Abstract
Background
Global immune activation and HLA alleles are each associated with the pathogenesis of human immunodeficiency virus (HIV) and hepatitis C virus .
Methods
We evaluated the relationship between 44 HLA class I and 28 class II alleles and percentages of activated CD8 (CD8+CD38+DR+) and CD4 (CD4+CD38+DR+) T cells in 586 women who were naive to highly active antiretroviral therapy. We used linear generalized estimating equation regression models, adjusting for race/ethnicity, age, HIV load, and hepatitis C virus infection and controlling for multiplicity using a false discovery rate threshold of 0.10.
Results
Ten HLA alleles were associated with CD8 and/or CD4 T-cell activation. Lower percentages of activated CD8 and/or CD4 T cells were associated with protective alleles B*57:03 (CD8 T cells, −6.6% [P = .002]; CD4 T cells, −2.7% [P = .007]), C*18:01 (CD8 T cells, −6.6%; P < .0008) and DRB1*13:01 (CD4 T cells, −2.7%; P < .0004), and higher percentages were found with B*18:01 (CD8 T cells, 6.2%; P < .0003), a detrimental allele. Other alleles/allele groups associated with activation included C*12:03, group DQA1*01:00, DQB1*03:01, DQB1*03:02, DQB1*06:02, and DQB1*06:03.
Conclusion
These findings suggest that a person’s HLA type may play a role in modulating T-cell activation independent of viral load and sheds light on the relationship between HLA, T-cell activation, immune control, and HIV pathogenesis.
Keywords: HIV, HCV, T-cell activation, HLA, immune activation
We found that specific HLA alleles predict CD8 and CD4 T-cell activation, independent of HLA associations with viral load. These findings suggest that host genetic variation could affect risk for end-organ diseases through an immune activation mechanism.
Global immune activation is a hallmark of human immunodeficiency virus (HIV) infection, leading to CD4 T-cell depletion, CD8 T-cell exhaustion and development of AIDS and non-AIDS conditions, especially in the presence of hepatitis C virus (HCV) coinfection [1–4]. The mechanisms involved in producing this hyperactive state are poorly understood.
HLA class I and II genes encode the major histocompatibility complex (MHC) proteins that present self and foreign peptides to CD8 and CD4 T cells, respectively [5]. Class I molecules (A, B, and C), present on nucleated cells, present intracellular-derived viral peptides on the surface of infected cells to cytotoxic CD8 T cells, stimulating their activation and clearance of virus-infected cells. They also interact with the innate immune system through natural killer cells and monocytes. Class II molecules (DRB1, DQA, and DQB), which are primarily expressed by antigen-presenting cells (eg, dendritic cells, macrophages), present exogenous antigen to CD4 T cells, stimulating their activation and expansion.
Variations in HLA genes are the most important host factor predicting outcomes including HIV load, rate of CD4 decline, and HIV disease progression [6]. Most notably, the B*57 allele group is overrepresented in elite controllers and is associated with slower HIV progression [7]. Other allele groups, such as C*04 and B*35-Px, are associated with more rapid HIV progression [8, 9].
Although HLA alleles and T-cell activation levels are each strongly linked to HIV pathogenesis, no study has evaluated the relationship between HLA and T-cell activation. We aimed to determine if there is a link between HLA and T-cell activation that could represent an independent heritable mechanism leading to accelerated HIV/HCV disease.
METHODS
Study Population
This study is nested within the Women’s Interagency HIV Study (WIHS), a prospective, multicenter cohort study of HIV-infected and high-risk HIV-uninfected women in the United States [10]. After institutional review board approval, participants provided consent and were enrolled from October 1994 through November 1995. Women were selected from 2 overlapping HIV and HIV/HCV coinfection WIHS initiatives evaluating T-cell activation and HLA on HIV disease [4, 11]. This included 586 HIV-infected women (229 HCV negative [39%], 64 HCV positive/RNA negative [11%], and 293 HCV positive/RNA positive [50%]), followed up through June 2003, with a median of 2 visits each (range, 1–9 visits).
We restricted the analysis to data collected before highly active antiretroviral therapy (HAART), because we were interested in the relationship between HLA genotype, HIV load, and T-cell activation before highly suppressive treatment. Participants who took antiretroviral drugs were receiving either monotherapy or non-HAART combination therapy, defined in our study period as either dual therapy (2 nucleoside reverse-transcriptase inhibitors [NRTIs], 1 NRTI and 1 nonnucleoside reverse-transcriptase inhibitor, or 1 NRTI and 1 protease inhibitor) or triple-NRTI therapy.
Flow Cytometry
Flow cytometry was performed at 2 National Institute of Allergy and Infectious Diseases–certified laboratories, in accordance with the AIDS Clinical Trials Group consensus protocol and using FACSCalibur and Cell Quest software (Becton Dickinson). Flow cytometry was performed on frozen (71% of samples) or fresh (29%) peripheral mononuclear cells. The analysis used the following fluorochrome-conjugated antibodies: anti-CD3, anti-CD4, anti-CD8, anti–HLA-DR, and anti-CD38 (Becton Dickinson or PharMingen). CD4 and CD8 T-cell activation was defined as the percentage of cells positive for both cell surface markers CD38 and HLA-DR (CD38+DR+).
Clinical Laboratory Testing
HIV-1 RNA was determined every 6 months (Organon Teknika and Roche Molecular Diagnostics). HCV antibody status was determined at baseline (using Abbott enzyme immunoassay 2.0 or 3.0), followed by HCV RNA testing for HCV antibody–positive women with polymerase chain reaction (Roche Molecular Diagnostics).
HLA Genotyping and Imputation
Our study used HLA data generated both by reference-standard HLA genotyping using sequence-specific oligonucleotide probes and by HLA imputation algorithms that we have assessed for accuracy in our multiracial parent cohort study [12]. Four-digit resolution HLA alleles were genotyped in 1587 WIHS women at the National Cancer Institute Frederick Laboratory, as described elsewhere [13, 14]. Because HLA class I and HLA-DRB1 genes were judged to be most relevant for studies of pathogens that are common in WIHS (eg, HIV, HCV, and human papillomavirus), most women had genotyping at the following loci: HLA-A (n = 1567; 46 unique alleles), HLA-B (n = 1537; 99 unique alleles), HLA-C (n = 1497; 37 unique alleles), and HLA-DRB1 (n = 1415; 50 unique alleles). Most of these women also had HLA-DQA1 (n = 1034; 13 unique alleles) and HLA-DQB1 (n = 1036; 18 unique alleles) typing.
