Abstract
The decrease of pH level in the water affects animals living in aquatic habitat, such as crustaceans. The molecular mechanisms enabling these animals to survive this environmental stress remain unknown. To understand the modulatory function of neuropeptides in crustaceans when encountering drops in pH level, we developed and implemented a multifaceted mass spectrometric platform to investigate the global neuropeptide changes in response to water acidification in the Atlantic blue crab, Callinectes sapidus. Neural tissues were collected at different incubation periods to monitor dynamic changes of neuropeptides under different stress conditions occurring in the animal. Neuropeptide families were found to exhibit distinct expression patterns in different tissues and even each isoform had its specific response to the stress. Circulating fluid in the crabs (hemolymph) was also analyzed after 2-hour exposure to acidification condition, and together with results from tissue analysis, enabled the discovery of neuropeptides participating in the stress accommodation process as putative hormones. Two novel peptide sequences were detected in the hemolymph that appeared to be involved in the stress-related regulation in the crabs. The data have been deposited to the ProteomeXchange with identifier PXD013536.
Keywords: Atlantic blue crab, Callinectes sapidus, neuropeptide, hemolymph, acidification, isotopic reductive dimethylation, mass spectrometry, LC-ESI-MS, MALDI-MS
Graphical Abstract
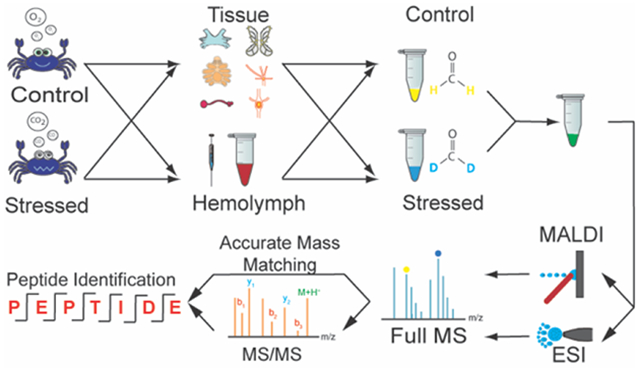
INTRODUCTION
Neuropeptides are the largest and most diverse group of endocrine signaling molecules in the nervous system. Recent studies have focused on characterization of neuropeptide signaling pathways1 and their regulatory roles in physiological processes, such as rhythmic motor pattern2,3, food intake4–6, molting7 and accommodating stress8–10. With a relatively simple, well-characterized nervous system, invertebrates have become a popular model system for neuropeptide-based studies11–16. The decapod crustacean nervous system consists of a central nervous system (including the brain), neuroendocrine system (e.g., pericardial organ and sinus glands) and stomatogastric nervous system (STNS), which contains 4 ganglia: paired commissural ganglia (CoG), unpaired stomatogastric ganglion (STG) and unpaired oesophageal ganglion (OG)17. Due to the crustacean open-circulatory system, all neuropeptide-rich tissues are bathed in fluid (i.e., hemolymph), functioning similarly as blood in mammals. Mature neuropeptides are secreted into the hemolymph17, after proteolytic processing from proneuropeptides, to achieve targeted modulation18. Thus, the study of neuropeptides in crustacean hemolymph may help to uncover their hormonal roles in regulating physiological processes, in addition to the tissue study 9,13,19.
As a species of marine animals, crustaceans are directly exposed to the water environment variation. Low pH stress is one of the most common stresses that crustaceans routinely experience in their habitats20. The global pH decreases in the seawater is mainly caused by the uptake of carbon dioxide (CO2) in the atmosphere. Usually, water holds more CO2 compared to oxygen (O2) due to chemical reactions21 which produce bicarbonate, carbonate and protons22, consequently causing acidification. Human activities have been considered to be the most significant contributor to CO2 release. As a principal sink of CO2, oceans have been acidified from approximately 8.25 to 8.1423 throughout this period, and their pH levels are now decreasing 50 times faster24 compared to the pre-industrial measurement25. It is predicted that by 2100, the pH levels of the oceans will decline by 0.15 to 0.31 units26, causing ecological and environmental problems 27–31.
Studies of a diverse range of crustacean species showed that acidified environments induce a decline in locomotive activities, decrease in growth rate, and dysregulation in gene expression32–38. The Atlantic blue crab, Callinectes sapidus, is a remarkably mobile crustacean whose foraging, reproducing, and survival from predators and stress are highly dependent on frequent locomotion. The animal has been found to get tired in low O2 (hypoxia) environments; however, the onset of fatigue was delayed when the animal was treated with additional CO2 at the same time39. Notably, the pH of the crustacean circulatory fluid (hemolymph) showed no distinguishable change from the passive in vitro response in the initial 2-3 hours40. The delay of CO2-induced response may be brought about by an increase in hemolymph HCO3- 40 or a CO2-specific effect on the blue crab’s hemocyanin, which exhibited increasing affinity to oxygen at low pH values, supporting locomotive ability39,41. Unfortunately, there is still no sufficient explanation of the molecular mechanism of the underlying changes and the regulation of physiological response to pH stress.
In this study, we performed quantitative investigation of neuropeptide changes in different neural tissues in Atlantic blue crab, Callinectes sapidus in response to low pH stress. Two complementary mass spectrometry (MS)-based methods, relying on either matrix-assisted laser desorption/ionization (MALDI) or electrospray ionization (ESI) coupled with liquid chromatography (LC) were both adopted and were evaluated by comparing the neuropeptide identifications and covered mass range. Crustacean neural tissues were collected at different time points (i.e., 2 hours (hrs) and 4 hrs) in order to monitor the dynamic changes of neuropeptides during development of the stress and the regulation of the adaptation process occurring in the nervous and neuroendocrine systems. In the circulating fluid, both known and newly discovered peptides were quantitatively measured to study their hormonal roles. This study provides a comprehensive analysis of crustacean neuropeptides in response to stress and will contribute to the future functional studies of neuropeptides in physiological regulation.
METHODS
Chemicals and Materials
Methanol (MeOH), acetonitrile (ACN), formic acid (FA), ammonium bicarbonate and glacial acetic acid were purchased from Fisher Scientific (Pittsburgh, PA). Borane pyridine, formaldehyde, and deuterium formaldehyde were from Sigma-Aldrich (St. Louis, MO). 2,5-dihydroxybenzoic acid (DHB) was obtained from Acros Organics (Morris Plains, NJ), and alpha-cyano-4-hydroxycinnamic acid (CHCA) was purchased from Sigma-Aldrich (St. Louis, MO). Acidified methanol was prepared as 90% methanol, 9% water and 1% glacial acetic acid (v/v/v). All water used in this study was doubly distilled on a Millipore filtration system (Burlington, MA) or Fisher HPLC grade.
Animals and Acidification Experiments
Female Atlantic blue crabs Callinectes sapidus were purchased from local markets (e.g., Midway Asian Market (Madison, WI) or the Louisiana Crawfish Company (Natchitoches, LA). All acquired crabs were maintained in recirculating artificial seawater, a salinity of 30 ppt, with a 12h:12h light/dark cycle, without food for at least one week in order to minimize the stress-related effects due to the transportation. Crabs at inactive conditions or with missing limbs were not eligible for the experiment. Crabs used in one experiment were selected from the same batch of purchased animals and were of similar body sizes, to ensure the baseline consistency in quantitative analysis (Figure S1). The selected crab was placed into a 10-gallon tank and was allowed to equilibrate at 18-20 °C for at least three days before experiments. Marine buffer purchased from Seachem Laboratories (Madison, GA) was added to avoid any deviation of initial pH level from 8.3, which is a normal pH for seawater. All housing parameters were the same between control and stressed groups. The acidified environment of the blue crabs’ native habitat is quite complicated20. Here in this study, the model was simplified and simulated by adding CO2 gas. A pH sensor (American Marine Pinpoint pH Monitor) and a dissolved oxygen (D.O.) sensor (American Marine Pinpoint Oxygen Monitor) were placed at the corner of the tank farthest from the CO2 gas tank tube. A moderate pH level (7.6-7.8) was achieved quickly by introducing CO2 via gas tank into the housing tank. The air saturation was at 50%−60%, which was considered as moderate hypoxic condition. It usually took a short time for the pH level to stabilize before the stress experiment started. D.O. level and pH level were monitored throughout the experiment. The crabs were kept in the acidified environment for the desired durations (2 or 4 hrs). A tarp was placed on the water surface before CO2 sparge in order to prevent water-air exchange for the experimental duration. Control and stressed crabs were cold-anesthetized on ice for 15-20 min followed by dissection, which usually took 45-50 min. Neuropeptide-rich organs, including brain, POs, SGs, TG, STG and OG, were dissected in chilled physiological saline (Composition: 440 mM NaCl, 11 mM KCl, 13 mM CaCl2, 26 mM MgCl2, 10 mM HEPES (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid) acid; pH 7.4, adjusted with NaOH). The dissection was carried out as described previously42. Hemolymph was collected by inserting a 25-gauge needle connected to a 1ml plastic syringe through the base of one of the legs. An aliquot of 0.75mL was withdrawn from the same crab before and after the low pH stress.
Tissue Extraction
Three animals’ tissues, either control or stressed, were pooled into one sample in order to minimize individual variability and manually homogenized using chilled acidified methanol. Undissolved large proteins were removed by centrifugation at 16,100 xg for 10 min. The pellet was washed twice with acidified methanol. Supernatants were combined, dried down and resuspended in 150 μl of 0.1% FA in water, followed by desalting using OMIX C18 Pipette Tips (Agilent Technologies, Santa Clara, CA). 150 μl 0.1% FA in 50% acetonitrile (v/v) was used to elute the peptides and the elution was dried down and subject to labeling steps.
Hemolymph Sample Preparation
0.75 mL of hemolymph was added to an equal volume of acidified methanol to extract peptides and precipitate large proteins. The samples were centrifuged at 16,100 xg for 10min. The pellet was washed twice by acidified methanol and the supernatants were combined, followed by ultrafiltration through 10kDa MWCO tube (rinsed by 0.1M NaOH and 50/50 MeOH/H2O, v/v) being centrifuged at 14,000 xg for 10min. Filtrate was concentrated to dryness using a SpeedVac concentrator (Thermo Fisher Scientific, Waltham, MA, USA) and was resuspended in 150 μl of 0.1% FA in water. Samples were then desalted by OMIX C18 pipette tips (Agilent Technologies, Santa Clara, CA) and eluted to 150 μl 0.1% FA in 50% acetonitrile (v/v). Elution was dried down and subject to labeling steps.
