Abstract
β-Lactam antibiotics can increase the resistance and virulence of individual intestinal microorganisms, which may affect host physiology and health. Klebsiella, a crucial gut inhabitant, has been confirmed to be resistant to most β-lactam antibiotics and contributes to the etiology of inflammatory bowel disease (IBD). In this study, the influence of amoxicillin (AMO) on Klebsiella and its role in colitis was investigated in an antibiotic cocktail (ABx) murine model. The results suggested that a 7-day AMO treatment significantly enriched the abundance of Klebsiella and enhanced serum resistance, antibiotic resistance, and biofilm formation ability of Klebsiella variicola (K. variicola) compared to the wild-type strain in the control group mice. Colonization of mice with the AMO-associated K. variicola could induce Th1 cells and inhibit Treg differentiation to promote inflammation in ABx murine model. In addition, inoculation of AMO-associated K. variicola in dextran sodium sulfate (DSS)-induced colitis murine model mice also confirmed that K. variicola colonization exacerbated inflammation as assessed by increased TNF-α, IFN-γ, IL-17a, and disease activity (DAI) levels; decreased colon length and bodyweight; and a disrupted Th1/Treg balance. The results of our study demonstrate that AMO enhances Klebsiella virulence in mice by disrupting the T cell equilibrium to exacerbate colitis, thereby providing a reference for proper antibiotic prescription.
Keywords: amoxicillin, colitis, Klebsiella variicola, proinflammation, Th1/Treg balance
Introduction
The gut microbiota has been recognized as being indispensable for maintaining host homeostasis via nutrient production and processing and immune system training (Samuli et al., 2012). Furthermore, the perturbation of the gut microbiota can promote infections, metabolic syndromes, and neurological and inflammation diseases (Menchén, 2009; Cristina et al., 2012; Rooks and Garrett, 2016; Sharon et al., 2016). Antibiotics indiscriminately kill or prevent the growth of both pathogenic and commensal bacteria, having a long-lasting impact on gut microbiota (Roberfroid et al., 2010; Maurice et al., 2013; Mikkelsen et al., 2016). Antibiotic exposure has been demonstrated to not only alter the composition and function of the gut microbiota and upregulate the expression of antibiotic resistance genes (ARGs) but also affect health by causing asthma, allergies, diarrhea, obesity, diabetes, and inflammatory bowel diseases (IBDs) in both early life and adulthood (Lewis and Pamer, 2017; Ni et al., 2017; Mccoy et al., 2018). While microbial disruption due to antibiotic has been linked to increased susceptibility to these diseases, a causal relationship has not yet been established. Amoxicillin (AMO), a β-lactam antibiotic, is one of the most widely used antibiotics worldwide due to its safety (Atli et al., 2016). Recent studies found that AMO disrupted the gut microbiota and induced ARGs that may increase the healthy risk (Korry et al., 2020; Wang et al., 2020). However, the impact of AMO on microbiome-associated diseases remains lacking. It was previously reported that penicillin-perturbed gut microbiota could be transferred to offspring, making them more susceptible to the development of colitis (Schulfer et al., 2018). In addition, ampicillin has been shown to affect the virulence and resistance of the gut microbiota by altering bacterial toxin production (Goneau et al., 2015). However, most studies have primarily focused on analyzing microbial composition and function disruption in response to antibiotic exposure (Cekanaviciute et al., 2017). An adequate understanding of the relationship between the effects of antibiotics toward specific bacteria and the paradigm of such alterations in associated disease pathogenesis remains lacking.
Klebsiella spp. are members of the intestinal flora and are among the most common intestinal opportunistic pathogens that are widely present in water, soil, and air, promoting their spread and causing infections among the immunocompromised population (Bengoechea and Sa Pessoa, 2019). These microbes can quickly acquire exogenetic genes through horizontal gene transfer from other environmental bacteria, and the resulting increase in virulence and antibiotic resistance increases their ability to cause infection in community and hospital settings (Tzouvelekis et al., 2012; Bengoechea and Sa Pessoa, 2019). Importantly, no less than one-third of Klebsiella pneumoniae (K. pneumoniae) isolates have been confirmed to be resistant to at least one antibiotic (Munoz-Price et al., 2013; Qin et al., 2014). In particular, β-lactam-resistant K. pneumoniae capable of carbapenemase production have spread across many countries and regions (Munoz-Price et al., 2013). Once exposed to these resistant pathogenic bacteria, incurable infections may occur (Patel et al., 2008). In vitro experiments have demonstrated that long-term low-dose antibiotic treatment can induce bacterial toxin production and biofilm formation and facilitate antibiotic resistance (Goneau et al., 2015). However, the effect of antibiotics on enhancing the antibiotic resistance and virulence of Klebsiella species in vivo remains unclear.
The intricate interactions between multidrug and hypervirulent Klebsiella species and the immune system may cause difficulties for therapeutic interventions (Bengoechea and Sa Pessoa, 2019). Germ-free animals bred in sterile environment was used for such comparative studies (Henrik et al., 2011). However, such a mouse model that displayed an immature and disordered immune system is expensive and requires specific infrastructure (Smith et al., 2007; Henrik et al., 2011). Generally accessible alternative broad-spectrum antibiotic administration against both Gram-positive (ampicillin and vancomycin) and Gram-negative (ampicillin and neomycin) aerobic, facultative strains and anaerobic (metronidazole), substantial deplete gut microbiota (antibiotic cocktail, ABx) in mice that possessed germ-like phenotype was therefore more used as an alternative model for germ-free animals to study host–microbe interaction (Rakoff-Nahoum et al., 2004; Henrik et al., 2011). It has been studied that Enterobacteriaceae are essential modulators of colitis severity (Sovran et al., 2018). Klebsiella can be recognized through receptors such as Toll-like receptors (TLRs) or nucleotide-binding and oligomerization domain-like receptors (NLRs). These multidrug and hypervirulent pathogenic bacteria further promote the differentiation of T cells and stimulate the production of cytokines, such as IL-1β, IL-18, TNF-α, IL-22, and IL-17, which promote inflammation (Happel et al., 2003; Takeuchi and Akira, 2010; Cai et al., 2012; Boxx and Cheng, 2016; Friedrich et al., 2019). Murine experiments have demonstrated that Th1 and Th17 cells play proinflammatory roles in many bacterial-induced intestinal diseases, while Tregs have been shown to have anti-inflammatory and tolerance-maintaining functions (Atarashi et al., 2017; Saleh et al., 2019). Recently, some studies have revealed that the ectopic colonization of oral Klebsiella strains in the intestine drives Th1 cell induction and inflammation through TLRs and IL-18 signaling of the dendritic cell-mediated pathway using germ-free mice (Atarashi et al., 2017). Although these studies have confirmed the role of Klebsiella in pathogenesis of inflammation, how the virulence and resistance of the pathogens affect the immune response during the pathogenesis of colitis remains poorly understood.
Therefore, in this study, it was hypothesized that antibiotic-dependent variations in specific bacteria in vivo would enhance the risk of colitis in mice. First, we investigated the influence of AMO on Klebsiella antibiotic resistance and virulence. Then, based on the results of these assays, an antibiotic cocktail (ABx; consisting of vancomycin, neomycin, colistin, and ampicillin) murine model was used to explore how the alterations in Klebsiella abundance and activity induce the host Th1/Treg immune reaction and exacerbate inflammation. In addition, the pathogenic role of immune system alterations caused by Klebsiella on a dextran sulfate sodium (DSS)-induced colitis murine model was assessed. These findings revealed that antibiotics enhanced Klebsiella virulence by altering Th1/Treg differentiation to exacerbate colitis.