Single-nucleotide polymorphisms were genotyped using an Illumina HumanOmni2.5-quad BeadChip system (Illumina) and allowing HLA imputation algorithms developed by other groups [15] to impute high-resolution HLA class I and II data for WIHS women who were not genotyped directly for HLA (n = 3353) [12, 16]. The HLA Genotype Imputation with Attribute Bagging (HIBAG) algorithm accurately imputed 89% of classic HLA alleles. Accuracy by HLA gene was 93% for HLA-A, 84% for HLA-B, 94% for HLA-C, 83% for HLA-DQA1, 91% for HLA-DQB1, and 88% for HLA-DRB1. Supplementary Table 1 shows the percentages by HLA alleles that were imputed.
Statistical Analysis
Descriptive data collected at WIHS enrollment included age, race, ethnicity, mode of HIV acquisition, antiretroviral therapy (ART), AIDS status, HIV load, HCV infection status (HCV antibody negative or positive, RNA positive or negative), and CD4 and CD8 T-cell counts. Longitudinal analysis of associations of HLA class I and II alleles with CD4 and CD8 T-cell activation levels was restricted to alleles with ≥5% prevalence. The generalized estimating equation (GEE) approach with exchangeable working correlation structure was used to model the repeated measures data. Multiple testing was accounted for by requiring a false discovery rate (FDR) of 0.1, according to the Benjamini-Hochberg method.
HLA alleles were individually modeled with each T-cell activation outcome, adjusting for select combinations of age, race/ethnicity, ART, HIV load, HCV infection status, and laboratory where activation data were generated. HIV load and ART were modeled as time-varying covariates using samples collected at the same visit as activation. If viral load (VL) was missing at a visit, we carried forward the data from a preceding visit within 1 year. Owing to rare frequencies of most alleles, a dominant genetic model was assumed, with alleles coded as prevalent (1 or 2 copies) or absent (0 copies). Adjustment for race/ethnicity included both self-reported and principal components of ancestry informative single-nucleotide polymorphisms to control for confounding by population substructure.
A natural log transformation was applied to CD4 T-cell activation and log (base 10) transformation to HIV load to reduce positive skewness. HCV infection status was included as an adjustment covariate because higher CD8 T-cell activation percentages have been associated with AIDS in the same population of coinfected women [4]. Possible statistical interactions between HCV infection status and HLA alleles were evaluated using analogous GEE models. For alleles with significant interactions with HCV infection status, stratified analyses were conducted.
For each combination of variant alleles that was observed in ≥10 woman, an allele-allele interaction term was evaluated in the GEE model. For alleles with significant associations with CD4 or CD8 T-cell activation, linkage disequilibrium (LD) was calculated using Pearson correlation coefficients. Alleles in LD were analyzed jointly in multiallelic GEE models. Differences in mean VL among women with and those without significant alleles were evaluated using GEE models adjusted for age, race/ethnicity, ART, HCV infection status, laboratory where activation data were generated, and CD4, CD8 and both CD4 and CD8 T-cell activation in separate models. Statistical analyses were performed using SAS (version 9.4; SAS Institute), Stata Statistical (version 14.1; StataCorp), and R (version 3.4.3) software.
RESULTS
Demographic and Clinical Characteristics of the Study Population
Demographic and clinical characteristics for the 586 HIV-infected and HAART-naive women are shown in Table 1. The majority of the women were non-Hispanic black (58%) or Hispanic (25%), with a history of injection drug use (45%) or heterosexual contact (35%) as the HIV risk factor. At enrollment, women had a median HIV RNA level of 15 050 copies/mL (range, 48–1 800 000 copies/mL), and 34% had CD4 T-cell counts >500/µL.
Table 1.
Baseline Demographic and Clinical Characteristics of Human Immunodeficiency Virus–Infected Women Naive to Highly Active Antiretroviral Treatment
| Characteristic | HIV-Infected Women, No. (%)a (N = 586) |
|---|---|
| Age, median (range), y | 37 (17–67) |
| Race/ethnicity | |
| White, non-Hispanic | 99 (17) |
| Black, non-Hispanic | 342 (58) |
| Hispanic | 145 (25) |
| HIV transmission risk factor | |
| Injection drug use | 263 (45) |
| Heterosexual contact | 207 (35) |
| Transfusion | 22 (4) |
| Not identified | 90 (15) |
| Data missing | 4 (1) |
| CD4 T-cell count, cells/μL | |
| ≤200 | 93 (16) |
| 201–350 | 142 (24) |
| 351–500 | 143 (24) |
| >500 | 202 (34) |
| Median (range) | 405 (2–1972)c |
| Data missing | 6 (1) |
| CD8 T-cell count | |
| ≤580/ μ L | 93 (16) |
| 581–820/ μ L | 164 (28) |
| 821–1140/ μ L | 167 (28) |
| >1140/ μ L | 154 (26) |
| CD8 T-cell count, median (range), cells/ μ L | 875 (91–3866) |
| CD8 T-cell data missing | 8 (1.4) |
| HIV load, median (range), copies/mL | 15 050 (48–1 800 000)b |
| Antiretroviral therapy | |
| No therapy | 372 (63) |
| Monotherapy | 164 (28) |
| Combination therapy | 50 (9) |
| HAART | 0 |
| HCV status | |
| HCV negative | 229 (39) |
| HCV positive (nonviremic) | 64 (11) |
| HCV positive (viremic) | 293 (50) |
| Ever had AIDS | |
| Yes | 51 (9) |
| No | 535 (91) |
Abbreviations: HAART, highly active antiretroviral therapy; HCV, hepatitis C virus; HIV, human immunodeficiency virus.
aData represent no. (%) of women unless otherwise specified.
bAmong 584 women with baseline HIV load data.
cAmong 580 women with baseline CD4 T-cell data.
Associations Between HLA Alleles and CD8 and CD4 T-Cell Activation
Among the 203 observed HLA alleles, 44 class I (15 HLA-A, 15 HLA-B, and 14 HLA-C) and 28 class II (6 HLA-DQA1, 8 HLA-DQB1, and 14 HLA-DRB1) alleles had ≥5% prevalence and were analyzed to identify associations with T-cell activation. Supplementary Table 2 shows the 72 HLA allele frequencies by race/ethnicity, median CD4 T-cell counts and VL, and median CD8+CD38+DR+ and CD4+CD38+DR+ percentages averaged over visits for the 586 women. Variability in median VL by allele ranged from 1200 copies/mL (for B*57:03, present in 7.3% of women) to 48 850 copies/mL (for B*1801, present in 7.2%).
