Abstract
The ability of the endosymbiont Wolbachia pipientis to restrict RNA viruses is presently being leveraged to curb global transmission of arbovirus-induced diseases. Past studies have shown that virus replication is limited early in arthropod cells colonized by the bacterium, although it is unclear if this phenomenon is replicated in mosquito cells that first encounter viruses obtained through a vertebrate blood meal. Furthermore, these cellular events neither explain how Wolbachia limits dissemination of viruses between mosquito tissues, nor how it prevents transmission of infectious viruses from mosquitoes to vertebrate host. In this study, we try to address these issues using an array of mosquito cell culture models, with an additional goal being to identify a common viral target for pathogen blocking. Our results establish the viral RNA as a cellular target for Wolbachia-mediated inhibition, with the incoming viral RNA experiencing rapid turnover following internalization in cells. This early block in replication in mosquito cells initially infected by the virus thus consequently reduces the production of progeny viruses from these same cells. However, this is not the only contributor to pathogen blocking. We show that the presence of Wolbachia reduces the per-particle infectivity of progeny viruses on naïve mosquito and vertebrate cells, consequently limiting virus dissemination and transmission, respectively. Importantly, we demonstrate that this aspect of pathogen blocking is independent of any particular Wolbachia-host association and affects viruses belonging to Togaviridae and Flaviviridae families of RNA viruses. Finally, consistent with the idea of the viral RNA as a target, we find that the encapsidated virion RNA is less infectious for viruses produced from Wolbachia-colonized cells. Collectively, our findings present a common mechanism of pathogen blocking in mosquitoes that establish a link between virus inhibition in the cell to virus dissemination and transmission.
Author summary
Viruses transmitted by arthropod vectors pose a significant global health risk. Incidence of diseases caused by these viruses can thus be reduced by implementing effective vector control strategies. This need is further exacerbated due to the lack of commercially available vaccines and antivirals. Presence of the intracellular bacteria Wolbachia pipientis is associated with virus inhibition in multiple mosquito vectors. Furthermore, Wolbachia is inherited transovarially and spreads across the vector population like a natural gene drive, making it an attractive vector control agent. In this study, we examine how the presence of the bacterium in arthropod cells prevents initial establishment of vertebrate cell derived viruses. Our results indicate rapid turnover of incoming viral RNA very early during infection in Wolbachia-colonized cells, thus establishing it as a cellular target for pathogen blocking. Additionally, upon evaluating how these events might further limit virus spread, we find that infectivity of progeny viruses belonging to multiple RNA virus families are reduced on a per-particle basis. This aspect of virus inhibition is independent of any particular Wolbachia-host association and affects how these viruses replicate in naïve mosquito and vertebrate cells, thus providing a collective basis of reduced virus dissemination and transmission in Wolbachia-colonized mosquitoes.
Introduction
The pathogen blocking ability of the arthropod endosymbiont Wolbachia pipientis makes it an exciting biocontrol agent that is currently being used to limit transmission of arboviruses around the world [1]. The importance of studying the underlying mechanism of pathogen blocking, however, is not only to determine the long-term feasibility of this strategy, but to also learn how this particular arthropod host-endosymbiont association enables the former to become refractory to a wide range of RNA viruses. The degree of virus inhibition varies between different Wolbachia-host associations and is dependent on the Wolbachia strain and the arthropod host species [2–5]. Collectively, three major aspects of pathogen blocking have been reported consistently across all associations: inhibition of viruses possessing positive-sense single-stranded RNA (+ ssRNA) genomes, limited virus dissemination, and lower virus transmission [2, 6–9]. Earlier studies have primarily concentrated on identifying cellular events that lead to virus inhibition in arthropod cells, including viral entry, genome replication and protein translation [3, 10–14]. Indeed, this approach has proven to be very useful in identifying important host determinants of pathogen blocking. However, too often results from these studies are limited to a single Wolbachia-host association, giving the impression that different Wolbachia-host permutations involve distinct mechanisms of inhibition [3, 10–13]. Furthermore, these studies do not address the question of how virus inhibition in Wolbachia-colonized cells ultimately limit virus dissemination within mosquito tissues, and transmission from mosquitoes to vertebrates. To this end, the goal of this study was to utilize an array of mosquito cell culture models representing different Wolbachia-host combinations to identify a common viral target for pathogen blocking and to determine the link between intracellular virus inhibition, restricted virus dissemination and virus transmission. Our results identify the viral RNA genome as a target for pathogen blocking in arthropods, which include both fruit flies and mosquitoes. In Wolbachia-colonized cells, viral RNA targeting occurs at multiple stages of the replication cycle, notably in the very early stages following virus internalization and genome delivery, that lead to a shortened half-life of the incoming viral RNA. Additionally, we demonstrate that viral RNA present within viruses produced from Wolbachia-colonized cells are less infectious in vertebrate cells, contributing to the overall reduced infectivity of the progeny viruses. We provide evidence that this aspect of pathogen blocking is likely independent of any particular Wolbachia strain and arthropod host association and affects members of at least two + ssRNA virus families. Finally, we show that two major aspects of pathogen blocking i.e. limited virus dissemination and transmission occur due to the inability of these less infectious viruses to propagate in naïve arthropod and vertebrate cells.
Results
Viral RNA is a shared target for multi-stage inhibition in Wolbachia-colonized cells
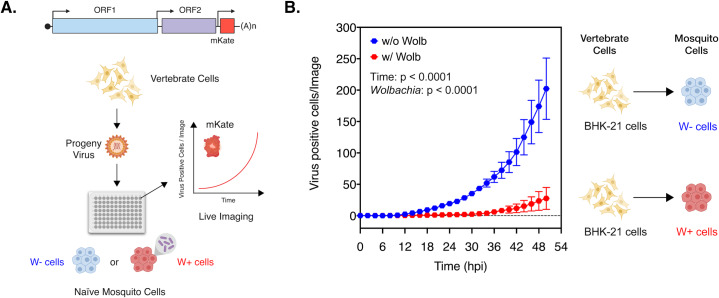
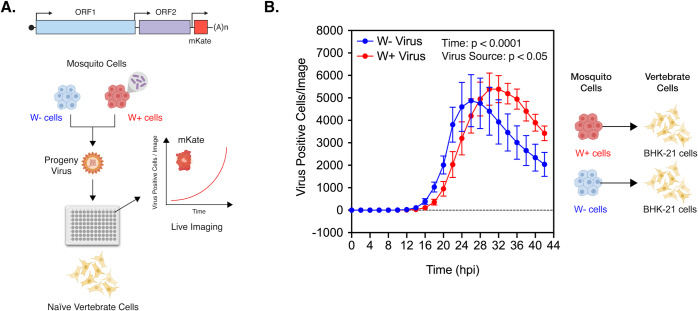
Presence of Wolbachia is associated with reduced viral gene expression in arthropod cells. This widely reported aspect of virus inhibition can be observed both in vivo, in Aedes aegypti mosquitoes colonized with wAlbB (S1A Fig; Welch-corrected unpaired two-tailed t-test, p = 0.0256, t = 2.989, df = 5.749) as well as in cell culture, Aedes albopictus cells colonized with wMel (S1B Fig; Welch-corrected unpaired two-tailed t-test, p < 0.0001, t = 55.23, df = 5.712) [10–12, 14]. However, this quantification represents total viral RNA accumulated over multiple rounds of virus replication in mosquito cells. To determine how inhibition occurs in the initial stages of infection in the vector i.e. during establishment of viruses acquired through a blood meal, we monitored the spread of vertebrate cell-derived viruses in naïve Aedes albopictus mosquito cells (C710 cells) colonized with and without Wolbachia derived from the planthopper host, Laodelphax striatellus (wStri). It should be noted that throughout this study, multiplicity of infection (MOI) was calculated in terms of viral genome equivalents (particles) instead of infectious units (PFUs), given that the former is independent and the latter is dependent on a single host/tissue cell line (e.g. vertebrate or arthropod) that the viruses are assayed on to determine virus “infectivity”. Spread of Chikungunya viruses derived from BHK-21 (Baby Hamster Kidney fibroblast cells) expressing a fluorescent reporter protein (CHIKV-mKate) was thus assessed in these mosquito cells following a synchronized infection at a low MOI (1 particle/cell) that involved virus adsorption at 4ºC. Cell monolayers were then extensively washed with 1XPBS to remove any unbound viruses and warm media (37ºC) was added to cells to initialize virus internalization and infection. Virus spread was then measured over 50 hours by quantifying mean virus-encoded fluorescent reporter expression observed over four distinct fields of view taken per well every 2 hours (Fig 1A). Two-way ANOVA was used to determine the effect of destination cell type (with or without wStri) and/or time on virus spread. Virus growth was significantly reduced over time in cells colonized with wStri compared to cells without the bacterium; Ordinary Two-way ANOVA with Sidak’s multiple comparisons test, Wolbachia: p < 0.0001, Time: p < 0.0001, Time X Wolbachia: p < 0.0001 (Fig 1).
Fig 1. Presence of Wolbachia reduces spread of vertebrate-derived viruses in naïve mosquito cells.
