Abstract
The microbial community of the yak (Bos grunniens) rumen plays an important role in surviving the harsh Tibetan environment where seasonal dynamic changes in pasture cause nutrient supply imbalances, resulting in weight loss in yaks during the cold season. A better understanding of rumen microbiota under different feeding regimes is critical for exploiting the microbiota to enhance feed efficiency and growth performance. This study explored the impact of different dietary energy levels on feed efficiency, rumen fermentation, bacterial community, and abundance of volatile fatty acid (VFA) transporter transcripts in the rumen epithelium of yaks. Fifteen healthy castrated male yaks were divided into three groups and fed with low (YL), medium (YM), and high energy (YH) levels diet having different NEg of 5.5, 6.2, and 6.9 MJ/kg, respectively. The increase in feed efficiency was recorded with an increase in dietary energy levels. The increase in dietary energy levels decreased the pH and increased the concentrations of acetate, propionate, butyrate, and valerate in yak rumens. The increase in the mRNA abundance of VFA transporter genes (MCT1, DRA, PAT1, and AE2) in the rumen epithelium of yaks was recorded as dietary energy level increased. High relative abundances of Firmicutes and Bacteroidetes were recorded with the increase in dietary energy levels. Significant population shifts at the genus level were recorded among the three treatments. This study provides new insights into the dietary energy-derived variations in rumen microbial community.
Keywords: yak, dietary energy, rumen microbiota, volatile fatty acid, feed efficiency, growth performance
Introduction
During recent years, the manipulation of rumen microbiota by altering diet to improve animal growth performance has gained increasing attention. The rumen is a complex environment containing diverse microbes that assist the host in digesting and utilizing feed energy. Rumen microbes produce various glucanases and xylanases to digest solid fiber by adhering to its surface, finally converting it into volatile fatty acids (VFAs), that is, acetate, propionate, and butyrate (Luo et al., 2001; Kamra, 2005). VFAs are the main source of energy in ruminants, as they provide 70–80% of the body’s energy needs and aid in growth and production performance. The VFAs produced in the rumen are absorbed and transported through the rumen epithelium into the blood by VFA transporters (Van Houtert, 1993; Yohe et al., 2019). So far, VFA transporters, including monocarboxylic acid transporters (MCT1), anion exchange carriers (AE2), downregulated in adenoma (DRA) proteins, and putative anion transporters (PAT1) are reported to be actively involved in VFA transportation through the rumen epithelium (Rajendran et al., 2000; Foltz et al., 2004).
Yak rumen microbiome coevolved with the host genome to adapt to extreme environmental conditions in the Qinghai Tibetan Plateau, where dynamic seasonal changes in the pasture cause nutrient supply imbalances, resulting in weight loss in yaks (Xue et al., 2005). Yaks greatly depend on metabolites produced by microbes in the rumen for their growth and survival (Saleem et al., 2013). Therefore, a better understanding of rumen microbiota under different feeding regimes is important for exploiting the microbiota to enhance feed efficiency (Jami and Mizrahi, 2012; Bannink et al., 2016). Feed efficiency is an important management tool in yak husbandry to improve economics of meat and milk production (Xu et al., 2017).
An increase in dietary energy levels displayed promising results in the performance of 3-year-old male yaks. Yaks showed average daily weight gains of 286.91, 446.75, and 770.42 g/day when fed with diets of 3.72, 4.52, and 5.32 MJ/kg, respectively (Hongze, 2015). A study conducted on yaks reported the difference in rumen microbiota and certain metabolites when fed different feed types divided into forage and concentrate groups (Liu et al., 2019). However, little information is available regarding the impact of different dietary energy levels on dynamic changes in the rumen fermentation and microbiota and their influence on transporter genes in the rumen epithelium of yaks. The objectives of this study were to evaluate the effect of three dietary energy levels on feed efficiency, rumen fermentation, and bacterial composition of yak. Moreover, we also studied the effects of different dietary energy levels on transcript abundance of VFA transporter genes in the rumen epithelium of yaks.
Materials and Methods
Ethics Statement
All procedures involving animal care and use were in strict accordance with the Guide for the Care and Use of Laboratory Animals, Lanzhou Institute of Husbandry and Pharmaceutical Sciences, CAAS, China [SCXK (Gan) 2014-0002]. After experiment completion, all yaks were transferred to a facility where they were humanely slaughtered by electrical stunning and exsanguination.
Animals, Diets, and Experimental Design
The feeding trial was conducted from February to May 2016 at Hongtu Yak Breeding Cooperatives of Tibetan Autonomous Prefecture in Gannan, Gansu Province, China. In brief, 15 adult castrated male yaks with similar body conditions were randomly divided into three treatment groups. Three diets of different net energy levels and a concentrate-to-forage ratio of 30:70 (DM basis) were formulated, containing similar roughage mixtures (40% microbial corn stalk silage, 40% oat silage, and 20% highland barley hay) and different energy concentrates: low (YL: 5.5 MJ/kg), medium (YM: 6.2 MJ/kg), and high energy levels (YH: 6.9 MJ/kg). The ingredients and nutrient compositions of the three diets are shown in Table 1 (Yang et al., 2017). The experiment lasted for 60 days after a 14-day adaptation period. The animals were fed ad libitum twice daily at 08:00–09:00 and 17:00–18:00 with a total of 2.45-kg concentrate and 5.75-kg roughage mixtures with free access to water. The orts were recorded to calculate average daily feed intake (ADFI), and ADFI was determined as the total feed intake/60 (Yang et al., 2017, 2019). Animals of each group were weighed before the morning feeding on the first and 60th days of the trial; feed efficiency was calculated as average daily gain/ADFI during the experimental period.
TABLE 1.
Ingredients and nutrient composition of three concentrates used during the experiment.
| Item | Treatment1 |
||
| YL | YM | YH | |
| Ingredient, g/kg of DM | |||
| Corn | 290 | 448 | 560 |
| Corn germ | 300 | 200 | 120 |
| Wheat bran | 40 | 40 | – |
| DDGS2 | 150 | 70 | 63 |
| Prickly ash seed | 100 | 20 | 40 |
| Cottonseed meal | 60 | 120 | 160 |
| Soybean meal | – | 50 | – |
| Salt | 8 | 8 | 8 |
| White stone powder | 20 | 20 | 20 |
| Dicalcium phosphate | 6 | 6 | 6 |
| Urea | 8 | – | 5 |
| Sodium bicarbonate | 10 | 10 | 10 |
| Premix3 | 8 | 8 | 8 |
| Nutrient composition, g/kg of DM | |||
| Crude protein | 165.3 | 167.4 | 172.1 |
| Crude fat | 37.3 | 41.8 | 55.7 |
| NEg4 (MJ/kg DM) | 55 | 62 | 69 |
| Neutral detergent fiber | 159.3 | 131.5 | 123.2 |
| Acid detergent fiber | 45.4 | 41.4 | 37.2 |
| Calcium | 6.4 | 8.4 | 7.5 |
| Phosphorus | 3.1 | 3.4 | 3.6 |
1LE, low energy level; ME, medium energy level; HE, high energy level. 2DDGS, distillers dried grains with solubles. 3Premix was provided per kilogram of total diet DM, and the composition was as follows: 22,520 IU of vitamin A, 1920 IU of vitamin D3, 18 IU of vitamin E, 0.36 IU of vitamin K3, 5.28 mg of vitamin B2, 0.008 mg of vitamin B12, 21.2 mg of D-calcium pantothenate, 9 mg of Cu, 132.8 mg of Zn, 240 mg of Fe, 8 mg of Mn, and 0.28 mg of Co. 4NEg, net energy for gain, is calculated according to Feeding Standard of Beef Cattle (NY/T 815-2004); others were measured values.
