Summary
Human antibody repertoire data captured through next-generation sequencing (NGS) has enabled deeper insights into B cell immunogenetics and paratope diversity. By analyzing large public NGS datasets, we map the landscape of non-canonical cysteines in human variable heavy-chain domains (VHs) at the repertoire level. We identify remarkable usage of non-canonical cysteines within the heavy-chain complementarity-determining region 3 (CDR-H3) and other CDRs and framework regions. Furthermore, our study reveals the diversity and location of non-canonical cysteines and their associated motifs in human VHs, which are reminiscent of and more complex than those found in other non-human species such as chicken, camel, llama, shark, and cow. These results explain how non-canonical cysteines strategically occur in the human antibodyome to expand its paratope space. This study will guide the design of human antibodies harboring disulfide-stabilized long CDR-H3s to access difficult-to-target epitopes and influence a paradigm shift in developability involving non-canonical cysteines.
Keywords: non-canonical cysteine, D gene, B-cell receptor, antibody repertoire, antibodyome, disulfide-bonded CDR-H3, next-generation sequencing, antibody library, antibody engineering, antibody discovery
Graphical Abstract
Highlights
-
•
NGS-based non-canonical cysteine landscape in human VHs
-
•
1 to 8 non-canonical cysteines and up to 30% in long CDR-H3s
-
•
An array of potential disulfide motifs adds paratope diversity
-
•
Non-canonical cysteines in human VHs are reminiscent of lower animals
Prabakaran and Chowdhury reveal the remarkable patterns of non-canonical cysteines in human antibody heavy chains (VHs) and their role in paratope diversification. These patterns mimic features observed separately in chicken, camel, llama, shark, and cow. These findings can help design and develop next-generation human antibodies and libraries.
Introduction
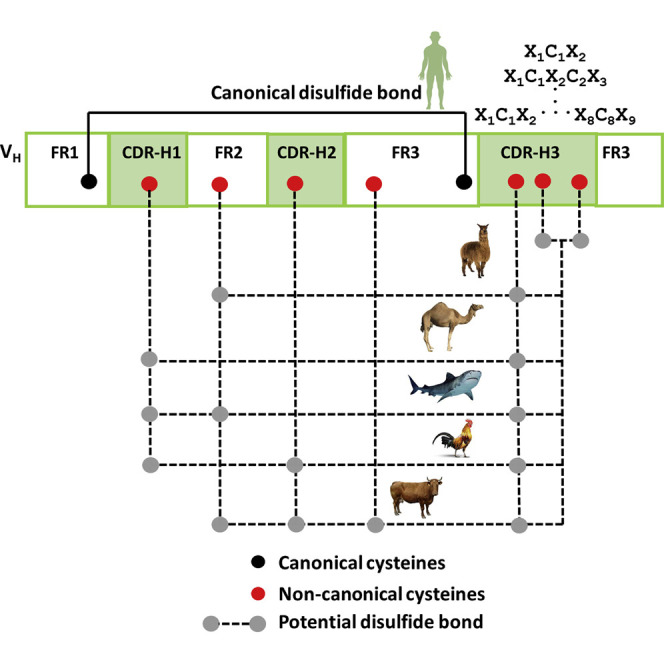
Cysteines in human antibodies play a fundamental structural role by forming intra- and inter-chain disulfide bonds (Frangione et al., 1969). They are found at the core in each of the immunoglobulin (Ig) domains and connect the polypeptide chains of an antibody molecule, which are encoded by variable gene (V gene) and constant gene (C gene) segments (Tonegawa, 1983) and referred as canonical cysteines. However, non-canonical cysteines are normally encoded by certain human diversity gene (D gene) segments, mainly IGHD2 and other D gene families. Non-canonical cysteines are thought to be less prevalent in human compared with species such as chicken (Wu et al., 2012), camel (Muyldermans et al., 1994), llama (Harmsen et al., 2000), shark (Feng et al., 2019; Stanfield et al., 2004), and cow (Haakenson et al., 2019; Saini et al., 1999; Wang et al., 2013). Non-canonical cysteines in these non-human species form various intra-heavy-chain complementarity-determining region 3 (CDR-H3) disulfide bonds and disulfide bonds between CDR-H3 and other CDRs or with framework regions (FRs). These non-canonical cysteines are vital in generating the diversity of antibody repertoires and distinct antigen-combining site structures and in mediating functions (Conroy et al., 2017; de los Rios et al., 2015; Dong et al., 2019; Finlay and Almagro, 2012). Lately, X-ray crystal structures of several human antibodies (Flyak et al., 2018; Kong et al., 2013; Lee et al., 2014; Sui et al., 2009; Wu et al., 2015; Ying et al., 2015) have revealed non-canonical cysteines to be mostly encoded by members of the IGDH2 family, forming disulfide motifs in CDR-H3s. Two previous studies have reported on the presence of non-canonical cysteines using a limited number of human antibody sequences (Chen et al., 2017; Zemlin et al., 2003) and suggested that the two-cysteine motifs found in human CDR-H3s, namely, CXnC, where Xn is the distance between the two cysteines in terms of amino acid (aa) length, could be useful as diversity elements. However, knowledge of the frequency and diversity of non-canonical cysteines and their associated motifs in human antibody VH repertoires has remained obscure.
Recently, next-generation sequencing (NGS) of human antibody repertoires has provided information on immunogenetic diversity with unprecedented depth and detail. More than 130 million annotated sequences of CDR-H3s from multiple individuals were made publicly available by two studies (Briney et al., 2019; DeWitt et al., 2016). Here, we analyzed these datasets to decipher the landscape and diversity of non-canonical cysteines in the human CDR-H3 repertoire. We determined the numbers, prevalence, and patterns with which non-canonical cysteines and potential disulfide motifs are present in CDR-H3s. We identified a remarkably high level of cysteine usage, mainly in long CDR-H3s, and various potential disulfide bonds that could form within CDR-H3s and possibly between CDR-H3s and other CDRs or FRs. These results may strongly suggest an evolutionary relationship between the variable heavy-chain domains (VHs) of human and those of other species (de los Rios et al., 2015). In particular, our analysis exposed the two-cysteine CXnC motifs (n = 4) that existed in more than 3 million unique CDR-H3s, of which several thousand were distinctive tetrapeptide motifs. In addition, we found that higher numbers of cysteines, three to eight, formed diverse and unique motifs that were not previously recognized in human CDR-H3s. We further reviewed the diverse structural mechanisms and functions of known human antibodies that dominantly use the disulfide motifs in their CDR-H3s to recognize various antigens. Altogether, this study discovered potential immunogenetic characteristics of human repertoire shaped by the presence of non-canonical cysteines and non-canonical disulfide bridges in VHs that are thought to be rare in or absent from humans. These identified non-canonical cysteines in CDR-H3s of expressed human repertoires can form a new and complex paratope space that one might explore to find solutions for challenging antigen targets. Furthermore, these results provide a deeper understanding of germline-encoded and somatic hypermutation (SHM)-generated non-canonical cysteine motifs in human antibody VH repertoires and have wide implications for the development of human antibody-based technologies.
Results
Immunogenetic Analysis Reveals High Frequency, Extensive Diversity, and Recurring Patterns of Non-canonical Cysteines
We surveyed the landscape of non-canonical cysteines in the expressed human antibody VH repertoires through analysis of two large NGS datasets; the circulating B cell populations of ten human subjects (Briney et al., 2019) (dataset A) and the circulating naive and memory B cells of three human donors (DeWitt et al., 2016) (dataset B). Dataset A contained annotated sequences of nearly 3 billion VH sequences representing more than 106.5 million unique antibodies from consensus clusters, with available in-frame VH and CDR-H3 sequences, Ig isotypes, IgG subtypes, and IGHV, IGHD, and IGHJ gene families (Briney et al., 2019). The frequency of non-canonical cysteines in CDR-H3s of dataset A ranged from 8.5% to 16.7%, with a median value 14.2%, as shown by the breakdown values for the number of non-canonical cysteines in Table S1. The number of non-canonical cysteines in CDR-H3s ranged from one to eight, which was unexpected in human, because the higher cysteine numbers are hallmarks of antibodies from chicken (Wu et al., 2012), shark (Feng et al., 2019), and cow (Wang et al., 2013). We identified more than 12 million unique VH sequences from IgM- and IgG-derived antibodies, which is 11.3% of the repertoire sequences that have at least one non-canonical cysteine in their CDR-H3s. The bivariate normal density plot of the non-canonical cysteine residue distribution showed that human CDR-H3s containing non-canonical cysteines associate with diverse IGHV germline families and Ig isotypes of circulating B cells (Figure S1). The IgM repertoire was found to contain most non-canonical cysteines, as well as the widest range in cysteine numbers, indicating that it has the largest pool of antibody sequences bearing diverse cysteine motifs in CDR-H3s. It was also observed that 1,298,403 unique cysteine-containing CDR-H3s associated with more than one germline IGHV gene or Ig isotype. The existence of IgD could not be reliably determined using the current dataset. However, because IgD is a spliced variant of the pre-mRNA that produces IgM (Gutzeit et al., 2018), and cysteine-containing CDR-H3s are quite abundant in IgM, it is reasonable to presume that cysteine-containing CDR-H3s are likely to be present in IgDs. However, this assumption, if validated, will imply that cysteine-containing CDR-H3s are prevalent across all Ig classes and IGHV germline families.
