Abstract
Purpose
The purpose of this study was to investigate the microbiome in the meibum, conjunctival sac, and eyelid skin in young and elderly healthy subjects, and analyze the effect that age, sex, and region have on microbiome composition.
Methods
This study involved 36 healthy subjects (young-age subjects: 9 men/9 women, age range: 20–35 years; elderly age subjects: 9 men/9 women, age range: 60–70 years). In all subjects, lower-eyelid meibum, lower conjunctival sac, and lower-eyelid skin specimens were collected from one eye, and then stored at –20°C. Taxonomic composition of the microbiome was obtained via 16S rRNA gene sequencing, and then analyzed.
Results
The meibum microbiome showed a high α-diversity (within-community diversity), particularly in the young subjects. However, in approximately 30% of the elderly subjects, a low-diversity microbiome dominated by Corynebacterium sp. or Neisseriaceae was observed. In the young subjects, the microbiome of the meibum resembled that of the conjunctival-sac, yet in the elderly subjects, the microbiome of the conjunctival-sac became more similar to that of the eyelid skin. The eyelid-skin microbiome was relatively simple, and was typically dominated by Propionibacterium acnes in the young subjects, or by Corynebacterium sp. or Neisseriaceae in the elderly subjects. In both age groups, no significant difference was seen between the men and women in regard to the meibum, conjunctival-sac, and eyelid-skin microbiome.
Conclusions
Our findings confirmed that the meibum of healthy adult-age subjects harbors highly diverse microbiota, and revealed that the meibum microbiome, especially the decrease of its diversity, alters with aging and may affect the homeostasis of the ocular surface.
Keywords: microbiome, meibomian gland, meibum, conjunctiva, eyelid, Propionibacterium acnes, coagulase-negative Staphylococcus, Staphylococcus epidermidis, Corynebacterium sp., meibomian gland dysfunction, meibomitis-related keratoconjunctivitis (MRKC), ocular surface
In humans, the overall condition of the meibomian glands reportedly has a strong influence on the health of the ocular surface, as well as related diseases.1 Meibomian glands are large modified sebaceous glands embedded in the tarsal plates that open on the skin of the eyelid just anterior to the mucocutaneous junction and secrete lipids (i.e. so-called “meibum”),2 onto the outer-most layer of the tear film.3 The secreted meibum plays an important role in the health of the ocular surface, as it prevents tear evaporation, stabilizes tear-film lubrication during the blinking process,4 and forms an optically smooth ocular surface, all of which help to assist in maintaining excellent visual acuity.5 Tear fluid is known to have antibacterial components, such as lactoferrin,6 IgA,7,8 and defensin,7 among others. Recently, it has been reported that meibum also has bactericidal effects and protects the ocular surface from microorganisms.9 Hence, meibomian gland dysfunction (MGD), a disorder in which both the quality and quantity of the meibum changes, can lead to tear-film instability combined with evaporative dry eye3,10–12 that can impair general visual acuity and alter the microbiota on the ocular surface. Due to the findings in the recent TFOS DEWS II report,13 as well as the findings in other studies,14,15 a general consensus has been reached that the ocular surface is a paucibacterial microbiome, yet is not sterile.
To date, there have been only a few studies focusing on meibum microbiota, and in all of those previous reports, conventional culture analysis was used. Meibomian glands were initially thought to be sterile.16 Scobee was the first to report that S. aureus is present in the meibomian gland.17 That report was followed by the findings in the Dougherty and McCulley study,18 in which the same species of bacteria (i.e. coagulase-negative Staphylococcus sp., Corynebacterium sp., and Propionibacterium acnes [P. acnes, recently renamed Cutibacterium acnes19]) as that in the lid margin were reportedly cultured from approximately 50% of the freshly expressed meibum of a normal subject. Thus, their findings indicated that commensal bacteria exist in the meibomian glands, yet did not indicate that the infection occurs in the meibomian glands. Recently, Zhang et al. confirmed that the predominant species isolated from the conjunctiva and meibomian gland secretion were S. epidermidis (aerobes) and P. acnes (anaerobes), and they reportedly discovered more complex bacterial flora in the patients with MGD than in the controls.20 Furthermore, meibomitis, an inflammatory form of MGD, is thought to be caused by bacterial ingrowth, and studies have reported the importance of using a systemic antimicrobial treatment to effectively eliminate the ocular surface inflammation in meibomitis cases.1,21–23
Since 2001, and especially over the past decade, a number of articles have been published regarding the analysis of the conjunctival microbiome via the use of 16S rRNA gene sequencing.14,15,24–31 However, and to the best of our knowledge, no previous studies have reported analyzing the detailed microbiome of meibum via 16S rRNA gene sequencing,32,33 so it has yet to be elucidated. Although one published study did investigate the meibomian gland microbiome via 16S rRNA gene sequencing for the strain once isolated from the culture of the meibum, some of the dominant anaerobic commensal bacteria, such as P. acnes, were not detected in that meibum.32
Thus, the purpose of this present study was to perform a comprehensive, yet “first step,” analysis of the microbiome of human meibum, conjunctival sac, and eyelid skin via 16S rRNA gene sequencing in order to specifically establish the baseline data of meibum obtained from healthy subjects.
