Abstract
Bacitracin is a broad-spectrum cyclic peptide antibiotic mainly produced by Bacillus, precursor amino acid supply served as the critical role during its synthesis. In this study, we systematically engineered branch-chain amino acid (BCAA) supply modules for bacitracin production. Firstly, we demonstrated that Ile and Leu acted as limiting precursors for bacitracin synthesis, and that BCAA synthetic pathways were strengthened via simultaneous overexpression of, feedback-resistance acetolactate synthase IlvBNfbr, 2-isopropylmalate synthetase LeuAfbr and BCAA aminotransferase YbgE. Using this approach, bacitracin yield from strain DW-BCAA2 was 892.54 U/mL, an increase of 18.32% compared with that DW2 (754.32 U/mL). Secondly, the BCAA permeases, YvbW and BraB, which have higher affinities for Leu and Ile transportation, respectively, were both identified as BCAA importers, with their overexpression improving intracellular BCAA accumulations and bacitracin yields. Finally, the leucine-responsive family regulator, lrpC was deleted to generate the final strain DW-BCAA6, with intracellular concentrations of Ile, Leu and Val increased by 2.26-, 1.90- and 0.72-fold, respectively. The bacitracin yield from DW-BCAA6 was 1029.83 U/mL, an increase of 36.52%, and is the highest bacitracin yield reported. Equally, concentrations of other byproducts including acetic acid, acetoin and 2,3-butanediol were all reduced. Taken together, we devised an efficient strategy for the enhanced production of bacitracin, and a promising B. licheniformis DW-BCAA6 strain was constructed for industrial production of bacitracin.
Keywords: Bacillus licheniformis, Bacitracin, Branch-chain amino acids, Branch-chain amino acid permease, lrpC
Highlights
-
•
Enhancing intracellular BCAA accumulations benefited bacitracin synthesis.
-
•
YvbW and BraB were both identified as BCAA importers in B. licheniformis.
-
•
Deleting lrp increased brnQ transcription and intracellular BCAA concentrations.
-
•
Bacitracin yield produced by DW-BCAA6 was the highest currently reported.
1. Introduction
Bacitracin is a cyclic peptide antibiotic mainly produced by Bacillus subtilis and Bacillus licheniformis, and is synthesized by non-ribosomal polypeptide synthetase (NRPS) with 11 kinds of amino acids (Orn, D-Phe, His, D-Asp, Asn, Lys, D-Glu, Cys, Leu, Ile and Val) as precursors. Bacitracin synthetase was encoded by gene cluster bacTABC, in which, bacA is used to activate and polymerize five kinds of amino acids (Ile, Cys, Leu, D-Glu and Ile) at bacitracin tail, bacB and bacC are responsible for activating and polymerizing seven kinds of amino acids in heptapeptide loop. However, the relationship between function of bacT and bacitracin synthesis is unclear. In addition, bacitracin is also composed of multiple components (bacitracin A, B1, B2, etc.), of which bacitracin A has the highest property (Li et al., 2018). Acting as a broad spectrum antibiotic, bacitracin inhibits gram-positive and some gram-negative bacteria by repressing bacterial cell wall synthesis. Additionally, this antibiotic is safe to use, has good stability and low-absorption rates (Fang et al., 2017; Rietkotter et al., 2008), and is therefore widely used in feed additives.
With the rapid development of synthetic biology and metabolic engineering, several strategies have been developed to enhance target metabolite production (Hao et al., 2013; Wohlleben et al., 2012; Zabala et al., 2013). For bacitracin, deleting branch chain amino acid (BCAA) transporter gene, yhdG and overexpressing the BCAA permease, BrnQ have been shown to improve intracellular BCAA accumulations, leading to 11% and 10.43% increases in bacitracin yields, respectively (Li et al., 2018; Zhu et al., 2018). The Lys and Orn synthetic pathways were also strengthened, which led to 28.95% and 16.5% increases of bacitracin yields (Wu et al., 2019; Yu et al., 2019). In addition, metabolic engineering of transcriptional factors and strengthening tricarboxylic acid cycle elements also improved precursor amino acid supplies for bacitracin production (Cai et al., 2019b; Liu et al., 2018). Based on these observations, insufficient BCAA supplies may be a bottleneck for efficient bacitracin production, however as yet, no studies have been performed to enhance bacitracin yield via the systematic engineering of BCAA supply modules.
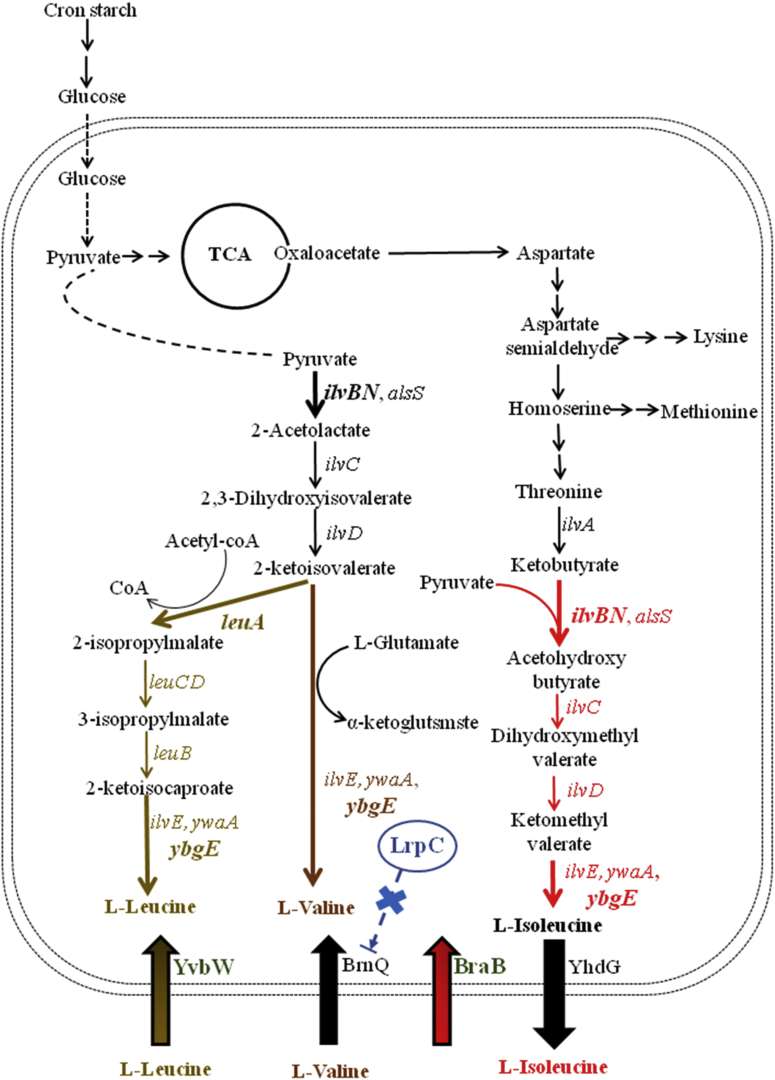
Compared to the synthetic pathways of other amino acids, the synthetic pathways for BCAAs are efficient and economical, since other pathways require more enzymes to synthesize a single kind of amino acid, while only eight enzymes (IlvA, IlvBN, IlvC, IlvD, YbgE, LeuA, LeuB and LeuCD) are required for BCAA syntheses (Fig. 2), and BCAA aminotransferase serves as the common enzyme for Ile, Leu and Val syntheses (Amorim and Blanchard, 2017). Recently, several metabolic engineering approaches have been conducted to improve BCAA production. Vogt et al. constructed a feedback-resistant 2-isopropylmalate synthetase, LeuAfbr for Leu production, with Leu yield further enhanced by deleting ltbR and iolR, reducing citrate synthetase activity and introducing a feedback-resistant IlvNfbr (Vogt et al., 2014). By overexpressing the feedback-resistant threonine dehydratase IlvAfbr and acetohydroxy acid synthetase IlvBNfbr, L-Ile titer produced by Corynebacterium glutamicum JH13-156 reached 30.7 g/L, an increase of 131.7% (Yin et al., 2012), and similarly, improving NADPH generation was also effective for L-Ile production (Shi et al., 2015). Additionally, Chen et al. showed that deletion of aceE, alaT, ilvA, and overexpression of ilvB, ilvN, ilvC and lrp, was beneficial for L-Val production in C. glutamicum (ATCC 13869), and authors also indicated that overexpression of BCAA transporter, BrnFE, promoted L-Val production (Chen et al., 2015). Moreover, L-Val yield was increased to 0.47 mmol/L by blocking three branched pathways (ilvA, leuA and ldh) in B. licheniformis by our group (Liang et al., 2015).