Dataset B included nearly 30 million unique CDR-H3s of human B cell receptor (BCR) sequences from the naive and memory repertoires of three donors (DeWitt et al., 2016). In this, 3,019,883 human CDR-H3s with at least one non-canonical cysteine residue were identified, i.e., 10.2% of the total number of repertoire sequences. The different numbers of non-canonical cysteines in dataset B are shown in Table S2. In this dataset, the maximum number of cysteines found was 7 and occurred only in one donor (D1). An interesting observation from this dataset was that naive population in all donors consistently showed 2% fewer cysteines in CDR-H3s than the memory population, suggesting that a subpopulation of cysteines in CDR-H3s might have been added through SHM. We further noticed that odd-numbered cysteines (one, three, or five) were 2%–5% more likely in the memory population (Table S2). In datasets A and B, most cysteine-containing CDR-H3s had either one or two cysteines, as previously reported (Chen et al., 2017; Zemlin et al., 2003). However, this analysis provided evidence for unusual diversity involving two non-canonical cysteines. It also showed the presence of three to eight non-canonical cysteines in several human CDR-H3s that had not been seen previously in human. These findings prompted a detailed analysis to discover the diversity of patterns and motifs associated with these non-canonical cysteines.
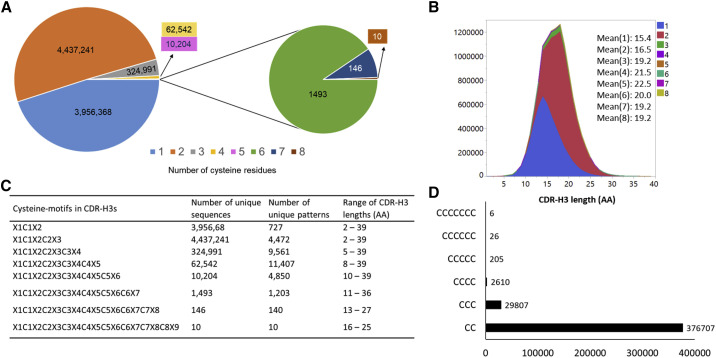
We observed ∼10% of CDR-H3s contained one or two non-canonical cysteines in the human VH repertoires (Tables S1 and S2). In the genome, 23 human germline D gene allele segments encode either one or two non-canonical cysteines; 10 of them are found in a 5′-3′ direct orientation and 13 are found in a 3′-5′ inverted orientation (Table S3A). Specifically, D gene segments in inverted orientations accounted for ∼2.5% of CDR-H3s containing non-canonical cysteines expressed in human VH repertoire (Table S3B). These results clearly showed the usage of inverted IGHD genes in the expressed human antibody repertoire, which has remained inconclusive so far because of the smaller datasets used in previous studies (Benichou et al., 2013; Ohm-Laursen et al., 2006). We also observed that D genes that do not encode non-canonical cysteines could be involved in the formation of CDR-H3s containing non-canonical cysteines, albeit at smaller percentages (Tables S3A and S3B). These might be due to mutational hotspots in D genes, base changes through junctional modification, or a combination of both. Therefore, we expected reasonable distribution of non-canonical cysteines upon VDJ recombination in CDR-H3s of human VH repertoires. We classified unique antibodies based on the number of non-canonical cysteines in CDR-H3s, ranging from one to eight, as shown in Figure 1 A. Among these, CDR-H3s containing two cysteines were the most prevalent, with 6,630,839 sequences (6.2%), followed by CDR-H3s containing a single cysteine, with 4,985,244 sequences (4.7%) (Table S1). Altogether, they accounted for 96.4% of the non-canonical cysteine-containing CDR-H3 repertoire. The higher numbers of non-canonical cysteines, three to eight, were in relatively low abundance, as indicated by their frequencies (Table S1). In particular, seven or eight non-canonical cysteines in CDR-H3s were determined to be a rare occurrence, because only 146 and 10, respectively, were found out of more than 106 million antibodies in dataset A. However, they were found in multiple individuals, implying that their occurrence is not sporadic.
Figure 1.
Analysis of Non-canonical Cysteines in Human CDR-H3s
(A) Pie charts showing the number of CDR-H3s with non-canonical cysteine residues ranging from 1 to 8 as identified in dataset A.
(B) Length distributions of CDR-H3s versus number of cysteines, along with their mean values. Color codes refer to the number of cysteines.
(C) Cysteine motifs observed in human CDR-H3s displaying a diverse pattern of number of cysteines and CDR-H3 length diversity. C1–C8, cysteine residues; X1–X9, number of other aas between cysteines and/or flanking them (see Figure S6).
(D) Number of unique CDR-H3s with contiguous cysteines occurring mostly as duplets and triplets and rarely up to septuplets found in human.
We further analyzed the relationship between the presence of non-canonical cysteines and the lengths of CDR-H3s (Figure 1B; Figure S2). We observed that the number of non-canonical cysteines found in CDR-H3s increased as the length of CDR-H3 increased, particularly for up to five cysteines (Figure 1B). The percentages of all antibodies, with and without non-canonical cysteines, in CDR-H3 lengths of 1–15 aa (average), 16–25 aa (long), and 26–39 aa (ultra-long) were 5.5%, 4.4%, and 0.1%, respectively, as observed in dataset A (Figure S2A). The percentages of antibodies containing non-canonical cysteines only, as observed in dataset A, were 8.7%, 18.9%, and 27.8% for CDR-H3s with lengths of 1–15 aa (average), 16–25 aa (long), and 26–39 aa (ultra-long), respectively (Figure S2B). Similar trends were observed for dataset B, for which the percentages of all antibodies with CDR-H3 lengths of 1–15 aa (average), 16–25 aa (long), and 26–39 aa (ultra-long) were 7.8%, 8.6%, and 0.3%, respectively. In contrast, the percentages of antibodies with non-canonical cysteines only were 5.4%, 15.9%, and 28.4% for average, long, and ultra-long CDR-H3s, respectively (Figures S2C and S2D).
To identify cysteine motifs and their unique patterns, we classified the cysteine motifs of CDR-H3s as tandem cysteines, C1 through C8, that are separated and/or flanked by different lengths of other aas, X1 through X9 (Figure 1C). Thus, a two-cysteine motif can be defined by the notation X1C1X2C2X3, where the two cysteines are separated by an X2 number of aas and flanked by X1 and X3 numbers of aas. Similarly, CDR-H3s with the three- and four-cysteine motifs are represented by X1C1X2C2X3C3X4 and X1C1X2C2X3C3X4C4X5. This analysis revealed multiple, contiguous cysteines that commonly occurred as duplets and triplets and rarely up to septuplets in CDR-H3s, which were previously unknown in human (Figure 1D). Until this point, these types of contiguous cysteines in CDR-H3s were observed only in antibodies derived from chicken (Wu et al., 2012), cows (Wang et al., 2013), and sharks (Feng et al., 2019). The higher frequency of cysteine-duplet occurrence (Figure 1D) could result from the predominate usage of the IGHD6-13∗01 germline gene in 3′-5′ inverted orientation reading frame 3 that encodes two contiguous cysteines. It is also possible that diversification events, such as through SHM (Brenner and Milstein, 1966), V(DD)J recombination (Briney et al., 2012), or combinations thereof, may lead to the formation of contiguous cysteines. Thus, these results revealed human CDR-H3s harbor a complex pattern of cysteines that can potentially create a diverse set of paratopes for unique antigen binding.
