Abstract
Glycopeptide antibiotics (GPAs) are considered drugs of “last resort” for the treatment of life-threatening infections caused by relevant Gram-positive pathogens (enterococci, staphylococci, and clostridia). Driven by the issue of the never-stopping evolution of bacterial antibiotic resistance, research on GPA biosynthesis and resistance is developing fast in modern “post-genomic” era. It is today widely accepted that resistance mechanisms emerging in pathogens have been acquired from the soil-dwelling antibiotic-producing actinomycetes, which use them to avoid suicide during production, rather than being orchestrated de novo by pathogen bacteria upon continued treatment. Actually, more and more genomes of GPA producers are being unraveled, carrying a broad collection of differently arranged GPA resistance (named van) genes. In the producer actinomycetes, van genes are generally associated with the antibiotic biosynthetic gene clusters (BGCs) deputed to GPA biosynthesis, being probably transferred/arranged together, favoring a possible co-regulation between antibiotic production and self-resistance. GPA BGC-associated van genes have been also found mining public databases of bacterial genomic and metagenomic sequences. Interestingly, some BGCs for antibiotics, seemingly unrelated to GPAs (e.g., feglymycin), carry van gene homologues. Herein, we would like to cover the recent advances on the distribution of GPA resistance genes in genomic and metagenomics datasets related to GPA potential/proved producer microorganisms. A thorough understanding of GPA resistance in the producing microorganisms may prove useful in the future surveillance of emerging mechanisms of resistance to this clinically relevant antibiotic class.
Keywords: antimicrobial resistance, glycopeptide antibiotics, van genes, glycopeptide producers, biosynthetic gene clusters
Gpa Mode of Action and Resistance Genes in Gram-Positive Pathogens
According to a recent report (WHO, 2017), drug-resistant infections will kill more people than cancer in just over three decades: by 2050, 10 million people are going to die every year due to antimicrobial resistance (AMR). Consequently, it is mandatory to stimulate discovery and development of novel antibiotics to counteract AMR (O’Neill, 2016). Glycopeptide antibiotics (GPAs) are frequently used to treat life-threatening infections caused by multidrug-resistant Gram-positive pathogens, such as Staphylococcus aureus, Enterococcus spp., and Clostridium difficile (for a review on their discovery and development, see Marcone et al., 2018; on their antimicrobial activity and clinical use, Zeng et al., 2016). GPAs inhibit bacterial cell wall synthesis in Gram-positive bacteria by binding to d-alanyl-d-alanine (d-Ala-d-Ala) dipeptide terminus of peptidoglycan (PG) precursors, sequestering the substrate from transpeptidation and transglycosylation reactions in the late extracellular stages of PG cross-linking. Thus, GPA action ultimately results in destabilizing cell wall integrity, causing bacterial cell death (Perkins and Nieto, 1974). Gram-negative microorganisms are intrinsically resistant to GPAs, because of their outer membrane, which prevents these molecules entering into the periplasm. In Gram-positive bacteria, the onset of vancomycin resistance was long-delayed in comparison to other antibiotic classes. The first vancomycin-resistant clinical isolate – an Enterococcus faecium strain – was reported in 1987, more than 30 years after the clinical introduction of vancomycin (Leclercq et al., 1988, Miller et al., 2016). Unfortunately, today a vast majority of E. faecium isolates harbor vancomycin resistance genes (van) (Vehreschild et al., 2019). The first vancomycin-resistant S. aureus (VRSA) isolate was reported in 2002 as a result of horizontal gene transfer from resistant enterococci (Bartley, 2002; Weigel et al., 2003); nowadays, 52 VRSA strains have been described worldwide (Cong et al., 2020).
The GPA resistance mechanisms in Gram-positive pathogens were intensively studied starting from the pioneering work published in the 1990s (Arthur et al., 1992, 1996). Gram-positive pathogens escape GPA action by reprogramming PG precursor biosynthesis, replacing the terminal d-Ala with d-lactate (d-Ala-d-Lac) or d-serine (d-Ala-d-Ser), thus reducing the affinity for cellular targets (Arthur et al., 1992, 1996; Courvalin, 2006). In enterococci, many different GPA-resistant phenotypes have been described according to their van gene operon organization (for a review, see Binda et al., 2014): in vanA, vanB, vanD, and vanM the key ligase determines the replacement of the terminus d-Ala with d-Lac, whereas in vanC, vanE, vanG, vanL, and vanN d-Ala. The d-Ala-d-Lac-type operons are located either on plasmids or on chromosomes, whereas the d-Ala-d-Ser-type ones are exclusively on the bacterial chromosome, except the case of vanN found on a plasmid in E. faecium. Operon expression could be inducible by GPAs (vanA, vanB, vanG, vanE, vanL, and vanM) or constitutive (vanC, vanD, and vanN) (Reynolds and Courvalin, 2005; Depardieu et al., 2007; Binda et al., 2014). The most clinically relevant manifestation of GPA resistance occurs in VanA enterococci and staphylococci, and in VanB enterococci. The first group is highly resistant to both vancomycin and teicoplanin, whereas the second group only to vancomycin. In both of them, resistance is mediated by the GPA-induced expression of the transposone-located vanHAX gene operon under the transcriptional control of the VanR/VanS two-component system (TCS). VanS is a membrane-associated sensor that in VanA bacteria is activated by the presence of either vancomycin or teicoplanin, whereas in VanB it is activated only by vancomycin. Consequently, VanB enterococci are sensitive to teicoplanin (Arthur et al., 1997, 1999; Arthur and Quintiliani, 2001). Activated VanS transfers a phosphoryl group to VanR, which is the response regulator that controls the co-transcription of the vanH, vanA, vanX, and vanY genes (Wright et al., 1993; Arthur et al., 1997, 1999; Arthur and Quintilliani, 2001). VanH is a dehydrogenase that reduces pyruvate to d-lactate; VanA is the key ligase that catalyzes the formation of the d-Ala-d-Lac resistant depsipeptide (Bugg et al., 1991; Arthur et al., 1992); VanX is a d,d-dipeptidase, which removes the intracellular pool of d-Ala-d-Ala produced by the native enterococcal ligase, ensuring that d-Ala-d-Lac is incorporated into PG precursors (Reynolds et al., 1994; Wu et al., 1995); and finally VanY has an ancillary role as a d,d-carboxypeptidase cleaving the last d-Ala from the residual pentapeptide PG precursors terminating in d-Ala-d-Ala (Arthur et al., 1998). Among the d-Ala-d-Ser-type operons, the better investigated was the vanC. It encodes for a racemase (VanT) that converts l-Ser to d-Ser, a ligase (VanC) that synthesizes d-Ala-d-Ser, and a bi-functional d,d-dipeptidase/d,d-carboxypeptidase (VanXYc) that cleaves the residual pools of d-Ala-d-Ala (Billot-Klein et al., 1994; Reynolds and Courvalin, 2005). In VanC phenotype, the TCS VanRcSc is located downstream the operon, but the resistance is constitutive due to mutations in the sensor VanSc (Healy et al., 2000; Hong et al., 2008; Koteva et al., 2010). VanC enterococci are intrinsically resistant to low levels of vancomycin, although they remain sensitive to teicoplanin.
