Abstract
This study was conducted to determine the effect of a fermented corn–soybean meal [fermented feed (FF)] on the gene expression of immunity in the blood, the level of secretory immunoglobulin A (sIgA), and mucosa-associated bacterial community in the duodenum and colon of grower-finisher pigs. In this study, crossbred barrows (Duroc × Landrace × Large White) were randomly assigned to either an unfermented corn–soybean diet (Ctrl) (n = 6) or an FF diet (n = 6), and then the following were examined: the expression of immunity using real-time reverse transcription polymerase-chain reaction in the blood, sIgA using enzyme-linked immunosorbent assay (ELISA), and changes in the bacterial community using Illumina Hiseq sequencing in the mucosa of the duodenum and colon. Compared with control pigs fed with a standard diet, the results showed that FF caused upregulation of the mRNA expression of Toll-like receptor 3 (TLR3), TLR4, TLR6, and TLR8 in the blood (P < 0.05). Moreover, sequencing of 16S rRNA genes in duodenal mucosa samples indicated that the FF diet had a lower proportion of Tenericutes (P < 0.05) in the duodenal mucosa-associated microbiota, and FF significantly increased the percentage of Rikenellaceae and Christensenellaceae but decreased the abundance of Lachnospiraceae (P < 0.05) in the colonic mucosa-associated microbiota. The ELISA results showed that FF significantly increased the concentration of sIgA in the colonic mucosa (P < 0.05). More importantly, our correlation analysis indicated that the gene expression of immunity in the blood and the concentration of sIgA was associated with colonic mucosa-associated microbiota. Our data provide new knowledge into the adaptation response of the intestine to fermented feeding in monogastric animals.
Keywords: fermented feed, gut health, immunity, mucosa-associated microbiota, pig
Introduction
Fermentation has been used to prepare healthy human foods for a long time (1, 2). In the livestock industry, fermented feed (FF) has generated special attention because it is associated with the improvement of nutritional quality (3), with the reduction of antinutritional factors (4). More importantly, FF also provides probiotics, beneficial microbiota, and metabolites for animals (5, 6). Furthermore, fermentation could increase amino acid and phosphorus digestibility in soybean meal (7). In recent years, research on finding alternatives to antibiotics in feed and reducing feed costs has continually promoted the development of FF for probiotics (8). Using FF could improve the growth performance of nursery (9) and weaned piglets (10). Meanwhile, FF has also been observed to improve the growth performance of their progeny and decrease diarrhea incidence (11). FF is beneficial to swine nutrition and has been successfully applied in several European countries such as Denmark. Also adding luster to its growing profile is the recent report that FF has beneficial effects on modulating pig gut microbiota (12, 13).
The gut microbiota of pigs strongly affects gastrointestinal and systemic health (14, 15). Using next-generation sequencing, extensive studies have been conducted to characterize the gut microbiota, primarily using fecal samples. However, little work has been done on the more proximal regions of the gut or on the microbiota living in the outer mucosal layer, which may differ from the fecal microbiome. Mucosa-associated microbiota, residing within the outer mucus layer, plays an important role in the intestine (16, 17). Researchers found that mucosa-associated microbiota is vital in exploring bacterial-triggered host immunity and gut–brain communication (18). In addition, although the effects of diet/dietary supplements on gut microbiota have become apparent, little research has been carried out on the effect of diet on mucosa-associated microbiota. Therefore, it is important to understand the effect of diet supplements on mucosa-associated microbiota (small intestine and large intestine) and serum immunity.
This study was part of a larger project investigating the effect of an FF meal on the whole-body health of pigs. The main study tested the hypothesis that a fermented complete commercial soybean diet will regulate gut health. Therefore, in this study, we mainly focused on the effect of FF on the mucosa-associated microbiota and gene expression of immunity in the blood. We hypothesized that feeding FF to pigs could affect the gene expression of immunity in the blood, the level of secretory immunoglobulin A (sIgA), and the mucosa-associated microbiota in the intestine of grower–finisher pigs. The potential links between colonic mucosa-associated microbiota modulation and gene expression of immunity and concentration of sIgA of FF intake are also discussed herein. These results are important for a better understanding of the composition of mucosa-associated microbiota in the intestine and how FF influences the mucosal health of the entire gastrointestinal tract of monogastric animals.
Materials and Methods
Ethics Statement
Animal protocols for this study were approved by the Animal Ethics Committee of Northwest A&F University (Yangling, Shaanxi, China).
Diets and Animals
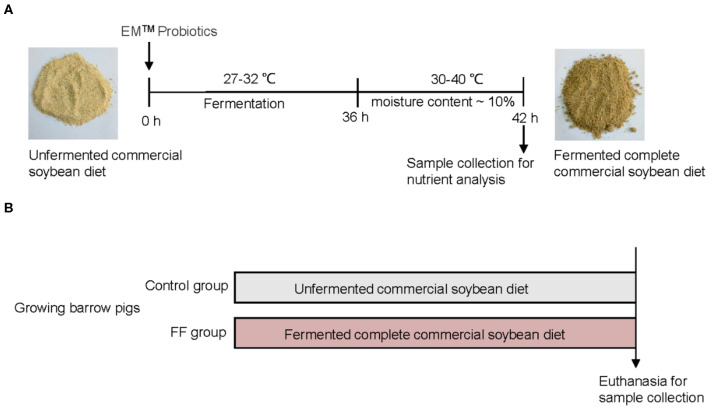
The preparation of FF was performed as previously described (19) and shown in Figure 1A. Briefly, the corn–soybean meals (Beijing Dabeinong, Beijing, China) were fermented with probiotics (Nongfukang, Zhengzhou, China) at 27–32°C for 36 h according to the manufacturer's instructions. After fermentation, the FF meal was dried until the moisture content was nearly 10%. The ingredients and nutrient composition of experimental meals are shown in Supplementary Table 1.
Figure 1.
Experimental design. (A) Preparation and composition of FF. (B) Experimental design.
A total of 48 growing barrow pigs [(Duroc × (Landrace × Large White)] were allocated randomly to two groups fed with either a corn–soybean diet or a fermented complete commercial soybean diet (Figure 1B). Pigs were housed in a controlled environment (four pigs per pen, six pens per group) where water and feed were provided ad-libitum. There were no significant differences in terms of body weights between the two groups (53.19 ± 2.17 vs. 54.60 ± 1.62 kg) at the beginning of the feeding trial. All pigs remained healthy and showed no signs of enteric or metabolic disturbances.