Materials and Methods
Bacterial Strain Identification and Inoculum Preparation
The 6- to 8-week wild-type specific-pathogen-free mice (C57BL/6J) from Huafukang Company (Beijing, China) were acclimated to animal facility for 1 week and then randomly divided into two groups that administered a single course of AMO (AMO-treated group, n = 10) or water (control group, n = 10) three times per day for 7 days. Mice were housed in standard specific-pathogen-free colonies at the Laboratory Animal Resource Center of Institute of Radiation Medicine, Chinese Academy of Medical Sciences and Peking Union Medical College following relating ethical approvals of Animal Ethics and Welfare Committee (Project IRM-DWLL-2016121). During antibiotic exposure experiment, mice were maintained on a 12-h light/dark cycle with free access to food and water. AMO dosage of 25.0 mg⋅kg–1 gavaged three times for mice per day was based on the therapeutic dose achieved peak effective bacteriostatic serum concentration in human children equal to that achieved in a similar serum concentration in mice (Andes and Craig, 1998; Fonseca et al., 2013; Nobel et al., 2015; Atli et al., 2016)Craig. Three aliquots of fresh fecal samples were randomly collected from the mice in these two groups and immediately added to PBS at a ratio of 1:9 (feces/PBS; W/V). Subsequently, 100 μL of the mixture was transferred to MIAC medium to promote Klebsiella growth as previously described (Atarashi et al., 2017). A total of 28 strains were obtained, from four to five colonies of each fecal sample (Supplementary Tables S2, S3). Subsequently, the total of 28 strains were identified by 16S rRNA gene sequencing using the full-length 16S-amplicon primers 27F (5′-AGAGTTTGATCCTGGCTCAG2-3′) and 1492R (5′-GGTTACCTTGTTACGACTT-3′) (Frank et al., 2008). Because 18 colonies matching Klebsiella account for 64% of all identified strains (K. variicola occupied 78% of the identified Klebsiella, Supplementary Tables S2, S3), K. variicola was chosen for further analysis. Representative K. variicola strains K6 and Z5 were thus picked from the control and AMO-treated groups, respectively. Both K. variicola strains (K6 and Z5) were grown in Luria-Bertani (LB) medium overnight at 37°C. Subsequently, 200 μL of bacterial suspensions in water (approximately 5 × 109 CFU mL–1) were administered to antibiotic cocktail (ABx) or dextran sodium sulfate (DSS)-treated mice by intragastric gavage.
Bacterial 16S rRNA Gene Sequencing
DNA was extracted from fecal samples collected from the AMO-treated and control groups using a Magen HiPure Stool DNA kit according to the manufacturer’s instructions. The V3–V4 region of the bacterial 16S rRNA gene was amplified and sequenced using an Illumina MiSeq platform at Majorbio Bio-pharm Technology Co., Ltd. (Shanghai, China). The data analysis was performed on I-sanger platform of Majorbio1. The sequenced 16S rRNA data were deposited in NCBI Sequence Read Archive under the accession number SAMN12429552–SAMN12429557.
Whole-Genome Sequencing
Genomic DNA was extracted from K. variicola strains Z5 and K6 using a QIAamp DNA kit (Qiagen). Whole-genome sequencing was performed using an Illumina HiSeq X10 PE150 platform. The average nucleotide identity (ANI) was calculated according to a previously established method (Goris et al., 2007)2. The sequence data were deposited at NCBI under the accession numbers JAAGKS000000000 and JAAGKT000000000.
Antimicrobial Susceptibility Testing
The minimum inhibitory concentrations (MICs) of K. variicola K6 and Z5 for chloramphenicol (chl), ampicillin (amp), kanamycin (kan), gentamicin (gen), neomycin (neo), imipenem (ipm), streptomycin (str), and tetracycline (tet) were determined as previously described (Clinical and Laboratory Standards Institute [CLSI], 2009). Briefly, K. variicola cell suspensions were prepared using an overnight culture at a concentration of 0.5 MCF (n = 3 for each bacteria). Then, 100-μL aliquots of the suspensions were added to 96-well microplates containing 100 μL of serially diluted antibiotics (512 mg⋅L–1 to 1 mg⋅L–1) prepared in MH (B)medium. The optical density at 600 nm (OD600) of each well was determined using an ELISA plate reader after 20 h of incubation at 37°C. The MIC values were reported as the concentration of antibiotics that inhibited 95% of bacterial growth in broth.
Serum Resistance Assays
Serum resistance assays were performed as previously described (Gu et al., 2018). Aliquots (25 μl) of bacterial suspensions (106 CFU⋅mL–1 in saline) were mixed in 96-well microplates with 75 μl of fresh, unheated human serum collected from healthy adult volunteers (n = 3 for each test). The microplates were incubated for 3 h at 37°C, and 100-μL aliquots collected at 60, 120, and 180 min were plated onto LB agar-solidified medium. The plates were incubated overnight at 37°C, and viable bacterial colonies were enumerated. Inactivated serum preincubated at 65°C for 30 min was used as a negative control. The enumerated cells were used to calculate the survival rate.
Biofilm Formation Assays
Biofilm assays were performed in 24-well plates following previously described methods with some modifications (Khalil et al., 2019). Briefly, polymers were cut into 0.5-cm pieces and then sterilized. Three overnight cultures of K. variicola as same dilutions (106CFU⋅mL–1) were used to inoculate 24-well plates containing five polymer pieces. The biofilm biomass was quantified after 48 h of growth at 37°C using crystal violet staining as described elsewhere (Khalil et al., 2019). Assays were performed in quintuplet, and OD570 values were analyzed using GraphPad eight by t test to evaluate the differences in biofilm formation between the strains.
Animal Treatment
All animal experiments were conducted in accordance with the guidelines for animal care and use of the Chinese Academy of Medical Sciences and Peking Union Medical College following related ethical approvals of the Animal Ethics and Welfare Committee (Project IRM-DWLL-2016121).
Six- to eight-week-old specific-pathogen-free male C57BL/6J mice (Huafukang Co., Ltd., Beijing, China) weighing 18–25 g were used in this study. The animals were allowed to acclimate for 1 week before the study began. All mice were bred in standard specific-pathogen-free colonies at the Institute of Radiation Medicine, Chinese Academy of Medical Sciences and Peking Union Medical College were allowed ad libitum access to sterile food and water and maintained on a 12-h light/dark cycle. Before treatment, all mice were randomly divided into six groups. The mice were then prepared for inoculation by the oral delivery of antibiotics (low aspartame-based sweetener added for good taste to mice) or water in the drinking water (1.0 g⋅L–1 neomycin, 1.0 g⋅L–1 ampicillin, 1.0 g⋅L–1 metronidazole, and 0.5 g⋅L–1 vancomycin) for 14 days. Such duration of 14 days antibiotic treatment was chosen based on bacteria absent (<1 CFU⋅mg–1 in 107-fold dilutions) from feces aerobes (37°C for 48 h) and anaerobes (37°C for 48 h in an anaerobic chamber) incubation plated on blood agar medium. The antibiotic cocktail was freshly prepared every 36 h. After 14 days, all antibiotic-treated mice were randomly divided into five groups: the Antibiotic group (administrated water, n = 10); the K6 group (administered the K. variicola strain K6 isolated from the feces of the control mice, n = 10); the Z5 group (administered the K. variicola Z5 strain isolated from the feces of the AMO-treated mice, n = 10); the K6D group (7 days DSS treatment and then administered the K. variicola strain K6 isolated from the feces of the control mice, n = 10); and the Z5D group (7 days DSS treatment and then administered the K. variicola strain Z5 isolated from the feces of the AMO-treated mice, n = 10). The K6D and Z5D groups were given 2.5% (wt/vol) DSS (Meilun Biotechnology Co., Ltd, Dalian, China) in drinking water ad libitum for an additional 7 days after the antibiotic treatment. In addition, the K6 and Z5 groups were administered the K6 and Z5 strains at 108 CFU for 7 days. After the DSS treatment, the K6D and Z5D groups were inoculated with K6 and Z5 for the following 7 days. The water-treated mice were used as a healthy control group without antibiotic treatment (received water throughout the experiment, n = 10). The control and antibiotic group mice were euthanized along with K6 and Z5 at 21 days or K6D and Z5D treatment at 27 days. The body weight of the mice was monitored daily, while colon length was measured at the end of the experiment. Disease activity was scored as previously described (Zhang et al., 2016).
