ABSTRACT
Members of Bifidobacterium are among the first microbes to colonise the human gut, and certain species are recognised as the natural resident of human gut microbiota. Their presence in the human gut has been associated with health-promoting benefits and reduced abundance of this genus is linked with several diseases. Bifidobacterial species are assumed to have coevolved with their hosts and include members that are naturally present in the human gut, thus recognised as Human-Residential Bifidobacteria (HRB). The physiological functions of these bacteria and the reasons why they occur in and how they adapt to the human gut are of immense significance. In this review, we provide an overview of the biology of bifidobacteria as members of the human gut microbiota and address factors that contribute to the preponderance of HRB in the human gut. We highlight some of the important genetic attributes and core physiological traits of these bacteria that may explain their adaptive advantages, ecological fitness, and competitiveness in the human gut. This review will help to widen our understanding of one of the most important human commensal bacteria and shed light on the practical consideration for selecting bifidobacterial strains as human probiotics.
Keywords: bifidobacteria, human health, natural inhabitants, physiological properties, genetic adaptation, ecological fitness
Bifidobacterial species that ubiquitously encounter in the human host, collectively known as the Human-Residential Bifidobacteria (HRB), possess unique genetic elements and superior physiological functions that contribute to their ecological fitness and competitiveness, and they are thus more beneficial to humans.
INTRODUCTION
Members of the genus Bifidobacterium are of substantial importance due to their purported health-promoting effects in human across their lifespan (O'Callaghan and van Sinderen 2016). Their presence in the human gastrointestinal tract is often associated with health benefits including the production of metabolites such as short-chain fatty acids and vitamins, immune system development and prevention of gut disorders (O'Callaghan and van Sinderen 2016). Recent advances in bifidobacterial research reveal that bifidobacterial strains have coevolved with their hosts and many physiological characteristics can be residential-origin dependent (Lamendella et al. 2008; Sun et al. 2015; Wong et al. 2018; Zou et al. 2019; Rodriguez and Martiny 2020). In this regard, some species of bifidobacteria have been identified as the natural inhabitants of the human host (Wong et al. 2018). Nevertheless, the reason why bifidobacterial species reside in the human gut across the life course, their adaptation and survival in the harsh environment of the human gastrointestinal tract and their impact on human health remain elusive. Thus, exploration of their human niche-specific adaptations and functional traits as members of human gut microbiota is of utmost significance.
This review aims at providing an overview of the ecological fitness of the bifidobacterial species that are commonly present in the human gut and elucidate how this bacterial group contribute to human health. Here we summarise current evidence on the colonisation, genetic adaptation and physiological characteristics of the human gut commensal bifidobacteria and address their roles as members of human gut microbiota. An increased understanding of the physiological attributes of this genus and their functions in the human host, which are of immense industrial value, will aid the selection of probiotic strains for human use.
COLONISATION OF BIFIDOBACTERIA IN THE HUMAN GUT
Bifidobacterium is one of the most abundant bacterial genera present in the healthy infant gut (Favier et al. 2002; Odamaki et al. 2016). The abundance of this genus in the human gastrointestinal tract substantially decrease after weaning and continue to decrease with age (Yatsunenko et al. 2012; Kato et al. 2017). During adulthood the levels decrease considerably but remain relatively stable; decreasing again in old age (Odamaki et al. 2016). Colonisation by bifidobacteria is believed to play pivotal roles in maintaining human health. It is associated with a range of beneficial health effects, involved in the maturation of the immune, digestive and metabolic systems and consequently protecting against the susceptibility to various diseases later in life (Turroni et al. 2008).
Initial bifidobacterial colonisation occurs from birth and is influenced by several extrinsic factors (Arboleya et al. 2016). It has become evident that bifidobacterial species are vertically transmitted from the mother and colonise the intestine of the infant at the very early stages of life (Makino 2018). Several studies have linked the transmission of bifidobacteria from the mother's vaginal tract, gastrointestinal tract, breast milk, placenta and amniotic fluid to the infant (Makino et al. 2013; Milani et al. 2015b; Collado et al. 2016; Ferretti et al. 2018; de Goffau et al. 2019).
The delivery mode, in particular, has been demonstrated to potently impact this initial colonisation, with vaginally born infants displaying an increased abundance of bifidobacteria compared to those born by caesarean section (Rutayisire et al. 2016; Reyman et al. 2019). More specifically, a study based on analysis of the gut microbiota of mothers and corresponding children demonstrated that vaginally delivered infants share at least one monophyletic strain belonging to the genus Bifidobacterium with their mothers, whereas the monophyletic strains were not observed among infants delivered by caesarean section, which is thus indicative of vertical transmission (Makino et al. 2013). Moreover, in recent comparative genome analysis, the strains of bifidobacteria isolated from vaginal and gut microbiomes were indistinguishable, thus enforcing the importance of the maternal vaginal microbiome as a source of bifidobacterial colonisation (Freitas and Hill 2018).
The maternal birth canal has therefore been implicated as an essential source of bacteria, including bifidobacteria, during the delivery process. Although the colonisation of the gut environment was traditionally thought to begin at birth; new evidence of the presence of bacteria in the uterine environment suggests a primary foetal colonisation (de Goffau et al. 2019). Recent studies also suggest that many foetuses are exposed to microbes through the amniotic fluid that was continuously swallowed from mid to late gestation in utero (Chu et al. 2017). However, the ‘in utero colonisation hypothesis’ remains subject to debate. It is argued that, due to methodological difficulties, current scientific evidence does not support the existence of microbiome within the healthy foetal milieu (Perez-Muñoz et al. 2017). A recent study also demonstrates that there was no evidence for the presence of bacteria in the human placenta (de Goffau et al. 2019). Therefore, conclusions remain unachievable, and more studies are needed in this area.
Also, during vaginal delivery, as the infants pass through the birth canal, their oral and nasal cavities are infiltrated with vaginal secretions (Torres-Alipi et al. 1990; Li et al. 2018). For this reason, the neonatal oral fluid at delivery appears to be a mother-to-child-transmission route for bifidobacteria. It is indeed true that bifidobacteria are present in the neonatal oral fluid at birth and the same strains are detected in both the oral fluid and faecal samples collected at one month after birth, suggesting neonatal oral fluid at delivery is an essential source of bacteria from both maternal and environmental ecosystems and may contribute to the early delivery of bacteria to the digestive tracts of neonates (Toda et al. 2019).
Furthermore, accumulating evidence suggests that human breast milk harbours a microbial community and represents a source of commensal bacteria for the neonates (Gueimonde et al. 2007; Jost et al. 2014; Ruiz, García-Carral and Rodriguez 2019). It has been reported that bifidobacterial strains were present in maternal milk, suggesting the possibility of breast milk as a transmission route of bifidobacteria from the mother to the infant gut (Jost et al. 2014; Makino et al. 2015; Kordy et al. 2020). In this context, feeding mode is considered to be some of the major forces that could impact the colonisation level and species composition of bifidobacteria in the infant gut. Breastfeeding is known to provide the infant with a broad spectrum of biologically active factors including, among others, human milk oligosaccharides (HMOs), that aid in the colonisation of bifidobacteria, as well as a set of commensal microbes to initially colonise the infant gut (Le Doare et al. 2018). Numerous studies comparing the infant gut microbiome of exclusively breast-fed versus formula-fed infants have reported that the guts of breast-fed infants are enriched with Bifidobacterium spp. whereas formula-fed infants have a lower abundance of beneficial bacteria (Wang et al. 2015; Lewis and Mills 2017).
Altogether, these studies provide clues of where and how bifidobacteria colonise the human gut. It is implicated that early life colonisation patterns and successions of bifidobacteria may contribute to the risk of developing health complications during neonatal stage or later in life, highlighting the importance of maintaining proper levels of bifidobacteria in the human gut (Arboleya et al. 2016).
GENERAL FEATURES OF BIFIDOBACTERIUM SPECIES
Bifidobacteria are Gram-positive, anaerobic, non-motile, non-spore-forming, polymorphic rods that belong to the family Bifidobacteriaceae, order Bifidobacteriales and phylum Actinobacteria. Bifidobacteria display a range of distinct cell forms, including curved, short and bifurcated Y shapes. The genomic DNA of bifidobacteria contains a high guanine-plus-cytosine content, with numerous genes involved in the metabolism of dietary and host-derived carbohydrates (Milani et al. 2014; Ventura et al. 2014). At present, the genus Bifidobacterium encompasses approximately 80 species, including four species (Bifidobacterium animalis, B. longum, B. pseudolongum and B. thermacidophilum) that are further divided into subspecies (Parte 2018; Sakanaka et al. 2020).
In addition to the presence in the human gut, bifidobacterial species also naturally occur in the gastrointestinal tract of animals as well as a few that are present in sewage, human vagina, oral cavity, breast milk, and foods (Ventura et al. 2007). Bifidobacteria are widely distributed in the gut of social animals (e.g. mammals, birds and insects), whose offspring are dependent on parental care (Turroni, Van Sinderen and Ventura 2011). Vertical transmission from mother to offspring appears to be a common ecological trademark of this bacterial genus, reflecting a clear evolutionary link between the parent, offspring/progeny and the bifidobacterial species present. A study has revealed that the clades of Bifidobacteriaceae arose via cospeciation with humans, chimpanzees, bonobos and gorillas over the past 15 million years, among which the species have been maintained exclusively within host lineages across hundreds of host generations (Moeller et al. 2016). After that, bifidobacteria have developed a diverse number of genetic strategies to adapt to their respective hosts and display differences in their ecological adaptation among species.
In general, bifidobacteria could be categorised into two major groups based on their residential origins; bifidobacterial species that are naturally encountered in the human gastrointestinal tract are referred to as Human-Residential Bifidobacteria (HRB), whereas other species which are the natural inhabitants of animals or environment as non-HRB (Sugahara, Odamaki and Xiao 2015; Wong et al. 2018) (Fig. 1). Among HRB, B. breve, B. longum subsp. infantis, B. longum subsp. longum and B. bifidum, which are the dominant species in the infant's intestines, are referred to as infant-type HRB (Turroni et al. 2012), whereas B. adolescentis, B. catenulatum, B. pseudocatenulatum, B. longum subsp. longum, etc., which are the dominant species in the adult intestines, are referred to as adult-type HRB (Ishikawa et al. 2013). However, there does not seem to be an absolute infant versus adult division of bifidobacterial (sub)species (Turroni et al. 2012, 2019). It is noted that HRB strains might not be adapted to particular habitats (gut/blood/human milk) and life stages (infant/adult) within humans, for which the phylogenetic and genomic traits are undisguisable from the strains studied (Freitas and Hill 2018; Rodriguez and Martiny 2020). After that, B. longum subsp. longum was referred to as species that predominantly inhabit both the infant and adult intestines (Odamaki et al. 2018). The ubiquitous distribution of B. longum subsp. longum species across the human lifespan was suggested to be associated with their genetic diversity which could enhance their adaptation and increase competitiveness in the gut environment, and at least partly due to extensive transmission across family members, a phenomenon that was shown not to be confined to mother-infant pairs (Odamaki et al. 2018, 2019).
Figure 1.
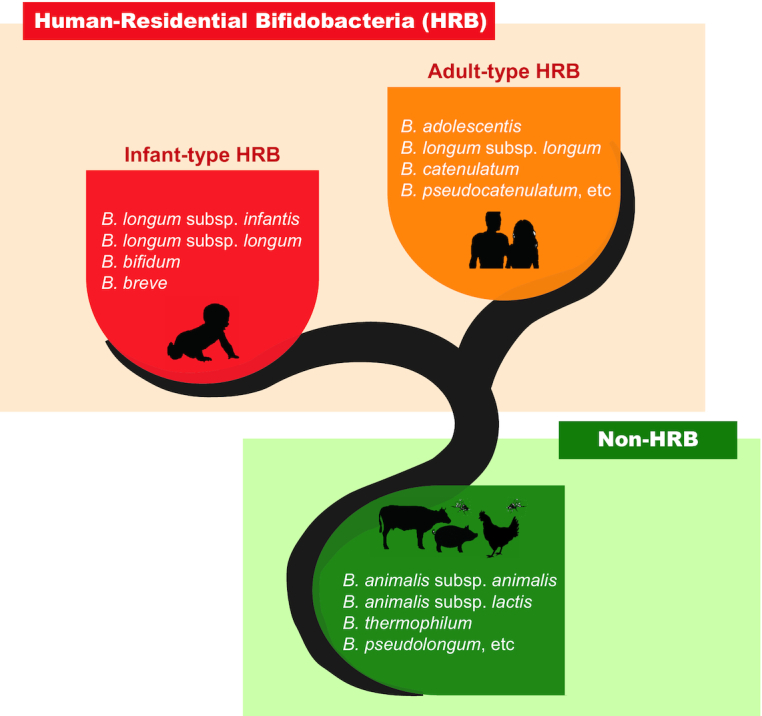
Distinctive differences in ecological distribution of bifidobacteria. Bifidobacterial species are distributed in a wide range of niches, encompassing the human intestine, the gastrointestinal tract of animals, human vagina, human oral cavity, breast milk, sewage and food. The species that naturally occur in the human host are referred to as Human-Residential Bifidobacteria (HRB). Among HRB, Bifidobacterium longum subsp. infantis, B. longum subsp. longum, B. bifidum and B. breve are recognised as the four exclusive members of the infant gut and are referred to as infant-type HRB. Meanwhile, bifidobacterial species that are predominantly present in the adult gut are referred to as adult-type HRB. Conversely, other species which are the natural inhabitants of animals or environment are referred to as non-HRB. The species of HRB and non-HRB display differences in their ecological adaptation.