Sample Collection
Upon reaching slaughter weight (~110 kg), one pig from each pen was randomly selected and fasted overnight (sampling number of pigs were six in each group). For sample collection, 1-ml whole blood samples were collected from the external jugular vein of pigs and then immediately mixed with 3-ml RNA Lock Reagent (Bioteke Co., Beijing, China). The mixture was kept at room temperature for 2 h and then stored at −80°C until use. Then, serum samples were carefully collected and centrifuged at 2,500 rpm for 15 min at 4°C; afterward, the samples were brought back to the laboratory for further analysis. Meanwhile, the pigs were anesthetized and slaughtered. Mucosal scrapings from the duodenum and colon were removed, quickly frozen, and then stored at −80°C.
Real-Time Quantitative Reverse Transcription-Polymerase Chain Reaction
Total RNA extraction from blood was performed using RNA Lock Reagent (Tiangen Biotech, Beijing, China) according to the manufacturer's instructions. Reverse transcription (RT) and quantitative RT polymerase chain reaction (qRT-PCR) were performed as previously described (20). All measurements were performed in triplicate. The qRT-PCR primers are shown in Supplementary Table 2.
Enzyme-Linked Immunosorbent Assay (ELISA) Analysis of Interleukin-10, Tumor Necrosis Factor-α, and sIgA
For serum interleukin-10 (IL-10) and tumor necrosis factor-alpha (TNF-α) measurements, serum was measured using a swine IL-10 and TNF-α ELISA kit (Abcam, Cambridge, UK) as per the manufacturer's protocols.
For duodenal mucosa and colonic mucosa sIgA measurements, the loop of the colon was obtained and gently rinsed with 0.9% saline; then, the digest was carefully removed to obtain the mucosa samples and immediately placed in 1.5-ml centrifuge tubes. The concentration of sIgA was measured using a swine IgA ELISA kit (Abcam) following the manufacturer's instructions. Each sample was tested in triplicate.
16S rRNA High-Throughput Sequencing of Mucosa-Associated Microbiota
Bacterial genomic DNA (gDNA) was extracted from the duodenal mucosa and colonic mucosa samples using the bacterial gDNA stool mini-kit (Qiagen, Hilden, Germany), and the quality of the DNA was verified with visual monitoring after 1% agarose gel electrophoresis. According to the V4–V5 hypervariable region of the 16S rRNA genes, universal primers with barcode (Supplementary Table 3) were used to amplify the DNA sequences, followed by generation of sequencing libraries using the TruSeq DNA PCR-free sample preparation kit. Next, the libraries were sequenced on an Illumina Hiseq platform (performed by Novogene Biological Information Technology Co., Beijing, China). Raw data were submitted to the National Center of Biotechnology Information Sequence Read Archive Database (https://www.ncbi.nlm.nih.gov/sra) with accession no. PRJNA607631.
The paired-end reads were merged using Fast Length Adjustment of Short (FLASH) reads software. After quality filtering using QIIME 1.7.02, effective reads were finally obtained by removing the chimera sequences using the UCHIME algorithm. The sequences were then aligned into the UPARSE software (Uparse v7.0.1001) of bacterial 16S rRNA genes, and the operational taxonomic units (OTUs) were identified as one cluster at the 97% similarity level. Next, MUSCLE software (Version 3.8.31) was used to obtain an abundance-based coverage estimator (ACE) and the Shannon index. The different relative abundances of bacterial taxa within different groups [false discovery rate (FDR) <0.05] from the inner to outer rings with phylum and family were measured.
Statistical Analysis
Statistical analyses were performed using SPSS software (v.20.0; IBM Corp., Armonk NY, USA), and values are presented as means ± standard error of the mean (SEM). The data of genes related to immunity, inflammatory factors, and sIgA concentrations were evaluated using Student's t-test. The data of duodenal mucosa and colonic mucosa microbiota were evaluated using non-parametric Kruskal–Wallis test followed by post-hoc test, and the P-values were corrected using FDR as previously described (21). FDR values <0.05 were considered significantly different. The correlations between colonic mucosa microbial composition and genes related to immunity in blood and concentration of sIgA in the colonic mucosa, which was significantly influenced by FF, were evaluated using Spearman's correlation test. The Pearson correlation coefficient was >0.5, and P < 0.05 were considered to be significantly different.
Results
Effect of FF on Expression of Genes Related to Immunity in the Blood and Serum Inflammatory Factor
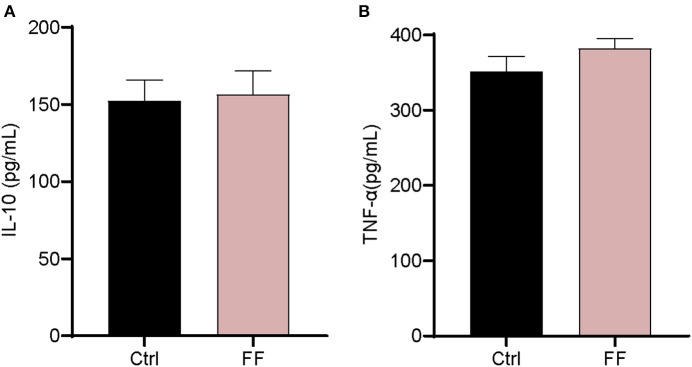
We first characterized the influence of FF on the mRNA expression of Toll-like receptors (TLRs) and two antimicrobial peptide-encoding genes in the blood. The results are presented in Figure 2. The mRNA abundance of TLR3, TLR4, TLR6, and TLR8 in FF-fed pigs was significantly increased (P < 0.05) compared with that in the control group. Furthermore, the impact of FF on serum inflammatory factors was determined. Cytokines, as a series of small peptide molecules, are vital in the modulation of immunity and inflammatory responses (22). As shown in Figure 3, we found no statistical difference between the two groups. These results indicate that FF administration increased the mRNA expression of TLRs in the blood.
Figure 2.