Sample Collection
At the end of the experiment, rumen fluid (20 ml) from yaks of each group was extracted via an oral stomach tube before the morning feeding on day 60. The tube was thoroughly cleaned using fresh warm water between sample collections, and 10–15 ml of the sample from each yak was always discarded to avoid contamination from saliva. The rumen fluid was immediately frozen in liquid nitrogen and then stored at −80°C for later analysis of VFA and bacterial diversity. Yaks were then transferred to a facility and humanely slaughtered by electrical stunning and exsanguination. The rumen was immediately separated and placed on dry ice; the rumen epithelial tissues of ∼10-cm2 size were excised from the caudoventral sac and washed with ice-cold phosphate buffer saline (pH = 7.0) to remove plant particles. The samples were snap frozen in liquid nitrogen and later stored at −80°C for total RNA isolation.
Determination of Rumen Fermentation Parameters
The rumen fluid pH was measured using a portable pH meter (Sartorius PB-10, Sartorius, Göttingen, Germany) during fluid collection. The cryopreserved rumen fluid sample was thawed at 4°C and thoroughly mixed by vortexing. Next, 10 ml of the rumen fluid was taken and centrifuged at 3000 × g for 10 min; then, 1 ml of supernatant was placed in a 1.5-ml centrifuge tube, and 0.2 ml of a metaphosphoric acid solution containing the internal standard 2-ethylbutyric acid was added. The sample was mixed, placed in an ice water bath for 30 min, and centrifuged at 10,000 × g at 4°C. The supernatant was placed in a new 1.5-ml centrifuge tube and stored in a 4°C refrigerator for testing. The VFA concentration was determined by gas chromatography (Varian 450, Agilent Technologies China, Co., Ltd., China). The gas chromatographic conditions and subsequent test procedures were conducted as described previously (Horwitz, 2010).
Rumen Bacterial Diversity Analysis
Genomic DNA was extracted from rumen fluid samples by the cetyltrimethylammonium bromide method (William et al., 2012). The DNA concentration was determined by NanoDrop 2000 (Thermo Fisher Scientific, Waltham, MA, United States), and its integrity was checked by 1.2% agarose gel electrophoresis. The extracted DNA was used for PCR amplification of the V4 region of the 16S rRNA gene using universal primer pairs (515F–806R) with barcodes. PCR amplification was performed using the Phusion® High-Fidelity PCR Master Mix with GC Buffer (New England BioLabs, Ipswich, MA, United States). The amplified products were subjected to 1.5% agarose gel electrophoresis and gel purified by using the QIAquick PCR Purification Kit to construct libraries. The libraries were constructed using the TruSeq® DNA PCR-Free Sample Preparation Kit (Illumina, San Diego, CA, United States) according to the manufacturer’s protocol. The constructed libraries were quantified by the Qubit dsDNA High-Sensitivity Assay Kit (Invitrogen, Carlsbad, CA, United States), and paired-end sequencing was performed using the Illumina HiSeq 2500 PE250 system (Illumina, San Diego, CA, United States) according to standard protocol. The sequencing data were analyzed by using the QIIME (Quantitative Insights Into Microbial Ecology, Version 1.7.0) pipeline. All sequence reads were trimmed and assigned to each sample based on their barcodes. High-quality sequences were clustered into operational taxonomic units (OTUs) at 97% identity using UCLUST software Version 7.11. The QIIME software was used to calculate Chao1, Shannon, Simpson, ACE, and Good’s coverage indices, while R software (Version 2.15.3) was used to construct weighted UniFrac distance-based principal coordinates analysis (PCoA) plot to illustrate significance differences between samples.
Analysis of the Transcripts Abundance of VFA Transporter Genes
Total RNA was isolated from the rumen epithelium by the TaKaRa MiniBEST Universal RNA Extraction Kit (code no. 9767; TaKaRa, Dalian, China) according to the manufacturer’s instructions. The quality and integrity of total RNA were determined by NanoDrop 2000 (Thermo Fisher Scientific, Waltham, MA, United States) and 1.2% agarose gel electrophoresis, respectively. The RNA samples were reverse transcribed using the PrimeScriptTM RT Reagent Kit (TaKaRa, Dalian, China) according to the manufacturer’s instructions. Primers were designed using the Primer Premier 5 software (PREMIER Biosoft International, San Francisco, CA, United States) for VFA transporter genes MCT1, DRA, PAT1, and AE2 (Supplementary Table S1). Quantitative real-time PCR (qRT-PCR) was performed in triplicate to determine the transcripts’ relative abundances using SYBR1 Premix Ex TaqTM II (TaKaRa, Dalian, China) relative to transcript levels of reference gene β-actin. Reactions were run on a fluorescence thermal cycler (CFX96, Bio-Rad, Hercules, CA, United States), and the program was as follows: 95°C for 30 s, 40 cycles of 95°C for 5 s and annealing at 60°C for 30 s, and a melting curve with a temperature increase of 0.5°C every 5 s starting at 65°C. qRT-PCR analysis for each studied gene was performed using cDNA from five biological replicates with three technical replicates per biological replicate. The threshold cycle (CT) resulting from qRT-PCR was analyzed using the 2–Δ Δ Ct method, and all data were normalized with the reference gene β-actin.
Statistical Analysis
The R software (Version 2.15.3) was used to perform variation tests among three treatments at various classification levels (phylum and genus) to obtain P-values. The lm function of the Estimability package in R was used to evaluate the linear and quadratic effects of the dietary energy levels on the bacterial abundance. Analysis of similarities (Anosim) was executed using the Anosim function of the R vegan package. The growth parameters, VFA concentration data, and abundance of transcripts of VFA transporter genes were processed by one-way analysis of variance using the least significant difference procedure to perform multiple comparisons in the SPSS software. Significance was declared at P < 0.05. Pearson correlations analysis was performed to calculate relations of VFA concentration and abundance of transcripts of VFA transporter genes with relative abundance of top bacterial genera having a relative abundance of ≥0.1% in at least one of the samples using GraphPad Prism 8.0.2, and a heatmap was generated by R software (Version 2.15.3). P-values were adjusted with false discovery rate, and the corrected P-values < 0.05 were regarded as statistically significant.
Data Availability
The data for this study have been deposited in the European Nucleotide Archive (ENA) at EMBL-EBI under accession number PRJEB342982.
Results
Yak Growth Performance
The growth performance of yaks has been previously reported (Yang et al., 2019). The final body weights of YM and YH group animals were significantly higher than those in the YL group (P < 0.05). However, no significant increase in the final body weight was observed between YM and YH groups (P < 0.05). Feed efficiency was significantly affected (P < 0.001) by an increased dietary energy level and was higher in the YM and YH groups than in the YL group; however, no significant difference was observed between YH and YM groups (Table 2).
TABLE 2.
Effects of dietary energy levels on growth performance of yak.
| Parameters | Treatment groups |
SEM | P-value | ||
| YL | YM | YH | |||
| Initial BW (kg) | 276.0 | 277.0 | 275.2 | 2.55 | 0.78 |
| Final BW (kg) | 313.4b | 330.0a | 326.8a | 4.35 | 0.037 |
| Average daily gain (kg/d) | 0.63 b | 0.88a | 0.86a | 0.034 | <0.001 |
| Average daily feed intake (ADFI) (kg/d) | 8.12a | 7.96 | 7.78b | 0.05 | 0.01 |
| Feed efficiency | 0.076a | 0.11b | 0.11b | 0.01 | <0.001 |
YL, low energy; YM, medium energy; YH, high energy; SEM, standard error of the mean. Values are presented as mean ± SEM. Means in a row with different small letter superscripts differ significantly (P < 0.05); those with the same letter superscripts present no difference (P > 0.05).