Meanwhile, non-HRB species encompass B. animalis subsp. animalis, B. animalis subsp. lactis, B. thermophilum, B. pseudolongum, etc., among which some species show a strict ecological adaptation to a particular animal gut (Lamendella et al. 2008). For instance, the non-HRB species B. magnum and B. cuniculi were ubiquitously found in rabbit faeces, B. pullorum and B. gallinarum in the chicken intestine, and B. longum subsp. suis in the piglet faeces (Biavati et al. 2000). Such variations were suggested to be related to the diet, age, and species of the host animals (Mitsuoka and Kaneuchi 1977). Bifidobacteria are also found in other atypical ecological niches that are either partly linked with the gut: sewage (e.g. B. minimum and B. thermacidophilum) (Biavati, Scardovi and Moore 1982; Dong et al. 2000) or completely different from that of the gut: food products (e.g. B. mongoliense from fermented mare's milk product and B. aquikefiri and B. tibiigranuli from water kefir) (Watanabe et al. 2009; Laureys et al. 2016; Eckel et al. 2019). Notably, a small population of B. animalis subsp. lactis, which is originated from animal gut and commonly incorporated as probiotics in dairy products, was also detected in human faeces (Turroni et al. 2009). It has been suggested that most of the strains of B. animalis subsp. lactis currently applied in commercial products were genetically indistinguishable (Xiao et al. 2010b; Milani et al. 2013). Its detection might attribute to human diets, such as intake of yoghurt, suggesting that B. animalis subsp. lactis is not a commensal of human gut microbiota (Kato et al. 2017). Altogether, the identification of bifidobacteria in these environments could be plausibly a consequence of ‘natural’ contaminations from human/animal gut origins and/or from accidental contaminations during the sampling procedures. Conversely, there could be a strong rationale behind the species of HRB being the natural inhabitant of the human gut. As will be discussed later, accumulating evidence suggests that the residential origins of bifidobacteria are associated with a number of significant differences in their physiological features and health-promoting functions in the human host (Xiao et al. 2010a; Odamaki et al. 2015; Sugahara et al. 2015; Sugahara, Odamaki and Xiao 2015; Minami et al. 2016; Sakurai et al. 2017, 2018; Wong et al. 2018, 2020; Sakurai, Odamaki and Xiao 2019).
NATURAL SELECTION OF INFANT-TYPE HRB BY HUMAN MILK
Human milk is regarded as the principal source of nutrition for infants. It contains a rich source of essential nutrients that nourishes the neonate, supporting proper growth and development. In addition, human milk also provides the neonate with its own microbiota as well as a wide array of bioactive molecules that indirectly can contribute to the establishment of infant gut microbiome (Le Doare et al. 2018). Breast-fed infants are characterised by a gut population dominated by the species of HRB, including B. bifidum, B. breve, B. longum subsp. longum, and B. longum subsp. infantis (Turroni et al. 2012; Makino et al. 2015). This predominance has been explained in part by the high amounts of a class of molecules called human milk oligosaccharides (HMOs) (Lawson et al. 2019). Intriguingly, these molecules are indigestible by the infant, and thus provide no direct nutritive value. Instead, HMOs are thought to selectively support the colonisation of HRB species capable of utilising these diverse substrates (Sela and Mills 2010; Asakuma et al. 2011; Katayama 2016). In addition to HMOs, human milk contains antibacterial compounds such as immunoglobulins, lactoferrin, lactoferricin, lysozyme, lactoperoxidase, free fatty acids, antimicrobial peptides and others (Field 2005). Among them, lysozyme has been reported to be another possible selective factor in human milk that could affect the colonisation of bifidobacterial species in the infant gut (Gagnon et al. 2004; Rada et al. 2010; Minami et al. 2016) (Fig 2).
Figure 2.
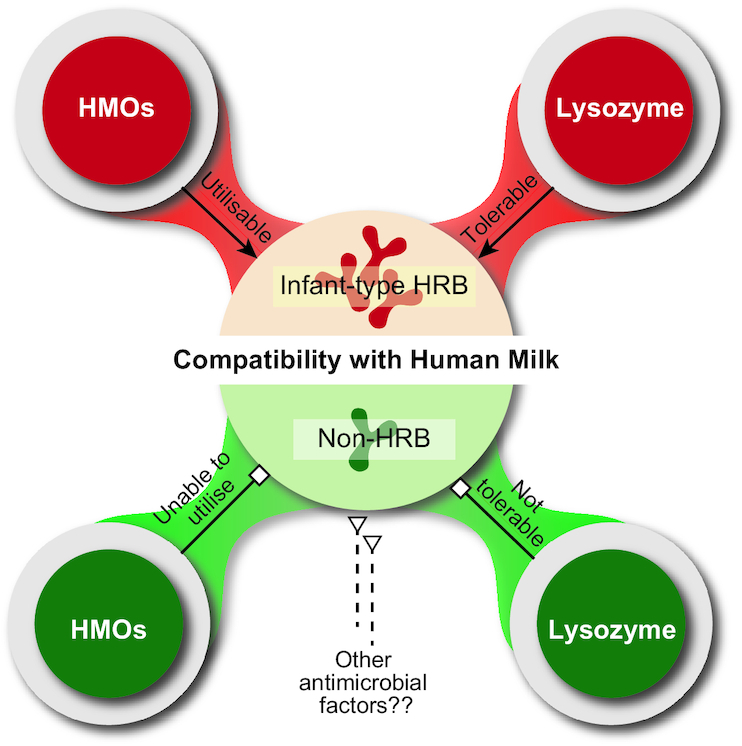
Differences in the compatibility with human milk among bifidobacterial species. Infant-type HRB species are adapted to utilise human milk oligosaccharides (HMOs), which are one of the most important component in the human milk that function to selectively support the colonisation of bifidobacteria in infant gut. Infant-type HRB are also highly tolerant to the antibacterial component present in the human milk called lysozyme. In contrary, non-HRB species are excluded by these selective factors present in the human milk. The species of non-HRB are lacking with the enzymatic arsenal dedicated for HMOs utilisation and are sensitive to human milk lysozyme.
HMO utilisation
HMOs are the third most abundant substantial component in human milk after lactose and lipids, reaching concentrations of up to 15 g/L in mature human milk (Kobata 2010). HMOs are complex carbohydrates composing of five monosaccharides: glucose (Glc), galactose (Gal), N-acetylglucosamine (GlcNAc), fucose (Fuc) and N-acetylneuraminic acid (NeuAc) or sialic acid (Kirmiz et al. 2018). HMOs are synthesised from a lactose (Galβ1–4Glc) backbone at the reducing end and are elongated by the repeats of β-1,3- or β-1,6-linked lacto-N-biose I (LNB) or N-acetyllactosamine (Chen 2015). The terminal lactose molecules can be further decorated with the addition of fucose and/or sialic acid residues (Bode and Jantscher-Krenn 2012). With varying degrees of polymerisation and multiple linkage isomers, more than 200 different structures of HMOs have been observed (Thomson, Medina and Garrido 2018). The overabundant examples of HMOs are lacto-N-tetraose (LNT), lacto-N-fucopentaose I (LNFP I), lacto-N-difucohexaose I (LNDFH I) and 2’-fucosyllactose (2’-FL), with three of the four HMOs are type I chains that exhibit the core structure of LNT (Kunz et al. 2000; Chen 2015). Degradation of LNT and its constituent component, LNB, has therefore been implicated as an essential step in HMO metabolism.
Utilisation of HMOs by bifidobacteria
Bifidobacterial species display differential ability to consume various HMOs, among which the infant-type HRB, namely, B. bifidum, B. breve, B. longum subsp. infantis and B. longum subsp. longum are adapted to utilise HMOs, while the ability was not present in many species of adult-type HRB and the non-HRB (LoCascio et al. 2007; Xiao et al. 2010a; Asakuma et al. 2011). More specifically, considerable differences in HMO utilisation among the four species of infant-type HRB were observed. B. longum subsp. infantis aggressively consumes LNB and almost all types of HMOs including fucosylated and sialylated molecules, and the consumption capability is highly and widely conserved in this subspecies (Xiao et al. 2010a; Asakuma et al. 2011; Garrido et al. 2015; Thomson, Medina and Garrido 2018). Meanwhile, the capability to consume HMOs is somewhat variable in B. bifidum strains, utilising almost all classes of HMOs and LNB (Xiao et al. 2010a; Asakuma et al. 2011; Garrido et al. 2015; Gotoh et al. 2018; Thomson, Medina and Garrido 2018). By contrast, the ability of the B. breve and B. longum subsp. longum strains to assimilate HMOs is limited. Most of the B. breve strains can utilise only LNT, lacto-N-neotetraose (LNnT), and LNB (Xiao et al. 2010a; Asakuma et al. 2011; Ruiz-Moyano et al. 2013; James et al. 2016; Thomson, Medina and Garrido 2018), whereas the majority of the B. longum subsp. longum strains can consume solely LNT and LNB, leaving other HMOs unmodified (Xiao et al. 2010a; Asakuma et al. 2011; Garrido et al. 2016; Thomson, Medina and Garrido 2018). Moreover, a few strains of these two (sub)species have been described to be capable of assimilating exceptionally other complex fucosylated HMOs, such as 2’-FL, 3-fucosyllactose (3-FL), lactodifucotetraose (LDFT) and LNFP I/II/III (Ruiz-Moyano et al. 2013; Garrido et al. 2016; Matsuki et al. 2016).
In addition, HMO utilisation capability by other strains of HRB (B. kashiwanohense and B. pseudocatenulatum) has been reported (Xiao et al. 2010a; Bunesova, Lacroix and Schwab 2016). B. kashiwanohense was found to consume preferably 2’-FL and 3-FL rather than LNT and LNnT (Bunesova, Lacroix and Schwab 2016; James et al. 2019). B. pseudocatenulatum was shown to be capable of utilising LNT (Matsuki et al. 2016). The ability to consume 2’-FL, 3-FL, LDFT and LNB has also been described in some strains of B. pseudocatenulatum (Xiao et al. 2010a; Matsuki et al. 2016). Nonetheless, the majority of adult-type HRB and non-HRB are unable to utilise HMOs. It is not surprising that such capability is absent and/or has been lost in some adult-type HRB and non-HRB that typically reside in the adult and animal intestines as well as other ecological niches.
Strategies in utilisation of HMOs by infant-type HRB
Infant-type HRB species have evolved two different strategies to degrade HMOs (LoCascio et al. 2009; Garrido, Barile and Mills 2012; Katayama 2016). The first strategy is oligosaccharide transporter-dependent, as observed in B. longum subsp. infantis, B. breve and most strains of B. longum subsp. longum, where a wider array of ATP-binding cassette (ABC) transporters (solute-binding proteins [SBPs]) are required for consumption of a broader set of HMOs; while the second is extracellular glycosidase-dependent, as observed in B. bifidum and certain strains of B. longum subsp. longum that contain the gene encoding lacto-N-biosidase (LnbX), where fewer transporters but cell wall-associated glycosyl hydrolases (GHs) are required for HMO utilisation. In the former case, intact HMOs are directly imported into the cells by ABC transporters where they are then hydrolysed intracellularly for consumption. In the latter case, HMOs are hydrolysed outside the cells by cell wall-associated GHs to liberate mono- and/or disaccharides. These liberated carbohydrates are subsequently catabolised inside the cells after their internalisation. This extracellular glycosidase-dependent strategy could then lead to cross-feeding of HMO degradants within Bifidobacterium community whereby the extracellularly liberated mono- and disaccharides may be consumed by other bifidobacterial species that are not able to consume the more complex HMOs (Gotoh et al. 2018). This has been observed in the case of sialyllactose-mediated cross-feeding between B. bifidum and B. breve (Egan et al. 2014b; Nishiyama et al. 2018). In these cases, B. breve, which is a non-sialyllactose consumer, can grow on the residual sialic acid that is liberated from sialyllactose by the extracellular sialidase of B. bifidum. Noteworthy, given that the abundance of extracellular glycosidase-dependent B. bifidum and LnbX-positive B. longum subsp. longum is generally lower in the infant gut microbiota (Matsuki et al. 2016; Yamada et al. 2017), it is suggested that most of the infant-type HRB employ a transporter-dependent strategy for HMO assimilation.
Comparative genomic analysis has exemplified the functional capabilities of infant-type HRB to assimilate HMOs. Infant-type HRB, including B. breve, B. bifidum, B. longum subsp. longum, and B. longum subsp. infantis, are equipped with sets of HMO-related genes, and such genetic elements are hardly detected in the genomes of the majority species of adult-type HRB and the non-HRB (Garrido et al. 2015; Odamaki et al. 2015; James et al. 2016, 2019; Sakanaka et al. 2020). The conservation and prevalence of the HMO utilisation genes were remarkably variable among infant-type HRB species. Infant-type HRB species employ different HMO metabolic pathways with different degrees of degradation and size limitations. It appears that HMO-related genes are highly conserved in the genome of B. longum subsp. infantis for which it contains an army of intracellular GHs and ABC transporters that are necessary for HMO utilisation (Sela et al. 2008; LoCascio et al. 2010). Notably, B. longum subsp. infantis internalises particular, intact small-mass HMOs, relying on the ABC transporters with defined specificity for individual HMO families (Garrido et al. 2011). In contrast, B. bifidum relies on a set of diverse cell wall-associated extracellular GHs including lacto-N-biosidase (LnbB) and 1,2-α-L-fucosidase that exhibit similar enzymatic affinities for HMOs compared to the intracellular enzymes from B. longum subsp. infantis (Wada et al. 2008; Ashida et al. 2009; Turroni et al. 2010; Kitaoka 2012; Sela et al. 2012). All genes required for extracellular HMO degradation are highly conserved across the species of B. bifidum (Gotoh et al. 2018; Sakanaka et al. 2020).