Histology and Cytokine Quantification
About 1-cm proximal colon sections located between ascending and transverse colon (1.5 cm from cecum) were harvested. Colon pieces were fixed in 10% neutral-buffered formalin, embedded in paraffin, and then stained with hematoxylin-eosin (H&E) for histological analysis. Histological analysis was performed in a blinded manner as previously described (Goneau et al., 2015). Serum samples were collected and stored at −80°C until further processing, and ELISA was performed using the serum samples according to the manufacturer’s instructions (ABclonal Inc., Wuhan, China).
Mouse Immune Cell Isolation and Flow Cytometry
Mesenteric lymph nodes, cervical lymph nodes, blood, and spleens were collected from the mice, and the samples were cut into small pieces and forced through a 100-μm cell strainer. Single-cell suspensions were stimulated with 40.5 ng⋅mL–1 of phorbol-12-myristate-13-acetate (PMA) and 700 ng⋅mL–1 of ionomycin in the presence of 2.5 μg⋅mL–1 of a protein transport inhibitor (Biolegend, #423303) for 6–8 h at 37°C. Then, the cells were incubated in 5% of fetal calf serum for 15 min and stained with anti-CD45-PerCP (Biolegend, #103130), anti-CD4-FITC (Biolegend, #100510), and anti-IFN-γ-PE/Cy7 (Biolegend, #505825). Aliquots of the cells were fixed and permeabilized with FoxP3/True-NuclearTM Transcription factor buffer set (Biolegend, #424401) and stained with anti-FoxP3-PE (Biolegend, #320007), anti-CD45-PerCP (Biolegend, #103130), and anti-CD4-FITC (Biolegend, #100510).
Statistics
Average values and standard deviations were prepared and calculated in GraphPad Prism version eight (San Diego, CA, United States). For all data displayed in graphs, the results are expressed as the mean ± SD (n = 3–7 mice for each group). Student’s t-test was used for two-group comparisons and one-way analysis of variance (ANOVA) followed by Turkey’s post hoc test was used for more than two groups. Differences with a P-value less than 0.05 were considered statistically significant.
Results
The Antibiotic Resistance and Virulence of K. variicola Are Enhanced in AMO-Treated Mice
The 16S rRNA gene was PCR amplified from mouse feces samples and sequenced to assess the influence of AMO on the microbiota. In this study, the mice treated for 7 days with AMO exhibited significantly reduced microbial community diversity compared to that observed in the control group (P < 0.05, Figure 1A). In agreement with these results, the structure of the gut microbiota was also disrupted. The principal component analysis (PCA) results showed that the microbial taxa in the AMO treatment group clustered separately from that of the control group (Figure 1B). More importantly, the relative abundance of Klebsiella (Proteobacteria) was increased by 41-fold in the AMO treatment group compared with the control group (Figure 1C). In addition, the abundance of human diseases categories (KEGG level 1) was significantly increased in the AMO group (Supplementary Figure S1A, P < 0.05). Further correlation analysis found that Klebsiella significantly correlated with enriched abundance of bacterial invasion of epithelial cells (KEGG level 3 of human diseases) (Supplementary Figure S1B, R > 0.9, P < 0.01). Because Klebsiella species has been proved to interfere with host inflammation and increases in their abundance can cause human diseases (Bengoechea and Sa Pessoa, 2019), this microbe was selected for further study.
FIGURE 1.
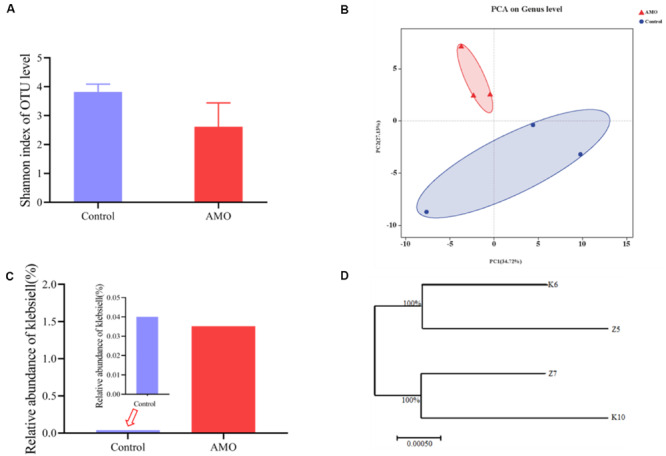
Effect of AMO on the mouse gut microbial community. (A) Diversity of microbiota after AMO treatment, assessed by Shannon index. (B) PCA plot based on the genus level between the AMO and control groups. (C) Relative percentage of Klebsiella between the AMO and control groups. (D) Cladogram of 4 representative Klebsiella isolated from AMO and control based on the full-length 16S rRNA gene sequencing results. Data presented as means ± SD (n = 3), difference at significance level *P < 0.05 (t-test) between two groups.
Culture-based methods and whole-genome sequencing were used to identify the isolated strains. Bacterial colonies grown on MacConkey inositol adonitol carbenicillin agar (MIAC) cultured from both the AMO treatment and control groups showed that OTUs mapped to Klebsiella were confirmed to be K. variicola. Two representative strains, designated as K. variicola K6 (isolated from the control group) and K. variicola Z5 (obtained from the AMO-treated group), were chosen based on the following criteria (Cekanaviciute et al., 2017): (1) they were present in both the control and AMO treatment groups to distinguish the antibiotic effect, (2) they have been reported to alter the immune response, (3) they are widely distributed pathogens associated with high morbidity and mortality, and (4) they were culturable to ensure their reproducibility. The multilocus sequence typing (MLST) analysis results using whole-genome sequencing data showed that two selected strains belonged to ST197. The ANI based on the genome sequencing data showed that the two strains have very high genomic similarity to each other (approximately 100%), suggesting that the two strains belong to the same species (Figure 1D, Supplementary Figure S2). These results indicated that K. variicola Z5 was the AMO-influenced strain corresponding to the wild-type strain K6, qualifying them for further study.
Both the antibiotic resistance and virulence of K. variicola significantly increased with the AMO treatment in the mouse intestine (Figures 2A,B). Antimicrobial susceptibility assay results showed that the MICs of imipenem and streptomycin were notably increased to 32 and 64 mg⋅L–1 in K. variicola Z5 compared to only 1 and 4 mg⋅L–1 in K. variicola K6, respectively (Figure 2C). Furthermore, the biofilm formation and serum resistance abilities of K. variicola Z5 were also significantly greater than those observed for K. variicola K6 (P < 0.01, Figures 2A,B).
FIGURE 2.
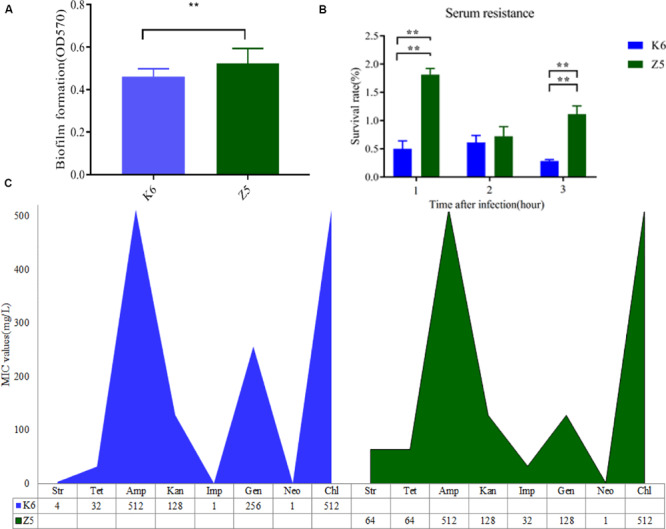
Virulence potential and antibiotic resistance profiles of K. variicola Z5 and K6. (A) Biofilm formation quantified by crystal violet staining and analysis at OD570 (n = 3). (B) Human serum resistance assays were performed using healthy human serum to assess the survival rate of 1 × 106 colony-forming units of each strain after 3 h (n = 3). (C) Antibiotic resistance characteristics of all strains using CLSI antimicrobial susceptibility testing standards (Clinical and Laboratory Standards Institute [CLSI], 2009). The different colors represent different strains, with the height of the column indicating the MIC value of the corresponding antibiotic (n = 3). Data presented as means ± SD (n = 3), difference at significance level *P < 0.05, **P < 0.01 (t-test) between two groups.