(A) Schematic representation of the experiment. CHIKV expressing mKate fluorescent protein from a second sub-genomic promoter was grown in BHK-21 cells. These progeny viruses were then used to infect naïve C710 cells with (depicted in red) and without (depicted in blue) Wolbachia (wStri strain) synchronously at an MOI of 1 particle/cell. Virus growth in cells, plated on a ninety-six-well plate, was measured in real time by imaging and quantifying the number of red cells (Virus Positive Cells/Image) expressing the virus encoded mKate protein over a period of 48 hours, using live cell imaging. (B) Color of the data points distinguish the two destination cell lines where virus replication was assayed on; blue represent C710 cells without Wolbachia while red represent C710 cells with Wolbachia. Y-axis label (Virus Positive Cells/Image) represent red cells expressing virus-encoded mKate fluorescent protein in a single field of view, four of which were averaged/sample at every two-hour time point collected over the course of infection. Two-way ANOVA with Sidak’s post-hoc test. Error bars represent standard error of mean (SEM) of biological replicates (n = 7–9).
Although our previous result validates the initial stage of virus inhibition in the vector, i.e. during virus establishment in Wolbachia-colonized mosquito tissues following an infectious blood meal, it is unknown how early this inhibition takes place at the cellular level in cells initially infected with the virus. Previous studies by other groups have indicated that inhibition of both alphaviruses and flaviviruses occur at an early stage of infection in Wolbachia-colonized arthropod cells [12–13]. Furthermore, data from the latter study indicate that viral RNA is susceptible to degradation immediately post-internalization [12]. Therefore, to determine whether viral RNA is degraded faster in Wolbachia-colonized cells following virion internalization, we asked if the incoming viral RNA half-life is altered between cells with and without Wolbachia. We also asked whether or not this event is exclusive to mosquito cells.
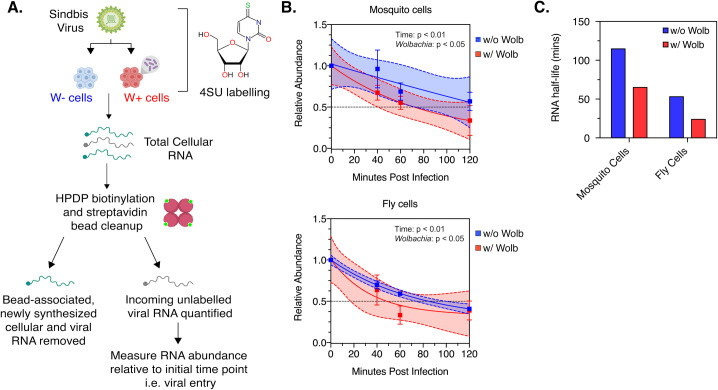
C710 Aedes albopictus cells with (wStri strain) or without Wolbachia (w/o wStri) and JW18 Drosophila melanogaster cells with (wMel strain) or without Wolbachia (w/o wMel) were grown overnight in media containing the cross-linkable nucleoside analog 4-thiouridine (4SU) at 50 μM (Fig 2A). In the cell, 4SU is converted to 4S-UTP before being incorporated into newly synthesized RNA, thus allowing labelling of all cellular RNA. Sindbis virus (SINV) derived from vertebrate BHK-21 cells grown in normal media, and therefore containing an unlabeled virion RNA genome, was then used to synchronously infect the 4SU-treated cells at an high MOI of 10 particles/cell. Following virus adsorption at 4ºC, cell monolayers were extensively washed with 1XPBS to remove any unbound viruses. Warm media (37ºC) containing 4SU was added to cells to initialize virus internalization and infection was then carried out under labelling conditions, thus allowing 4S-UTP incorporation into newly synthesized host and viral RNA, leaving the incoming viral RNA as the only unlabeled RNA species in the cell. All 4SU-labelled RNA was separated from the total RNA pool following biotinylation and streptavidin cleanup. Next, levels of unlabeled RNA at each time point was measured relative to that present at 0 minutes post internalization, which represent initial viral genome delivery. RNA half-life was extrapolated using non-linear, exponential decay model (Fig 2B). We observed steady monophasic decay of the incoming viral RNA in both mosquito and fly cells, either in the presence or absence of Wolbachia. Two-way ANOVA with Sidak’s post-hoc test revealed an effect of both time and Wolbachia on the relative abundance of incoming viral RNA; C710 mosquito cells, Time: p = 0.004, Wolbachia (wStri) = 0.0469, JW18 fly cells, Time: p = 0.0042, Wolbachia (wMel) = 0.021. (Fig 2B). However, in each case mean half-life of viral RNA was reduced in Wolbachia-colonized cells. In mosquito cells, RNA half-life was reduced approximately 1.8-fold; One-phase decay, 115.2 minutes (w/o wStri), 65.58 minutes (w/ wStri). In comparison, half-life was reduced approximately 2.2-fold in fly cells; 53.7 minutes (w/o wMel), 24.4 minutes (w/ wMel) (Fig 2C). Based on these results, we conclude that the incoming viral RNA undergoes faster turnover in Wolbachia-colonized cells, reducing the cellular pool of viral RNA very early in the replication cycle. Abrogation of both viral RNA synthesis and protein expression have been previously demonstrated in the presence of Wolbachia [10–14]. However, our current data alongside observations made by Thomas et.al, further support the idea of viral RNA as a cellular target for pathogen blocking at multiple steps of the viral infection cycle [13].
Fig 2. Incoming viral RNA is degraded quicker in Wolbachia-colonized cells.
Half-life of incoming viral RNA was assessed following infection of C710 Aedes albopictus cells colonized with and without Wolbachia (wStri strain) and JW18 Drosophila melanogaster cells with and without Wolbachia (wMel strain). (A) Schematic representation of the experiment. Sindbis virus derived from vertebrate BHK-21 cells was used to synchronously infect C710 or JW18 cells pre-labelled with 50 μM 4-thiouridine (4SU) for 12 hours. Infection was carried out under labelling conditions. At indicated times post infection, total RNA was extracted from cells, biotinylated and streptavidin beads were used to isolate incoming unlabeled viral RNA from 4SU-labelled cellular and newly synthesized RNA. Viral RNA was quantified at each time-point using quantitative RT-PCR relative to viral RNA detected at the start of infection (0h). (B) Relative abundance of incoming viral RNA in cells colonized with and without Wolbachia over 120 minutes post-infection were determined by qRT-PCR analysis as described in Materials and Methods. Non-linear, exponential regression analyses was performed to determine the viral RNA decay profile (represented by solid lines). Dashed lines represent the 95% CI of the aforementioned regression. Two-way ANOVA with Sidak’s post-hoc test. Error bars represent standard error of mean (SEM) of biological replicates (n = 3) (C) Viral RNA half-lives were estimated from data showed in (B).
Progeny viruses generated from Wolbachia colonized cells are less infectious
It is unclear how the aforementioned cellular events lead to reduced virus dissemination within the mosquito vector as well as reduced transmission into vertebrate hosts. In our previous study, we reported reduced infectivity of Sindbis viruses derived from Wolbachia-colonized fly cells on vertebrate BHK-21 cells [12]. In light of this result, we wondered whether these progeny viruses are compromised in their ability to infect and propagate in naïve vertebrate and arthropod cells. We reasoned that lower production of total viruses from wStri-colonized C710 mosquito cells in combination with their inability to propagate in naïve C710 mosquito cells might explain why virus dissemination is reduced in mosquitoes carrying Wolbachia. Additionally, the observed loss in transmission could occur as a result of these viruses being unable to spread and kill vertebrate cells. But given that Wolbachia-mediated reduction of virus dissemination and transmission has been observed against multiple RNA viruses across multiple Wolbachia-host associations, we first wanted to expand our previous findings to determine whether our previous observation regarding the loss in per-particle infectivity is limited either to any particular virus or certain Wolbachia-host associations [2, 6–9, 12].
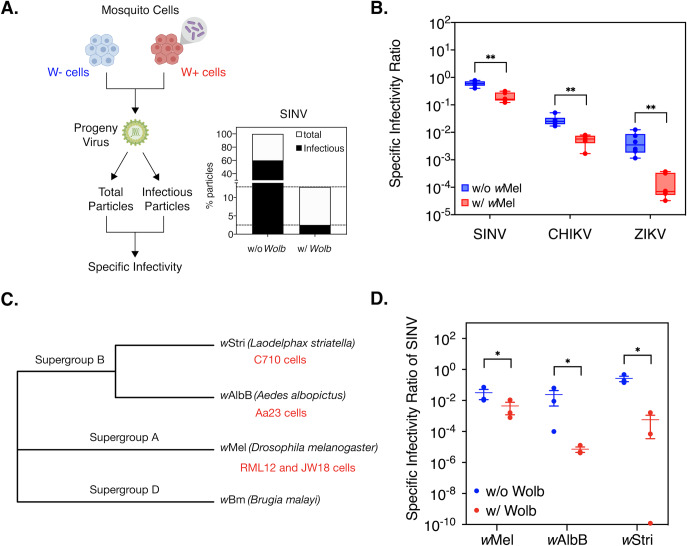
Viruses relevant to our study i.e. alphaviruses and flaviviruses are not known to form empty virions [15]. Thus, all virus particles produced from cells, regardless of infectivity, contain viral RNA cargo that can be measured using quantitative PCR. We therefore quantified viral genome copies present in the cell supernatant as a proxy for total virus particles released following infection in an arthropod host i.e. Aedes albopictus-derived or Drosophila melanogaster-derived cells with and without Wolbachia. Infectious viruses present in the same cell supernatant were assayed by quantifying plaque-forming units on vertebrate cells. Following independent quantification these attributes in tandem, we then calculated per-particle infectivity or specific infectivity ratio (SI) of progeny viruses as the ratio of infectious virus to total virus (Fig 3A) [12, 16].