Methods
Subjects
The protocols used for the experiments in this study were approved by the Institutional Review Board of Kyoto Prefectural University of Medicine and the Kyoto City Hospital Organization, Kyoto, Japan, and in accordance with the tenets set forth in the Declaration of Helsinki, written informed consent was obtained from all subjects prior to their participation in the study.
This study involved 36 healthy volunteer subjects comprised of 9 young women (mean age: 25.9 ± 5.2 SD years), 9 young men (mean age: 31.8 ± 3.8 years), 9 elderly women (mean age: 64.0 ± 2.9 years), and 9 elderly men (mean age: 65.4 ± 2.6years), and all subjects were native Japanese (i.e. of Asian ethnicity). The subjects in the young female group were healthy premenopausal women with a regular 28 to 30-day menstrual cycle (duration: 6–7 days), and they were seen in the follicular phase (i.e. before ovulation). The following subjects in both age groups were excluded from the study: tobacco smokers, contact lens wearers, and subjects with any eye and/or systemic disease, or who were taking medication at the time of the study.
Sample Collection
From one eye in each subject, microbiota at the following three sites were collected via single-use, clean, and sterile 3-mm diameter cotton swabs (JCB Industry Ltd., Tokyo, Japan): 1) a lower-eyelid skin sample from 3 mm below the eyelash line, 2) a lower conjunctival-sac sample, and 3) a lower-eyelid meibum sample. The specimens were collected in the following order. First, the skin microbiota was collected by rubbing with a swab moistened with sterile TET buffer (10 mM Tris-Cl [pH 8.0], 1 mM EDTA, 0.1% Tween 20). Next, the conjunctival-sac microbiota was collected by rubbing with a dry swab. The meibum sample was then collected via strict adherence to the following precautions in order to avoid any possible bacterial contamination from surrounding tissues. First, the lid margin was sterilized by use of 10% povidone-iodine, cleaned with sterile saline applied to a swab, and then wiped with a dry swab. The meibum was then squeezed out of the eyelid margin by use of a Yoshitomi Meibomian Gland Compressor (T.M.I. Co., Ltd., Saitama, Japan) under a surgical microscope, collected by use of a Daviel cataract spoon, and transferred to a dry swab. Each of the microbiota-sample swabs was then cut, with the head of the swab then being placed into a DNase-free Eppendorf Tube (Thermo Fisher Scientific, Waltham, MA, USA) and immediately stored at –20°C for later analysis.
DNA Extraction and Sequencing
DNA was isolated from each obtained specimen using a DNeasy PowerSoil Kit (Qiagen, Venlo, The Netherlands) according to the manufacturer's instructions. The V1-V2 region of the 16S rRNA gene was then amplified by polymerase chain reaction (PCR) using the following primers; 27modF: 5′-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGGRGTTTGATYMTGGCTCAG-3′ and 338R: 5′- GTCTCGTGGGCTCGGAGATGTGTATAAGAG
ACAGTGCTGCCTCCCGTAGGAGT, where R indicates purine, M indicates A or C, and underlined resides correspond to Illumina (Illumina Inc., San Diego, CA, USA) adapters. Dual index barcodes were added to the obtained PCR amplicon. Finally, those barcoded libraries were equimolarly pooled and paired-end sequenced (2 × 301 bp) on a MiSeq (Illumina) using a MiSeq Reagent kit version 3 (Illumina) for 600 cycles.