Rumen VFA Concentrations and Transcript Abundances of VFA Transporter Genes
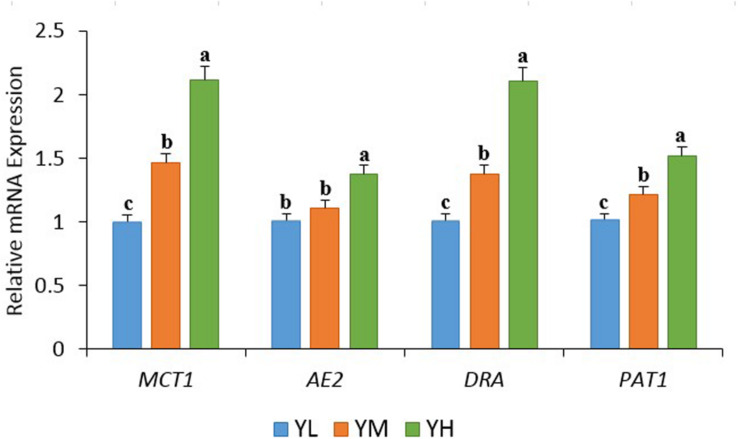
The increase in dietary energy level significantly decreased the pH of yak rumen fluid (P < 0.05), and significant increases in the concentrations of acetate, propionate, butyrate, valerate, and total VFA were observed (P < 0.05) (Table 3). The influence of different dietary energy levels was investigated by examining the transcript abundance of VFA transporter genes (Figure 1). The increase in dietary energy levels significantly enhanced (P < 0.05) the mRNA abundance of MCT1, DRA, and PAT1 in the yak rumen epithelium. Abundance of AE2 transcripts showed no difference between YL and YM groups (P > 0.05) but was significantly higher in the YH group (P < 0.05).
TABLE 3.
Effects of different dietary energy levels on rumen VFA concentrations of yak.
| Parameters | Treatment groups |
SEM | P-value | ||
| YL | YM | YH | |||
| pH | 6.90a | 6.66b | 6.47c | 0.05 | <0.001 |
| Total VFA (mM) | 51.4c | 60.9b | 72.1a | 2.41 | <0.001 |
| Acetate (mM) | 37.4c | 44.9b | 51.9a | 1.71 | <0.001 |
| Propionate (mM) | 8.7c | 9.7b | 11.4a | 0.36 | 0.001 |
| Butyrate (mM) | 4.02c | 4.98b | 7.11a | 0.45 | <0.01 |
| Isobutyrate (mM) | 0.32 | 0.33 | 0.37 | 0.01 | 0.08 |
| Valerate (mM) | 0.26c | 0.31b | 0.44a | 0.02 | <0.01 |
| Isovalerate (mM) | 0.66 | 0.66 | 0.77 | 0.02 | 0.09 |
YL, low energy; YM, medium energy; YH, high energy; SEM, standard error of the mean. Values are presented as mean ± SEM. Means in a row with different small letter superscripts differ significantly (P < 0.05); those with the same letter superscripts present no difference (P > 0.05).
FIGURE 1.
The transcript abundances of VFA transporter genes in the rumen epithelium of yak among three dietary energy treatments. Within a panel, means without a common letter differ (P < 0.05). MCT1, monocarboxylate transporter 1; DRA, downregulated in adenoma; AE2, anion exchanger 2; PAT1, putative anion transporter 1.
Bacterial 16S rRNA Gene Sequencing
In total, 911,966 raw reads (average 59,192 sequences per sample) were obtained from bacterial 16S rRNA sequencing of 15 samples. After quality filtering, 895,658 sequences were obtained, of which 7783 were chimeric reads that were removed from further analysis. The mean sequence read length was 253 bp. The non-chimeric reads were clustered into 2662 OTUs at 97% similarity levels, of which 2118 were identified as low-abundance OTUs (79.6%) having less than 10 reads, and 749 OTUs were present in all three treatment groups (28%). The qualified sequences were clustered at 97% similarity levels. Good’s coverage for all samples was more than 99%. The sequencing depth to describe the OTU-level bacterial diversity was evaluated by a rarefaction curve of observed species of all samples (Supplementary Figure S1). The curves of all samples reached a plateau, indicating that a sufficient number of sequences had been generated to investigate bacterial diversity in the rumen. OTU analysis identified 1828 common OTUs in three groups, while 314, 67, and 69 unique OTUs were discovered in the YL, YM, and YH groups, respectively.
Of the 26 identified bacterial phyla in the rumen samples, Firmicutes, Bacteroidetes, Tenericutes, and Lentisphaerae showed high relative abundance among the three groups. The mean relative abundances of Firmicutes and Bacteroidetes were 48.4 ± 5.7 and 39.4 ± 5.4%, respectively, accounting for 87.8% of the total phyla (Supplementary Figure S2), and were higher in YM and YL groups, respectively. A high Firmicutes: Bacteroidetes F:B ratio (1.41) was recorded in the YM group, followed by YH (1.27) and YL (1.12); however, these changes were not significant among the three groups (Table 4). Other phyla, that is, Proteobacteria, Chloroflexi, Fibrobacteres, and Spirochaetes had relative abundances of ≥0.1% in all groups. These predominant phyla displayed different relative abundances and compositions among the three groups (Supplementary Figure S2). Different dietary energy levels had little influence on relative abundances of the major phyla; however, Tenericutes (P = 0.04), Lentisphaerae (P = 0.02), Spirochaetes (P = 0.02), and SHA-109 (P = 0.02) showed significant differences. Lentisphaerae, Spirochaetes, and SHA-109 were linearly increased as the diet energy level changed from low to high, while Tenericutes showed a quadratic relationship (Table 4).
TABLE 4.
Main phylum of the rumen samples from different dietary treatment groups of yak.
| Phylum | Treatment groups |
SEM | P-value | ||
| YL | YM | YH | |||
| Firmicutes | 0.46 | 0.51 | 0.48 | 0.014 | 0.52 |
| Bacteroidetes | 0.43 | 0.36 | 0.38 | 0.014 | 0.16 |
| Tenericutes | 0.03ab | 0.05a | 0.02b | 0.004 | 0.04 |
| Lentisphaerae | 0.026b | 0.03b | 0.04a | 0.003 | 0.02 |
| Proteobacteria | 0.015 | 0.012 | 0.012 | 0.001 | 0.72 |
| Chloroflexi | 0.004 | 0.002 | 0.007 | 0.001 | 0.31 |
| Fibrobacteres | 0.005 | 0.006 | 0.004 | 0.0008 | 0.70 |
| Spirochaetes | 0.003b | 0.004b | 0.008a | 0.0008 | 0.02 |
| SHA-109 | 0.0004b | 0.0005b | 0.001a | 0.0002 | 0.02 |
| F:B | 1.12 | 1.41 | 1.27 | 0.082 | 0.39 |
YL, low energy; YM, medium energy; YH, high energy; SEM, standard error of mean. Means in a row with different small letter superscripts differ significantly (P < 0.05).