Reductive Dimethylation Labeling
Purified samples were dissolved into 10 μl 0.1% FA in water and labeled with formaldehyde-H2 (1% v/v) and formaldehyde-D2 (1% v/v), for control and stressed sample respectively. Borane pyridine (30mM) was added to the samples, which were then incubated for 15 min in a 37 °C water bath. Ammonium bicarbonate (100mM) was added to quench the reaction. For each biological replicate, one heavy labeled (stressed, formaldehyde-D2) sample and one light labeled (control, formaldehyde-H2) sample were mixed in equal volume, then dried down and re-dissolved in 15 μl 0.1% FA in water. A 10 μl aliquot was subjected to further desalting process using C18 Ziptip Pipette Tips (Merck Millipore Ltd, Tullagreen, Carrightwohill, Co. Cork, IRL) before being loaded to LC-ESI-MS, and the remaining 5 μl was subjected to MALDI-MS analysis.
Mass Spectrometry Analysis
For ESI-MS analysis, desalted sample in 10 μl 0.1% FA was subjected to online separation with a Waters nano-Acquity Ultra Performance LC system equipped with a self-packed column (150mm length of 1.7μm C18 with a 3μm C18 cap) connected to a Q-Exactive quadrupole Orbitrap mass spectrometer (Thermo Fisher Scientific, Bremen, Germany). A 120-min gradient was used at flow rate of 0.3μl/min, starting from 100% A (0.1% formic acid in water) and 0% B (0.1% formic acid in ACN), increasing to 10% B at 1 min, 35% B at 90 min, 95% B at 92 min, and remaining for 10 min , and then dropping back to 0% at 105 min. Typical mass spectrometry conditions were as listed: spray voltage, 2.1 kV; no sheath and auxiliary gas flow; heated capillary temperature, 275 °C. Data was collected in data-dependent mode, with top 15 abundant precursor ions selected for HCD fragmentation with the following settings: Full-MS, resolution, 70,000; AGC, 1e6; maximum injection time, 250ms; scan range, m/z 200-2000; dd-MS2, resolution, 17,500; AGC, 2e5; maximum injection time, 120ms; loop count, 15; isolation window, m/z 2.0; fixed first mass, m/z 100.0; normalized collision energy, 30.
For MALDI-MS, samples were separated into two fractions and spotted on MALDI target plate with DHB (2,5-dihydroxybenzoic acid, 150mg/ml, 50% MeOH/50% H2O/0.1% FA) and CHCA (α-cyano-4-hydroxycinnamic acid, 10mg/ml, 84% acetonitrile/ 13% ethanol/0.003% trifluoroacetic acid) before analysis by MALDI-LTQ-Orbitrap XL mass spectrometer (Thermo Scientific Bremen, Germany) at mass range of m/z 200-2000.
Data Processing
LC-ESI-MS raw data was analyzed by PEAKS Studio 7 (Bioinformatics Solution Inc., Waterloo, ON, Canada). Variable post-translational modifications (PTMs) were selected as C-terminal amidation, methionine oxidation, pyroglutamation, dehydration, dimethylation-H2 and dimethylation-D2. Parent mass error tolerance was 50ppm and fragment mass error tolerance was 0.02 Da. No enzyme cleavage was specified in de novo sequencing and database search. An in-house crustacean neuropeptide database was used. Quantitative analysis was conducted with a mass tolerance of 0.2 Da and retention time range of 1.0 min. Peptide spectrum matches (PSMs) with a −10logP value cutoff of 20 in PEAKS were considered for data filtration and validation.
MALDI-MS raw data was imported to a self-coded Java program to filter peak pairs that had mass increments of 28.0313 and 32.0564 Da from exact masses of NPs in database with mass error tolerance at ±5ppm. Peak intensities of heavy (stressed) and light (control) labeled samples were calculated for fold changes in neuropeptide expression.
In both instrument methods, 5 biological replicates were analyzed for the brain, PO, SG, TG, STG, OG at 2hr stress, brain, SG at 4hr stress. Four biological replicates were analyzed for the PO, TG and STG at 4hr stress and also hemolymph. Each biological replicate consisted of 3 technical replicates. Student’s t-test was applied (unpaired t-test for tissue analysis, paired t-test for hemolymph analysis) and a p-value smaller than 0.05 was considered to be a statistically significant change.
RESULTS AND DISCUSSION
Neuropeptidome analysis by MALDI-MS vs. ESI-MS
As an essential and indispensable analysis method for complex samples, mass spectrometry (MS) has been widely used in proteomics43–45, peptidomics46–48 and lipidomics49,50,51studies across species. Especially in the last several decades, the advent of ESI and MALDI have revolutionized the analysis of large biomolecules. MALDI is featured with relatively high tolerance to contaminants and salts, which allows for simple sample preparation, leading to less sample loss. It has been successfully applied to crustacean neuropeptide profiling52,53 and stress-induced response study9,11. However, MALDI mostly produces singly-charged ions, which often have limited fragmentation efficiency to generate sufficient sequence information. The simplified sample preparation of MALDI also leads to analyte suppression issues, which is detrimental to the detection of low level of endogenous neuropeptides in the crustacean54. ESI coupled with LC offers better separation followed by more efficient fragmentation due to the production of multiply-charged ions. In this study, we adopted both methods in order to facilitate a more comprehensive neuropeptidome analysis. Furthermore, results from MALDI-MS were collected using both CHCA and DHB as matrices. These two matrices yielded complementary identifications of neuropeptides, which allowed better coverage of the neuropeptidome. Also, the expression level changes of neuropeptides detected in both matrices exhibited high consistency (data not shown), supporting the reproducibility and reliability of the results.
To compare the coverage contributed by each ionization method, MALDI-MS and ESI-MS were both applied to analyze brain, PO, SG, TG, STG, and OG at 2hr (Supplemental Information, Tables S1–6) and 4hr time points (Supplemental Information, Tables S7–11). With singly-charged ions, MALDI usually performed better in the detection of neuropeptides at the low mass range, whereas ESI facilitated detection at a higher mass range due to its capacity for producing multi-charged ions. In the POs treated with 2hr stress (Figures 1a, b), the mass of neuropeptides detected by ESI-MS could be up to 3.8 kDa, whereas MALDI-MS covered mass range up to 2 kDa. Differences in mass distribution could be observed throughout the experiments. As an example, after 2hr stress, the molecular weights of neuropeptides in the TG (Figure 1c) detected by MALDI-MS showed predominant occupancy in low mass while ESI-MS contributed to the detection of a greater number of larger peptides.
Figure 1.
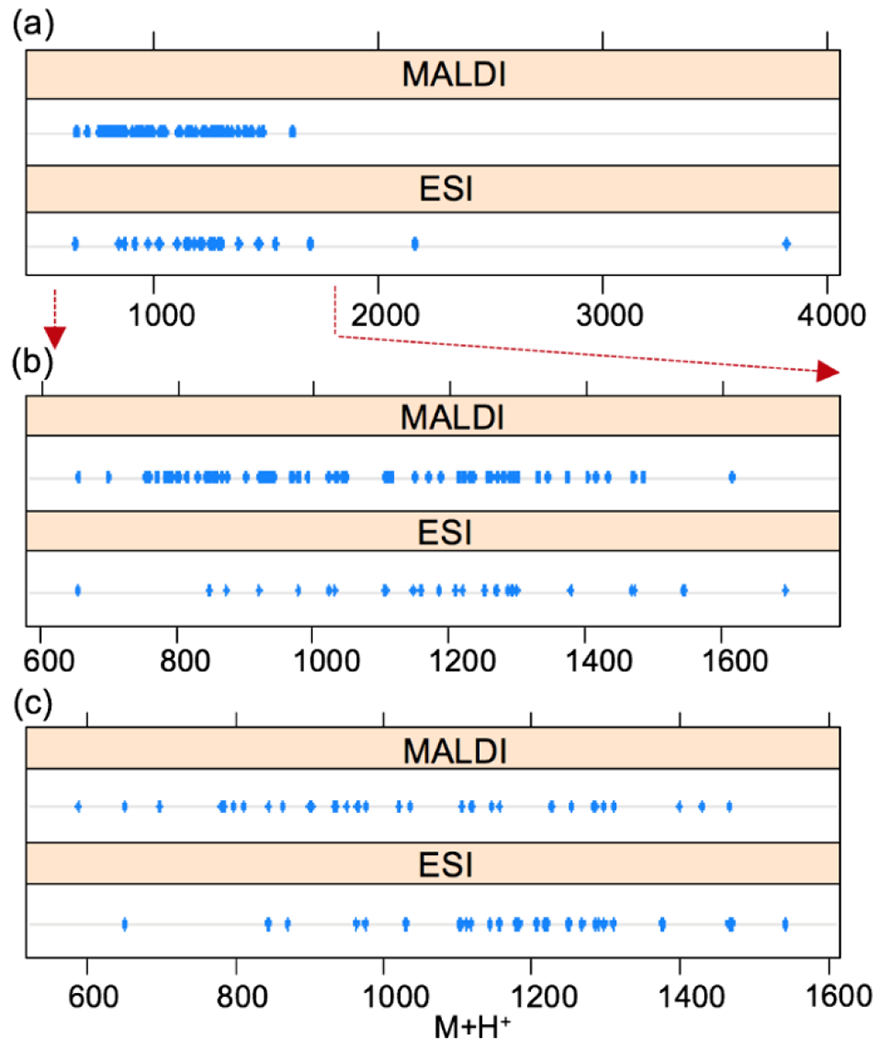
Mass distribution of neuropeptides detected in response to 2 hr acidification stimulus in the PO (a) and the TG (c). Low mass range of neuropeptides in the PO was enlarged (b).