Fig. 2.
Metabolic engineering of BCAA synthetic, transportation and regulation modules for enhanced production of bacitracin in B. licheniformis DW2.
B. licheniformis DW2 is an industrial strain used for bacitracin production (Wang et al., 2017). Several strategies have been developed to improve bacitracin yield in our laboratory (Cai et al., 2019b). In this study, we sought to improve bacitracin yield via systematically engineering BCAA supply modules, including strengthening BCAA synthetic pathways, identifying and modifying BCAA transporters and manipulating the Leu-responsive family regulator, lrpC. We demonstrated that strengthening BCAA supplies was efficient in enhancing bacitracin production, and we provided a promising strain for industrial production of bacitracin.
2. Materials and methods
2.1. Strains, plasmids and cultivation conditions
The strains and plasmids used in this research were provided in Table 1. B. licheniformis DW2 (CCTCC M2011344) acts as the original strain for constructing recombinants. The vector T2(2)-Ori was applied for gene deletion, promoter replacement and integration expression in B. licheniformis, and pHY300PLK serves as a E. coli-Bacillus shuttle expression vector for gene expression. The primers used for strain construction were listed in Table S1 (seeing in Supplementary Materials).
Table 1.
The strains and plasmids used in this research.
| Strains and plasmids | Relevant properties | Source of reference |
|---|---|---|
| Strains | ||
| Escherichia coli DH5α | supE44 ΔlacU169 (f 80 lacZΔM15) hsd R17 recA1 gyrA96 thi1 relA1 | This study |
| Bacillus licheniformis DW2 | Wide-type CCTCC M2011344 | CCTCC |
| DW2/pHY-ilvA | DW2 harboring IlvA expression vector pHY-ilvA | This study |
| DW2/pHY-ilvBN | DW2 harboring IlvBN expression vector pHY-ilvBN | This study |
| DW2/pHY-ilvCD | DW2 harboring IlvCD expression vector pHY-ilvCD | This study |
| DW2/pHY-leuA | DW2 harboring LeuA expression vector pHY-leuA | This study |
| DW2/pHY-leuB | DW2 harboring LeuB expression vector pHY-leuB | This study |
| DW2/pHY-leuC | DW2 harboring LeuC expression vector pHY-leuC | This study |
| DW2/pHY-leuD | DW2 harboring LeuD expression vector pHY-leuD | This study |
| DW2/pHY-300 | DW2 harboring pHY300PLK, as control strain | This study |
| DW2/pHY-ilvBNfbr | DW2 harboring IlvBNfbr expression vector pHY-ilvBNfbr | This study |
| DW2/pHY-leuAfbr | DW2 harboring LeuAfbr expression vector pHY-leuAfbr | This study |
| DW2-ilvBNfbr | IlvBNfbr integration overexpression strain | This study |
| DW2-leuAfbr | LeuAfbr integration overexpression strain | This study |
| DW-BCAA1 | IlvBNfbr and LeuAfbr integration overexpression strain | This study |
| DW-BCAA1/pHY300 | DW-BCAA1 harboring pHY300PLK | This study |
| DW-BCAA1/pHY-IlvE | DW-BCAA1 harboring IlvE expression vector pHY-IlvE | This study |
| DW-BCAA1/pHY-YbgE | DW-BCAA1 harboring YbgE expression vector pHY-YbgE | This study |
| DW-BCAA1/pHY-YwaA | DW-BCAA1 harboring YwaA expression vector pHY-YwaA | This study |
| DW-BCAA2 | Overexpression of YbgE via promoter replacement in DW-BCAA1 | This study |
| DW2△yvbW | Deleting yvbW in DW2 | This study |
| DW2/pHY-YvbW | DW2 harboring YvbW expression vector pHY-YvbW | This study |
| DW2△BraB | Deleting braB in DW2 | This study |
| DW2/pHY-BraB | DW2 harboring BraB expression vector pHY-BraB | This study |
| DW-BCAA3 | Overexpression of YvbW via promoter replacement in DW-BCAA2 | This study |
| DW-BCAA4 | Overexpression of BraB via promoter replacement in DW-BCAA2 | This study |
| DW-BCAA5 | Simultaneously overexpressing YvbW and BraB via promoter replacement in DW-BCAA2 | This study |
| DW-BCAA6 | Deleting regulator gene lrpC in DW-BCAA5 | This study |
| Plasmids | ||
| pHY300PLK | E. coli and B. s shuttle vector; Ampr, Tetr | Lab collection |
| T2(2)-Ori | Bacillus knockout vector; Kanr | Lab collection |
| pHY-ilvA | IlvA expression vcctor based on pHY300PLK | This study |
| pHY-ilvBN | IlvBN expression vcctor based on pHY300PLK | This study |
| pHY-ilvCD | IlvCD expression vcctor based on pHY300PLK | This study |
| pHY-leuA | LeuA expression vcctor based on pHY300PLK | This study |
| pHY-leuB | LeuB expression vcctor based on pHY300PLK | This study |
| pHY-leuC | LeuC expression vcctor based on pHY300PLK | This study |
| pHY-leuD | LeuD expression vcctor based on pHY300PLK | This study |
| pHY-ilvBNfbr | IlvBNfbr expression vcctor based on pHY300PLK | This study |
| pHY-leuAfbr | LeuAfbr expression vcctor based on pHY300PLK | This study |
| pHY-IlvE | IlvE expression vcctor based on pHY300PLK | This study |
| pHY-YbgE | YbgE expression vcctor based on pHY300PLK | This study |
| pHY-YwaA | YwaA expression vcctor based on pHY300PLK | This study |
| T2-::ilvBNfbr | T2(2)-Ori-ilvBNfbr(A + tnrA + B); to overexpress ilvBNfbr | This study |
| T2-::leuAfbr | T2(2)-Ori-leuAfbr(A + tnrA + B); to overexpress leuAfbr | This study |
| T2-PbacA-PybgE | T2(2)-Ori-PybgE(A + PbacA + B); to replace the promoter of ybgE by PbacA | This study |
| T2-PbacA-PyvbW | T2(2)-Ori-PyvbW(A + PbacA + B); to replace the promoter of yvbW by PbacA | This study |
| T2-PbacA-PbraB | T2(2)-Ori-PbraB(A + PbacA + B); to replace the promoter of braB by PbacA | This study |
| T2-lrpC | T2(2)-Ori-lrpC(A + B); to delete lrpC | This study |
B. licheniformis and E. coli were cultivated at 37 °C, and Luriae-Bertani (LB) medium served as the basic medium for strain cultivation, when necessary, adding with corresponding antibiotic (20 mg/L kanamycin or 20 mg/L tetracycline). The medium for bacitracin production contains (g/L) 100 soybean meal, 45 corn starch, 1 (NH4)2SO4, and 6 CaCO3, with natural pH. The seed was cultivated in a 250 mL flask containing 20 mL LB medium to OD600 4.0–4.5, and transferred (1 mL) into fermentation medium, and then cultivated at 230 rpm for 48 h.
As for identifying the transportation modules of BCAA permeases, YvbW and BraB, the medium E (ME medium, pH 7.2) containing (g/L) 20 glucose, 20 sodium glutamate, 10 sodium citrate, 7 NH4Cl, 0.5 K2HPO4·3H2O, 0.5 MgSO4·7H2O, 0.04 FeCl3·6H2O, 0.104 MnSO4·H2O, 0.15 CaCl2·2H2O was applied (Birrer et al., 1994).
2.2. Construction of gene overexpression strain
The construction procedure of gene overexpression strain was referred to our previously reported method (Cai et al., 2017), and IlvB overexpression strain was served as an example. Briefly, P43 promoter from B. subtilis 168, gene ilvB and amyL terminator from B. licheniformis DW2, were amplified with corresponding primers, and fused by Splicing Overlap Extension (SOE)-PCR. The fused fragment was inserted into pHY300PLK at restriction sites EocRI/XbaI, diagnostic PCR and DNA sequence confirmed that IlvB expression vector pHY-ilvB was constructed successfully. Then, pHY-ilvB was electro-transferred into B. licheniformis DW2 to attain IlvB overexpression strain, named as DW2/pHY-ilvB. Similarly, other genes (ilvA, ilvCD, leuA, leuB, leuC, leuD, ilvE, ybgE, ywaA) overexpression strains were constructed according to the same method, and DW2/pHY300 was obtained to serve as the control.