Additional variants of these van gene operons were found in other Gram-positive pathogens including Listeria spp., streptococci, clostridia (Biavasco et al., 1996; Poyart et al., 1997; Peltier et al., 2013), and also in nonpathogenic Gram-positives, including Bacillus circulans, Oerskovia spp., Corynebacterium spp., and Streptomyces coelicolor (Power et al., 1995; Fontana et al., 1997; Hong et al., 2004). A novel vancomycin vanF operon (vanYFZFHFFXF) was described in Paenibacillus popilliae, an environmental bacteria used as biopesticide to counteract beetle larvae that caused milky disease in Japan (Patel et al., 2000; Ahmed and Baptiste, 2018). The dissemination of GPA resistance more recently reached zoonotic pathogens such as the emergent Streptococcus suis, where the low level of vancomycin-resistance is due to the presence of a vanG-like operon (Huang et al., 2018). Herein, we focus our attention on van genes distribution and function in the GPA-producing actinomycetes, which are considered the putative primary source of the variety of GPA-resistant determinants occurring in environmental bacteria and pathogens (Marshall et al., 1998; Beltrametti et al., 2007; Marcone et al., 2010, 2014; Schäberle et al., 2011).
Updating The Glycopeptide Resistance Paradigm For The Gpa-Producing Strains: Van Genes And Their Organization In Known And Putative Gpa Bgcs
Actinomycetes are Gram-positive soil-dwelling bacteria, which produce about two-thirds of the naturally derived antibiotics with clinical use (Bérdy, 2012; Barka et al., 2015), including GPAs (Nicolaou et al., 1999). Clinically relevant GPAs are produced by Amycolatopsis orientalis (vancomycin), Actinoplanes teichomyceticus (teicoplanin), and Nonomuraea gerenzanensis (dalbavancin precursor – A40926) (Zeng et al., 2016; Marcone et al., 2018). GPA producers require self-resistance mechanisms to avoid suicide during antibiotic production and, like in pathogens, such resistance is due to van genes, whose description dates back to the end of the 1990s, one decade later than in pathogens (Marshall et al., 1997, 1998). Sequence and operon structure similarities of van genes between pathogens and GPA-producers are significant (Hong et al., 2008; Binda et al., 2014). The intriguing aspect is that in GPA producers, van genes are usually located within the GPA biosynthetic gene clusters (BGCs) deputed to the antibiotic biosynthesis (Pootoolal et al., 2002; Beltrametti et al., 2007; Marcone et al., 2010, 2014; Schäberle et al., 2011). In the last two decades, multiple novel GPA BGCs from actinomycetes were sequenced and annotated, and each of them (with few exceptions, see below) contains van genes (Figure 1).
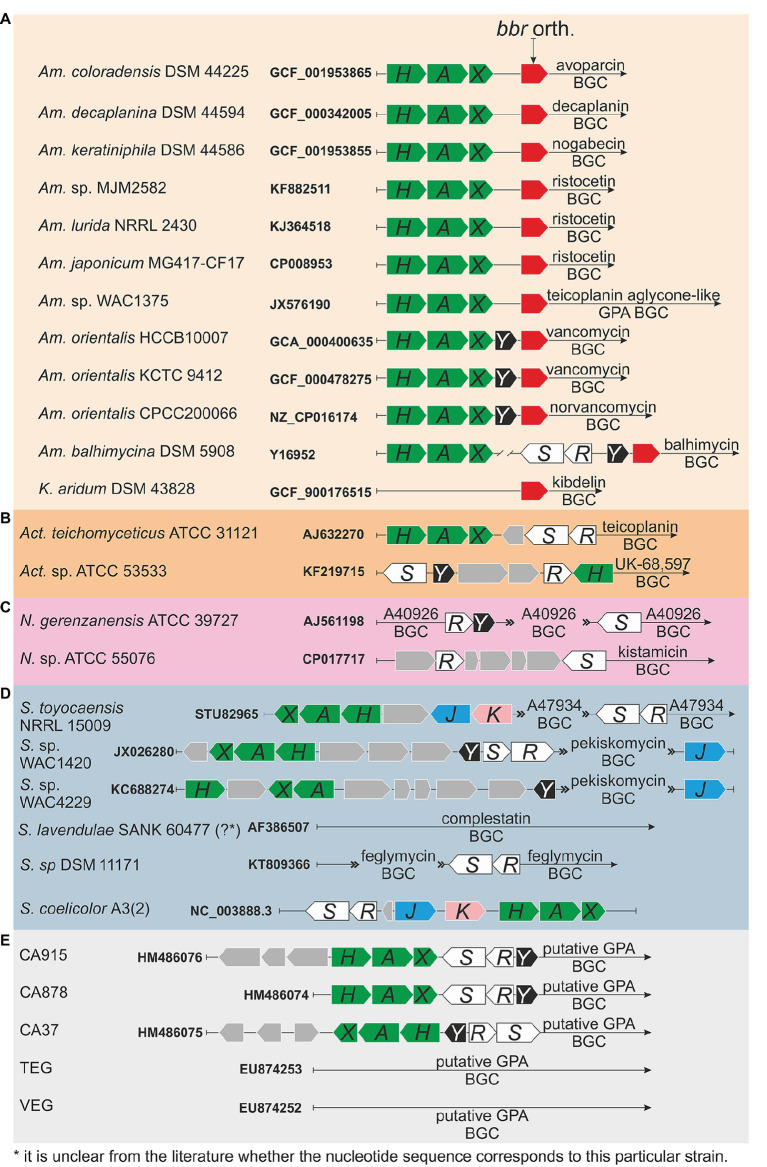
Figure 1(Continued)FIGURE 1.
Organization of van genes orthologues in glycopeptide antibiotic (GPA) biosynthetic gene clusters (BGCs) of producers and metagenomics sequences: (A) family Pseudonocardiaceae: Amycolatopsis coloradensis DSM 44225 (GCF_001953865) – avoparcin producer (Kunstmann et al., 1968; Labeda, 1995); Amycolatopsis decaplanina DSM 44594 (GCF_000342005, Kaur et al., 2013) – decaplanin producer (Sanchez et al., 1992); Amycolatopsis keratiniphila subsp. nogabecina DSM 44586 (=Amycolatopsis keratiniphila subsp. nogabecina FH1893, GCF_001953855) – nogabecin producer (Wink et al., 2003); Amycolatopsis sp. MJM2582 (KF882511, Truman et al., 2014), Amycolatopsis lurida NRRL2430 (KJ364518, Truman et al., 2014), Amycolatopsis japonicum MG417-CF17 (CP008953, Spohn et al., 2014) – ristocetin producers; Amycolatopsis sp. WAC4169 (JX576190, Thaker et al., 2013) – producer of teicoplanin aglycone-like GPA; Amycolatopsis orientalis HCCB10007 (=Am. keratiniphila, GCA_000400635, Xu et al., 2014) and Amycolatopsis orientalis KCTC 9412 (=Am. orientalis DSM 40040, GCF_000478275, Jeong et al., 2013) – vancomycin producers; Amycolatopsis orientalis CPCC200066 (=Am. orientalis B-37, NZ_CP016174, Lei et al., 2015) – norvancomycin producer; Amycolatopsis balhimycina DSM 5908 (Y16952, Shawky et al., 2007) – balhimycin producer (Nadkarni et al., 1994; Wink et al., 2003); Kibdelosporangium aridum DSM 43828 (GCF_900176515) – the producer of kibdelins (Shearer et al., 1985), (B) genus Actinoplanes (please see main text for more details), (C) genus Nonomuraea (please see main text for more details), (D) genus Streptomyces (please see main text for more details); although S. coelicolor is not a GPA producer, organization of S. coelicolor van gene orthologues is also given, and (E) metagenomics sequences (please see main text for more details).