The differences in distribution between MALDI- and ESI-MS were also found among neuropeptide families. After 2hr incubation in low pH environment, in the brain tissue, more neuropeptides from allatostatin B-type (AST-B) and tachykinin families were detected by ESI-MS (Figure 2b) than that by MALDI-MS (Figure 2a). The responses of several peptides from the crustacean hyperglycemic hormone precursor-related peptide (CPRP) family to the stress were found in the brain using MALDI-MS; nevertheless, none of them was detected in the brain using ESI-MS. Interestingly, more orcokinin isoforms at a higher mass range within their family were detected by ESI-MS than by MALDI-MS (Figure 2c). The reason for the differences in family distribution could be attributed by the fact that some peptides ionized better with the assistance of MALDI matrix, whereas some other peptides would generate multiply-charged ions more easily, resulting in preferential detection by either ionization method.
Figure 2.
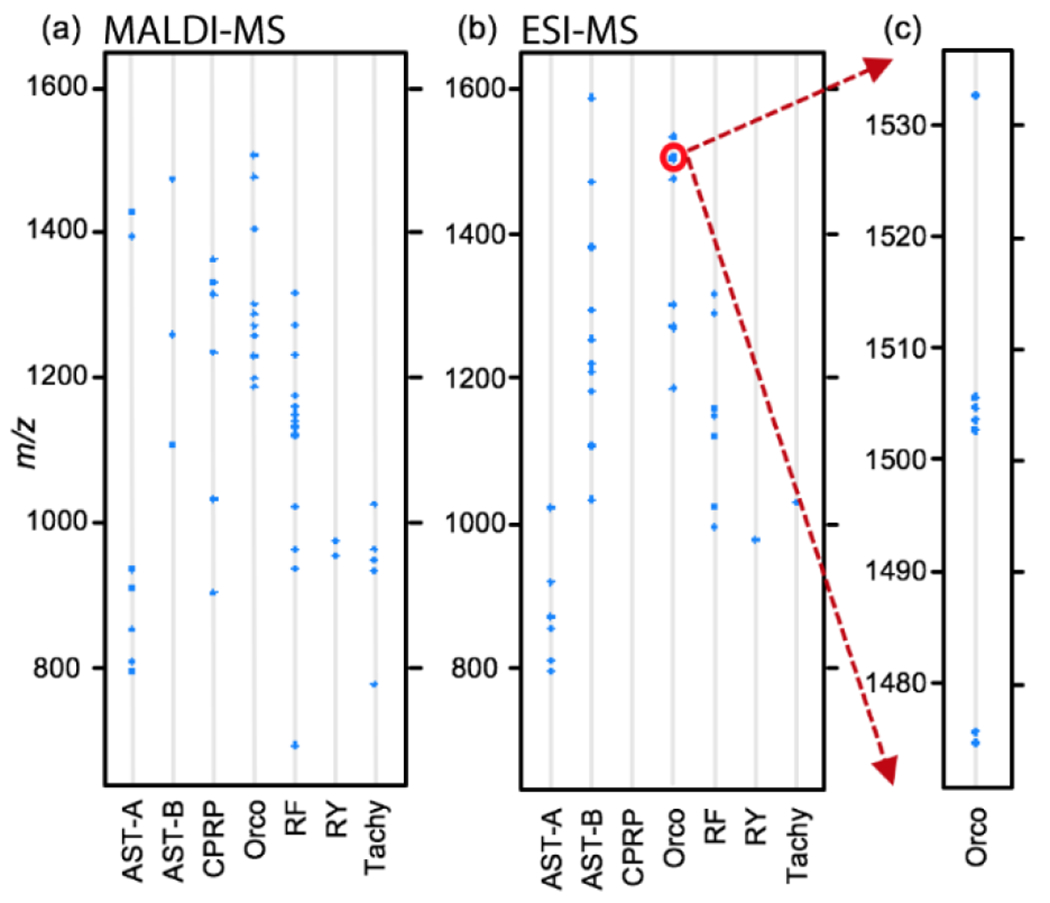
Neuropeptide families observed in the brain in response to 2 hr low pH stress using (a) MALDI-MS and (b) ESI-MS. Mass range between 1460-1540 in the orcokinin family detected by ESI-MS was zoomed in (c). AST-A, A-type allatostatins; AST-B, B-type allatostatins; Orco, Orcokinin; RF, RFamide; RY, RYamide; Tachy, Tachykinin.
As stated above, ESI and MALDI together contributed to a broadened coverage of the neuropeptidome in our study. Furthermore, consistency in expression level changes should be demonstrated in the overlaps of MALDI- and ESI-MS analysis. As shown in Figure 3, most neuropeptides that were detected in both MALDI and ESI exhibited similar trends in response to 2 hr acidification stress. However, several outliers were found, such as AST-B GNWNKFQGSWamide (m/z 1222.5752) in SG, AST-B AWSNLGQAWamide (m/z 1031.5057) and RFamide QDLDHVFLRFamide (m/z 1288.6797) in PO. The discrepancy may result from different data analysis methods applied to the two datasets. When identifying neuropeptides from MALDI-MS data using accurate mass matching, we searched through the in-house database which contained exact masses calculated by neuropeptide sequences (pyroglutamine (pQ) and C-terminus amidation were involved in calculation if indicated in sequences). Other PTMs (e.g., methionine oxidation and dehydration) were included in PEAKS search against ESI-MS data. Some neuropeptides with C-terminus amidation were also accompanied by their non-amidation isoforms. PEAKS applied statistical methods to determine the intensity ratio from multiple isoforms and charge states of a neuropeptide. This result suggests that peptides with different modifications may play distinct roles in physiological regulation.
Figure 3.
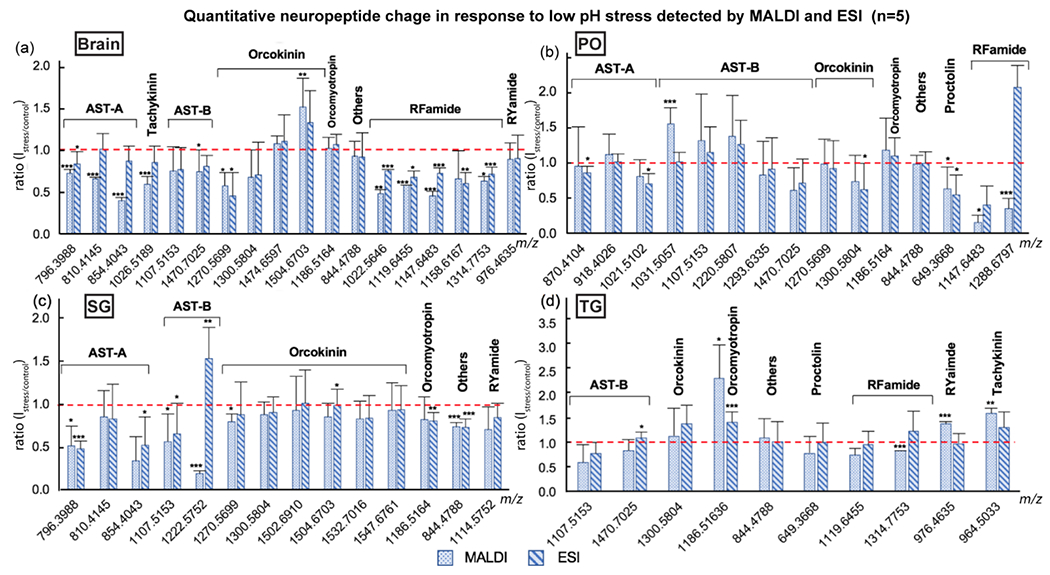
Neuropeptide changes in the (a) brain, (b) pericardial organ (PO), (c) sinus gland (SG) and (d) thoracic ganglia (TG) upon 2 hr acidified incubation (n=5), acquired by MALDI-MS and ESI-MS. Ratios were calculated by dividing the heavy-labeled groups (stressed, pH 7.6-7.8) by the light-labeled (control, pH 8.3). X-axis, exact mass of neuropeptides (M+H+); Y-axis, peak intensity ratio of stress over control; Error bar, standard deviation; Red dash line indicated no change in peptide levels. Student’s t-test was applied to evaluate the significance of neuropeptide level changes upon stress compared to control groups. *p<0.05; **p<0.005; ***p<0.001.
Neuropeptide changes in neuronal tissues upon 2hr water acidification stress
Neuropeptides can be classified into different families according to the common sequence motif they shared. Distinct motifs among neuropeptide families are expected to give rise to their different functional roles in physiological modulation14,15,55–58. In this study, several neuropeptide families were found, throughout the crustacean neuronal tissues, to respond to the low pH stress. RFamides in the brain (Figure 3a), proctolin in the PO (Figure 3b), and allatostatin A-type (AST-A) in the SG (Figure 3c) were all found to be down-regulated, while the expression levels of tachykinin and orcomyotropin in the TG (Figure 3d) were up-regulated. The diversity in the response further illustrated that each neuropeptide family contributed differently to the adaptation to the environmental stress.
We also discovered that isoforms in a neuropeptide family, although having similar sequences, may still differ from each other in their reactions to the stimulus. For example, in the brain, orcokinin NFDEIDRSGFA (m/z 1270.5699) decreased significantly (MALDI: s/c, 0.57; p, 0.0475; ESI: s/c, 0.45; p, 0.0240;) due to the acidification; however, another isoform NFDEIDRSSFGFA (m/z 1504.6703) exhibited significant increasing trend (MALDI: s/c, 1.50; p, 0.0042; ESI: s/c, 1.31;). In the PO, level of AST-B AGWSSMRGAWamide (m/z 1107.5153; MALDI: s/c, 1.30; ESI: s/c, 1.14;) and SGDWSSLRGAWamide (m/z 1220.5807; MALDI: s/c, 1.37; ESI: s/c, 1.26;) were elevated when the animal suffered from low pH stress. In contrast, the expression level of VPNDWAHFRGSWamide (m/z 1470.7025) was reduced (MALDI: s/c, 0.61; ESI: s/c, 0.7;). The disparity of response within the same neuropeptide family suggested that the modulatory function of a single isoform may differ from each other. Each isoform likely has its unique role in physiological regulation 59,60.