On the other hand, the species of B. breve shows high conservation of genes encoding for the SBP of ABC transporters (LNnT-BP (NahS) and GNB/LNB-BP (GltA)) and intracellular enzymes (GH 42 LNT β-1,3-galactosidase, GH 20 β-N-acetylglucosaminidase, GH 2 β-1,4-galactosidase and GH 112 GNB/LNB phosphorylase) that are necessary for the degradation of LNT, LNnT and LNB (James et al. 2016; Sakanaka et al. 2020). It was found that, although this species commonly possesses an intracellular GH95 α-fucosidase, the prevalence of the fucosyllactose transporters (FL1-BP and FL2-BP) is remarkably low (Sakanaka et al. 2019), and thus exhibit strain-dependent differences in the capability to utilise fucosylated HMO. Indeed, only a few strains of B. breve have been shown to contain GH29 α-fucosidase and the ABC transporter system and are capable of assimilating 2’-FL and larger fucosylated HMOs such as LNFP I/II (Ruiz-Moyano et al. 2013; Matsuki et al. 2016). Moreover, it has been described that the strain B. breve UCC2003 utilises LNT and LNnT through two different mechanisms, which resemble the degradation pathway of B. longum subsp. infantis (James et al. 2016). B. breve UCC2003 was also shown to possess functional pathways to internalise and utilise L-fucose (James et al. 2019).
The strains of B. longum subsp. longum who are recognised as LNT/LNB consumers possess the SBP of ABC transporter (GNB/LNB-BP (GltA)) as well as LNT- and LNB-degrading intracellular enzymes including GH42 LNT β-1,3-galactosidase, GH20 β-N-acetylglucosaminidase, GH2 β-1,4-galactosidase and GH 112 GNB/LNB phosphorylase (Suzuki et al. 2008; Garrido et al. 2016; Sakanaka et al. 2020). Moreover, certain strains of B. longum subsp. longum also contain extracellular GH136 lacto-N-biosidase (LnbX) enzyme in its genome (Sakurama et al. 2013). Noteworthy, strains containing LnbX mainly degrade LNT to LNB and lactose, and subsequently import LNB with the SBP of ABC transporters (GNB/LNB-BP (GltA)) and use GNB/LNB phosphorylase (LnpA) for intracellular phosphorolysis, whereas LnbX-negative strains hydrolyse LNT inside the cell with the enzymes LNT β-1,3-galactosidase, β-N-acetylglucosaminidase, and β-1,4-galactosidase (Sakurama et al. 2013; Odamaki et al. 2015). Recent studies have also found that specific strains of B. longum subsp. longum are able to utilise 2’-FL and 3-FL as the unique carbon sources (Garrido et al. 2016; Arboleya et al. 2018). These strains were found to possess a novel gene cluster devoted to the utilisation of fucosylated HMOs, including genes encoding an α-fucosidase enzyme, the ABC transporter system that is responsible for import of fucosylated molecules, and components for fucose metabolism (Garrido et al. 2016; Arboleya et al. 2018).
Interestingly, a recent report has highlighted that some strains of HRB could employ a unique adaptation strategy to assimilate fucosylated HMOs (Sakanaka et al. 2019). The study identified two functionally distinct but overlapping fucosyllactose transporters (FL transporter-1 and -2) that are highly conserved in B. longum subsp. infantis. FL transporter-2 is capable of taking up 2’-FL, 3-FL, LDFT, and LNFP I, whereas FL transporter-1 can import only the former two molecules. The distinct specificities between FL transporter-1 and -2 (the homologous ABC transporters) may be determined by the partially different FL-binding sites of the corresponding SBPs (Sakanaka et al. 2019). The homologs of FL transporter-1 or -2 are also sporadically distributed among the infant-type HRB species (B. longum subsp. longum and B. breve) as well as a few strains of B. pseudocatenulatum, B. kashiwanohense and B. longum subsp. suis (James et al. 2019). It appears that the different capabilities of bifidobacteria to assimilate the fucosylated HMOs are mostly attributed to conservation profiles of FL transporter-1 and/or -2 in bifidobacteria, emphasising the pivotal role of these transporters in dictating the preference of HMO uptake (Garrido et al. 2016; Matsuki et al. 2016; Sakanaka et al. 2019). Furthermore, the SBP homolog gene of FL transporter-2 was found to be positively and actively associated with the abundance of Bifidobacterium in the gut of breast-fed infants, suggesting the FL transporter-2 may function as a pivotal fitness factor involved in the dominance and adaptation of bifidobacterial species, particularly infant-type HRB, in the infant gut microbiota (Sakanaka et al. 2019). Another recent report also reinforced the importance of fucosylated HMO metabolism for the successful bifidobacterial establishment in the breast-fed infant gut (James et al. 2019). The study revealed that the four gene homologs (FumFDCE) involved in fucose/fucosylated HMO utilisation pathway are distributed in Bifidobacterium strains typically found in infants, including B. breve and B. longum subsp. infantis (James et al. 2019). Collectively, these findings provide intriguing insights into the regulatory networks behind HMO utilisation. It is implicated that infant-type HRB has undergone specific genetic adaptation to and strict co-evolution with the infant host. In addition to the implications to host-bacterial coevolution, this wide array of responses to HMOs may impact probiotics design and applications, where the residential origin and strain-level differences in substrate utilisation require additional considerations.
Tolerance to lysozyme
A recent study has demonstrated that bifidobacterial species of different residential origins exhibit differential growth characteristics in human milk (Minami et al. 2016). The ability to grow in human milk was found to be highly conserved in infant-type HRB strains of B. longum subsp. infantis and B. breve, while the growth characteristics of B. longum subsp. longum and B. bifidum were shown to be strain-dependent; some strains grew and some retained their inoculated cell numbers. In particular, the tested strains of adult-type HRB and non-HRB were unable to grow in human milk and had reduced viable cell numbers after incubation. Given the fact that human milk also contains an essential amount of lactose (approximately 7% by weight) (Ballard and Morrow 2013), a carbohydrate source that can be assimilated by bifidobacteria, it seems likely and has indeed been shown to be true that nutrient deficiency may not be the reason for the repressed growth and that the presence of lysozyme in human milk is related to the inability of the strains of adult-type HRB and non-HRB to grow in human milk (Minami et al. 2016).
Lysozyme is a ubiquitous antibacterial enzyme that is present in large quantities in human milk (up to 400 µg/mL), which is approximately 1000–3000-fold higher concentration than the bovine milk (Prieur 1986). In addition to human milk, lysozyme is naturally present in almost all tissues and biological fluids, such as tears, saliva, sweat, and mucus (Field 2005). It is therefore plausible that lysozyme susceptibility might represent a threat for bifidobacteria to survive in the human body. Studies have shown that bifidobacterial species that are the natural inhabitants of human intestines (HRB) are more resistant to lysozyme compared to those that are commonly encountered in the animal intestines (non-HRB) (Rada et al. 2010; Rockova et al. 2012; Minami et al. 2016). More specifically, infant-type HRB strains were tolerant to high concentrations of lysozyme, and lysozyme tolerance was positively correlated with their growth in human milk. In contrast, the tested strains of adult-type HRB displayed intermediate tolerance, while non-HRB were relatively sensitive to lysozyme, which would, in turn, led to their failure to grow in human milk (Minami et al. 2016). One study examining the tolerance mechanisms of HRB strains against lysozyme has revealed that differences of lysozyme susceptibility between HRB and non-HRB are independent of the peptidoglycan-degrading property of lysozyme (Sakurai et al. 2017). Instead, the tolerance to lysozyme among some HRB strains is due to their resistance to the cationic properties of lysozyme that are associated with the cytotoxic activity (Sakurai et al. 2017). This finding provides a fundamental insight into how HRB are protected against lysozyme action, which would, in turn, contribute to their colonisation in the human host. Further research is needed to elucidate the specific mechanisms in HRB against lysozyme hydrolysis. Altogether, these findings suggest that the presence of lysozyme in human milk could act as another crucial selective factor that prevents the colonisation of non-HRB in the guts of breast-fed infants.
Bifidobacterial human milk compatibility and colonisation of infants
As aforementioned, bifidobacteria of different residential origins display genotypic and phenotypic variations in their compatibility with human milk. Importantly, not all bifidobacteria are equally adapted to consume HMOs and tolerant to human milk lysozyme, which suggests that certain strains of bifidobacteria (infant-type HRB) are better able to colonise a breast-fed infant. This concept has been addressed in a clinical study investigating the gut microbial composition of premature infants — infants who often have a delayed bifidobacterial colonisation in comparison to term infants (Underwood et al. 2013). Premature infants receiving milk were given the probiotic B. longum subsp. infantis and B. animalis subsp. lactis. The results demonstrated that B. longum subsp. infantis, a member of infant-type HRB, colonised better than B. animalis subsp. lactis, a non-HRB strain, in premature infants exposed to both formula and human milk feeding. This study suggests a close linkage between HMOs and human milk lysozyme (in this case, as part of whole human milk) and colonisation of the gut, specific to certain strains of (infant-type) HRB but not non-HRB (Underwood et al. 2013; Minami et al. 2016; Lawson et al. 2019).
Furthermore, HRB species (B. longum subsp. longum, B. breve and B. bifidum) but not the non-HRB species (B. animalis) were more frequently detected in the gut microbiota of breast-fed infants (Tannock et al. 2013). In contrast, non-HRB species were more abundant in formula-fed infants than in breast-fed infants, indicating the presence of HMOs and lysozyme in the human milk, but not the infant formulas, contributed to the different abundancy of bifidobacterial species in infant gut microbiota (Tannock et al. 2013; Minami et al. 2016). Another study evaluating the early gut microbial composition of healthy neonates also revealed that the colonisation of the non-HRB species, B. animalis subsp. lactis, was dictated by the type of feeding (Martin et al. 2016). Noteworthy, B. animalis subsp. lactis was more frequently detected in the gut microbiota of formula-fed infants and its colonisation was relatively weak in breast-fed infants, suggesting that B. animalis subsp. lactis might not be a common member of the infant gut microbiota. In contrast, the species of infant-type HRB such as B. breve or B. longum subsp. infantis were confirmed as early infant gut colonisers and their colonisation was not affected by the mode of delivery and type of feeding (Martin et al. 2016). Taken together, these data clearly demonstrated the superiority of infant-type HRB – bifidobacterial species of high compatibility with human milk – to colonise the infant gut, highlighting that certain infant-type HRB strains are more beneficial and could be a better probiotic candidate for human use, especially in infants. In this regard, there must be a good reason why human milk naturally selected the species of HRB, particularly the infant-type HRB, for the developing infants. It is implicated that HRB are better adapted to the intestinal environment of the human host and could contribute to better human health.
PHYSIOLOGICAL FUNCTIONS OF HUMAN-RESIDENTIAL BIFIDOBACTERIA (HRB)
Carbohydrate metabolism
Comparative genomic studies of bifidobacterial species have uncovered specific genetic strategies of the members of bifidobacteria to establish and persevere in the human gastrointestinal tract (Milani et al. 2014, 2015a; Odamaki et al. 2015). It appears that currently known bifidobacterial taxa have undergone a substantial number of gene acquisition events during the evolutionary process where such acquired genes – a specific genetic repertoire that encodes glycosyl hydrolases (GHs) – are related to carbohydrate metabolism. After that, adaptation to habitats rich in complex carbohydrates, as such in the human gut, is seen as the main driving force responsible for speciation among members of the genus Bifidobacterium (Milani et al. 2016).
Even though bifidobacteria are not a dominant member of the adult gut microbiota, their functional biological roles in the metabolism of dietary and host-derived glycans cannot be neglected. Bifidobacteria display saccharolytic behaviour and their ability to colonise and survive in the gastrointestinal tract is largely dependent on their carbohydrate metabolic capabilities (Milani et al. 2016). According to the Carbohydrate Active Enzymes (CAZy) classification, the pan-genome of the Bifidobacterium genus is among the largest predicted glycobiomes of the known gut commensals, with a large proportion of annotated genes encode an enzymatic arsenal involved in carbohydrate metabolism, including GHs, glycosyltransferases, and carbohydrate esterases (Milani et al. 2015a). The bifidobacterial glycobiome is enriched in enzymes belonging to the GH13 family, which are the typical enzymes for degradation of a wide range of complex carbohydrates, such as starch, glycogen, and related substrates (e.g. amylose, amylopectin, pullulan, maltodextrin and cyclomaltodextrin), as well as palatinose, stachyose, raffinose and melibiose (Milani et al. 2015a). Notably, all these sugars are dominant glycans found in the adult mammalian diet. Moreover, part of the predicted glycobiome of bifidobacteria is extracellular, allowing them to degrade carbohydrates whose direct uptake is not possible due to the size and complexity of the glycan.
Intriguingly, it appears that the occurrence of HRB in the human gut is supported by their metabolic abilities about various complex, host-indigestible carbohydrates either (in)directly derived from the host (i.e. mucin and HMOs) or the diet (Milani et al. 2014, 2015a, 2016). Analyses of the genome sequences of the various type strains representing HRB and non-HRB revealed an abundant presence of genes related to plant-derived carbohydrate metabolism in the genomes of HRB, especially the adult-type HRB, B. adolescentis and B. longum subsp. longum. The genome of B. adolescentis is particularly enriched with a much larger set of GH 13 enzymes, which include amylase, pullulanase, and cyclomaltodextrinase, than other HRB species, exemplifying their capabilities to metabolise various dietary starch and starch-like oligo/polysaccharides such as amylopectin, pullulan, maltotriose, and maltodextrin (Duranti et al. 2014, 2016). Such genetic architecture may reflect an adaptation of B. adolescentis species to the adult diet, which often contains high amounts of plant-derived glycans, and thus highlighting the superiority of this HRB species to persist in the gastrointestinal tract of adult human beings. Moreover, the analyses of fermentation profiles of B. adolescentis strains have further exemplified their preference for the utilisation of different sugars (e.g. galactose, mannose and glucose), as well as plant-derived carbohydrates that are typically present in the human diet, such as starch (Duranti et al. 2014).