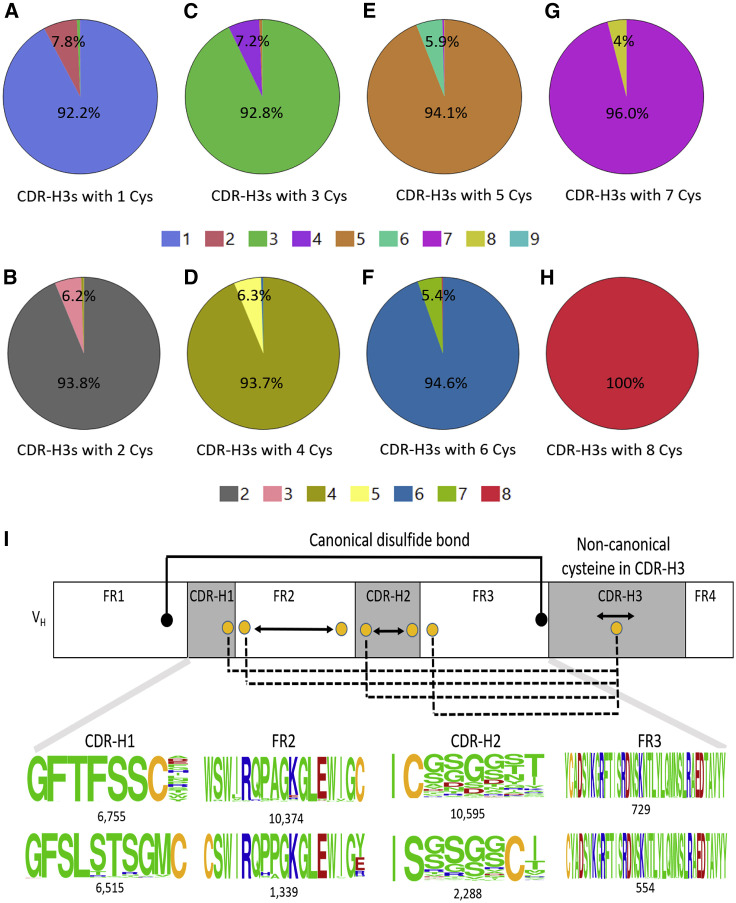
As we observed earlier, CDR-H3s containing two cysteines were the most prevalent. Although CDR-H3s with even numbers of cysteines might allow for the formation of intra-CDR-H3 disulfide bonds, an unpaired cysteine found in CDR-H3 could potentially form a disulfide bond with a free cysteine in other parts of the VH region. For example, a non-canonical cysteine that naturally exists in the germline-encoded CDR-H1 of the IGHV2-70∗01 lineage (Lefranc et al., 1999) could be available for a free cysteine in CDR-H3 to form an inter-CDR disulfide bond. More strikingly, we observed that 13,389 VH sequences of the IGHV2-70∗01 germline origin was associated with CDR-H3s containing non-canonical cysteines, ranging from one to four. Markedly, we found that 72% of those CDR-H3s contained free cysteines, suggesting potential disulfide bonds between non-canonical cysteines of CDR-H3s and germline-encoded cysteines of CDR-H1s. Likewise, a SHM-generated cysteine could appear anywhere in the antibody variable regions that may also mediate a non-canonical disulfide bond with the unpaired cysteine in CDR-H3. To this end, we calculated the frequencies of VH sequences that have non-canonical cysteines in FR1, CDR-H1, FR2, CDR-H2, or FR3, in addition to that found in CDR-H3s using dataset A, because it had full-length VH sequences as shown in Figure 2 . In these pie charts, the major arcs represent the percentage of VH sequences that have non-canonical cysteines only at their CDR-H3s. The smaller arcs show the percentage of VH sequences that have non-canonical cysteines both at their CDR-H3s and in other parts of VH regions, such as FR1, CDR-H1, FR2, CDR-H2, or FR3. For instance, Figure 2A shows that 7.8% of VH sequences, as represented by the smaller arc in the pie chart, contain a single free cysteine in CDR-H3s but have additional non-canonical cysteines elsewhere in the VH. The same holds true for the other pie charts in Figures 2B–2G. In Figure 2A, we identified 313,492 VHs that had single cysteines in CDR-H3s and another cysteine elsewhere in their VHs. Sequence analysis of these VHs revealed the locations of these non-canonical cysteine within and outside CDR-H3s. In the variable region, these cysteines are found in one of the following positions: at the end of CDR-H1, on one or the other end of CDR-H2 and FR2, and at the beginning of FR3 (Figure 2I). Moreover, we observed that these non-canonical cysteines at those specific locations in CDR-H1, FR2, CDR-H2, or FR3 are generally near CDR-H3s, as seen in the three-dimensional structures of antibodies, suggesting potential for disulfide bond formation. These results suggest that human antibodies, like those in chicken, camel, llama, shark, and cow, probably use non-canonical cysteines for paratope diversification. It is also conceivable that like the other non-human species mentioned earlier, humans use non-canonical disulfide bonds to stabilize antibodies. The potential for disulfide bond formation between CDR-H3s and other parts of VH regions, such as CDR-H1, FR2, CDR-H2, and FR3, in human antibodies is quite analogous to non-canonical disulfide bonds existing in antibody repertoires of chicken, camel, llamas, and shark (de los Rios et al., 2015; Finlay and Almagro, 2012). What is remarkable is that although each of these non-human species has unique non-canonical disulfides, because of the number and location of the non-canonical cysteines, humans appear to have exceptionally unique features in their antibodyome compared with these other species. It is therefore tantalizing to imagine that the paratope space of humans is larger than previously thought.
Figure 2.
Frequencies and Locations of Non-canonical Cysteines in Human CDR-H3s and Other Parts of VHs as Observed in Dataset A
(A–H) Pie charts with the larger arcs showing the frequencies of VHs with CDR-H3 non-canonical cysteines, one through eight that are represented by (A) through (H), respectively. The smaller arcs show the frequencies for VHs with non-canonical cysteines observed at both CDR-H3 and other parts of VHs, namely, other heavy-chain CDRs and FRs. Therefore, VHs in smaller arcs could potentially have various disulfide bonds between those non-canonical cysteines in CDR-H3s and any of the non-canonical cysteines from CDRs or FRs, which are reminiscent of disulfide bonds discovered in antibodies from chicken, camel, llama, shark, and cow.
(I) Schematic depicting the locations of non-canonical cysteines (yellow) in CDR-H1, FR2, CDR-H2, and FR3 of human VHs. In FR2 and CDR-H2, non-canonical cysteine can occur at either end. Their locations are shown by the WebLogos. The number of sequences contributing to each logo is given. A single cysteine in CDR-H3s was simultaneously found with an unpaired cysteine in other regions of VHs, indicating potentially diverse non-canonical disulfide bonds in human VHs (dotted lines).
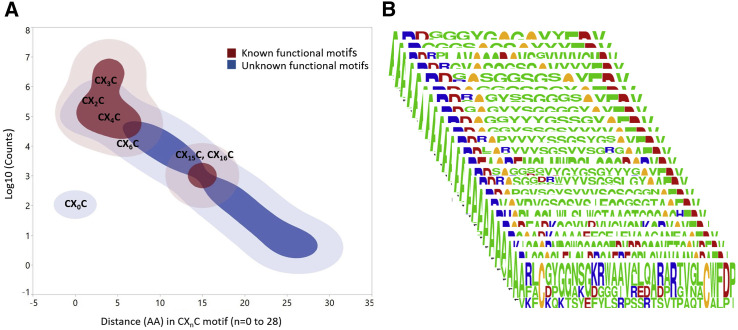
CXnC Motifs Display an Extraordinary Sequence Diversity
The two-cysteine motif, CXnC, in human CDR-H3s was the most prevalent type of non-canonical cysteine motif identified and found in 6,630,839 and 2,270,432 unique human antibodies in datasets A and B, respectively (Tables S1 and S2). These sequences contain a range of CXnC motifs in terms of aa length and diversity between cysteines and potentially forming intra-CDR-H3 disulfide bonds. Previously, a handful of structural studies showed that disulfide-containing CDR-H3 motifs in human antibodies play important roles in antigen recognition (Almagro et al., 2012; Doria-Rose et al., 2014; Flyak et al., 2018; Kong et al., 2013; Lee et al., 2014; Wu et al., 2015; Ying et al., 2015). This prompted us to analyze the diversity of the CXnC motifs within CDR-H3s of human antibody repertoires and correlate those motifs to known antigen binding modes and functions. Furthermore, previous sequence analyses using smaller datasets showed that the two cysteines had Xn separation of 0–12 aa only, although most of them were generally separated by 4 aa (Chen et al., 2017; Zemlin et al., 2003). However, our analysis exposed a range of Xn values up to 28 aa and associated cysteines motifs in CDR-H3s at the repertoire level. In Figure 3 A, a contour plot in red highlights human antibodies containing CXnC motifs (n = 2, 3, 4, 6, 15, and 16) associated with known structures and functions. The contour plot in blue in Figure 3A represents antibodies containing other CXnC motifs existing in the human antibody repertoire but for which no functional activities are known. We observed remarkable diversity of CXnC motifs within CDR-H3s, comprising 4,472 unique patterns of the type X1C1X2C2X3 (Figure 1C), for which high-frequency, two-cysteine motifs with different Xn values of 1 to 24 aa were selected and shown as WebLogos in Figure 3B.
Figure 3.
CXnC Motifs in Human CDR-H3s
(A) Contour plot showing the categories of CXnC motifs in human CDR-H3s with a distance of 0 to 28 aa (CXnC, n = 0–28). The relative abundance of the lengths (Xn, aa distance) between two cysteines is plotted against count values of log10 for 4,279,148 CXnC motifs in dataset A. Shown in red are examples of the lengths with Xn = 2, 3, 4, 6, 15, and 16 that have been reported to exhibit functional activities with known structures (Figure S5). Shown in blue are the other CXnC motifs for which no functional activities have yet been ascribed.
(B) Diverse CXnC motifs observed in the present study, showing a high-frequency motif selected as an example for each Xn value, are shown in stacked WebLogos.