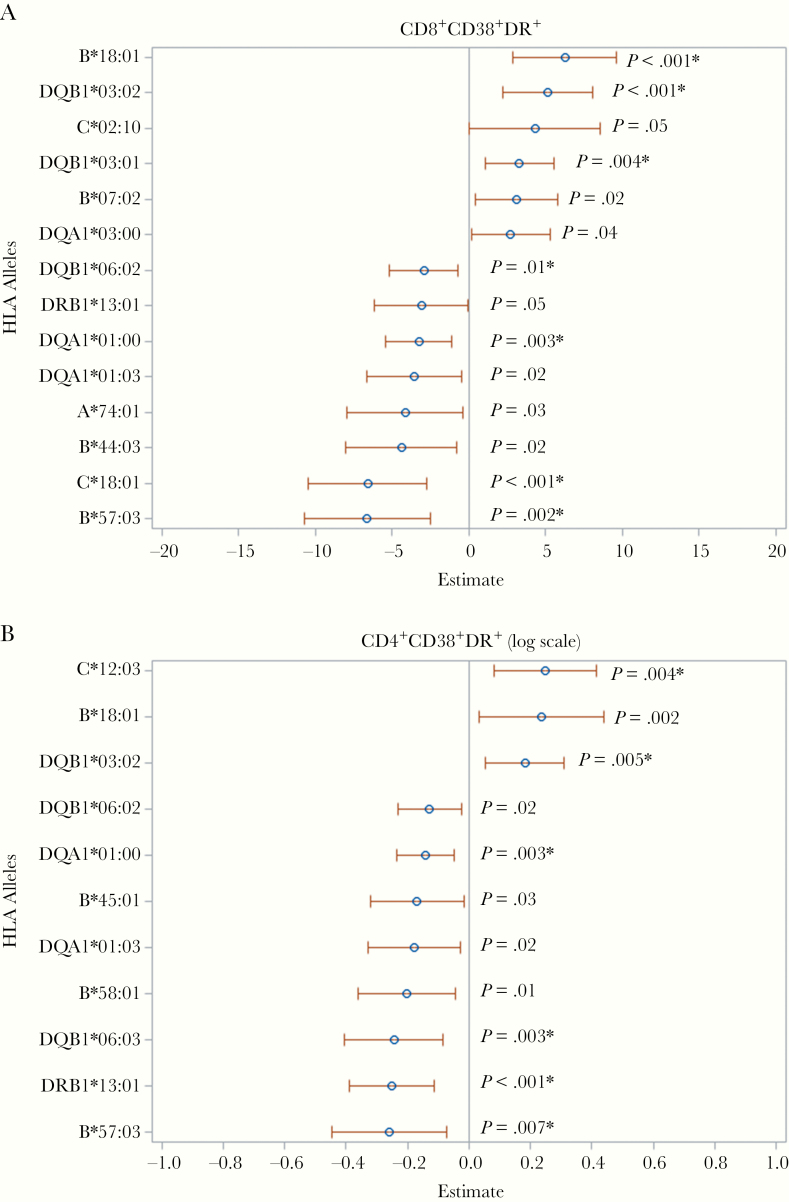
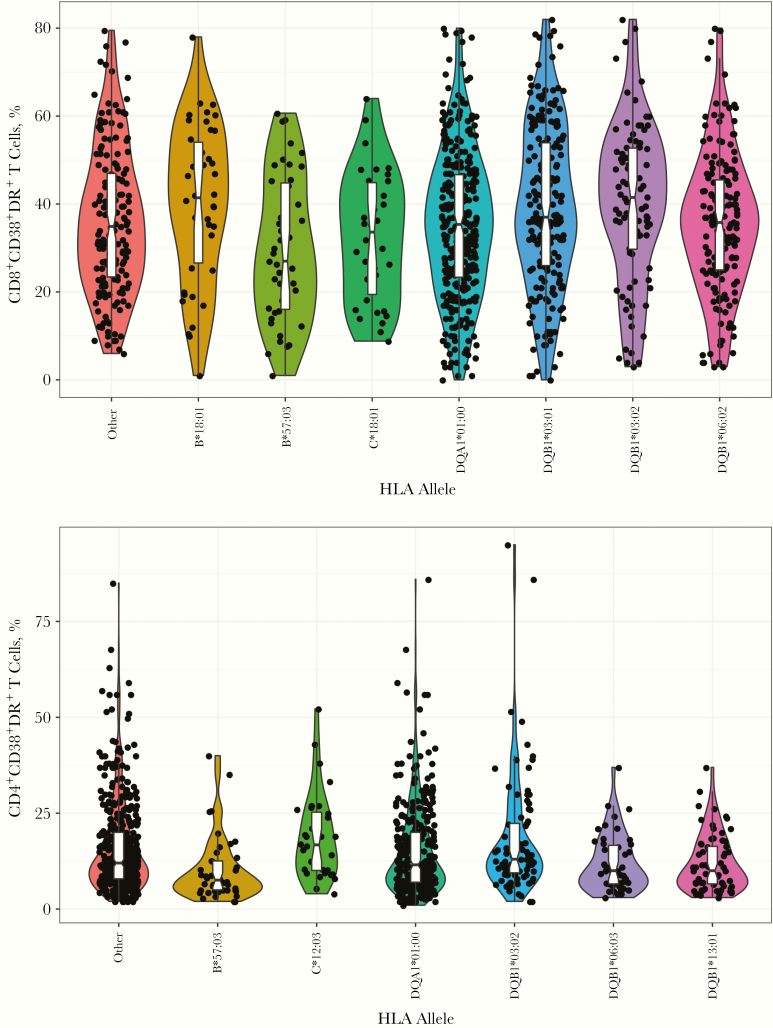
Adjusted models for CD8 and CD4 T-cell activation for each of the 72 alleles are shown in Supplementary Table 3A and 3B; 14 alleles achieved the nominal .05 significance level for association with CD8 T-cell activation (Figure 1A), and after multiplicity adjustment, 7 achieved a 0.10 FDR (B*18:01, B*57:03, C*18:01, DQA1*01:00, DQB1*03:01, DQB1*03:02, and DQB1*06:02). For CD4 T-cell activation, 11 alleles achieved the nominal .05 significance level for association, and 6 achieved the 0.10 FDR (B*57:03, C*12:03, DQA1*01:00, DQB1*03:02, DQB1*06:03, and DRB1*13:01) (Figure 1B). The violin plots in Figure 2 show the distributions of CD8+CD38+DR+ and CD4+CD38+DR+ T-cell proportions among participants with each of the associated (FDR, <0.10) HLA alleles.
Figure 1.
Relationship between significant HLA alleles and CD8 (A) or CD4 (B) activation. Significant (P ≤ .05) associations of HLA alleles with percentage of CD8+CD38+DR+ (A) and log percentage of CD4+CD38+DR+ (B) T cells in multivariate linear models, with adjustments for race/ethnicity, principal components, age, laboratory data source, antiretroviral therapy, human immunodeficiency virus load, and hepatitis C virus status. The β estimate (with 95% confidence interval) is shown for each allele. *P ≤ .01 after adjustment for multiple tests (false discovery rate, <0.10).
Figure 2.
Violin plots showing percentages of CD8 and CD4 T-cell activation for each patient by HLA allele. Distributions of percentages of CD8+CD38+DR+ and CD4+CD38+DR+ T cells are shown by allele variant group. For plots of the allele variants, each point represents a unique subject. Activation levels at multiple visits were averaged for each subject. The “Other” group includes subjects with none of the allele variants.