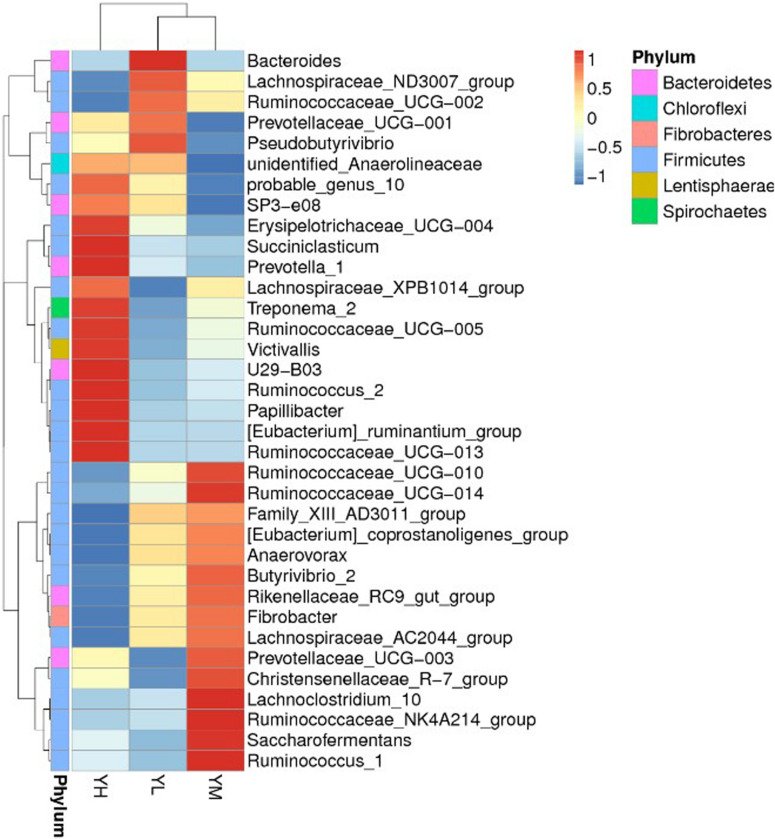
At the genus level, 274 genera were identified in all samples, of which Christensenellaceae_R-7_group, Rikenellaceae_RC9_gut_group, Prevotella_1, Ruminococcaceae_NK4A214_group, and Prevotellaceae_UCG-003 were the most abundant. The mean relative abundances of Christensenellaceae_R-7_group and Rikenellaceae_RC9_gut_group were 12.4 ± 4 and 10.9 ± 3.2%, respectively, accounting for 23.3% of the total genera (Figure 2). A heatmap was constructed to examine the relative abundance of different genera in the three treatment groups (Figure 2). The relative abundances of Bacteroides and Pseudobutyrivibrio were higher in YL than in YM and YH groups. High relative abundances of Rikenellaceae_RC9_gut_group, Ruminococcaceae, Ruminococcus, Lachnoclostridium, and Saccharofermentans were recorded in the YM group, while Erysipelotrichaceae, U29-B03, Succiniclasticum, Prevotella, Treponema, Victivallis, and Papillibacter were the most abundant in the YH group.
FIGURE 2.
Heatmap showing the rumen bacterial composition at genus level in yak fed different dietary energy levels. YL, low energy; YM, medium energy; YH, high energy.
Bacterial Diversity Analysis
Significant differences were found in the bacterial communities at the OTU level between YH and YL (Anosim, P < 0.05) groups and YH and YM (P < 0.01) groups, while no significant difference was observed between the YM and YL (P > 0.05) groups. For alpha diversity analysis, we calculated the Shannon diversity, Simpson, ACE, and Chao1 indices for each treatment. Alpha diversity analysis showed no significant (P > 0.05) difference in the rumen bacterial community as dietary energy levels increased (Table 5). The rumen bacterial diversity of each group tended to be stable, but YM group diversity indices were slightly higher than those of the YL and YH groups.
TABLE 5.
Alpha diversity indices for different dietary treatment groups of yak.
| Indices | Treatment groups |
SEM | P-value | ||
| YL | YM | YH | |||
| Shannon | 8.09 | 8.18 | 8.17 | 0.03 | 0.49 |
| Simpson | 0.99 | 0.99 | 0.99 | 0.0006 | 0.05 |
| Chao1 | 1721.6 | 1732.4 | 1624.3 | 32.50 | 0.35 |
| ACE | 1720.4 | 1736.6 | 1646.5 | 27.07 | 0.37 |
YL, low energy; YM, medium energy; YH, high energy; SEM, standard error of the mean.
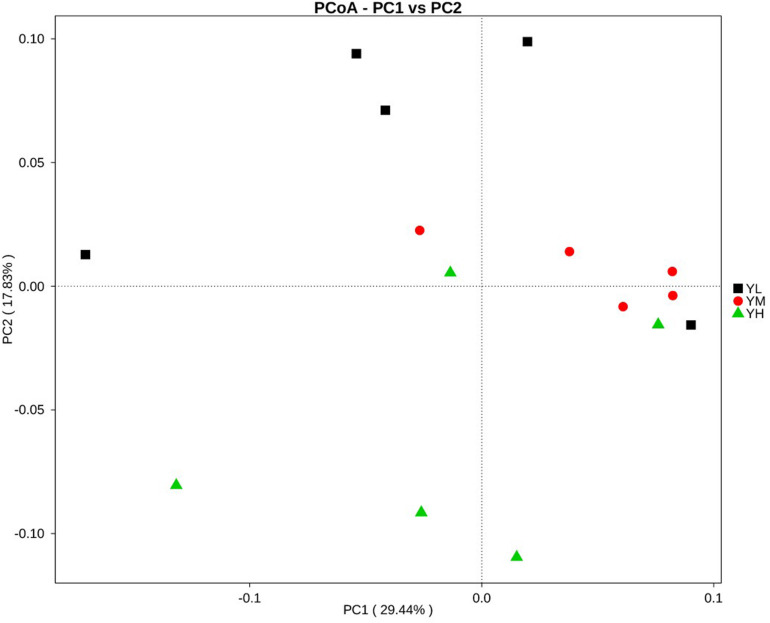
Principal coordinates analysis for beta diversity showed that the bacterial composition and structure under different energy diets were significantly different, as indicated by the first two principal component scores that accounted for 29.44 and 17.83% of the total variations (Figure 3).
FIGURE 3.
Principal coordinate analysis (PCoA) for three dietary energy treatments. X-axis, first principal component, and Y-axis, second principal component. Different colors represent different groups.