Among the neuronal tissues collected in this study, many peptides were found in common (Figure 4c). Interestingly, it was found that an identical neuropeptide may exhibit different expression trend in response to low pH stress in different tissues. Orcokinin NFDEIDRSSFGF (m/z 1433.6332) detected by MALDI-MS (Figure 4b), was significantly reduced in the SG (s/c, 0.64; p, 0.0085;) and OG (s/c, 0.35; p, 0.0083;), while its expression level was elevated by nearly one and half-fold in the TG (s/c, 1.45; p, 0.0262;). In the PO, this neuropeptide remained unaffected (s/c, 1.05; p, 0.7688;) by the acidification. Similar phenomenon was also observed in the ESI-MS studies (Figure 4a). The level of AST-B AWSNLGQAWamide (m/z 1031.5057) was reduced in the STG (s/c, 0.75; p, 0.0179;), increased in the SG (s/c, 1.55; p, 0.0505;), yet kept approximately the same in other tissues (brain: s/c, 0.91; p, 0.1918; PO: s/c, 1,01; p, 0.9879; TG: s/c, 0.96; p, 0.5494;). It was noted that neuropeptides detected in both STG and OG followed the same trend in response to the stress. The disparity and the consistency in neuropeptide expression in different tissues may reflect that, under environmental stress, the role of a neuropeptide could be excitatory or inhibitory depending on the location of the neuropeptide and the position of the tissue within the nervous system. POs release neuropeptides directly into the circulatory system in the region of heart; thus, the dynamic changes of neuropeptides in the PO in response to the stress may indicate their modulation on cardiac activity. The TG has extended nerve cords connecting to the limbs of crabs, therefore, it is reasonable to consider that the TG, to some degree, controls the movements of crustacean. It has been shown that blue crabs got fatigued in a shorter period and their pull force decreased faster in the addition of CO2 at 50% air saturation treatment when compared to 100% air saturation group39. Hence, neuropeptide changes in the TG may provide clues to unveil the physiological functions in locomotion control during the low pH stress. The STG contains the motor neurons which manage the movements of the muscles of pyloric regions of the stomach16. The OG contains descending modulatory neurons that can influence activity of the STG2. Accordingly, neuropeptides in the STG and the OG may act as modulators in pyloric rhythms61. The multiple directions of a single neuropeptide being regulated under environmental stress may elucidate the multi-functions and distinct roles of the neuropeptide in different neuronal regions.
Figure 4.
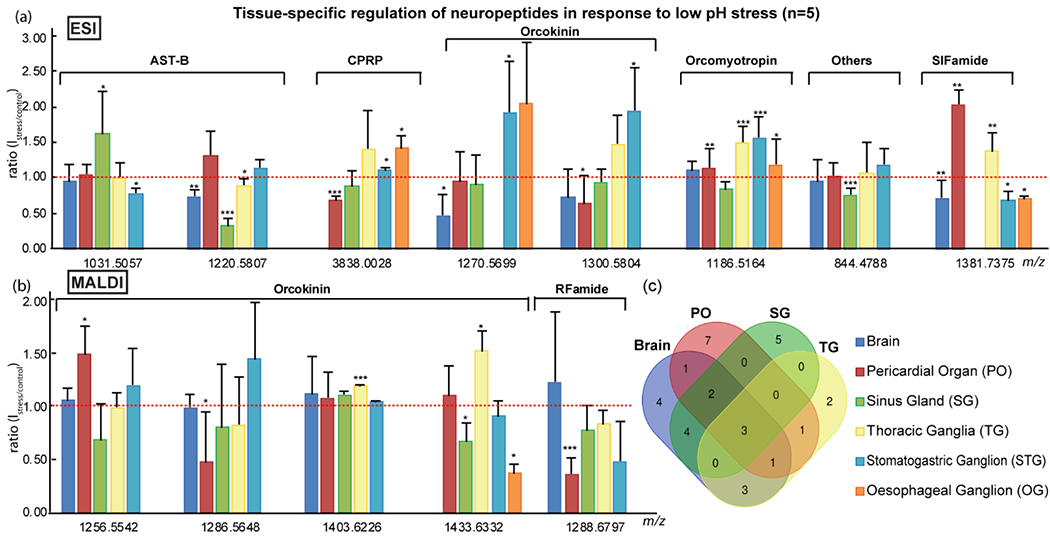
Expression level changes of single neuropeptide isoforms across various tissues under 2hr low pH stress (n=5) analyzed by ESI-MS (a) and MALDI-MS (b). Venn diagram (c) for the numbers of neuropeptides detected after 2 hr low pH stress in four major neural tissues (Brain, PO, SG and TG). Only neuropeptides identified by both MALDI- and ESI-MS were included in this graph. X-axis, exact mass of neuropeptides (M+H+); Y-axis, peak intensity ratio of stress over control; Error bar, standard deviation; Red dash line indicated no change in peptide levels. Student’s t-test was applied to evaluate the significance of neuropeptide level changes upon stress compared to control groups. *p<0.05; **p<0.005; ***p<0.001.
Another possible explanation to the distinct trends of identical neuropeptide in six different tissues could be that, in additional to their local functions, neuropeptides were also released from neurons and neuroendocrine tissues as circulating hormones55,62–64. For example, after 2hr incubation in acidified conditions, SIFamide GYRKPPFNGSIFamide (m/z 1381.7375) was measured to be significantly reduced in the brain (ESI: s/c, 0.69; p, 0.0037;), STG (ESI: s/c, 0.65; p, 0.0464;) and OG (ESI: s/c, 0.67; p, 0.0400;), meanwhile its expression level was up-regulated in the PO (ESI: s/c, 1.94; p, 0.0050;) and the TG (ESI: s/c, 1.31; p, 0.0030;). This may indicate that SIFamide was released from the brain, STG and OG, followed by transporting via circulating fluid, and exerted influence on the PO and TG. Neuropeptides in the brain were found to be unaffected by temperature elevation9. In this study, orcokinin NFDEIDRSGFG (m/z 1256.5542, s/c, 1.01; p, 0.9373;), NFDEIDRSSFG (m/z 1286.5648, s/c, 0.94; p, 0.4126;) and NFDEIDRSGFGF (m/z 1403.6226, s/c, 1.07; p, 0.8089;), detected by MALDI-MS, orcomyotropin FDAFTTGFGHS (m/z 1186.5164, MALDI: s/c, 1.01; p, 0.8771; ESI: s/c, 1.06; p, 0.1406;), and others HL/IGSL/IYRamide (m/z 844.4788, MALDI: s/c, 0.91; p, 0.1922; ESI: s/c, 0.91; p, 0.2409;), detected by both MALDI-MS and ESI-MS, remained constant after exposure to the low pH stress. While it might be tempting to conclude that neuropeptides in the brain were irrelevant to the adaptation of acidification, this may also suggest that the fluctuation of neuropeptide levels in the brain was balanced out by those in the peripheral neuronal organs. For example, orcokinin NFDEIDRSGFG (m/z 1256.5542) exhibited statistically significant increase in the PO (MALDI: s/c, 1.42; p, 0.0115;) and decrease in the SG (MALDI: s/c, 0.65;), respectively, while in the TG (MALDI: s/c, 0.95; p, 0.3165;) and STG (MALDI: s/c, 1.14; p, 0.5366;), the orcokinin level had minimal change. The opposite trends of this orcokinin isoform in surrounding tissues may contribute to the relatively stable expression level in the brain, since it acts as a central control of the entire nervous system.
Stress development analysis under 2hr and 4hr incubation
In order to investigate how the low pH stress developed in the crustacean and how the neuropeptides modulated the accommodation to the stimulus, we also collected tissue from crabs incubated for 4 hrs in an acidified environment. The neuropeptide expression changes were calculated against the control groups using the same method applied to 2 hr stress. The stress-to-control ratio of the same peptide was compared between 2 hr and 4 hr stress incubation (Figure 5a). Several neuropeptides, such as AST-B TSWGKFQGSWamide (m/z 1182.5691) in the TG, orcokinin NFDEIDRSGFGFV (m/z 1502.6910) in the STG, and SIFamide GYRKPPFNGSIFamide (m/z 1381.7375) in the TG all significantly increased at 2 hr measured point (s/c, 1.23; p, 0.0165; s/c, 1.21; p, 0.0059; s/c, 1.31; p, 0.0030, respectively) but dropped remarkably after 4hr incubation (s/c, 0.81; p, 0.0009; s/c, 0.40; p, 0.0032; s/c, 0.60; p, 0.0025, respectively). This observation may suggest that the regulation of these neuropeptides was different when the animals encountered with acute or long-term stress. The decrease of peptide expression level may indicate either degradation by protease within the tissue or released externally to other targets, being involved in physiological processes. Further studies concerning the circulating fluid will be needed to prove the secretion of peptides and the targeted function of these peptides as hormones. Similarly, AST-B NDWSKFGQSWamide (m/z 1253.5698) in the PO and orcokinin DFDEIDRSSFGFA (m/z 1505.6543) in the SG recovered to normal levels (s/c, 0.93; p, 0.2407; s/c, 0.92; p, 0.0625, respectively) by the end of the 4 hr stress duration after a dramatic change (s/c, 0.74; p, 0.0239; s/c, 0.82; p, 0.0120, respectively) during the halfway point. Neuropeptides were produced or used up to regulate the stress at the beginning. As the self-adjustment of the animal brought the physiological environment towards normal condition, the level of peptide also recovered close to normal level. Some other neuropeptides, like AST-B (m/z 1294.6175), Orcokinin (m/z 1270.5699) and RYamide (m/z 977.4476) in the PO, remained unaffected in the first two hours (s/c, 1.00; p, 0.7233; s/c, 0.91; p, 0.3444; s/c, 0.96; p, 0.1473, respectively) but deviated from normal level significantly (s/c, 0.73; p, 0.0248; s/c, 2.03; p, 0.0037; s/c, 0.71; p, 0.0460, respectively) by the end of the 4hr incubation, exhibiting that the initiation of stress-induced response of these neuropeptides was delayed for at least two hours. Studies on blue crab locomotion have revealed that the onset of fatigue would be delayed because of the additional CO2 to hypoxia induced by N2 only39. This could likely be explained by the CO2-specific effect on blue crab hemocyanin, the affinity to oxygen of which would largely increase to overcome the negative effect due to acidification 39,41. In another circulating fluid study, a lag for the initiation of any active response to the hemolymph pH change was also found when the blue crab was put in water bubbled with CO240. Here, the neuropeptidomic analysis provided deeper insight into the mechanisms of how crustaceans accommodated to low pH stress at the molecular level and facilitated functional discovery of neuropeptides.