Direction and Magnitude of Significant HLA Allele Associations With CD8 T-Cell Activation
Differences in mean CD8+CD38+DR+ T-cell percentages were estimated using adjusted GEE models where β is the difference in CD8+CD38+DR+ T-cell percentages between women with or without each HLA allele with a positive β value indicating increased activation and a negative value indicating decreased activation. (Table 2) Women with B*18:01 (β = 6.2% [95% confidence interval (CI), 2.9–9.6]; FDR, 0.02), DQB1*03:01 (β = 3.3% [1.0–5.5]; FDR, 0.05), or DQB1*03:02 (β = 5.1% [95% CI, 2.2–8.0]; FDR, 0.02), had higher CD8 T-cell activation percentages than women without these alleles. On the other hand, women with B*57:03 (β = −6.6% [95% CI, −10.8 to −2.5]; FDR, 0.03), C*18:01 (β = −6.6% [−10.5 to −2.7]; FDR, 0.02), group DQA1*01:00 (β = −3.3% [−5.4 to −1.1]; FDR, 0.04), or DQB1*06:02 (β = −2.9% [−5.2 to −0.7]; FDR, 0.10), had lower CD8 T-cell activation percentages than those without these alleles.
Table 2.
Association of CD8 T-Cell Activation Levels (CD8+CD38+DR+) With HLA Alleles Among Human Immunodeficiency Virus–Infected Women Naive to Highly Active Antiretroviral Therapya
| Allele | β (95% CI)b | Mean Differencec | P Value | FDRd |
|---|---|---|---|---|
| B*18:01 | 6.2 (2.9–9.6) | 6.2 | <.0003 | 0.02 |
| B*57:03 | −6.6 (−10.8 to −2.5) | −6.6 | .002 | 0.03 |
| C*18:01 | −6.6 (−10.5 to −2.7) | −6.6 | <.0008 | 0.02 |
| DQA1*01:00 | −3.3 (−5.4 to −1.1) | −3.3 | .003 | 0.04 |
| DQB1*03:01 | 3.3 (1.0–5.5) | 3.3 | .004 | 0.05 |
| DQB1*03:02 | 5.1 (2.2–8.0) | 5.1 | <.0006 | 0.02 |
| DQB1*06:02 | −2.9 (−5.2 to −0.7) | −2.9 | .01 | 0.10 |
Abbreviations: CI, confidence interval; FDR, false discovery rate.
aSample size of 586 women with a total of 1177 study visits. Samples with missing HLA data were imputed (see Supplementary Table 1 for percentage of data imputed by HLA type). The FDR was controlled at 0.10.
bThis value (β) is the difference in percentage of activated CD8 T cells between individuals with the allele and those without the allele, estimated using generalized estimating equation models. Models were controlled for age, race, principal components, laboratory data source, antiretroviral therapy, human immunodeficiency virus RNA level, and hepatitis C virus status.
cThe expected difference in mean percentage of activated CD8 T cells between individuals with the allele and those without the allele (covariates at mean or reference level).
dFDR computed using the Benjamini and Hochberg method.
Direction and Magnitude of Significant HLA Allele Associations With CD4 T-Cell Activation
For CD4 T-cell activation, (Table 3), β estimates are in a logarithmic scale; hence, we report expected mean differences in raw scale between women with or without each allele. The mean CD4+CD38+DR+ percentage was higher in women with alleles C*12:03 or DQB1*03:02 (3.3% and 2.3% [both FDR, 0.06], respectively), and lower in those with B*57:03 (−2.7%; FDR, 0.07), group DQA1*01:00 (−1.7%; FDR, 0.06), DQB1*06:03 (−2.6%; FDR, 0.06), or DRB1*13:01 (−2.7%; FDR, 0.03), compared with women without these alleles.
Table 3.
Association of CD4 T-Cell Activation Levels (Log of CD4+CD38+DR+) With HLA Alleles Among Human Immunodeficiency Virus–Infected Women Naive to Highly Active Antiretroviral Therapya
| Allele | β (95% CI), Log Scaleb | Mean Difference, Raw Scalec | P Value | FDRd |
|---|---|---|---|---|
| B*57:03 | −0.26 (−0.45 to −0.07) | −2.7 | .007 | 0.07 |
| C*12:03 | 0.25 (0.08–0.42) | 3.3 | .004 | 0.06 |
| DQA1*01:00 | −0.14 (−0.24 to −0.05) | −1.7 | .003 | 0.06 |
| DQB1*03:02 | 0.18 (0.05–0.31) | 2.3 | .005 | 0.06 |
| DQB1*06:03 | −0.24 (−0.41 to −0.08) | −2.6 | .003 | 0.06 |
| DRB1*13:01 | −0.25 (−0.39 to −0.11) | −2.7 | <.0004 | 0.03 |
Abbreviations: CI, confidence interval; FDR, false discovery rate.
aSample size of 580 women with a total of 1161 study visits. Samples with missing HLA data were imputed (see Supplementary Table 1 for percentage of data imputed by HLA type). The FDR was controlled at 0.10.
bThis value (β) is the difference in percentage of activated CD4 T cells in individuals with the allele as compared to individuals without the allele, estimated using generalized estimating equation models. Models were controlled for age, race, principal components, laboratory data source, antiretroviral therapy, human immunodeficiency virus RNA, and hepatitis C virus status.
cThe expected difference in mean percentage of activated CD4 T cells between individuals with and those without the allele (covariates at mean or reference level).
dFDR computed using the Benjamini and Hochberg method.
HLA Associations With Immune Control (HIV Load) After Adjustment for T-Cell Activation
Adjusted models evaluating immune control for women with or without significant alleles are shown in Table 4. Women with B*57:03 had significantly lower VL after adjustment for age, race, ART, and HCV status (model 1), and the differences remained significant with adjustments for CD4 (model 2), CD8 (model 3), or both CD4 and CD8 (model 4) T-cell activation (all P < .0001) than those without this allele. VLs were also marginally lower with C*18:01 (P = .05) and higher with B*18:01 (P = .03), but only before adjustment for activation. No differences in VL were found for the other alleles. However, after adjustment for activation, VL was lower for DQB1*03:02.
Table 4.