Sequence Processing and Taxonomic Classification
Processing and analysis of the sequenced reads were conducted via the use of the QIIME analysis tool package version 1.9.0 (www.quiime.org).34 The acquired paired-end reds were merged into a single read by USEARCH version 8.0.1623 (www.drive5.com),35 and the adaptor sequences were trimmed by Cutadapt version 1.8.1 (www.cutadapt.readthedocs.io/en/v1.8.1/index.html).36 Next, high-quality reads that satisfy the following criteria were extracted using the USEARCH tool: (1) the expected number of errors in the read, which is calculated based on sequencing quality scores, does not exceed 0.5, (2) the length is 250 bp or longer, and (3) the sequence is not considered as a chimera of different species. Using the resultant high-quality reads, operational taxonomy units (OTUs) were generated by clustering the reads with 97% or higher similarity. The seed sequence of each OTU was then chosen and used for taxonomic classification by using UCLUST taxonomy assigner (www.drive5.com)35 and Greengenes reference sequence database (gg_13_8_otus; www.greengenes.secondgenome.com).37 Based on these data, the taxonomic composition of each microbiota was determined. The Shannon index and weighted UniFrac metrics38 were also calculated in QIIME with taxonomic abundance profiles at the species and OTU levels, respectively.
Data Analysis and Statistical Test
The statistical environment R39 was used for the principal coordinate analysis (PCoA) and statistical tests.
Results
The Microbiome of Healthy Subjects
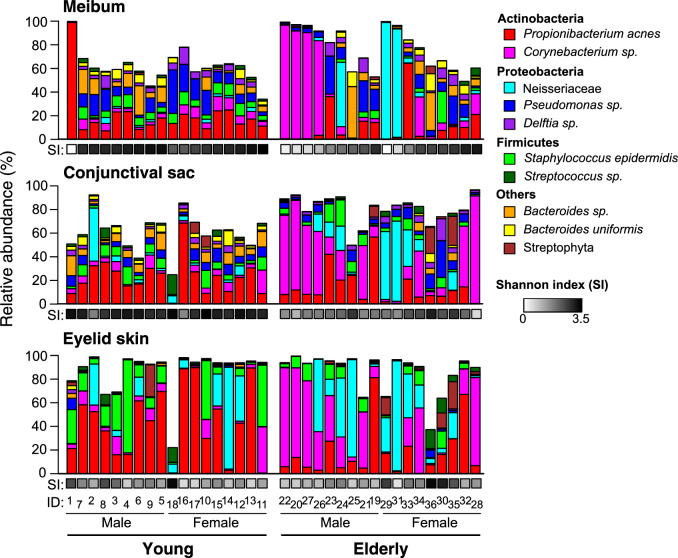
The bacterial composition of each sample revealed by 16S rRNA analysis is shown in Figure 1, and the major 10 taxa, whose mean relative abundance is >1%, are depicted. It has been reported that, typically, the microbiome of human skin is relatively simple, and is dominated by Propionibacterium and Staphylococcus genera.40 Consistently, the eyelid-skin samples collected from the young subjects showed low α-diversity, or within-community diversity, index (Shannon index), and contained P. acnes and Staphylococcus epidermidis (S. epidermidis) as the dominant species, whose mean relative abundance was 43% and 18%, respectively. Interestingly, the meibum and conjunctival-sac samples obtained from the young subjects exhibited microbiomes distinct from that of skin. They were characterized by high α-diversity index, and consisted of a large number of bacteria species. Typically, the most abundant species is P. acnes or Pseudomonas sp. for meibum and P. acnes for the conjunctival sac, whose mean relative abundance are 22% or less. We also found that the microbiomes in the elderly subjects were different from those in the young subjects. On the eyelid skin, the community diversity was once-again low, yet Corynebacterium sp. and the Neisseriaceae family (mostly a species that has no genus or species level affiliation) were the predominant taxa in many cases. The meibum and conjunctival sac microbiota were divided into high- and low-diversity types in the elderly subjects. In the low-diversity microbiota, either that of Corynebacterium sp. or the Neisseriaceae family was the most abundant taxon.
Figure 1.
The composition of bacterial microbiota in the samples collected at the meibum, conjunctival sac, and eyelid skin sites. The 10 most abundant taxa are shown. Shannon index (SI), a measure of species diversity in a sample, is indicated by the gray-scale color. The subjects were sorted by SI of the meibum sample in ascending order, within each group of young male, young female, elderly male, and elderly female subjects. ID, subject identification number.