Colonization of Mice With AMO-Associated K. variicola Induces Inflammation
ABx-treated mice were inoculated with AMO-associated K. variicola Z5 and K6 strains to assess the effects of these species on inflammation during the AMO treatment of mice (Figure 3A). Control group treated with water throughout the experiment was set to healthy control in that its body weight shows normal growth. Compared to the healthy control, mice treated with multiple antibiotics lost a little weight with no detectable distress or pain within 14 days, which allowed us to further compare the specific organism’s effect (Figures 3B,C, P > 0.05). Consistent with the enhanced serum resistance and biofilm formation, K. variicola Z5 administration induced an increase in inflammation activity in ABx-treated mice that were characterized by significant decreases in weight, shorter colon lengths, and increase in higher disease activity index (DAI) values compared with those observed in the K. variicola K6, control, and antibiotic groups (P < 0.05, Figures 3B–D). Specifically, K. variicola Z5 significantly increased the serum levels of the proinflammatory cytokine IL-17a and mildly increased those of the inflammation markers TNF-α and IFN-γ (P > 0.05) (Supplementary Figure S3). In addition, the levels of the anti-inflammatory cytokine IL-10 were also increased in the K. variicola Z5 group (Supplementary Figure S3). Mice administered K. variicola Z5 exhibited more severe colonic damage that was revealed by occurrence of loosely arranged muscle fibers in muscle layers and inflammatory cell infiltration in mucous layer compared with that observed in the control group based on the results of histological structure analyses (Figure 4). In contrast, the mice administered K. variicola K6 exhibited almost no inflammation as assessed by observations of an insignificant variation in colon length, DAI value, serum IL-10, TNF-α, and IFN-γ levels and histological damage compared with that observed in the control mice (P > 0.05, Figures 3B–D, Supplementary Figure S3). Thus, these results indicated that K. variicola Z5 was responsible for the observed alterations in inflammation cytokine levels and colon histological structure.
FIGURE 3.
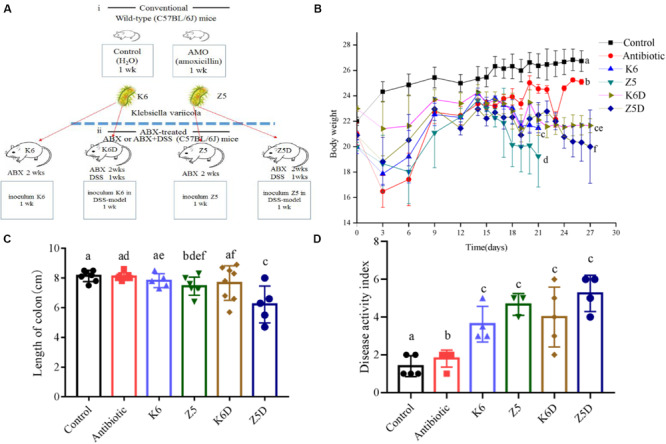
Increased proinflammation in colonized AMO-resistant K. variicola in ABx-treated or DSS-induced colitis mice. (A) Experimental design for colonization with each K. variicola strain in the ABx model. (B) Weight loss was reported as the mean ± SD from each group. (C) The length of the colon measured during treatment. (D) Disease activity index. Different letters indicate significant differences (P < 0.05, one-way ANOVA) between different groups (mean ± SD, n = 3–8 mice per group).
FIGURE 4.
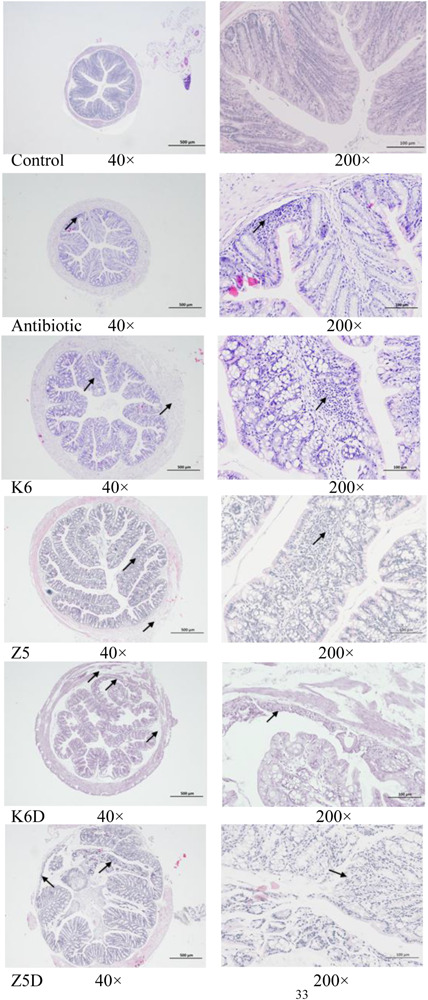
Histopathological analysis of the effects of AMO-resistant K. variicola on mouse colons in the ABx-treated or DSS-induced colitis mouse model. Representative H&E staining of the proximal colon 7 days after treatment. The arrow indicates lesions. The scale bar is labeled in the image.
AMO-Associated K. variicola Induces Th1 Cells and Inhibits Treg Differentiation
AMO associated K. variicola Z5 induced inflammation by modulating Th1/Treg differentiation. In the ABx-treated mice colonized with K. variicola Z5, T cell differentiation was assessed in multiple peripheral lymphoid tissues, including the spleens, blood, mesenteric lymph nodes (MLNs) and cervical lymph nodes (CLNs). Upon evaluation of the antibiotic and control group at 21 and 27 days, we found that multiple antibiotics did not significantly change the Th1 or Treg after antibiotic cessation (Supplementary Figure S4, P > 0.05). That may mean that neither 14 days of multiple antibiotic treatment nor 7 days (from 14 days ABx) or 14 days (from 21 days ABx) of microbial re-established after antibiotics within the gut were sufficient to have an impact on Th1 or Treg. We deem the 21- and 27-day samples of control and antibiotic as two combined whole groups in the following discussion. The results showed that K. variicola Z5 significantly increased the proportion of Th1 cells in the spleens, blood, MLNs, and CLNs compared to that observed in the control group (P < 0.05), potentially exacerbating inflammation (Figures 5A,B). No apparent induction of Th1 cells was observed in the K. variicola K6 group (P > 0.05, Figure 5B). Treg differentiation in the spleen, MLNs, CLNs, and blood were also detected (Figures 6A,B). The results demonstrated that colonization of mice with AMO-associated K. variicola Z5 inhibited Treg differentiation in the CLNs and MLNs compared to that observed in the control group (P < 0.05, Figure 6B). In contrast, the proportions of Tregs from the spleens, CLNs, MLNs, and blood of mice in the K. variicola K6 group were almost equal to those observed in the control mice (P > 0.05, Figure 6B).
FIGURE 5.