2.3. Gene deletion in B. licheniformis
The procedure for constructing gene deletion strain of B. licheniformis was referred to our previously reported method (Cai et al., 2018), and BCAA permease gene, yvbW deletion strain was served as an example. In brief, the upstream and downstream homogenous arms of yvbW were amplified, fused by SOE-PCR. After digested by SacI/XbaI, the fragments were then inserted into T2(2)-Ori. Plasmid extraction and DNA sequence confirmed that yvbW deletion vector was constructed successfully, named as T2-yvbW.
Then, T2-yvbW was electro-transferred into B. licheniformis DW2, and the positive transformant was picked and cultivated in LB medium with 20 mg/L kanamycin at 45 °C, and sub-cultured for three generations. The single-cross transformant was verified, and then cultivated at 37 °C without antibiotic addition for several generations. The kanamycin-sensitive colonies were picked, diagnostic PCR and DNA sequence were applied to confirm that yvbW deficient strain was constructed successfully, named as DW2△yvbW. Similarly, another BCAA permease gene, braB, was successfully deleted in DW2 by the same method, named as DW2△braB.
2.4. Construction of promoter replacement strain
The construction procedure of promoter PybgE replacement strain was accorded to the previous research (Qiu et al., 2014). Briefly, the up-stream and down-stream homogenous arms of promoter PybgE, bacitracin synthetase operon promoter PbacA (Shi et al., 2019), were amplified by corresponding primers, after fused by SOE-PCR, the fused fragment was inserted into T2(2)-Ori to form the promoter replacement vector T2-PbacA-PybgE. Then, T2-PbacA-PybgE was electro-transferred into B. licheniformis, and promoter replacement strain was attained via homologous double crossover, the same as that of gene deletion.
2.5. Construction of gene integration overexpression strain
To construct the strain which overexpressing multiple genes together, individual genes mediated by P43 promoter were integrated into chromosome of B. licheniformis, following to previously reported protocol (Wu et al., 2019), and verified by diagnostic PCR and DNA sequence.
2.6. Analytical methods
Bacitracin yield was measured by Agilent 1260 high performance liquid chromatography (HPLC) equipped with C18 column (ZORBAX SB-C18), and cell biomass was measured by dilution coating. The concentrations of extracellular and intracellular amino acids were determined by Agilent 7890B gas chromatography (GC), equipped with Trace TR-5 column (30 m × 0.25 mm × 0.25 μm) and flame ionizationdetector (FID) at 280 °C (Wu et al., 2019). The concentrations of byproducts acetic acid, acetoin and 2,3-butanediol were determined by GC (Zhan et al., 2018). The activities of acetolactate synthetase and 2-isopropylmalate synthetase were measured via according to previous researches (Vogt et al., 2014; Yin et al., 2012). Gene transcriptional levels of recombinant strains were measured by RT-qPCR, comparing with those of control strains after being normalized to reference gene 16S rDNA (Cai et al., 2017), and primers used for RT-qPCR were listed in Table S2 (seeing in Supplementary Materials).
2.7. Statistical analysis
All data were conducted to analyze the variance at P < 0.05 and P < 0.01, and a t-test was applied to compare the mean values using the software package, Statistica 6.0. Each experiment was repeated at least three times, and data are represented as the means of three replicates, and bars represented the standard deviations, ∗, P < 0.05; and ∗∗, P < 0.01 indicated the significance levels between experimental and control groups (Cai et al., 2018).
3. Results and discussion
3.1. The effects of exogenous BCAA addition on bacitracin production
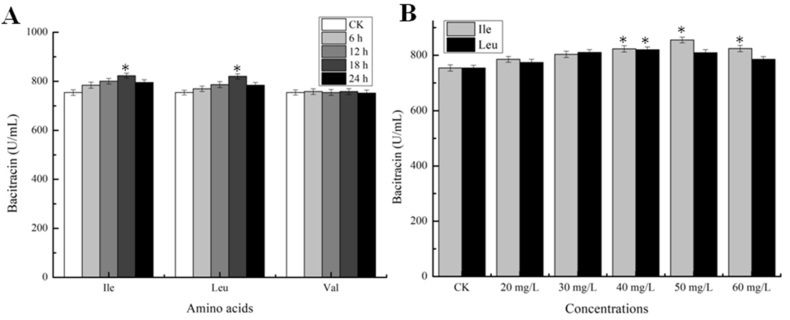
Acting as precursor amino acids of bacitracin, BCAA supplies may serve as critical roles on bacitracin synthesis. Firstly, to evaluate the important role of BCAA supplies on bacitracin production, 40 mg/L of Ile, Leu and Val were individually added into bacitracin fermentation medium at 6, 12, 18, 24 h in separate experiments. Our data suggested that Ile and Leu additions benefited bacitracin synthesis, however, Val addition had no effect on its production (Fig. 1A). Then, different Ile and Leu concentrations were tested at 18 h, with 50 mg/L Ile and 40 mg/L Leu, bacitracin yields were increased by 13.40% and 8.75%, respectively, compared with the control group. These data indicated that Ile and Leu may act as the limiting precursors for bacitracin synthesis.
Fig. 1.
Effects of Ile, Leu and Val additions on bacitracin production. A: Effects of 40 mg/L Ile, Leu and Val addition at different time points on bacitracin production, respectively. B: Effects of different concentrations of Ile and Leu additions at 18 h on bacitracin production, respectively. Each experiment was repeated at least three times, and data are represented as the means of three replicates and bars represent the standard deviations, ∗, P < 0.05; and ∗∗, P < 0.01 indicate the significance levels between recombinant strains and control strain.
3.2. Strengthening BCAA synthetic pathways improved bacitracin yields
3.2.1. The effects of overexpressing BCAA synthetic pathway genes on bacitracin production
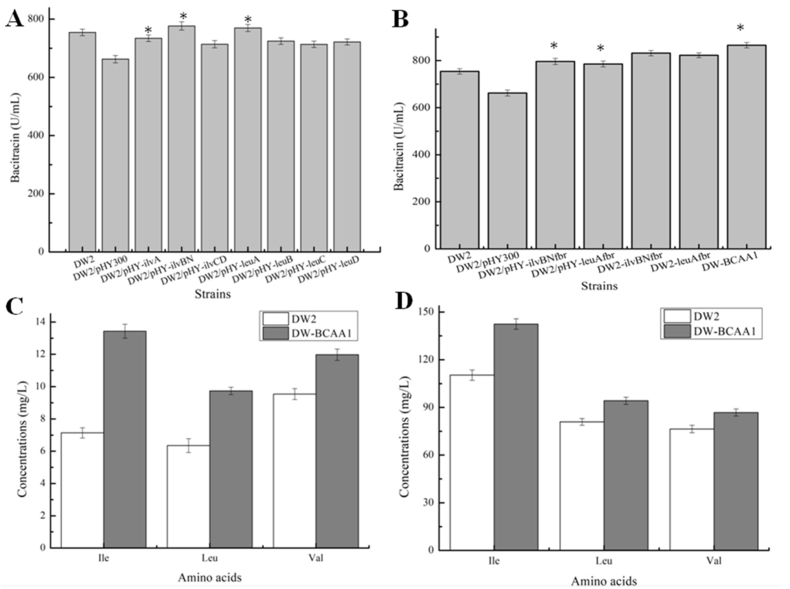
A schematic diagram of BCAA synthetic pathways in B. licheniformis DW2 is shown in Fig. 2. The following genes, ilvA, ilvBN, ilvCD, leuA, leuB, leuC and leuD were overexpressed to evaluate the effects of BCAA synthetic pathway enhancement on bacitracin production. Based on our results (Fig. 3A), overexpression of these genes enhanced bacitracin yields to 10.85%, 17.22%, 7.76%, 16.15%, 9.36%, 7.70% and 8.91%, respectively, higher than that of DW2/pHY300 (662.43 U/mL). Of these recombinant strains, the IlvBN and LeuA overexpression strains DW2/pHY-ilvBN and DW2/pHY-leuA showed the best performances, and agreed with previous researches showing that IlvBN and LeuA were critical in Ile and Leu production, respectively (Vogt et al., 2014; Yin et al., 2012). Similarly, strengthening IlvBN and LeuA expression also benefited cell growth (Fig. S1), as cell biomasses of DW2/pHY-ilvBN and DW2/pHY-leuA showed 9.32% and 9.30% higher than that of DW2/pHY300, respectively.