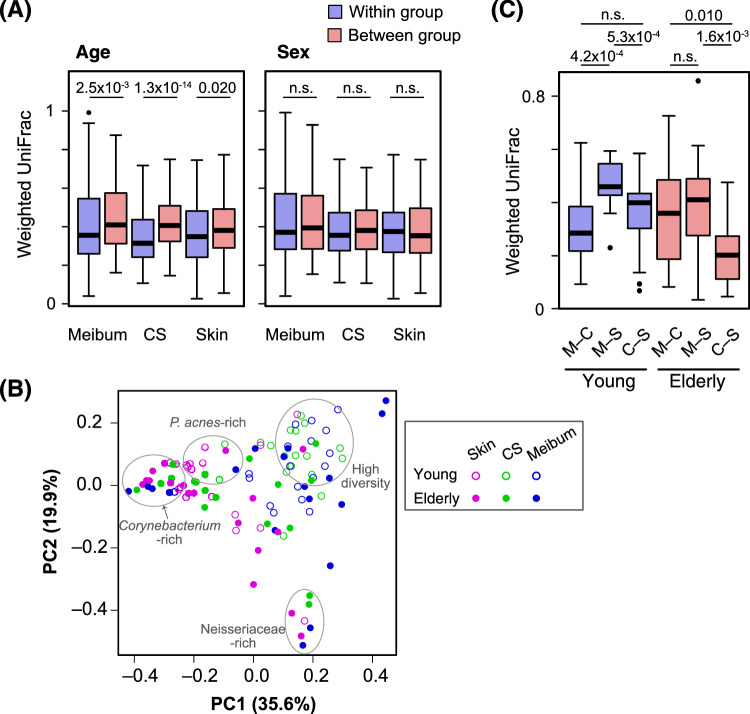
Next, we assessed how interpersonal variation is affected by the age and sex of the subjects. Weighted UniFrac38 was used to measure the distance, or the degree of dissimilarity, between a pair of samples. At all of the sample collection sites, the weighted UniFrac pairwise distances between different age groups showed significantly higher values than those within the same age group (Fig. 23A). In contrast, the sex of the subject was found to have little impact on bacterial communities, because no significant difference in weighted UniFrac pairwise distances was found between the within-group and between-group comparisons (Fig. 2A).
Figure 2.
Analyses based on the weighted UniFrac pairwise distance between samples. (A) Distribution of the pairwise distances. All pairs of the samples from either of meibum, conjunctival sac (CS), or eyelid skin (skin) were divided into “within the same age groups” and “between the different age groups” (left) or “within the same sex groups” and “between the different sex groups” (right). The weighted UniFrac distances of each set of pairs are depicted in the box plot. Blue, pairs within the same age/sex groups; red, pairs between the different age/sex groups. The numbers above the box plots are P values of the Mann-Whitney U test. n.s., not significant (or P > 0.05). (B) A principal coordinate analysis plot based on the weighted UniFrac distance. Samples representing each of the collection sites, and subject age are shown by distinct symbols. Clusters of the samples with characteristic microbiomes are indicated. (C) Weighted UniFrac distances of the samples collected at the three different sites of the same subject. M–C, sample pairs between the meibum and the CS; M–S, sample pairs between the meibum and the skin; C–S, sample pairs between the CS and the skin. The numbers above the box plots are P values of the Wilcoxon signed-rank test. n.s., not significant (or P > 0.05).
Figure 3.
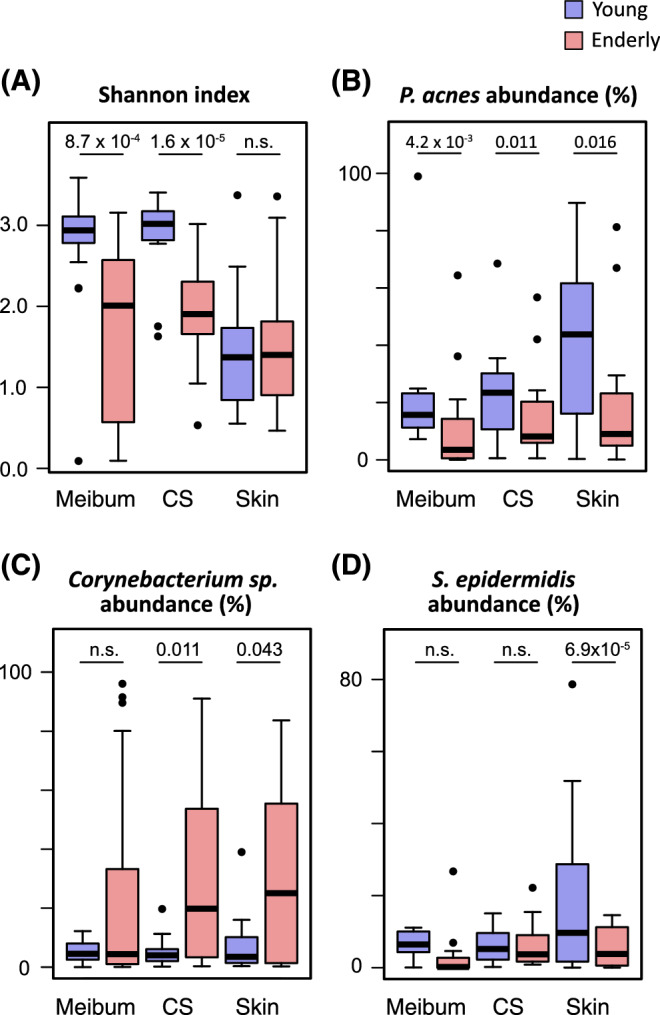
Microbiomes collected at either the meibum, conjunctival sac (CS), or eyelid skin (skin) sites were compared between the young subjects (blue) and the elderly subjects (red) in the Shannon index (A) and relative abundance of P. acnes (B), Corynebacterium sp. (C), or S. epidermidis (D). The numbers above the box plots are the P values of the Mann-Whitney U test. n.s., not significant (or P > 0.05).