Viral Load Compared by HLA Allele Among Human Immunodeficiency Virus–Infected Women Naive to Highly Active Antiretroviral Therapya
| Allele | Prev- alent | Women, No. (%) | Model 1 | Model 2 | Model 3 | Model 4 | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| HIV Load Mean (95 CI)b | P Valuec | HIV Load Mean (95 CI)b | P Valuec | HIV Load, Mean (95 CI)b | P Valuec | HIV Load, Mean (95 CI)b | P Valuec | |||
| HLA class I | ||||||||||
| B*18:01 | Yes | 42 (7.2) | 9453 (4431–20 169) | .03 | 6867 (3465–13 608) | .39 | 6648 (3491–12 659) | .32 | 6109 (3243–11 506) | .62 |
| No | 544 (92.8) | 4353 (2966–6388) | 5152 (3635–7300) | 4877 (3436–6921) | 5250 (3757–7336) | |||||
| B*57:03 | Yes | 43 (7.3) | 681 (328–1414) | <.001 | 1396 (721–2702) | <.001 | 1422 (735–2750) | <.001 | 1779 (936–3380) | <.0001 |
| No | 543 (92.7) | 5559 (3832–8063) | 5930 (4200–8372) | 5604 (3968–7916) | 5832 (4189–8119) | |||||
| C*12:03 | Yes | 32 (5.5) | 4063 (1745–9459) | .74 | 5387 (2637–11 005) | .96 | 3199 (1570–6518) | .17 | 4056 (2058–7994) | .39 |
| No | 554 (94.5) | 4673 (3185–6855) | 5295 (3739–7498) | 5155 (3641–7298) | 5410 (3876–7551) | |||||
| C*18:01 | Yes | 34 (5.8) | 2210 (927–5268) | .05 | 3950 (1957–7973) | .34 | 3503 (1675–7327) | .28 | 4339 (2205–8539) | .50 |
| No | 552 (94.2) | 4947 (3405–7186) | 5434 (3853–7664) | 5167 (3654–7307) | 5416 (3884–7552) | |||||
| HLA class II | ||||||||||
| DQA1*01:00 | Yes | 330 (56.3) | 4788 (3187–7193) | .67 | 5839 (4058–8402) | .12 | 5515 (3838–7924) | .11 | 5959 (4205–8444) | .054 |
| No | 256 (43.7) | 4415 (2866–6802) | 4541 (3072–6712) | 4277 (2880–6351) | 4416 (3030–6436) | |||||
| DQB1*03:01 | Yes | 200 (34.1) | 5176 (3319–8071) | .43 | 5100 (3367–7726) | .75 | 5266 (3465–8003) | .70 | 5219 (3480–7826) | .86 |
| No | 386 (65.9) | 4431 (2965–6622) | 5386 (3768–7701) | 4927 (3450–7036) | 5372 (3820–7554) | |||||
| DQB1*03:02 | Yes | 88 (15) | 3750 (2118–6642) | .35 | 3470 (2091–5759) | .03 | 3414 (2112–5519) | .04 | 3388 (2129–5389) | .01 |
| No | 498 (85) | 4819 (3275–7090) | 5710 (4017–8116) | 5388 (3780–7678) | 5792 (4122–8138) | |||||
| DQB1*06:02 | Yes | 168 (28.7) | 4490 (2829–7128) | .82 | 5773 (3787–8803) | .51 | 5613 (3689–8542) | .36 | 6159 (4110–9230) | .23 |
| No | 418 (71.3) | 4708 (3162–7009) | 5112 (3570–7320) | 4777 (3335–6843) | 5003 (3546–7059) | |||||
| DQB1*06:03 | Yes | 47 (8) | 4888 (2301–10 383) | .88 | 6402 (3262–12 565) | .54 | 7140 (3605–14 142) | .26 | 7650 (3979–14 706) | .22 |
| No | 539 (92) | 4623 (3153–6778) | 5237 (3703–7406) | 4911 (3465–6959) | 5208 (3731–7271) | |||||
| DRB1*13:01 | Yes | 71 (12.1) | 3930 (2152–7175) | .51 | 5314 (3144–8981) | .99 | 5956 (3490–10 164) | .44 | 6425 (3869–10 668) | .37 |
| No | 515 (87.9) | 4723 (3218–6931) | 5298 (3743–7500) | 4933 (3478–6997) | 5227 (3741–7304) |
Abbreviations: CI, confidence interval; HIV, human immunodeficiency virus.
aThe analysis included clinical data at all visits with T-cell activation data. Model 1 controlled for age, race, antiretroviral therapy (ART), hepatitis C virus (HCV) status, and laboratory data source; model 2, for age, race, ART, HCV status, laboratory data source, and CD4 T-cell activation; model 3, for age, race, ART, HCV status, laboratory data source and CD8 T-cell activation; and model 4, for age, race, ART, HCV status, laboratory data source, and CD4 and CD8 T-cell activation.
bAdjusted means estimated using generalized estimating equation models. The HIV load was log-transformed for analysis, and resulting mean estimates and 95% CIs were back-transformed.
c P values for comparison between group in which the allele was prevalent and group in which it was not.
HCV Infection, HLA, and T-Cell Activation
We next evaluated potential effect modification between HCV infection and HLA alleles, because prior WIHS research found an association between CD8 T-cell activation and development of AIDS among coinfected women [4]. We observed that HCV infection modifies the effect of DQB1*03:01 on CD8 T-cell activation (P = .008; data not shown), but HCV did not modify other allele associations (all P ≥ .05; data not shown). In a stratified analysis, only HCV antibody–positive nonviremic and viremic women with DQB1*03:01 had significantly higher CD8 T-cell percentages (8.6% [P = .01] and 5.6% [P < .0007], respectively) than those without the allele (Supplementary Table 4). This was not found for women with HIV infection only.
Evaluation of Multiallelic Effects
Pairwise LD was estimated for alleles significantly associated with CD4 or CD8 T-cell activation (Supplementary Figure 1). Allele pairs with LD, suggested by a P value ≤ .05 by Pearson correlation coefficient and ≥10 women possessing both alleles (Supplementary Table 5), were considered in multiallelic adjusted models of CD4 or CD8 T-cell activation. No evidence was found for statistical interactions between pairs of alleles in LD. Allelic effect estimates from multiallelic models were similar to those from single-allele models (Supplementary Table 6).
DISCUSSION
Understanding the pathogenesis of HIV infection, including the role of genetics, is essential not only for the development of new strategies to treat and prevent HIV infection, but also for vaccine development. The delicate balance between innate and adaptive immunity that maintains steady-state control of infectious diseases is closely linked to MHC diversity. This well-orchestrated system, however, is hampered by HIV, as it leads to alterations in CD8 and CD4 T-cell immune responses and finally CD8 T-cell anergy and CD4 depletion.
To our knowledge, this is the first comprehensive study to evaluate associations between HLA alleles and CD8 and CD4 T-cell activation in persons living with HIV (PLWH). We identified 10 class I and class II alleles associated with significantly higher or lower levels of activated CD4 and/or CD8 T cells independent of VL. Most alleles identified had been reported to be associated with HIV or HCV disease [13, 14, 17–21]. Some alleles were associated with significantly lower (B*57:03 and group DQA1*01:00) or higher (DQB1*03:02) percentages of activated CD8 and CD4 T cells than those without these alleles. Importantly, the study suggests that the increased longevity of PLWH with B*57:03 may be due to both lower HIV load and lower levels of T-cell activation. These findings, if replicated, have implications for understanding the role of genetics on T-cell activation and HIV/HCV pathogenesis.