Figure 5.
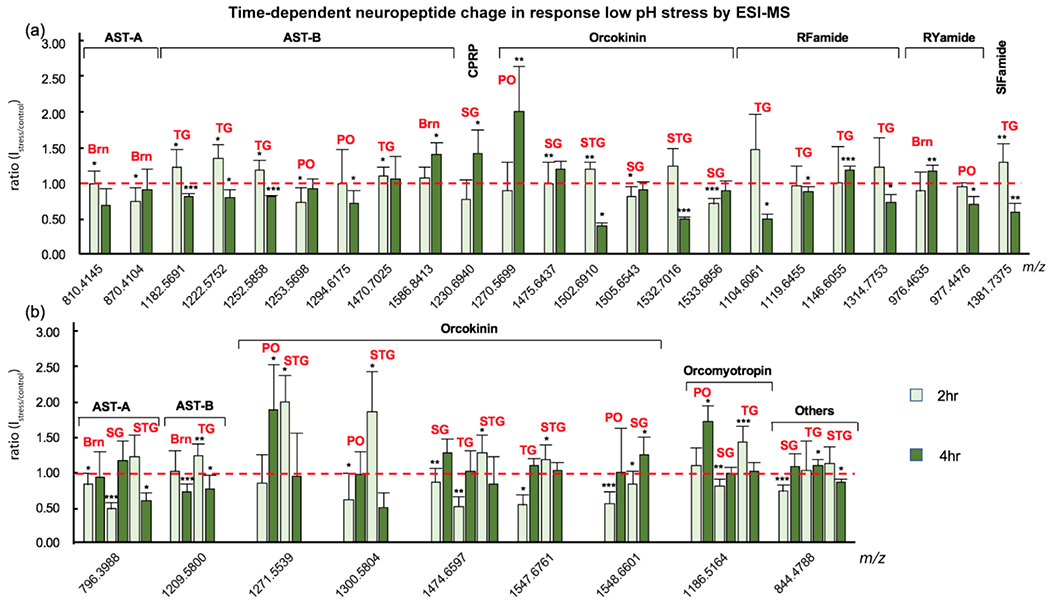
Neuropeptide changes identified by ESI-MS at different time points in a single tissue (a) indicating the stress-related function of the peptide and in multiple tissues (b), indicating the hormonal role of the peptide when pH declined. X-axis, exact mass of neuropeptides (M+H+); Y-axis, peak intensity ratio of stress over control; Error bar, standard deviation; Red dash line indicated no change in peptide levels. Student’s t-test was applied to evaluate the significance of neuropeptide level changes upon stress compared to control groups. *p<0.05; **p<0.005; ***p<0.001. Brn, brain.
Stress development was also monitored across different tissues (Figure 5b). Again, tissue-specific response was observed. For instance, at 2 hr time point, orcokinin DFDEIDRSGFA (m/z 1271.5539) was at standard level in the PO (s/c, 0.84; p, 0.1274;), but was up-regulated in the STG (s/c, 2.00; p, 0.0156;). When it came to the end of 4 hr stress treatment, the level of this orcokinin peptide in the STG returned to normal level (s/c, 0.94; p, 0.5291;) while elevated in the PO significantly (s/c, 1.88; p, 0.0078;). The transmission pathway could likely be described as: when the animal initially encountered the stimulus, the orcokinin was generated and accumulated in the STG; with time development and self-accommodation, the neuropeptide was released from the STG and transported to the PO, indicating the cardiac activity as a modulation target in this period.
Quantification of neuropeptide levels in circulating system
In the crustacean, hemolymph fills the hemocoel of the body and surrounds all the organs and cells. When encountered with acidification, several physiological aspects are affected by the stress, such as immune system and acid-base balance65–67. Neuropeptides are secreted upon stimuli from neuronal tissue into hemolymph and exert influence on target tissue, which provides critical information for mapping the peptidomic regulatory pathway. In the ESI-MS analysis (Table 1), CPRP RSAEGLGRMGRLLASLKSDTVTPLRGFEGETGHPLE (m/z 3838.0028) remained at constant level in the hemolymph after 2 hr exposure to low pH stress, while it was either up or down-regulated in the PO, SG and TG. Donors and receptors delivered and received similar amount of peptides, yielding a relatively stable concentration in the hemolymph during stress incubation. CPRP is a circulating hormone with unknown function63. Our study may reveal potential regulatory role of this CPRP in pH stress adaptation. Similarly, proctolin RYLPT (m/z 649.3668) was found to have stable expression level in the brain and the TG, while decreased level in the PO, SG and STG, yet dramatically elevated in the OG. Its relative concentration in the hemolymph was increased significantly, suggesting its secretion into the circulating system and possible hormonal role.
Table 1.
Neuropeptide changes in the hemolymph in response to 2hr low pH stress.
| Source | Family | Mass | Sequence | Aves/c | Stds/c | Tissue | Aves/c | Stds/c |
|---|---|---|---|---|---|---|---|---|
| ESI | AST-B | 997.5003 | AWSALHGAWamide | 0.76 | 0.22 | - | - | - |
| Others | 1254.8144 | AVLLPKKTEKK | 0.29*** | 0.06 | - | - | - | |
| CPRP | 3838.0028 | RSAEGLGRMGRLL ASLKSDTVTPLRG FEGETGHPLE |
1.00 | 0.32 | PO*** SG TG |
0.656 0.840 1.345 |
0.05 0.21 0.53 |
|
| MALDI | Proctolin | 649.3668 | RYLPT | 2.02 | 0.66 | Brain PO* SG TG STG OG** |
1.07 0.63 0.68 0.79 0.89 4.75 |
0.12 0.31 0.37 0.34 0.16 0.76 |
| RFamide | 966.5268 | DRNFLRFamide | 0.04* | 0.02 | PO TG |
0.38 0.69 |
0.38 0.38 |
|
| SIFamide | 1161.6527 | RKPPFNGSIFamide | 0.03*** | 0.02 | - | - | - | |
| Actin | 1209.6837 | LRVAPEESPVL | 0.61 | 0.31 | - | - | - | |
| RFamide | 1253.6538 | HDSPHVFLRFamide | 0.12* | 0.04 | - | - | - |
p<0.05;
p<0.005;
p<0.001
In general, we expect to observe opposite trends of neuropeptide level changes within the neuroendocrine or secretory tissue and circulating fluid assuming hormonal role of secreted peptides. However, RFamide DRNFLRFamide (m/z 966.5268) detected by MALDI-MS and others such as AVLLPKKTEKK (m/z 1254.8144) observed via ESI-MS were measured to be down-regulated in both tissue and hemolymph. One possible explanation would be that normally the source of peptides in hemolymph was the secretion from tissues. However, when the animal was exposed to stress, less neuropeptides were produced in neurons or the produced neuropeptides were enzymatically degraded, therefore, less neuropeptides was released into the circulating system or a particular function of the peptide was suppressed due to stress.
Two novel peptides were found to be up-regulated due to the decrease of pH, Pep1599 LTEELANQEELLAK (m/z 1599.8356) and Pep1835 ANNVLSGGTTMYPGLADR (m/z 1835.8836) (Figure 6). With dimethyl derivatization, the fragmentation was relatively complete, accounting for most of the b- and y- ions (Figures 7a–d). In the isotopic labeling, two methyl groups were added to primary amine groups, such as N-terminus and lysine residue (K). For Pep1835, dimethyl labeling on the N-terminus resulted in mass increment of 28.0313 Da and 32.0564 Da, by two CH2- or two CD2-, respectively. In the tandem mass spectrum, a 4.0251 Da mass shift was observed in all b- ions, while y- ions showed good alignments (Figure 7f). For Pep1599, isotopic formaldehyde reacted with both the N-terminus and the lysine on the C-terminus, yielding mass increment by 56.0626 Da and 64.1128 Da. Therefore, a 4.0251 Da mass shift occurred on both b- and y- ions (Figure 7e).
Figure 6.

Mass spectra (full scan) of two novel peptides detected in the hemolymph using ESI-MS. Blue circle, control; Red circle, 2hr acidification stress.
Figure 7.
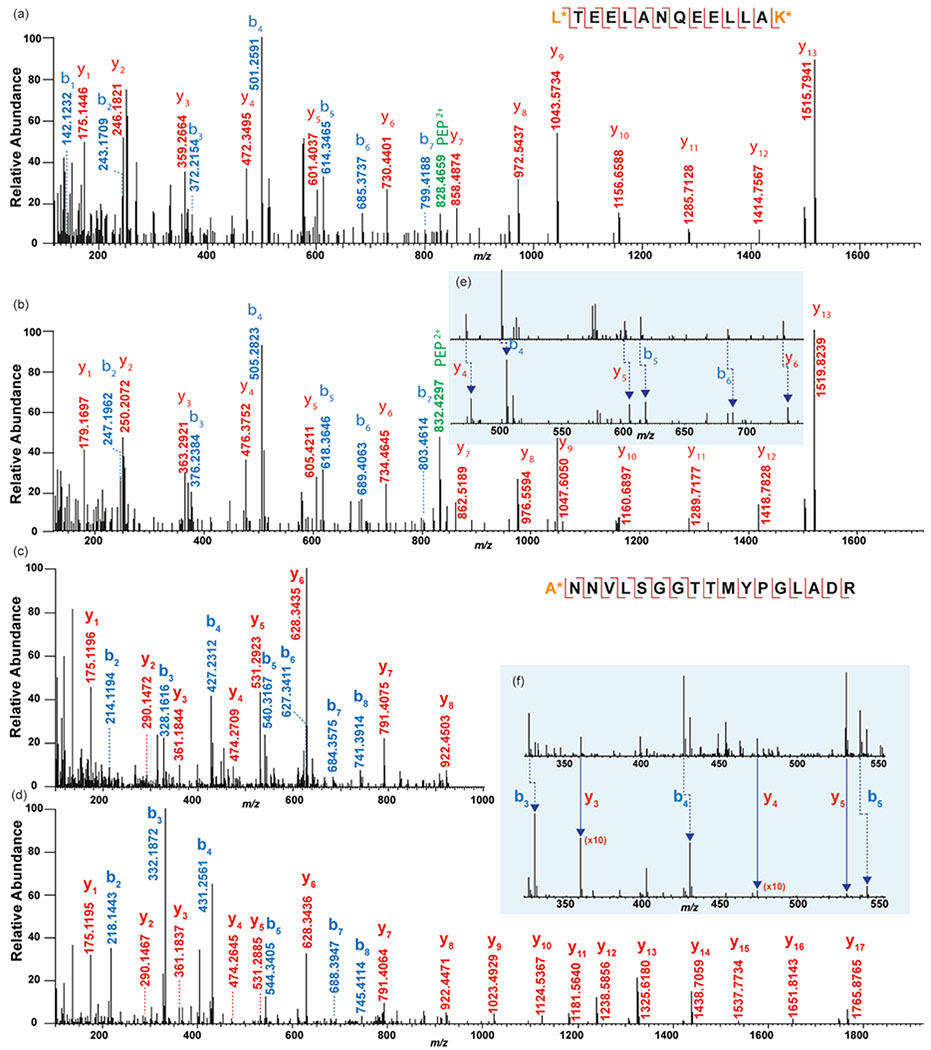
Tandem mass spectra of novel peptides (LTEELANQEELLAK and ANNVLSGGTTMYPGLADR) acquired from ESI-MS for control (a)(c) and 2 hr stressed (b)(d) hemolymph samples. Dimethyl labeling was applied for quantification. Amino acid residues with asterisks showed the position of dimethyl groups added. Mass difference caused by isotopic labeling was illustrated in (e)(f).