Correlation of Bacterial Genera With VFA Concentrations and Transcript Abundances of VFA Transporter Genes
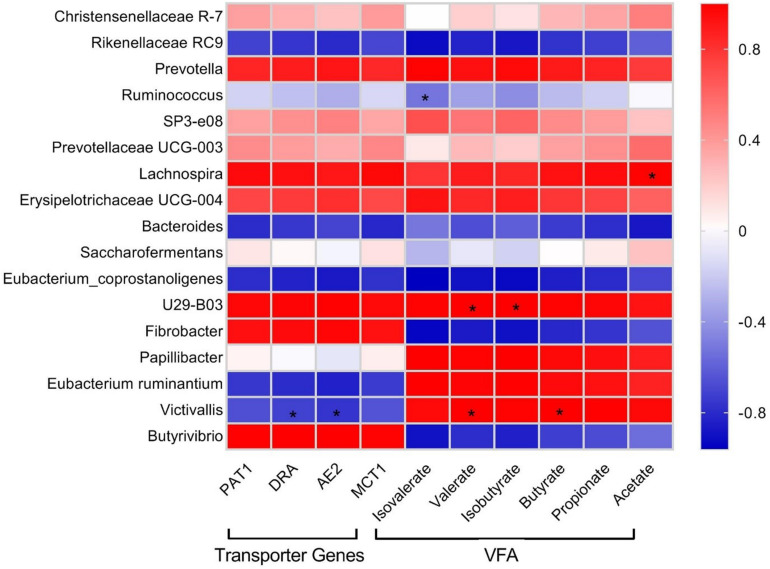
Pearson correlation analysis was performed in order to check the relationships of the relative abundance of selected bacterial genera identified by 16S rRNA sequencing with rumen VFA concentration and abundance of VFA transporter transcripts (Figure 4). Erysipelotrichaceae UCG-004, Lachnospira, Prevotellaceae UCG-003, SP3-e08, Prevotella, U29-B03, and Christensenellaceae_R-7 showed positive correlations with VFA and VFA transporter genes. Papillibacter displayed a strong positive correlation with VFA, weak positive correlation with PAT-1 and MCT-1, and weak negative correlation with DRA and AE2. Eubacterium ruminantium and Victivallis showed strong positive correlations with VFA and strong negative correlations with VFA transporter genes. Saccharofermentans showed weak positive correlations with acetate, propionate, butyrate, PAT-1, DRA, and MCT-1 and weak negative correlations with isobutyrate, valerate, isovalerate, and AE2. Butyrivibrio and Fibrobacter displayed strong negative correlations with both VFA and VFA transporter genes. Eubacterium_coprostanoligenes, Bacteroides, Ruminococcus, and Rikenellaceae_RC9 exhibited negative correlations with VFA and VFA transporter genes.
FIGURE 4.
Pearson correlation coefficients for microbial genera with rumen VFA concentrations and transcript abundances of VFA transporter genes in yak. Color intensity represents correlation coefficient. Significant difference (P < 0.05) is represented by an asterisk (*).
Discussion
Feed efficiency is an important trait for yak husbandry and economic income. Feed efficiency is influenced by many factors such as management practices, physiological mechanism, nutrition, host genetics, and rumen microbiota. Rumen microbiota provides feed energy to the animal through fermentation in the form of VFA. Studies have shown that rumen microbiota can be manipulated by diet to enhance feed efficiency of animals (Jiao et al., 2015; Yang et al., 2017). So, in this study, we evaluated feed efficiency, rumen bacterial community, and microbial fermentation of yak fed with different dietary energy levels. Moreover, we also checked the effects of different dietary energy levels on VFA transporter genes in the rumen epithelium of yak.
We observed significant increases in acetate, propionate, butyrate, and valerate concentrations as dietary energy levels increased; however, no significant increase in the concentrations of isobutyrate and isovalerate was observed. Among all VFAs, acetate, propionate, and butyrate are particularly known to be associated with animal feed efficiency. Acetate and butyrate are used for fat synthesis and energy supply, while propionate is the main source of glucose for ruminants (Den Besten et al., 2013). We recorded high feed efficiency in yaks from YM and YH groups, which could be explained by a significant increase in concentrations of VFA with the resulting increase of metabolizable energy content of the feed (Yang et al., 2019). High concentrations of acetate, propionate, and valerate in rumen of high milk-producing Holstein dairy cows were reported (Xue et al., 2019). High concentrations of butyrate and a tendency for greater total VFA concentration and acetate have been reported in efficient animals (Guan et al., 2008). A study conducted on beef cattle reported the association of concentrations of rumen VFA and feed efficiency (Li et al., 2019). Increase in concentrations of rumen VFA is known to be associated with lower rumen pH and decrease in animal performance (Koike and Kobayashi, 2001; Klieve et al., 2003). We recorded a decline in rumen pH with an increase in VFA concentrations, but no significant difference in feed efficiency was recorded between YM and YH groups. One reason could be that rumen pH in this study did not cross the threshold value for rumen acidosis that is associated with low feed efficiency in ruminants (Krause and Oetzel, 2006). To maintain rumen pH, VFAs are transported from the rumen to the blood through VFA transporters. The increase in VFA concentrations in yak rumen presumably enhanced the mRNA expression of transporter genes that are involved in VFA absorption and transportation, that is, MCT1, AE2, DRA, and PAT1, in the rumen epithelium. The increases in transcripts abundances of MCT1, AE2, DRA, and PAT1 were also reported in goats fed a high-energy diet (Yan et al., 2014). The concentrations of different VFAs in the rumen associate with the composition of microbial community (Griswold et al., 1999). We then evaluated the differences in rumen bacterial community of yak and their association in improving feed efficiency. In this study, alpha diversity indices did not differ significantly between treatments. Studies have shown host specificity of rumen bacterial community, which might have played an important role in this study (Wiener et al., 2006; Clemmons et al., 2019; Li et al., 2019). However, we recorded significant differences in beta diversity supporting the variations between the microbial communities of each group. Of the 26 bacterial phyla identified in yak rumens, Firmicutes and Bacteroidetes showed high relative abundances. The presence of these phyla has also been reported in the rumen of different ruminants, indicating their importance in the rumen (Ley et al., 2008). These two phyla are considered as an important microbial parameter to assess the energy requirements of ruminants (Xue et al., 2017). Firmicutes are known to be involved in cellulose, hemicellulose, starch, and oligosaccharide degradation as well as in acetate, propionate, and butyrate production. Moreover, Bacteroidetes are known for their role in oligosaccharide hydrolysis and acetate and propionate production. Yaks fed with different feed types showed high relative abundances of Bacteroidetes and Firmicutes in forage and concentrate groups, respectively (Liu et al., 2019). Grazing yaks with low feed efficiency displayed high Bacteroidetes and less Firmicutes abundances in rumen (Zou et al., 2019). The high relative abundance of Firmicutes, increasing the Firmicutes-to-Bacteroidetes ratio, was reported to be associated with efficient feed utilization in animals (Ley et al., 2005; Turnbaugh et al., 2006; Myer et al., 2015). In this study, the high abundance of Firmicutes in the rumen suggests that this shift might have played an important role in affecting feed efficiency. The lack of significant differences in the major phyla between groups might indicate the importance of variations in microbial communities at the genus level. Many of the changes were identified within the phylum Firmicutes at the genus level, which included Christensenellaceae_R-7_group, Ruminococcaceae_NK4A214_group, Ruminococcaceae, Ruminococcus, Lachnoclostridium, Saccharofermentans, Erysipelotrichaceae, Succiniclasticum, and Papillibacter. We recorded significant differences in the relative abundances of phyla Lentisphaerae and Tenericutes with the increase in dietary energy levels. The phylum Lentisphaerae is reported to be associated with changes in feed efficiency, and Tenericutes is essential for rumen homeostasis and health of the host (Mao et al., 2013; Guan et al., 2017). However, little data have been presented on the role of these phyla in improving feed efficiency in the rumen, and the significance of their role in the rumen remains to be determined.