Fig 3. Presence of Wolbachia in mosquito cells reduces progeny virus infectivity in vertebrate cells.
RML12 mosquito cells with and without Wolbachia (wMel strain) were infected with alphaviruses Sindbis (SINV), Chikungunya (CHIKV) or flavivirus Zika (ZIKV) at an MOI of 10 particles/cell. Viral supernatants were harvested at 48 hours post infection. Infectious virus titer was quantified by performing plaque assays on BHK-21 (SINV, CHIKV) and Vero (ZIKV) cells. Total virus particles were determined by quantifying viral genome copies present in the supernatant using qRT-PCR. Reported specific infectivity (SI) ratios were calculated as total infectious virus titer divided by total particles produced per mL of viral supernatant. (A) Percentage of total SINV particles produced from cells with and without Wolbachia that are infectious on BHK-21 cells. (B) Specific Infectivity Ratios of progeny SINV, CHIKV and ZIKV viruses (n = 6). Welch’s t-test performed on log-transformed values. (C) Maximum-likelihood tree representing the phylogenetic relationship between the Wolbachia strains used in the study was generated using MEGA X, using a MUSCLE alignment of concatenated sequences of multi-locus typing (MLST) genes (coxA, gatB, ftsZ, hcpA, fbpA). Sequences from Wolbachia strain wBm, native to the filarial nematode Brugia malayi, was used as a distant outgroup. Corresponding host cell lines used in this study are indicated in red below their respective Wolbachia strain tip labels. (D) Specific Infectivity of progeny SINV derived from Aedes albopictus cells colonized with non-native (wMel and wStri) and native (wAlbB) Wolbachia strains. Cells were infected with virus at an MOI of 0.1 particles/cell and infectious virus titer produced after 48 hours was quantified via plaque assays on BHK-21 cells. Error bars represent standard error of mean (SEM) of biological replicates (n = 3). One-way ANOVA with Tukey’s post-hoc test performed on log-transformed values. *P < 0.05, **P < 0.01.
First, we assessed the ability of a single Wolbachia strain-type, wMel, to inhibit multiple RNA viruses across two different arthropod host cell types, mosquito (Aedes albopictus RML12 cells) and the native fruit fly (Drosophila melanogaster JW18 cells). Mosquito cells were challenged with a panel of arboviruses, including alphaviruses Sindbis (SINV) and Chikungunya (CHIKV) and one flavivirus, Zika (ZIKV) to determine the breadth of this phenotype against multiple arboviruses. In all cases, viruses grown in the presence of Wolbachia (W+ viruses) exhibited a lower SI ratio than viruses grown in cells without the endosymbiont (W- viruses). Degree to which infectivity (SI) was reduced varied depending on the virus type; Welch-corrected unpaired two-tailed t-test on log-transformed values, SINV: 3-fold reduction (p = 0.000059, df = 10, t-ratio = 6.627), CHIKV: 5-fold reduction (t-test, p = 0.000071, df = 10, t-ratio = 6.6.471), ZIKV: 32-fold reduction (t-test, p = 0.000144, df = 10, t-ratio = 5.934) (Fig 3A and 3B, S2 Fig). We obtained similar results when we tested the virus panel in Drosophila melanogaster cells with and without Wolbachia (wMel); Welch-corrected unpaired two-tailed t-test on log-transformed values, SINV (p = 0.0045, df = 9.458, t-ratio = 3.698), CHIKV (t-test, p = 0.013204, df = 10, t-ratio = 3.006), ZIKV (t-test, p = 0.033939, df = 7, t-ratio = 2.629) (S3 Fig). Presence of the same Wolbachia genotype in different arthropod hosts thus leads to reduced infectivity of arboviruses belonging to different virus families, indicating that this phenotype is independent of virus type and any specific Wolbachia-host association.
We next examined whether different Wolbachia strains are capable of reducing progeny virus infectivity in the context of a single arthropod host (Aedes albopictus) cell type. A panel of three Aedes albopictus derived mosquito cells were obtained, each colonized by a distinct Wolbachia strain, including Wolbachia strain wAlbB (Aa23 cells) derived from the native Aedes albopictus and non-native strains wStri (C710 cells) derived from a planthopper, Laodelphax striatellus and wMel (RML12 cells), derived from Drosophila melanogaster [17]. Phylogenetic analyses of these different Wolbachia strains classify them into two distinct clades, with wAlbB and wStri representing Supergroup B and wMel Supergroup A (Fig 3C). Historically, the wMel strain has been used successfully in Wolbachia-mediated vector control efforts [4, 7]. However, both Supergroup B strains wAlbB and wStri are promising candidates that are capable of limiting arbovirus dissemination, given their strong pathogen blocking phenotype in cell culture [10].
All cells, regardless of the Wolbachia strain present within, were challenged with SINV at a very low MOI of 0.1 particle/cell to additionally investigate the effect of high (MOI = 10 particle/cell) versus low (MOI = 0.1 particle/cell) starting virus titers on infection outcome in the presence of Wolbachia. We found starting MOI to influence progeny SINV infectivity for both W- and W+ viruses, as a lower starting MOI = 0.1 particle/cell (Fig 3D) resulted in overall lower SI ratios at 48 hours post infection relative to higher starting MOI = 10 particles/cell in the same host-Wolbachia background (RML12-wMel) (Fig 3B). However, we found SI of progeny W+ SINV to be reduced in each host-Wolbachia combination tested, irrespective of the Wolbachia strain present (Fig 3D; One-way ANOVA on log-transformed values with Tukey’s post-hoc test; Wolbachia, p = 0.00245, wMel, p = 0.0474, wAlbB, p = 0.0257, wStri, p = 0.04953). Extent to which SINV infectivity was reduced seemingly varied between cells colonized with different Wolbachia strains, which could be a result of either the genotype of the cells, Wolbachia strains or both. We observed differences in relative Wolbachia titers between the three cell lines. Notably, titers of both Supergroup B strains (wAlbB, wStri), that had a more pronounced effect on SINV (Fig 3D), were on average, slightly higher than the Supergroup A wMel strain (RML12 cells) (S4 Fig; Kruskal-Wallis of multivariate comparisons with Dunn’s post-hoc test; RML12-wMel vs C710-wStri, p = 0.3803, RML12-wMel vs Aa23-wAlbB, p = 0.0038, Aa23-wAlbB vs C710-wStri, p = 0.3803). It is unclear whether these moderate differences in endosymbiont titer are in any way correlated with the extent to which virus infectivity is reduced, given the considerable variation in Wolbachia titers across passaged cell lines. Based on these results, we conclude that presence of pathogen-blocking Wolbachia strains are commonly associated with a concomitant reduction in progeny virus infectivity.
Progeny viruses spread poorly in naïve mosquito cells
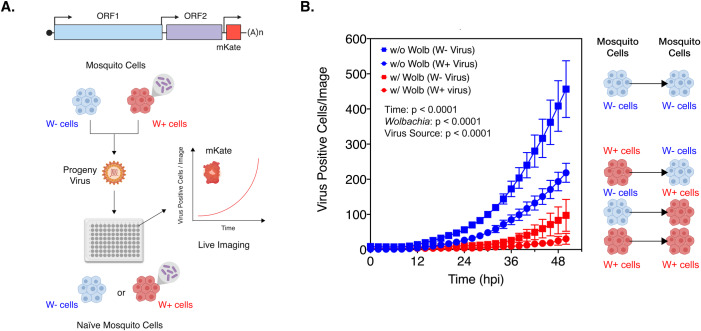
We next monitored the ability of progeny viruses derived from mosquito cells colonized with (W+ virus) and without (W- virus) Wolbachia to propagate in naïve mosquito cells. CHIKV expressing a fluorescent reporter protein (CHIKV-mKate) was grown in C710 Aedes albopictus cells with and without Wolbachia (wStri strain), purified and subsequently used to infect naïve mosquito cells at equal MOIs (MOI = 5 particles/cell) following a synchronous infection. Virus infection and spread was then monitored over a period of 50 hours using a live-cell imaging system by quantifying mean virus-encoded fluorescent reporter expression observed over four distinct fields of view taken per well every 2 hours (Fig 4A). Three-way ANOVA was used to determine the effects of virus source (Source i.e. W- or W+ virus), destination cell type (with or without Wolbachia) and/or time, on virus spread. Our results show significant effects of all three variables on virus growth, both on their own as well as in combination with each other; Three-way ANOVA with Tukey’s post-hoc test, Source: p < 0.0001, Wolbachia: p < 0.0001, Time: p < 0.0001, Source X Time: p < 0.0001, Source X Wolbachia: p < 0.0001, Wolbachia X Time: p < 0.0001, Source X Time X Wolbachia: p = 0.0016 (Fig 4B). As expected, W- virus grew more poorly in naïve cells with Wolbachia compared to those without (Fig 4B, square symbols). However, W+ viruses (Fig 4B, circle symbols) exhibited reduced growth in naïve cells both in the presence and absence of Wolbachia (wStri strain). Importantly, these results indicate that reduced infectivity of W+ viruses might contribute to their inability to spread to new mosquito cells regardless of whether or not these cells are colonized by Wolbachia. Assuming that the mechanism of cellular inhibition of virus replication is cell-autonomous, this model might help explain why limited virus dissemination occurs within tissues with both high and low Wolbachia densities. As evident from the results from Fig 4B, our data is consistent with previous observations made by Koh et.al, which reported loss in infectivity of DENV isolates generated following serial passages in wMel-colonized Aag2 cells [6]. Importantly, loss in progeny virus infectivity is not limited to cell culture models. Indeed, as reported by Dutra et.al, in 2016, ZIKV viruses isolated from salivary gland secretions of wMel-colonized mosquitoes fail to establish systemic infections in naïve mosquitoes following intrathoracic injection, demonstrating a quantifiable loss in infectivity in vivo [7].