Thus far, the majority of GPA BGCs were found in members of the genus Amycolatopsis (Adamek et al., 2018), which belongs to the Pseudonocardiaceae family. Besides the vancomycin producers, other Amycolatopsis spp. produce avoparcin, decaplanin, nogabecin, ristocetin, teicoplanin aglycone-like GPA, norvancomycin, balhimycin, and chloroeremomycin. In their corresponding BGCs, van genes were found just upstream the genes coding for the StrR-like pathway-specific regulators (orthologues of bbr from balhimycin BGC, Figure 1). We excluded from this comparison the chloroeremomycin BGC from Amycolatopsis orientalis PA-42867 (?) (van Wageningen et al., 1998), which apparently was not completely covered with sequencing. Thus, three patterns for the organization of van genes are recognizable (Figure 1) in Pseudonocardiaceae GPA producers. In the producers of avoparcin, decaplanin, nogabecin, ristocetin, and teicoplanin-like aglycone GPA, the GPA BGCs carry vanHAX orthologues, but not vanY or vanRS orthologues. In vancomycin and norvancomycin producers, vanY orthologues are clustered with vanHAX ones. Balhimycin producer – Amycolatopsis balhimycina – possesses a BGC with vanRS (vlnRAbSAb) and vanY (vanYAb) orthologues, but vanHAX orthologues (vanHAbAAbXAb) were actually found 2 kbp away from balhimycin BGC (Schäberle et al., 2011; Frasch et al., 2015). Indeed, no cluster-situated van genes were found sequencing the genome of Kibdelosporangium aridum – the producer of kibdelins (Shearer et al., 1986) – which also belongs to Pseudonocardiaceae family (Figure 1).
Other known GPA BGCs are from Actinoplanes spp. (family Micromonosporaceae) and Nonomuraea spp. (family Streptosporangiaceae). Act. teichomyceticus and Actinoplanes sp. ATCC 53533 produce teicoplanin (Bardone et al., 1978) and UK-68,597 (Skelton et al., 1990), respectively. Teicoplanin BGC (named tei) contains vanHAX and vanRS orthologues (tei7-6-5 and tei2-3, respectively) organized in two separate operons, but none vanY orthologue (Figure 1; Li et al., 2004; Yushchuk et al., 2020b). In contrast, UK-68,597 BGC contains a vanH, not contiguous vanR and vanS, and a vanY orthologue (Figure 1; Yim et al., 2014). In the genus Nonomuraea, N. gerenzanenis ATCC 39727 and Nonomuraea sp. ATCC 55076 produce A40926 (Goldstein et al., 1987) and the type V glycopeptide kistamicin (Naruse et al., 1993), respectively. In A40926 BGC (named dbv) (Sosio et al., 2003), vanR and vanS homologues (dbv6 and dbv22, respectively) are not contiguous and GPA resistance is due to the expression of vanY orthologue (dbv7, Figure 1; Marcone et al., 2010, 2014; Binda et al., 2012; ). No vanHAXY genes are present in kistamicin BGC, although kistamicin BGC contains homologues of vanS and vanR named kisG and kisB (Nazari et al., 2017).
GPAs are also produced by few Streptomyces species (Figure 1). Interestingly, functional van genes were also found in S. coelicolor, which is not a GPA producer (Hong et al., 2004). A47934 BGC from Streptomyces toyocaensis NRRL 15009 contains vanHStAStXSt and vanRStSSt operons, together with staO and staP orthologues to S. coelicolor vanJ and vanK, respectively (Pootoolal et al., 2002). Pekiskomycin BGC from Streptomyces sp. WAC1420 contains vanY, vanJ as well as vanHAX and vanRS homologues, but pekiskomycin BGC from Streptomyces sp. WAC4229 lacks vanRS homologues (Thaker et al., 2013). No homologues of van genes were found in complestatin (type V GPA) BGC from S. lavendulae SANK 60477 (?) (Chiu et al., 2001), although this antibiotic possesses a moderate antibacterial activity. However, complestatin was shown to inhibit the fatty acid biosynthesis in Gram-positive bacteria (Kwon et al., 2015), therefore the producer may require no cell wall remodeling for complestatin self-resistance. Finally, feglymycin BGC from Streptomyces sp. DSM11171 (Figure 1) encodes for a 13-mer peptide antibiotic acting on bacterial cell wall biosynthesis by inhibiting MurA and MurC. Albeit the structure and the mode of action of feglymycin differs from the ones of GPAs, feglymycin BGC shares a high level of similarity with GPA BGCs (Gonsior et al., 2015; Yushchuk et al., 2020a), including the presence of vanRS-like genes – fegM and fegN.
To conclude, CA915, CA37, and CA878 GPA BGCs (Banik et al., 2010), which were sequenced from metagenomics samples, contain vanHAX, vanY and vanRS homologues, whereas none van gene was found in other metagenome-derived GPA BGCs as TEG and VEG (Banik and Brady, 2008; Figure 1).
Updating On What Is Known About The In Vivo Function Of Van Genes In Gpa-Producing Strains
Although van genes were found in multiple GPA BGCs, only for few of them the function was experimentally proven. Balhimycin resistance in Am. balhimycina is likely the most deeply investigated model among GPA producers (Figure 2A). vanHAbAAbXAb, that is located outside the BGC (Figure 1), was shown to be constitutively expressed through all the periods of growth and during balhimycin production (Schäberle et al., 2011). Deletion of vanHAbAAbXAb genes makes Am. balhimycina significantly more sensitive to its own product, decreasing its MIC from 5 to 0.25 mg/ml, and causing an earlier expression of the BGC-situated vanYAb (Frasch et al., 2015). However, vanYAb itself does not play a decisive role in GPA-resistance since its deletion did not alter the GPA resistance phenotype (Frasch et al., 2015). Double vanHAbAAbXAb and vanYAb knocked-out mutants showed the same GPA resistance phenotype as ΔvanHAbAAbXAb. PG precursors ending in d-Ala-d-Lac were still found in the single ΔvanHAbAAbXAb and in the double ΔvanHAbAAbXAb ΔvanYAb mutants together with d-Ala-d-Ala ending PG precursors and tetrapeptides (Frasch et al., 2015). The residual GPA resistance in these mutants is probably due to an accessory Ddl1Ab, a putative d-Ala-d-Lac ligase encoded in the genome of Am. balhimycina, which shares 72% of amino acid sequence identity with VanAAb (Frasch et al., 2015). Ddl1Ab might add d-Lac to the tetrapeptide PG precursors generated by the d,d-carboxypetidase VanYAb (although the presence of some other d-Ala-d-Ala carboxypeptidases encoded in the genome cannot be completely ruled out considering the resistant phenotype of the ΔvanYAb mutant). In the absence of VanHAb, d-Lac for this reaction is probably obtained from the primary metabolic pool. Expression of vanHAbAAbXAb was demonstrated to be independent from the BGC-situated regulator vlnRAb (Kilian et al., 2016). However, VlnRAb is important for the activation of the BGC-situated vanYAb expression (Kilian et al., 2016). Heterologous expression of vlnRAbSAb in S. coelicolor ΔvanRS mutants indicated that both VlnRAb and VlnSAb are active and able to replace their counterparts VanR and VanS, which in S. coelicolor control the expression of vanHAX genes in response to vancomycin (Hong et al., 2004), restoring resistance to both balhimycin and teicoplanin in the complemented strains (Kilian et al., 2016). Overall, it seems that the BGC-situated vlnRAbSAb-vanYAb regulatory circuit is functional, but does not play a major role in balhimycin resistance, which is mostly determined by vanHAbAAbXAb expression. It would be interesting to test GPA resistance in ddl1Ab knocked-out mutant generated in Am. balhimycina ΔvanHAbAAbXAb ΔvanYAb to better understand the role of this accessory ligase and its connection with the d,d-carboxypetidase activity of VanYAb (or of some other still-unknown carboxypeptidases).