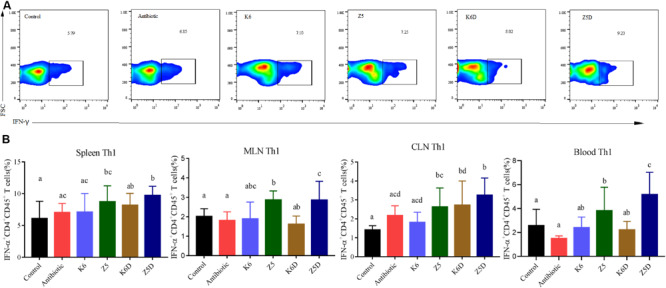
Th1 cell induction and its role in DSS-induced colitis in mice model. (A) Data represent flow plots and quantification of Th1 cell portion of spleen. (B) In the spleen, MLN, CLN, and blood in each group. Different letters indicate significant differences (P < 0.05, one-way ANOVA) between different groups (mean ± SD, n = 3–7 mice per group).
FIGURE 6.
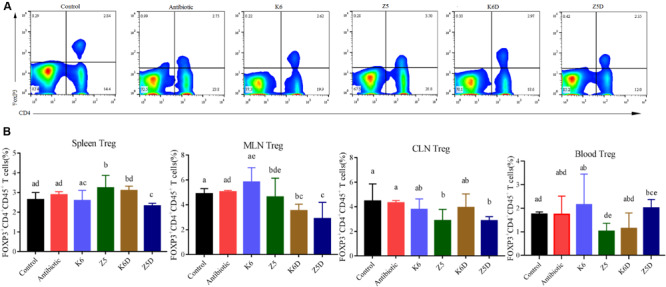
Treg inhibition and its role in DSS-induced colitis in mice. (A,B) Flow plots and quantification of Tregs in the spleen and in the spleens, MLNs, CLNs, and blood in each group. Different letters indicate significant differences (P < 0.05, one-way ANOVA) between different groups (mean ± SD, n = 3–7 mice per group).
AMO-Associated K. variicola Exacerbates Disease Severity in DSS-Induced Colitis
Colonization of mice with K. variicola Z5 recapitulated Th1/Treg differentiation and exacerbated DSS-induced colitis severity. Remarkably, the disease symptom DAI value was significantly increased, while the body weights and colon lengths of mice were notably decreased in the Z5D group (DSS model monocolonized with AMO-resistant K. variicola Z5) compared with that observed in the K6D group (DSS model monocolonized with wild K. variicola K6) or control group (P < 0.05, Figures 3B–D). Furthermore, the histological results showed enhanced colitis phenotypes with a loss of histological structure, necrosis of some intestinal gland cells, abscission of epithelial cell, and marked inflammatory cell infiltration in the mucous layers of mice in the Z5D group compared with the moderately injured K6D or control mice (Figure 4). In addition, the DSS-associated colitis immune alterations were also more obvious in the Z5D group compared with the K6D or control groups, as shown by increased serum levels of proinflammation cytokines, such as TNF-α, IFN-γ, and IL-17a (P < 0.05, Supplementary Figure S3).
The induction of Th1 cells and the inhibition of Tregs was recapitulated in a DSS-colitis model. The Z5D treatment group mice exhibited significantly increased Th1 cell induction in the spleens, blood, MLNs, and CLNs compared to that observed in mice from the K6D and control groups (P < 0.05, Figure 5B). Furthermore, decreased Treg proportion was also observed in the spleens, blood, MLNs, and CLNs (P < 0.05, Figure 6B), suggesting the inhibitory effect of AMO-associated K. variicola on lymphocyte differentiation in the DSS model (Figure 6B). The lack of Th1 induction and Treg inhibition was observed in both the K6D and control groups, which was consistent with the unaltered colitis symptoms (Figures 5, 6).
Discussion
Recently, numerous studies have reported that antibiotics reduce the taxonomic richness, diversity, and evenness of the intestinal microbial community, which may increase the risk of disease development, such as IBD, asthma, and obesity, both in the host and even in their offspring (Khan et al., 2016; Mikkelsen et al., 2016; MacPherson et al., 2018; Schulfer et al., 2018). However, it is also important to explore how antibiotics influence specific microbes to affect host functions. Klebsiella has been shown to disrupt the immune system and induce inflammation, resulting in an increased susceptibility to IBD (Bengoechea and Sa Pessoa, 2019). Nonetheless, studies on the influence of initial antibiotic use on Klebsiella within the gut are rare. Our results showed that AMO disruption of the gut microbiota was accompanied by an enhancement of the virulence and antibiotic resistance of the opportunistic pathogen K. variicola. These results also showed that AMO associated K. variicola can exacerbate the DSS-induced colitis by inducing Th1 cells and inhibiting Treg differentiation in the mice.
AMO-Associated Alterations in Intestinal Klebsiella Within the Gut
Identifying the mechanism, especially identifying the response of individual species, during antibiotic disruption is crucial for the development of precise therapies or microbial-associated dysbiosis preventative methods. Consistent with previous investigations of antibiotics, AMO treatment significantly reduced the diversity and perturbed the structure of the gut microbiota (Nobel et al., 2015; Schubert et al., 2015). Notably, in our study, the results of a genus-level analysis revealed that these responses were primarily attributed to a sharp increase in Klebsiella abundance. Reduced microbial diversity and a perturbed gut microbiota have been confirmed to be associated with immune disorder by several studies (Manichanh et al., 2006; Schulfer et al., 2018). Since AMO treatment induces microbiota disruption and is subsequently susceptible to diseases including colitis and gastrointestinal pathogen infection (Roubaud-Baudron et al., 2019), we would study the critical role of specific species. Nonetheless, due to limitations in 16S rRNA sequencing, alterations in specific bacteria among perturbed microbial communities cannot be evaluated at the species level and precisely identified within complicated bacterial community networks. The use of culture-based bacterial isolation methods enabled us to dissect the causal relationship between the microbial community and host well-being. Intriguingly, we found that the increased abundance of Klebsiella positively matches the bacterial invasion of epithelial cells. Furthermore, the observed enrichment of bacteria in the genus Klebsiella has been proved to affect the host’s immune system, and infections like Crohn’s disease and urinary tract infections (Atarashi et al., 2017; Cekanaviciute et al., 2017; Ni et al., 2017; Schirmer et al., 2019). Thus, in this study, Klebsiella strains were isolated from the AMO treatment and control groups as potential candidates to study the link between gut microbiota and colitis. Antimicrobial susceptibility, biofilm formation, and serum resistance test results revealed the enhanced antibiotic resistance and virulence of the AMO treatment group strain Z5, characteristics that are responsible for the hypervirulent potential of pathogens (Holt et al., 2015; Lam et al., 2018; Uzoechi and Abu-Lail, 2019). Although studies have shown that subinhibitory concentrations of antibiotics increase the biofilm formation of E. coli in vitro (Goneau et al., 2015), the rarely reported effects of AMO on enhancing the biofilm production and antibiotic resistance of the gut inhabitant Klebsiella in vivo were observed in our study. These findings suggest that alterations in K. variicola due to AMO treatment may promote inflammation.
AMO Treatment-Associated Alterations in Strain Z5 That Promote Mild Inflammation May Be Self-Regulated
The association of biofilm formation with the virulence of pathogens has been confirmed to be correlated with inflammation, including a decrease in E-cadherin levels, an increase in IL-6 levels and activation of the STAT3 pathway (Tomkovich et al., 2019). Increased biofilm formation by bacteria is associated with increased inflammation (Goneau et al., 2015). Combined with the observed enhancement in virulence (as assessed by biofilm formation and serum resistance tests) of AMO-associated Klebsiella, our study also showed that K. variicola Z5 could induce inflammation. The strain Z5-colonized mice also exhibited enhanced colitis in a DSS-induced colitis model, whereas an inflammation phenotype was observed in the K. variicola K6-colonized or control mice. Thus, these results demonstrate that AMO-induced alterations in K. variicola may contribute to its immune evasion and an increased risk of colitis in vivo.