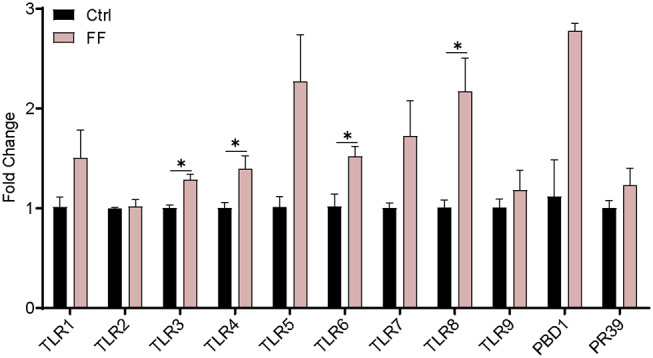
Effect of FF on the expression of genes related to blood immunity. The Ctrl group was designed as 1-fold change. Ctrl, pigs fed with normal commercial feed; FF, pigs fed with fermented meal. Data are presented as mean ± SEM (n = 6). *P < 0.05.
Figure 3.
Effect of FF on the serum cytokine levels. (A) IL-10; (B) TNF-α. Ctrl: pigs fed with normal commercial feed; FF: pigs fed with fermented meal. Data are presented as mean ± SEM (n = 6).
Effect of FF on Composition of Intestinal Mucosal-Associated Microbiota
We used bacterial 16S rRNA sequencing based on V4–V5 hypervariable regions to analyze the impact of FF supplementation on duodenal mucosa- and colonic mucosa-associated microbiota. The mean ± SEM values of duodenal mucosa and colonic mucosa for each sample were 59,762 ± 4,560 and 51,272 ± 2,109, respectively. Using a pairwise identity threshold of 97% packing sequence, each sample showed an average operating taxon of 892 ± 119 (duodenal mucosa) and 909 ± 24 (colonic mucosa). Meanwhile, FF exhibited no statistical difference in terms of diversity of microbiota, as evidenced by similar richness estimators (ACE and Chao 1 index) and diversity indices (Shannon and Simpson indices), compared with control subjects (Table 1).
Table 1.
Diversity estimation of the 16S.
| Items | Duodenal mucosa | Colonic mucosa | ||||
|---|---|---|---|---|---|---|
| Ctrl | FF | P-value | Ctrl | FF | P-value | |
| Ace | 932.58 ± 86.36 | 1,502.61 ± 207.95 | 0.142 | 1,066.42 ± 46.22 | 993.77 ± 32.24 | 0.226 |
| Chao 1 | 927.22 ± 82.17 | 1,517.51 ± 204.76 | 0.148 | 1,063.58 ± 51.49 | 992.19 ± 37.17 | 0.287 |
| Shannon | 6.97 ± 0.15 | 4.84 ± 0.66 | 0.075 | 7.47 ± 0.19 | 7.25 ± 0.26 | 0.516 |
| Simpson | 0.96 ± 0.04 | 0.80 ± 0.06 | 0.128 | 0.98 ± 0.01 | 0.97 ± 0.02 | 0.455 |
Ctrl, pigs fed with normal commercial feed; FF, pigs fed with fermented meal. Data are presented as the mean ± SEM (n = 6).
Next, we observed that Firmicutes and Bacteroidetes were the most predominant phyla in the duodenal and colonic mucosa of the control pigs at the phylum level (Tables 2, 3). The FF diet contained a lower proportion of Tenericutes (P < 0.05) in the duodenal mucosa-associated microbiota. Meanwhile, the FF diet had a higher proportion of Proteobacteria (P < 0.01) and a higher proportion of Acidobacteria (P < 0.05), but a lower proportion of Tenericutes in the colonic mucosa (P < 0.05).
Table 2.
Relative abundance of microbial phylum (percentage) in the duodenal mucosa-associated microbiota of pigs in the FF and control groups.
| Phylum | Ctrl | FF | P-value |
|---|---|---|---|
| Firmicutes | 59.15 ± 12.17 | 57.72 ± 8.16 | 0.939 |
| Proteobacteria | 1.03 ± 0.10 | 21.45 ± 9.43 | 0.175 |
| Bacteroidetes | 32.47 ± 9.96 | 7.91 ± 2.16 | 0.136 |
| Spirochaetes | 1.22 ± 0.34 | 0.34 ± 0.12 | 0.123 |
| Unclassified phylum | 4.19 ± 3.22 | 8.80 ± 6.66 | 0.665 |
| Tenericutes | 1.40 ± 0.28 | 0.45 ± 0.13 | 0.022 |
| Actinobacteria | 0.12 ± 0.03 | 2.23 ± 0.69 | 0.088 |
| Chloroflexi | 0.00 ± 0.01 | 0.72 ± 0.34 | 0.180 |
| Cyanobacteria | 0.24 ± 0.06 | 0.06 ± 0.02 | 0.116 |
| Acidobacteria | 0.02 ± 0.02 | 0.24 ± 0.12 | 0.238 |
| Verrucomicrobia; | 0.14 ± 0.08 | 0.08 ± 0.02 | 0.552 |
Ctrl, pigs fed with normal commercial feed; FF, pigs fed with fermented meal. Data are presented as mean ± SEM (control n = 4, FF n = 6).
Table 3.
Relative abundance of microbial phylum (percentage) in the colonic mucosa-associated microbiota of pigs in the FF and control groups.
| Phylum | Ctrl | FF | P-value |
|---|---|---|---|
| Firmicutes | 47.98 ± 5.46 | 50.54 ± 3.83 | 0.709 |
| Proteobacteria | 1.43 ± 0.13 | 0.87 ± 0.06 | 0.007 |
| Bacteroidetes | 44.10 ± 5.53 | 42.25 ± 17.25 | 0.789 |
| Spirochaetes | 1.84 ± 0.37 | 3.26 ± 0.75 | 0.132 |
| Unclassified phylum | 0.47 ± 0.04 | 0.72 ± 0.39 | 0.354 |
| Tenericutes | 3.47 ± 0.55 | 1.66 ± 0.29 | 0.022 |
| Actinobacteria | 0.19 ± 0.03 | 0.34 ± 0.10 | 0.222 |
| Planctomycetes | 0.04 ± 0.03 | 0.18 ± 0.11 | 0.286 |
| Verrucomicrobia | 0.27 ± 0.08 | 0.06 ± 0.04 | 0.043 |
| Cyanobacteria | 0.21 ± 0.09 | 0.11 ± 0.07 | 0.349 |
| Euryarchaeota | 0.14 ± 0.07 | 0.12 ± 0.05 | 0.841 |
Ctrl, pigs fed with normal commercial feed; FF, pigs fed with fermented meal. Data are presented as mean ± SEM (n = 6).