Pep1599 and Pep1835 do not belong to any known neuropeptide family based on our current knowledge. It has been reported that in blue crabs, hemolymph proteins (mainly hemocyanin) are involved in pH adjustment39,41. As for other species, hemolymph protein levels altered under hypercapnia in both shallow-water crabs and deep-sea crabs68. In brine shrimp, proteins related to metabolic process, stress response, immune defense, cytoskeletal and signal transduction were examined to change once hatched and raised in acidification environments69. Therefore, we performed a BLAST search against the protein database using the sequence of Pep1599 and Pep1835. There was no match of any known proteins found with Pep1599, indicating this peptide could be produced via cleavage from an unidentified protein related to pH stress. Pep1835 was searched to align with actin in several other crustacean species, such as Daphnia magna and Homarus americanus. Another actin fragment detected in this study was LRVAPEESPVL (m/z 1209.6838), which was found to be down-regulated in MALDI-MS analysis. This actin fragment was found to increase in the hemolymph of Jonah crab, Cancer borealis, upon feeding53. Actin typically participates in vital cellular process, such as muscle contraction and cell signaling. Our current study may reveal its additional function related to stress adaptation in crustaceans.
CONCLUSION
In this study, we quantitatively analyzed the influence of low pH stress on neuropeptide changes in Atlantic blue crab, Callinectes sapidus, both in tissue and body fluid. Different ionization methods were adopted and the results were combined to provide more comprehensive coverage. It has been demonstrated that neural organs, brain, PO, SG, TG, STG and OG, were involved in stress-related modulation. Each neuropeptide family and each isoform within the family has distinct functions depending on its sequence and the tissue from which it belongs. The discovery of identical neuropeptides in multiple tissues and the circulating fluid also indicated those peptides as hormonal molecules, being released from neuroendocrine organs upon stimulus, traveling through the circulatory system, and exerting effects on targeted organs. The time course study of low pH stress revealed dynamic changes of neuropeptides, indicating the pathway of self-adjustment and adaptation in the animal over time. Collectively, our study offers a more comprehensive view of the neuropeptidome as key molecular players in the physiological regulation in response to environmental stress in crustacean model system. Future directions of this study should focus on extending to more severe pH change and longer incubation period. The neuropeptidome analysis under only hypoxia stress should be investigated to facilitate better understanding of the CO2-induced low pH stress.
Supplementary Material
Table S1. Neuropeptide level changes in the brain in response to 2hr low pH stress (n=5)
Table S2. Neuropeptide level changes in the PO in response to 2hr low pH stress (n=5)
Table S3. Neuropeptide level changes in the SG in response to 2hr low pH stress (n=5)
Table S4. Neuropeptide level changes in the TG in response to 2hr low pH stress (n=5)
Table S5. Neuropeptide level changes in the STG in response to 2hr low pH stress (n=5)
Table S6. Neuropeptide level changes in the OG in response to 2hr low pH stress (n=5)
Table S7. Neuropeptide level changes in the brain in response to 4hr low pH stress (n=5)
Table S8. Neuropeptide level changes in the PO in response to 4hr low pH stress (n=4)
Table S9. Neuropeptide level changes in the SG in response to 4hr low pH stress (n=5)
Table S10. Neuropeptide level changes in the TG in response to 4hr low pH stress (n=4)
Table S11. Neuropeptide level changes in the STG in response to 4hr low pH stress (n=4)
Figure S1. MALDI-MS spectra of labeled brain tissues at different ratios.
ACKNOWLEDGEMENT
This work has been supported in part by the National Science Foundation grant (CHE-1710140) and National Institutes of Health (NIH) grants (1R01DK071801, 1R56DK071801, R01NS029436). The MALDI-Orbitrap and Q-Exactive Orbitrap instruments were purchased through an NIH shared instrument grant (NCRRS10RR029531). A.R.B. would like to acknowledge the NIH-General Medical Sciences NRSA Fellowship (1F31GM119365-01). K.D. acknowledges a predoctoral fellowship supported by the NIH, under Ruth L. Kirschstein National Research Service Award T32 HL 007936 from the National Heart Lung and Blood Institute to the University of Wisconsin-Madison Cardiovascular Research Center and the National Institutes of Health-General Medical Sciences F31 National Research Service Award (1F31GM126870-01A1) for funding. Y.L. and Z.L. would like to acknowledge the Li Lab for their support. L.L. acknowledges a Vilas Distinguished Achievement Professorship and Charles Melbourne Johnson Distinguished Chair Professorship with funding provided by the Wisconsin Alumni Research Foundation and University of Wisconsin-Madison School of Pharmacy.
ABBREVIATIONS
- PO
pericardial organ
- SG
sinus gland
- TG
thoracic ganglia
- STG
stomatogastric ganglion
- OG
oesophageal ganglion
- CHH
crustacean hyperglycemic hormone
- CPRP
CHH precursor-related peptide
- AST-A
A-type allatostatins
- AST-B
B-type allatostatins; s/c, stress/control
- MALDI
matrix-assisted laser desorption/ionization
- ESI
electrospray ionization
- LC
liquid chromatography
- MS
mass spectrometry
Footnotes
Data deposition
Representative mass spectrometry data have been deposited to the ProteomeXchange Consortium70 via the PRIDE partner repository with the dataset identifier PXD013536.
Supporting information
The supporting Information is available free of charge on the ACS Publication website.
The authors declare no competing financial interest.
REFERENCES
- (1).Hook V; Bandeira N Neuropeptidomics Mass Spectrometry Reveals Signaling Networks Generated by Distinct Protease Pathways in Human Systems. J. Am. Soc. Mass Spectrom 2015, 26 (12), 1970–1980. 10.1007/s13361-015-1251-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Nusbaum MP; Beenhakker MP A Small-Systems Approach to Motor Pattern Generation. Nature 2002, 417 (6886), 343–350. 10.1038/417343a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Blitz DM; Christie AE; Marder E; Nusbaum MP Distribution and Effects of Tachykinin-like Peptides in the Stomatogastric Nervous System of the Crab, Cancer Borealis. J. Comp. Neurol 1995, 354 (2), 282–294. 10.1002/cne.903540209. [DOI] [PubMed] [Google Scholar]
- (4).Turrigiano GG; Selverston AI A Cholecystokinin-like Hormone Activates a Feeding-Related Neural Circuit in Lobster. Nature 1990, 344 (6269), 866–868. 10.1038/344866a0. [DOI] [PubMed] [Google Scholar]
- (5).Schmerberg CM; Li L Mass Spectrometric Detection of Neuropeptides Using Affinity-Enhanced Microdialysis with Antibody-Coated Magnetic Nanoparticles. Anal. Chem 2013, 85 (2), 915–922. 10.1021/ac302403e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Schmerberg CM; Liang Z; Li L Data-Independent MS/MS Quantification of Neuropeptides for Determination of Putative Feeding-Related Neurohormones in Microdialysate. ACS Chem. Neurosci 2015, 6 (1), 174–180. 10.1021/cn500253u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Nakatsuji T; Lee CY; Watson RD Crustacean Molt-Inhibiting Hormone: Structure, Function, and Cellular Mode of Action. Comp. Biochem. Physiol., Part A Mol. Integr. Physiol 2009, 152 (2), 139–148. 10.1016/j.cbpa.2008.10.012. [DOI] [PubMed] [Google Scholar]
- (8).Hiremagalur B; Kvetnansky R; Nankova B; Fleischer J; Geertman R; Fukuhara K; Viskupic E; Sabban EL Stress Elicits Trans-Synaptic Activation of Adrenal Neuropeptide Y Gene Expression. Brain Res. Mol. Brain Res 1994, 27 (1), 138–144. [DOI] [PubMed] [Google Scholar]