Recent studies, including genome-wide association studies, have confirmed that polymorphisms in class I alleles are the most closely linked to immune control and HIV and HCV disease [14, 20, 22–27]. In our study, women with HLA B*57:03 had the lowest percentages of activated CD4 and CD8 T cells and the lowest VLs, even after adjustment for activation, suggesting the independent effect of this allele on maintaining low levels of activation and immune control. B*57:03 has been shown to have high functional avidity, polyfunctionality, proliferative capacity, and cytotoxicity of CD8 T cells to effectively kill HIV-infected cells and suppress viral replication [28]. B*57:03 has a conserved epitope for Gag with broad p24 Gag responses that correlate with disease protection [28–30]. However, selective pressure can lead to reduced fitness that may also result in development of protective CD8 responses [31]. Furthermore, the low levels of CD4 T-cell activation associated with B*57:03 may also affect HIV integration and replication, resulting in low VL and low HIV reservoirs [32].
We found that when an allele was associated with both CD4 and CD8 T-cell activation, the direction of activation was the same. For instance, women with B*57:03, present in 7.3% of the cohort and observed in multiple studies to be a protective allele [14, 20, 22–27], had a 6.6% lower percentage of activated CD8 T cells and a 2.7% lower percentage of activated CD4 T cells than women without this allele. The DQA1*01:00 allele group (present in 56.3%) was also associated with lower percentages of activated CD8 and CD4 T cells, whereas DQB1*03:02 (15%) had higher percentages of both. These results are not surprising given the cross-talk between antigen-presenting cells, CD4 and CD8 T cells and other populations that communicate through secretion of cytokines resulting in their activation [33]. Interestingly, our previous work demonstrated that concurrent hyperactivation of CD4 and CD8 T cells predicts AIDS, whereas women with low levels of activation and high levels of resting T cells had slower progression [34].
Of interest, we found that, except for B*57:03 and B*18:01, alleles with significant associations with T-cell activation were not associated with VL. In nonhuman primates, simian immunodeficiency virus (SIV) infection in its natural host is not associated with disease, despite high levels of SIV replication, whereas experimental SIV in rhesus macaques leads to simian AIDS [35]. Lack of chronic immune activation is one of the distinguishing features of nonpathogenic SIV. In our study, C*18:01 was associated with 6.6% lower CD8 T-cell activation levels, similar to B*57:03, but there were only borderline differences in VL between those with and those without the allele. Although this allele is in LD with HLA B*57:03 (17 women had both alleles), each allele remained significant in our multiallelic adjusted model (Supplementary Table 6).
B*18:01, which has a negative impact on HIV disease and HCV disease [17, 21], was associated with significantly higher levels of CD8 T-cell activation. CD4 T-cell activation was also higher, but this association did not pass FDR correction (Supplementary Table 3B). This allele is a member of the B*44 supertype that shares overlapping peptide-binding capacities for HIV and other viruses [36]. Another allele in the B*44 supertype is B*44:03, which was associated with lower activation levels but did not achieve 0.1 FDR. Differing binding affinities of peptides of HIV, HCV, hepatitis B virus, and herpesviruses, such as cytomegalovirus or Epstein-Barr virus (which may cross-react with self-peptides), may also play a role in activation of CD8 T cells [36], because most PLWH are infected with these herpesviruses.
Most of the alleles identified have been associated with autoimmune diseases [37], suggesting that the potential protective or deleterious effects of certain HLA alleles associated with autoimmune diseases may affect immune activation in the setting of HIV. For instance, HLA B*44 has been shown to be protective against some autoimmune diseases (eg, progressive multiple sclerosis [38] and diabetes [39]) and to modulate immune responses to vaccines [40]. In contrast, the HLA B*18 allele group is associated with progressive type 1 diabetes and response to a number of self-peptides [39, 41]. Thus, the differences in CD8 T-cell activation levels noted between women with and those without B*18:01 may potentially reflect differences in regulation of the immune response to HIV and other viral infections, such as HCV, cytomegalovirus, or Epstein-Barr virus infection, or to self-antigens [42]. Irrespective of its origin, high CD8 T-cell activation in women with B*18:01 represents an example of a putatively high-risk (eg, increased risk of end-organ diseases) subpopulation.
Our study found that the protective allele DRB1*13:01 was associated with lower percentages of activated CD4 T cells, but no differences in VL were found between those with and those without this allele. DRB1*13 allele group has been shown to be protective, not only for HIV, but also for HCV, hepatitis B virus, and other infections and some autoimmune diseases (eg, rheumatoid arthritis and systemic lupus erythematosus) suggesting a similar mechanism of protection [43–45]. In contrast, higher percentages of activated CD4 T cells were found with C*12:03, which is associated with Stevens-Johnson syndrome [46], but VLs did not differ between those with and those without this allele.
In our study, 50% of women were HIV/HCV coinfected, allowing us to evaluate the effect modification of HCV on HLA and T-cell activation. Like HIV, measures of HCV pathogenesis have strong associations with variation in HLA genes in WIHS women and in global cohorts. We found that women with DQB1*03:01 (present in 34.1%) had higher CD8 T-cell activation percentages among those with HIV/HCV coinfection, but not among HIV monoinfected women (Supplementary Table 4). Effect modification was not observed for any other significant allele. DQB1*03:01 has been associated with HCV clearance, lower HCV loads, and lack of progression of liver fibrosis and hepatocellular carcinoma in those infected only with HCV [11, 19, 47]. Furthermore, this allele was associated with decreased neutralizing antibody responses in the recent HIV vaccine RV44 trial [48]. Taken together, the increasing volume of significant associations of DQB1*03:01 in HCV-infected populations with a variety of phenotypes including in this study merits renewed attention to this allele in HIV/HCV pathogenesis. Interestingly, women with DQB1*03:02, a protective allele for HIV and HCV disease [49], had increased activation, suggesting that there may be differences in HIV and HCV peptide binding in the setting of coinfection [19].
Another potential mechanism that may affect level of activation is the variation in cell surface MHC protein expression [27]. HIV can modulate the expression of MHC class I and II on cell surfaces as a mechanism to evade immune control (eg, down-regulation of mature class I and II by HIV nef and up-regulation of the HLA class II Ii chain on antigen-presenting cells) [50]. Furthermore, cytokines, such as interferon γ, induce the expression of MHC molecules on a variety of tissues, such as endothelial and epithelial cells, thus affecting T-cell activation.
There are some limitations to our study. First, although the sample size was large, it may not be large enough to detect associations between activation and rare HLA alleles that were present in <5% of the population. Second, the population included only women and may not be generalizable to men. Third, not all races and ethnicities were represented, and thus results may not be applicable to other populations worldwide. Finally, these studies need to be reproduced in independent populations in among PLWH who are receiving effective ART treatment.