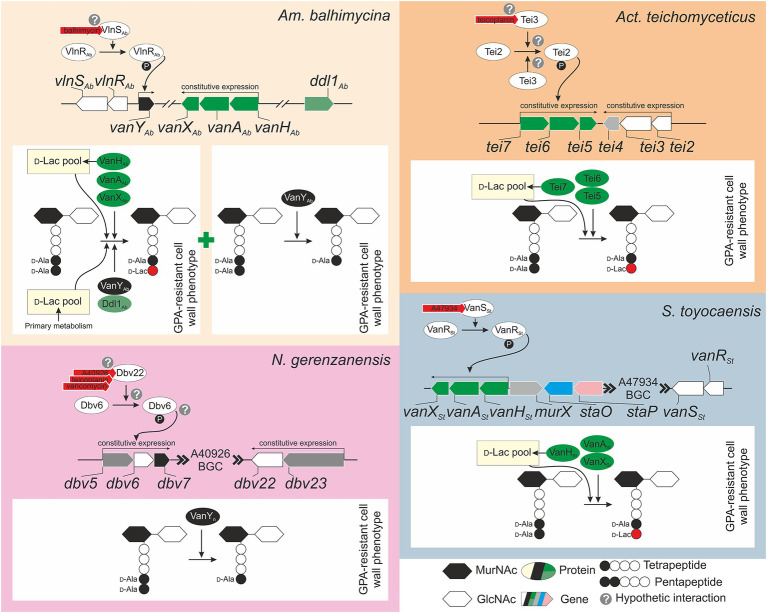
Figure 2.
Knowledge-based conceptual schemes for the GPA self-resistance mechanisms and their regulation in model GPA-producers: (A) Am. balhimycina, the producer of balhimycin, (B) Act. teichomyceticus, the producer of teicoplanin, (C) N. gerenzanensis, the producer of A40926, and (D) S. toyocaensis, the producer of A47934. Please refer to main text for more details on genes involved in GPA self-resistance. Please note that tei4 in Act. teichomyceticus, dbv5 and dbv23 in N. gerenzanensis, and murX in S. toyocaensis, colored in light gray, are not van gene orthologues, but they are reported since they are close to van genes or part of their operons.
Differently from Am. balhimycina, in Act. teichomyceticus vanHAX orthologues – tei7-6-5 – are located within the tei BGC together with vanRS orthologues – tei2-3 (Figure 2B). tei7-6-5 expression determines the production of PG precursors ending in d-Ala-d-Lac, conferring a GPA-resistant phenotype to Act. teichomyceticus (Beltrametti et al., 2007; Binda et al., 2018). Interestingly, the expression of tei7-6-5 operon is constant during the growth curve and in teicoplanin production conditions (Beltrametti et al., 2007; Yushchuk et al., 2019) and the VanX d,d-dipeptidase activity was detectable in cellular extracts independently from the addition of teicoplanin (Binda et al., 2018). One probable reason for the constitutive expression of tei7-6-5 is the non-inducibility of the sensor histidine kinase Tei3, due to its point mutations previously known to confer a constitutive kinase activity to S. coelicolor VanS (Beltrametti et al., 2007). Also, the expression of vanRS orthologues – tei2-3 – was also found constitutive under teicoplanin production conditions (Yushchuk et al., 2019) and these genes are co-expressed with tei4 – coding for a dehydrofolate reductase with no obvious role in teicoplanin-resistance (Yushchuk et al., 2020b). Moreover, tei2-3-4 expression is independent from tei cluster-encoded transcriptional regulators – Tei15* and Tei16* (Yushchuk et al., 2019, 2020b). Constitutive expression of tei2-3-4 could be granted by tei2 promoter, which was shown to be highly active in Act. teichomyceticus, starting from the very early stage of spore germination (Yushchuk et al., 2020b). More investigations are required for a complete understanding of teicoplanin-resistance in Act. teichomyceticus. Study of the Tei3 properties is among the most interesting tasks.
In N. gerenzanensis producing the teicoplanin-like A40926, vanHAX orthologues were not found neither in the BGC nor in the genome (D’Argenio et al., 2016). The only known mechanism of resistance relies on the action of VanYn, whose coding gene (dbv7) is within the dbv BGC (Figure 2C) and whose knockout abolishes the resistance phenotype (Marcone et al., 2010, 2014; Figure 2C). VanYn is a d,d-carboxypeptidase that cleaves the last d-Ala from pentapeptide PG precursors generating tetrapeptides, drastically reducing GPA affinity for cellular targets (Binda et al., 2012). A l,d-transpeptidase (Ldt) then uses the tetrapeptide acyl donors supplied by VanYn to synthetize the mature cell wall (Hugonnet et al., 2014). The role of this protein and its features that assimilate/distinguish it from enterococcal VanY and from VanYAb were investigated in detail (Marcone et al., 2010, 2014; Binda et al., 2012, 2013). Less clear is the regulatory circuit governing dbv7 expression. Direct VanYn carboxypeptidase activity measurement in N. gerenzanensis growing with the addition of different GPAs, unambiguously showed that VanYn activity is induced by vancomycin, teicoplanin, and A40926 (Binda et al., 2018). vanRS homologues – dbv6 and dbv22 – are present in the dbv BGC, but the knockout of dbv6 did not exert any influence on A40926 production and growth of N. gerenzanensis (Lo Grasso et al., 2015). Unfortunately, the GPA resistance phenotype of this mutant was not described. On the other side, transcriptional analysis of dbv genes indicated that the expression of dbv5-6-7 and dbv23-22 operons is rather constitutive (Alduina et al., 2007). Although the presence of other GPA-sensitive TCS beyond the borders of A40926 BGC cannot be ruled out, role of Dbv6 and Dbv22 in N. gerenzanensis A40926 self-resistance merits further investigations.
Finally, S. toyocaensis possesses, perhaps, the most straightforward resistance mechanism among all the investigated GPA producers (Figure 2D), reminding the situation in S. coelicolor (Hong et al., 2004). The BGC-located vanHStAStXSt operon was shown to be crucial for A47934 resistance and vanASt knockout made S. toyocaensis completely sensitive to A47934 (Pootoolal et al., 2002). At the same time, vanHAX-genes from the vancomycin producer Am. orientalis C329.2 were able to restore A47934 resistance phenotype in the knocked-out mutant (Pootoolal et al., 2002). Functions of VanRSt and VanSSt (both present in the A47934 BGC, Figures 1, 2D) were also studied in detail, showing that VanSSt has a remarkable specificity for A47934 and it is unable to sense teicoplanin or vancomycin (Koteva et al., 2010; Novotna et al., 2016). Moreover, also the interaction between VanRSt and VanSSt was found to be very specific, since VanRSt could not be phosphorylated by a non-cognate sensor-histidine kinase (Novotna et al., 2016). The roles (if there are any) of staP and staO (orthologues of S. coelicolor vanK and vanJ) in S. toyocaensis A47934 self-resistance were not investigated, thus the importance of these auxiliary resistance genes remains to be proved.
Outlook
Soil GPA producers are considered the putative source of GPA resistance determinants, which might had been recruited and differently combined in pathogens. The goal of this mini review is to update the knowledge on the occurrence and role of van genes in producing microorganisms. It emerges that more in silico, in vitro, and in vivo investigations on their function and regulation are required to shed light on the intriguing issue of their origin and role. Overall, a detailed phylogenetic analysis would be useful to illuminate the evolution of GPA-resistant determinants in GPA producers and from them to pathogens. A recent pioneering work on the reconstruction of GPA BGC phylogeny (Waglechner et al., 2019) reported on the possible origin and evolution of GPA cluster-situated van-genes. According to these authors, vanA had likely originated within Amycolatopsis genus, whereas vanH, vanX, and vanRS within Actinoplanes; and vanY probably originated within genus Nonomuraea and it was then distributed among GPA BGCs by multiple transfer events. Combination of these genes in pathogens is today determining the urgent clinical need for new drugs to combat multi-drug resistant Gram-positive pathogens.