The similarity of the individual samples was visualized via PCoA of the weighted UniFrac distance matrix (Fig. 2B). The meibum and conjunctival-sac samples were basically distributed separately from the skin samples, indicating a different microbiome structure between meibum/conjunctival-sac and skin. However, several samples derived from the meibum, the conjunctival sac, and the skin of the elderly subjects formed clusters, and those clusters corresponded to a Corynebacterium- or Neisseriaceae-rich microbiome. This finding is consistent with the emergence of the low-diversity meibum/conjunctival-sac microbiota in the elderly subjects (Fig. 1).
We also evaluated the similarity of bacterial community among the three sites of the same individual, using the weighted UniFrac distance (Fig. 2C). In the young subjects, the meibum and conjunctival-sac microbiomes were most similar (i.e. showed the smallest weighted UniFrac value), whereas the meibum and skin microbiomes were most distant. However, in the elderly subjects, the distance between the conjunctival sac and the skin is closer than that between the meibum and the conjunctival sac.
Our findings indicated that the meibum, conjunctival-sac, and eyelid-skin microbiota in the young subjects differed from that in the elderly subjects, and we validated that indication via the statistical test results. At the meibum and conjunctival-sac sites, the mean of the Shannon α-diversity index was greatly reduced upon aging, and this change was found to be statistically significant (Fig. 3A). The mean of the relative abundance of P. acnes was also significantly reduced in the elderly subjects at all of the tested sites (i.e. meibum, conjunctival sac, and eyelid skin) (Fig. 3B). Among the other abundant taxa, Corynebacterium sp. showed considerable increase in the mean relative abundance at the conjunctival sac and skin (Fig. 3C), whereas S. epidermidis showed significant decrease at the skin in the elderly subjects (Fig. 3D).
Discussion
To the best of our knowledge, this present study is the first to investigate meibum microbiomes in detail and analyze their differences in relation to aging and sex in comparison with that of the adjacent conjunctival-sac and eyelid-skin microbiomes via the use of the 16S rRNA gene sequencing method. Our findings revealed that the pure meibum in healthy subjects harbors highly diverse microbiota (i.e., >10 taxa) that usually cannot be recovered by a conventional culture technique. Furthermore, our findings show that there is a clear difference between young and elderly subjects in regard to the microbiome of the meibum, the conjunctival sac, and the eyelid skin.
In this present study, the Shannon α-diversity index of the meibum and conjunctival sac significantly decreased with aging. This finding is consistent with the previous reports in regard to the conjunctival microbiome,41,42 whereas another report showed no age difference29 or with a higher diversity in elderly subjects.43 One of the possible reasons for the difference in the Shannon index findings among the studies could be due to the different sequencing methods that were used (i.e. 16S genome sequence41,42 versus whole genome sequence43) and/or the study design (i.e. the subjects’ age setting and the sample collection of each group, as Ozkan et al.40 mentioned). Conversely, our eyelid-skin microbiome data was consistent with that of the skin microbiome, which is relatively simple and dominated by Propionibacterium and Staphylococcus genera.40 In fact, a low α-diversity with P. acnes and S. epidermidis was seen in the young subjects, and with Corynebacterium sp. or the Neisseriaceae family in the elderly subjects. Interestingly, no other changes in the microbiota associated with aging have been reported in other areas of the skin. Thus, whether or not this age-related change is a general feature of the eyelid needs further investigation. Corynebacterium sp. is reportedly noninflammatory under a steady-state condition; however, it could be an activator of skin immunity by expressing mycolic acid, which is required to mediate IL-23-dependent responses.44
Interestingly, our microbiota findings indicated that the conjunctival-sac microbiome is close to that of meibum; yet distinct from that of the eyelid skin in the young, whereas it becomes more similar to that of the eyelid skin in the elderly. This might be explained by the fact that human microbiota, although personalized, varies systemically across specific body environments (habitats) and time45 (i.e. bacteria in the eyelid skin was intrapersonally transplanted to the conjunctiva upon aging by rubbing the eyelids with the fingers).