At the family level, we observed that BS11_gut_group and Rhodospirillaceae tended to decrease in terms of relative abundance after FF administration in the duodenal mucosa samples (0.05 < P < 0.10) (Table 4). At the same time, the results showed that fermented feeding significantly increased the proportion of Rikenellaceae and Christensenellaceae, whereas the abundance of Lachnospiraceae, Streptococcaceae, Veillonellaceae, and Succinivibrionaceae decreased in the colonic mucosa samples (P < 0.05) (Table 5). Collectively, the FF had a minimal effect on duodenal mucosa microbiota and shaped the colonic mucosa microbiota.
Table 4.
Relative abundance (percentage) for the top 30 most abundant family in the duodenal mucosa-associated microbiota of pigs in the FF and control groups.
| Items | Ctrl | FF | P-value |
|---|---|---|---|
| Lactobacillaceae | 24.85 ± 19.52 | 23.33 ± 12.33 | 0.959 |
| Prevotellaceae | 15.09 ± 4.58 | 2.77 ± 0.83 | 0.114 |
| Clostridiaceae_1 | 9.85 ± 6.44 | 2.79 ± 0.96 | 0.439 |
| Ruminococcaceae | 8.29 ± 2.33 | 6.43 ± 2.27 | 0.656 |
| Bacteroidales_S24-7_group | 7.96 ± 2.49 | 2.08 ± 0.58 | 0.148 |
| Lachnospiraceae | 6.36 ± 1.78 | 2.90 ± 1.00 | 0.228 |
| Unclassified family | 6.19 ± 3.45 | 10.56 ± 6.90 | 0.665 |
| Rikenellaceae | 3.52 ± 1.24 | 0.79 ± 0.22 | 0.169 |
| Veillonellaceae | 2.30 ± 0.97 | 5.43 ± 1.68 | 0.247 |
| Porphyromonadaceae | 2.11 ± 0.77 | 0.87 ± 0.26 | 0.285 |
| Bacteroidales_RF16_group | 1.94 ± 0.54 | 0.53 ± 0.16 | 0.121 |
| Streptococcaceae | 1.76 ± 0.64 | 0.46 ± 0.27 | 0.202 |
| Peptostreptococcaceae | 1.70 ± 0.57 | 3.06 ± 1.73 | 0.577 |
| Erysipelotrichaceae | 1.32 ± 0.51 | 2.91 ± 1.63 | 0.495 |
| Spirochaetaceae | 1.22 ± 0.34 | 0.33 ± 0.12 | 0.123 |
| Bacteroidaceae | 0.90 ± 0.34 | 0.17 ± 0.05 | 0.182 |
| Acidaminococcaceae | 0.86 ± 0.24 | 0.35 ± 0.11 | 0.187 |
| Christensenellaceae | 0.78 ± 0.24 | 1.18 ± 0.45 | 0.562 |
| Family_XIII | 0.51 ± 0.19 | 0.51 ± 0.10 | 0.988 |
| Anaeroplasmataceae | 0.19 ± 0.08 | 0.01 ± 0.01 | 0.161 |
| Mycoplasmataceae | 0.18 ± 0.10 | 0.00 ± 0.00 | 0.229 |
| Clostridiales_vadinBB60_group | 0.17 ± 0.06 | 0.05 ± 0.02 | 0.181 |
| P-2534-18B5_gut_group | 0.17 ± 0.06 | 0.03 ± 0.01 | 0.164 |
| Alcaligenaceae | 0.16 ± 0.04 | 0.09 ± 0.03 | 0.351 |
| BS11_gut_group | 0.15 ± 0.05 | 0.01 ± 0.01 | 0.086 |
| Peptococcaceae | 0.13 ± 0.05 | 0.06 ± 0.02 | 0.395 |
| Campylobacteraceae | 0.12 ± 0.04 | 0.47 ± 0.29 | 0.398 |
| Rhodospirillaceae | 0.10 ± 0.03 | 0.02 ± 0.01 | 0.099 |
| Helicobacteraceae | 0.09 ± 0.02 | 0.22 ± 0.15 | 0.523 |
| Succinivibrionaceae | 0.09 ± 0.03 | 0.00 ± 0.00 | 0.131 |
| Neisseriaceae | 0.08 ± 0.03 | 0.05 ± 0.02 | 0.583 |
Ctrl, pigs fed with normal commercial feed; FF, pigs fed with fermented meal. Data are presented as mean ± SEM (control n = 4, FF n = 6).
Table 5.
Relative abundance (percentage) for the top 30 most abundant family in the colonic mucosa-associated microbiota of pigs in the FF and control groups.
| Items | Ctrl | FF | P-value |
|---|---|---|---|
| Prevotellaceae | 25.76 ± 5.88 | 13.43 ± 2.67 | 0.098 |
| Ruminococcaceae | 17.80 ± 2.51 | 22.44 ± 2.01 | 0.180 |
| Lachnospiraceae | 11.61 ± 0.76 | 8.62 ± 0.72 | 0.017 |
| Bacteroidales_S24-7_group | 7.89 ± 1.50 | 10.15 ± 1.01 | 0.245 |
| Streptococcaceae | 5.22 ± 1.68 | 0.05 ± 0.02 | 0.027 |
| Unclassified family | 5.38 ± 0.75 | 4.61 ± 0.49 | 0.159 |
| Rikenellaceae | 4.53 ± 0.41 | 10.38 ± 1.61 | 0.014 |
| Clostridiaceae_1 | 3.16 ± 0.97 | 1.40 ± 0.21 | 0.134 |
| Bacteroidales_RF16_group | 2.46 ± 0.56 | 2.29 ± 0.61 | 0.843 |
| Veillonellaceae | 2.15 ± 0.36 | 0.35 ± 0.18 | 0.003 |
| Porphyromonadaceae | 1.84 ± 0.35 | 3.75 ± 0.87 | 0.083 |
| Spirochaetaceae | 1.84 ± 0.37 | 3.26 ± 0.75 | 0.132 |
| Peptostreptococcaceae | 1.71 ± 0.40 | 1.70 ± 0.29 | 0.985 |
| Acidaminococcaceae | 1.47 ± 0.08 | 1.23 ± 0.29 | 0.446 |
| Erysipelotrichaceae | 1.41 ± 0.17 | 0.95 ± 0.12 | 0.057 |
| Christensenellaceae | 1.35 ± 0.40 | 3.43 ± 0.63 | 0.022 |
| Family_XIII | 0.66 ± 0.07 | 0.91 ± 0.13 | 0.141 |
| Bacteroidaceae | 0.60 ± 0.16 | 0.72 ± 0.13 | 0.566 |
| Lactobacillaceae | 0.60 ± 0.32 | 8.24 ± 5.47 | 0.221 |
| Clostridiales_vadinBB60_group | 0.42 ± 0.11 | 0.26 ± 0.03 | 0.221 |
| Anaeroplasmataceae | 0.36 ± 0.08 | 0.18 ± 0.06 | 0.109 |
| P-2534-18B5_gut_group | 0.27 ± 0.06 | 0.14 ± 0.03 | 0.118 |
| Campylobacteraceae | 0.26 ± 0.08 | 0.12 ± 0.03 | 0.140 |
| Bacteroidales_BS11_gut_group | 0.25 ± 0.13 | 0.65 ± 0.32 | 0.290 |
| Succinivibrionaceae | 0.22 ± 0.07 | 0.02 ± 0.02 | 0.037 |
| Alcaligenaceae | 0.19 ± 0.04 | 0.11 ± 0.05 | 0.276 |
| Fibrobacteraceae | 0.14 ± 0.05 | 0.04 ± 0.03 | 0.127 |
| Peptococcaceae | 0.13 ± 0.02 | 0.09 ± 0.01 | 0.123 |
| Unidentified_thermoplasmatales | 0.12 ± 0.07 | 0.04 ± 0.04 | 0.401 |
| Enterobacteriaceae | 0.11 ± 0.06 | 0.31 ± 0.10 | 0.125 |
| Coriobacteriaceae | 0.10 ± 0.02 | 0.12 ± 0.02 | 0.587 |
Ctrl, pigs fed with normal commercial feed; FF, pigs fed with fermented meal. Data are presented as the mean ± SEM (n = 6).