- (9).Chen R; Xiao M; Buchberger A; Li L Quantitative Neuropeptidomics Study of the Effects of Temperature Change in the Crab Cancer Borealis. J Proteome Res 2014, 13 (12), 5767–5776. 10.1021/pr500742q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Hook V; Podvin S; Bundey R; Toneff T; Ziegler M Profiles of Secreted Neuropeptides and Catecholamines Illustrate Similarities and Differences in Response to Stimulation by Distinct Secretagogues. Mol Cell Neurosci 2015, 68, 177–185. 10.1016/j.mcn.2015.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Zhang Y; Buchberger A; Muthuvel G; Li L Expression and Distribution of Neuropeptides in the Nervous System of the Crab Carcinus Maenas and Their Roles in Environmental Stress. Proteomics 2015, 15 (23–24), 3969–3979. 10.1002/pmic.201500256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Li L; Kelley WP; Billimoria CP; Christie AE; Pulver SR; Sweedler JV; Marder E Mass Spectrometric Investigation of the Neuropeptide Complement and Release in the Pericardial Organs of the Crab, Cancer Borealis. J. Neurochem 2003, 87 (3), 642–656. [DOI] [PubMed] [Google Scholar]
- (13).Liang Z; Schmerberg CM; Li L Mass Spectrometric Measurement of Neuropeptide Secretion in the Crab, Cancer Borealis, by in Vivo Microdialysis. Analyst 2015, 140 (11), 3803–3813. 10.1039/c4an02016b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Dircksen H; Skiebe P; Abel B; Agricola H; Buchner K; Muren JE; Nässel DR Structure, Distribution, and Biological Activity of Novel Members of the Allatostatin Family in the Crayfish Orconectes Limosus. Peptides 1999, 20 (6), 695–712. [DOI] [PubMed] [Google Scholar]
- (15).Glantz RM; Miller CS; Nässel DR Tachykinin-Related Peptide and GABA-Mediated Presynaptic Inhibition of Crayfish Photoreceptors. J. Neurosci 2000, 20 (5), 1780–1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Maynard DM; Dando MR The Structure of the Stomatogastric Neuromuscular System in Callinectes Sapidus, Homarus Americanus and Panulirus Argus (Decapoda Crustacea). Philos. Trans. R. Soc. Lond., B, Biol. Sci 1974, 268 (892), 161–220. 10.1098/rstb.1974.0024. [DOI] [PubMed] [Google Scholar]
- (17).Marder E; Bucher D Understanding Circuit Dynamics Using the Stomatogastric Nervous System of Lobsters and Crabs. Annu. Rev. Physiol 2007, 69, 291–316. 10.1146/annurev.physiol.69.031905.161516. [DOI] [PubMed] [Google Scholar]
- (18).Hook V; Funkelstein L; Lu D; Bark S; Wegrzyn J; Hwang S-R Proteases for Processing Proneuropeptides into Peptide Neurotransmitters and Hormones. Annu. Rev. Pharmacol. Toxicol 2008, 48, 393–423. 10.1146/annurev.pharmtox.48.113006.094812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Behrens HL; Chen R; Li L Combining Microdialysis, NanoLC-MS, and MALDI-TOF/TOF To Detect Neuropeptides Secreted in the Crab, Cancer Borealis. Anal. Chem 2008, 80 (18), 6949–6958. 10.1021/ac800798h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Cochran RE; Burnett LE Respiratory Responses of the Salt Marsh Animals, Fundulus Heteroclitus, Leiostomus Xanthurus, and Palaemonetes Pugio to Environmental Hypoxia and Hypercapnia and to the Organophosphate Pesticide, Azinphosmethyl. Journal of Experimental Marine Biology and Ecology 1996, 195 (1), 125–144. 10.1016/0022-0981(95)00102-6. [DOI] [Google Scholar]
- (21).Burnett LE The Challenges of Living in Hypoxic and Hypercapnic Aquatic Environments. Integr Comp Biol 1997, 37 (6), 633–640. 10.1093/icb/37.6.633. [DOI] [Google Scholar]
- (22).Turley C; Gattuso J-P Future Biological and Ecosystem Impacts of Ocean Acidification and Their Socioeconomic-Policy Implications. Current Opinion in Environmental Sustainability 2012, 4 (3), 278–286. 10.1016/j.cosust.2012.05.007. [DOI] [Google Scholar]
- (23).Jacobson MZ Studying Ocean Acidification with Conservative, Stable Numerical Schemes for Nonequilibrium Air-Ocean Exchange and Ocean Equilibrium Chemistry. J. Geophys. Res 2005, 110 (D07302). 10.1029/2004JD005220. [DOI] [Google Scholar]
- (24).Hönisch B; Ridgwell A; Schmidt DN; Thomas E; Gibbs SJ; Sluijs A; Zeebe R; Kump L; Martindale RC; Greene SE; et al. The Geological Record of Ocean Acidification. Science 2012, 335 (6072), 1058–1063. 10.1126/science.1208277. [DOI] [PubMed] [Google Scholar]
- (25).Hönisch B; Hemming NG Surface Ocean PH Response to Variations in PCO2 through Two Full Glacial Cycles. Earth and Planetary Science Letters 2005, 236 (1), 305–314. 10.1016/j.epsl.2005.04.027. [DOI] [Google Scholar]
- (26).Mora C; Wei CL; Rollo A; Amaro T; Baco AR; Billett D; Bopp L; Chen Q; Collier M; Danovaro R; et al. Biotic and Human Vulnerability to Projected Changes in Ocean Biogeochemistry over the 21st Century. PLOS Biology 2013, 11 (10), e1001682 10.1371/journal.pbio.1001682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Galloway JN Acidification of the World: Natural and Anthropogenic. Water, Air, & Soil Pollution 2001, 130 (1), 17–24. 10.1023/A:1012272431583. [DOI] [Google Scholar]
- (28).Riebesell U; Zondervan I; Rost B; Tortell PD; Zeebe RE; Morel FMM Reduced Calcification of Marine Plankton in Response to Increased Atmospheric CO2. Nature 2000, 407 (6802), 364–367. 10.1038/35030078. [DOI] [PubMed] [Google Scholar]
- (29).Langdon C; Atkinson MJ Effect of Elevated PCO2 on Photosynthesis and Calcification of Corals and Interactions with Seasonal Change in Temperature/Irradiance and Nutrient Enrichment. J. Geophys. Res 2005, 110 (C09S07). 10.1029/2004JC002576. [DOI] [Google Scholar]
- (30).Kuffner IB; Andersson AJ; Jokiel PL; Rodgers KS; Mackenzie FT Decreased Abundance of Crustose Coralline Algae Due to Ocean Acidification. Nature Geosci. 2008, 1 (2), 114–117. 10.1038/ngeo100. [DOI] [Google Scholar]
- (31).Gazeau F; Quiblier C; Jansen JM; Gattuso J-P; Middelburg JJ; Heip CHR Impact of Elevated CO2 on Shellfish Calcification. Geophys. Res. Lett 2007, 34 (L07603). 10.1029/2006GL028554. [DOI] [Google Scholar]
- (32).Allison V; Dunham DW; Harvey HH Low PH Alters Response to Food in the Crayfish Cambarus Bartoni. Can. J. Zool 1992, 70 (12), 2416–2420. 10.1139/z92-324. [DOI] [Google Scholar]
- (33).Walther K; Anger K; Pörtner H-O Effects of ocean acidification and warming on the larval development of the spider crab Hyas araneus from different latitudes (54° vs. 79°N). Mar Eco Prog Ser 2010, 417, 159–170. https://doi.org/doi: 10.3354/meps08807. [DOI] [Google Scholar]
- (34).Zheng C; Jeswin J; Shen K; Lablche M; Wang K; Liu H Detrimental Effect of CO2-Driven Seawater Acidification on a Crustacean Brine Shrimp, Artemia Sinica. Fish Shellfish Immunol. 2015, 43 (1), 181–190. 10.1016/j.fsi.2014.12.027. [DOI] [PubMed] [Google Scholar]
- (35).Carter HA; Ceballos-Osuna L; Miller NA; Stillman JH Impact of Ocean Acidification on Metabolism and Energetics during Early Life Stages of the Intertidal Porcelain Crab Petrolisthes Cinctipes. J. Exp. Biol 2013, 216 (8), 1412–1422. 10.1242/jeb.078162. [DOI] [PubMed] [Google Scholar]
- (36).Glover CN; Wood CM Physiological Characterisation of a PH- and Calcium-Dependent Sodium Uptake Mechanism in the Freshwater Crustacean, Daphnia Magna. J. Exp. Biol 2005, 208 (Pt 5), 951–959. 10.1242/jeb.01426. [DOI] [PubMed] [Google Scholar]
- (37).Harms L; Frickenhaus S; Schiffer M; Mark FC; Storch D; Held C; Pörtner H-O; Lucassen M Gene Expression Profiling in Gills of the Great Spider Crab Hyas Araneus in Response to Ocean Acidification and Warming. BMC Genomics 2014, 15, 789 10.1186/1471-2164-15-789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Lehtonen MP; Burnett LE Effects of Hypoxia and Hypercapnic Hypoxia on Oxygen Transport and Acid-Base Status in the Atlantic Blue Crab, Callinectes Sapidus, During Exercise. J Exp Zool A Ecol Genet Physiol 2016, 325 (9), 598–609. 10.1002/jez.2054. [DOI] [PubMed] [Google Scholar]
- (39).Stover KK; Burnett KG; McElroy EJ; Burnett LE Locomotory Fatigue during Moderate and Severe Hypoxia and Hypercapnia in the Atlantic Blue Crab, Callinectes Sapidus. Biol. Bull 2013, 224 (2), 68–78. 10.1086/BBLv224n2p68. [DOI] [PubMed] [Google Scholar]
- (40).Cameron JN Effects of Hypercapnia on Blood Acid-Base Status, NaCl Fluxes, and Trans-Gill Potential in Freshwater Blue Crabs,Callinectes Sapidus. J Comp Physiol B 1978, 123 (2), 137–141. 10.1007/BF00687841. [DOI] [Google Scholar]
- (41).MANGUM CP; BURNETF LE The CO2 Sensitivity of the Hemocyanins and Its Relationship to Cl - Sensitivity. Biol. Bull 1986, 171, 248–263. 10.2307/1541921. [DOI] [Google Scholar]
- (42).Kutz KK; Schmidt JJ; Li L In Situ Tissue Analysis of Neuropeptides by MALDI FTMS In-Cell Accumulation. Anal. Chem 2004, 76 (19), 5630–5640. 10.1021/ac049255b. [DOI] [PubMed] [Google Scholar]
- (43).Tiss A; Smith C; Menon U; Jacobs I; Timms JF; Cramer R A Well-Characterised Peak Identification List of MALDI MS Profile Peaks for Human Blood Serum. Proteomics 2010, 10 (18), 3388–3392. 10.1002/pmic.201000100. [DOI] [PubMed] [Google Scholar]