It should be noted that the diversity of the meibum/conjunctival-sac microbiome in elderly subjects is very low, and is occupied by either Corynebacterium sp. or Neisseriaceae. Corynebacterium sp., which reportedly has been found in the conjunctiva via the use of 16S RNA gene sequencing methods,27–29 is a causative bacterium of conjunctivitis, keratitis, and others, and greater attention is now being paid to its resistance to antimicrobial agents in the elderly.46 In fact, alteration of the microbiome due to the aging process might be the underlying background of these diseases.
In contrast to the age difference of the subjects, our study found that sex had no major impact on the microbiome at all three sites, which is consistent with previous findings.29,41
It is widely known that there is evidence indicating that the loss of gut microbiota diversity can occur and does affect the aging process,47 and that dysbiosis often drives infection and inflammation.48 Compared with S. aureus and S. epidermidis, P. acnes and Corynebacterium sp. reportedly produce relatively low lipase activity.49 Thus, a dysbiosis of meibum and/or conjunctiva could degrade meibomian lipids, results in meibomitis,50 unstable tear film,51 and ocular surface inflammation.23 Antimicrobial agents, such as minocycline and azithromycin, may contribute to a recovery from a dysbiosis of the ocular surface. Therefore, any change of the microbiome in meibum and conjunctiva could lead to a shift in ocular surface health, thus resulting in a diseased condition.
It should be noted that this present study did have some limitations. First, it should be noted that one limitation was that there was a possible risk of contamination. However, in all subjects, the meibum samples were obtained after careful cleaning of the eyelid border where the meibomian gland orifices are located, and far enough away from the conjunctival sac as well as from the eyelid skin. Second, although the sample size was not large, the microbiome, which was recurrently detected in each region and in each age group, proved the clear age-related difference. However, in order for the results to be deemed more conclusive, further study involving a larger sample size is needed. Third, the ethnicity of the subjects might have had an impact on the divergence of the microbiome.
In conclusion, the findings in this study confirmed diverse microbiota in the meibum of healthy human subjects, and revealed that it alters with aging, especially in regard to the decrease of its diversity. In the young subjects, the microbiome of the meibum closely resembled that of the conjunctival sac, yet in the elderly subjects, the microbiome of the conjunctival sac was found to have become more similar to that of the eyelid skin. Our findings and observations may possibly indicate one of the causes in the change of the meibum lipid composition in elderly subjects and patients with MGD.
Acknowledgments
The authors thank John Bush for editing the manuscript. This research was supported in part by grants from the Japanese Ministry of Education, Culture, Sports, Science and Technology (No. 16K11295 and No. 19K09996). The authors alone are responsible for the content and writing of the paper.
Disclosure: T. Suzuki, None; T. Sutani, None; H. Nakai, None; K. Shirahige, None; S. Kinoshita, None
References
- 1. Suzuki T, Teramukai S, Kinoshita S. Meibomian glands and ocular surface inflammation. Ocul Surf. 2015; 13: 133–149. [DOI] [PubMed] [Google Scholar]
- 2. Nicolaides N, Kaitaranta JK, Rawdah TN, Macy JI, Boswell FM 3rd, Smith RE. Meibomian gland studies: comparison of steer and human lipids. Invest Ophthalmol Vis Sci. 1981; 20: 522–536. [PubMed] [Google Scholar]
- 3. Knop E, Knop N, Millar T, Obata H, Sullivan DA. The international workshop on meibomian gland dysfunction: report of the subcommittee on anatomy, physiology, and pathophysiology of the meibomian gland. Invest Ophthalmol Vis Sci. 2011; 52: 1938–1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bron AJ, Benjamin L, Snibson GR. Meibomian gland disease. Classification and grading of lid changes. Eye (Lond). 1991; 5(Pt 4): 395–411. [DOI] [PubMed] [Google Scholar]
- 5. Montes-Mico R. Role of the tear film in the optical quality of the human eye. J Cataract Refract Surg. 2007; 33: 1631–1635. [DOI] [PubMed] [Google Scholar]