Cytokines, including IL-10, IL-6, IL-8, IL-1β, TNF-α, and IFN-γ, are produced in response to host–microbe interactions to defend the host and may be important for IBD development (Friedrich et al., 2019). The IL-10 cytokine family has been shown to facilitate the gut antimicrobial defense and promote mucosal barrier integrity and repair by activating STAT3 (Shouval et al., 2014), while IL-17, TNF-α, and IFN-γ induce colitis by disrupting the mucosal barrier function of the immune system (Leppkes et al., 2009; Friedrich et al., 2019). Studies have shown that the β-lactam antibiotic oxacillin stimulates the expression of lipoprotein-like (lpl) genes in methicillin-resistant Staphylococcus aureus (MRSA), which promotes the production of the proinflammation cytokines IL-6 and TNF-α in macrophages to trigger a TLR2-dependent immune response in mice (Shang et al., 2018). In agreement with these results, we showed that AMO treatment-associated changes in K. variicola also induce the production of proinflammatory cytokines such as cytokine IL-17a, TNF-α, and IFN-γ. Unlike previous studies, the levels of the anti-inflammatory cytokine IL-10 in mice treated with AMO-resistant K. variicola were also increased. This anti-inflammatory cytokine is important in suppressing IFN-γ and TNF-α by limiting macrophage activity, the production of which in mice was shown to be promoted by Bacteroides fragilis to inhibit colitis through the suppression of IL-17a production (Mazmanian et al., 2008). Such an increase in IL-10 may be due to antigen-specific Th1 cells, which have been shown to promote IL-10 production mediated by GPR43 (Sun et al., 2018). T-effector cell production of IL-10 was shown to be a self-regulated mechanism to prevent Th1 cell-induced inflammation (Sun et al., 2018), indicating that inflammation caused by AMO-resistant K. variicola may occur via the disruption of IL-10 levels. However, once K. variicola infection occurs, especially early on, the increased antibiotic resistance and virulence of this microbe caused by AMO administration may result in it being more difficult to treat. A recent study demonstrated that the multiple sequence types of extended-spectrum β-lactamases K. variicola isolates in intensive care unit present a large challenge for therapy (Giuffrè et al., 2016). Therefore, further attention needs to be paid to Klebsiella with respect to clinical AMO prescription.
AMO-Associated Klebsiella Exacerbates Colitis by Regulating the Th1/Treg Balance
CD4 T cells play an essential role in the host immune system, regulating immune responses against a wide variety of pathogens via the differentiation of naïve CD4 T cells into Th1/Th2/Th17 cells and Tregs (Knochelmann et al., 2018). The balance of T cells, especially Th1/Th17 cells and Tregs, has emerged as having a prominent role in IBD (Knochelmann et al., 2018). Previous investigations revealed that antibiotics can significantly promote the development of colitis in IL10–/– mice by perturbing the gut microbiota, indicating that antibiotics reshape the intestinal microbiota and can affect host physiology and health (Schulfer et al., 2018). Combined with the enhanced virulence of K. variicola observed in our study, we observed that AMO-resistant K. variicola promotion of Th1 cell induction and Treg inhibition triggers inflammation. These results are in agreement with those of a study by Atarashi, who showed that ectopic Klebsiella colonization of the gut of IBD patient induces an increase in the proportion of Th1 cells (Atarashi et al., 2017). In contrast, the opposite results were observed for K. variicola K6, which proved the effect of AMO on bacterial virulence in altering the Th1/Treg equilibrium. These results all demonstrate that antibiotic treatment altering individual bacterial species can disrupt the host T cell balance. However, in contrast to the immunomodulatory effect of AMO-associated K. variicola, whether the susceptibility of the host to variations in T cells play a crucial role in colitis remains to be determined. In this study, we showed that the AMO-associated strain Z5 exacerbated colitis in a DSS-induced mouse model. Human and mouse studies have proved that the host immune response, including with respect to T cell participation, influences the severity and outcome of colitis (Feghaly et al., 2013; Saleh et al., 2019). Notably, a recent study showed that the effect of colitis-associated inflammation on CD4 T cells affects the severity and mortality of Clostridium difficile infections (Saleh et al., 2019). Therefore, the ability of AMO-resistant Klebsiella to perturb the Th1/Treg balance may explain the pathogenesis of K. variicola in proinflammation and colitis. Once Klebsiella gains access to the host, it is recognized by immune cells through pattern recognition receptors (PRRs) to induce an immune response. TLR4, TLR2, and TLR9 signaling have been shown to play an important role in Klebsiella infection recognition and defense. Mice deficient in TLR9 were failed to produce Th1 cells to clear bacteria (Urvashi et al., 2007; Ivin et al., 2017). Furthermore, TLR–/– mice showed increased mortality when inoculated with Klebsiella due to an increase in the bacterial population in the lungs (Urvashi et al., 2007). In addition, TLR4–/– × TLR2–/– mice were also observed to be more susceptible to Klebsiella infection (Bengoechea and Sa Pessoa, 2019). However, the mechanism by which Klebsiella interacts with the immune system requires further study.
Perspectives
Our study has limitations. While we demonstrate that AMO increased Klebsiella correlation with bacterial invasion of epithelial cells that verified to induce Th1 and inhibit Treg causing inflammation, the causality needs more vigorous discussion. First, more samples were not taken, including the association between microbiota and bacterial function, the comparative examination of colonic pathology, and the effect of K. variicola on inflammation due to costs and operational errors. Thus, the statistical analysis in this study only gives a glimpse of the virulence between control and AMO-associated K. variicola that warrants more sample size. Second, the antibiotic cocktail mouse model did not involve sterile mice such that we cannot exclude the possibility of recovery and expanded inhibition of commensal bacteria, especially the effect of other Gram-negative bacteria on T cell differentiation and inflammation. For example, Acinetobacter increased Th1, thus exacerbating inflammation (Cekanaviciute et al., 2017). Spatiotemporal K. variicola and its influence throughout the intestine was not established. Germ-free monocolonized mice of K. variicola were urgently needed to compare the colonization difference between small intestine and colon, especially in the mucosal layer to quantify the inflammatory response. Lastly, although we sequenced K6 and Z5, in-depth data mining was not well-performed. Genetic level of variation may well be explained by the virulence of the strains. Molecular biological validation to systemic in vivo and in vitro studies is required to definitively establish causal relationships between AMO and its perturbed K. variicola on host inflammation.
Conclusion
Antibiotic exposure causes individual bacterial species (K. variicola) to have enhanced resistance and virulence. K. variicola was shown to induce Th1 cells and inhibit Treg differentiation, which triggered proinflammation and induced mild disruption of the colon histological structure. Furthermore, these alterations aggravated the development of DSS-induced colitis, indicating their role in the pathogenesis of colitis. These results demonstrate that increased attention needs to be paid to the variation in the virulence of individual pathogens in response to antibiotic treatment and its role in promoting diseases, supporting the need for appropriate antibiotic use to limit clinical Klebsiella-associated inflammation.
Data Availability Statement
This Whole Genome Shotgun project has been deposited at NCBI SRA SRR9891530–SRR9891532 and SRR9891535–SRR9891537.
Ethics Statement
The animal study was reviewed and approved by Chinese Academy of Medical Sciences and Peking Union Medical College.
Author Contributions
YL, DM, QW, and HL conceived and coordinated the study. HL carried out the experiment, analyzed the data, and drafted the manuscript. LL and YZ carried out the laboratory work. ZC, LL, RD, and QW revised the manuscript. YL, DM, ZC, and QW provided the financial support. All authors read and approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank the Institute of Radiation Medicine, Chinese’s Academy of Medical Sciences and Peking Union Medical College for keeping and caring for the animals. And express the sincerest thank to Meng Yuan at Nankai University and Yujun Yan at Institute of Radiation Medicline for animal care and methodological guidance.