Another important sign of bifidobacterial adaptation to the human gut is the ability of the HRB species, B. longum subsp. longum, to utilise a wide variety of plant-derived dietary carbohydrates. B. longum subsp. longum was shown to be genetically well equipped with a substantial number of GHs (e.g. xylanase, arabinosidase, galactosidase, neopullanase isomaltase, inulinase, glucosidase, hexosaminidase, and mannosidase), oligosaccharide transporters and proteins with a cell-surface anchor motif, reflecting the genetic strategies employed by this species to survive and compete in its ecological niche (Schell et al. 2002; Odamaki et al. 2015; Fujita et al. 2019). Noteworthy, a large number of GH 43 and 51 family members, which are enzymes responsible for the breakdown of arabinofuranoside and xylan, were found to be specific to B. longum subsp. longum as well as B. adolescentis and other adult-type HRB (Odamaki et al. 2015; Duranti et al. 2016; Komeno et al. 2019). The presence of such genetic elements for utilisation of plant-derived carbohydrates, which are assumed to be not introduced into infant gut before weaning, provides an advantage for this species to colonise in the human intestine. It appears that B. longum subsp. longum might play a key role in infant's digestion during weaning where a non-milk diet containing complex carbohydrates are being introduced for the first time. Consequently, such genetic architecture may explain why only B. longum subsp. longum predominantly inhabit in both the infant and adult intestines (Pacheco et al. 2015; Odamaki et al. 2018).
Moreover, B. breve is another HRB species that deserves a special mention as it appears to possess the ability to metabolise a wide range of α/β-glucose- and α/β-galactose-containing carbohydrates, corroborating its niche-specific adaptation to both infant and adult guts (Pokusaeva, Fitzgerald and van Sinderen 2011; Bottacini et al. 2018). Specific strains of B. breve encode various carbohydrate-modifying enzymes including α-glucosidases (belonging to the GH13 and GH31 families) that are involved in hydrolysis of α-glucosidic linkages usually present in di-, oligo- and polysaccharides (e.g. maltose, starch and related α-glycans) (Pokusaeva et al. 2009; Kelly et al. 2016), β-glucosidases (belonging to the GH1 and GH3 families) that are involved in assimilation of a variety of glycan substrates such as cellobiose and cellodextrin (Pokusaeva et al. 2011; Bottacini et al. 2018) and β-galactosidases (belonging to the GH2 and GH42 families) that are involved in degradation of lactose, galactan and galacto-oligossacharides (O'Connell Motherway, Fitzgerald and van Sinderen 2011; O'Connell Motherway et al. 2013). It has been found that certain B. breve strains were able to utilise starch, pullulan and amylopectin (Ryan, Fitzgerald and van Sinderen 2006). In addition, the strain B. breve UCC2003 is reported to encode various carbohydrate-modifying enzymes including amylopullulanase, endogalactanase, β-fructofuranosidase, β-1,4-glucosidase, α-1,6-glucosidases, and ribokinase) that allow it to metabolise various glycans such as sucrose, fructose, starch, amylopectin, panose, cellobiose, and ribose (Ryan, Fitzgerald and van Sinderen 2005; Motherway et al. 2008; O'Connell Motherway et al. 2009; Pokusaeva et al. 2010; O'Connell Motherway, Fitzgerald and van Sinderen 2011; Pokusaeva, Fitzgerald and van Sinderen 2011; Kelly et al. 2016). Given that B. breve is particularly abundant in early life where starch-containing foods, fruits and cereals or vegetables are among the first digestible dietary carbohydrates introduced at weaning (Stephen et al. 2012), the presence of such genetic elements for hydrolysis of glycosidic linkages may aid in their colonisation and persistence in the (infant) gut. Extensive research on carbohydrate utilisation in B. breve using a combination of comparative genomics, gene-trait matching (genotype-phenotype association) and functional genomics have also revealed that bifidobacterial mutualism and carbohydrate syntrophy occurs in the infant's gut wherein the species of B. breve may, perhaps, co-operatively, cross-feed with other (bifido)bacterial species like B. bifidum or B. longum subsp. longum in order to sustain growth on the various plant-derived carbohydrates (Pokusaeva, Fitzgerald and van Sinderen 2011; Egan et al. 2014b; Bottacini et al. 2018).
Furthermore, one study examining the metabolic capabilities of probiotic strains of bifidobacteria and lactic acid bacteria has revealed the superiority of HRB strains to assimilate galactooligosaccharide (GOS), a group of prebiotic compounds that are widely used in infant nutrition to stimulate growth of beneficial gut bacteria (Böger et al. 2019). HRB strains tested (B. adolescentis DSM 20 083, B. breve DSM 20 091, B. bifidum DSM 20 456, B. longum subsp. infantis DSM 20 088) grew well and were more efficient for GOS utilisation than non-HRB strains (B. lactis W51 and W52), with a distinct degree of polymerisation and different glycosidic linkage compositions. Noteworthy, most of the branched GOS were only consumed by HRB strains of B. breve, B. adolescentis, and B. longum subsp. infantis (Böger et al. 2019). This study suggests that HRB strains particularly B. breve and B. longum subsp. infantis are most suitable for symbiotic mixtures with GOS.
On the other hand, several species of non-HRB (B. animalis subsp. lactis, B. pseudolongum subsp. pseudolongum, B. pseudolongum subsp. globusum, and B. thermophilum) were found lacking the sets of genes encoding enzymes involved in the metabolism of arabinoxylan oligosaccharides (Rivière et al. 2014). Notably, the species of B. animalis subsp. lactis –a common probiotic agent used in dairy products and dietary supplements – can only metabolise a very minimal number of carbohydrates (Milani et al. 2013, 2015a). It is therefore plausible that the genetic architecture of non-HRB for plant-derived carbohydrates metabolism is associated with their corresponding hosts, revealing a co-evolution host–microbe profile. Nonetheless, such genetic elements may have undergone massive genome decay as a result of its industrial exploitation, which has involved long-term cultivation of B. animalis subsp. lactis on synthetic media (Milani et al. 2013).
HRB species have also been shown to be capable of utilising mucin as a nutrient source in order to colonise and survive within the host intestinal lining (Kiyohara et al. 2012; Katayama 2016). Mucins are host-derived glycans, secreted by intestinal goblet cells that essentially coat the surface of the intestinal mucosa. The main monosaccharides components are N-acetylglucosamine, N-acetylgalactosamine, and galactose, and these glycoproteins are decorated with fucose, sialic acid, and sulphate groups (Tailford et al. 2015). It has been reported that most strains of B. longum subsp. longum and B. bifidum are equipped with the gene homologs encoding endo-α-N-acetylgalactosaminidase and are thus capable of hydrolysing the core structures of mucin-type oligosaccharides, Galβ1–3GalNAcα-O-Ser/Thr (or T-antigen) to Galβ1–3GalNAc (Fujita et al. 2005). Also, infant-type HRB strains of B. longum subsp. infantis, B. longum subsp. longum, and B. breve have also been shown to possess the gene homolog encoding α-N-acetylgalactosaminidase (NagBb) that hydrolyses GalNAcα-O-Ser/Thr (Tn antigen) from the core-3-mucin-type O-glycans (GlcNAcβ1–3GalNAcα-O-Ser/Thr) (Odamaki et al. 2015). In particular, the strain B. breve UCC2003, which is a versatile bacterium from a metabolic perspective, was also shown to contain sulfatase-encoding gene clusters that allow it to metabolise N-acetylglucosamine-6-sulfate (GlcNAc-6-S), but apparently not on GlcNAc, as a sole carbon source (Pokusaeva, Fitzgerald and van Sinderen 2011; Egan et al. 2016). Interestingly, it is reported that B. breve UCC2003 produces an intracellular sulfatase enzymes and appears to employs a cross-feeding strategy with other members of the gut microbiota in order to gain access to mucin-derived sulphated monosaccharides (Egan et al. 2014a, 2016). Nonetheless, non-HRB strains of B. animalis were found lacking these genetic elements. These findings reflect the fitness and competitiveness of HRB strains, particularly the infant-type HRB, to survive and persist in the human intestine, where the glycans produced by the host serve as a carbon source for these bifidobacterial species.
Furthermore, these genetic data were substantiated by the analysis of mucin degradation of infant-type HRB, which highlighted the superior capability of the strains of B. bifidum D119 and L22, B. breve NCIMB8807 and B. longum subsp. longum NCIMB8809 to utilise mucin as a carbon source and grow well in mucin-containing defined medium (Ruas-Madiedo et al. 2008). In contrast, B. animalis and B. pseudocatenulatum strains, which are lacking mucin-utilisation-related genes (afcA and engBF glycosidase genes), were not able to utilise mucin and had minimal growth (Ruas-Madiedo et al. 2008). It is therefore implicated that HRB strains, particularly infant-type, are more competent to assimilate mucin, survive, and persist in the human gastrointestinal tract.
These findings suggest that HRB possess adaptive advantages and competitiveness to colonise in the human gut over the non-HRB that are less specialised with the human gut environment. Although the carbohydrates assimilation capability of bifidobacteria is more likely to be dependent on strain-specificity, the driving force of residential origin cannot be neglected. For instance, the HRB strains that are capable of utilising non-digestible glycans would aid in human digestion of glycans with valuable nutrient values which would otherwise be lost from the body and unused as waste to the outside environment.
Production of folate
In addition to carbohydrate metabolisms and adaptation to a wide variety of carbohydrates, bifidobacterial species that are naturally residing in the human gut (HRB) also possess interesting feature concerning folate biosynthetic capabilities. Folate, also known as vitamin B9 or pteroyl-L-glutamate, is water-soluble vitamin that plays a role as a cofactor. It is required for efficient DNA replication, repair and methylation, and synthesis of nucleotides, vitamins and certain amino acids (Jacob 2000; Lucock 2000). For this key role in the cell cycle, various fast proliferating cells such as leukocytes, erythrocytes, and enterocytes require high levels of folate for growth (Jacob 2000).
Humans are auxotroph for folate; they cannot synthesise on their own and must obtain it exogenously through the diet (e.g. leafy green vegetables, yeast extracts, liver, beans) or indigenous folate-synthesising bacteria (Strozzi and Mogna 2008; Aufreiter et al. 2009). Folate deficiency has been associated with malformation of the neural tube during embryonic development and increased risk of megaloblastic anaemia, certain forms of cancer, and cardiovascular diseases (Rayburn, Stanley and Garrett 1996; Choi and Mason 2000; Stanger 2002). Moreover, this vitamin is particularly important for some population groups, including the elderly, children, and pregnant women.
The folate molecule contains a pterin moiety originating from 6-hydroxymethyl-7,8-dihydropterin pyrophosphate (DHPPP), bound to a unit of para-aminobenzoic acid (pABA). The de novo biosynthesis process of folate necessitates both DHPPP and pABA, for which the synthesis of these precursors requires the three basic building blocks: guanosine triphosphate (GTP), chorismate, and glutamate. DHPPP is formed from GTP in four consecutive steps, while pABA can be produced from chorismate in the shikimate pathway (Rossi, Amaretti and Raimondi 2011; Andlid, D'Aimmo and Jastrebova 2018). Subsequently, the DHPPP and pABA are joined together with the formation of a C-N bond, catalysed by the enzyme dihydropteroate synthase (DHPS, EC 2.5.1.15) to form 7,8-dihydropteroate (DHP). Further, DHP is reduced to the biologically active cofactor tetrahydrofolate (THF) (Rossi, Amaretti and Raimondi 2011; Andlid, D'Aimmo and Jastrebova 2018).
Bifidobacteria have been shown to possess the folate synthesising machinery, albeit not all species have the complete enzymatic armoury (Rossi, Amaretti and Raimondi 2011; LeBlanc et al. 2013). Comparative genomic analysis has provided intriguing insights into the genes and corresponding enzymes presumptively involved in bifidobacterial folate biosynthesis. Based on the available genome information, all sequenced bifidobacterial species have thus far shown to harbour the required genes for the shikimate pathway and thus can produce chorismate (Barrangou et al. 2009; Ventura et al. 2009; Turroni et al. 2010). However, not all species are capable of converting chorismate into pABA. Although all species harbour pabA gene encoding ADCS enzyme, only the HRB species of B. adolescentis and B. dentium possess the pabC gene encoding for ADCL enzyme and can, therefore, synthesise pABA de novo (Rossi, Amaretti and Raimondi 2011; Andlid, D'Aimmo and Jastrebova 2018). The other species would require pABA supplementation in order to accomplish folate production.
Moreover, all sequenced HRB species (B. adolescentis, B. dentium, and B. longum subsp. longum) harbour a cluster of fol genes for DHPPP biosynthesis and are predicted to carry out the condensation of DHPPP and pABA (Rossi, Amaretti and Raimondi 2011). The non-HRB species of B. animalis subsp. lactis were, however, found missing the key genes (folE and folBK) needed for DHPPP biosynthesis and the folP gene encoding DHPS, enzyme involves in the condensation of DHPPP and pABA (Rossi, Amaretti and Raimondi 2011; LeBlanc et al. 2013; Milani et al. 2014), and are therefore auxotroph for folates even if pABA is present in the environment. Collectively, it is conceivable that the HRB species of B. adolescentis and B. dentium can produce folate de novo, B. longum subsp. longum requires pABA supplementation, and the non-HRB species of B. animalis requires folate supplementation.
Furthermore, many studies have assessed the capabilities of bifidobacterial species to produce folate and their possible contribution to the folate intake of the host (Pompei et al. 2007a; D'Aimmo et al. 2012, 2014; Sugahara et al. 2015). It appears that the mere ability to synthesise folate is restricted to certain species/strains of bifidobacteria, and to a large extent, their capabilities are also highly associated with the residential origins, leading to distinct differences between HRB and non-HRB in folate production (Sugahara et al. 2015). For instance, while most HRB strains were capable of producing high levels of folate, non-HRB strains produce the least amount or nothing (autotroph) (Sugahara et al. 2015). Interestingly, the folate production capacity in bifidobacteria is reported to be correlated with the phylogenetic lineage. Several HRB species isolated from non-human primates (B. adolescentis and B. dentium) were capable of producing a high level of total folate, whereas the non-HRB species isolated from non-primates (B. animalis subsp. animalis, B. animalis subsp. lactis, B. pseudolongum subsp. globosum, B. asteroides, B. coryneforme, and B. indicum) were incapable (D'Aimmo et al. 2014). It has become evident that folate production is characteristic of HRB but not of non-HRB. Adaptations to habitats that frequently contain external folate, mainly the animal intestines, have presumably led to reduced selection pressure for de novo folate biosynthesis and are most likely rendering many strains and possibly whole species of non-HRB to have lost their ability to synthesise folate (Andlid, D'Aimmo and Jastrebova 2018).