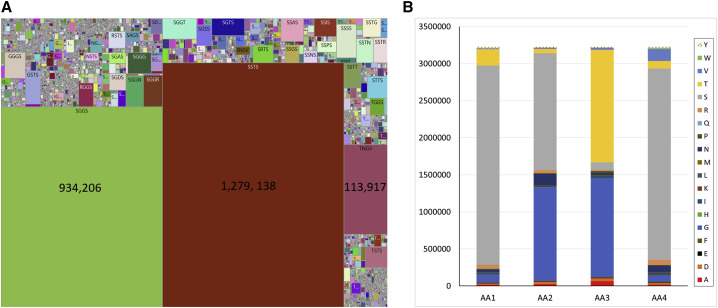
The CX4C motif accounted for nearly 75% of all CXnC motifs as observed in dataset A. The higher frequency of the CX4C motif could be largely determined by diverse IGHD2 germline families that mostly encode two cysteines separated by 4 aa, including SSTS, SGGS, TNGV, and TGGV (Table S3A). We found IGHD2 families of more than 19% in dataset A (Table S3B). Nonetheless, we identified 34,266 unique tetrapeptides embedded between the two cysteine residues of CX4C motifs, with occurrences up to 1,279,138. An aerial view of those tetrapeptides within CX4C motifs is shown by a Treemap chart in Figure 4 A. The IGHD2 germline-encoded tetrapeptides had a high prevalence of SSTS (1,279,138), SGGS (943,206), and TNGV (113,917) across dataset A. However, the other IGHD2 germline encoding the TGGV tetrapeptide had a lower frequency of 3,182. Notably, 118 high-frequency tetrapeptides appearing at least 1,000 times within CX4C motifs of human CDR-H3s in dataset A were identified (Figure S3). Furthermore, we calculated position-specific aa compositions of the tetrapeptides within the CX4C motif and found three germline-encoded aas (S, G, and T) predominate, as shown in Figure 4B. Specifically, the usage of S was found to be 83%, 49%, and 80% at positions aa1, aa2, and aa4, respectively, whereas G/S and G/T were found to be 88% at both position aa2 and position aa3. All other 19 aas appeared at lower frequencies. Apart from the CX4C motifs, the IGHD2-21 gene encoded the CX3C motif containing the germline tripeptide GGD, which resulted in the formation of 385,746 unique CDR-H3s, as observed in dataset A. This adds to the diversity of potential disulfide-bonded CXnC motifs. Thus, IGHD2 germline diversity, along with sequence lengths and diversities of the aas that separate and/or flank the cysteines and potential SHM, gave rise to 3,226,652 unique CDR-H3s in dataset A.
Figure 4.
Landscape of CX4C Motifs in Human CDR-H3s
(A) Treemap chart showing an aerial view of 34,266 unique tetrapeptides embedded between the two cysteines in CX4C motifs, ranging in frequencies from 1 to 1,279,138, as found in dataset A. The top three peptides with higher prevalence are shown with numbers in the Treemap chart. These are the only D gene germline-encoded products. There were 118 high-frequency tetrapeptides that are not D germline encoded but appeared more than 1,000 times in CX4C motifs of human CDR-H3s (see Figure S3).
(B) Histogram of position-specific frequency of aa usage (counts) within the CX4C motif as found in 3,226,652 unique human CDR-H3s.
CXnC Motifs Play a Determining Role in the Structure and Function of Antibodies
To understand structural and functional aspects of non-canonical cysteines, we performed analysis of the Protein Data Bank (PDB) and investigated immunogenetic origin of disulfide motifs in CDR-H3s of human antibodies and the role they play in interactions with antigens. We identified twelve human antibodies in complexes with various antigens (Figure S4) and twenty-five in apo forms (Figure S5) that had CXnC motifs within their CDR-H3s. These structurally characterized human antibodies containing CDR-H3 disulfide motifs showed a spectrum of functional activities and epitope specificity. These antibodies use the disulfide motifs of CDR-H3s, engaging in different binding modes, to target diverse viral antigens, including HIV, influenza, HCV, RSV, MERS CoV, and HCMV and other human proteins such as lecithin cholesterol acyltransferase (LCAT), Tau peptide, BLyS receptor 3 (BR3), celiac disease-specific gluten peptide, and L-rhamnose of Streptococcus pneumoniae (Figure S4). These antibodies used prominently expressed IGHV genes such as 1-69, 1-2, 3-23, 3-30, 3-36, 4-31, 4-59, and 5-51, pairing with both IGLV and IGKV genes (Figure S5). In these crystal structures, CDR-H3s, with lengths ranging from 16 to 38 aa, presented various disulfide motifs, including CX2C, CX3C, CX4C, CCX5CX4C, CX6C, CX15C, and CX16C.
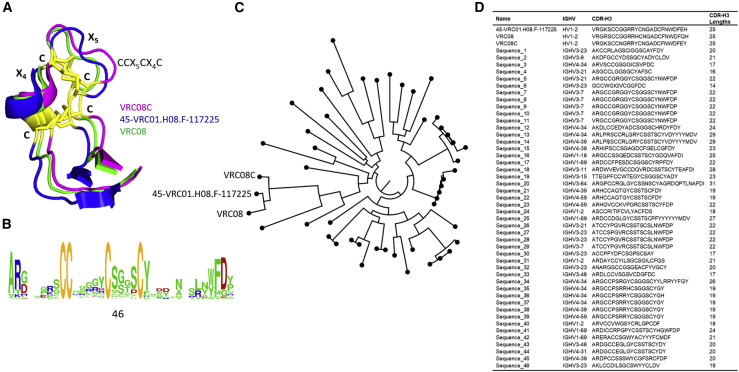
Next, we analyzed the immunogenetic origins of non-canonical cysteine motifs, including D gene usage, junctional modification, D-D fusion, and potential SHM, with specific examples as observed in the PDB. In Figures S5A–S5G, of all heavy chains of anti-HIV VRC-class antibodies, four of them originate from the HV1-2 gene family and three originate from the HV3-30 family. Among those anti-HIV antibodies, VRC08C, 45-VRC01.H08.F-117225, and VRC08 contain CCX5CX4C motifs, which form a pair of disulfide bonds in the 25-aa-long CDR-H3s (Figure 5 A) and create distinct binding surfaces for antigen binding (Wu et al., 2015). We identified a four-cysteine motif of the same CCX5CX4C type in 46 CDR-H3s in dataset A as depicted in Figure 5B. A circular phylogenetic tree showing the relationship among CDR-H3s bearing CCX5CX4C motifs from 46 human antibodies and VRC01-class antibodies is illustrated in Figure 5C. We further identified that the 46 VHs had diverse IGHV germline lineages, as well as CDR-H3 lengths ranging from 14 to 31 aa (Figure 5D). We could predict that the CCX5CX4C motif may have originated from IGHD6-13 and IGHD2 family genes, because these D genes encode contiguous double cysteines and CX4C motifs, respectively. Furthermore, we were able to identity 12 unique CDR-H3s containing the CCX5CX4C motif in dataset B (data not shown). In addition, we looked at a third dataset that consisted of NGS-derived B cell repertoire from three individuals to find high-frequency shared clonotypes (Soto et al., 2019) that were curated and included in the cAb-Rep database (Guo et al., 2019). Our analysis of these datasets also yielded 44 CDR-H3 sequences that contain the CCX5CX4C motif. Thus, these results confirmed the prevalence of the CCX5CX4C motif at the repertoire level from multiple individuals in different datasets.
Figure 5.
CCX5CX4C Cysteine Motifs in Human CDR-H3s
(A) Aligned 3D structures of CDR-H3s with CCX5CX4C motifs, forming a conserved double-disulfide bond (yellow), in three anti-HIV-1 broadly neutralizing VRC01-class antibodies (blue, green, and magenta) are shown.
(B) WebLogo generated from a group of 46 CDR-H3s presenting a 4-cysteine motif, CCX5CX4C, as found in dataset A, which is similar to that found in VRC01-class antibodies.
(C) Circular phylogram showing the relationship among CDR-H3s of 46 human antibodies with different IGHV germline lineages and the three VRC01-class antibodies derived from the IGHV1-2 lineage.
(D) CDR-H3s containing CCX5CX4C motifs in 46 human antibodies identified in dataset A are shown with IGHV germline families and aa lengths. Anti-HIV VRC01-class antibodies use the IGHV1-2 germline with CDR-H3 lengths of 25 aa, whereas those 46 human antibodies were found to have diverse IGHV germline usages and CDR-H3 lengths in the range of 14–31 aa.