Fig. 3.
Effects of strengthening BCAA synthetic pathways on bacitracin production. A: Effects of overexpression of ilvA, ilvBN, ilvCD, leuA, leuB, leuC, leuD on bacitracin yields, B: overexpression of the feedback resistant IlvBNfbr and LeuAfbr improved bacitraicn production, C: The concentrations of intracellular BCAAs, D: The concentrations of extracellular BCAAs. Each experiment was repeated at least three times, and data are represented as the means of three replicates and bars represent the standard deviations, ∗, P < 0.05; and ∗∗, P < 0.01 indicate the significance levels between recombinant strains and control strain.
3.2.2. Construction of the feedback-resistant IlvBNfbr and LeuAfbr and their effects on bacitracin production
Generally, genes in amino acid synthetic pathways are feedback inhibited by the increasing target concentrations (Amorim and Blanchard, 2017). Previously, to eliminate feedback inhibition by Ile, Yin et al. constructed the feedback-resistant IlvAfbr and IlvBNfbr, to further improve Ile production in C. glutamicum (Yin et al., 2012). Also, similar tactics were applied to modify LeuA, to resist the inhibition effects of Leu (Vogt et al., 2014). In this study, three amino acid exchanges (P167S, Q413E and F557W) were introduced to ilvBN to construct ilvBNfbr, and the relative feedback resistant LeuAfbr was generated by two amino acid changes i.e. T425H and G428D. The strains DW2/pHY-ilvBNfbr and DW2/pHY-leuAfbr were constructed, and the corresponding bacitracin yields were 796.26 U/mL and 785.33 U/mL, slightly higher than those produced by DW2/pHY-ilvBN and DW2/pHY-leuA strains (Fig. 3B), respectively. Additionally, acetolactate synthase activity in DW2/pHY-ilvBNfbr and 2-isopropylmalate synthase activity in DW2/pHY-leuAfbr were enhanced by 54.46% and 41.23%, compared with those of DW2/pHY-ilvBN and DW2/pHY-leuA, respectively (Table 2), which positively correlated with bacitracin yields (Fig. 3B).
Table 2.
The activities of acetolactate synthetase and 2-isopropylmalate synthetase in IlvBN and LeuA overexpression strains.
| Strains | Acetolactate synthetase (U/mL) | 2-isopropylmalate synthetase (U/mL) |
|---|---|---|
| DW2/pHY300 | 14.32 | 9.53 |
| DW2/pHY-ilvBN | 31.42 | – |
| DW2/pHY-leuA | – | 16.42 |
| DW2/pHY-ilvBNfbr | 48.53 | – |
| DW2/pHY-leuAfbr | – | 23.19 |
Furthermore, to eliminate the repression effects of free plasmids on cell growth and bacitracin synthesis (Zhu et al., 2019), ilvBNfbr and leuAfbr driven by P43 promoter were integrated into B. licheniformis DW2 chromosome to generate the recombinant strains, DW2-ilvBNfbr and DW2-leuAfbr. The bacitracin yields of which reached 831.54 U/mL and 822.43 U/mL, 10.24% and 9.44% higher than that of DW2 (754.32 U/mL), respectively. Furthermore, B. licheniformis DW-BCAA1 was generated by simultaneously overexpressing IlvBNfbr and LeuAfbr, and bacitracin yield reached 865.15 U/mL, an increase of 14.69% when compared to DW2. Moreover, the intracellular concentrations of Ile, Leu and Val in DW2-BCAA1 reached 13.43 mg/L, 9.74 mg/L and 11.98 mg/L, increased by 88.10%, 53.39% and 25.58%, respectively. Equally, the extracellular concentrations of Ile, Leu and Val were respectively increased by 29.06%, 16.41% and 13.56%, because of the enhancements of BCAA synthetic pathways.
3.2.3. Strengthening BCAA aminotransferase improved bacitracin production
As the last enzyme in the BCAA synthetic pathways (Amorim and Blanchard, 2017; Berger et al., 2003), BCAA aminotransferase has critical roles in BCAA syntheses (Feng et al., 2018). In this work, three BCAA aminotransferases, IlvE from E. coli, and YbgE and YwaA from B. licheniformis, were overexpressed in DW-BCAA1 strain, generating BCAA aminotransferase overexpression strains, DW-BCAA1/pHY-IlvE, DW-BCAA1/pHY-YbgE and DW-BCAA1/pHY-YwaA, respectively. Our experimental results showed that BCAA aminotransferase overexpression improved bacitracin yields, i.e. 8.62%, 14.66% and 11.02%, compared to DW-BCAA1/pHY300 (759.76 U/mL) (Fig. 4A), respectively.
Fig. 4.
Effects of BCAA aminotransferase overexpressions on bacitracin production. A: Effects of ilvE, ywaA and ybgE overexperessions on bacitracin yields, B: Effects of replacing the promoter of ybgE by PbacA on bacitracin production, C: The concentrations of intracellular BCAAs, D: The concentrations of extracellular BCAAs. Each experiment was repeated at least three times, and data are represented as the means of three replicates and bars represent the standard deviations, ∗, P < 0.05; and ∗∗, P < 0.01 indicate the significance levels between recombinant strains and control strain.
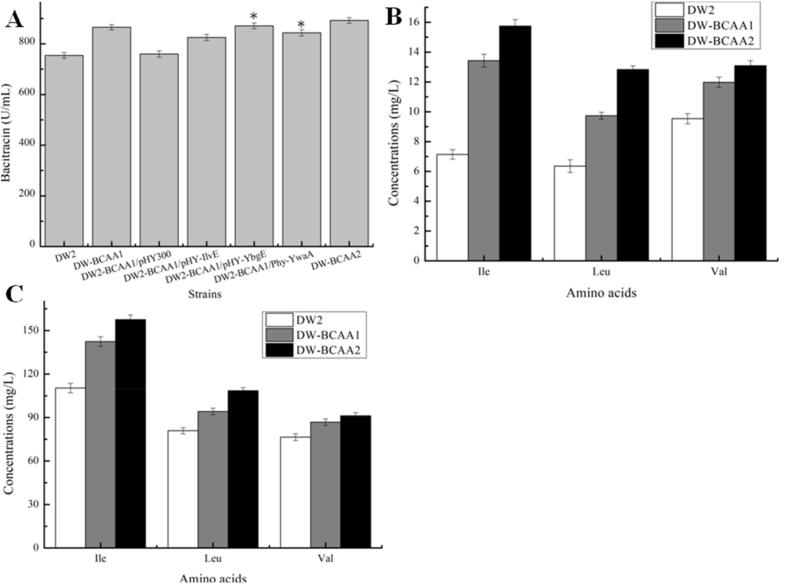
Furthermore, the ybgE promoter in B. licheniformis BCAA1 was replaced by PbacA, a strong promoter previously confirmed in our group (Shi et al., 2019), to generate DW-BCAA2. Fermentation results showed that bacitracin yield produced by DW-BCAA2 reached 892.54 U/mL, an increase of 18.32% and 3.17%, compared with DW2 and DW-BCAA1, respectively. In addition, intracellular and extracellular BCAA concentrations were all significantly enhanced (Fig. 4B and C). Previously, the BCAA aminotransferase activity of YbgE was much greater than YwaA, and it displayed higher affinity for Ile and Leu in B. subtilis, consistently, our results agreed with previous reports (Berger et al., 2003; Brinsmade et al., 2014).
3.3. Engineering BCAA permeases, YvbW and BraB improved bacitracin production
Amino acid transporters play important roles in the distribution and production of relative amino acids (Wendisch, 2019; Xie et al., 2012). Previously, Lys transporter LysE was shown to be a Lys exporter in C. glutamicum, with LysE overexpression significantly enhancing Lys yields (Naerdal et al., 2017). Overexpression of BCAA transporter, BrnFE was effective for Val production in C. glutamicum (Chen et al., 2015). The fermentation medium for bacitracin production contains 100 g/L soybean meal, which generates considerable amounts of free amino acids in fermentation broth (Cai et al., 2019a), therefore, engineering amino acid transporters may be an effective approach for bacitracin production. In our previous research, we observed that engineering Lys transporters, LysE, LysP and YvsH was beneficial for bacitracin synthesis, via improving intracellular Lys accumulation (Wu et al., 2019). As for BCAAs, deletion of BCAA exporter gene, yhdG, improved intracellular BCAA accumulations and bacitracin yield (Li et al., 2018), and overexpression of another BCAA importer, BrnQ, also benefited bacitracin synthesis (Zhu et al., 2018). Here, yvbW and braB were annotated as BCAA permease genes in B. licheniformis DW2, however, the transport modes are not yet resolved, nor are their effects on bacitracin production. Here, we sought to clarify the transportation modes of YvbW and BraB, and then manipulating their actions to improve intracellular BCAA accumulation for bacitracin production.