Effect of FF on sIgA Concentration in Intestinal Mucosa of Pigs
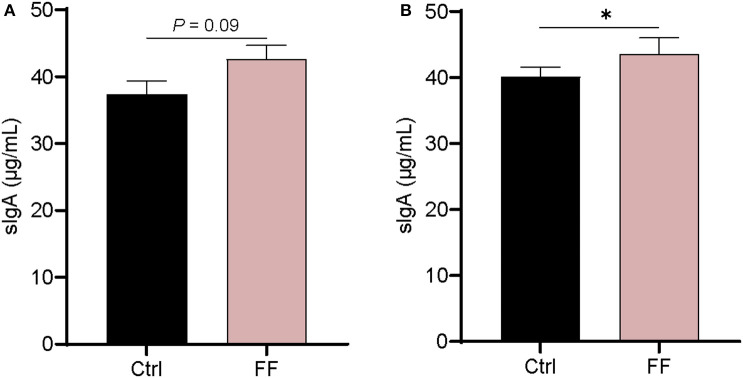
IgA, as the most enriched immunoglobulin in mammals, is mainly secreted from mucous membranes (23). It is reported that mucosa-associated microbiota is critically important for intestinal production and secretion of sIgA (24). As shown in Figure 4, there was no significant difference in the concentration of sIgA in the duodenal mucosa. In the colon mucosa, the concentration of sIgA in FF-fed pigs was significantly increased compared with that in control animals (P < 0.05). Overall, the impact of FF on sIgA concentration in the intestinal mucosa of pigs mainly occurred in the colonic mucosa.
Figure 4.
Effect of FF on the content of sIgA in the colonic mucosa. Ctrl, pigs fed with normal commercial feed; FF, pigs fed with fermented meal. Data are presented as mean ± SEM (n = 6). *P < 0.05. (A) Duodenal mucosa; (B) Colonic mucosa.
Correlations Between Colonic Mucosa-Associated Microbiota and Immunity-Related Genes and Colonic sIgA
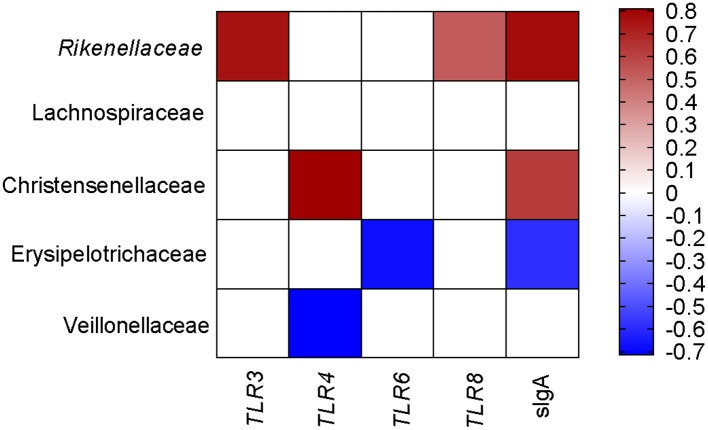
To comprehensively analyze the relationship between pig blood immune-related genes, sIgA, and colonic mucosa-associated microbiota, the Spearman correlation coefficient was calculated to generate the correlation matrix. As shown in Figure 5, we observed that TLR3, TLR8, and sIgA were positively associated with the proportion of Rikenellaceae. The gene expression of TLR4 was positively associated with the proportion of Christensenellaceae but negatively related to the proportion of Veillonellaceae. sIgA was positively associated with the proportion of Christensenellaceae but negatively related to the proportion of Erysipelotrichaceae. Meanwhile, the gene expression of TLR6 was negatively associated with the proportion of Erysipelotrichaceae. Taken together, these findings showed that colonic mucosa microbiota was associated with the expression of immunity-related genes and colonic sIgA.
Figure 5.
Correlation analysis between colonic microbial composition and genes related to blood immunity and sIgA, which were significantly influenced by the FF diet in the colonic of pigs compared to the Ctrl diet. Ctrl, pigs fed with normal commercial feed; FF, pigs fed with fermented meal. The color represents a significant correlation (P < 0.05), and the intensity of the colors represents the degree of association. Red represents significantly positive correlation and blue represents significantly negative correlation (n = 6).
Discussion
Over the past several decades, antibiotic resistance and antibiotic residues have become a growing problem worldwide (25, 26). FF, a widely investigated diet that has been used as an antibiotic growth promoter (27), was recently shown to drive down feed prices through the use of industrial by-products in livestock (28, 29). In this study, we compared the effects of FF and unfermented corn–soybean diet on blood immunity, serum inflammatory factor, and intestinal (duodenum and colon) mucosa-associated microbiota composition in grower–finisher pigs. Our data indicated that fermented feeding modulated the expression of the blood gene and selectively modified the mucosa-associated microbiota composition.