Funding. This work was supported by the Key Projects of the National Natural Science Foundation of China (41831287), the China National Science Fund for Distinguished Young Scholars (41525013), the National Natural Science Foundation of China (31870351, 31670509, 41807483, and 41703088), the Key Projects of Tianjin Natural Science Foundation (19JCZDJC40800), and the Key Projects of Research and Development of Hebei Province (19273707D).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.01256/full#supplementary-material
References
- Andes D., Craig W. A. (1998). In vivo activities of amoxicillin and amoxicillin-clavulanate against Streptococcus pneumoniae: application to breakpoint determinations. Antimicrob. Agents Chemother. 42 2375–2379. 10.1128/aac.42.9.2375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atarashi K., Suda W., Luo C., Kawaguchi T., Motoo I., Narushima S., et al. (2017). Ectopic colonization of oral bacteria in the intestine drives TH1 cell induction and inflammation. Science 358 359–365. 10.1126/science.aan4526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atli O., Demir-Ozkay U., Ilgin S., Aydin T. H., Akbulut E. N., Sener E. (2016). Evidence for neurotoxicity associated with amoxicillin in juvenile rats. Hum. Exp. Toxicol. 35 866–876. 10.1177/0960327115607948 [DOI] [PubMed] [Google Scholar]
- Bengoechea J. A., Sa Pessoa J. (2019). Klebsiella pneumoniae infection biology: living to counteract host defences. FEMS Microbiol. Rev. 43 123–144. 10.1093/femsre/fuy043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boxx G. M., Cheng G. (2016). The roles of type I interferon in bacterial infection. Cell Host Microbe 19 760–769. 10.1016/j.chom.2016.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai S., Batra S., Wakamatsu N., Pacher P., Jeyaseelan S. (2012). NLRC4 inflammasome-mediated production of IL-1β modulates mucosal immunity in the lung against gram-negative bacterial infection. J. Immunol. 188 5623–5635. 10.4049/jimmunol.1200195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cekanaviciute E., Yoo B. B., Runia T. F., Debelius J. W., Singh S., Nelson C. A., et al. (2017). Gut bacteria from multiple sclerosis patients modulate human T cells and exacerbate symptoms in mouse models. Proc. Natl. Acad. Sci. U.S.A. 114 10713–10718. 10.1073/pnas.1711235114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute [CLSI] (2009). Performance Standards for Antimicrobial Susceptibility Testing of Anaerobic Bacteria: Informational Supplement. Wayne, PA: Clinical and Laboratory Standards Institute (CLSI). [Google Scholar]
- Cristina M., Ana G. C., María V., Javier S. (2012). Cellular and molecular basis of intestinal barrier dysfunction in the irritable bowel syndrome. Gut Liver 6 305–315. 10.5009/gnl.2012.6.3.305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feghaly R. E., El Stauber J. L., Tarr P. I., Haslam D. B. (2013). Intestinal inflammatory biomarkers and outcome in pediatric clostridium difficile infections. J. Pediatr. 163 1697.e2–1704.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca W., Hoppu K., Rey L. C., Amaral J., Qazi S. (2013). Comparing pharmacokinetics of amoxicillin given twice or three times per day to children older than 3 months with pneumonia. Antimicrob. Agents Chemother. 47 997–1001. 10.1128/aac.47.3.997-1001.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank J. A., Reich C. I., Sharma S., Weisbaum J. S., Wilson B. A., Olsen G. J. (2008). Critical evaluation of two primers commonly used for amplification of bacterial 16S rRNA genes. Appl. Environ. Microbiol. 74 2461–2470. 10.1128/aem.02272-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich M., Pohin M., Powrie F. (2019). Cytokine networks in the pathophysiology of inflammatory bowel disease. Immunity 50 992–1006. 10.1016/j.immuni.2019.03.017 [DOI] [PubMed] [Google Scholar]
- Giuffrè M., Geraci D. M., Bonura C., Saporito L., Graziano G., Insinga V., et al. (2016). The increasing challenge of multidrug-resistant gram-negative bacilli: results of a 5-year active surveillance program in a neonatal intensive care unit. Medicine 95:e3016. 10.1097/md.0000000000003016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goneau L. W., Hannan T. J., MacPhee R. A., Schwartz D. J., Macklaim J. M., Gloor G. B., et al. (2015). Subinhibitory antibiotic therapy alters recurrent urinary tract infection pathogenesis through modulation of bacterial virulence and host immunity. mBio 6:e00356-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goris J., Konstantinidis K. T., Klappenbach J. A., Goris J., Konstantinidis K. T., Klappenbach J. A., et al. (2007). DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 57 81–91. 10.1099/ijs.0.64483-0 [DOI] [PubMed] [Google Scholar]
- Gu D., Dong N., Zheng Z., Lin D., Chen S. (2018). A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a chinese hospital: a molecular epidemiological study. Lancet Infect. Dis. 18 37–46. 10.1016/s1473-3099(17)30489-9 [DOI] [PubMed] [Google Scholar]
- Happel K. I., Mingquan Z., Erana Y., Quinton L. J., Euan L., Ramsay A. J., et al. (2003). Cutting edge: roles of Toll-like receptor 4 and IL-23 in IL-17 expression in response to Klebsiella pneumoniae infection. J. Immunol. 170 4432–4436. 10.4049/jimmunol.170.9.4432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrik R. D., Alexander E., Anders S., Vedrana G., Lars J. F., Peter G., et al. (2011). Depletion of murine intestinal microbiota: effects on gut mucosa and epithelial gene expression. PLos One 6:e17996. 10.1371/journal.pone.0017996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt K. E., Wertheim H., Zadoks R. N., Baker S., Whitehouse C. A., Dance D., et al. (2015). Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc. Natl. Acad. Scie. U.S.A. 112 E3574–E3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivin M., Dumigan A., de Vasconcelos F. N., Ebner F., Borroni M., Kavirayani A., et al. (2017). Natural killer cell-intrinsic type I IFN signaling controls Klebsiella pneumoniae growth during lung infection. PLoS Pathog. 13:e1006696. 10.1371/journal.ppat.1006696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil M. A. F., Hager R., Abd-El Reheem F., Mahmoud E. E., Samir T., Moawad S. S., et al. (2019). A study of the virulence traits of carbapenem-resistant klebsiella pneumoniae isolates in a galleria mellonella model. Microb. Drug Resist. 25 1063–1071. 10.1089/mdr.2018.0270 [DOI] [PubMed] [Google Scholar]
- Khan I., Azhar E. I., Abbas A. T., Kumosani T., Barbour E. K., Raoult D., et al. (2016). Metagenomic analysis of antibiotic-induced changes in gut microbiota in a pregnant rat model. Front.Pharmacol. 7:104. 10.3389/fphar.2016.00104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knochelmann H. M., Dwyer C. J., Bailey S. R., Amaya S. M., Elston D. M., Mazza-McCrann J. M., et al. (2018). When worlds collide: Th17 and Treg cells in cancer and autoimmunity. Cell Mol. Immunol. 15 458–469. 10.1038/s41423-018-0004-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korry B. J., Cabral D. J., Belenky P. (2020). Metatranscriptomics reveals antibiotic-induced resistance gene expression in the murine gut microbiota. Front. Microbiol. 11:322. 10.3389/fmicb.2020.00322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam M. M., Wick R. R., Wyres K. L., Gorrie C. L., Judd L. M., Jenney A. W., et al. (2018). Genetic diversity, mobilisation and spread of the yersiniabactin-encoding mobile element ICEKp in Klebsiella pneumoniae populations. Microb. Genomics 4:e000196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leppkes M., Becker C., Ivanov I. I., Hirth S., Wirtz S., Neufert C., et al. (2009). RORγ-Expressing Th17 cells induce murine chronic intestinal inflammation via redundant effects of IL-17A and IL-17F. Gastroenterology 136 257–267. 10.1053/j.gastro.2008.10.018 [DOI] [PubMed] [Google Scholar]