Heatmap analysis revealed significant differences in the relative abundance of specific genera between different dietary energy groups. The relative abundance of genus Bacteroides was higher in the YL group. The relative abundance of Bacteroides in the rumen is associated with dietary crude fiber content, and their main role is to degrade hemicellulose (Thomas et al., 2011). YL group yaks utilized more roughage, so Bacteroides were abundant in their rumen and presumably conducted fiber decomposition. A high relative abundance of Bacteroides was also reported in the rumens of goats fed with a low-concentrate diet (Hua et al., 2017). The relative abundances of Rikenellaceae_RC9_gut_group, Ruminococcaceae, Ruminococcus, Lachnoclostridium, and Saccharofermentans were higher in the YM group. Rikenellaceae_RC9_gut_group are known to indicate animal health and associated with obesity (Clarke et al., 2013). Rikenellaceae_RC9_gut_group are known for their role in producing acetate and propionate as fermentation end products (Holman and Gzyl, 2019). Studies showed the involvement of Ruminococcaceae in fiber degradation and biohydrogenation in the rumen (Gagen et al., 2015; Opdahl et al., 2018). Diet supplemented with linseed oil reported the increase in relative abundance of genus Ruminococcus in the rumen of Yanbian Yellow Cattle (Li et al., 2015). Ruminococcus is also known as an important genus involved in acetate production (Jeyanathan et al., 2014). In this study, Ruminococcus showed a negative correlation with acetate, although a high relative abundance of Ruminococcus was identified in the YM group. The increase in the relative abundance of Ruminococcus might be stimulated by resistant starch and crude fat present in the diet due to its characteristic amylolytic and lipolytic activities (Krause et al., 2003; Ferrario et al., 2017). In the YH group, Erysipelotrichaceae, U29-B03, Succiniclasticum, Prevotella, Treponema, Victivallis, and Papillibacter were the most dominant genera. Treponema is mainly involved in the degradation process of soluble carbohydrates (Stanton and Canale-Parola, 1980), and Succiniclasticum is a starch-degrading bacterium that degrades dietary starch, produces acetic acid, and succinic acid, and converts succinic acid to propionic acid (Fernando et al., 2010; Huws et al., 2015). In beef cow rumens, Treponema and Ruminobacter showed high abundances when a high-grain diet was provided, which suggested that these bacteria were present in response to adaptation to the high-grain diet (Chen et al., 2011). Prevotella, another important genus in the rumen, utilizes hemicellulose and plays important roles in protein metabolism and starch degradation (Griswold et al., 1999; Ramšak et al., 2000). The replacement of Ruminococcus and Butyrivibrio by Prevotella has been broadly reported during adaptations of rumen microbiota to high-energy diets (Callaway et al., 2010; Fernando et al., 2010; Pitta et al., 2014; Derakhshani et al., 2017). The high relative abundance of Prevotella was also reported in a high-energy-diet group compared to groups fed medium- and low-energy diets in white Cashmere goats (Xufeng, 2015). In cows, Prevotella abundance was significantly increased when the diet was switched from low grain to high grain (Fernando et al., 2010). An in vitro study conducted with sheep rumen fluid reported the increase in Prevotella with supplementation with plant and marine oils (Vargas et al., 2017). In this study, YH yaks had the highest rumen starch contents; furthermore, the high abundance of Prevotella and rapid starch degradation in the rumen, which increased acetic acid and propionic acid yields, indicated a positive correlation of Prevotella with VFA production. Dietary energy levels clearly influenced the rumen bacterial community and increased the relative abundances of non-structural carbohydrate-degrading bacteria. Here, we studied rumen bacterial community by using rumen fluid which lacks undigested solid fiber particles. Whole rumen content including liquid and solid fractions should be studied to better explore the bacterial composition.
Correlation analysis indicated a cluster of bacteria positively correlated with VFA and transcript abundance of VFA transporters mainly belonging to Firmicutes and Bacteroidetes, including Erysipelotrichaceae UCG-004, Lachnospira, Prevotellaceae UCG-003, SP3-e08, Prevotella, U29-B03, and Christensenellaceae_R-7, signifying their importance in VFA synthesis and energy generation. Christensenellaceae_R-7 and Rikenellaceae_RC9, the dominant genera in this study, displayed positive and negative correlations with all ruminal VFAs, respectively. We also identified that E. ruminantium and Victivallis displayed positive correlations with VFAs and negative correlation with VFA transporter genes. The correlation analysis in this study was based on a combined dataset; the lack of significant correlations between some bacterial taxa with VFA and their transporter genes does not suggest that those bacterial taxa are unimportant. More work is needed to explore the correlations, because a small number of species might have a strong impact on rumen fermentation parameters (Hanage, 2014; Patra and Yu, 2014; Shabat et al., 2016). It must be taken into account that although the variations in genera and their putative functions in the rumen could be correlated with the observed differences in the phenotypes, their contribution in feed efficiency is still not clear and require more study.
Conclusion
In conclusion, the results presented here provide new information regarding the effects of different dietary energy levels on feed efficiency, rumen fermentation, transcript abundance of VFA transporters, and microbial communities. The increase in dietary energy levels enhanced the feed efficiency of yaks. Increase in the concentration of VFAs and transcript abundance of VFA transporters was also recorded with an increase in dietary energy levels. The dietary energy levels affected the microbial diversity, and the increase in energy levels increased the relative abundance of Lentisphaerae and Tenericutes at the phylum level and increased the relative abundance of Erysipelotrichaceae, U29-B03, Succiniclasticum, Prevotella, Treponema, Victivallis, and Papillibacter at the genus level. These findings are of great importance for the targeted improvement of nutrient levels in ruminants.
Data Availability Statement
The datasets generated for this study can be found in the http://www.ebi.ac.uk/ena/data/view/PRJEB34298.
Ethics Statement
The animal study was reviewed and approved by guidelines for the Care and Use of Laboratory Animals, Lanzhou Institute of Husbandry and Pharmaceutical Sciences, Chinese Academy of Agricultural Sciences, China. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author Contributions
XD, PY, and RL designed the research. AA, CY, QK, JZ, CL, ZL, and MD conducted the experiment and collected the data. AA and CY analyzed the data. AA wrote the first draft of manuscript text. XD, CY, QK, JZ, and JH revised the manuscript. All authors read and approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding. This work was supported by a grant from the international cooperation and exchange program of the National Natural Science Foundation of China (No. 31461143020) and international cooperation projects in Gansu province (No. 1504WKCA053). None of the funders had any role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; or preparation, review, or approval of the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.00625/full#supplementary-material
References