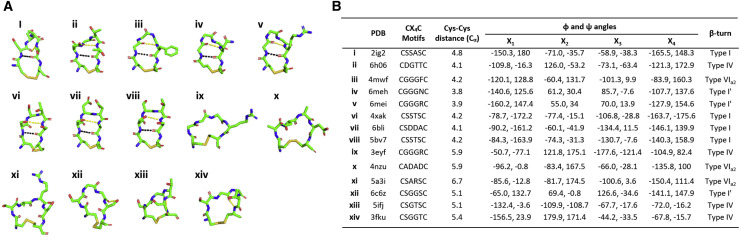
In the case of CX2C motifs (Figures S5A, S5H, and S5Y), as well as for the CX6C motif (Figure S5U), the germline origins could only have been attributed to SHM events, because IGHD germline segments do not encode two cysteines separated by either a dipeptide or a hexapeptide. Similarly, CX15C and CX16C motifs, shown in Figures S5E–S5G, were thought to have originated from P- and N-nucleotide addition and point mutations, as previously explained (Doria-Rose et al., 2014). The CX3C motif (Figure S5R) containing the tripeptide sequence GGD was found to have a clear IGHD2-21 germline origin, because the tripeptide sequence is fully germline encoded (Table S3A). 14 of 25 human antibodies containing non-canonical cysteines with known 3D structures have the CX4C motif in CDR-H3s (Figures S5I–S5Q, S5S, S5T, and S5V–S5X). We performed a detailed structural analysis of the 6 residue loops constituting the CX4C motif and found that it has wide structural diversity stabilized with disulfide bonds and other hydrogen bonds in some instances (Figure 6 A). Here, structural conformations of CX4C motifs were defined by dihedral angles, ϕ and ψ, of central residues of the tetrapeptides and disulfide bonds (Figure 6B), which resemble 4-residue β turns observed in protein structures (de Brevern, 2016; Venkatachalam, 1968). We further identified IGHD2 family genes as the main germline gene for CX4C motifs, because tetrapeptides flanked by two cysteines had sequence matches or similarities with corresponding germline residues of IGHD2 genes (Figure 6B). Furthermore, we noted from the structural details of twenty-five human antibodies that CDR-H3s exhibit unique conformations tethered by disulfide-bonded cysteines and mediate antibody-antigen interactions through various binding modes and novel mechanisms of action. In general, CDR-H3s with cysteine motifs CX2C, CX3C, CX4C, and CX6C, were found to adopt β-hairpin folds as stabilized by disulfide bridges. In addition, they mostly appeared protruding and oriented themselves either pointing toward or bending away from the light chains (Figure S5), thereby creating a repertoire of structurally diverse and stable paratopes. Finally, these results suggested that human antibodies containing non-canonical cysteine motifs with diverse patterns might be potentially useful to recognize a range of antigens through molecular mimicry mechanisms, such as the conserved extracellular cysteine-tethered loops found in the Cys-loop ligand-gated ion channel receptors (Thompson et al., 2010) and cysteine noose domains in anti-viral proteins (Lee et al., 2018).
Figure 6.
Structural Conformation of CX4C Motifs Observed in CDR-H3s of Human Antibodies
(A) CX4C motifs in CDR-H3s of human antibodies forming different types of β turns are shown in a stick representation. Black dots denote hydrogen bonds mimicking i → i + 3 classical hydrogen bonds found in classical β turns, and yellow dots represent other hydrogen bonds existing between backbone atoms or backbone and side-chain atoms.
(B) PDB codes, sequence, disulfide bond distances, and dihedral angles for tetrapeptides of CX4C motifs, along with β turn types, are given.
Multiple Non-canonical Cysteine Motifs Exist and Reveal Immunogenetic Mechanisms
To find out whether human antibodies have multiple cysteines in CDR-H3s, we used dataset A to analyze CDR-H3s of varied lengths that contain between three and six cysteines. To enable this analysis, we defined a pattern as an exact arrangement of cysteine residues within CDR-H3s that are interspersed and/or flanked by given number of aas. The aa compositional diversities of the interspersing and flanking segments increase the uniqueness of CDR-H3s. In this manner, we found a range of patterns defined by the distinct number of aas that separate and/or flank cysteines, ranging from 1 to 8, which are given in Figure S6. The variations in the number of aas for three- and four-cysteine motifs were found to be larger compared with other cysteine motifs, leading to a higher number of unique patterns for these categories (Figure 1C). Specifically, for the three-cysteine motif X1C1X2C2X3C3X, we identified 9,561 unique patterns (324,991 unique sequences); for the four-cysteine motif X1C1X2C2X3C3X4C4X5, 11,407 unique patterns were found (62,542 unique sequences). Furthermore, we identified exceptional five- and six-cysteine motifs within CDR-H3s of human antibodies, which had 4,850 and 1,203 unique patterns, respectively (10,204 and 1,493 unique sequences). The high-frequency motifs containing three to six non-canonical cysteines from a subset of CDR-H3s with diverse patterns selected from dataset A are shown using sequence logos in Figure 7 . The free cysteines available from any of these one- to seven-cysteine motifs in CDR-H3s may form disulfide bonds between CDR-H3 and other parts of VHs (CDR-H1, FR2, CDR-H2, or FR3), as shown in Figure 2, leading to more complex cysteine patterns. The diversity of multiple cysteine patterns may have evolved from several sources, including IGHD germline-encoded cysteines, SHM, and D-D fusion. To assess the extent of D-D fusion occurring in four-cysteine motifs of CDR-H3s, we calculated the frequencies of co-existing germline-encoded two-cysteine motifs that could be formed by potential V(DD)J recombination (Briney et al., 2012). The potential frequencies of D-D fusion involving either of the two IGHD germlines that encode two-cysteine motifs (CC, CX3C, and CX4C), as observed in dataset A, is shown in Figure S7A. We identified the diverse D-D fusions in many human CDR-H3s containing the four-cysteine motifs. Some of those high-frequency, four-cysteine motifs formed by the D-D fusion are shown by selected sequence logos (Figure S7B). These depict relevant D germline-encoded cysteine motifs, along with changes in other aas induced by SHM.
Figure 7.
Three- to Six-Cysteine Motifs Observed in a Subset of Human CDR-H3s Showing Diverse Patterns
(A–D) Relative abundance of different aas between and flanking conserved cysteines in (A) three-cysteine, (B) four-cysteine, (C) five-cysteine, and (D) six-cysteine motifs within a selected sets of high-frequency CDR-H3s (dataset A) are displayed as WebLogos. The number of CDR-H3s contributing to each pattern is given underneath each sequence logo.
Discussion
Detailed immunogenetics analysis of the available NGS data from the largest single collection of BCRs (Briney et al., 2019), comprising of nearly 3 billion antibody VH sequences, brought to light the landscape and diversity of non-canonical cysteines in human VH repertoire. This has been incomprehensible until now. We identified more than 12 million unique VH sequences containing non-canonical cysteines with exceptionally diverse motifs and potential disulfide loops of varying in size and composition involving CDR-H3s. These findings have implications for understanding how non-canonical cysteines strategically occur in the human antibodyome and the role they can play to expand the paratope space that were thought to be rare or absent. These results will trigger future studies on the design and development of novel human antibodies that can potentially access epitopes generally considered inaccessible.
This study revealed the potential for the formation of intra-CDR-H3 disulfide bonds and those between CDR-H3 and other CDRs or FRs of human VHs. It also showed the range of intervening aas within any two non-canonical cysteines that exist in CDR-H3s. In addition, the present analysis uncovered various three-, four-, five-, and six-cysteine motifs in human CDR-H3s. We could expect that some of these CDR-H3s with multiple non-canonical cysteines form the extended β-hairpin structures stabilized by single- and double-disulfide bonds as seen in other proteins (Gunasekaran et al., 1997). The contiguous double cysteines observed with a large prevalence in the expressed human antibody repertoire might form a new class of antibodies with vicinal disulfides, which might provide biological roles through allosteric function or binding to sugar or multiring ligand moieties (Carugo et al., 2003; de Araujo et al., 2013; Richardson et al., 2017). The new information of patterns and motifs associated with non-canonical cysteines in CDR-H3s of human antibodies can guide the designs of novel cysteine nooses and cysteine-containing long CDR-H3 libraries. The diverse non-canonical cysteine motifs in human CDR-H3s may also be useful for computational design of new antibodies. Furthermore, the finding of certain non-canonical cysteine-enriched VHs can be used to trace the phylogeny of broadly neutralizing anti-HIV antibodies that have disulfide-bonded CDR-H3s for identifying putative templates for the B cell-lineage immunogen design (Haynes et al., 2012).