- 6. Jensen OL, Gluud BS, Birgens HS. The concentration of lactoferrin in tears of normals and of diabetics. Acta Ophthalmol (Copenh). 1986; 64: 83–87. [DOI] [PubMed] [Google Scholar]
- 7. Vinding T, Eriksen JS, Nielsen NV. The concentration of lysozyme and secretory IgA in tears from healthy persons with and without contact lens use. Acta Ophthalmol (Copenh). 1987; 65: 23–26. [DOI] [PubMed] [Google Scholar]
- 8. Haynes RJ, Tighe PJ, Dua HS. Antimicrobial defensin peptides of the human ocular surface. Br J Ophthalmol. 1999; 83: 737–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mudgil P. Antimicrobial role of human meibomian lipids at the ocular surface. Invest Ophthalmol Vis Sci. 2014; 55: 7272–7277. [DOI] [PubMed] [Google Scholar]
- 10. Bron AJ, Tiffany JM. The contribution of meibomian disease to dry eye. Ocul Surf. 2004; 2: 149–165. [DOI] [PubMed] [Google Scholar]
- 11. Nelson JD, Shimazaki J, Benitez-del-Castillo JM, et al.. The international workshop on meibomian gland dysfunction: report of the definition and classification subcommittee. Invest Ophthalmol Vis Sci. 2011; 52: 1930–1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bron AJ, Tiffany JM, Gouveia SM, Yokoi N, Voon LW. Functional aspects of the tear film lipid layer. Exp Eye Res. 2004; 78: 347–360. [DOI] [PubMed] [Google Scholar]
- 13. Craig JP, Nichols KK, Akpek EK, et al.. TFOS DEWS II Definition and Classification Report. Ocul Surf. 2017; 15: 276–283. [DOI] [PubMed] [Google Scholar]
- 14. Huang Y, Yang B, Li W. Defining the normal core microbiome of conjunctival microbial communities. Clin Microbiol Infect. 2016; 22: 643.e647–643.e612. [DOI] [PubMed] [Google Scholar]
- 15. de Paiva CS, Jones DB, Stern ME, et al.. Altered mucosal microbiome diversity and disease severity in Sjogren Syndrome. Sci Rep. 2016; 6: 23561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gifford H. Notes of ophthalmic bacteriology, partly with reference to asepsis. Arch Ophthalmol. 1898; 27: 616–664. [Google Scholar]
- 17. Scobee RG. The role of the meibomian glands in recurrent conjunctivitis. Am J Ophthalmol. 1942; 25: 184–192. [Google Scholar]
- 18. Dougherty JM, McCulley JP. Comparative bacteriology of chronic blepharitis. Br J Ophthalmol. 1984; 68: 524–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Scholz CFP, Kilian M. “The natural history of cutaneous propionibacteria,and reclassification of selected species within the genus Propionibacterium to the proposed novel genera Acidipropionibacterium gen. nov., Cutibacterium gen. nov. and Pseudopropionibacterium gen. nov”. Int J Syst Evol Microbiol. 2016; 66: 4422–4432. [DOI] [PubMed] [Google Scholar]
- 20. Zhang SD, He JN, Niu TT, et al.. Bacteriological profile of ocular surface flora in meibomian gland dysfunction. Ocul Surf. 2017; 15: 242–247. [DOI] [PubMed] [Google Scholar]
- 21. Suzuki T, Mitsuishi Y, Sano Y, Yokoi N, Kinoshita S. Phlyctenular keratitis associated with meibomitis in young patients. Am J Ophthalmol. 2005; 140: 77–82. [DOI] [PubMed] [Google Scholar]
- 22. Suzuki T. Meibomitis-related keratoconjunctivitis: implications and clinical significance of meibomian gland inflammation. Cornea. 2012; 31 suppl 1: S41–S44. [DOI] [PubMed] [Google Scholar]
- 23. Suzuki T. Inflamed Obstructive Meibomian Gland Dysfunction Causes Ocular Surface Inflammation. Invest Ophthalmol Vis Sci. 2018; 59: DES94–DES101. [DOI] [PubMed] [Google Scholar]
- 24. Schabereiter-Gurtner C, Maca S, Rolleke S, et al.. 16S rDNA-based identification of bacteria from conjunctival swabs by PCR and DGGE fingerprinting. Invest Ophthalmol Vis Sci. 2001; 42: 1164–1171. [PubMed] [Google Scholar]
- 25. Graham JE, Moore JE, Jiru X, et al.. Ocular pathogen or commensal: a PCR-based study of surface bacterial flora in normal and dry eyes. Invest Ophthalmol Vis Sci. 2007; 48: 5616–5623. [DOI] [PubMed] [Google Scholar]