Some in vivo studies further confirmed the capabilities of HRB to produce high folate levels and to act as an endogenous source of this vitamin by elucidating its release inside the intestines of the murine model and human subject (Pompei et al. 2007b; Strozzi and Mogna 2008; Sugahara et al. 2015). In an in vivo study using germ-free mice administered with a single strain of bifidobacteria, the levels of folate were found to be higher in mice administered with the HRB strain of B. longum subsp. longum than the non-HRB strain of B. animalis subsp. lactis. This was accompanied by a significant improvement in the haematological indicators related to anaemia in germ-free mice colonised with HRB strain (Sugahara et al. 2015). The study suggests that folate-producing HRB strain could be more beneficial than those of the non-HRB strain in protecting the human host against folate deficiency health complications such as anaemia. Furthermore, in a human pilot study, daily consumption of the HRB strains (B. adolescentis DSMZ 18 352, B. adolescentis DSMZ 18 350, and B. pseudocatenulatum DSMZ 18 353) for 30 days resulted in a significant higher folate content in the intestinal lumen, indicating these HRB strains are capable of producing folate and providing an endogenous source of this vitamin to the human host (Strozzi and Mogna 2008).
The species of HRB have thus far been shown to harbour a functioning folate biosynthesis machinery and exhibit a greater capacity for folate production than the non-HRB. It seems possible that HRB species have been favoured during the evolution with the human host wherein they can both colonise the human gut and produce folate (symbiotic coevolution). Whether this is true or not is unknown but the findings show a great promise for high-folate producing HRB strains supplementation in improving human folate status. Folate-producing HRB strains, either incorporated in functional foods or as trophic probiotics, would help in intestinal homeostasis of vitamins and confer beneficial effects on human health.
Degradation of food-derived opioid peptides
Food-derived opioid peptides are substances generated from enzymatic hydrolysis of dietary proteins, including milk, vegetable, cereal, and meat/poultry that are having morphine-like activities (Teschemacher 2003). Due to structural similarity to endogenous opioids, food-derived opioid peptides could be recognised by opioid receptors that are found primarily in the central nervous system and gastrointestinal tract, and display opioid-like molecular and physiological activities (Liu and Udenigwe 2019). The first identified and widely studied milk casein-derived opioid peptide was β-casomorphin-7 (BCM-7; Tyr-Pro-Phe-Pro-Gly-Pro-Ile) released from β-casein of bovine or human milk during technological processes and/or enzymatic digestion in the intestine (Brantl et al. 1981). Studies have suggested that BCM-7 may pass through human intestinal barriers, and could, in turn, influence on nervous, digestive and immune functions by altering the gene expression of µ-opioid receptors located on cell surfaces of these systems (Liu and Udenigwe 2019). BCMs -4, -5, and -6, which are derived by sequential removal of three, two or one amino acid residues, respectively from the C-terminus of BCM-7, have also been demonstrated to possess opioid agonist effects (Brantl et al. 1981). Also, gluten exorphin and gliadorphin-7 (or gliadinomorphin-7), derived from the gluten found in wheat flour, rye, barley and oats, were acknowledged as substances with opioid activity (Pruimboom and De Punder 2015).
Food-derived opioid peptides have received much attention in human health. Some food-derived opioid peptides have been reported to exert beneficial health bioactivities in healthy humans (Rutherfurd-Markwick 2012; Liu and Udenigwe 2019), however, some studies implicate that they may have adverse health effects in susceptible people. Milk β-casein-derived BCM-7, for instance, has been demonstrated to be associated with many negative health outcomes, including autism, type I diabetes, sudden infant death syndrome, and atopic dermatitis (Wasilewska et al. 2011; Fiedorowicz et al. 2014; Sokolov et al. 2014; Chia et al. 2017). BCMs have been reported to be a causal trigger to apnoea in sudden infant death syndrome for which penetration of BCMs into the infant's immature central nervous system may inhibit the respiratory centre in the brainstem, thereby leading to abnormal ventilator responses, apnoea, and death (Sun et al. 2003). Several other studies provide direct evidence that BCM-7 is positively correlated with childhood mental disorders and apparent-life threatening events in infants wherein an elevated level of BCM-7 was detected in the urine of autistic children (Sokolov et al. 2014) and the sera of infants with apnoea (Wasilewska et al. 2011). In addition, exclusion of gluten from the diet can restore regular gut mucosa functions and mitigate gastrointestinal inflammation (Jianqin et al. 2015) and celiac disease (Anania et al. 2017; Brietzke et al. 2018), both of which could lead to the development of other autoimmune disorders like type I diabetes (Serena et al. 2015; Krzewska and Ben-Skowronek 2016).
There is also evidence that patients who have celiac disease, an autoimmune disorder triggered by gluten ingestion in genetically predisposed individuals, had a decreased representation of bifidobacteria species in their gut microbiota as compared to those of the healthy subjects (Golfetto et al. 2014). Several attempts have been made to supplement some bifidobacterial strains as a probiotics therapy for celiac disease (Smecuol et al. 2013; Klemenak et al. 2015; Quagliariello et al. 2016; Pinto-Sánchez et al. 2017). For instance, in a double-blind, randomised, placebo-controlled trial, administration of B. longum subsp. infantis NLS-SS to celiac disease patients for three weeks resulted in symptomatic improvement (Smecuol et al. 2013). Similarly, administration of two probiotic strains, B. breve B632 and BR03, suppressed tumour necrosis factor-alpha (TNF-α) production in celiac disease children on a gluten-free diet and such positive effect was reversed after three months of trial where probiotics supplementation had ceased (Klemenak et al. 2015). Moreover, some bifidobacterial strains (B. longum subsp. longum IATA-ES1 and B. bifidum IATA-ES2) have also been shown to degrade gliadin peptides into inactive peptides, thereby reducing the cytotoxic and inflammatory effects of gluten peptides (Laparra and Sanz 2010; Cinova et al. 2011).
Noteworthy, many of the strains exerting positive effects on gluten-mediated disorders are the commensal bacteria of the human gut and belong to HRB, albeit some non-HRB strain (B. animalis subsp. lactis) was also reported to have a positive effect (Lindfors et al. 2008). It is therefore implicated that the capability to degrade the potentially harmful food-derived opioid peptides could be a specific physiological function of HRB, explaining at least in part why HRB naturally inhabit the human gastrointestinal tract across the lifespan and how they contribute to human health. Indeed, HRB species were found to possess more potent degradative capabilities for human milk- and bovine milk-derived BCM-7 as well as wheat gluten-derived α-gliadorphin-7 (Sakurai et al. 2018). Specifically, infant-type HRB strains were reported to exert a relatively higher dipeptidyl peptidase IV — a proline-specific enzyme that can hydrolyse opioid peptides — activity than the other strains wherein certain strains of B. longum subsp. infantis and B. bifidum demonstrated the greatest degradative capabilities for all three food-derived opioid peptides. In contrast, the strains of non-HRB were inactive for the hydrolysis of food-derived opioid peptides (Sakurai et al. 2018). Taken together, these findings provide some clues on the possible role of HRB as prominent members of the human gut microbiota. It is implicated that HRB species could aid in the degradation of the potentially harmful food-derived opioid peptides, particularly the strains of infant-type HRB as of which these peptides are more potent to infants with immature gastrointestinal system, thereby contributing to host health.
Production of aromatic lactic acids
Another vital feature of HRB species is their capability to produce tryptophan-derived indoles. Gut bacterial tryptophan metabolites have increasingly been recognised as an essential mediator in host physiology and may contribute to intestinal and systemic homeostasis (Tremaroli and Bäckhed 2012; Agus, Planchais and Sokol 2018). For instance, microbial tryptophan-derived indoles, including indole-3-acetic acid (IAA), indole-3-aldehyde (IAld), indole-3-lactic acid (ILA) and indole-3-propionic acid (IPA), have been reported to act as ligands of aryl hydrocarbon receptor (AhR) and may potentially improve gut barrier function and regulate gut mucosal immune responses (Hubbard et al. 2015; Cervantes-Barragan et al. 2017; Dodd et al. 2017; Wilck et al. 2017). ILA and IPA have also been identified as antioxidants and as free-radical scavengers (Chyan et al. 1999; Karbownik et al. 2006; Suzuki et al. 2013). In this regard, it seems likely that the production of tryptophan-derived indoles could be the precise mechanism deploys by HRB species in host-microbial interactions, contributing to their beneficial effects on human health.
This concept has been demonstrated in a recent study investigating the anti-inflammatory effect of ILA — a breastmilk tryptophan metabolite secreted by B. longum subsp. infantis — in the immature intestine (Meng et al. 2020). The study found that this tryptophan metabolite produced by an infant-type HRB strain B. longum subsp. infantis ATCC 15 697 interacts with AhR and reduces the inflammatory interleukin-8 (IL-8) response in immature but not mature intestinal enterocytes (Meng et al. 2020). Studies have shown that ILA is involved in host immune homeostasis via AhR activation, leading to reprogramming of immunoregulatory T cells and inflammatory T cells (Cervantes-Barragan et al. 2017; Lanz et al. 2017; Wilck et al. 2017). Such function would be beneficial for healthy growth, including the immune development and maturation, in infants.
Interestingly, in another recent report, ILA was found as the only tryptophan metabolite produced by bifidobacterial species; no others, including IPA, IAA, and IAld, were produced (Sakurai, Odamaki and Xiao 2019). Noteworthy, the species of infant-type HRB displayed the highest capacity for ILA production among which the strains of B. longum subsp. longum, B. longum subsp. infantis, B. breve, and B. bifidum, produced higher levels of ILA than other strains (Sakurai, Odamaki and Xiao 2019). Numerous studies have also demonstrated that infant-type HRB are particularly superior in producing ILA (Aragozzini et al. 1979; Russell et al. 2013; Ehrlich et al. 2018). Nevertheless, the exact biological role of ILA; how and why only limited species (infant-type HRB) are allowed to harbour in the human infant gut, remains unclear. Very recently, in a preprints, Laursen et al. 2020 demonstrated that aromatic lactate dehydrogenase, an enzyme which was explicitly found in breastmilk-promoted Bifidobacterium species (infant-type HRB), is involved in the formation of aromatic lactic acid metabolites including ILA from aromatic amino acids. They also showed that stool concentrations of aromatic lactic acids are determined by the abundance of breast milk-promoted Bifidobacterium species harbouring the aromatic lactate dehydrogenase enzyme. ILA was pointed as a relevant early life AhR agonist and could be associated with intestinal and systemic homeostasis. ILA has been reported to have antimicrobial activity (Shigeno et al. 2018) in addition to hydrogen peroxide production as a by-product during tryptophan deamination (Dieuleveux, Lemarinier and Gueguen 1998), suggesting ILA production by infant-type HRB may contribute to their preponderance and competitiveness in the infant intestines. It is therefore implicated that bifidobacteria-derived ILA may have conserved roles in human health development. Taken together, these findings shed lights on the mechanism by which HRB, especially the infant-type, exert their adaptive abilities and beneficial properties in the human gut.
CONCLUDING REMARKS
Through more than a century after its discovery, evidence has indicated that bifidobacteria have coevolved with their respective hosts. Conversely, bifidobacterial species that naturally occur in the human gut, better known as HRB, have adapted to the human host and possess many unique physiological characteristics. This organism is seen as an essential member of the human gut microbiota across the lifespan, and is associated with human health. The acquisition of HRB species commences from birth, for which the bacteria are vertically transmitted from the mother's vaginal tract, gastrointestinal tract, breast milk, placenta and amniotic fluid to the infant. The studies, to date, have revealed that the persistence of infant-type HRB in the infant gut is attributed to their competitive abilities with regards to human milk (i.e. their ability to metabolise HMOs and tolerance to lysozyme). Noteworthy, infant-type HRB are naturally selected by human milk; they are genetically equipped and adapted to assimilate HMOs, explaining why the species of infant-type HRB have such a close relationship to the breast-fed infant. With a progressive reduction of breastfeeding and an increase of solid food intake, the species of HRB are then shifted from infant-type to adult-type. During adulthood, although their abundance is relatively low, HRB may still impact host overall health through their metabolic and physiological activities. In particular, adult-type HRB appear to possess a large amount of the gut-associated enzymatic arsenal dedicated to the metabolism of dietary plant polysaccharides and host-derived carbohydrates, thus enforcing the importance of HRB in facilitating the metabolism of complex carbohydrates that are not digested by human intestinal enzymes or by other gut bacteria. Besides, it has become clear that certain strains of HRB, particularly infant-type HRB, have a better functional capacity in various physiological activities, including the production of folate and ILA as well as degradation of food-derived opioid peptides. The studies discussed here shed lights on the role of HRB, particularly infant-type HRB in most of the cases, as members of human gut microbiota across the lifespan. Despite such findings, many potential physiological characteristics of HRB have yet to be explored. Furthermore, we still lack a comprehensive understanding of how HRB species interact with the human host and contribute to human health. More studies are thus needed to provide in-depth insight into the evolutionary scenario of this critical human gut commensal bacteria, which would, in turn, facilitate the selection of strains for application as human probiotics.
ACKNOWLEDGEMENTS
The authors are grateful to all the researchers whom we cited in this review for their significant and valuable research. In particular, the authors would like express their deep gratitude to Dr Takane Katayama, Kyoto University, Dr Mikiyasu Sakanaka, Technical University of Denmark and Dr Francois Bernier, Next Generation Science Institute, Morinaga Milk Industries Co., Ltd. for their critical review of this manuscript.