Fig 4. Progeny viruses derived from Wolbachia colonized cells replicate poorly in naïve mosquito cells.
(A) Schematic representation of the experiment. CHIKV expressing mKate fluorescent protein from a second sub-genomic promoter was grown in C710 Aedes albopictus cells in the presence (W+ virus) or absence (W- virus) of Wolbachia (wStri strain). These progeny viruses were then used to infect naïve C710 cells with (depicted in red) and without (depicted in blue) Wolbachia (wStri strain). synchronously at an MOI of 5 particles/cell. Virus growth in cells, plated on a ninety-six-well plate, was measured in real time by imaging and quantifying the number of red cells (Virus Positive Cells/Image) expressing the virus encoded mKate protein over a period of 48 hours, using live cell imaging. (B) Color of the data points distinguish the two destination cell lines where virus replication was assayed on; blue represent C710 cells without Wolbachia while red represent C710 cells with Wolbachia. Shape of the data points refer to the nature of the progeny viruses used to initiate infection; squares represent W- viruses, grown in C710 cells without Wolbachia, while circles represent W+ viruses, grown in C710 cells with Wolbachia. Y-axis label (Virus Positive Cells/Image) represent red cells expressing virus-encoded mKate fluorescent protein in a single field of view, four of which were averaged/sample at every two-hour time point collected over the course of infection. Three-way ANOVA with Tukey’s post-hoc test. Error bars represent standard error of mean (SEM) of biological replicates (n = 9).
Progeny viruses spread poorly in naïve vertebrate cells
We further assessed the infectivity of W+ progeny viruses to spread in vertebrate cells using live-cell imaging to validate our earlier results (Fig 3B). As before, purified progeny W+ and W- viruses derived from mosquito cells colonized with (W+ virus) and without (W- virus) Wolbachia (wStri strain) was subsequently used to infect naïve BHK-21 cells at an MOI of 5 particles/cell. Infection was synchronized as before and virus spread was measured over 42 hours by quantifying mean virus-encoded fluorescent reporter expression observed over four distinct fields of view taken per well every 2 hours (Fig 5A). Two-way ANOVA was used to determine the effect of virus source (Source i.e. W- or W+ virus) and/or time on virus spread over the course of the infection. Spread of W- viruses were consistently faster compared to W+ viruses, with peak number of W- virus positive cells observed at 26 hours post infection compared to 30 hours post infection for W+ viruses; Ordinary Two-way ANOVA with Tukey’s post-hoc test, Source: p = 0.0261, Time: p < 0.0001, Time X Source: p = 0.0068 (Fig 5B). Interestingly, peak number of virus positive cells was higher for W+ viruses relative to W- viruses between 28 and 42 hours. However, this is due to a delay in cell death in W+ infected cell populations, thus allowing greater and prolonged expression of virus encoded reporter (S5 Fig). In comparison, cells infected with W- viruses succumb early to infection, resulting in a faster loss in reporter activity between 26 and 42 hours (Fig 5B, S5 Fig).
Fig 5. Progeny viruses derived from Wolbachia colonized cells replicate poorly in naïve vertebrate cells.
(A) Schematic representation of the experiment. CHIKV expressing mKate fluorescent protein from a second sub-genomic promoter was grown in C710 Aedes albopictus cells in the presence (W+ virus) or absence (W- virus) of Wolbachia (wStri strain). These progeny viruses were then used to infect naïve vertebrate BHK-21 cells synchronously at an MOI of 5 particles/cell. Virus growth in cells, plated on a ninety-six-well plate, was measured in real time by imaging and quantifying the number of red cells expressing the virus encoded mKate protein over a period of 42 hours using live-cell imaging. (B) Color of the data points distinguish the progeny viruses used to initiate infection in BHK-cells; blue represent progeny viruses derived from C710 cells without Wolbachia (W- virus) while red represent progeny viruses derived from C710 cells with Wolbachia (W+ virus). Y-axis label (Virus Positive Cells/Image) represent red cells expressing virus-encoded mKate fluorescent protein in a single field of view, four of which were averaged/sample at every two-hour time point collected over the course of infection. Two-way ANOVA with Tukey’s post-hoc test. Error bars represent standard error of mean (SEM) of biological replicates (n = 3–4).
Encapsidated viral RNA within progeny viruses are less infectious
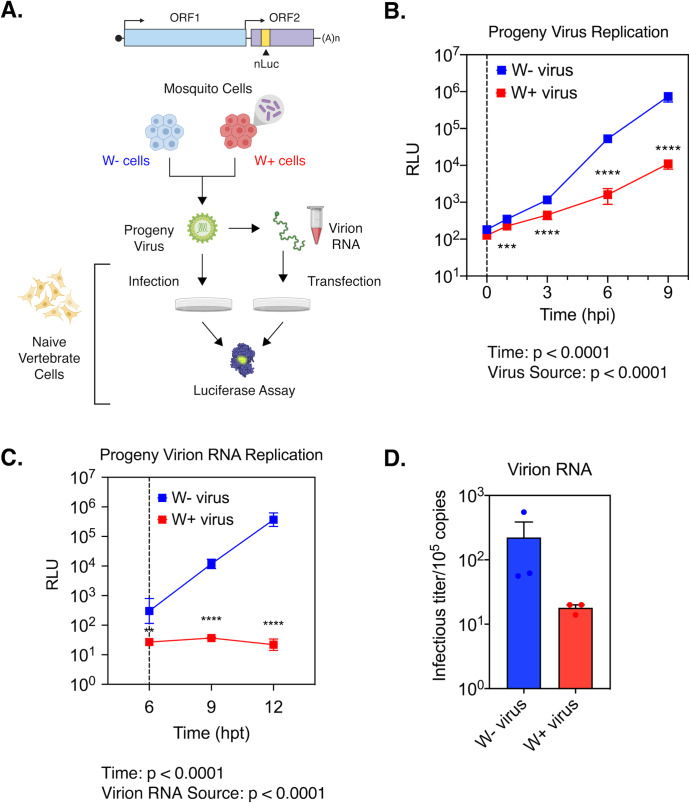
While reduced infectivity of progeny viruses explains limited virus dissemination and transmission in Wolbachia-colonized mosquitoes, it is not evident from our previous data why W+ viruses are less infectious. Simplistically, two key factors can result in the observed loss in virus infectivity; compromised virion structure and dysfunctional viral RNA. The former might impair virus attachment and entry to cells, while the latter would prevent efficient virus replication post entry. We reasoned that if W+ viruses exhibit structural defects that impair their ability to bind and enter cells, direct delivery of the encapsidated viral RNA into the cell should bypass this blockade and thus allow genome replication comparable to W- virus derived RNA. Therefore, we isolated encapsidated virion RNA from W+ and W- viruses produced from wMel-colonized RML12 mosquito cells and assessed their ability to replicate in naïve vertebrate BHK-21 cells following transfection. For this replication assay, we used SINV with nanoluciferase reporter fused to the second open reading frame (SINV-nLuc) and measured luciferase activity as a proxy for viral structural protein expression over time (Fig 6A). First, we used progeny viruses at an MOI of 5 particles/cell to a establish a synchronized infection in BHK-21 cells as described earlier and measured viral replication over a period of 9 hours post infection. We found W+ virus replication to be significantly reduced relative to W- viruses over time, reaffirming our earlier results regarding poor infectivity of W+ viruses on vertebrate cells (Fig 5B); Two-way ANOVA with Tukey’s post-hoc test, Time: p < 0.0001, Wolbachia: p < 0.0001, Time X Wolbachia, p < 0.0001 (Fig 6B). Next, we isolated virion encapsidated RNA from W+ and W- viruses and transfected naïve BHK-21 cells with equal, approximately 105 viral genome copies (Fig 6A). As before, we used luciferase reporter activity as a proxy for virus replication over time and found significantly reduced reporter activity in cells transfected with W+ virus-derived RNA; Two-way ANOVA with Tukey’s post-hoc test, Time: p < 0.0001, Wolbachia: < 0.0001, Time X Wolbachia, p < 0.0001 (Fig 6C). This reduction in reporter activity was more severe compared to the reduction observed in our previous experiments where infections were initiated with W+ viruses (Fig 6B). We have previously demonstrated that interactions between Sindbis virus capsid protein and the viral RNA (SINV C: R interaction) are important in regulating the function of the incoming viral RNA in vertebrate cells [16]. Absence of critical SINV C:R interaction sites that impact association of the viral RNA with the capsid proteins lead to increased degradation of the incoming viral RNA, reducing the half-life by almost 2.5 -fold. As SINV C:R interactions are absent during our transfection experiments, it is possible that virion RNA derived from W+ viruses are turned over at a faster rate, causing the observed reduction in replication of W+ virus-derived RNA (Fig 6C). Finally, to test whether reduced replication of W+ virion RNA result in the production of fewer infectious units, we quantified plaque-forming units following transfection of 105 copies of virion isolated RNA into BHK-21 cells and found W+ virion RNA to produce 10-times fewer infectious units relative to W- virion RNA after 48 hours (Fig 6D). We obtained similar results upon transfecting BHK-21 cells with virion RNA isolated from SINV-nLuc W- and W+ viruses derived from JW18 fly cells colonized with Wolbachia (wMel strain); Two-tailed Welch’s t-test, p = 0.0196, t = 4.883, df = 2.774 (S6 Fig). Taken together, these results suggest that encapsidated viral RNA present within W+ viruses are deficient in their ability to replicate in naïve vertebrate cells, ultimately resulting in the formation of fewer infectious units.