Corn and soybean meal account for about 70% of global feed consumption. However, the utility of conventional corn and soybean meal mixed is limited by several antinutrient factors that inhibit nutrient bioavailability and impair animal health (30). Solid-state fermentation has provided a solution, which has been widely used in China, to improve the nutritional quality of feed (31). In the present study, we used a complete fermented corn–soybean meal (100% fermented), which may increase manufacturing costs in conventional swine production. Hence, further studies specifically focusing on different inclusion levels of FF are warranted to explore the appropriate dose of supplementary feeds to help increase income in swine production.
It has been reported that TLRs play a vital role in immunity (32). One previous study showed that the expression of TLRs may reflect the status of disease resistance of pigs (33, 34). In our study, we found that the mRNA abundance of TLR3, TLR4, TLR6, and TLR8 in FF-fed pigs was significantly higher (P < 0.05), indicating that the FF diet could influence the gene expression of TLRs. Interestingly, we found no significant difference between the two antimicrobial peptides (PBD-1 and PR-39), whereas the mRNA abundances of PBD-1 and PR39 in the duodenum were significantly increased (P < 0.05) in our previous study (19). In the same study, we had observed high concentrations of immunoglobulins in the serum samples, and intestinal inflammation was not detected, indicating that the gene expression of both groups was in normal ranges. Furthermore, it is essential to develop an in vivo pathogenic challenge model as a further study to confirm that the FF diet enhances immune performance.
There are extensive studies on the beneficial effects of FF on pig gut microbiota, but so far only a few studies have focused on mucosa-associated microbiota. Nevertheless, researchers had found that colonic mucosa-associated microbiota plays an important role in host metabolism and immune homeostasis (35, 36). In the present study, we found that FF had no significant effect on the number of OTUs, whereas the abundance of certain phyla was significantly affected. The proportion of Proteobacteria and Acidobacteria were significantly increased (P < 0.05), whereas the proportion of Tenericutes was decreased in the colonic mucosa (P < 0.05) in response to FF meal. We further analyzed the influence of the FF diet on colonic mucosa-associated microbiota at the family level. It has been reported that the increased proportion of Christensenellaceae (a butyrate producer) was beneficial for the sow's health (37), and our data found that FF diet significantly increased the proportion of Christensenellaceae. In addition, a previous study found that the enhanced ratio of Christensenellaceae might promote a lean host phenotype in pigs (38). Meanwhile, the family of Rikenellaceae may be related to anaerobic metabolism (39). One study has shown that Erysipelotrichaceae was related to the digestion of protein and energy in dogs (40). In the present study, the significantly decreased proportion of Erysipelotrichaceae may provide benefit to the pig's gut health. In addition, some species of Veillonellaceae, as an opportunistic pathogen for animals, could be responsible for polymicrobial infections (41). Interestingly, in the present study, the proportion of Lachnospiraceae was significantly decreased in FF-fed pigs compared with that in the control group. In short, the FF diet shaped the colonic mucosa-associated microbiota.
Then we determined the concentration of sIgA in the duodenal mucosa and colonic mucosa. It has been reported that sIgA is the most abundant colonic antibody antigen known as “immune exclusion” (42). The result was consistent with previous studies (43, 44), which reported that diet could promote intestinal sIgA secretion through intestinal microbiota and could be beneficial to the animal's health (45). Furthermore, we analyzed the correlation between colonic mucosa-associated microbiota and sIgA, and the results indicated that sIgA was influenced by the intestinal mucosa-associated microbiota. Our data showed that FF is beneficial to swine gut health. Moreover, the tight junction in the gut should be taken into consideration in further studies as well as in potential practical applications.
FF meal altered the blood immunity and composition of colonic mucosa-associated microbiota in pigs. The colonic mucosa-associated microbiota was closely associated with the expression of blood immunity-related genes. For instance, Christensenellaceae, a bacterial family known to be abundant in the intestinal ecosystem, was positively correlated with lipid metabolism (46). Furthermore, our data showed that Christensenellaceae was also positively correlated with TLR4 expression in the blood and sIgA in the colonic mucosa. These lines of evidence suggest that the beneficial effect of FF might be attributable to the higher relative abundances of some beneficial bacteria on the colonic health of grower–finisher pigs. Further studies are necessary to confirm the roles of these gut bacteria in regulating swine health.
Conclusion
Taken together, the findings of this study revealed that feeding complete fermented corn–soybean meal (100% fermented) to grower–finisher pigs could increase the expression of genes related to immunity in the blood, and changed the concentration of sIgA in the colonic mucosa and modified the microbiota community in the colonic mucosa. These findings highlight the positive role of FF diet in pig health. Furthermore, it would be interesting to explore the best dose for feed supplements, which we would like to focus on in our next study.
Data Availability Statement
The datasets generated for this study can be found in the NCBI Repository [accession number: PRJNA607631].
Ethics Statement
Animal protocols for this study were approved by the Animal Ethics Committee of Northwest A&F University (Yangling, Shaanxi, China).
Author Contributions
XS and JL designed the study. JL, HC, XuZ, XiZ, XL, and JH helped performed the experiments and took samples. MZ and ZW analyzed the data. JL and MZ wrote and revised the manuscript. MZ, XL, JH, GY, and XS modified the manuscript. All authors reviewed the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the people in XL's team and XS's team for their excellent support during this experiment.