3.3.1. Identification of transportation modes of the BCAA permeases, YvbW and BraB in B. licheniformis DW2
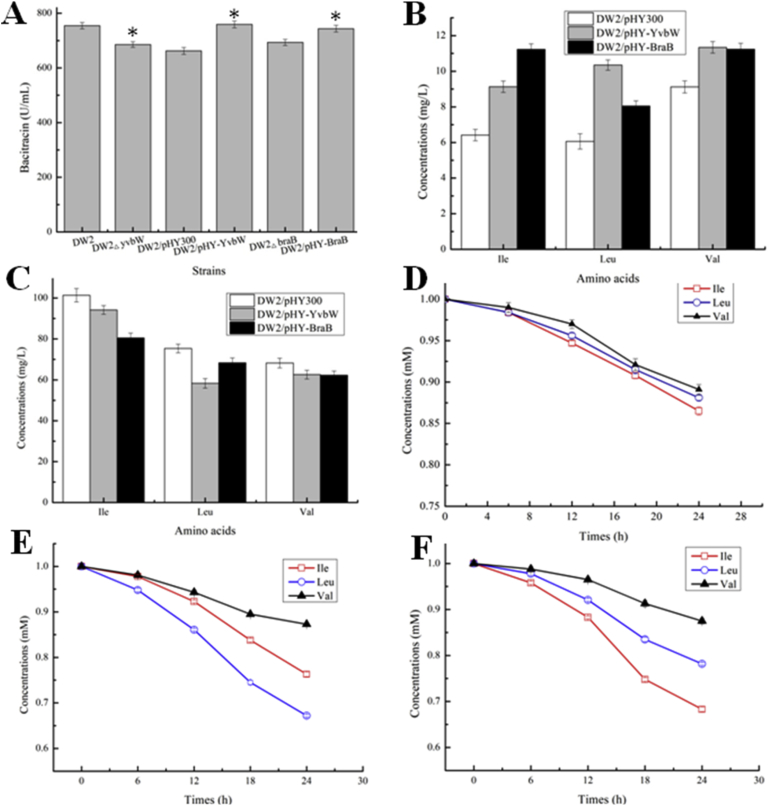
The genes, yvbW and braB were knocked out in DW2, generating the mutant strains DW2△yvbW and DW2△braB. Gene overexpression strains DW2/pHY-yvbW and DW2/pHY-braB were also constructed. Our experimental results showed that bacitracin yields were decreased by 7.95% and 6.48% in yvbW and braB deletion strains, respectively, while YvbW and BraB overexpressions promoted bacitracin syntheses (Fig. 5A). Meantime, intracellular BCAA concentrations were increased and extracellular BCAA concentrations were decreased in yvbW and braB overexpression strains, respectively (Fig. 5B and C). Therefore, our results indicated YvbW and BraB may function as BCAA importers in B. licheniformis DW2.
Fig. 5.
Identification of BCAA permeases, YvbW and BraB, in B. licheniformis DW2. A: Effects of overexpression and deletion of yvbW and braB on bacitracin production, B: The concentrations of intracellular BCAAs, C: The concentrations of extracellular BCAAs, D: The concentrations of extracellular BCAAs of DW2/pHY300, E: The concentrations of extracellular BCAAs of DW2/pHY-YvbW, F: The concentrations of extracellular BCAAs of DW2/pHY-BraB. Each experiment was repeated at least three times, and data are represented as the means of three replicates and bars represent the standard deviations, ∗, P < 0.05; and ∗∗, P < 0.01 indicate the significance levels between recombinant strains and control strain.
To test this hypothesis, all strains were cultivated in ME medium for 24 h, after which cells were washed and transferred into ME medium containing 1 mM Ile, Leu or Val. After this period, extracellular BCAA concentrations were measured for another 24 h. Our data revealed that concentrations of extracellular BCAAs dropped significantly in YvbW and BraB overexpression strains, when compared to DW2/pHY300 (Fig. 5D–F). The import rates of extracellular BCAAs were reduced in gene deletion strains (Fig. S2). Moreover, YvbW appeared more suitable for Leu import, and BraB may have higher affinity for Ile import, however, both showed limited capabilities for Val import. Taken together, these results confirmed that YvbW and BraB acted as BCAA importers in DW2, suggesting they were efficient permeases for Leu and Ile import, respectively. In B. subtilis, the BCAA transporters, BcaP (YhdG in B. licheniformis DW2), BraB and BrnQ were implicated in BCAA uptake, with BcaP appearing to be the most efficient permease for Ile and Val import (Belitsky et al., 2015). Additionally, these transporters, as well as the BCAA synthetase gene cluster, ilv-leu, were regulated by CodY in B. subtilis (Belitsky et al., 2015). Moreover, YvbW is assumed to be a BCAA permease with high affinity for Leu transportation in B. subtilis (Wels et al., 2008), however, no studies have yet to test and verify this mechanism. Our results confirmed that YvbW served as a BCAA importer in B. licheniformis DW2, and acted as the most efficient permease for Leu uptake.
3.3.2. Engineering the BCAA permeases, YvbW and BraB to improve bacitracin production
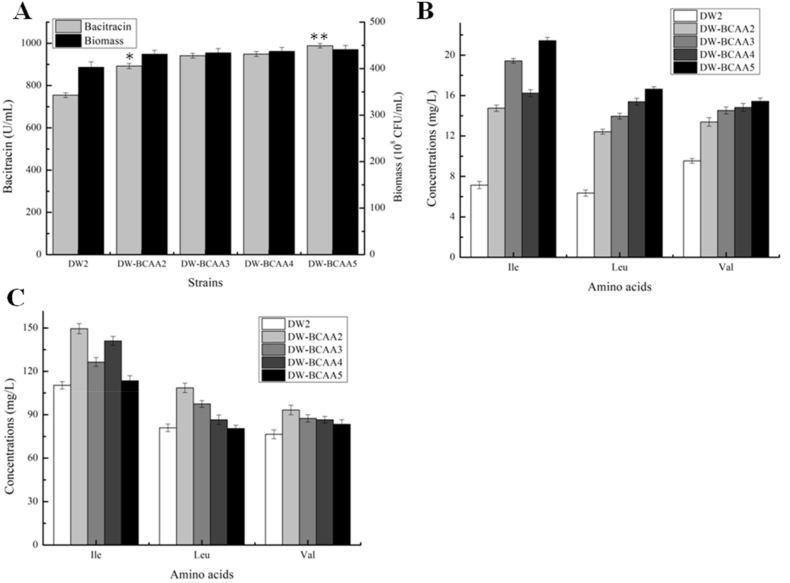
To further improve intracellular BCAA accumulations for bacitracin production, YvbW and BraB were overexpressed in DW-BCAA2, by replacing their promoters with PbacA, resulting in strains DW-BCAA3 and DW-BCAA4, respectively. After experimental analyses, our results (Fig. 6A) suggested that bacitracin yields produced by DW-BCAA3 and DW-BCAA4 were 941.42 U/mL and 949.12 U/mL, increased by 24.80% and 25.83% when compared with DW2, respectively. Additionally, a further strain DW2-BCAA5 was generated by a combinatorial overexpression strategy (overexpression of YvbW and BraB simultaneously), and bacitracin yield of which reached 988.52 U/mL, increased by 31.05% and 10.75% compared with those of DW2 and DW-BCAA2, respectively (Fig. 6A). Additionally, intracellular concentrations of Ile, Leu and Val were 21.43 mg/L, 16.64 mg/L and 15.43 mg/L, respectively, increased by 2.00-, 1.62- and 0.62-fold compared to DW-BCAA2 cells, while concentrations of extracellular BCAAs were all decreased (Fig. 6B and C).
Fig. 6.
Engineering BCAA permeases, YvbW and BraB, for enhancement production of bacitracin. A: Strengthening YvbW and BraB expressions improved bacitracin production. B: The concentrations of intracellular BCAAs, C: The concentrations of extracellular BCAAs. Each experiment was repeated at least three times, and data are represented as the means of three replicates and bars represent the standard deviations, ∗, P < 0.05; and ∗∗, P < 0.01 indicate the significance levels between recombinant strains and control strain.