- Bannink A., Van Lingen H. J., Ellis J. L., France J., Dijkstra J. (2016). The contribution of mathematical modeling to understanding dynamic aspects of rumen metabolism. Front. Microbiol. 7:1820. 10.3389/fmicb.2016.01820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaway T., Dowd S., Edrington T., Anderson R., Krueger N., Bauer N., et al. (2010). Evaluation of bacterial diversity in the rumen and feces of cattle fed different levels of dried distillers grains plus solubles using bacterial tag-encoded FLX amplicon pyrosequencing. J. Anim. Sci. 88 3977–3983. 10.2527/jas.2010-2900 [DOI] [PubMed] [Google Scholar]
- Chen Y., Penner G. B., Li M., Oba M., Guan L. L. (2011). Changes in bacterial diversity associated with epithelial tissue in the beef cow rumen during the transition to a high-grain diet. Appl. Environ. Microbiol. 77 5770–5781. 10.1128/aem.00375-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke S. F., Murphy E. F., O’sullivan O., Ross R. P., O’toole P. W., Shanahan F., et al. (2013). Targeting the microbiota to address diet-induced obesity: a time dependent challenge. PLoS ONE 8:e65790. 10.1371/journal.pone.0065790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemmons B. A., Voy B. H., Myer P. R. (2019). Altering the gut microbiome of cattle: considerations of host-microbiome interactions for persistent microbiome manipulation. Microb. Ecol. 77 523–536. 10.1007/s00248-018-1234-9 [DOI] [PubMed] [Google Scholar]
- Den Besten G., Van Eunen K., Groen A. K., Venema K., Reijngoud D.-J., Bakker B. M. (2013). The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J. Lipid Res. 54 2325–2340. 10.1194/jlr.r036012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derakhshani H., Tun H. M., Cardoso F. C., Plaizier J. C., Khafipour E., Loor J. (2017). Linking peripartal dynamics of ruminal microbiota to dietary changes and production parameters. Front. Microbiol. 7:2143 10.3389/fmicb.2016.02143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernando S. C., Purvis H., Najar F., Sukharnikov L., Krehbiel C., Nagaraja T., et al. (2010). Rumen microbial population dynamics during adaptation to a high-grain diet. Appl. Environ. Microbiol. 76 7482–7490. 10.1128/aem.00388-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrario C., Statello R., Carnevali L., Mancabelli L., Milani C., Mangifesta M., et al. (2017). How to feed the mammalian gut microbiota: bacterial and metabolic modulation by dietary fibers. Front. Microbiol. 8:1749 10.3389/fmicb.2017.01749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foltz M., Boll M., Raschka L., Kottra G., Daniel H. (2004). A novel bifunctionality: PAT1 and PAT2 mediate electrogenic proton/amino acid and electroneutral proton/fatty acid symport. FASEB J. 18 1758–1760. 10.1096/fj.03-1387fje [DOI] [PubMed] [Google Scholar]
- Gagen E. J., Padmanabha J., Denman S. E., Mcsweeney C. S. (2015). Hydrogenotrophic culture enrichment reveals rumen Lachnospiraceae and Ruminococcaceae acetogens and hydrogen-responsive Bacteroidetes from pasture-fed cattle. FEMS Microb. Lett. 362:104. [DOI] [PubMed] [Google Scholar]
- Griswold K. E., White B. A., Mackie R. I. (1999). Diversity of extracellular proteolytic activities among Prevotella species from the rumen. Curr. Microbiol. 39 187–194. 10.1007/s002849900443 [DOI] [PubMed] [Google Scholar]
- Guan L. L., Nkrumah J. D., Basarab J. A., Moore S. S. (2008). Linkage of microbial ecology to phenotype: correlation of rumen microbial ecology to cattle’s feed efficiency. FEMS Microbiol. Lett. 288 85–91. 10.1111/j.1574-6968.2008.01343.x [DOI] [PubMed] [Google Scholar]
- Guan Y., Yang H., Han S., Feng L., Wang T., Ge J. (2017). Comparison of the gut microbiota composition between wild and captive sika deer (Cervus nippon hortulorum) from feces by high-throughput sequencing. AMB Express 7:212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanage W. P. (2014). Microbiology: microbiome science needs a healthy dose of scepticism. Nat. News 512 247. 10.1038/512247a [DOI] [PubMed] [Google Scholar]
- Holman D. B., Gzyl K. E. (2019). A meta-analysis of the bovine gastrointestinal tract microbiota. FEMS Microbiol. Ecol. 95. [DOI] [PubMed] [Google Scholar]
- Hongze W. (2015). Effects of Dietary Energy Levels on Performance, rumen Fermentation and Intramuscular fat Metabolism of Fattening Calves. Ya’an: Sichuan Agricultural University. [Google Scholar]
- Horwitz W. (2010). Official Methods of Analysis of AOAC International: Agricultural Chemicals, Contaminants, Drugs/Edited by William Horwitz, Vol. 1 Gaithersburg, MD: AOAC International, 1997. [Google Scholar]
- Hua C., Tian J., Tian P., Cong R., Luo Y., Geng Y., et al. (2017). Feeding a high concentration diet induces unhealthy alterations in the composition and metabolism of ruminal microbiota and host response in a goat model. Front. Microbiol 8:138 10.3389/fmicb.2017.00138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huws S. A., Edwards J. E., Creevey C. J., Rees Stevens P., Lin W., Girdwood S. E., et al. (2015). Temporal dynamics of the metabolically active rumen bacteria colonizing fresh perennial ryegrass. FEMS Microb. Ecol. 92:137. [DOI] [PubMed] [Google Scholar]
- Jami E., Mizrahi I. (2012). Composition and similarity of bovine rumen microbiota across individual animals. PLoS ONE 7:e33306. 10.1371/journal.pone.0033306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeyanathan J., Martin C., Morgavi D. (2014). The use of direct-fed microbials for mitigation of ruminant methane emissions: a review. Animal 8 250–261. 10.1017/s1751731113002085 [DOI] [PubMed] [Google Scholar]
- Jiao J. Z., Huang J. Y., Zhou C., Tan Z. L. (2015). Taxonomic identification of ruminal epithelial bacterial diversity during rumen development in goats. Appl. Environ. Microbiol. 81 3502–3509. 10.1128/AEM.00203-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamra D. N. (2005). Rumen Microbial Ecosystem. Berlin: Springer Science & Business Media. [Google Scholar]
- Klieve A. V., Hennessy D., Ouwerkerk D., Forster R. J., Roderick I. M., Attwood G. T. (2003). Establishing populations of Megasphaera elsdenii YE 34 and Butyrivibrio fibrisolvens YE 44 in the rumen of cattle fed high grain diets. J. Appl. Microbiol. 95 621–630. 10.1046/j.1365-2672.2003.02024.x [DOI] [PubMed] [Google Scholar]
- Koike S., Kobayashi Y. (2001). Development and use of competitive PCR assays for the rumen cellulolytic bacteria: Fibrobacter succinogenes, Ruminococcus albus and Ruminococcus flavefaciens. FEMS Microbiol. Lett. 204 361–366. 10.1111/j.1574-6968.2001.tb10911.x [DOI] [PubMed] [Google Scholar]
- Krause D. O., Denman S. E., Mackie R. I., Morrison M., Rae A. L., Attwood G. T., et al. (2003). Opportunities to improve fiber degradation in the rumen: microbiology, ecology, and genomics. FEMS Microbiol. Rev. 27 663–693. 10.1016/s0168-6445(03)00072-x [DOI] [PubMed] [Google Scholar]
- Krause K. M., Oetzel G. R. (2006). Understanding and preventing subacute ruminal acidosis in dairy herds: a review. Anim. Feed Sci. Technol. 126 215–236. 10.1016/j.anifeedsci.2005.08.004 [DOI] [Google Scholar]