In conclusion, specific HLA alleles predict CD8 and CD4 T-cell activation, independent of HLA associations with VL. These findings suggest that host genetic variation could affect risk for end-organ diseases through an immune activation mechanism. Whether variation in activation levels in the presence of different HLA alleles is related to response to HIV or other viral peptides and/or self-antigens needs further investigation. These results have implications for potential therapeutic interventions and HIV vaccine studies.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. We thank all the women who participated in this study over so many years.
Study sites. Data in this manuscript were collected by the Women’s Interagency HIV Study (WIHS) at the following sites (principal investigators; National Institutes of Health [NIH] grant no.): University of Alabama at Birmingham WIHS (Mirjam-Colette Kempf and Deborah Konkle-Parker; U01-AI-103401), Atlanta WIHS (Ighovwerha Ofotokun and Gina Wingood; U01-AI-103408), Bronx WIHS (Kathryn Anastos; U01-AI-035004), Brooklyn WIHS (Howard Minkoff and Deborah Gustafson; U01-AI-031834), Chicago WIHS (Mardge Cohen and Audrey French; U01-AI-034993), Metropolitan Washington WIHS (Seble Kassaye; U01-AI-034994), Miami WIHS (Margaret Fischl and Lisa Metsch; U01-AI-103397), University of North Carolina WIHS (Adaora Adimora; U01-AI-103390), Connie Wofsy Women’s HIV Study, Northern California (Ruth Greenblatt, Bradley E. Aouizerat, and Phyllis C. Tien; U01-AI-034989), WIHS Data Management and Analysis Center (Stephen Gange and Elizabeth T. Golub; U01-AI-042590), and Southern California WIHS (Joel Milam; U01-HD-032632) (WIHS I–WIHS IV).
Disclaimer. The contents of this publication are solely the responsibility of the authors and do not represent the official views of the NIH.
Financial support. This work was supported by the National Institute of Allergy and Infectious Diseases: R56AI052065, R01AI052065, and the National Institute on Drug Abuse R01DA044111 (Andrea Kovacs, PI). This work was supported by the National Institute of Allergy and Infectious Diseases, the Eunice Kennedy Shriver National Institute of Child Health and Human Development, the National Cancer Institute, the National Institute on Drug Abuse, the National Institute of Mental Health, the National Institute of Dental and Craniofacial Research, the National Institute on Alcohol Abuse and Alcoholism, the National Institute on Deafness and other Communication Disorders, and the NIH’s Office of Research on Women’s Health, and the NIH (grants UL1-TR000004 [to University of California, San Francisco, Clinical & Translational Science Institute], UL1-TR000454 [to Atlanta Clinical & Translational Science Alliance], and P30-AI-050410 [to University of North Carolina Center for AIDS Research]).
Potential conflicts of interest. All authors: No reported conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Hazenberg MD, Otto SA, van Benthem BH, et al. Persistent immune activation in HIV-1 infection is associated with progression to AIDS. AIDS 2003; 17:1881–8. [DOI] [PubMed] [Google Scholar]
- 2. Giorgi JV, Hultin LE, McKeating JA, et al. Shorter survival in advanced human immunodeficiency virus type 1 infection is more closely associated with T lymphocyte activation than with plasma virus burden or virus chemokine coreceptor usage. J Infect Dis 1999; 179:859–70. [DOI] [PubMed] [Google Scholar]
- 3. Papagno L, Spina CA, Marchant A, et al. Immune activation and CD8+ T-cell differentiation towards senescence in HIV-1 infection. PLoS Biol 2004; 2:E20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kovacs A, Karim R, Mack WJ, et al. Activation of CD8 T cells predicts progression of HIV infection in women coinfected with hepatitis C virus. J Infect Dis 2010; 201:823–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Neefjes J, Jongsma ML, Paul P, Bakke O. Towards a systems understanding of MHC class I and MHC class II antigen presentation. Nat Rev Immunol 2011; 11:823–36. [DOI] [PubMed] [Google Scholar]
- 6. Martin MP, Carrington M. Immunogenetics of HIV disease. Immunol Rev 2013; 254:245–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pereyra F, Addo MM, Kaufmann DE, et al. Genetic and immunologic heterogeneity among persons who control HIV infection in the absence of therapy. J Infect Dis 2008; 197:563–71. [DOI] [PubMed] [Google Scholar]
- 8. Gao X, Nelson GW, Karacki P, et al. Effect of a single amino acid change in MHC class I molecules on the rate of progression to AIDS. N Engl J Med 2001; 344:1668–75. [DOI] [PubMed] [Google Scholar]
- 9. Leslie A, Matthews PC, Listgarten J, et al. Additive contribution of HLA class I alleles in the immune control of HIV-1 infection. J Virol 2010; 84:9879–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Adimora AA, Ramirez C, Benning L, et al. Cohort profile: the Women’s Interagency HIV Study (WIHS). Int J Epidemiol 2018; 47:393–4i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kuniholm MH, Gao X, Xue X, et al. The relation of HLA genotype to hepatitis C viral load and markers of liver fibrosis in HIV-infected and HIV-uninfected women. J Infect Dis 2011; 203:1807–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kuniholm MH, Xie X, Anastos K, et al. Human leucocyte antigen class I and II imputation in a multiracial population. Int J Immunogenet 2016; 43:369–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kuniholm MH, Kovacs A, Gao X, et al. Specific human leukocyte antigen class I and II alleles associated with hepatitis C virus viremia. Hepatology 2010; 51:1514–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kuniholm MH, Gao X, Xue X, et al. Human leukocyte antigen genotype and risk of HIV disease progression before and after initiation of antiretroviral therapy. J Virol 2011; 85:10826–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Leslie S, Donnelly P, McVean G. A statistical method for predicting classical HLA alleles from SNP data. Am J Hum Genet 2008; 82:48–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zheng X, Shen J, Cox C, et al. HIBAG—HLA genotype imputation with attribute bagging. Pharmacogenomics J 2014; 14:192–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Goulder PJ, Walker BD. HIV and HLA class I: an evolving relationship. Immunity 2012; 37:426–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kiepiela P, Leslie AJ, Honeyborne I, et al. Dominant influence of HLA-B in mediating the potential co-evolution of HIV and HLA. Nature 2004; 432:769–75. [DOI] [PubMed] [Google Scholar]
- 19. Lee MH, Huang YH, Chen HY, et al. Human leukocyte antigen variants and risk of hepatocellular carcinoma modified by hepatitis C virus genotypes: A genome-wide association study. Hepatology 2018; 67:651–661. [DOI] [PubMed] [Google Scholar]