Previously, several amino acid transporters were engineered for target metabolite production, i.e. engineering LysE for Lys production and BrnFE for Val production (Chen et al., 2015; Naerdal et al., 2017). Here, high levels of free amino acids degraded from soybean meal remained in the fermentation broth (Cai et al., 2019a), and how to effectively utilize these free amino acids is a key factor for high-level production of bacitracin. Because of this, engineering amino acid transporters has proven to be effective for bacitracin synthesis (Wu et al., 2019), and bacitracin yields were significantly enhanced by overexpressing the BCAA permeases, BraB and YvbW in this study, and we suggested that this strategy could be applied to the production of other metabolites.
3.4. Deletion of the leucine-responsive regulator gene, lrpC improved bacitracin production
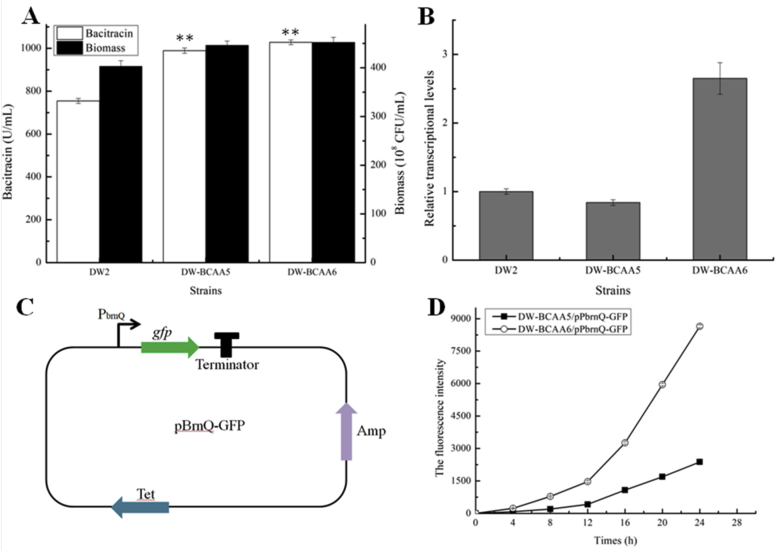
The leucine-responsive regulator LrpC, which is present in bacteria and archaea, is a negative regulator in several physiological processes (Beloin et al., 2000; Lopez-Torrejon et al., 2006). Similarly, genes in the BCAA synthetic (ilvGMEDA, ilvIH and leuABCD) and transportation pathways (ilvJ and ygaZH) are regulated by regulator LrpC in E. coli (Park et al., 2007). Chip-on-chip analysis confirmed that LrpC directly regulated BCAA transporter BrnFE in C. glutamicum (Lange et al., 2012). In addition, deleting the lrp family regulator, SACE_lrp strengthened BCAA accumulations, which further improved erythromycin production in Saccharopolyspora erythraea (Liu et al., 2017). In this study, lrpC was deleted in DW-BCAA5, resulting in DW-BCAA6. From our experimental results, bacitracin yield produced by DW-BCAA6 reached 1027.78 U/mL, an increase of 36.25%, when compared to DW2 (Fig. 7A). Equally, the transcriptional levels of BCAA permease, brnQ, were enhanced 1.65-fold (Fig. 7B). Furthermore, we also constructed a green fluorescent protein (GFP) expression vector, which driven by promoter PbrnQ (Fig. 7C), and then transferred into DW-BCAA5 and DW-BCAA6 respectively. Based on our results of Fig. 7D, green fluorescence intensities were significantly enhanced in DW-BCAA6, indicating the increased PbrnQ transcription in lrpC deletion strain. Collectively, these results suggested that PbrnQ was negatively regulated by regulator LrpC. Since BrnQ has been shown to be a BCAA importer in B. licheniformis (Zhu et al., 2018), the increased BrnQ expression benefited intracellular BCAA accumulations, which promoted bacitracin synthesis in lrpC deletion strain. Meantime, lrpC deletion had no effect on ilvA, ilvB and ilvCD transcription in BCAA synthetic pathways (Fig. S3).
Fig. 7.
Effects of lrpC deletion on bacitracin synthesis. A: Deletion of lrpC improved bacitracin yield, B: The transcriptional level of BCAA transporter gene brnQ, C: The schematic diagram of GFP expression vector mediated by promoter PbrnQ, D: The green fluorescence intensities of recombinant strains. Each experiment was repeated at least three times, and data are represented as the means of three replicates and bars represent the standard deviations, ∗, P < 0.05; and ∗∗, P < 0.01 indicate the significance levels between recombinant strains and control strain.
Additionally, BCAA addition experiments were conducted in DW-BCAA6, our results in Fig. S4 suggested that addition of Ile, Leu or Val could not further improve bacitracin yield of DW-BCAA6, suggested that intracellular BCAAs were adequate for bacitracin synthesis via our metabolic engineering approaches.
3.5. Fermentation parameters for B. licheniformis DW2 and DW-BCAA6
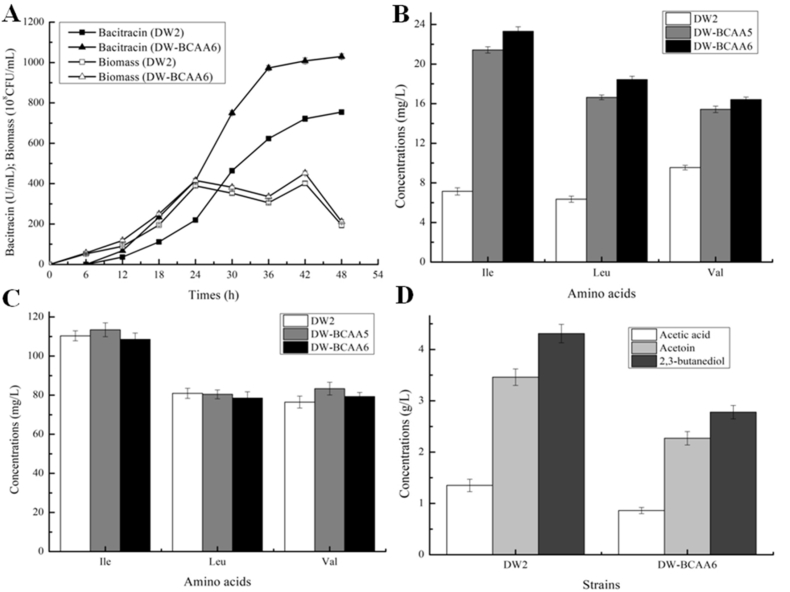
We assessed several outputs of B. licheniformis DW2 and DW-BCAA6 in terms of bacitracin production. As shown in Fig. 8A, bacitracin yields produced by DW-BCAA6 were higher than DW2 throughout the fermentation process, with the maximum bacitracin yield for DW-BCAA6 was increased to 1029.83 U/mL, a 36.52% increase compared to DW2. Strengthening BCAA supplies was also beneficial for fatty acid synthesis and cell growth (Bentley et al., 2016), and the maximum cell biomass for DW-BCAA6 was 452.42 × 108 CFU/mL, 12.73% higher than that of DW2 (401.33 × 108 CFU/mL). In addition to the critical role of BCAAs on metabolite synthesis and cell growth, the strategies outlined in this research could be extended to the efficient production of heterologous proteins and other metabolites in future work. The bacitracin yield per DW-BCAA6 cell was 2.28 × 10−8 U/CFU, an increase of 21.11% compared to DW2. Furthermore, the concentrations of intracellular Ile, Leu and Val were 23.31 mg/L, 18.43 mg/L and 16.41 mg/L in DW-BCAA6, increased by 2.26-, 1.90- and 0.72-fold compared with DW2 (7.14 mg/L, 6.35 mg/L and 9.54 mg/L) (Fig. 8B and C), respectively. Of three amino acids (Ile, Leu and Val), Ile had the largest increase ratio (2.26-fold) in DW-BCAA6, compared to Leu (1.90-fold) and Val (0.72-fold). Since Ile acts as a critical constituent amino acid of bacitracin A, the most active component in bacitracin family (Li et al., 2018), in this study, bacitracin A proportion of DW-BCAA6 was 56.37%, increased by 24.62% compared with DW2 (45.27%), indicated that strengthening Ile supply not only improve bacitracin yield, but also bacitracin A potency is enhanced in DW-BCAA6, and which is beneficial for the increase of bacitracin effectiveness. Additionally, the intracellular concentrations of other precursor amino acids (Asn, Cys and Lys) were also significantly enhanced in DW-BCAA6 cells, compared with those of DW2 (Fig. S5). In addition, since acetic acid, acetoin and 2,3-butanediol were the main byproducts during Bacillus fermentation (Zhan et al., 2018), our results implied that the yields of acetic acid, acetoin and 2,3-butanediol by-products were significantly decreased, when compared with DW2 (Fig. 8D).