Fig 6. RNA encapsidated within progeny viruses derived from Wolbachia colonized cells are less infectious.
(A) Schematic of experiments performed using Sindbis Virus (SINV) carrying a translationally fused nanoluciferase (nLuc) gene in the second open reading frame (ORF2). (B) Sindbis nLuc reporter viruses (SINV-nLuc) derived from RML12 mosquito cells with (W+ virus) or without (W- virus) Wolbachia (wMel strain) were subsequently used to synchronously infect naïve BHK-21 cells at equivalent MOIs of 5 particles/cell (n = 6). Cell lysates were collected at indicated times post infection and luciferase activity (RLU) was measured and used as a proxy to quantify viral replication. Two-way ANOVA with Tukey’s post-hoc test. Vertical dashed line indicates the first time point collected following removal of the initial inoculum. (C) Replication kinetics of virion encapsidated RNA isolated from progeny SINV-nLuc viruses derived from RML12 mosquito cells with (W+ virus) or without (W- virus) Wolbachia (wMel strain) was determined by measuring luciferase activity (RLU) following transfection of 105 copies of virion encapsulated RNA into BHK-21 cells (n = 6). Two-way ANOVA with Tukey’s post-hoc test. Vertical dashed line at 6 hours post transfection (hpt) indicates the first time point collected following removal of the initial transfection mixture. (D) Infectious titer generated from the aforementioned virion RNAs was determined by counting the number of plaques produced after 48 hours post transfection into BHK-21 cells. Error bars represent standard error of mean (SEM) of biological replicates (n = 3). **P < 0.01, ***P < 0.001, ****P < 0.0001.
Considering that viral RNA and protein synthesis is abrogated in arthropod cells in the presence of Wolbachia, we conclude based on the data presented above that the viral RNA serves as a shared target for endosymbiont-mediated inhibition in arthropod cells.
Discussion
Uptake of viruses occur as the mosquito takes an infectious bloodmeal from a vertebrate animal. As the blood meal is digested, these viruses infect midgut cells before escaping the midgut barrier and disseminating to other mosquito tissues, which become persistently infected. Therefore, it is important to note that the viruses initially establishing infection in the vector are of vertebrate origin, while those that undergo dissemination and eventually transmission, are derived from mosquito cells. Prior studies have shown that initial infection of viruses in mosquito midguts is reduced in the presence of Wolbachia, suggesting an inability of vertebrate-derived viruses to establish infection [8, 18]. In this study, we show that presence of Wolbachia restricts vertebrate cell-derived virus growth in naïve mosquito cells. While past studies have shown inhibition of viral RNA and protein synthesis in Wolbachia-colonized cells, it is still unclear how early virus inhibition occurs in cells that are initially infected [13–14]. Using Semilki Forest Viruses (SFV) carrying a translationally fused luciferase reporter, Rainey et.al. demonstrated reduced expression of reporter genes at 7 hours post infection in fly (JW18) cells colonized with Wolbachia (wMel strain), suggesting that inhibition occurs early in the infection process [14]. Importantly, the authors also observed reduced luciferase expression following transfection of an in vitro transcribed virus replicon reporter, which indicates that inhibition of virus replication is independent of virus entry into Wolbachia-colonized cells and therefore might involve an intracellular viral target [13–14]. More recently, Thomas et.al. reported that Wolbachia-mediated inhibition of mosquito cell (C6/36) derived DENV-2 in wMel-colonized Aag-2 cells involve degradation of the viral RNA [13]. Interestingly, the authors observe reduced abundance of viral RNA in Aag2-wMel cells as early as 1h post infection, following synchronized binding (4ºC) and internalization (25ºC) of DENV-2 viruses. It is important to note that this reduction likely represent a loss of incoming viral RNA, due to low amounts of nascently synthesized viral RNA being present in the cell this early in infection. Here using metabolic labelling, we determined the half-life of the incoming vertebrate cell-derived viral RNA to show that virus inhibition occurs very early during infection, with the incoming viral RNA undergoing faster turnover in Wolbachia-colonized cells. This likely involves one or more cellular RNA degradation pathways, including perhaps orthologs of the RNA exonuclease Xrn1, whose role in pathogen blocking of flaviviruses has been demonstrated in Aedes aegypti derived Aag2-wMel cells [13]. Taken together, our results agree with past observations of this phenomenon involving other viruses belonging to both Togaviridae (SFV) and Flaviviridae (DENV-2) families, suggesting perhaps a common mechanism of action across different cell types and RNA viruses [13–14].
Presence of Wolbachia in mosquitoes has been shown to also reduce the rate of virus infection in different mosquito tissues. These include tissues proximal to the midgut like Malpighian tubules and fat bodies, as well as distal tissues like salivary glands, implying limited virus dissemination in the presence of the endosymbiont [8]. Interestingly, the degree to which virus inhibition occurs in these tissues is not correlated with either Wolbachia-density or its role in innate immunity. In fact, viral RNA levels are dramatically reduced in the mosquito head, which is poorly colonized by Wolbachia [8]. These results are confounding, given that the mechanism of pathogen blocking is thought to be cell-autonomous [19]. In addition, virus transmission is demonstrably reduced in Wolbachia-colonized mosquitoes [2, 7–9]. It is important to note that in this case, viruses transmitted to a vertebrate animal following a secondary bloodmeal are produced in the mosquito salivary gland tissues. In Wolbachia-colonized mosquitoes, reduced transmission is thought to likely occur as a result of reduced virus growth in salivary gland tissues. Interestingly, however, previous studies have found that although salivary gland secretions from Wolbachia-colonized mosquitoes contain detectable levels of Zika viral genome copies (ZIKV, Flaviviridae), these levels far exceed the actual number of infectious viruses that can be assayed on either vertebrate or mosquito cells, implying a reduction in per-particle infectivity [7]. Similar results have also been obtained in cell culture for dengue virus (DENV, Flavivirus) [20].
We have previously shown reduced per-particle infectivity of Sindbis virus grown in Drosophila melanogaster cells colonized with wMel on vertebrate BHK-21 cells [12]. In this study, we provide evidence that this attribute of pathogen blocking is present in Aedes albopictus mosquito cells, colonized with both native (wAlbB) and non-native (wMel and wStri) Wolbachia strains. Additionally, Wolbachia reduces the infectivity of progeny viruses belonging to two distinct families of + ssRNA viruses, Togaviridae (SINV, CHIKV) and Flaviviridae (ZIKV) in both mosquito and fly cells. Using growth assays we clearly demonstrate the inability of progeny W+ viruses to replicate in naïve vertebrate cells, thus linking virus infectivity to loss in transmission. Importantly, we demonstrate that reduced infectivity of these progeny viruses also limits their ability to grow in naïve mosquito cells. Remarkably, limited virus growth is independent of whether or not these naïve cells are colonized with Wolbachia, thus providing an explanation for why viral RNA levels are reduced in tissues with low Wolbachia titers. Therefore, while it is unclear whether virus production is abrogated completely from Wolbachia-colonized cells, as Wolbachia infection rates are known vary significantly between different cell lines and across passages, it is possible that the small proportion of infectious W+ virus progeny is produced from sub-populations of cells with very low/no Wolbachia.
While further investigation is required to determine whether these results are limited to single virus type, Wolbachia strain and/or host cell species, we expect this phenotype to remain consistent in Wolbachia-host associations where pathogen blocking have been reported previously. However, it would also be interesting to explore this phenotype in particular Wolbachia-host combinations that lack pathogen blocking abilities against susceptible viruses e.g. Dengue Virus (DENV, Flaviviridae) in Aedes aegypti mosquitoes with wPip, West Nile Virus (WNV, Flaviviridae) in Culex tarsalis mosquitoes systemically infected with wAlbB and Cricket Paralysis Virus (CrPV, Dicistroviridae) in Drosophila pandora infected with wPanCI [21–23].