Contributor Information
Chyn Boon Wong, Next Generation Science Institute, Morinaga Milk Industry Co., Ltd., 5-1-83, Higashihara, Zama, Kanagawa, 252–8583 Japan.
Toshitaka Odamaki, Next Generation Science Institute, Morinaga Milk Industry Co., Ltd., 5-1-83, Higashihara, Zama, Kanagawa, 252–8583 Japan.
Jin-zhong Xiao, Next Generation Science Institute, Morinaga Milk Industry Co., Ltd., 5-1-83, Higashihara, Zama, Kanagawa, 252–8583 Japan.
Conflict of interest
The authors, CBW, TO, and J-ZX are employees of Morinaga Milk Industry Co., Ltd., which has several probiotic products marketed worldwide. This does not alter our adherence to FEMS Microbiology Review policies on sharing data and materials.
REFERENCES
- Agus A, Planchais J, Sokol H. Gut microbiota regulation of tryptophan metabolism in health and disease. Cell Host Microbe. 2018;23:716–24. [DOI] [PubMed] [Google Scholar]
- Anania C, Pacifico L, Olivero Fet al.. Cardiometabolic risk factors in children with celiac disease on a gluten-free diet. World J Clin Pediatr. 2017;6:143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andlid TA, D'Aimmo MR, Jastrebova J. Folate and Bifidobacteria. The Bifidobacteria and Related Organisms. Cambridge, Massachusetts, United States: Academic Press Inc., Elsevier, 2018, 195–212. [Google Scholar]
- Aragozzini F, Ferrari A, Pacini Net al.. Indole-3-lactic acid as a tryptophan metabolite produced by Bifidobacterium spp. Appl Environ Microbiol. 1979;38:544–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arboleya S, Bottacini F, O'Connell-Motherway Met al.. Gene-trait matching across the Bifidobacterium longum pan-genome reveals considerable diversity in carbohydrate catabolism among human infant strains. BMC Genomics. 2018;19:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arboleya S, Watkins C, Stanton Cet al.. Gut bifidobacteria populations in human health and aging. Front Microbiol. 2016;7:1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asakuma S, Hatakeyama E, Urashima Tet al.. Physiology of consumption of human milk oligosaccharides by infant gut-associated bifidobacteria. J Biol Chem. 2011;286:34583–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashida H, Miyake A, Kiyohara Met al.. Two distinct α-L-fucosidases from Bifidobacterium bifidum are essential for the utilization of fucosylated milk oligosaccharides and glycoconjugates. Glycobiology. 2009;19:1010–7. [DOI] [PubMed] [Google Scholar]
- Aufreiter S, Gregory JF III, Pfeiffer CMet al.. Folate is absorbed across the colon of adults: evidence from cecal infusion of 13C-labeled [6S]-5-formyltetrahydrofolic acid. Am J Clin Nutr. 2009;90:116–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballard O, Morrow AL. Human milk composition: nutrients and bioactive factors. Pediatr Clin. 2013;60:49–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrangou R, Briczinski EP, Traeger LLet al.. Comparison of the complete genome sequences of Bifidobacteriumanimalis subsp. lactis DSM 10140 and Bl-04. J Bacteriol. 2009;191:4144–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biavati B, Scardovi V, Moore WEC. Electrophoretic patterns of proteins in the genus Bifidobacterium and proposal of four new species. Int J Syst Evol Microbiol. 1982;32:358–73. [Google Scholar]
- Biavati B, Vescovo M, Torriani Set al.. Bifidobacteria: history, ecology, physiology and applications. Ann Microbiol. 2000;50:117–32. [Google Scholar]
- Bode L, Jantscher-Krenn E. Structure-function relationships of human milk oligosaccharides. Adv Nutr. 2012;3:383S–91S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottacini F, Morrissey R, Esteban-Torres Met al.. Comparative genomics and genotype-phenotype associations in Bifidobacterium breve. Sci Rep. 2018;8:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brantl V, Teschemacher H, Bläsig Jet al.. Opioid activities of β-casomorphins. Life Sci. 1981;28:1903–9. [DOI] [PubMed] [Google Scholar]
- Brietzke E, Cerqueira RO, Mansur RBet al.. Gluten related illnesses and severe mental disorders: a comprehensive review. Neurosci Biobehav Rev. 2018;84:368–75. [DOI] [PubMed] [Google Scholar]
- Bunesova V, Lacroix C, Schwab C. Fucosyllactose and L-fucose utilization of infant Bifidobacterium longum and Bifidobacterium kashiwanohense. BMC Microbiol. 2016;16:248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böger M, van Leeuwen SS, Lammerts van Bueren Aet al.. Structural identity of galactooligosaccharide molecules selectively utilized by single cultures of probiotic bacterial strains. J Agric Food Chem. 2019;67:13969–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervantes-Barragan L, Chai JN, Tianero MDet al.. Lactobacillus reuteri induces gut intraepithelial CD4+ CD8αα+ T cells. Science (80-). 2017;357:806–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. Human milk oligosaccharides (HMOS): structure, function, and enzyme-catalyzed synthesis. Advances in Carbohydrate Chemistry and Biochemistry. Vol 72, Cambridge, Massachusetts, United States: Academic Press Inc., Elsevier, 2015, 113–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia JSJ, McRae JL, Kukuljan Set al.. A1 beta-casein milk protein and other environmental pre-disposing factors for type 1 diabetes. Nutr Diabetes. 2017;7:e274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S-W, Mason JB. Folate and carcinogenesis: an integrated scheme. J Nutr. 2000;130:129–32. [DOI] [PubMed] [Google Scholar]
- Chu DM, Ma J, Prince ALet al.. Maturation of the infant microbiome community structure and function across multiple body sites and in relation to mode of delivery. Nat Med. 2017;23:314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chyan Y-J, Poeggeler B, Omar RAet al.. Potent neuroprotective properties against the Alzheimer β-amyloid by an endogenous melatonin-related indole structure, indole-3-propionic acid. J Biol Chem. 1999;274:21937–42. [DOI] [PubMed] [Google Scholar]
- Cinova J, De Palma G, Stepankova Ret al.. Role of intestinal bacteria in gliadin-induced changes in intestinal mucosa: study in germ-free rats. PLoS One. 2011;6:e16169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collado MC, Rautava S, Aakko Jet al.. Human gut colonisation may be initiated in utero by distinct microbial communities in the placenta and amniotic fluid. Sci Rep. 2016;6:23129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Aimmo MR, Mattarelli P, Biavati Bet al.. The potential of bifidobacteria as a source of natural folate. J Appl Microbiol. 2012;112:975–84. [DOI] [PubMed] [Google Scholar]
- D'Aimmo MR, Modesto M, Mattarelli Pet al.. Biosynthesis and cellular content of folate in bifidobacteria across host species with different diets. Anaerobe. 2014;30:169–77. [DOI] [PubMed] [Google Scholar]
- de Goffau MC, Lager S, Sovio Uet al.. Human placenta has no microbiome but can contain potential pathogens. Nature. 2019;572:329–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieuleveux V, Lemarinier S, Gueguen M. Antimicrobial spectrum and target site of D-3-phenyllactic acid. Int J Food Microbiol. 1998;40:177–83. [DOI] [PubMed] [Google Scholar]
- Dodd D, Spitzer MH, Van Treuren Wet al.. A gut bacterial pathway metabolizes aromatic amino acids into nine circulating metabolites. Nature. 2017;551:648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X, Xin Y, Jian Wet al.. Bifidobacterium thermacidophilumsp. nov., isolated from an anaerobic digester. Int J Syst Evol Microbiol. 2000;50:119–25. [DOI] [PubMed] [Google Scholar]
- Duranti S, Milani C, Lugli GAet al.. Evaluation of genetic diversity among strains of the human gut commensal Bifidobacterium adolescentis. Sci Rep. 2016;6:23971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duranti S, Turroni F, Lugli GAet al.. Genomic characterization and transcriptional studies of the starch-utilizing strain Bifidobacterium adolescentis 22 L. Appl Environ Microbiol. 2014;80:6080–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckel VPL, Ziegler L-M, Vogel RFet al.. Bifidobacterium tibiigranuli sp. nov. isolated from homemade water kefir. Int J Syst Evol Microbiol. 2019:ijsem003936. [DOI] [PubMed] [Google Scholar]
- Egan M, Jiang H, Motherway MOet al.. Glycosulfatase-encoding gene cluster in Bifidobacterium breve UCC2003. Appl Environ Microbiol. 2016;82:6611–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan M, Motherway MO, Kilcoyne Met al.. Cross-feeding byBifidobacterium breve UCC2003 during co-cultivation with Bifidobacterium bifidum PRL2010 in a mucin-based medium. BMC Microbiol. 2014a;14:282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan M, Motherway MO, Ventura Met al.. Metabolism of sialic acid byBifidobacterium breveUCC2003. Appl Environ Microbiol. 2014b;80:4414–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich AM, Henrick B, Pacheco Aet al.. Bifidobacterium grown on human milk oligosaccharides produce tryptophan metabolite Indole-3-lactic acid that significantly decreases inflammation in intestinal cells in vitro. FASEB J. 2018;32:lb359. [Google Scholar]
- Favier CF, Vaughan EE, De Vos WMet al.. Molecular monitoring of succession of bacterial communities in human neonates. Appl Environ Microbiol. 2002;68:219–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferretti P, Pasolli E, Tett Aet al.. Mother-to-infant microbial transmission from different body sites shapes the developing infant gut microbiome. Cell Host Microbe. 2018;24:133–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiedorowicz E, Kaczmarski M, Cieślińska Aet al.. β-casomorphin-7 alters μ-opioid receptor and dipeptidyl peptidase IV genes expression in children with atopic dermatitis. Peptides. 2014;62:144–9. [DOI] [PubMed] [Google Scholar]
- Field CJ. The immunological components of human milk and their effect on immune development in infants. J Nutr. 2005;135:1–4. [DOI] [PubMed] [Google Scholar]
- Freitas AC, Hill JE. Bifidobacteria isolated from vaginal and gut microbiomes are indistinguishable by comparative genomics. PLoS One. 2018;13:e0196290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita K, Oura F, Nagamine Net al.. Identification and molecular cloning of a novel glycoside hydrolase family of core 1 type O-glycan-specific endo-α-N-acetylgalactosaminidase from Bifidobacterium longum. J Biol Chem. 2005;280:37415–22. [DOI] [PubMed] [Google Scholar]
- Fujita K, Sakamoto A, Kaneko Set al.. Degradative enzymes for type II arabinogalactan side chains in Bifidobacterium longum subsp. longum. Appl Microbiol Biotechnol. 2019;103:1299–310. [DOI] [PubMed] [Google Scholar]
- Gagnon M, Kheadr EE, Le Blay Get al.. In vitro inhibition of Escherichia coli O157: H7 by bifidobacterial strains of human origin. Int J Food Microbiol. 2004;92:69–78. [DOI] [PubMed] [Google Scholar]
- Garrido D, Barile D, Mills DA. A molecular basis for bifidobacterial enrichment in the infant gastrointestinal tract. Adv Nutr. 2012;3:415S–21S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido D, Kim JH, German JBet al.. Oligosaccharide binding proteins from Bifidobacterium longum subsp. infantis reveal a preference for host glycans. PLoS One. 2011;6:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido D, Ruiz-Moyano S, Kirmiz Net al.. A novel gene cluster allows preferential utilization of fucosylated milk oligosaccharides in Bifidobacterium longum subsp. longum SC596. Sci Rep. 2016;6:35045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido D, Ruiz-Moyano S, Lemay DGet al.. Comparative transcriptomics reveals key differences in the response to milk oligosaccharides of infant gut-associated bifidobacteria. Sci Rep. 2015;5:13517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golfetto L, de Senna FD, Hermes Jet al.. Lower bifidobacteria counts in adult patients with celiac disease on a gluten-free diet. Arq Gastroenterol. 2014;51:139–43. [DOI] [PubMed] [Google Scholar]
- Gotoh A, Katoh T, Sakanaka Met al.. Sharing of human milk oligosaccharides degradants within bifidobacterial communities in faecal cultures supplemented with Bifidobacterium bifidum. Sci Rep. 2018;8:13958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gueimonde M, Laitinen K, Salminen Set al.. Breast milk: a source of bifidobacteria for infant gut development and maturation? Neonatology. 2007;92:64–6. [DOI] [PubMed] [Google Scholar]
- Hubbard TD, Murray IA, Bisson WHet al.. Adaptation of the human aryl hydrocarbon receptor to sense microbiota-derived indoles. Sci Rep. 2015;5:12689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa E, Matsuki T, Kubota Het al.. Ethnic diversity of gut microbiota: species characterization of Bacteroides fragilis group and genus Bifidobacteriumin healthy Belgian adults, and comparison with data from Japanese subjects. J Biosci Bioeng. 2013;116:265–70. [DOI] [PubMed] [Google Scholar]
- Jacob RA. Folate, DNA methylation, and gene expression: factors of nature and nurture. 2000. [DOI] [PubMed]
- James K, Bottacini F, Contreras JISet al.. Metabolism of the predominant human milk oligosaccharide fucosyllactose by an infant gut commensal. Sci Rep. 2019;9:1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James K, Motherway MO, Bottacini Fet al.. Bifidobacterium breve UCC2003 metabolises the human milk oligosaccharides lacto-N-tetraose and lacto-N-neo-tetraose through overlapping, yet distinct pathways. Sci Rep. 2016;6:38560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jianqin S, Leiming X, Lu Xet al.. Effects of milk containing only A2 beta casein versus milk containing both A1 and A2 beta casein proteins on gastrointestinal physiology, symptoms of discomfort, and cognitive behavior of people with self-reported intolerance to traditional cows’ milk. Nutr J. 2015;15:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jost T, Lacroix C, Braegger CPet al.. Vertical mother–neonate transfer of maternal gut bacteria via breastfeeding. Environ Microbiol. 2014;16:2891–904. [DOI] [PubMed] [Google Scholar]