Footnotes
Funding. This study was supported by the Agricultural Special Fund Project in Shaanxi Province (NYKJ-2018-YL01) and the Technical Innovation Guidance Special (Foundation) Project of Shaanxi Province (2017ZKC07-114).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2020.00246/full#supplementary-material
References
- 1.Sanchez Mainar M, Stavropoulou DA, Leroy F. Exploring the metabolic heterogeneity of coagulase-negative staphylococci to improve the quality and safety of fermented meats: a review. Int J Food Microbiol. (2017) 247:24–37. 10.1016/j.ijfoodmicro.2016.05.021 [DOI] [PubMed] [Google Scholar]
- 2.Shiferaw Terefe N, Augustin MA. Fermentation for tailoring the technological and health related functionality of food products. Crit Rev Food Sci Nutr. 4:1–27. 10.1080/10408398.2019.1666250 [DOI] [PubMed] [Google Scholar]
- 3.Xu B, Zhu L, Fu J, Li Z, Wang Y, Jin M. Overall assessment of fermented feed for pigs: a series of meta-analyses. J Anim Sci. (2019) 97:4810–21. 10.1093/jas/skz350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang C, Shi C, Zhang Y, Song D, Lu Z, Wang Y. Microbiota in fermented feed and swine gut. Appl Microbiol Biotechnol. (2018) 102:2941–8. 10.1007/s00253-018-8829-4 [DOI] [PubMed] [Google Scholar]
- 5.Parvez S, Malik KA, Ah Kang S, Kim HY. Probiotics and their fermented food products are beneficial for health. J Appl Microbiol. (2006) 100:1171–85. 10.1111/j.1365-2672.2006.02963.x [DOI] [PubMed] [Google Scholar]
- 6.Liu P, Zhao J, Guo P, Lu W, Geng Z, Levesque CL, et al. Dietary corn bran fermented by Bacillus Subtilis MA139 decreased gut cellulolytic bacteria and microbiota diversity in finishing pigs. Front Cell Infect Microbiol. (2017) 7:526. 10.3389/fcimb.2017.00526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shi C, Zhang Y, Yin Y, Wang C, Lu Z, Wang F, et al. Amino acid and phosphorus digestibility of fermented corn-soybean meal mixed feed with Bacillus subtilis and Enterococcus faecium fed to pigs. J Anim Sci. (2017) 95:3996–4004. 10.2527/jas.2017.1516 [DOI] [PubMed] [Google Scholar]
- 8.Thacker PA. Alternatives to antibiotics as growth promoters for use in swine production: a review. J Anim Sci Biotechnol. (2013) 4:35. 10.1186/2049-1891-4-35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng SS, Li Y, Geng SJ, Hu LS, Fu XF, Han XY. Effects of dietary fresh fermented soybean meal on growth performance, ammonia and particulate matter emissions, and nitrogen excretion in nursery piglets. J Zhejiang Univ Sci B. (2017) 18:1083–92. 10.1631/jzus.B1700180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu J, Gao M, Zhang R, Sun Z, Wang C, Yang F, et al. Effects of soybean meal fermented by L. Plantarum, B subtilis and S cerevisieae on growth, immune function and intestinal morphology in weaned piglets. Microb Cell Fact. (2017) 16:191. 10.1186/s12934-017-0809-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang C, Lin C, Su W, Zhang Y, Wang F, Wang Y, et al. Effects of supplementing sow diets with fermented corn and soybean meal mixed feed during lactation on the performance of sows and progeny. J Anim Sci. (2018) 96:206–14. 10.1093/jas/skx019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jeong JS, Park JW, Lee SI, Kim IH. Apparent ileal digestibility of nutrients and amino acids in soybean meal, fish meal, spray-dried plasma protein and fermented soybean meal to weaned pigs. Anim Sci J. (2016) 87:697–702. 10.1111/asj.12483 [DOI] [PubMed] [Google Scholar]
- 13.Tufarelli V, Crovace AM, Rossi G, Laudadio V. Effect of a dietary probiotic blend on performance, blood characteristics, meat quality and faecal microbial shedding in growing-finishing pigs. S Afr J Anim Sci. (2017) 47:875–82. 10.4314/sajas.v47i6.15 [DOI] [Google Scholar]
- 14.Mann E, Dzieciol M, Metzler-Zebeli BU, Wagner M, Schmitz-Esser S. Microbiomes of unreactive and pathologically altered ileocecal lymph nodes of slaughter pigs. Appl Environ Microbiol. (2014) 80:193–203. 10.1128/AEM.03089-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang L, Wu W, Lee Y-K, Xie J, Zhang H. Spatial heterogeneity and co-occurrence of mucosal and luminal microbiome across swine intestinal tract. Front Microbiol. (2018) 9:48. 10.3389/fmicb.2018.00048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tlaskalova-Hogenova H, Stepankova R, Kozakova H, Hudcovic T, Vannucci L, Tuckova L, et al. The role of gut microbiota (commensal bacteria) and the mucosal barrier in the pathogenesis of inflammatory and autoimmune diseases and cancer: contribution of germ-free and gnotobiotic animal models of human diseases. Cell Mol Immunol. (2011) 8:110–20. 10.1038/cmi.2010.67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burrough ER, Arruda BL, Plummer PJ. Comparison of the luminal and mucosa-associated microbiota in the colon of pigs with and without swine dysentery. Front Vet Sci. (2017) 4:139. 10.3389/fvets.2017.00139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thaiss CA, Zmora N, Levy M, Elinav E. The microbiome and innate immunity. Nature. (2016) 535:65–74. 10.1038/nature18847 [DOI] [PubMed] [Google Scholar]
- 19.Lu J, Zhang X, Liu Y, Cao H, Han Q, Xie B, et al. Effect of fermented corn-soybean meal on serum immunity, the expression of genes related to gut immunity, gut microbiota, and bacterial metabolites in grower-finisher pigs. Front Microbiol. (2019) 10:2620. 10.3389/fmicb.2019.02620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang J, Ge J, Cao H, Zhang X, Guo Y, Li X, et al. Leptin promotes white adipocyte browning by inhibiting the Hh signaling pathway. Cells. (2019) 8:372. 10.3390/cells8040372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Benjamini Y, Hochberg Y. On the adaptive control of the false discovery rate in multiple testing with independent statistics. J Edu Behav Stat. (2000) 25:60–83. 10.3102/1076998602500106014584715 [DOI] [Google Scholar]
- 22.Cook AD, Christensen AD, Tewari D, McMahon SB, Hamilton JA. Immune cytokines and their receptors in inflammatory pain. Trends Immunol. (2018) 39:240–55. 10.1016/j.it.2017.12.003 [DOI] [PubMed] [Google Scholar]