Author Contributions
EB and OY collected data and papers and co-wrote the review. OY prepared the figures. FM and EB supervised the work.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Consorzio Italbiotec for supporting EB.
Footnotes
Funding. This work was supported by grant “Fondo di Ateneo per la Ricerca” 2017, 2018 to FM, and Consorzio Italbiotec to EB.
References
- Adamek M., Alanjary M., Sales-Ortells H., Goodfellow M., Bull A. T., Winkler A., et al. (2018). Comparative genomics reveals phylogenetic distribution patterns of secondary metabolites in Amycolatopsis species. BMC Genomics 19:426. 10.1186/s12864-018-4809-4, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed M. O., Baptiste K. E. (2018). Vancomycin-resistant enterococci: a review of antimicrobial resistance mechanisms and perspectives of human and animal health. Microb. Drug Resist. 24, 590–606. 10.1089/mdr.2017.0147, PMID: [DOI] [PubMed] [Google Scholar]
- Alduina R., Piccolo L. L., Dalia D., Ferraro C., Gunnarsson N., Donadio S., et al. (2007). Phosphate-controlled regulator for the biosynthesis of the dalbavancin precursor A40926. J. Bacteriol. 189, 8120–8129. 10.1128/JB.01247-07, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthur M., Depardieu F., Cabanié L., Reynolds P., Courvalin P. (1998). Requirement of the VanY and VanX d,d-peptidases for glycopeptide resistance in enterococci. Mol. Mirobiol. 30, 819–830. 10.1046/j.1365-2958.1998.01114.x, PMID: [DOI] [PubMed] [Google Scholar]
- Arthur M., Depardieu F., Courvalin P. (1999). Regulated interactions between partner and non-partner sensors and response regulators that control glycopeptide resistance gene expression in enterococci. Microbiologica 145, 1849–1858. 10.1099/13500872-145-8-1849, PMID: [DOI] [PubMed] [Google Scholar]
- Arthur M., Depardieu F., Gerbaud G., Galimand M., Leclercq R., Courvalin P. (1997). The VanS sensor negatively controls VanR-mediated transcriptional activation of glycopeptide resistance genes of Tn1546 and related elements in the absence of induction. J. Bacteriol. 179, 97–106. 10.1128/JB.179.1.97-106.1997, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthur M., Molinas C., Bugg T. D., Wright G. D., Walsh C. T., Courvalin P. (1992). Evidence for in vivo incorporation of d-lactate into peptidoglycan precursors of vancomycin-resistant enterococci. Antimicrob. Agents Chemother. 36, 867–869. 10.1128/aac.36.4.867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthur M., Quintiliani R. (2001). Regulation of VanA‐ and VanB-type glycopeptide resistance in enterococci. Antimicrob. Agents Chemother. 45, 375–381. 10.1128/aac.45.2.375-381.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthur M., Reynolds P., Courvalin P. (1996). Glycopeptide resistance in enterococci. Trends Microbiol. 4, 401–407. 10.1016/0966-842X(96)10063-9, PMID: [DOI] [PubMed] [Google Scholar]
- Banik J. J., Brady S. F. (2008). Cloning and characterization of new glycopeptide gene clusters found in an environmental DNA megalibrary. Proc. Natl. Acad. Sci. U. S. A. 105, 17273–17277. 10.1073/pnas.0807564105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banik J. J., Craig J. W., Calle P. Y., Brady S. F. (2010). Tailoring enzyme-rich environmental DNA clones: a source of enzymes for generating libraries of unnatural natural products. J. Am. Chem. Soc. 132, 15661–15670. 10.1021/ja105825a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardone M. R., Paternoster M., Coronelli C. (1978). Teichomycins, new antibiotics from Actinoplanes teichomyceticus nov. sp. II. Extraction and chemical characterization. J. Antibiot. 31, 170–177. 10.7164/antibiotics.31.170, PMID: [DOI] [PubMed] [Google Scholar]
- Barka E. A., Vatsa P., Sanchez L., Gavetas-Vaillant N., Jacquard C., Meier-Kolthoff J. P., et al. (2015). Taxonomy, physiology, and natural products of Actinobacteria. Microbiol. Mol. Biol. Rev. 80, 1–43. 10.1128/MMBR.00019-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartley J. (2002). First case of VRSA identified in Michigan. Infect. Control Hosp. Epidemiol. 23:480. 10.1017/s0195941700082333, PMID: [DOI] [PubMed] [Google Scholar]
- Beltrametti F., Consolandi A., Carrano L., Bagatin F., Rossi R., Leoni L., et al. (2007). Resistance to glycopeptide antibiotics in the teicoplanin producer is mediated by van gene homologue expression directing the synthesis of a modified cell wall peptidoglycan. Antimicrob. Agents Chemother. 51, 1135–1141. 10.1128/AAC.01071-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bérdy J. (2012). Thoughts and facts about antibiotics: where we are now and where we are heading. J. Antibiot. 65, 385–395. 10.1038/ja.2012.27, PMID: [DOI] [PubMed] [Google Scholar]
- Biavasco F., Giovanetti E., Miele A., Vignaroli C., Facinelli B., Varaldo P. E. (1996). In vitro conjugative transfer of VanA vancomycin resistance between Enterococci and Listeriae of different species. Eur. J. Clin. Microbiol. Infect. Dis. 15, 50–59. 10.1007/BF01586185, PMID: [DOI] [PubMed] [Google Scholar]
- Billot-Klein D., Blanot D., Gutmann L., van Heijenoort J. (1994). Association constants for the binding of vancomycin and teicoplanin to N-acetyl-d-alanyl-d-alanine and N-acetyl-d-alanyl-d-serine. Biochem. J. 304, 1021–1022. 10.1042/bj3041021, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binda E., Cappelletti P., Marinelli F., Marcone G. L. (2018). Specificity of induction of glycopeptide antibiotic resistance in the producing actinomycetes. Antibiotics 7, 36–46. 10.3390/antibiotics7020036, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binda E., Marcone G. L., Berini F., Pollegioni L., Marinelli F. (2013). Streptomyces spp. as efficient expression system for a d,d-peptidase/d,d-carboxypeptidase involved in glycopeptide antibiotic resistance. BMC Biotechnol. 13:24. 10.1186/1472-6750-13-24, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binda E., Marinelli F., Marcone G. L. (2014). Old and new glycopeptide antibiotics: action and resistance. Antibiotics 3, 572–594. 10.3390/antibiotics3040572, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binda E., Marcone G. L., Pollegioni L., Marinelli F. (2012). Characterization of VanYn, a novel 485 d,d-peptidase/d,d-carboxypeptidase involved in glycopeptide antibiotic resistance in Nonomuraea sp. ATCC 39727. FEBS J. 279, 3203–3213. 10.1111/j.1742-4874658.2012.08706.x, PMID: [DOI] [PubMed] [Google Scholar]