Finally, we demonstrate that loss in virus infectivity occur at the level of the encapsidated virion RNA and affects its ability to replicate following direct delivery into vertebrate cells. Further work is required to determine the exact fate of W+ virus derived viral RNA in vertebrate cells in terms of its stability, localization and translation. Given the absence of information regarding potential structural differences between viruses derived from mosquito cells with and without Wolbachia, our data does not exclude the possibility of an additional block in progeny virus binding that might exacerbate this inhibitory effect. One notable limitation of our experimental system is the use of a limited number of mammalian epithelial cell lines as a generalized vertebrate model of infection. While cultured fibroblast and epithelial cells like BHK-21 and Vero, respectively, are physiologically relevant and have been regularly used towards the study of alphavirus and flavivirus pathogenesis, further studies using a diverse set of tissue-specific cell lines is required to fully understand the effect of Wolbachia on virus transmission [24].
Notably, our findings support the hypothesis of viral RNA being the target for pathogen blocking. Wolbachia’s ability to restrict viruses is seemingly limited to positive-sense single-stranded RNA (+ssRNA) viruses, as inhibition of neither negative-sense single-stranded RNA (-ssRNA) nor DNA viruses have been observed either in the field, or under laboratory conditions. It is therefore likely that factor(s) regulating virus inhibition affect this particular genomic feature shared by all susceptible viruses [3, 25–26]. We have previously reported the role of Drosophila melanogaster RNA methyltransferase Dnmt2 as a host determinant of pathogen blocking [12]. Interestingly, the mosquito ortholog of Dnmt2 has also been shown to play an important role in pathogen blocking in mosquitoes, albeit in a manner opposite to flies [27]. Furthermore, loss of Dnmt2 levels in fly cells significantly increase viral RNA replication and progeny virus infectivity, implying an antiviral mechanism of action that might involve targeting of the viral RNA. Given the biological role of Dnmt2 as an RNA cytosine methyltransferase, future studies will also focus on exploring the possibility of changes in the methylation profile of viral RNA in the presence and absence of Wolbachia.
Materials and methods
Insect and Mammalian cell culture
RML12 Aedes albopictus cells with and without Wolbachia wMel (a generous gift from Dr. Seth Bordenstein, Vanderbilt University) were grown at 24 ºC in Schneider’s insect media (Sigma-Aldrich) supplemented with 10% (v/v) heat-inactivated fetal bovine serum (Corning), 1% (v/v) each of L-Glutamine (Corning), non-essential amino acids (Corning) and penicillin-streptomycin-antimycotic (Corning). C710 Aedes albopictus cells with and without Wolbachia wStri (a generous gift from Dr. Horacio Frydman, Boston University) were grown in 1X Minimal Essential Medium (Corning) supplemented with 5% (v/v) heat-inactivated fetal bovine serum (Corning), 1% (v/v) each of L-Glutamine (Corning), non-essential amino acids (Corning) and penicillin-streptomycin-antimycotic (Corning). Aa23 Aedes albopictus cells with and without Wolbachia wAlbB (a generous gift from Dr. Horacio Frydman, Boston University) were grown at 24 ºC in Schneider’s insect media (Sigma-Aldrich) supplemented with 20% (v/v) heat-inactivated fetal bovine serum (Corning), 1% (v/v) each of L-Glutamine (Corning), non-essential amino acids (Corning) and penicillin-streptomycin-antimycotic (Corning). JW18 Drosophila melanogaster cells with and without Wolbachia wMel were grown at 24 ºC in Shields and Sang M3 insect media (Sigma-Aldrich) supplemented with 10% (v/v) heat-inactivated fetal bovine serum, 1% (v/v) each of L-Glutamine (Corning), non-essential amino acids (Corning) and penicillin-streptomycin-antimycotic (Corning). Aedes albopictus C636 cells and mammalian BHK-21 (Baby Hamster Kidney Fibroblast) and Vero (African Green Monkey Kidney Epithelial) cells were grown at either 28ºC (C6/36) or 37 ºC (BHK/Vero) under 5% ambient CO2 in 1X Minimal Essential Medium (Corning) supplemented with 10% (v/v) heat-inactivated fetal bovine serum (Corning), 1% (v/v) each of L-Glutamine (Corning), non-essential amino acids (Corning) and penicillin-streptomycin-antimycotic (Corning).
Virus infection in cells and progeny virus production
Virus stocks were generated from Aedes albopictus derived cells (RML12, C710, Aa23) or Drosophila melanogaster derived cells (JW18) with or without Wolbachia (RML12-wMel, C710-wStri, Aa23-wAlbB, JW18-wMel) by infecting naïve cells with virus at an MOI of 10 particles/cell. In all cases, serum-free media was used for downstream virus purification. Media containing virus was collected 5 days post-infection for alphaviruses SINV (SINV-nLuc), CHIKV (CHIKV18125-capsid-mKate), and 7 days post-infection for flavivirus ZIKV (MR766 Uganda Strain). Virus stocks were subsequently purified and concentrated by ultracentrifugation (43K for 2.5 h) over a 27% (w/v) sucrose cushion dissolved in HNE buffer (20mM HEPES, 0.15M NaCl, 0.1mM EDTA). Viral pellets were stored and aliquoted in HNE buffer before being used for all subsequent experiments. Viral titers were determined using standard plaque assay on vertebrate cells (BHK-21 cells for SINV, CHIKV and Vero cells for ZIKV). Cells were fixed either 48 hours post infection (SINV, CHIKV) or 72 hours post infection (ZIKV) using 10% (v/v) formaldehyde and stained with crystal violet to visualize plaques. Virus particles were determined by quantifying viral genome copies via quantitative RT-PCR using primers listed in the primer table (S1 Table) and standard curves comprised of linearized infectious clone sequences containing either full-length viral genomes (SINV, CHIKV) or partial Env gene (ZIKV).
Mosquito rearing and infectious blood meals
Aedes aegypti mosquitoes either -infected and -uninfected with Wolbachia (wAlbB strain) (generously provided by Dr. Zhiyong Xi, Michigan State University, USA), were reared in an insect incubator (Percival Model I-36VL, Perry, IA, USA) at 28°C and 75% humidity with 12 h light/dark cycle. Four to six-day old mated female mosquitoes were allowed to feed for 1h on approximately 108 PFUs of SINV (TE12-untagged) containing citrated rabbit blood (Fisher Scientific DRB030) supplemented with 1mM ATP (VWR) and 10% sucrose using a Hemotek artificial blood feeding system (Hemotek, UK) maintained under constant temperature of 37°C. Engorged mosquitoes were then isolated and reared at 28°C in the presence of male mosquitoes. Infected mosquitoes were harvested 5–7 days post blood meal before being snap frozen in liquid nitrogen and stored at −80°C before further processing. Samples for qPCR and qRT-PCR were homogenized in TRiZOL (Sigma Aldrich) reagent and further processed for nucleic acid extractions using manufacturer’s protocols.
Virion RNA extraction and transfection
Virion encapsidated RNA was extracted from viruses (SINV-nLuc) were purified over a 27% sucrose cushion using TRiZOL reagent (Sigma Aldrich) using manufacturer’s protocol. Post extraction, RNAs were DNase (RQ1 RNase-free DNase, NEB) treated using manufacturer’s protocol to remove cellular contaminants and viral RNA copies were quantified via quantitative RT-PCR using primers probing for SINV nsP1 and E1 genomic regions (Supplementary S1 Table) and a standard curve comprised of linearized SINV infectious clone containing the full-length viral genome. To determine infectivity or replication kinetics of Sindbis virion RNA, equal copies of virion isolated RNA (105 copies), quantified using qRT-PCR, were transfected into BHK-21 cells in serum-free Opti-MEM (Gibco). To maximize production of infectious units, equal mass (1 μg) of virion (SINV-nLuc) isolated RNA derived from JW18 fly cells was transfected into BHK-21 cells. Transfection was carried out for 6 hours before the transfection inoculum was removed and overlay was applied. Cells were fixed 48 hours post transfection using 10% (v/v) formaldehyde and stained with crystal violet to visualize plaques.
Live cell imaging
Growth of fluorescent reporter viruses in mosquito (C710) and vertebrate (BHK-21) cells were monitored using Incucyte live cell analysis system (Essen Biosciences, USA). Cells were grown under standard conditions as described earlier under 5% ambient CO2 either at 37ºC for vertebrate BHK-21 cells and 27ºC for Aedes albopictus C710 cells. Cells were plated to 75–80% confluency in 96-well plates to allow distinct separation between adjacent cells and preserve cell shape for optimal automated cell counting. Cells per well were imaged and averaged across four distinct fields of view, each placed in one quarter of the well, every 2 hours over the course of the infection. For every sample, total fluorescence generated by cells expressing the red fluorescent reporter mKate was calculated and normalized by the cell number. A manual threshold was set to minimize background signal via automated background correction at the time of data collection. Following acquisition, data was analyzed real-time using the native Incucyte Base Analysis Software.
Incoming viral genomic RNA half-life assay
C710 Aedes albopictus cells with (w/ wStri) or without Wolbachia (w/o wStri) and JW18 Drosophila melanogaster cells with (w/ wMel) or without Wolbachia (w/o wMel) were grown overnight (~12h) in media supplemented with 50 μM nucleoside analog 4-thiouridine (Sigma). These 4SU-treated cells were synchronously infected with Sindbis virus (SINV) derived from vertebrate BHK-21 cells at an MOI of 10 particles/cell. At indicated times post infection, tissue culture supernatants were removed, and cell monolayers were washed with 1XPBS prior to RNA extraction using TRiZOL reagent (Sigma Aldrich) using manufacturer’s protocols. A total of 1μg of total RNA per sample was biotinylated using HPDP-Biotin (Pierce) and all 4SU-labelled RNA was separated from the total RNA pool using streptavidin cleanup and levels of unlabeled RNA at each time point was measured relative to that present at 0 minutes post internalization. RNA half-life was extrapolated using non-linear, exponential decay model as described previously [16, 28].