Fig. 8.
Fermentation processes of B. licheniformis DW2 and DW-BCAA6. A: Bacitracin yields and cell biomasses of DW2 and DW-BCAA6 during bacitracin production, B: The concentrations of intracellular BCAAs, C: The concentrations of extracellular BCAAs, D: The concentrations of main byproducts, acetic acid, acetoin and 2,3-butanediol. Each experiment was repeated at least three times, and data are represented as the means of three replicates and bars represent the standard deviations, ∗, P < 0.05; and ∗∗, P < 0.01 indicate the significance levels between recombinant strains and control strain.
4. Conclusions
Precursor amino acid supplies serve critical roles in bacitracin production. In this research, Ile and Leu were confirmed as limiting amino acids for bacitracin synthesis, suggesting that strengthening BCAA synthetic pathways could improve bacitracin yield. Furthermore, the BCAA transporters, YvbW and BraB were identified as BCAA importers, and overexpression of these importers enhanced intracellular BCAA accumulations and bacitracin yields. Additionally, deletion of the leucine-responsive regulator, lrpC improved BrnQ expression, and further improved intracellular BCAA supplies and bacitracin production. Finally, bacitracin yields produced by DW-BCAA6 reached 1029.83 U/mL, an increase of 36.52% when compared with DW2. This is the highest bacitracin yield currently reported in the literature. This study has outlined an efficient strain improvement strategy to improve bacitracin yield, and a promising B. licheniformis DW-BCAA6 strain was generated for the industrial production of bacitracin.
CRediT authorship contribution statement
Dongbo Cai: Conceptualization, Investigation, Writing - original draft. Jiang Zhu: Investigation. Yang Li: Formal analysis. Lingfeng Li: Investigation. Meng Zhang: Formal analysis. Zhi Wang: Methodology. Hanbo Yang: Resources. Junhui Li: Resources. Zhifan Yang: Conceptualization, Writing - review & editing. Shouwen Chen: Conceptualization, Supervision, Funding acquisition.
Declaration of competing interest
The author Junhui Li was employed by Lifecome Biochemistry Co. Ltd in China, and the remaining authors have a commercial interest in bacitracin production with Lifecome Biochemistry Co. Ltd.
Acknowledgements
This work was supported by the National Key Research and Development Program of China (2018YFA0900300), the Technical Innovation Special Fund of Hubei Province (2018ACA149), and the Natural Science Foundation of Hubei Province of China (2019CFB319), China Postdoctoral Science Foundation (2018M642802) and Open Funding Project of State Key Laboratory of Biocatalysis and Enzyme Engineering (SKLBEE2018005).
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.mec.2020.e00136.
Contributor Information
Zhifan Yang, Email: mel212@126.com.
Shouwen Chen, Email: sailyangzhf@hubu.edu.cn.
Appendix A. Supplementary data
The following are the supplementary data related to this article:
References
- Amorim Franco T.M., Blanchard J.S. Bacterial branched-chain amino acid biosynthesis: structures, mechanisms, and drugability. Biochemistry. 2017;56:5849–5865. doi: 10.1021/acs.biochem.7b00849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belitsky B.R., Brinsmade S.R., Sonenshein A.L. Intermediate levels of Bacillus subtilis CodY activity are required for derepression of the branched-chain amino acid permease. BraB. PLoS Genet. 2015;11 doi: 10.1371/journal.pgen.1005600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beloin C., Exley R., Mahe A.L., Zouine M., Cubasch S., Le Hegarat F. Characterization of LrpC DNA-binding properties and regulation of Bacillus subtilis lrpC gene expression. J. Bacteriol. 2000;182:4414–4424. doi: 10.1128/jb.182.16.4414-4424.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley G.J., Jiang W., Guaman L.P., Xiao Y., Zhang F. Engineering Escherichia coli to produce branched-chain fatty acids in high percentages. Metab. Eng. 2016;38:148–158. doi: 10.1016/j.ymben.2016.07.003. [DOI] [PubMed] [Google Scholar]
- Berger B.J., English S., Chan G., Knodel M.H. Methionine regeneration and aminotransferases in Bacillus subtilis, Bacillus cereus, and Bacillus anthracis. J. Bacteriol. 2003;185:2418–2431. doi: 10.1128/JB.185.8.2418-2431.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birrer G.A., Cromwick A.M., Gross R.A. Gamma-poly(glutamic acid) formation by Bacillus licheniformis 9945a: physiological and biochemical studies. Int. J. Biol. Macromol. 1994;16:265–275. doi: 10.1016/0141-8130(94)90032-9. [DOI] [PubMed] [Google Scholar]
- Brinsmade S.R., Alexander E.L., Livny J., Stettner A.I., Segre D., Rhee K.Y., Sonenshein A.L. Hierarchical expression of genes controlled by the Bacillus subtilis global regulatory protein CodY. Proc. Natl. Acad. Sci. U. S. A. 2014;111:8227–8232. doi: 10.1073/pnas.1321308111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai D., Chen Y., He P., Wang S., Mo F., Li X., Wang Q., Nomura C.T., Wen Z., Ma X., Chen S. Enhanced production of Poly-gamma-glutamic acid by improving ATP supply in metabolically engineered Bacillus licheniformis. Biotechnol. Bioeng. 2018;115:2541–2553. doi: 10.1002/bit.26774. [DOI] [PubMed] [Google Scholar]
- Cai D., He P., Lu X., Zhu C., Zhu J., Zhan Y., Wang Q., Wen Z., Chen S. A novel approach to improve poly-γ-glutamic acid production by NADPH Regeneration in Bacillus licheniformis WX-02. Sci. Rep. 2017;7 doi: 10.1038/srep43404. 43404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai D., Zhang B., Rao Y., Li L., Zhu J., Li J., Ma X., Chen S. Improving the utilization rate of soybean meal for efficient production of bacitracin and heterologous proteins in the aprA-deficient strain of Bacillus licheniformis. Appl. Microbiol. Biotechnol. 2019;103:4789–4799. doi: 10.1007/s00253-019-09804-0. [DOI] [PubMed] [Google Scholar]
- Cai D., Zhu J., Zhu S., Lu Y., Zhang B., Lu K., Li J., Ma X., Chen S. Metabolic engineering of main transcription factors in carbon, nitrogen, and phosphorus metabolisms for enhanced production of bacitracin in Bacillus licheniformis. ACS Synth. Biol. 2019;8:866–875. doi: 10.1021/acssynbio.9b00005. [DOI] [PubMed] [Google Scholar]
- Chen C., Li Y., Hu J., Dong X., Wang X. Metabolic engineering of Corynebacterium glutamicum ATCC13869 for L-valine production. Metab. Eng. 2015;29:66–75. doi: 10.1016/j.ymben.2015.03.004. [DOI] [PubMed] [Google Scholar]
- Fang C., Nagy-Staron A., Grafe M., Heermann R., Jung K., Gebhard S., Mascher T. Insulation and wiring specificity of BceR-like response regulators and their target promoters in Bacillus subtilis. Mol. Microbiol. 2017;104:16–31. doi: 10.1111/mmi.13597. [DOI] [PubMed] [Google Scholar]
- Feng L.Y., Xu J.Z., Zhang W.G. Improved l-leucine production in Corynebacterium glutamicum by optimizing the aminotransferases. Molecules. 2018;23 doi: 10.3390/molecules23092102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao F., Wu Q., Xu Y. Precursor supply strategy for tetramethylpyrazine production by Bacillus subtilis on solid-state fermentation of wheat bran. Appl. Biochem. Biotechnol. 2013;169:1346–1352. doi: 10.1007/s12010-012-0083-0. [DOI] [PubMed] [Google Scholar]