- Bugg T. D., Wright G. D., Dutka-Malen S., Arthur M., Courvalin P., Walsh C. T. (1991). Molecular basis for vancomycin resistance in Enterococcus faecium BM4147: biosynthesis of a depsipeptide peptidoglycan precursor by vancomycin resistance proteins VanH and VanA. Biochemist 30, 10408–10415. 10.1021/bi00107a007, PMID: [DOI] [PubMed] [Google Scholar]
- Chiu H.-T., Hubbard B. K., Shah A. N., Eide J., Fredenburg R. A., Walsh C. T., et al. (2001). Molecular cloning and sequence analysis of the complestatin biosynthetic gene cluster. Proc. Natl. Acad. Sci. U. S. A. 98, 8548–8553. 10.1073/pnas.151246498, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong Y., Yang S., Rao X. (2020). Vancomycin resistant Staphylococcus aureus infections: a review of case updating and clinical features. J. Adv. Res. 21, 169–176. 10.1016/j.jare.2019.10.005, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courvalin P. (2006). Vancomycin resistance in gram-positive cocci. Clin. Infect. Dis. 42, S25–S34. 10.1086/491711, PMID: [DOI] [PubMed] [Google Scholar]
- D’Argenio V., Petrillo M., Pasanisi D., Pagliarulo C., Colicchio R., Talà A., et al. (2016). The complete 12Mb genome and transcriptome of Nonomuraea gerenzanensis with new insights into its duplicated “magic” RNA polymerase. Sci. Rep. 6, 1–13. 10.1038/s41598-016-0025-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depardieu F., Podglajen I., Leclercq R., Collatz E., Courvalin P. (2007). Modes and modulations of antibiotic resistance gene expression. Clin. Microbiol. Rev. 20, 79–114. 10.1128/CMR.00015-06, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontana R., Ligozzi M., Pedrotti C., Padovani E. M., Cornaglia G. C. (1997). Vancomycin-resistant Bacillus circulans carrying the vanA gene responsible for vancomycin resistance in enterococci. Eur. J. Clin. Microbiol. Infect. Dis. 16, 473–474. 10.1007/BF02471915, PMID: [DOI] [PubMed] [Google Scholar]
- Frasch H.-J., Kalan L., Kilian R., Martin T., Wright G. D., Stegmann E. (2015). Alternative pathway to a glycopeptide-resistant cell wall in the balhimycin producer Amycolatopsis balhimycina. ACS Infect. Dis. 1, 243–252. 10.1021/acsinfecdis.5b00011, PMID: [DOI] [PubMed] [Google Scholar]
- Goldstein B. P., Selva E., Gastaldo L., Berti M., Pallanza R., Ripamonti F., et al. (1987). A40926, a new glycopeptide antibiotic with anti-Neisseria activity. Antimicrob. Agents Chemother. 31, 1961–1966. 10.1128/aac.31.12.1961, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonsior M., Mühlenweg A., Tietzmann M., Rausch S., Poch A., Süssmuth R. D. (2015). Biosynthesis of the peptide antibiotic feglymycin by a linear nonribosomal peptide synthetase mechanism. Chembiochem 16, 2610–2614. 10.1002/cbic.201500432, PMID: [DOI] [PubMed] [Google Scholar]
- Healy V. L., Lessard I. A., Roper D. I., Knox J. R., Walsh C. T. (2000). Vancomycin resistance in enterococci: reprogramming of the d-ala-d-ala ligases in bacterial peptidoglycan biosynthesis. Chem. Biol. 7, R109–R119. 10.1016/S1074-5521(00)00116-2, PMID: [DOI] [PubMed] [Google Scholar]
- Hong H. J., Hutchings M. I., Buttner M. J. (2008). Vancomycin resistance VanS/VanR two-component systems. Adv. Exp. Med. Biol. 631, 200–213. 10.1007/978-0-387-78885-2_14 [DOI] [PubMed] [Google Scholar]
- Hong H. J., Hutchings M. I., Neu J. M., Wright G. D., Paget M. S. B., Buttner M. J. (2004). Characterization of an inducible vancomycin resistance system in Streptomyces coelicolor reveals a novel gene (vanK) required for drug resistance. Mol. Microbiol. 52, 1107–1121. 10.1111/j.1365-2958.2004.04032.x, PMID: [DOI] [PubMed] [Google Scholar]
- Huang J., Chen L., Li D., Wang M., Du F., Gao Y., et al. (2018). Emergence of a vanG-n carrying and multidrug resistant ICE in zoonotic pathogen Streptococcus suis. Vet. Microbiol. 222, 109–113. 10.1016/j.vetmic.2018.07.008, PMID: [DOI] [PubMed] [Google Scholar]
- Hugonnet J.-E., Haddache N., Veckerlé C., Dubost L., Marie A., Shikura N., et al. (2014). Peptidoglycan cross-linking in glycopeptide-resistant actinomycetales. Antimicrob. Agents Chemother. 58, 1749–1756. 10.1128/aac.02329-13, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong H., Sim Y. M., Kim H. J., Lee D.-W., Lim S.-K., Lee S. J. (2013). Genome sequence of the vancomycin-producing Amycolatopsis orientalis subsp. orientalis strain KCTC 9412T. Genome Announc. 1, e00408–e00413. 10.1128/genomea.00408-13, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur N., Kumar S., Bala M., Raghava G. P. S., Mayilraj S. (2013). Draft genome sequence of Amycolatopsis decaplanina strain DSM 44594T. Genome Announc. 1, e00138–e00113. 10.1128/genomea.00138-13, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilian R., Frasch H.-J., Kulik A., Wohlleben W., Stegmann E. (2016). The VanRS homologous two-component system VnlRSAb of the glycopeptide producer Amycolatopsis balhimycina activates transcription of the vanHAXSc genes in Streptomyces coelicolor, but not in A. balhimycina. Microb. Drug Resist. 22, 499–509. 10.1089/mdr.2016.0128, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koteva K., Hong H.-J., Wang X. D., Nazi I., Hughes D., Naldrett M. J., et al. (2010). A vancomycin photoprobe identifies the histidine kinase VanSsc as a vancomycin receptor. Nat. Chem. Biol. 6, 327–329. 10.1038/nchembio.350, PMID: [DOI] [PubMed] [Google Scholar]
- Kunstmann M. P., Mitscher L. A., Porter J. N., Shay A. J., Darken M. A. (1968). LL-AV290, a new antibiotic. I. Fermentation, isolation, and characterization. Antimicrob. Agents Chemother. 8, 242–245. PMID: [PubMed] [Google Scholar]
- Kwon Y.-J., Kim H.-J., Kim W.-G. (2015). Complestatin exerts antibacterial activity by the inhibition of fatty acid synthesis. Biol. Pharm. Bull. 38, 715–721. 10.1248/bpb.b14-00824, PMID: [DOI] [PubMed] [Google Scholar]
- Labeda D. P. (1995). Amycolatopsis coloradensis sp., nov., the avoparcin (LL-AV290)-producing strain. Int. J. Syst. Evol. Microbiol. 45, 124–127. 10.1099/00207713-45-1-124 [DOI] [Google Scholar]
- Leclercq R., Derlot R., Duval E. J., Courvalin P. (1988). Plasmid-mediated resistance to vancomycin and teicoplanin in Enterococcus faecium. N. Engl. J. Med. 319, 157–161. 10.1056/NEJM198807213190307, PMID: [DOI] [PubMed] [Google Scholar]
- Lei X., Yuan F., Shi Y., Li X., Wang L., Hong B. (2015). Draft genome sequence of norvancomycin-producing strain Amycolatopsis orientalis CPCC200066. Genome Announc. 3, e00296–e00215. 10.1128/genomea.00296-15, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T., Huang F., Haydock S., Mironenko T., Leadlay P., Spencer J. (2004). Biosynthetic gene cluster of the glycopeptide antibiotic teicoplanin characterization of two glycosyltransferases and the key acyltransferase. Chem. Biol. 11, 107–119. 10.1016/j.chembiol.2004.01.001, PMID: [DOI] [PubMed] [Google Scholar]