- (44).Blonder J; Goshe MB; Moore RJ; Pasa-Tolic L; Masselon CD; Lipton MS; Smith RD Enrichment of Integral Membrane Proteins for Proteomic Analysis Using Liquid Chromatography−Tandem Mass Spectrometry. J. Proteome Res 2002, 1 (4), 351–360. 10.1021/pr0255248. [DOI] [PubMed] [Google Scholar]
- (45).Lombard‐Banek C; Moody SA; Nemes P Single‐Cell Mass Spectrometry for Discovery Proteomics: Quantifying Translational Cell Heterogeneity in the 16‐Cell Frog (Xenopus) Embryo. Angew Chem Int Ed Engl 2016, 55 (7), 2454–2458. 10.1002/anie.201510411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Sköld K; Svensson M; Kaplan A; Björkesten L; Aström J; Andren PE A Neuroproteomic Approach to Targeting Neuropeptides in the Brain. Proteomics 2002, 2 (4), 447–454. . [DOI] [PubMed] [Google Scholar]
- (47).Yew JY; Dikler S; Stretton AO De Novo Sequencing of Novel Neuropeptides Directly from Ascaris Suum Tissue Using Matrix-Assisted Laser Desorption/Ionization Time-of-Flight/Time-of-Flight. Rapid Commun. Mass Spectrom 2003, 17 (24), 2693–2698. 10.1002/rcm.1240. [DOI] [PubMed] [Google Scholar]
- (48).Livnat I; Tai H-C; Jansson ET; Bai L; Romanova EV; Chen T; Yu K; Chen S; Zhang Y; Wang Z; et al. A D-Amino Acid-Containing Neuropeptide Discovery Funnel. Anal. Chem 2016, 88 (23), 11868–11876. 10.1021/acs.analchem.6b03658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Guan M; Zhang Z; Li S; Liu J; Liu L; Yang H; Zhang Y; Wang T; Zhao Z Silver Nanoparticles as Matrix for MALDI FTICR MS Profiling and Imaging of Diverse Lipids in Brain. Talanta 2018, 179, 624–631. 10.1016/j.talanta.2017.11.067. [DOI] [PubMed] [Google Scholar]
- (50).Wörmer L; Lipp JS; Schröder JM; Hinrichs K-U Application of Two New LC–ESI–MS Methods for Improved Detection of Intact Polar Lipids (IPLs) in Environmental Samples. Organic Geochemistry 2013, 59, 10–21. 10.1016/j.orggeochem.2013.03.004. [DOI] [Google Scholar]
- (51).Niehoff A-C; Kettling H; Pirkl A; Chiang YN; Dreisewerd K; Yew JY Analysis of Drosophila Lipids by Matrix-Assisted Laser Desorption/Ionization Mass Spectrometric Imaging. Anal. Chem 2014, 86 (22), 11086–11092. 10.1021/ac503171f. [DOI] [PubMed] [Google Scholar]
- (52).Ye H; Hui L; Kellersberger K; Li L Mapping of Neuropeptides in the Crustacean Stomatogastric Nervous System by Imaging Mass Spectrometry. J. Am. Soc. Mass Spectrom 2013, 24 (1), 134–147. 10.1007/s13361-012-0502-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Chen R; Ma M; Hui L; Zhang J; Li L Measurement of Neuropeptides in Crustacean Hemolymph via MALDI Mass Spectrometry. J. Am. Soc. Mass Spectrom 2009, 20 (4), 708–718. 10.1016/j.jasms.2008.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Li Q; Zubieta J-K; Kennedy RT Practical Aspects of in Vivo Detection of Neuropeptides by Microdialysis Coupled Off-Line to Capillary LC with Multi-Stage MS. Anal Chem 2009, 81 (6), 2242–2250. 10.1021/ac802391b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Skiebe P; Schneider H Allatostatin Peptides in the Crab Stomatogastric Nervous System: Inhibition of the Pyloric Motor Pattern and Distribution of Allatostatin-like Immunoreactivity. J. Exp. Biol 1994, 194 (1), 195–208. [DOI] [PubMed] [Google Scholar]
- (56).Cruz-Bermúdez ND; Marder E Multiple Modulators Act on the Cardiac Ganglion of the Crab, Cancer Borealis. J. Exp. Biol 2007, 210 (16), 2873–2884. 10.1242/jeb.002949. [DOI] [PubMed] [Google Scholar]
- (57).Dickinson PS; Stemmler EA; Barton EE; Cashman CR; Gardner NP; Rus S; Brennan HR; McClintock TS; Christie AE Molecular, Mass Spectral, and Physiological Analyses of Orcokinins and Orcokinin Precursor-Related Peptides in the Lobster Homarus Americanus and the Crayfish Procambarus Clarkii. Peptides 2009, 30 (2), 297–317. 10.1016/j.peptides.2008.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Fu Q; Tang LS; Marder E; Li L Mass Spectrometric Characterization and Physiological Actions of VPNDWAHFRGSWamide, a Novel B Type Allatostatin in the Crab, Cancer Borealis. J. Neurochem 2007, 101 (4), 1099–1107. 10.1111/j.1471-4159.2007.04482.x. [DOI] [PubMed] [Google Scholar]
- (59).Chung JS; Zmora N Functional Studies of Crustacean Hyperglycemic Hormones (CHHs) of the Blue Crab, Callinectes Sapidus - the Expression and Release of CHH in Eyestalk and Pericardial Organ in Response to Environmental Stress. FEBS J. 2008, 275 (4), 693–704. 10.1111/j.1742-4658.2007.06231.x. [DOI] [PubMed] [Google Scholar]
- (60).Nusbaum MP; Blitz DM Neuropeptide Modulation of Microcircuits. Curr Opin Neurobiol 2012, 22 (4), 592–601. 10.1016/j.conb.2012.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Christie AE; Stemmler EA; Dickinson PS Crustacean Neuropeptides. Cell. Mol. Life Sci 2010, 67 (24), 4135–4169. 10.1007/s00018-010-0482-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Li L; Pulver SR; Kelley WP; Thirumalai V; Sweedler JV; Marder E Orcokinin Peptides in Developing and Adult Crustacean Stomatogastric Nervous Systems and Pericardial Organs. J. Comp. Neurol 2002, 444 (3), 227–244. 10.1002/cne.10139. [DOI] [PubMed] [Google Scholar]
- (63).Wilcockson DC; Chung SJ; Webster SG Is Crustacean Hyperglycaemic Hormone Precursor-Related Peptide a Circulating Neurohormone in Crabs? Cell Tissue Res. 2002, 307 (1), 129–138. 10.1007/s00441-001-0469-8. [DOI] [PubMed] [Google Scholar]
- (64).Ma M; Wang J; Chen R; Li L Expanding the Crustacean Neuropeptidome Using a Multifaceted Mass Spectrometric Approach. J. Proteome Res 2009, 8 (5), 2426–2437. 10.1021/pr801047v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Holman JD; Burnett KG; Burnett LE Effects of Hypercapnic Hypoxia on the Clearance of Vibrio Campbellii in the Atlantic Blue Crab, Callinectes Sapidus Rathbun. Biol. Bull 2004, 206 (3), 188–196. 10.2307/1543642. [DOI] [PubMed] [Google Scholar]
- (66).Spicer JI; Raffo A; Widdicombe S Influence of CO2-Related Seawater Acidification on Extracellular Acid–Base Balance in the Velvet Swimming Crab Necora Puber. Mar Biol 2007, 151 (3), 1117–1125. 10.1007/s00227-006-0551-6. [DOI] [Google Scholar]
- (67).Tanner CA; Burnett LE; Burnett KG The Effects of Hypoxia and PH on Phenoloxidase Activity in the Atlantic Blue Crab, Callinectes Sapidus. Comp. Biochem. Physiol., Part A Mol. Integr. Physiol 2006, 144 (2), 218–223. 10.1016/j.cbpa.2006.02.042. [DOI] [PubMed] [Google Scholar]
- (68).F. Pane E; P. Barry J Extracellular Acid-Base Regulation during Short-Term Hypercapnia Is Effective in a Shallow-Water Crab, but Ineffective in a Deep-Sea Crab. Marine Ecology-progress Series - MAR ECOL-PROGR SER 2007, 334, 1–9. 10.3354/meps334001. [DOI] [Google Scholar]
- (69).Chang XJ; Zheng CQ; Wang YW; Meng C; Xie XL; Liu HP Differential Protein Expression Using Proteomics from a Crustacean Brine Shrimp (Artemia Sinica) under CO2-Driven Seawater Acidification. Fish Shellfish Immunol. 2016, 58, 669–677. 10.1016/j.fsi.2016.10.008. [DOI] [PubMed] [Google Scholar]
- (70).Perez-Riverol Y; Csordas A; Bai J; Bernal-Llinares M; Hewapathirana S; Kundu DJ; Inuganti A; Griss J; Mayer G; Eisenacher M; et al. The PRIDE Database and Related Tools and Resources in 2019: Improving Support for Quantification Data. Nucleic Acids Res. 2019, 47 (D1), D442–D450. 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Neuropeptide level changes in the brain in response to 2hr low pH stress (n=5)
Table S2. Neuropeptide level changes in the PO in response to 2hr low pH stress (n=5)
Table S3. Neuropeptide level changes in the SG in response to 2hr low pH stress (n=5)
Table S4. Neuropeptide level changes in the TG in response to 2hr low pH stress (n=5)
Table S5. Neuropeptide level changes in the STG in response to 2hr low pH stress (n=5)
Table S6. Neuropeptide level changes in the OG in response to 2hr low pH stress (n=5)
Table S7. Neuropeptide level changes in the brain in response to 4hr low pH stress (n=5)
Table S8. Neuropeptide level changes in the PO in response to 4hr low pH stress (n=4)
Table S9. Neuropeptide level changes in the SG in response to 4hr low pH stress (n=5)
Table S10. Neuropeptide level changes in the TG in response to 4hr low pH stress (n=4)
Table S11. Neuropeptide level changes in the STG in response to 4hr low pH stress (n=4)
Figure S1. MALDI-MS spectra of labeled brain tissues at different ratios.