- Lange C., Mustafi N., Frunzke J., Kennerknecht N., Wessel M., Bott M., Wendisch V.F. Lrp of Corynebacterium glutamicum controls expression of the brnFE operon encoding the export system for L-methionine and branched-chain amino acids. J. Biotechnol. 2012;158:231–241. doi: 10.1016/j.jbiotec.2011.06.003. [DOI] [PubMed] [Google Scholar]
- Li Y., Wu F., Cai D., Zhan Y., Li J., Chen X., Chen H., Chen S., Ma X. Enhanced production of bacitracin by knocking out of amino acid permease gene yhdG in Bacillus Licheniformis DW2. Sheng Wu Gong Cheng Xue Bao. 2018;34(6):916–927. doi: 10.13345/j.cjb.170500. [DOI] [PubMed] [Google Scholar]
- Liang C., Huo Y., Qi G., Wei X., Wang Q., Chen S. Enhancement of L-valine production in Bacillus licheniformis by blocking three branched pathways. Biotechnol. Lett. 2015;37:1243–1248. doi: 10.1007/s10529-015-1783-7. [DOI] [PubMed] [Google Scholar]
- Liu J., Chen Y., Wang W., Ren M., Wu P., Wang Y., Li C., Zhang L., Wu H., Weaver D.T., Zhang B. Engineering of an Lrp family regulator SACE_Lrp improves erythromycin production in Saccharopolyspora erythraea. Metab. Eng. 2017;39:29–37. doi: 10.1016/j.ymben.2016.10.012. [DOI] [PubMed] [Google Scholar]
- Liu Z., Yu W., Nomura C.T., Chen S., Yang Y., Wang Q. Increased flux through the TCA cycle enhances bacitracin production by Bacillus licheniformis DW2. Appl. Microbiol. Biotechnol. 2018;102(16):6935–6946. doi: 10.1007/s00253-018-9133-z. [DOI] [PubMed] [Google Scholar]
- Lopez-Torrejon G., Martinez-Jimenez M.I., Ayora S. Role of LrpC from Bacillus subtilis in DNA transactions during DNA repair and recombination. Nucleic Acids Res. 2006;34:120–129. doi: 10.1093/nar/gkj418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naerdal I., Netzer R., Irla M., Krog A., Heggeset T.M.B., Wendisch V.F., Brautaset T. l-lysine production by Bacillus methanolicus: genome-based mutational analysis and l-lysine secretion engineering. J. Biotechnol. 2017;244:25–33. doi: 10.1016/j.jbiotec.2017.02.001. [DOI] [PubMed] [Google Scholar]
- Park J.H., Lee K.H., Kim T.Y., Lee S.Y. Metabolic engineering of Escherichia coli for the production of L-valine based on transcriptome analysis and in silico gene knockout simulation. Proc. Natl. Acad. Sci. U. S. A. 2007;104:7797–7802. doi: 10.1073/pnas.0702609104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Y., Xiao F., Wei X., Wen Z., Chen S. Improvement of lichenysin production in Bacillus licheniformis by replacement of native promoter of lichenysin biosynthesis operon and medium optimization. Appl. Microbiol. Biotechnol. 2014;98:8895–8903. doi: 10.1007/s00253-014-5978-y. [DOI] [PubMed] [Google Scholar]
- Rietkotter E., Hoyer D., Mascher T. Bacitracin sensing in Bacillus subtilis. Mol. Microbiol. 2008;68:768–785. doi: 10.1111/j.1365-2958.2008.06194.x. [DOI] [PubMed] [Google Scholar]
- Shi F., Li K., Li Y. Comparative proteome analysis of global effect of POS5 and zwf-ppnK overexpression in L-isoleucine producing Corynebacterium glutamicum ssp. lactofermentum. Biotechnol Lett. 2015;37:1063–1071. doi: 10.1007/s10529-015-1768-6. [DOI] [PubMed] [Google Scholar]
- Shi J., Zhan Y., Zhou M., He M., Wang Q., Li X., Wen Z., Chen S. High-level production of short branched-chain fatty acids from waste materials by genetically modified Bacillus licheniformis. Bioresour. Technol. 2019;271:325–331. doi: 10.1016/j.biortech.2018.08.134. [DOI] [PubMed] [Google Scholar]
- Vogt M., Haas S., Klaffl S., Polen T., Eggeling L., van Ooyen J., Bott M. Pushing product formation to its limit: metabolic engineering of Corynebacterium glutamicum for L-leucine overproduction. Metab. Eng. 2014;22:40–52. doi: 10.1016/j.ymben.2013.12.001. [DOI] [PubMed] [Google Scholar]
- Wang D., Wang Q., Qiu Y., Nomura C.T., Li J., Chen S. Untangling the transcription regulatory network of the bacitracin synthase operon in Bacillus licheniformis DW2. Res. Microbiol. 2017;168:515–523. doi: 10.1016/j.resmic.2017.02.010. [DOI] [PubMed] [Google Scholar]
- Wels M., Groot Kormelink T., Kleerebezem M., Siezen R.J., Francke C. An in silico analysis of T-box regulated genes and T-box evolution in prokaryotes, with emphasis on prediction of substrate specificity of transporters. BMC Genom. 2008;9:330. doi: 10.1186/1471-2164-9-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendisch V.F. Metabolic engineering advances and prospects for amino acid production. Metab. Eng. 2019;58:17–34. doi: 10.1016/j.ymben.2019.03.008. [DOI] [PubMed] [Google Scholar]
- Wohlleben W., Mast Y., Muth G., Rottgen M., Stegmann E., Weber T. Synthetic biology of secondary metabolite biosynthesis in actinomycetes: engineering precursor supply as a way to optimize antibiotic production. FEBS Lett. 2012;586:2171–2176. doi: 10.1016/j.febslet.2012.04.025. [DOI] [PubMed] [Google Scholar]
- Wu F., Cai D., Li L., Li Y., Yang H., Li J., Ma X., Chen S. Modular metabolic engineering of lysine supply for enhanced production of bacitracin in Bacillus licheniformis. Appl. Microbiol. Biotechnol. 2019;103:8799–8812. doi: 10.1007/s00253-019-10110-y. [DOI] [PubMed] [Google Scholar]
- Xie X., Xu L., Shi J., Xu Q., Chen N. Effect of transport proteins on L-isoleucine production with the L-isoleucine-producing strain Corynebacterium glutamicum YILW. J. Ind. Microbiol. Biotechnol. 2012;39:1549–1556. doi: 10.1007/s10295-012-1155-4. [DOI] [PubMed] [Google Scholar]
- Yin L., Hu X., Xu D., Ning J., Chen J., Wang X. Co-expression of feedback-resistant threonine dehydratase and acetohydroxy acid synthase increase L-isoleucine production in Corynebacterium glutamicum. Metab. Eng. 2012;14:542–550. doi: 10.1016/j.ymben.2012.06.002. [DOI] [PubMed] [Google Scholar]
- Yu W., Li D., Jia S., Liu Z., Nomura C.T., Li J., Chen S., Wang Q. Systematic metabolic pathway modification to boost L-ornithine supply for bacitracin production in Bacillus licheniformis DW2. Appl. Microbiol. Biotechnol. 2019;103:8383–8392. doi: 10.1007/s00253-019-10107-7. [DOI] [PubMed] [Google Scholar]
- Zabala D., Brana A.F., Florez A.B., Salas J.A., Mendez C. Engineering precursor metabolite pools for increasing production of antitumor mithramycins in Streptomyces argillaceus. Metab. Eng. 2013;20:187–197. doi: 10.1016/j.ymben.2013.10.002. [DOI] [PubMed] [Google Scholar]
- Zhan Y., Sheng B., Wang H., Shi J., Cai D., Yi L., Yang S., Wen Z., Ma X., Chen S. Rewiring glycerol metabolism for enhanced production of poly-gamma-glutamic acid in Bacillus licheniformis. Biotechnol. Biofuels. 2018;11:306. doi: 10.1186/s13068-018-1311-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Cai D., Xu H., Liu Z., Zhang B., Wu F., Li J., Chen S. Enhancement of precursor amino acid supplies for improving bacitracin production by activation of branched chain amino acid transporter BrnQ and deletion of its regulator gene lrp in Bacillus licheniformis. Synth Syst Biotechnol. 2018;3:236–243. doi: 10.1016/j.synbio.2018.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S., Cai D., Liu Z., Zhang B., Li J., Chen S., Ma X. Enhancement of bacitracin production by NADPH generation via overexpressing glucose-6-phosphate dehydrogenase zwf in Bacillus licheniformis. Appl. Biochem. Biotechnol. 2019;187:1502–1514. doi: 10.1007/s12010-018-2894-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.