Viral replication assays
Quantification of viral protein translation was performed using cellular lysates following synchronized infections with reporter viruses (SINV-nLuc), or transfections with virion-derived RNA from the aforementioned viruses. At indicated times post infection, samples were collected and homogenized in 1X Cell Culture Lysis Reagent (Promega). Samples were mixed with NanoGlo luciferase reagent (Promega), incubated at room temperature for 3 minutes before luminescence was recorded using a Synergy H1 microplate reader (BioTech instruments).
Real-time quantitative PCR and RT-PCR analyses
Total DNA and RNA from samples were extracted using TRiZOL reagent (Sigma Aldrich) using manufacturer’s protocols. Complementary DNA (cDNA) was synthesized using MMuLV Reverse Transcriptase (NEB) with random hexamer primers (Integrated DNA Technologies). Negative (no RT or no gDNA) controls were performed for each target. Quantitative PCR or RT-PCR analyses were performed using Brilliant III SYBR green QPCR master mix (Bioline) with gene-specific primers according to the manufacturer's protocol and with the Applied Bioscience StepOnePlus qPCR machine (Life Technologies). All primer sets, except for ZIKV_MR766 Env and CHIKV_18125 E2 were designed based on information present in existing literature [15, 20]. For primers designed specifically for this study, primer amplification efficiency was calculated using standard curves (see S1 Table for more details). The expression levels were normalized to the endogenous 18S rRNA expression using the delta-delta comparative threshold method (ΔΔCT) (S1 Table).
Phylogenetic analyses
MUSCLE alignment was performed on concatenated Wolbachia sequences comprising five MLST genes coxA, gatB, ftsZ, hcpA and fbpA. The resulting alignment was used to perform Maximum-likelihood analyses in MEGA X with a bootstrap value set to 1000. The resulting newick (.nwk) file was used to construct a phylogenetic tree using FigTree v1.4.4.
Statistical analyses of experimental data
All statistical analyses were conducted using GraphPad Prism 8 (GraphPad Software Inc., San Diego, CA).
Supporting information
(A) Viral RNA levels were quantified in individual adult female Aedes aegypti mosquitoes with and without Wolbachia (wAlbB strain) using qRT-PCR at 7-days post infectious blood meal with SINV. Error bars represent standard error of mean (SEM) of biological replicates (n = 6). Welch’s t-test performed on log-transformed values. *P < 0.05 (B) Viral RNA replication in mosquito cells colonized with Wolbachia. RML12 mosquito cells with or without Wolbachia (wMel strain) were infected with SINV at an MOI of 10 particles/cell. Total cellular RNA was harvested 48 hours post infection and assayed for viral RNA levels using quantitative RT-PCR. Error bars represent standard error of mean (SEM) of biological replicates (n = 6). Welch’s t-test performed on log-transformed values. ****P < 0.0001.
(TIF)
Mean percentage of total CHIKV (A) and ZIKV (B) particles produced from Aedes albopictus RML12 cells colonized with (w/ Wolb) or without (w/o Wolb) Wolbachia (wMel strain) (n = 6). Corresponding specific infectivity ratios are presented in Fig 3B.
(TIF)
Welch’s t-test performed on log-transformed values. Error bars represent standard error of mean (SEM) of biological replicates (n = 5–6). *P < 0.05; **P < 0.01.
(TIF)
Kruskal-Wallis test of multivariate comparisons with Dunn’s post-hoc test. Error bars represent standard error of mean (SEM) of biological replicates (n = 4). **P < 0.01, ns = non-significant.
(TIF)
Micrograph images presented here represent of one out of four fields of view collected per replicate/time point (n = 6) every 4 hours (A-L). For each set, left and right images represent BHK-21 cells infected with W- virus (derived from C710 cells without wStri) and W+ virus (derived from C710 cells with wStri) respectively. Infected cell populations are visible as red cells, outlines of which are masked in blue to allow automated quantification using Incucyte Base Analysis Software (quantified in Fig 5).
(TIF)
Viruses were derived from JW18 fly cells with (W+ virus) or without (W- virus) Wolbachia (wMel strain). 1 μg of total virion RNA was transfected into BHK-21 cells and infectious titer was determined by counting the number of plaques produced after 48 hours post transfection. Details of the assay can be found in Materials and Methods. Error bars represent standard error of mean (SEM) of biological replicates (n = 3). Welch’s t-test on log-transformed data. *P < 0.05.
(TIF)
All primers were used at a final concentration of 10μM for quantitative RT-PCR reactions. All primer sets, except for ZIKV_MR766 Env and CHIKV_18125 E2 were designed based on information present in existing literature [15, 20]. For primer sets designed specifically for this study, primer amplification efficiency was carried out using standard curves. (CHIKV_18125 E2: Slope = -0.69, Amplification factor = 4.90, Efficiency ~ 100%, ZIKV_MR766 Env: Slope = -0.761, Amplification factor = 5.77, Efficiency ~ 100%).
(DOCX)
Acknowledgments
We thank Lauren Nease for proofreading the revised version of the manuscript. We thank all members of the IU Virology Labs for fostering ideas through helpful discussions and for sharing their instruments. We thank our colleagues outside of IU for their generosity in sharing reagents, including Dr. Horacio Frydman, Boston University for providing us Aa23 and C710 Aedes albopictus cells, Dr. Seth Bordenstein, Vanderbilt University for providing us RML12 Aedes albopictus cells and finally, Dr. Zhiyong Xi, Michigan State University for providing us Aedes aegypti mosquitoes.
Data Availability
All relevant data are within the manuscript and its Supporting Information files except for the Incucyte data. These full datasets have been made available at doi:10.5061/dryad.w0vt4b8n9.
Funding Statement
ILGN and RWH received funding from the National Institutes of Health grant R21 AI121849 (https://www.nih.gov), and from the Indiana University Office for the Vice Provost for Research’s Faculty Research Support Program (https://research.iu.edu/funding-proposals/funding/opportunities/support-program/index.html). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Ferguson NM. Challenges and opportunities in controlling mosquito-borne infections. Nature. 2018. July;559(7715):490–497. 10.1038/s41586-018-0318-5 Epub 2018 Jul 25. . [DOI] [PubMed] [Google Scholar]
- 2.Mousson L, Zouache K, Arias-Goeta C, Raquin V, Mavingui P, Failloux AB. The native Wolbachia symbionts limit transmission of dengue virus in Aedes albopictus. PLoS Negl Trop Dis. 2012;6(12):e1989 10.1371/journal.pntd.0001989 Epub 2012 Dec 27. ; PMCID: PMC3531523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lindsey ARI, Bhattacharya T, Newton ILG, Hardy RW. Conflict in the Intracellular Lives of Endosymbionts and Viruses: A Mechanistic Look at Wolbachia-Mediated Pathogen-blocking. Viruses. 2018. March 21;10(4):141 10.3390/v10040141 ; PMCID: PMC5923435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frentiu FD, Zakir T, Walker T, Popovici J, Pyke AT, van den Hurk A et al. Limited dengue virus replication in field-collected Aedes aegypti mosquitoes infected with Wolbachia. PLoS Negl Trop Dis. 2014. February 20;8(2):e2688 10.1371/journal.pntd.0002688 ; PMCID: PMC3930499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moreira LA, Iturbe-Ormaetxe I, Jeffery JA, Lu G, Pyke AT, Hedges LM et al. A Wolbachia symbiont in Aedes aegypti limits infection with dengue, Chikungunya, and Plasmodium. Cell. 2009. December 24;139(7):1268–78. 10.1016/j.cell.2009.11.042 . [DOI] [PubMed] [Google Scholar]