- Ley R. E., Bäckhed F., Turnbaugh P., Lozupone C. A., Knight R. D., Gordon J. I. (2005). Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. U.S.A. 102 11070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley R. E., Hamady M., Lozupone C., Turnbaugh P. J., Ramey R. R., Bircher J. S., et al. (2008). Evolution of mammals and their gut microbes. Science 320 1647–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Li C., Chen Y., Liu J., Zhang C., Irving B., et al. (2019). Host genetics influence the rumen microbiota and heritable rumen microbial features associate with feed efficiency in cattle. Microbiome 7:92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. Z., Park B. K., Shin J. S., Choi S. H., Smith S. B., Yan C. G. (2015). Effects of dietary linseed oil and propionate precursors on ruminal microbial community, composition, and diversity in yanbian yellow cattle. PLoS ONE 10:e0126473. 10.1371/journal.pone.0126473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Wu H., Liu S., Chai S., Meng Q., Zhou Z. (2019). Dynamic alterations in yak rumen bacteria community and metabolome characteristics in response to feed type. Front. Microbiol. 10:1116 10.3389/fmicb.2019.01116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y., Pfister P., Leisinger T., Wasserfallen A. (2001). The Genome of archaeal prophage ΨM100 encodes the lytic enzyme responsible for autolysis of Methanothermobacter wolfeii. J. Bacteriol. 183 5788–5792. 10.1128/jb.183.19.5788-5792.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao S., Zhang R., Wang D., Zhu W. (2013). Impact of subacute ruminal acidosis (SARA) adaptation on rumen microbiota in dairy cattle using pyrosequencing. Anaerobe 24 12–19. 10.1016/j.anaerobe.2013.08.003 [DOI] [PubMed] [Google Scholar]
- Myer P. R., Smith T. P., Wells J. E., Kuehn L. A., Freetly H. C. (2015). Rumen microbiome from steers differing in feed efficiency. PLoS ONE 10:e0129174. 10.1371/journal.pone.0129174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opdahl L., Gonda M., St-Pierre B. (2018). Identification of uncultured bacterial species from firmicutes, bacteroidetes and candidatus Saccharibacteria as candidate cellulose utilizers from the rumen of beef cows. Microorganisms 6 17. 10.3390/microorganisms6010017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patra A. K., Yu Z. (2014). Effects of vanillin, quillaja saponin, and essential oils on in vitro fermentation and protein-degrading microorganisms of the rumen. Appl. Microb. Biotechnol. 98 897–905. 10.1007/s00253-013-4930-x [DOI] [PubMed] [Google Scholar]
- Pitta D., Kumar S., Vecchiarelli B., Shirley D., Bittinger K., Baker L., et al. (2014). Temporal dynamics in the ruminal microbiome of dairy cows during the transition period. J. Anim. Sci. 92 4014–4022. 10.2527/jas.2014-7621 [DOI] [PubMed] [Google Scholar]
- Rajendran V. M., Black J., Ardito T. A., Sangan P., Alper S. L., Schweinfest C., et al. (2000). Regulation of DRA and AE1 in rat colon by dietary Na depletion. Am. J. Physiol. Gastroint. Liver Physiol. 279 G931–G942. [DOI] [PubMed] [Google Scholar]
- Ramšak A., Peterka M., Tajima K., Martin J. C., Wood J., Johnston M. E. A., et al. (2000). Unravelling the genetic diversity of ruminal bacteria belonging to the CFB phylum. FEMS Microbiol. Ecol. 33 69–79. 10.1111/j.1574-6941.2000.tb00728.x [DOI] [PubMed] [Google Scholar]
- Saleem F., Bouatra S., Guo A. C., Psychogios N., Mandal R., Dunn S. M., et al. (2013). The bovine ruminal fluid metabolome. Metabolomics 9 360–378. 10.1007/s11306-012-0458-9 [DOI] [Google Scholar]
- Shabat S. K. B., Sasson G., Doron-Faigenboim A., Durman T., Yaacoby S., Miller M. E. B., et al. (2016). Specific microbiome-dependent mechanisms underlie the energy harvest efficiency of ruminants. ISME J. 10:2958. 10.1038/ismej.2016.62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanton T., Canale-Parola E. (1980). Treponema bryantii sp. nov., a rumen spirochete that interacts with cellulolytic bacteria. Arch. Microbiol. 127 145–156. 10.1007/bf00428018 [DOI] [PubMed] [Google Scholar]
- Thomas F., Hehemann J.-H., Rebuffet E., Czjzek M., Michel G. (2011). Environmental and gut bacteroidetes: the food connection. Front. Microbiol. 2:93 10.3389/fmicb.2011.00093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh P. J., Ley R. E., Mahowald M. A., Magrini V., Mardis E. R., Gordon J. I. (2006). An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444 1027–1031. [DOI] [PubMed] [Google Scholar]
- Van Houtert M. (1993). The production and metabolism of volatile fatty acids by ruminants fed roughages: a review. Anim. Feed Sci. Technol. 43 189–225. 10.1016/0377-8401(93)90078-x [DOI] [Google Scholar]
- Vargas J. E., Andrés S., Snelling T. J., López-Ferreras L., Yánez-Ruíz D. R., García-Estrada C., et al. (2017). Effect of sunflower and marine oils on ruminal microbiota, in vitro fermentation and digesta fatty acid profile. Front. Microbiol. 8:1124 10.3389/fmicb.2017.01124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiener G., Jianlin H., Ruijun L. (2006). The Yak. Bangkok: FAO Regional Office for Asia and the Pacific. [Google Scholar]
- William S., Feil H., Copeland A. (2012). Bacterial genomic DNA isolation using CTAB. Sigma 50 6876. [Google Scholar]
- Xu T., Xu S., Hu L., Zhao N., Liu Z., Ma L., et al. (2017). Effect of dietary types on feed intakes, growth performance and economic benefit in tibetan sheep and yaks on the qinghai-tibet plateau during cold season. PLoS ONE 12:e0169187. 10.1371/journal.pone.0169187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue B., Zhao X., Zhang Y. (2005). Seasonal changes in weight and body composition of yak grazing on alpine-meadow grassland in the Qinghai-Tibetan plateau of China. J. Anim. Sci. 83 1908–1913. 10.2527/2005.8381908x [DOI] [PubMed] [Google Scholar]
- Xue D., Chen H., Zhao X., Xu S., Hu L., Xu T., et al. (2017). Rumen prokaryotic communities of ruminants under different feeding paradigms on the Qinghai-Tibetan Plateau. Syst. Appl. Microbiol. 40 227–236. 10.1016/j.syapm.2017.03.006 [DOI] [PubMed] [Google Scholar]
- Xue M. Y., Sun H. Z., Wu X. H., Guan L. L., Liu J. X. (2019). Assessment of rumen bacteria in dairy cows with varied milk protein yield. J. Dairy Sci. 102 5031–5041. 10.3168/jds.2018-15974 [DOI] [PubMed] [Google Scholar]
- Xufeng H. (2015). Study on the Effects of Daily Age and Diet Ratio on the Rumen Microflora of Shanbei White Cashmere Goat. Xianyang: Northwest A&F University. [Google Scholar]
- Yan L., Zhang B., Shen Z. (2014). Dietary modulation of the expression of genes involved in short-chain fatty acid absorption in the rumen epithelium is related to short-chain fatty acid concentration and pH in the rumen of goats. J. Dairy Sci. 97 5668–5675. 10.3168/jds.2013-7807 [DOI] [PubMed] [Google Scholar]
- Yang C., Liu J., Wu X., Bao P., Long R., Guo X., et al. (2017). The response of gene expression associated with lipid metabolism, fat deposition and fatty acid profile in the longissimus dorsi muscle of Gannan yaks to different energy levels of diets. PLoS ONE 12:e0187604. 10.1371/journal.pone.0187604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C., Zhang J., Ahmad A. A., Bao P., Guo X., Long R., et al. (2019). Dietary energy levels affect growth performance through growth hormone and insulin-like growth factor 1 in Yak (Bos grunniens). Animals 9:39. 10.3390/ani9020039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yohe T. T., Schramm H., White R. R., Hanigan M. D., Parsons C. L. M., Tucker H. L. M., et al. (2019). Form of calf diet and the rumen. II: impact on volatile fatty acid absorption. J. Dairy Sci. 102 8502–8512. 10.3168/jds.2019-16450 [DOI] [PubMed] [Google Scholar]
- Zou H., Hu R., Wang Z., Shah A. M., Zeng S., Peng Q., et al. (2019). Effects of nutritional deprivation and re-alimentation on the feed efficiency, blood biochemistry, and rumen microflora in yaks (Bos grunniens). Animals 9:807. 10.3390/ani9100807 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data for this study have been deposited in the European Nucleotide Archive (ENA) at EMBL-EBI under accession number PRJEB342982.
The datasets generated for this study can be found in the http://www.ebi.ac.uk/ena/data/view/PRJEB34298.