Although comparatively rare in humans, the presence of high cysteine numbers and motifs in CDR-H3s, as consistently observed in multiple individuals, is reminiscent of that found in chicken, camel, llama, shark, and cow. Knowledge of non-canonical cysteines and better understanding of their roles in creating genetic diversity and distinct paratope surfaces of these non-human species have led to the development of several promising antibody-discovery platform technologies (Feng et al., 2019; Gjetting et al., 2019; Könitzer et al., 2017; Muyldermans, 2013; Muyldermans and Smider, 2016). In contrast, for human, it was generally held that such genetic and structural diversity did not exist. This consequently thwarted the development of potential human antibody therapeutics that could capitalize on non-canonical cysteines in human CDR-H3s. With this new information, the multifaceted roles of cysteines in the conformational stabilization of peptides and proteins (Góngora-Benítez et al., 2014) may be applicable to human antibodies with disulfide-bonded CDR-H3s. Intra-CDR-H3 disulfide bridges in humanized antibodies isolated from chicken showed normal expression and stability similar to those from fully human antibodies in clinical development (Gjetting et al., 2019). In addition, aggregation-resistant human VHs selected by an in vitro method were found to have intra- and inter-CDR-H3 disulfide bonds (Arbabi-Ghahroudi et al., 2009). More importantly, human antibodies containing disulfide-bonded CDR-H3s can form pre-configured, rigid structures that may not incur a large entropic penalty for interacting with protein antigens (Goldenzweig and Fleishman, 2018). Therefore, one can envisage, using newly found knowledge from this analysis, the design of antibodies, or human antibody libraries, with long disulfide-bonded CDR-H3s to bind recessed or concave epitopes. Furthermore, human antibodies containing multiple disulfide bridges and ultra-long CDR-H3s identified in this study could mimic some disulfide-rich ligands, receptors, and ion channel inhibitors (Góngora-Benítez et al., 2014; Osbourn, 1997). Moreover, it is important to appreciate that the redox potential and metal binding capacity of cysteine motifs (Miseta and Csutora, 2000) in CDR-H3s could make them suitable for catalytic functions (Chmura et al., 2001; Pollack et al., 1989), because it is being realized that protective and pathogenic catalytic antibodies occur naturally (Bowen et al., 2017). Another possible role for cysteine-containing CDR-H3s may be to form a covalent antigen-antibody complex to serve immunoregulatory function (Taylor et al., 1979). A similar idea had been previously proposed for αβ T cell receptors (TCRs) containing a central CDR3 cysteine, whereby an inter-TCR disulfide bond between central CDR3 cysteines or a disulfide bond between the CDR3 cysteine and a cysteine in the peptide-major histocompatibility complex (MHC) is hypothesized to induce strong TCR signaling (Wirasinha et al., 2018). Finally, some cysteine motifs in CDR-H3s of human antibody sequences with certain aa lengths and compositions may form predictable, canonical structures that might help define new H3 rules (Shirai et al., 1996). Overall, these results provide the fundamental framework for understanding the role of non-canonical cysteines in shaping the complex paratope diversity in human antibody VH repertoires. They can serve as a guiding resource to enable novel designs of human antibody libraries and in silico disulfide-engineered antibodies and ultimately lead to the discovery and development of a new class of human antibodies.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Deposited Data | ||
| Dataset A | GitHub | http://www.github.com/briney/grp_paper |
| Dataset B | Dryad | https://datadryad.org/stash/dataset/doi:10.5061/dryad.35ks2 |
| Software and Algorithms | ||
| JMP 14.2.0 | SAS Institute Inc. | https://www.jmp.com/en_us/home.html |
| MOE 2018.0101 | Chemical Computing Group | https://www.chemcomp.com/ |
| CLC Main Workbench 8.1 | QIAGEN | https://www.qiagen.com/us/ |
| WebLogo server | University of California, Berkeley | https://weblogo.berkeley.edu/ |
| Protein Data Bank | Research Collaboratory for Structural Bioinformatics PDB | http://www.rcsb.org/ |
| IgBLAST | The National Center for Biotechnology Information | https://www.ncbi.nlm.nih.gov/igblast/ |
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to lead contact, Dr. Ponraj Prabakaran (prabakaran.ponraj@sanofi.com).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
This study did not generate any unique datasets or code.
Experimental Model and Subject Details
NGS datasets of human antibody VH repertoires available from two different studies involving ten and three individuals were downloaded at GitHub (http://www.github.com/briney/grp_paper) and Dryad (https://datadryad.org/stash/dataset/doi:10.5061/dryad.35ks2) respectively. These refer to datasets A and B, respectively, from the ten and three individuals (Tables S1 and S2). To analyze the dataset A, all unique in-frame VH amino acid sequences with annotated IGHV, IGHD and IGHJ germline gene families, CDR-H3s and isotype information were extracted using UNIX scripts. All IGHV, IGHD and IGHJ germline gene families in the dataset A were previously annotated using Abstar (version 0.3.3, https://github.com/briney/abstar/releases) and deposited into GitHub public repository. All IGHD gene segments in a total of 106,521,023 sequences were annotated except for 88,526 (dataset A). This is probably because inferring the D gene usages could be impossible or misleading under certain circumstances such as for shorter IGHD segments (ex. 3 aas) and ambiguities existing due to junctional modification, D-D fusion and multiple SHM events. The CDR-H3 aa lengths as well as cysteine numbers in CDR-H3s and VHs were further calculated from the extracted data. Human IGHD germline segments encoding non-canonical cysteines and IGHD segments expressed in human VH repertoire were curated (Table S3A). The total numbers of CDR-H3s and those containing non-canonical cysteines involving different IGHD gene segments were calculated (Table S3B). The IGHD germline segments encoding CC, CX3C, CX4C motifs for the potential creation of four-cysteine motifs due to possible V(DD)J recombinations were inferred (Figure S7A). JMP SQL was used to retrieve the selected CDR-H3s with D-D fusions and shown with WebLogos (Figure S7B). It should be noted that annotated CSV and JSON files as downloaded from GitHub contained the consensus sequences from nearly 3 billion VHs. However, most of the clusters contained only a single sequence and many of the consensus sequences could be identified in the raw datasets.
Method Details
Computational analysis and identification of non-canonical cysteine motifs
The extracted information from the dataset A containing CDR-H3 sequences with non-canonical cysteines, CDR-H3 lengths, cysteine numbers, and names of IGHV, IGHD and IGHJ germline gene families were imported into JMP to create Data Tables, enabling the analysis of large datasets. For analysis of dataset B, productive and unique CDR-H3 amino acid sequences were extracted from both memory and naive B cell receptors for which cysteine numbers were calculated and imported into JMP. Frequency distributions for the numbers of non-canonical cysteines in datasets A and B were calculated using JMP and shown in Tables S1 and S2 respectively. The bivariate normal density analysis for IGHV germline genes and cysteine numbers for different antibody isotypes from the dataset A was performed with 90% coverage using JMP Query Builder (Figure S1). Specific SQL queries were created and executed for generating data needed for analyzing the frequency distribution of cysteine numbers in CDR-H3s (Figure 1A) and VHs (Figures 2A–2H) as well as CDR-H3 AA lengths (Figure 1B). CDR-H3s were grouped into 3 different length categories such as average (up to 15 aa), long (16-25 aa) and ultra-long (26 – 39 aa) for analyzing the influence of non-canonical cysteines. Frequency plots were made of CDR-H3 length categories using the JMP and calculated the percentages of all CDR-H3s as well as those that contain non-canonical cysteine only for datasets A and B (Figure S2). We created a dataset of 313,492 VHs which had a single cysteine in CDR-H3s and contained single cysteines elsewhere in their VHs. We used scripting and JMP analysis for identifying the cysteine containing motifs in CDR-H1, FR2, CDR-H2 and FR3 and used WebLogos for depicting the locations of non-canonical cysteines (Figure 2I). To further analyze possible cysteine containing motifs and their unique patterns in CDR-H3s, we searched for all tandem cysteines, either interspersed with other amino acids or contiguous, as occurred in the dataset A by using JMP scripts (Figures 1C and 1D). The distance between two cysteines in terms of AA length (Xn) for all observed CXnC motifs in CDR-H3s were calculated. A contour plot, having the x axis with number of AAs and the y axis with number of motifs on a log10 scale, was generated using JMP. Mapping of CXnC motifs based on known functional information from the PDB was carried out (Figure 3A). WebLogos were created and stacked to depict diverse CXnC motifs with a range of Xn values observed in dataset A (Figure 3B). To study the diversity of CX4C motifs, all tetrapeptides in between the two cysteines occurring in the CDR-H3s, CX4C, were extracted by using JMP SQL query. Frequencies of tetrapeptides and position-specific AA composition were calculated, and the results were visualized with a Treemap chart and a histogram respectively (Figures 4A and 4B). Top ranking unique CX4C motifs in CDR-H3s that occurred at least more than 1000 times were selected and drawn with a Treemap chart using JMP (Figure S3). Atomic coordinates were downloaded for antibodies containing non-canonical cysteines and their complexes from the PDB for further analysis and visualizations (Figures S4 and S5). The frequency distributions for number of aas that separate, and flank cysteines were calculated and visualized using boxplots using JMP graph builder (Figure S6). The potential D-D fusions in CDR-H3s were queried using SQL by selecting germline-encoded non-canonical cysteine motifs and illustrated with sequence logos (Figure S7).
Structural analysis of CDR-H3 disulfide motifs
The MOE Antibody Database as implemented in version 2018.01 was used to identify the 3D structures of antibodies that contain disulfide motifs in their CDR-H3s. By selecting the MOE internal database of Antibody Project Search panel and CDR_H3 sequence along with expression, $CDR_H3 and prosite “C,” we obtained a list of all PDB codes with other information such as resolution, species and CDR lengths for structurally characterized antibodies containing cysteine motifs in CDR-H3s. We further selected antibodies of human origin possessing the intra-disulfide bonded CDR-H3s in the scFv and Fab formats. The germline IGHV and IGLV/IGLK gene families were annotated using the NCBI IgBLAST. Structural analysis of antibodies and complexes was performed to examine conformations of CDR-H3s, particularly, cysteine motifs, and the role of CDR-H3s in antigen-antibody interactions using MOE and PyMOL. The CDR-H3s bearing CCX5CX4C motifs identified in 46 antibodies from this analysis were aligned and the circular phylogram was constructed using Neighbor-Joining method within the CLC Main Workbench.