- 26. Dong Q, Brulc JM, Iovieno A, et al.. Diversity of bacteria at healthy human conjunctiva. Invest Ophthalmol Vis Sci. 2011; 52: 5408–5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shin H, Price K, Albert L, Dodick J, Park L, Dominguez-Bello MG. Changes in the Eye Microbiota Associated with Contact Lens Wearing. MBio. 2016; 7: e00198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Doan T, Akileswaran L, Andersen D, et al.. Paucibacterial Microbiome and Resident DNA Virome of the Healthy Conjunctiva. Invest Ophthalmol Vis Sci. 2016; 57: 5116–5126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ozkan J, Nielsen S, Diez-Vives C, Coroneo M, Thomas T, Willcox M. Temporal Stability and Composition of the Ocular Surface Microbiome. Sci Rep. 2017; 7: 9880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cavuoto KM, Mendez R, Miller D, Galor A, Banerjee S. Effect of clinical parameters on the ocular surface microbiome in children and adults. Clin Ophthalmol. 2018; 12: 1189–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ozkan J, Coroneo M, Willcox M, Wemheuer B, Thomas T. Identification and visualization of a distinct microbiome in ocular surface conjunctival tissue. Invest Ophthalmol Vis Sci. 2018; 59: 4268–4276. [DOI] [PubMed] [Google Scholar]
- 32. Jiang X, Deng A, Yang J, et al.. Pathogens in the Meibomian gland and conjunctival sac: microbiome of normal subjects and patients with Meibomian gland dysfunction. Infect Drug Resist. 2018; 11: 1729–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Asao K, Hashida N, Ando S, et al.. Conjunctival dysbiosis in mucosa-associated lymphoid tissue lymphoma. Sci Rep. 2019; 9: 8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Caporaso JG, Kuczynski J, Stombaugh J, et al.. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010; 7: 335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010; 26: 2460–2461. [DOI] [PubMed] [Google Scholar]
- 36. Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet Journal. 2011; 17: 10–12. [Google Scholar]
- 37. DeSantis TZ, Hugenholtz P, Larsen N, et al.. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006; 72: 5069–5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lozupone CA, Hamady M, Kelley ST, Knight R. Quantitative and qualitative beta diversity measures lead to different insights into factors that structure microbial communities. Appl Environ Microbiol. 2007; 73: 1576–1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ihaka R, Gentleman R. R: A language for data analysis and graphics. J Comp Graph Stat. 1996; 5: 299–314. [Google Scholar]
- 40. Grice EA, Kong HH, Conlan S, et al.. Topographical and temporal diversity of the human skin microbiome. Science. 2009; 324: 1190–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhou Y, Holland MJ, Makalo P, et al.. The conjunctival microbiome in health and trachomatous disease: a case control study. Genome Med. 2014; 6: 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ozkan J, Willcox M, Wemheuer B, Wilcsek G, Coroneo M, Thomas T. Biogeography of the human ocular microbiota. Ocul Surf. 2019; 17: 111–118. [DOI] [PubMed] [Google Scholar]
- 43. Wen X, Miao L, Deng Y, et al.. The influence of age and sex on ocular surface microbiota in healthy adults. Invest Ophthalmol Vis Sci. 2017; 58: 6030–6037. [DOI] [PubMed] [Google Scholar]
- 44. Ridaura VK, Bouladoux N, Claesen J, et al.. Contextual control of skin immunity and inflammation by Corynebacterium. J Exp Med. 2018; 215: 785–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. Bacterial community variation in human body habitats across space and time. Science. 2009; 326: 1694–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Deguchi H, Kitazawa K, Kayukawa K, et al.. The trend of resistance to antibiotics for ocular infection of Staphylococcus aureus, coagulase-negative staphylococci, and Corynebacterium compared with 10-years previous: a retrospective observational study. PLoS One. 2018; 13: e0203705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. O'Toole PW, Jeffery IB. Gut microbiota and aging. Science. 2015; 350: 1214–1215. [DOI] [PubMed] [Google Scholar]
- 48. Levy M, Kolodziejczyk AA, Thaiss CA, Elinav E. Dysbiosis and the immune system. Nat Rev Immunol. 2017; 17: 219–232. [DOI] [PubMed] [Google Scholar]
- 49. Flanagan JL, Khandekar N, Zhu H, et al.. Glycerol monolaurate inhibits lipase production by clinical ocular isolates without affecting bacterial cell viability. Invest Ophthalmol Vis Sci. 2016; 57: 544–550. [DOI] [PubMed] [Google Scholar]
- 50. McCulley JP, Dougherty JM, Deneau DG. Classification of chronic blepharitis. Ophthalmology. 1982; 89: 1173–1180. [DOI] [PubMed] [Google Scholar]
- 51. Arciniega JC, Nadji EJ, Butovich IA. Effects of free fatty acids on meibomian lipid films. Exp Eye Res. 2011; 93: 452–459. [DOI] [PMC free article] [PubMed] [Google Scholar]