- Lo Grasso L., Maffioli S., Sosio M., Bibb M., Puglia A. M., Alduina R. (2015). Two master switch regulators trigger A40926 biosynthesis in Nonomuraea sp. strain ATCC 39727. J. Bacteriol. 197, 2536–2544. 10.1128/JB.00262-15, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcone G. L., Beltrametti F., Binda E., Carrano L., Foulston L., Hesketh A., et al. (2010). Novel mechanism of glycopeptide resistance in the A40926 producer Nonomuraea sp. ATCC 39727. Antimicrob. Agents Chemother. 54, 2465–2472. 10.1128/AAC.00106-10, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcone G. L., Binda E., Berini F., Marinelli F. (2018). Old and new glycopeptide antibiotics: from product to gene and back in the post-genomic era. Biotechnol. Adv. 36, 534–554. 10.1016/j.biotechadv.2018.02.009, PMID: [DOI] [PubMed] [Google Scholar]
- Marcone G. L., Binda E., Carrano L., Bibb M., Marinelli F. (2014). Relationship between glycopeptide production and resistance in the actinomycete Nonomuraea sp. ATCC 39727. Antimicrob. Agents Chemother. 58, 5191–5201. 10.1128/aac.02626-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall C. G., Broadhead G., Leskiw B. K., Wright G. D. (1997). d-Ala-d-Ala ligases from glycopeptide antibiotic-producing organisms are highly homologous to the enterococcal vancomycin-resistance ligases VanA and VanB. Proc. Natl. Acad. Sci. U. S. A. 94, 6480–6483. 10.1073/pnas.94.12.6480, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall C. G., Lessard I. A. D., Park I.-S., Wright G. D. (1998). Glycopeptide antibiotic resistance genes in glycopeptide-producing organisms. Antimicrob. Agents Chemother. 42, 2215–2220. 10.1128/AAC.42.9.2215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller W. R., Murray B. E., Rice L. B., Arias C. A. (2016). Vancomycin-resistant enterococci: therapeutic challenges in the 21st century. Infect. Dis. Clin. North Am. 30, 415–439. 10.1016/j.idc.2016.02.006, PMID: [DOI] [PubMed] [Google Scholar]
- Nadkarni S. R., Patel M. V., Chatterjee S., Vijayakumar E. K. S., Desikan K. R., Blumbach J., et al. (1994). Balhimycin, a new glycopeptide antibiotic produced by Amycolatopsis sp. Y-86,21022. Taxonomy, production, isolation and biological activity. J. Antibiot. 47, 334–341. 10.7164/antibiotics.47.334, PMID: [DOI] [PubMed] [Google Scholar]
- Naruse N., Oka M., Konishi M., Oki T. (1993). New antiviral antibiotics, Kistamicins A and B. II. Structure determination. J. Antibiot. 46, 1812–1818. 10.7164/antibiotics.46.1812 [DOI] [PubMed] [Google Scholar]
- Nazari B., Forneris C. C., Gibson M. I., Moon K., Schramma K. R., Seyedsayamdost M. R. (2017). Nonomuraea sp. ATCC 55076 harbours the largest actinomycete chromosome to date and the kistamicin biosynthetic gene cluster. Med. Chem. Commun. 8, 780–788. 10.1039/C6MD00637J, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolaou K. C., Boddy C. N. C., Bräse S., Winssinger N. (1999). Chemistry, biology, and medicine of the glycopeptide antibiotics. Angew. Chem. Int. Ed. Engl. 38, 2096–2152. [DOI] [PubMed] [Google Scholar]
- Novotna G. B., Kwun M. J., Hong H.-J. (2016). In vivo characterization of the activation and interaction of the VanR-VanS two-component regulatory system controlling glycopeptide antibiotic resistance in two related Streptomyces species. Antimicrob. Agents Chemother. 60, 1627–1637. 10.1128/aac.01367-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill J. (ed.). (2016). Tackling drug-resistant infections globally: final report and recommendations. The review on antimicrobial resistance. Available at: http://amr-review.org/sites/default/files/160518_Final%20paper_with%20cover.pdf
- Patel R., Piper K., Cockerill F. R., Steckelberg J. M., Yousten A. A. (2000). The biopesticide Paenibacillus popilliae has a vancomycin resistance gene cluster homologous to the enterococcal VanA vancomycin resistance gene cluster. Antimicrob. Agents Chemother. 44, 705–709. 10.1128/aac.44.3.705-709.2000, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltier J., Courtin P., Meouche E. I., Catel-Ferreira M., Chapot-Chartier M. P., Lemée L., et al. (2013). Genomic and expression analysis of the vanG-like gene cluster of Clostridium difficile. Microbiologica 159, 1510–1520. 10.1099/mic.0.065060-0, PMID: [DOI] [PubMed] [Google Scholar]
- Perkins H. R., Nieto M. (1974). The chemical basis for the action of the vancomycin group of antibiotics. Ann. N. Y. Acad. Sci. 235, 348–363. 10.1111/j.1749-6632.1974.tb43276.x, PMID: [DOI] [PubMed] [Google Scholar]
- Pootoolal J., Thomas M. G., Marshall C. G., Neu J. M., Hubbard B. K., Walsh C. T., et al. (2002). Assembling the glycopeptide antibiotic scaffold: the biosynthesis of from Streptomyces toyocaensis NRRL15009. Proc. Natl. Acad. Sci. U. S. A. 99, 8962–8967. 10.1073/pnas.102285099, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power E. G. M., Abdulla Y. H., Talsania H. G., Spice W., Aathithan S., French G. L. (1995). vanA genes in vancomycin-resistant clinical isolates of Oerskovia turbata and Arcanobacterium (Corynebacterium) haemolyticum. J. Antimicrob. Chemother. 36, 595–606. 10.1093/jac/36.4.595, PMID: [DOI] [PubMed] [Google Scholar]
- Poyart C., Pierre C., Quesne G., Pron B., Berche P., Trieu-Cuot P. (1997). Emergence of vancomycin resistance in the genus Streptococcus: characterization of a vanB transferable determinant in Streptococcus bovis. Antimicrob. Agents Chemother. 41, 24–29. PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds P. E., Courvalin P. (2005). Vancomycin resistance in enterococci due to synthesis of precursors terminating in d-alanyl-d-serine. Antimicrob. Agents Chemother. 49, 21–25. 10.1128/AAC.49.1.21-25.2005, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds P. E., Depardieu F., Dutka-Malen S., Arthur M., Courvalin P. (1994). Glycopeptide resistance mediated by enterococcal transposon Tn1546 requires production of VanX for hydrolysis of d-alanyl-d-alanine. Mol. Microbiol. 13, 1065–1070. 10.1111/j.1365-2958.1994.tb00497.x, PMID: [DOI] [PubMed] [Google Scholar]