- Karbownik M, Stasiak M, Zygmunt Aet al.. Protective effects of melatonin and indole-3-propionic acid against lipid peroxidation, caused by potassium bromate in the rat kidney. Cell Biochem Funct Cell Biochem its Modul by Act agents or Dis. 2006;24:483–9. [DOI] [PubMed] [Google Scholar]
- Katayama T. Host-derived glycans serve as selected nutrients for the gut microbe: human milk oligosaccharides and bifidobacteria. Biosci Biotechnol Biochem. 2016;80:621–32. [DOI] [PubMed] [Google Scholar]
- Kato K, Odamaki T, Mitsuyama Eet al.. Age-related changes in the composition of gut Bifidobacteriumspecies. Curr Microbiol. 2017;74:987–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly ED, Bottacini F, O'Callaghan Jet al.. Glycoside hydrolase family 13 α-glucosidases encoded by Bifidobacterium breve UCC2003; a comparative analysis of function, structure and phylogeny. Int J Food Microbiol. 2016;224:55–65. [DOI] [PubMed] [Google Scholar]
- Kirmiz N, Robinson RC, Shah IMet al.. Milk glycans and their interaction with the infant-gut microbiota. Annu Rev Food Sci Technol. 2018;9:429–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitaoka M. Bifidobacterial enzymes involved in the metabolism of human milk oligosaccharides. Adv Nutr. 2012;3:422S–9S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyohara M, Nakatomi T, Kurihara Set al.. α-N-acetylgalactosaminidase from infant-associated bifidobacteria belonging to novel glycoside hydrolase family 129 is implicated in alternative mucin degradation pathway. J Biol Chem. 2012;287:693–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemenak M, Dolinšek J, Langerholc Tet al.. Administration of Bifidobacterium breve Decreases the Production of TNF-α in Children with Celiac Disease. Dig Dis Sci. 2015;60:3386–92. [DOI] [PubMed] [Google Scholar]
- Kobata A. Structures and application of oligosaccharides in human milk. Proc Japan Acad Ser B. 2010;86:731–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komeno M, Hayamizu H, Fujita Ket al.. Two novel α-L-arabinofuranosidases from Bifidobacterium longum subsp. longum belonging to glycoside hydrolase family 43 cooperatively degrade arabinan. Appl Environ Microbiol. 2019;85:e02582–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kordy K, Gaufin T, Mwangi Met al.. Contributions to human breast milk microbiome and enteromammary transfer of Bifidobacterium breve. PLoS One. 2020;15:e0219633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzewska A, Ben-Skowronek I. Effect of associated autoimmune diseases on type 1 diabetes mellitus incidence and metabolic control in children and adolescents. Biomed Res Int. 2016;2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunz C, Rudloff S, Baier Wet al.. Oligosaccharides in human milk: structural, functional, and metabolic aspects. Annu Rev Nutr. 2000;20:699–722. [DOI] [PubMed] [Google Scholar]
- Lamendella R, Santo Domingo JW, Kelty Cet al.. Bifidobacteria in feces and environmental waters. Appl Environ Microbiol. 2008;74:575–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanz TV, Becker S, Mohapatra SRet al.. Suppression of Th1 differentiation by tryptophan supplementation in vivo. Amino Acids. 2017;49:1169–75. [DOI] [PubMed] [Google Scholar]
- Laparra JM, Sanz Y. Bifidobacteria inhibit the inflammatory response induced by gliadins in intestinal epithelial cells via modifications of toxic peptide generation during digestion. J Cell Biochem. 2010;109:801–7. [DOI] [PubMed] [Google Scholar]
- Laureys D, Cnockaert M, De Vuyst Let al.. Bifidobacterium aquikefiri sp. nov., isolated from water kefir. Int J Syst Evol Microbiol. 2016;66:1281–6. [DOI] [PubMed] [Google Scholar]
- Laursen MF, Sakanaka M, von Burg Net al.. Breastmilk-promoted bifidobacteria produce aromatic lactic acids in the infant gut. bioRxiv. 2020. [Google Scholar]
- Lawson MAE, O'Neill IJ, Kujawska Met al.. Breast milk-derived human milk oligosaccharides promote Bifidobacterium interactions within a single ecosystem. ISME J. 2019:14:635–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBlanc JG, Milani C, De Giori GSet al.. Bacteria as vitamin suppliers to their host: a gut microbiota perspective. Curr Opin Biotechnol. 2013;24:160–8. [DOI] [PubMed] [Google Scholar]
- Le Doare K, Holder B, Bassett Aet al.. Mother's milk: A purposeful contribution to the development of the infant microbiota and immunity. Front Immunol. 2018;9:361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis ZT, Mills DA. Differential establishment of bifidobacteria in the breastfed infant gut. Intestinal Microbiome: Functional Aspects in Health and Disease. Vol 88, Karger AG, Basel: Karger Publishers, 2017, 149–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Wang J, Wu Let al.. The impacts of delivery mode on infant's oral microflora. Sci Rep. 2018;8:11938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindfors K, Blomqvist T, Juuti‐Uusitalo Ket al.. Live probioticBifidobacterium lactisbacteria inhibit the toxic effects induced by wheat gliadin in epithelial cell culture. Clin Exp Immunol. 2008;152:552–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Udenigwe CC. Role of food‐derived opioid peptides in the central nervous and gastrointestinal systems. J Food Biochem. 2019;43:e12629. [DOI] [PubMed] [Google Scholar]
- LoCascio RG, Desai P, Sela DAet al.. Broad conservation of milk utilization genes in Bifidobacterium longum subsp. infantis as revealed by comparative genomic hybridization. Appl Environ Microbiol. 2010;76:7373–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LoCascio RG, Ninonuevo MR, Freeman SLet al.. Glycoprofiling of bifidobacterial consumption of human milk oligosaccharides demonstrates strain specific, preferential consumption of small chain glycans secreted in early human lactation. J Agric Food Chem. 2007;55:8914–9. [DOI] [PubMed] [Google Scholar]
- LoCascio RG, Niñonuevo MR, Kronewitter SRet al.. A versatile and scalable strategy for glycoprofiling bifidobacterial consumption of human milk oligosaccharides. Microb Biotechnol. 2009;2:333–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucock M. Folic acid: nutritional biochemistry, molecular biology, and role in disease processes. Mol Genet Metab. 2000;71:121–38. [DOI] [PubMed] [Google Scholar]
- Makino H, Kushiro A, Ishikawa Eet al.. Mother-to-infant transmission of intestinal bifidobacterial strains has an impact on the early development of vaginally delivered infant's microbiota. PLoS One. 2013;8:e78331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino H, Martin R, Ishikawa Eet al.. Multilocus sequence typing of bifidobacterial strains from infant's faeces and human milk: are bifidobacteria being sustainably shared during breastfeeding? Benef Microbes. 2015;6:563–72. [DOI] [PubMed] [Google Scholar]
- Makino H. Bifidobacterial strains in the intestines of newborns originate from their mothers. Biosci microbiota, food Heal. 2018;37:79–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin R, Makino H, Yavuz ACet al.. Early-life events, including mode of delivery and type of feeding, siblings and gender, shape the developing gut microbiota. PLoS One. 2016;11:e0158498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuki T, Yahagi K, Mori Het al.. A key genetic factor for fucosyllactose utilization affects infant gut microbiota development. Nat Commun. 2016;7:11939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng D, Sommella E, Salviati Eet al.. Indole-3-lactic acid, a metabolite of tryptophan, secreted by Bifidobacterium longum subspecies infantis is anti-inflammatory in the immature intestine. Pediatr Res. 2020:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milani C, Duranti S, Lugli GAet al.. Comparative genomics ofBifidobacterium animalis subsp. lactis reveals a strict monophyletic bifidobacterial taxon. Appl Environ Microbiol. 2013;79:4304–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milani C, Lugli GA, Duranti Set al.. Bifidobacteria exhibit social behavior through carbohydrate resource sharing in the gut. Sci Rep. 2015a;5:15782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milani C, Lugli GA, Duranti Set al.. Genomic encyclopedia of type strains of the genus Bifidobacterium. Appl Environ Microbiol. 2014;80:6290–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milani C, Mancabelli L, Lugli GAet al.. Exploring vertical transmission of bifidobacteria from mother to child. Appl Environ Microbiol. 2015b;81:7078–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milani C, Turroni F, Duranti Set al.. Genomics of the genus Bifidobacterium reveals species-specific adaptation to the glycan-rich gut environment. Appl Environ Microbiol. 2016;82:980–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minami J, Odamaki T, Hashikura Net al.. Lysozyme in breast milk is a selection factor for bifidobacterial colonisation in the infant intestine. Benef Microbes. 2016;7:53–60. [DOI] [PubMed] [Google Scholar]
- Mitsuoka T, Kaneuchi C. Ecology of the bifidobacteria. Am J Clin Nutr. 1977;30:1799–810. [DOI] [PubMed] [Google Scholar]
- Moeller AH, Caro-Quintero A, Mjungu Det al.. Cospeciation of gut microbiota with hominids. Science (80-). 2016;353:380–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motherway MO, Fitzgerald GF, Neirynck Set al.. Characterization of ApuB, an extracellular type II amylopullulanase from Bifidobacterium breve UCC2003. Appl Environ Microbiol. 2008;74:6271–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama K, Nagai A, Uribayashi Ket al.. Two extracellular sialidases from Bifidobacterium bifidumpromote the degradation of sialyl-oligosaccharides and support the growth of Bifidobacterium breve. Anaerobe. 2018;52:22–8. [DOI] [PubMed] [Google Scholar]
- O'Callaghan A, van Sinderen D. Bifidobacteria and their role as members of the human gut microbiota. Front Microbiol. 2016;7:925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell Motherway M, Fitzgerald GF, van Sinderen D. Metabolism of a plant derived galactose‐containing polysaccharide byBifidobacterium breve UCC2003. Microb Biotechnol. 2011;4:403–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell Motherway M, Kinsella M, Fitzgerald GFet al.. Transcriptional and functional characterization of genetic elements involved in galacto‐oligosaccharide utilization by Bifidobacterium breveUCC 2003. Microb Biotechnol. 2013;6:67–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell Motherway M, O'Driscoll J, Fitzgerald GFet al.. Overcoming the restriction barrier to plasmid transformation and targeted mutagenesis inBifidobacterium breve UCC2003. Microb Biotechnol. 2009;2:321–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odamaki T, Bottacini F, Kato Ket al.. Genomic diversity and distribution of Bifidobacterium longumsubsp. longum across the human lifespan. Sci Rep. 2018;8:85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odamaki T, Bottacini F, Mitsuyama Eet al.. Impact of a bathing tradition on shared gut microbe among Japanese families. Sci Rep. 2019;9:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odamaki T, Horigome A, Sugahara Het al.. Comparative genomics revealed genetic diversity and species/strain-level differences in carbohydrate metabolism of three probiotic bifidobacterial species. Int J Genomics. 2015;2015:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odamaki T, Kato K, Sugahara Het al.. Age-related changes in gut microbiota composition from newborn to centenarian: a cross-sectional study. BMC Microbiol. 2016;16:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacheco AR, Barile D, Underwood MAet al.. The impact of the milk glycobiome on the neonate gut microbiota. Annu Rev Anim Biosci. 2015;3:419–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parte AC. LPSN–List of Prokaryotic names with Standing in Nomenclature (bacterio. net), 20 years on. Int J Syst Evol Microbiol. 2018;68:1825–9. [DOI] [PubMed] [Google Scholar]
- Perez-Muñoz ME, Arrieta M-C, Ramer-Tait AEet al.. A critical assessment of the “sterile womb” and “in utero colonization” hypotheses: implications for research on the pioneer infant microbiome. Microbiome. 2017;5:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto-Sánchez MI, Smecuol EC, Temprano MPet al.. Bifidobacterium infantisNLS super strain reduces the expression of α-defensin-5, a marker of innate immunity, in the mucosa of active celiac disease patients. J Clin Gastroenterol. 2017;51:814–7. [DOI] [PubMed] [Google Scholar]
- Pokusaeva K, Fitzgerald GF, van Sinderen D. Carbohydrate metabolism in Bifidobacteria. Genes Nutr. 2011;6:285–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokusaeva K, Neves AR, Zomer Aet al.. Ribose utilization by the human commensal Bifidobacterium breve UCC2003. Microb Biotechnol. 2010;3:311–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokusaeva K, O'Connell-Motherway M, Zomer Aet al.. Cellodextrin utilization by Bifidobacterium breve UCC2003. Appl Environ Microbiol. 2011;77:1681–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokusaeva K, O'Connell-Motherway M, Zomer Aet al.. Characterization of two novel α-glucosidases from Bifidobacterium breveUCC2003. Appl Environ Microbiol. 2009;75:1135–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pompei A, Cordisco L, Amaretti Aet al.. Administration of folate-producing bifidobacteria enhances folate status in Wistar rats. J Nutr. 2007b;137:2742–6. [DOI] [PubMed] [Google Scholar]
- Pompei A, Cordisco L, Amaretti Aet al.. Folate production by bifidobacteria as a potential probiotic property. Appl Environ Microbiol. 2007a;73:179–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieur DJ. Tissue specific deficiency of lysozyme in ruminants. Comp Biochem Physiol B. 1986;85:349–53. [DOI] [PubMed] [Google Scholar]
- Pruimboom L, De Punder K. The opioid effects of gluten exorphins: asymptomatic celiac disease. J Heal Popul Nutr. 2015;33:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quagliariello A, Aloisio I, Bozzi Cionci Net al.. Effect of Bifidobacterium breveon the intestinal microbiota of coeliac children on a gluten free diet: a pilot study. Nutrients. 2016;8:660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada V, Splichal I, Rockova Set al.. Susceptibility of bifidobacteria to lysozyme as a possible selection criterion for probiotic bifidobacterial strains. Biotechnol Lett. 2010;32:451–5. [DOI] [PubMed] [Google Scholar]