- 20. Kim AY, Kuntzen T, Timm J, et al. Spontaneous control of HCV is associated with expression of HLA-B 57 and preservation of targeted epitopes. Gastroenterology 2011; 140:686–96 e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Frías M, Rodríguez-Cano D, Cuenca-López F, et al. HLA-B18 as a risk factor of short-term progression to severe liver fibrosis in HIV/HCV co-infected patients with absent or minimal fibrosis: implications for timing of therapy. Pharmacogenomics J 2017; 17:551–5. [DOI] [PubMed] [Google Scholar]
- 22. Migueles SA, Sabbaghian MS, Shupert WL, et al. HLA B*5701 is highly associated with restriction of virus replication in a subgroup of HIV-infected long term nonprogressors. Proc Natl Acad Sci U S A 2000; 97:2709–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fellay J, Shianna KV, Ge D, et al. A whole-genome association study of major determinants for host control of HIV-1. Science 2007; 317:944–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kaslow RA, Carrington M, Apple R, et al. Influence of combinations of human major histocompatibility complex genes on the course of HIV-1 infection. Nat Med 1996; 2:405–11. [DOI] [PubMed] [Google Scholar]
- 25. Pereyra F, Jia X, McLaren PJ, et al. International HIV Controllers Study. The major genetic determinants of HIV-1 control affect HLA class I peptide presentation. Science 2010; 330:1551–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Stephens HA. Immunogenetic surveillance of HIV/AIDS. Infect Genet Evol 2012; 12:1481–91. [DOI] [PubMed] [Google Scholar]
- 27. Apps R, Qi Y, Carlson JM, et al. Influence of HLA-C expression level on HIV control. Science 2013; 340:87–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lécuroux C, Sáez-Cirión A, Girault I, et al. Both HLA-B*57 and plasma HIV RNA levels contribute to the HIV-specific CD8+ T cell response in HIV controllers. J Virol 2014; 88:176–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Serwanga J, Shafer LA, Pimego E, et al. Host HLA B*allele-associated multi-clade Gag T-cell recognition correlates with slow HIV-1 disease progression in antiretroviral therapy-naïve Ugandans. PLoS One 2009; 4:e4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kloverpris HN, Stryhn A, Harndahl M, et al. HLA-B*57 micropolymorphism shapes HLA allele-specific epitope immunogenicity, selection pressure, and HIV immune control. J Virol 2012; 86:919–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pohlmeyer CW, Buckheit RW 3rd, Siliciano RF, Blankson JN. CD8+ T cells from HLA-B*57 elite suppressors effectively suppress replication of HIV-1 escape mutants. Retrovirology 2013; 10:152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Descours B, Avettand-Fenoel V, Blanc C, et al. ; ALT ANRS CO15 Study Group Immune responses driven by protective human leukocyte antigen alleles from long-term nonprogressors are associated with low HIV reservoir in central memory CD4 T cells. Clin Infect Dis 2012; 54:1495–503. [DOI] [PubMed] [Google Scholar]
- 33. Mangalam AK, Taneja V, David CS. HLA class II molecules influence susceptibility versus protection in inflammatory diseases by determining the cytokine profile. J Immunol 2013; 190:513–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Karim R, Mack WJ, Stiller T, et al. Association of HIV clinical disease progression with profiles of early immune activation: results from a cluster analysis approach. AIDS 2013; 27:1473–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chahroudi A, Bosinger SE, Vanderford TH, Paiardini M, Silvestri G. Natural SIV hosts: showing AIDS the door. Science 2012; 335:1188–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sidney J, Southwood S, Pasquetto V, Sette A. Simultaneous prediction of binding capacity for multiple molecules of the HLA B44 supertype. J Immunol 2003; 171:5964–74. [DOI] [PubMed] [Google Scholar]
- 37. Lenz TL, Deutsch AJ, Han B, et al. Widespread non-additive and interaction effects within HLA loci modulate the risk of autoimmune diseases. Nat Genet 2015; 47:1085–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Healy BC, Liguori M, Tran D, et al. HLA B*44: protective effects in MS susceptibility and MRI outcome measures. Neurology 2010; 75:634–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Valdes AM, Erlich HA, Noble JA. Human leukocyte antigen class I B and C loci contribute to type 1 diabetes (T1D) susceptibility and age at T1D onset. Hum Immunol 2005; 66:301–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ovsyannikova IG, Jacobson RM, Vierkant RA, Pankratz VS, Poland GA. HLA supertypes and immune responses to measles-mumps-rubella viral vaccine: findings and implications for vaccine design. Vaccine 2007; 25:3090–100. [DOI] [PubMed] [Google Scholar]
- 41. Hickman HD, Luis AD, Buchli R, et al. Toward a definition of self: proteomic evaluation of the class I peptide repertoire. J Immunol 2004; 172:2944–52. [DOI] [PubMed] [Google Scholar]
- 42. Münz C, Lünemann JD, Getts MT, Miller SD. Antiviral immune responses: triggers of or triggered by autoimmunity? Nat Rev Immunol 2009; 9:246–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Höhler T, Gerken G, Notghi A, et al. HLA-DRB1*1301 and *1302 protect against chronic hepatitis B. J Hepatol 1997; 26:503–7. [DOI] [PubMed] [Google Scholar]
- 44. Julg B, Moodley ES, Qi Y, et al. Possession of HLA class II DRB1*1303 associates with reduced viral loads in chronic HIV-1 clade C and B infection. J Infect Dis 2011; 203:803–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Thursz MR, Kwiatkowski D, Allsopp CE, Greenwood BM, Thomas HC, Hill AV. Association between an MHC class II allele and clearance of hepatitis B virus in the Gambia. N Engl J Med 1995; 332:1065–9. [DOI] [PubMed] [Google Scholar]
- 46. Wakamatsu TH, Ueta M, Tokunaga K, et al. Human leukocyte antigen class I genes associated with Stevens-Johnson syndrome and severe ocular complications following use of cold medicine in a Brazilian population. JAMA Ophthalmol 2017; 135:355–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Duggal P, Thio CL, Wojcik GL, et al. Genome-wide association study of spontaneous resolution of hepatitis C virus infection: data from multiple cohorts. Ann Intern Med 2013; 158:235–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Paris R, Bejrachandra S, Thongcharoen P, et al. ; Thai AIDS Vaccine Evaluation Group HLA class II restriction of HIV-1 clade-specific neutralizing antibody responses in ethnic Thai recipients of the RV144 prime-boost vaccine combination of ALVAC-HIV and AIDSVAX(®) B/E. Vaccine 2012; 30:832–6. [DOI] [PubMed] [Google Scholar]
- 49. Rallón N, Restrepo C, Vicario JL, et al. Human leucocyte antigen (HLA)-DQB1*03:02 and HLA-A*02:01 have opposite patterns in their effects on susceptibility to HIV infection. HIV Med 2017; 18:587–94. [DOI] [PubMed] [Google Scholar]
- 50. Schindler M, Würfl S, Benaroch P, et al. Down-modulation of mature major histocompatibility complex class II and up-regulation of invariant chain cell surface expression are well-conserved functions of human and simian immunodeficiency virus nef alleles. J Virol 2003; 77:10548–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.