Quantification and Statistical Analysis
JMP (version 14.2.0, SAS Institute Inc., Cary, NC) statistical software was used for data analysis, statistical calculations and generating all plots. Ribbon diagrams of antibodies and their complexes were made using PyMOL Molecular Graphics System (version 2.2.3 Schrödinger, LLC). Analysis of antibody structural database, as built from the Protein Data Bank (https://www.rcsb.org), was performed using MOE (Version 2018.01, Chemical Computing Group). Phylogenetic tree was generated with CLC Main Workbench (version 8.1). Sequence logos for cysteine containing CDR-H3s were generated by WebLogo server (version 2.8.2, https://weblogo.berkeley.edu/logo.cgi).
Acknowledgments
We thank Sarah Tao, Carlos Garcia-Echeverria, Brian Mackness, and Anusuya Ramasubramanian for their contributions to improving the manuscript. We thank Maria Wendt and Katarina Radosevic for their comments on this work. We acknowledge the significant contributions of researchers whose work formed the basis for this expert analysis.
Author Contributions
P.P. and P.S.C. performed background research and designed the study. P.P. collected data, wrote scripts, and performed all computational analysis. P.S.C. helped formulate the hypothesis, data analysis, and interpretation. P.P. and P.S.C. discussed the results and drafted the manuscript.
Declaration of Interests
The authors are employees of Sanofi.
Published: June 30, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2020.107831.
Contributor Information
Ponraj Prabakaran, Email: prabakaran.ponraj@sanofi.com.
Partha S. Chowdhury, Email: partha.chowdhury@sanofi.com.
Supplemental Information
References
- Almagro J.C., Raghunathan G., Beil E., Janecki D.J., Chen Q., Dinh T., LaCombe A., Connor J., Ware M., Kim P.H. Characterization of a high-affinity human antibody with a disulfide bridge in the third complementarity-determining region of the heavy chain. J. Mol. Recognit. 2012;25:125–135. doi: 10.1002/jmr.1168. [DOI] [PubMed] [Google Scholar]
- Arbabi-Ghahroudi M., To R., Gaudette N., Hirama T., Ding W., MacKenzie R., Tanha J. Aggregation-resistant VHs selected by in vitro evolution tend to have disulfide-bonded loops and acidic isoelectric points. Protein Eng. Des. Sel. 2009;22:59–66. doi: 10.1093/protein/gzn071. [DOI] [PubMed] [Google Scholar]
- Benichou J., Glanville J., Prak E.T.L., Azran R., Kuo T.C., Pons J., Desmarais C., Tsaban L., Louzoun Y. The restricted DH gene reading frame usage in the expressed human antibody repertoire is selected based upon its amino acid content. J. Immunol. 2013;190:5567–5577. doi: 10.4049/jimmunol.1201929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen A., Wear M., Casadevall A. Antibody-Mediated Catalysis in Infection and Immunity. Infect. Immun. 2017;85:e00202-17. doi: 10.1128/IAI.00202-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S., Milstein C. Origin of antibody variation. Nature. 1966;211:242–243. doi: 10.1038/211242a0. [DOI] [PubMed] [Google Scholar]
- Briney B.S., Willis J.R., Hicar M.D., Thomas J.W., 2nd, Crowe J.E., Jr. Frequency and genetic characterization of V(DD)J recombinants in the human peripheral blood antibody repertoire. Immunology. 2012;137:56–64. doi: 10.1111/j.1365-2567.2012.03605.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briney B., Inderbitzin A., Joyce C., Burton D.R. Commonality despite exceptional diversity in the baseline human antibody repertoire. Nature. 2019;566:393–397. doi: 10.1038/s41586-019-0879-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carugo O., Cemazar M., Zahariev S., Hudáky I., Gáspári Z., Perczel A., Pongor S. Vicinal disulfide turns. Protein Eng. 2003;16:637–639. doi: 10.1093/protein/gzg088. [DOI] [PubMed] [Google Scholar]
- Chen L., Duan Y., Benatuil L., Stine W.B. Analysis of 5518 unique, productively rearranged human VH3-23∗01 gene sequences reveals CDR-H3 length-dependent usage of the IGHD2 gene family. Protein Eng. Des. Sel. 2017;30:603–609. doi: 10.1093/protein/gzx027. [DOI] [PubMed] [Google Scholar]
- Chmura A.J., Orton M.S., Meares C.F. Antibodies with infinite affinity. Proc. Natl. Acad. Sci. USA. 2001;98:8480–8484. doi: 10.1073/pnas.151260298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conroy P.J., Law R.H., Caradoc-Davies T.T., Whisstock J.C. Antibodies: From novel repertoires to defining and refining the structure of biologically important targets. Methods. 2017;116:12–22. doi: 10.1016/j.ymeth.2017.01.003. [DOI] [PubMed] [Google Scholar]
- de Araujo A.D., Herzig V., Windley M.J., Dziemborowicz S., Mobli M., Nicholson G.M., Alewood P.F., King G.F. Do vicinal disulfide bridges mediate functionally important redox transformations in proteins? Antioxid. Redox Signal. 2013;19:1976–1980. doi: 10.1089/ars.2013.5365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Brevern A.G. Extension of the classical classification of β-turns. Sci. Rep. 2016;6:33191. doi: 10.1038/srep33191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de los Rios M., Criscitiello M.F., Smider V.V. Structural and genetic diversity in antibody repertoires from diverse species. Curr. Opin. Struct. Biol. 2015;33:27–41. doi: 10.1016/j.sbi.2015.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeWitt W.S., Lindau P., Snyder T.M., Sherwood A.M., Vignali M., Carlson C.S., Greenberg P.D., Duerkopp N., Emerson R.O., Robins H.S. A Public Database of Memory and Naive B-Cell Receptor Sequences. PLoS ONE. 2016;11:e0160853. doi: 10.1371/journal.pone.0160853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J., Finn J.A., Larsen P.A., Smith T.P.L., Crowe J.E., Jr. Structural Diversity of Ultralong CDRH3s in Seven Bovine Antibody Heavy Chains. Front. Immunol. 2019;10:558. doi: 10.3389/fimmu.2019.00558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doria-Rose N.A., Schramm C.A., Gorman J., Moore P.L., Bhiman J.N., DeKosky B.J., Ernandes M.J., Georgiev I.S., Kim H.J., Pancera M., NISC Comparative Sequencing Program Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies. Nature. 2014;509:55–62. doi: 10.1038/nature13036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng M., Bian H., Wu X., Fu T., Fu Y., Hong J., Fleming B.D., Flajnik M.F., Ho M. Construction and next-generation sequencing analysis of a large phage-displayed VNAR single-domain antibody library from six naive nurse sharks. Antib. Ther. 2019;2:1–11. doi: 10.1093/abt/tby011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlay W.J., Almagro J.C. Natural and man-made V-gene repertoires for antibody discovery. Front. Immunol. 2012;3:342. doi: 10.3389/fimmu.2012.00342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flyak A.I., Ruiz S., Colbert M.D., Luong T., Crowe J.E., Jr., Bailey J.R., Bjorkman P.J. HCV Broadly Neutralizing Antibodies Use a CDRH3 Disulfide Motif to Recognize an E2 Glycoprotein Site that Can Be Targeted for Vaccine Design. Cell Host Microbe. 2018;24:703–716.e3. doi: 10.1016/j.chom.2018.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frangione B., Milstein C., Pink J.R. Structural studies of immunoglobulin G. Nature. 1969;221:145–148. doi: 10.1038/221145a0. [DOI] [PubMed] [Google Scholar]
- Gjetting T., Gad M., Fröhlich C., Lindsted T., Melander M.C., Bhatia V.K., Grandal M.M., Dietrich N., Uhlenbrock F., Galler G.R. Sym021, a promising anti-PD1 clinical candidate antibody derived from a new chicken antibody discovery platform. MAbs. 2019;11:666–680. doi: 10.1080/19420862.2019.1596514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldenzweig A., Fleishman S.J. Principles of Protein Stability and Their Application in Computational Design. Annu. Rev. Biochem. 2018;87:105–129. doi: 10.1146/annurev-biochem-062917-012102. [DOI] [PubMed] [Google Scholar]
- Góngora-Benítez M., Tulla-Puche J., Albericio F. Multifaceted roles of disulfide bonds. Peptides as therapeutics. Chem. Rev. 2014;114:901–926. doi: 10.1021/cr400031z. [DOI] [PubMed] [Google Scholar]
- Gunasekaran K., Ramakrishnan C., Balaram P. Beta-hairpins in proteins revisited: lessons for de novo design. Protein Eng. 1997;10:1131–1141. doi: 10.1093/protein/10.10.1131. [DOI] [PubMed] [Google Scholar]