- 23.Macpherson AJ, Yilmaz B, Limenitakis JP, Ganal-Vonarburg SC. IgA function in relation to the intestinal microbiota. Annu Rev Immunol. (2018) 36:359–81. 10.1146/annurev-immunol-042617-053238 [DOI] [PubMed] [Google Scholar]
- 24.Kubinak JL, Petersen C, Stephens WZ, Soto R, Bake E, O'Connell RM, et al. MyD88 signaling in T cells directs IgA-mediated control of the microbiota to promote health. Cell Host Microbe. (2015) 17:153–63. 10.1016/j.chom.2014.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu YG, Johnson TA, Su JQ, Qiao M, Guo GX, Stedtfeld RD, et al. Diverse and abundant antibiotic resistance genes in Chinese swine farms. Proc Natl Acad Sci USA. (2013) 110:3435–40. 10.1073/pnas.1222743110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang H, Paruch L, Chen X, van Eerde A, Skomedal H, Wang Y, et al. Antibiotic application and resistance in swine production in China: current situation and future perspectives. Front Vet Sci. (2019) 6:136. 10.3389/fvets.2019.00136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Plumed-Ferrer C, von Wright A. Fermented pig liquid feed: nutritional, safety and regulatory aspects. J Appl Microbiol. (2009) 106:351–68. 10.1111/j.1365-2672.2008.03938.x [DOI] [PubMed] [Google Scholar]
- 28.Wang J, Han Y, Zhao J, Zhou Z, Fan H. Consuming fermented distillers' dried grains with solubles (DDGS) feed reveals a shift in the faecal microbiota of growing and fattening pigs using 454 pyrosequencing. J Integr Agric. (2017) 16:900–10. 10.1016/S2095-3119(16)61523-X [DOI] [Google Scholar]
- 29.Mamuad LL, Kim SH, Biswas AA, Yu Z, Cho KK, Kim SB, et al. Rumen fermentation and microbial community composition influenced by live Enterococcus Faecium supplementation. AMB Express. (2019) 9:123. 10.1186/s13568-019-0848-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feng J, Liu X, Xu Z, Lu Y, Liu Y. Effect of fermented soybean meal on intestinal morphology and digestive enzyme activities in weaned piglets. Dig Dis Sci. (2007) 52:1845. 10.1007/s10620-006-9705-0 [DOI] [PubMed] [Google Scholar]
- 31.Wang C, Shi C, Su W, Jin M, Xu B, Hao L, et al. Dynamics of the physicochemical characteristics, microbiota, and metabolic functions of soybean meal and corn mixed substrates during two-stage solid-state fermentation. mSystems. (2020) 5:e00501-00519. 10.1128/mSystems.00501-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cen X, Liu S, Cheng K. The role of toll-like receptor in inflammation and tumor immunity. Front Pharmacol. (2018) 9:878. 10.3389/fphar.2018.00878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cheng C, Sun WK, Liu R, Wang RM, Chen YH, Wang Y, et al. Comparison of gene expression of toll-like receptors and antimicrobial peptides in immune organs and tissues between yorkshire and tibetan pigs. Anim Genet. (2015) 46:272–9. 10.1111/age.12286 [DOI] [PubMed] [Google Scholar]
- 34.Hu J, Yang D, Wang H, Li C, Zeng Y, Chen W. CpG oligodeoxynucleotides induce differential cytokine and chemokine gene expression profiles in dapulian and landrace pigs. Front Microbiol. (2016) 7:1992. 10.3389/fmicb.2016.01992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun Y, Zhou L, Fang L, Su Y, Zhu W. Responses in colonic microbial community and gene expression of pigs to a long-term high resistant starch diet. Front Microbiol. (2015) 6:877. 10.3389/fmicb.2015.00877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu J, Bian G, Sun D, Zhu W, Mao S. Starter feeding supplementation alters colonic mucosal bacterial communities and modulates mucosal immune homeostasis in newborn lambs. Front Microbiol. (2017) 8:429. 10.3389/fmicb.2017.00429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xiong Y, Pang J, Lv L, Wu Y, Li N, Huang S, et al. Effects of maternal supplementation with rare earth elements during late gestation and lactation on performances, health, and fecal microbiota of the sows and their offspring. Animals. (2019) 9:738. 10.3390/ani9100738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu C, Zhou J, Li Y, Zhang D, Wang Z, Li Y, et al. Structural modulation of gut microbiota in Bama minipigs in response to treatment with a “growth-promoting agent,” salbutamol. Appl Microbiol Biotechnol. (2017) 101:5809–18. 10.1007/s00253-017-8329-y [DOI] [PubMed] [Google Scholar]
- 39.Graf J. The Family Rikenellaceae. The Prokaryotes: Other Major Lineages of Bacteria and The Archaea. Berlin, Heidelberg: Springer; (2014). p. 857–9. 10.1007/978-3-642-38954-2_134 [DOI] [Google Scholar]
- 40.Bermingham EN, Maclean P, Thomas DG, Cave NJ, Young W. Key bacterial families (Clostridiaceae, Erysipelotrichaceae and Bacteroidaceae) are related to the digestion of protein and energy in dogs. PeerJ. (2017) 5:e3019. 10.7717/peerj.3019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marchandin H, Jumas-Bilak E. The Family Veillonellaceae. The Prokaryotes: Firmicutes and Tenericutes. Berlin, Heidelberg: Springer; (2014). p. 433–53. 10.1007/978-3-642-30120-9_361 [DOI] [Google Scholar]
- 42.Palm NW, de Zoete MR, Cullen TW, Barry NA, Stefanowski J, Hao L, et al. Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell. (2014) 158:1000–10. 10.1016/j.cell.2014.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xiao D, Tang Z, Yin Y, Zhang B, Hu X, Feng Z, et al. Effects of dietary administering chitosan on growth performance, jejunal morphology, jejunal mucosal sIgA, occludin, claudin-1 and TLR4 expression in weaned piglets challenged by enterotoxigenic Escherichia Coli. Int Immunopharmacol. (2013) 17:670–6. 10.1016/j.intimp.2013.07.023 [DOI] [PubMed] [Google Scholar]
- 44.Wu M, Xiao H, Liu G, Chen S, Tan B, Ren W, et al. Glutamine promotes intestinal SIgA secretion through intestinal microbiota and IL-13. Mol Nutr Food Res. (2016) 60:1637–48. 10.1002/mnfr.201600026 [DOI] [PubMed] [Google Scholar]
- 45.Hand TW. All bacteroides are equal but some are more equal than others for the induction of IgA. Cell Host Microbe. (2019) 27:319–21. 10.1016/j.chom.2020.02.011 [DOI] [PubMed] [Google Scholar]
- 46.Ghazalpour A, Cespedes I, Bennett BJ, Allayee H. Expanding role of gut microbiota in lipid metabolism. Curr Opin Lipidol. (2016) 27:141–7. 10.1097/MOL.0000000000000278 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated for this study can be found in the NCBI Repository [accession number: PRJNA607631].