- Rayburn WF, Stanley JR, Garrett ME. Periconceptional folate intake and neural tube defects. J Am Coll Nutr. 1996;15:121–5. [DOI] [PubMed] [Google Scholar]
- Reyman M, van Houten MA, van Baarle Det al.. Impact of delivery mode-associated gut microbiota dynamics on health in the first year of life. Nat Commun. 2019;10:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivière A, Moens F, Selak Met al.. The ability of bifidobacteria to degrade arabinoxylan oligosaccharide constituents and derived oligosaccharides is strain dependent. Appl Environ Microbiol. 2014;80:204–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockova S, Rada V, Nevoral Jet al.. Inter-species differences in the growth of bifidobacteria cultured on human milk oligosaccharides. Folia Microbiol (Praha). 2012;57:321–4. [DOI] [PubMed] [Google Scholar]
- Rodriguez CI, Martiny JBH. Evolutionary relationships among bifidobacteria and their hosts and environments. BMC Genomics. 2020;21:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi M, Amaretti A, Raimondi S. Folate production by probiotic bacteria. Nutrients. 2011;3:118–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruas-Madiedo P, Gueimonde M, Fernández-García Met al.. Mucin degradation by Bifidobacterium strains isolated from the human intestinal microbiota. Appl Environ Microbiol. 2008;74:1936–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Moyano S, Totten SM, Garrido DAet al.. Variation in consumption of human milk oligosaccharides by infant gut-associated strains ofBifidobacterium breve. Appl Environ Microbiol. 2013;79:6040–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz L, García-Carral C, Rodriguez JM. Unfolding the human milk microbiome landscape in the omics era. Front Microbiol. 2019;10:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell WR, Duncan SH, Scobbie Let al.. Major phenylpropanoid‐derived metabolites in the human gut can arise from microbial fermentation of protein. Mol Nutr Food Res. 2013;57:523–35. [DOI] [PubMed] [Google Scholar]
- Rutayisire E, Huang K, Liu Yet al.. The mode of delivery affects the diversity and colonization pattern of the gut microbiota during the first year of infants’ life: a systematic review. BMC Gastroenterol. 2016;16:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherfurd-Markwick KJ. Food proteins as a source of bioactive peptides with diverse functions. Br J Nutr. 2012;108:S149–57. [DOI] [PubMed] [Google Scholar]
- Ryan SM, Fitzgerald GF, van Sinderen D. Screening for and identification of starch-, amylopectin-, and pullulan-degrading activities in bifidobacterial strains. Appl Environ Microbiol. 2006;72:5289–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan SM, Fitzgerald GF, van Sinderen D. Transcriptional regulation and characterization of a novel β-fructofuranosidase-encoding gene from Bifidobacterium breveUCC2003. Appl Environ Microbiol. 2005;71:3475–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakanaka M, Gotoh A, Yoshida Ket al.. Varied Pathways of Infant Gut-Associated Bifidobacterium to Assimilate Human Milk Oligosaccharides: Prevalence of the Gene Set and Its Correlation with Bifidobacteria-Rich Microbiota Formation. Nutrients. 2020;12:71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakanaka M, Hansen ME, Gotoh Aet al.. Evolutionary adaptation in fucosyllactose uptake systems supports bifidobacteria-infant symbiosis. Sci Adv. 2019;5:eaaw7696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai T, Hashikura N, Minami Jet al.. Tolerance mechanisms of human-residential bifidobacteria against lysozyme. Anaerobe. 2017;47:104–10. [DOI] [PubMed] [Google Scholar]
- Sakurai T, Odamaki T, Xiao J. Production of Indole-3-Lactic Acid by Bifidobacterium Strains Isolated fromHuman Infants. Microorganisms. 2019;7:340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai T, Yamada A, Hashikura Net al.. Degradation of food-derived opioid peptides by bifidobacteria. Benef Microbes. 2018;9:675–82. [DOI] [PubMed] [Google Scholar]
- Sakurama H, Kiyohara M, Wada Jet al.. Lacto-N-biosidase encoded by a novel gene of Bifidobacterium longum subspecies longum shows unique substrate specificity and requires a designated chaperone for its active expression. J Biol Chem. 2013;288:25194–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schell MA, Karmirantzou M, Snel Bet al.. The genome sequence of Bifidobacterium longum reflects its adaptation to the human gastrointestinal tract. Proc Natl Acad Sci. 2002;99:14422–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sela DA, Chapman J, Adeuya Aet al.. The genome sequence of Bifidobacterium longumsubsp. infantis reveals adaptations for milk utilization within the infant microbiome. Proc Natl Acad Sci. 2008;105:18964–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sela DA, Garrido D, Lerno Let al.. Bifidobacterium longum subsp. infantis ATCC 15697 α-fucosidases are active on fucosylated human milk oligosaccharides. Appl Environ Microbiol. 2012;78:795–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sela DA, Mills DA. Nursing our microbiota: molecular linkages between bifidobacteria and milk oligosaccharides. Trends Microbiol. 2010;18:298–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serena G, Camhi S, Sturgeon Cet al.. The role of gluten in celiac disease and type 1 diabetes. Nutrients. 2015;7:7143–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigeno Y, Zhang H, Banno Tet al.. Gut microbiota development in mice is affected by hydrogen peroxide produced from amino acid metabolism during lactation. FASEB J. 2018;33:3343–52. [DOI] [PubMed] [Google Scholar]
- Smecuol E, Hwang HJ, Sugai Eet al.. Exploratory, randomized, double-blind, placebo-controlled study on the effects of Bifidobacterium infantis natren life start strain super strain in active celiac disease. J Clin Gastroenterol. 2013;47:139–47. [DOI] [PubMed] [Google Scholar]
- Sokolov O, Kost N, Andreeva Oet al.. Autistic children display elevated urine levels of bovine casomorphin-7 immunoreactivity. Peptides. 2014;56:68–71. [DOI] [PubMed] [Google Scholar]
- Stanger O. Physiology of folic acid in health and disease. Curr Drug Metab. 2002;3:211–23. [DOI] [PubMed] [Google Scholar]
- Stephen A, Alles M, De Graaf Cet al.. The role and requirements of digestible dietary carbohydrates in infants and toddlers. Eur J Clin Nutr. 2012;66:765–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strozzi GP, Mogna L. Quantification of folic acid in human feces after administration of Bifidobacterium probiotic strains. J Clin Gastroenterol. 2008;42:S179–84. [DOI] [PubMed] [Google Scholar]
- Sugahara H, Odamaki T, Hashikura Net al.. Differences in folate production by bifidobacteria of different origins. Biosci microbiota, food Heal. 2015;34:87–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugahara H, Odamaki T, Xiao J. Genotypic and phenotypic evaluation revealed the appropriateness of human-residential bifidobacteria for human use. 2015.
- Sun Z, Zhang W, Guo Cet al.. Comparative genomic analysis of 45 type strains of the genus Bifidobacterium: a snapshot of its genetic diversity and evolution. PLoS One. 2015;10:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Zhang Z, Wang Xet al.. Relation of β-casomorphin to apnea in sudden infant death syndrome. Peptides. 2003;24:937–43. [DOI] [PubMed] [Google Scholar]
- Suzuki R, Wada J, Katayama Tet al.. Structural and thermodynamic analyses of solute-binding protein from Bifidobacterium longumspecific for core 1 disaccharide and lacto-N-biose I. J Biol Chem. 2008;283:13165–73. [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Kosaka M, Shindo Ket al.. Identification of antioxidants produced by Lactobacillus plantarum. Biosci Biotechnol Biochem. 2013:121006. [DOI] [PubMed] [Google Scholar]
- Tailford LE, Crost EH, Kavanaugh Det al.. Mucin glycan foraging in the human gut microbiome. Front Genet. 2015;6:81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tannock GW, Lawley B, Munro Ket al.. Comparison of the compositions of the stool microbiotas of infants fed goat milk formula, cow milk-based formula, or breast milk. Appl Environ Microbiol. 2013;79:3040–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teschemacher H. Opioid receptor ligands derived from food proteins. Curr Pharm Des. 2003;9:1331–44. [DOI] [PubMed] [Google Scholar]
- Thomson P, Medina DA, Garrido D. Human milk oligosaccharides and infant gut bifidobacteria: molecular strategies for their utilization. Food Microbiol. 2018;75:37–46. [DOI] [PubMed] [Google Scholar]
- Toda K, Hisata K, Satoh Tet al.. Neonatal oral fluid as a transmission route for bifidobacteria to the infant gut immediately after birth. Sci Rep. 2019;9:8692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Alipi BI, Fragoso-Ramírez JA, Martínez-Limón AJet al.. Bacterial colonization of the oral cavity in the newborn. Bol Med Hosp Infant Mex. 1990;47:78–83. [PubMed] [Google Scholar]
- Tremaroli V, Bäckhed F. Functional interactions between the gut microbiota and host metabolism. Nature. 2012;489:242. [DOI] [PubMed] [Google Scholar]
- Turroni F, Bottacini F, Foroni Eet al.. Genome analysis of Bifidobacterium bifidum PRL2010 reveals metabolic pathways for host-derived glycan foraging. Proc Natl Acad Sci. 2010;107:19514–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turroni F, Duranti S, Milani Cet al.. Bifidobacterium bifidum: A Key Member of the Early Human Gut Microbiota. Microorganisms. 2019;7:544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turroni F, Foroni E, Pizzetti Pet al.. Exploring the diversity of the bifidobacterial population in the human intestinal tract. Appl Environ Microbiol. 2009;75:1534–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turroni F, Peano C, Pass DAet al.. Diversity of bifidobacteria within the infant gut microbiota. PLoS One. 2012;7:e36957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turroni F, Ribbera A, Foroni Eet al.. Human gut microbiota and bifidobacteria: from composition to functionality. Antonie Van Leeuwenhoek. 2008;94:35–50. [DOI] [PubMed] [Google Scholar]
- Turroni F, Van Sinderen D, Ventura M. Genomics and ecological overview of the genus Bifidobacterium. Int J Food Microbiol. 2011;149:37–44. [DOI] [PubMed] [Google Scholar]
- Underwood MA, Kalanetra KM, Bokulich NAet al.. A comparison of two probiotic strains of bifidobacteria in premature infants. J Pediatr. 2013;163:1585–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura M, Canchaya C, Tauch Aet al.. Genomics of Actinobacteria: tracing the evolutionary history of an ancient phylum. Microbiol Mol Biol Rev. 2007;71:495–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura M, Turroni F, Lugli GAet al.. Bifidobacteria and humans: our special friends, from ecological to genomics perspectives. J Sci Food Agric. 2014;94:163–8. [DOI] [PubMed] [Google Scholar]
- Ventura M, Turroni F, Zomer Aet al.. The Bifidobacterium dentium Bd1 genome sequence reflects its genetic adaptation to the human oral cavity. PLos Genet. 2009;5:e1000785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada J, Ando T, Kiyohara Met al.. Bifidobacterium bifidum lacto-N-biosidase, a critical enzyme for the degradation of human milk oligosaccharides with a type 1 structure. Appl Environ Microbiol. 2008;74:3996–4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Li M, Wu Set al.. Fecal microbiota composition of breast-fed infants is correlated with human milk oligosaccharides consumed. J Pediatr Gastroenterol Nutr. 2015;60:825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasilewska J, Sienkiewicz-Szłapka E, Kuźbida Eet al.. The exogenous opioid peptides and DPPIV serum activity in infants with apnoea expressed as apparent life threatening events (ALTE). Neuropeptides. 2011;45:189–95. [DOI] [PubMed] [Google Scholar]
- Watanabe K, Makino H, Sasamoto Met al.. Bifidobacterium mongoliense sp. nov., from airag, a traditional fermented mare's milk product from Mongolia. Int J Syst Evol Microbiol. 2009;59:1535–40. [DOI] [PubMed] [Google Scholar]
- Wilck N, Matus MG, Kearney SMet al.. Salt-responsive gut commensal modulates TH17 axis and disease. Nature. 2017;551:585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong CB, Sugahara H, Odamaki Tet al.. Different physiological properties of human-residential and non-human-residential bifidobacteria in human health. Benef Microbes. 2018;9. DOI: 10.3920/BM2017.0031. [DOI] [PubMed] [Google Scholar]
- Wong CB, Tanaka A, Kuhara Tet al.. Potential Effects of Indole-3-Lactic Acid, a Metabolite of Human Bifidobacteria, on NGF-induced Neurite Outgrowth in PC12 Cells. Microorganisms. 2020;8:398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J, Takahashi S, Nishimoto Met al.. Distribution of in vitro fermentation ability of lacto-N-biose I, a major building block of human milk oligosaccharides, in bifidobacterial strains. Appl Environ Microbiol. 2010a;76:54–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J, Takahashi S, Odamaki Tet al.. Antibiotic susceptibility of bifidobacterial strains distributed in the Japanese market. Biosci Biotechnol Biochem. 2010b;74:336–42. [DOI] [PubMed] [Google Scholar]
- Yamada C, Gotoh A, Sakanaka Met al.. Molecular insight into evolution of symbiosis between breast-fed infants and a member of the human gut microbiome Bifidobacteriumlongum. Cell Chem Biol. 2017;24:515–24. [DOI] [PubMed] [Google Scholar]
- Yatsunenko T, Rey FE, Manary MJet al.. Human gut microbiome viewed across age and geography. Nature. 2012;486:222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Y, Xue W, Luo Get al.. 1,520 reference genomes from cultivated human gut bacteria enable functional microbiome analyses. Nat Biotechnol. 2019;37:179–85. [DOI] [PMC free article] [PubMed] [Google Scholar]