- Guo Y., Chen K., Kwong P.D., Shapiro L., Sheng Z. cAb-Rep: A Database of Curated Antibody Repertoires for Exploring Antibody Diversity and Predicting Antibody Prevalence. Front. Immunol. 2019;10:2365. doi: 10.3389/fimmu.2019.02365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutzeit C., Chen K., Cerutti A. The enigmatic function of IgD: some answers at last. Eur. J. Immunol. 2018;48:1101–1113. doi: 10.1002/eji.201646547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haakenson J.K., Deiss T.C., Warner G.F., Mwangi W., Criscitiello M.F., Smider V.V. A Broad Role for Cysteines in Bovine Antibody Diversity. Immunohorizons. 2019;3:478–487. doi: 10.4049/immunohorizons.1900058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmsen M.M., Ruuls R.C., Nijman I.J., Niewold T.A., Frenken L.G., de Geus B. Llama heavy-chain V regions consist of at least four distinct subfamilies revealing novel sequence features. Mol. Immunol. 2000;37:579–590. doi: 10.1016/s0161-5890(00)00081-x. [DOI] [PubMed] [Google Scholar]
- Haynes B.F., Kelsoe G., Harrison S.C., Kepler T.B. B-cell-lineage immunogen design in vaccine development with HIV-1 as a case study. Nat. Biotechnol. 2012;30:423–433. doi: 10.1038/nbt.2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong L., Giang E., Nieusma T., Kadam R.U., Cogburn K.E., Hua Y., Dai X., Stanfield R.L., Burton D.R., Ward A.B. Hepatitis C virus E2 envelope glycoprotein core structure. Science. 2013;342:1090–1094. doi: 10.1126/science.1243876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Könitzer J.D., Pramanick S., Pan Q., Augustin R., Bandholtz S., Harriman W., Izquierdo S. Generation of a highly diverse panel of antagonistic chicken monoclonal antibodies against the GIP receptor. MAbs. 2017;9:536–549. doi: 10.1080/19420862.2016.1276683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee P.S., Ohshima N., Stanfield R.L., Yu W., Iba Y., Okuno Y., Kurosawa Y., Wilson I.A. Receptor mimicry by antibody F045-092 facilitates universal binding to the H3 subtype of influenza virus. Nat. Commun. 2014;5:3614. doi: 10.1038/ncomms4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Klenow L., Coyle E.M., Golding H., Khurana S. Protective antigenic sites in respiratory syncytial virus G attachment protein outside the central conserved and cysteine noose domains. PLoS Pathog. 2018;14:e1007262. doi: 10.1371/journal.ppat.1007262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefranc M.P., Giudicelli V., Ginestoux C., Bodmer J., Müller W., Bontrop R., Lemaitre M., Malik A., Barbié V., Chaume D. IMGT, the international ImMunoGeneTics database. Nucleic Acids Res. 1999;27:209–212. doi: 10.1093/nar/27.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miseta A., Csutora P. Relationship between the occurrence of cysteine in proteins and the complexity of organisms. Mol. Biol. Evol. 2000;17:1232–1239. doi: 10.1093/oxfordjournals.molbev.a026406. [DOI] [PubMed] [Google Scholar]
- Muyldermans S. Nanobodies: natural single-domain antibodies. Annu. Rev. Biochem. 2013;82:775–797. doi: 10.1146/annurev-biochem-063011-092449. [DOI] [PubMed] [Google Scholar]
- Muyldermans S., Smider V.V. Distinct antibody species: structural differences creating therapeutic opportunities. Curr. Opin. Immunol. 2016;40:7–13. doi: 10.1016/j.coi.2016.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muyldermans S., Atarhouch T., Saldanha J., Barbosa J.A., Hamers R. Sequence and structure of VH domain from naturally occurring camel heavy chain immunoglobulins lacking light chains. Protein Eng. 1994;7:1129–1135. doi: 10.1093/protein/7.9.1129. [DOI] [PubMed] [Google Scholar]
- Ohm-Laursen L., Nielsen M., Larsen S.R., Barington T. No evidence for the use of DIR, D-D fusions, chromosome 15 open reading frames or VH replacement in the peripheral repertoire was found on application of an improved algorithm, JointML, to 6329 human immunoglobulin H rearrangements. Immunology. 2006;119:265–277. doi: 10.1111/j.1365-2567.2006.02431.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osbourn K.J. 1997. Cysteine noose antibody libraries, means for their production and uses thereof. International patent application publication WO/1999/023222, filed October 30, 1998, and published May 14, 1999. [Google Scholar]
- Pollack S.J., Nakayama G.R., Schultz P.G. Design of catalytic antibodies. Methods Enzymol. 1989;178:551–568. doi: 10.1016/0076-6879(89)78039-3. [DOI] [PubMed] [Google Scholar]
- Richardson J.S., Videau L.L., Williams C.J., Richardson D.C. Broad Analysis of Vicinal Disulfides: Occurrences, Conformations with Cis or with Trans Peptides, and Functional Roles Including Sugar Binding. J. Mol. Biol. 2017;429:1321–1335. doi: 10.1016/j.jmb.2017.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini S.S., Allore B., Jacobs R.M., Kaushik A. Exceptionally long CDR3H region with multiple cysteine residues in functional bovine IgM antibodies. Eur. J. Immunol. 1999;29:2420–2426. doi: 10.1002/(SICI)1521-4141(199908)29:08<2420::AID-IMMU2420>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- Shirai H., Kidera A., Nakamura H. Structural classification of CDR-H3 in antibodies. FEBS Lett. 1996;399:1–8. doi: 10.1016/s0014-5793(96)01252-5. [DOI] [PubMed] [Google Scholar]
- Soto C., Bombardi R.G., Branchizio A., Kose N., Matta P., Sevy A.M., Sinkovits R.S., Gilchuk P., Finn J.A., Crowe J.E., Jr. High frequency of shared clonotypes in human B cell receptor repertoires. Nature. 2019;566:398–402. doi: 10.1038/s41586-019-0934-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanfield R.L., Dooley H., Flajnik M.F., Wilson I.A. Crystal structure of a shark single-domain antibody V region in complex with lysozyme. Science. 2004;305:1770–1773. doi: 10.1126/science.1101148. [DOI] [PubMed] [Google Scholar]
- Sui J., Hwang W.C., Perez S., Wei G., Aird D., Chen L.M., Santelli E., Stec B., Cadwell G., Ali M. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat. Struct. Mol. Biol. 2009;16:265–273. doi: 10.1038/nsmb.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor R.B., Tite J.P., Manzo C. Immunoregulatory effects of a covalent antigen-antibody complex. Nature. 1979;281:488–490. doi: 10.1038/281488a0. [DOI] [PubMed] [Google Scholar]
- Thompson A.J., Lester H.A., Lummis S.C. The structural basis of function in Cys-loop receptors. Q. Rev. Biophys. 2010;43:449–499. doi: 10.1017/S0033583510000168. [DOI] [PubMed] [Google Scholar]
- Tonegawa S. Somatic generation of antibody diversity. Nature. 1983;302:575–581. doi: 10.1038/302575a0. [DOI] [PubMed] [Google Scholar]
- Venkatachalam C.M. Stereochemical criteria for polypeptides and proteins. V. Conformation of a system of three linked peptide units. Biopolymers. 1968;6:1425–1436. doi: 10.1002/bip.1968.360061006. [DOI] [PubMed] [Google Scholar]
- Wang F., Ekiert D.C., Ahmad I., Yu W., Zhang Y., Bazirgan O., Torkamani A., Raudsepp T., Mwangi W., Criscitiello M.F. Reshaping antibody diversity. Cell. 2013;153:1379–1393. doi: 10.1016/j.cell.2013.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirasinha R.C., Singh M., Archer S.K., Chan A., Harrison P.F., Goodnow C.C., Daley S.R. αβ T-cell receptors with a central CDR3 cysteine are enriched in CD8αα intraepithelial lymphocytes and their thymic precursors. Immunol. Cell Biol. 2018;96:553–561. doi: 10.1111/imcb.12047. [DOI] [PubMed] [Google Scholar]
- Wu L., Oficjalska K., Lambert M., Fennell B.J., Darmanin-Sheehan A., Ní Shúilleabháin D., Autin B., Cummins E., Tchistiakova L., Bloom L. Fundamental characteristics of the immunoglobulin VH repertoire of chickens in comparison with those of humans, mice, and camelids. J. Immunol. 2012;188:322–333. doi: 10.4049/jimmunol.1102466. [DOI] [PubMed] [Google Scholar]
- Wu X., Zhang Z., Schramm C.A., Joyce M.G., Kwon Y.D., Zhou T., Sheng Z., Zhang B., O’Dell S., McKee K., NISC Comparative Sequencing Program Maturation and Diversity of the VRC01-Antibody Lineage over 15 Years of Chronic HIV-1 Infection. Cell. 2015;161:470–485. doi: 10.1016/j.cell.2015.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying T., Prabakaran P., Du L., Shi W., Feng Y., Wang Y., Wang L., Li W., Jiang S., Dimitrov D.S., Zhou T. Junctional and allele-specific residues are critical for MERS-CoV neutralization by an exceptionally potent germline-like antibody. Nat. Commun. 2015;6:8223. doi: 10.1038/ncomms9223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemlin M., Klinger M., Link J., Zemlin C., Bauer K., Engler J.A., Schroeder H.W., Jr., Kirkham P.M. Expressed murine and human CDR-H3 intervals of equal length exhibit distinct repertoires that differ in their amino acid composition and predicted range of structures. J. Mol. Biol. 2003;334:733–749. doi: 10.1016/j.jmb.2003.10.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate any unique datasets or code.