- 6.Koh C, Audsley MD, Di Giallonardo F, Kerton EJ, Young PR et al. Sustained Wolbachia-mediated blocking of dengue virus isolates following serial passage in Aedes aegypti cell culture. Virus Evol. 2019. June 8;5(1):vez012 10.1093/ve/vez012 ; PMCID: PMC6555872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dutra HL, Rocha MN, Dias FB, Mansur SB, Caragata EP, Moreira LA. Wolbachia Blocks Currently Circulating Zika Virus Isolates in Brazilian Aedes aegypti Mosquitoes. Cell Host Microbe. 2016. June 8;19(6):771–4. 10.1016/j.chom.2016.04.021 Epub 2016 May 4. ; PMCID: PMC4906366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Amuzu HE, McGraw EA. Wolbachia-Based Dengue Virus Inhibition Is Not Tissue-Specific in Aedes aegypti. PLoS Negl Trop Dis. 2016. November 17;10(11):e0005145 10.1371/journal.pntd.0005145 ; PMCID: PMC5113870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ant TH, Herd CS, Geoghegan V, Hoffmann AA, Sinkins SP. The Wolbachia strain wAu provides highly efficient virus transmission blocking in Aedes aegypti. PLoS Pathog. 2018. January 25;14(1):e1006815 10.1371/journal.ppat.1006815 ; PMCID: PMC5784998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schultz MJ, Tan AL, Gray CN, Isern S, Michael SF, Frydman HM et al. Wolbachia wStri Blocks Zika Virus Growth at Two Independent Stages of Viral Replication. mBio. 2018. May 22;9(3):e00738–18. 10.1128/mBio.00738-18 ; PMCID: PMC5964347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Geoghegan V, Stainton K, Rainey SM, Ant TH, Dowle AA, Larson T et al. Perturbed cholesterol and vesicular trafficking associated with dengue blocking in Wolbachia-infected Aedes aegypti cells. Nat Commun. 2017. September 13;8(1):526 10.1038/s41467-017-00610-8 ; PMCID: PMC5597582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bhattacharya T, Newton ILG, Hardy RW. Wolbachia elevates host methyltransferase expression to block an RNA virus early during infection. PLoS Pathog. 2017. June 15;13(6):e1006427 10.1371/journal.ppat.1006427 ; PMCID: PMC5472326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thomas S, Verma J, Woolfit M, O'Neill SL. Wolbachia-mediated virus blocking in mosquito cells is dependent on XRN1-mediated viral RNA degradation and influenced by viral replication rate. PLoS Pathog. 2018. March 1;14(3):e1006879 10.1371/journal.ppat.1006879 ; PMCID: PMC5833283.Bottom of Form [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rainey SM, Martinez J, McFarlane M, Juneja P, Sarkies P, Lulla A et al. Wolbachia Blocks Viral Genome Replication Early in Infection without a Transcriptional Response by the Endosymbiont or Host Small RNA Pathways. PLoS Pathog. 2016. April 18;12(4):e1005536 10.1371/journal.ppat.1005536 Erratum in: PLoS Pathog. 2016 May;12(5):e1005639. ; PMCID: PMC4835223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wengler G. The mode of assembly of alphavirus cores implies a mechanism for the disassembly of the cores in the early stages of infection. Brief review. Arch Virol. 1987;94(1–2):1–14. 10.1007/BF01313721 . [DOI] [PubMed] [Google Scholar]
- 16.Sokoloski KJ, Nease LM, May NA, Gebhart NN, Jones CE, Morrison TE et al. Identification of Interactions between Sindbis Virus Capsid Protein and Cytoplasmic vRNA as Novel Virulence Determinants. PLoS Pathog. 2017. June 29;13(6):e1006473 10.1371/journal.ppat.1006473 ; PMCID: PMC5507600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fallon AM, Baldridge GD, Higgins LA, Witthuhn BA. Wolbachia from the planthopper Laodelphax striatellus establishes a robust, persistent, streptomycin-resistant infection in clonal mosquito cells. In Vitro Cell Dev Biol Anim. 2013. January;49(1):66–73. 10.1007/s11626-012-9571-3 Epub 2012 Dec 28. ; PMCID: PMC3630504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Salazar MI, Richardson JH, Sánchez-Vargas I, Olson KE, Beaty BJ. Dengue virus type 2: replication and tropisms in orally infected Aedes aegypti mosquitoes. BMC Microbiol. 2007. January 30;7:9 10.1186/1471-2180-7-9 ; PMCID: PMC1797809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nainu F, Trenerry A, Johnson KN. Wolbachia-mediated antiviral protection is cell-autonomous. J Gen Virol. 2019. November;100(11):1587–1592. 10.1099/jgv.0.001342 Epub 2019 Oct 10. . [DOI] [PubMed] [Google Scholar]
- 20.Teramoto T, Huang X, Armbruster PA, Padmanabhan R. Infection of Aedes albopictus Mosquito C6/36 Cells with the wMelpop Strain of Wolbachia Modulates Dengue Virus-Induced Host Cellular Transcripts and Induces Critical Sequence Alterations in the Dengue Viral Genome. J Virol. 2019. July 17;93(15):e00581–19. 10.1128/JVI.00581-19 ; PMCID: PMC6639269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dodson BL, Hughes GL, Paul O, Matacchiero AC, Kramer LD, Rasgon JL. Wolbachia enhances West Nile virus (WNV) infection in the mosquito Culex tarsalis. PLoS Negl Trop Dis. 2014. July 10;8(7):e2965 10.1371/journal.pntd.0002965 ; PMCID: PMC4091933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fraser JE, O'Donnell TB, Duyvestyn JM, O'Neill SL, Simmons CP, Flores HA. Novel phenotype of Wolbachia strain wPip in Aedes aegypti challenges assumptions on mechanisms of Wolbachia-mediated dengue virus inhibition. bioRxiv. 2020. January 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Asselin AK, Villegas-Ospina S, Hoffmann AA, Brownlie JC, Johnson KN. Contrasting Patterns of Virus Protection and Functional Incompatibility Genes in Two Conspecific Wolbachia Strains from Drosophila pandora. Appl Environ Microbiol. 2019. February 20;85(5):e02290–18. 10.1128/AEM.02290-18 ; PMCID: PMC6384105.Bottom of Form [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Atkins GJ. The pathogenesis of alphaviruses. ISRN Virology. 2012. December 4;2013. [Google Scholar]
- 25.Teixeira L, Ferreira A, Ashburner M. The bacterial symbiont Wolbachia induces resistance to RNA viral infections in Drosophila melanogaster. PLoS Biol. 2008. December 23;6(12):e2 10.1371/journal.pbio.1000002 ; PMCID: PMC2605931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Glaser RL, Meola MA. The native Wolbachia endosymbionts of Drosophila melanogaster and Culex quinquefasciatus increase host resistance to West Nile virus infection. PLoS One. 2010. August 5;5(8):e11977 10.1371/journal.pone.0011977 ; PMCID: PMC2916829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang G, Hussain M, O'Neill SL, Asgari S. Wolbachia uses a host microRNA to regulate transcripts of a methyltransferase, contributing to dengue virus inhibition in Aedes aegypti. Proc Natl Acad Sci U S A. 2013. June 18;110(25):10276–81. 10.1073/pnas.1303603110 Epub 2013 Jun 3. ; PMCID: PMC3690878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dölken L, Ruzsics Z, Rädle B, Friedel CC, Zimmer R, Mages J et al. High-resolution gene expression profiling for simultaneous kinetic parameter analysis of RNA synthesis and decay. RNA. 2008. September;14(9):1959–72. 10.1261/rna.1136108 Epub 2008 Jul 24. ; PMCID: PMC2525961. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Viral RNA levels were quantified in individual adult female Aedes aegypti mosquitoes with and without Wolbachia (wAlbB strain) using qRT-PCR at 7-days post infectious blood meal with SINV. Error bars represent standard error of mean (SEM) of biological replicates (n = 6). Welch’s t-test performed on log-transformed values. *P < 0.05 (B) Viral RNA replication in mosquito cells colonized with Wolbachia. RML12 mosquito cells with or without Wolbachia (wMel strain) were infected with SINV at an MOI of 10 particles/cell. Total cellular RNA was harvested 48 hours post infection and assayed for viral RNA levels using quantitative RT-PCR. Error bars represent standard error of mean (SEM) of biological replicates (n = 6). Welch’s t-test performed on log-transformed values. ****P < 0.0001.
(TIF)
Mean percentage of total CHIKV (A) and ZIKV (B) particles produced from Aedes albopictus RML12 cells colonized with (w/ Wolb) or without (w/o Wolb) Wolbachia (wMel strain) (n = 6). Corresponding specific infectivity ratios are presented in Fig 3B.
(TIF)
Welch’s t-test performed on log-transformed values. Error bars represent standard error of mean (SEM) of biological replicates (n = 5–6). *P < 0.05; **P < 0.01.
(TIF)
Kruskal-Wallis test of multivariate comparisons with Dunn’s post-hoc test. Error bars represent standard error of mean (SEM) of biological replicates (n = 4). **P < 0.01, ns = non-significant.
(TIF)
Micrograph images presented here represent of one out of four fields of view collected per replicate/time point (n = 6) every 4 hours (A-L). For each set, left and right images represent BHK-21 cells infected with W- virus (derived from C710 cells without wStri) and W+ virus (derived from C710 cells with wStri) respectively. Infected cell populations are visible as red cells, outlines of which are masked in blue to allow automated quantification using Incucyte Base Analysis Software (quantified in Fig 5).
(TIF)
Viruses were derived from JW18 fly cells with (W+ virus) or without (W- virus) Wolbachia (wMel strain). 1 μg of total virion RNA was transfected into BHK-21 cells and infectious titer was determined by counting the number of plaques produced after 48 hours post transfection. Details of the assay can be found in Materials and Methods. Error bars represent standard error of mean (SEM) of biological replicates (n = 3). Welch’s t-test on log-transformed data. *P < 0.05.
(TIF)
All primers were used at a final concentration of 10μM for quantitative RT-PCR reactions. All primer sets, except for ZIKV_MR766 Env and CHIKV_18125 E2 were designed based on information present in existing literature [15, 20]. For primer sets designed specifically for this study, primer amplification efficiency was carried out using standard curves. (CHIKV_18125 E2: Slope = -0.69, Amplification factor = 4.90, Efficiency ~ 100%, ZIKV_MR766 Env: Slope = -0.761, Amplification factor = 5.77, Efficiency ~ 100%).
(DOCX)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files except for the Incucyte data. These full datasets have been made available at doi:10.5061/dryad.w0vt4b8n9.