- Sanchez M. L., Wenzel R. P., Jones R. N. (1992). In vitro activity of decaplanin (M86-1410), a new glycopeptide antibiotic. Antimicrob. Agents Chemother. 36, 873–875. 10.1128/aac.36.4.873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäberle T. F., Vollmer W., Frasch H.-J., Hüttel S., Kulik A., Röttgen M., et al. (2011). Self-resistance and cell wall composition in the glycopeptide producer Amycolatopsis balhimycina. Antimicrob. Agents Chemother. 55, 4283–4289. 10.1128/aac.01372-10, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shawky R. M., Puk O., Wietzorrek A., Pelzer S., Takano E., Wohlleben W., et al. (2007). The border sequence of the balhimycin biosynthesis gene cluster from Amycolatopsis balhimycina contains bbr, encoding a StrR-like pathway-specific regulator. J. Mol. Microbiol. Biotechnol. 13, 76–88. 10.1159/000103599, PMID: [DOI] [PubMed] [Google Scholar]
- Shearer M. C., Actor P., Bowie B. A., Grappel S. F., Nash C. H., Newman D. J., et al. (1985). Aridicins, novel glycopeptide antibiotics. I. Taxonomy, production and biological activity. J. Antibiot. 38, 555–560. 10.7164/antibiotics.38.555, PMID: [DOI] [PubMed] [Google Scholar]
- Shearer M. C., Giovenella A. J., Grappel S. F., Hedde R. D., Mehta R. J., Oh Y. K., et al. (1986). Kibdelins, novel glycopeptide antibiotics. I. Discovery, production, and biological evaluation. J. Antibiot. 39, 1386–1394. 10.7164/antibiotics.39.1386, PMID: [DOI] [PubMed] [Google Scholar]
- Skelton N. J., Williams D. H., Monday R. A., Ruddock J. C. (1990). Structure elucidation of the novel glycopeptide antibiotic UK-68,597. J. Org. Chem. Res. 55, 3718–3723. 10.1021/jo00299a008 [DOI] [Google Scholar]
- Sosio M., Stinchi S., Beltrametti F., Lazzarini A., Donadio S. (2003). The gene cluster for the biosynthesis of the glycopeptide antibiotic A40926 by Nonomuraea species. Chem. Biol. 10, 541–549. 10.1016/S1074-5521(03)00120-0, PMID: [DOI] [PubMed] [Google Scholar]
- Spohn M., Kirchner N., Kulik A., Jochim A., Wolf F., Muenzer P., et al. (2014). Overproduction of ristomycin A by activation of a silent gene cluster in Amycolatopsis japonicum MG417-CF17. Antimicrob. Agents Chemother. 58, 6185–6196. 10.1128/aac.03512-14, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaker M. N., Wang W., Spanogiannopoulos P., Waglechner N., King A. M., Medina R., et al. (2013). Identifying producers of antibacterial compounds by screening for antibiotic resistance. Nat. Biotechnol. 31, 922–927. 10.1038/nbt.2685, PMID: [DOI] [PubMed] [Google Scholar]
- Truman A. W., Kwun M. J., Cheng J., Yang S. H., Suh J.-W., Hong H.-J. (2014). Antibiotic resistance mechanisms inform discovery: identification and characterization of a novel Amycolatopsis strain producing ristocetin. Antimicrob. Agents Chemother. 58, 5687–5695. 10.1128/aac.03349-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vehreschild M. J. G. T., Haverkamp M., Biehl L. M., Lemmen S., Fätkenheuer G. (2019). Vancomycin-resistant enterococci (VRE): a reason to isolate? Infection 47, 7–11. 10.1007/s15010-018-1202-9, PMID: [DOI] [PubMed] [Google Scholar]
- van Wageningen A. V., Kirkpatrick P. N., Williams D. H., Harris B. R., Kershaw J. K., Lennard N. J., et al. (1998). Sequencing and analysis of genes involved in the biosynthesis of a vancomycin group antibiotic. Chem. Biol. 5, 155–162. 10.1016/S1074-5521(98)90060-6, PMID: [DOI] [PubMed] [Google Scholar]
- Waglechner N., McArthur A. G., Wright G. D. (2019). Phylogenetic reconciliation reveals the natural history of glycopeptide antibiotic biosynthesis and resistance. Nat. Microbiol. 4, 1862–1871. 10.1038/s41564-019-0531-5, PMID: [DOI] [PubMed] [Google Scholar]
- Weigel L. M., Clewell D. B., Gill S. R., Clark N. C., McDougal L. K., Flannagan S. E., et al. (2003). Genetic analysis of a high-level vancomycin-resistant isolate of Staphylococcus aureus. Science 302, 1569–1571. 10.1126/science.1090956, PMID: [DOI] [PubMed] [Google Scholar]
- WHO (2017). Antibacterial agents in clinical development: an analysis of the antibacterial clinical development pipeline, including tuberculosis. World Health Organization. WHO/EMP/IAU/2017.11
- Wink J. M., Kroppenstedt R. M., Ganguli B. N., Nadkarni S. R., Schumann P., Seibert G., et al. (2003). Three new antibiotic producing species of the genus Amycolatopsis, Amycolatopsis balhimycina sp. nov., A. tolypomycina sp. nov., A. vancoresmycina sp. nov., and description of Amycolatopsis keratiniphila subsp. keratiniphila subsp. nov. and A. keratiniphila subsp. nogabecina subsp. nov. Syst. Appl. Microbiol. 26, 38–46. 10.1078/072320203322337290, PMID: [DOI] [PubMed] [Google Scholar]
- Wright G. D., Holman T. R., Walsh C. T. (1993). Purification and characterization of VanR and the cytosolic domain of VanS: a two-component regulatory system required for vancomycin resistance in Enterococcus faecium BM4147. Biochemist 32, 5057–5063. 10.1021/bi00070a013, PMID: [DOI] [PubMed] [Google Scholar]
- Wu Z., Wright G. D., Walsh C. T. (1995). Overexpression, purification, and characterization of VanX, a d,d-dipeptidase which is essential for vancomycin resistance in Enterococcus faecium BM4147. Biochemist 34, 2455–2463. 10.1021/bi00008a008, PMID: [DOI] [PubMed] [Google Scholar]
- Xu L., Huang H., Wei W., Zhong Y., Tang B., Yuan H., et al. (2014). Complete genome sequence and comparative genomic analyses of the vancomycin-producing Amycolatopsis orientalis. BMC Genomics 15:363. 10.1186/1471-2164-15-363, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yim G., Kalan L., Koteva K., Thaker M. N., Waglechner N., Tang I., et al. (2014). Harnessing the synthetic capabilities of glycopeptide antibiotic tailoring enzymes: characterization of the UK-68,597 biosynthetic cluster. Chembiochem 15, 2613–2623. 10.1002/cbic.201402179, PMID: [DOI] [PubMed] [Google Scholar]
- Yushchuk O., Homoniuk V., Ostash B., Marinelli F., Fedorenko V. (2020b). Genetic insights into the mechanism of teicoplanin self-resistance in Actinoplanes teichomyceticus. J. Antibiot. 73, 255–259. 10.1038/s41429-019-0274-9 [DOI] [PubMed] [Google Scholar]
- Yushchuk O., Horbal L., Ostash B., Marinelli F., Wohlleben W., Stegmann E., et al. (2019). Regulation of teicoplanin biosynthesis: refining the roles of tei cluster-situated regulatory genes. Appl. Microbiol. Biotechnol. 103, 4089–4102. 10.1007/s00253-019-09789-w, PMID: [DOI] [PubMed] [Google Scholar]
- Yushchuk O., Ostash B., Truman A. W., Marinelli F., Fedorenko V. (2020a). Teicoplanin biosynthesis: unraveling the interplay of structural, regulatory, and resistance genes. Appl. Microbiol. Biotechnol. 104, 3279–3291. 10.1007/s00253-020-10436-y [DOI] [PubMed] [Google Scholar]
- Zeng D., Debabov D., Hartsell T. L., Cano R. J., Adams S., Schuyler J. A., et al. (2016). Approved glycopeptide antibacterial drugs: mechanism of action and resistance. Cold Spring Harb. Perspect. Med. 6:a026989. 10.1101/cshperspect.a026989 [DOI] [PMC free article] [PubMed] [Google Scholar]