- Lewis B. B., Pamer E. G. (2017). Microbiota-based therapies for clostridium difficile and antibiotic-resistant enteric infections. Annu. Rev. Microbiol. 71 157–178. 10.1146/annurev-micro-090816-093549 [DOI] [PubMed] [Google Scholar]
- MacPherson C. W., Mathieu O., Tremblay J., Champagne J., Nantel A., Girard S.-A., et al. (2018). Gut bacterial microbiota and its resistome rapidly recover to basal state levels after short-term amoxicillin-clavulanic acid treatment in healthy adults. Sci. Rep. 8:11192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manichanh C., Rigottier-Gois L., Bonnaud E., Gloux K., Dore J. (2006). Reduced diversity of faecal microbiota in Crohn’s disease revealed by a metagenomic approach. Digest World Core Med. J. 55 205–211. 10.1136/gut.2005.073817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurice C. F., Haiser H. J., Turnbaugh P. J. (2013). Xenobiotics shape the physiology and gene expression of the active human gut microbiome. Cell 152 39–50. 10.1016/j.cell.2012.10.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazmanian S. K., Round J. L., Kasper D. L. (2008). A microbial symbiosis factor prevents intestinal inflammatory disease. Nature 453 620–625. 10.1038/nature07008 [DOI] [PubMed] [Google Scholar]
- Mccoy K. D., Ignacio A., Geuking M. B. (2018). Microbiota and Type 2 immune responses. Curr. Opin. Immunol. 54 20–27. 10.1016/j.coi.2018.05.009 [DOI] [PubMed] [Google Scholar]
- Menchén L. (2009). [Cellular and molecular bases of intestinal barrier dysfunction induced by experimental stress]. Gastroenterol. Hepatol. 32 (Suppl. 2), 55–61. [DOI] [PubMed] [Google Scholar]
- Mikkelsen K. H., Allin K. H., Knop F. K. (2016). Effect of antibiotics on gut microbiota, glucose metabolism and body weight regulation: a review of the literature. Diabetes Obesity Metab. 18 444–453. 10.1111/dom.12637 [DOI] [PubMed] [Google Scholar]
- Munoz-Price L. S., Poirel L., Bonomo R. A., Schwaber M. J., Daikos G. L., Cormican M., et al. (2013). Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect. Dis. 13 785–796. 10.1016/s1473-3099(13)70190-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni J., Wu G. D., Albenberg L., Tomov V. T. (2017). Gut microbiota and IBD: causation or correlation? Nat. Rev. Gastroenterol. Hepatol. 14 573–584. 10.1038/nrgastro.2017.88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobel Y. R., Cox L. M., Kirigin F. F., Bokulich N. A., Yamanishi S., Teitler I., et al. (2015). Metabolic and metagenomic outcomes from early-life pulsed antibiotic treatment. Nat. Commun. 6:7486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel G., Huprikar S., Factor S. H., Jenkins S. G., Calfee D. P. (2008). Outcomes of carbapenem-resistant Klebsiella pneumoniae infection and the impact of antimicrobial and adjunctive therapies. Infect. Control Hospital Epidemiol. 29 1099–1106. 10.1086/592412 [DOI] [PubMed] [Google Scholar]
- Qin S., Fu Y., Zhang Q., Qi H., Wen J. G., Xu H., et al. (2014). High incidence and endemic spread of NDM-1-positive Enterobacteriaceae in henan province. China. Antimicrob. Agents Chemother. 58 4275–4282. 10.1128/aac.02813-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakoff-Nahoum S., Paglino J., Eslami-Varzaneh F., Edberg S., Medzhitov R. (2004). Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell 118 229–241. 10.1016/j.cell.2004.07.002 [DOI] [PubMed] [Google Scholar]
- Roberfroid M., Gibson G. R., Hoyles L., McCartney A. L., Rastall R., Rowland I., et al. (2010). Prebiotic effects: metabolic and health benefits. Br. J. Nutr. 104 S1–S63. [DOI] [PubMed] [Google Scholar]
- Rooks M. G., Garrett W. S. (2016). Gut microbiota, metabolites and host immunity. Nat. Rev. Immunol. 16 341–352. 10.1038/nri.2016.42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roubaud-Baudron C., Ruiz V. E., Swan A. M., Vallance B. A., Ozkul C., Pei Z., et al. (2019). Long-term effects of early-life antibiotic exposure on resistance to subsequent bacterial infection. mBio 10:e02820-19. 10.1128/mBio.02820-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh M. M., Frisbee A. L., Leslie J. L., Buonomo E. L., Cowardin C. A., Ma J. Z., et al. (2019). Colitis-induced Th17 cells increase the risk for severe subsequent clostridium difficile Infection. Cell Host Microbe 25 756–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuli R., Raakel L., Seppo S., Erika I. (2012). Microbial contact during pregnancy, intestinal colonization and human disease. Nat. Rev. Gastroenterol. Hepatol. 9 565–576. 10.1038/nrgastro.2012.144 [DOI] [PubMed] [Google Scholar]
- Schirmer M., Garner A., Vlamakis H., Xavier R. J. (2019). Microbial genes and pathways in inflammatory bowel disease. Nat. Rev. Microbiol. 17 497–511. 10.1038/s41579-019-0213-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert A. M., Sinani H., Schloss P. D. (2015). Antibiotic-induced alterations of the murine gut microbiota and subsequent effects on colonization resistance against Clostridium difficile. MBio 6:e00974-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulfer A. F., Battaglia T., Alvarez Y., Bijnens L., Ruiz V. E., Ho M., et al. (2018). Intergenerational transfer of antibiotic-perturbed microbiota enhances colitis in susceptible mice. Nat. Microbiol. 3 234–242. 10.1038/s41564-017-0075-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang W., Rao Y., Ying Z., Yi Y., Hu Q., Zhen H., et al. (2018). β-lactam antibiotics stimulate the pathogenicity of methicillin-resistant Staphylococcus aureus Via SarA-controlled tandem lipoprotein expression. mBio 10:e00880-19. 10.1128/mBio.00880-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon G., Sampson T. R., Geschwind D. H., Mazmanian S. K. (2016). The central nervous system and the gut microbiome. Cell 167 915–932. 10.1016/j.cell.2016.10.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shouval D. S., Biswas A., Goettel Jeremy A., Mccann K., Conaway E., Redhu Naresh S., et al. (2014). Interleukin-10 receptor signaling in innate immune cells regulates mucosal immune tolerance and anti-inflammatory macrophage function. Immunity 40 706–719. 10.1016/j.immuni.2014.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith K., Mccoy K. D., Macpherson A. J. (2007). Use of axenic animals in studying the adaptation of mammals to their commensal intestinal microbiota. Semin. Immunol. 19 59–69. 10.1016/j.smim.2006.10.002 [DOI] [PubMed] [Google Scholar]
- Sovran B., Planchais J., Jegou S., Straube M., Sokol H. (2018). Enterobacteriaceae are essential for the modulation of colitis severity by fungi. Microbiome 6:152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun M., Wu W., Chen L., Yang W., Huang X., Ma C., et al. (2018). Microbiota-derived short-chain fatty acids promote Th1 cell IL-10 production to maintain intestinal homeostasis. Nat. Commun. 9:3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi O., Akira S. (2010). Pattern recognition receptors and inflammation. Cell 140 805–820. 10.1016/j.cell.2010.01.022 [DOI] [PubMed] [Google Scholar]
- Tomkovich S., Dejea C. M., Winglee K., Drewes J. L., Chung L., Housseau F., et al. (2019). Human colon mucosal biofilms from healthy or colon cancer hosts are carcinogenic. J. Clin. Investigat. 130 1699–1712. 10.1172/jci124196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzouvelekis L. S., Markogiannakis A., Psichogiou M., Tassios P. T., Daikos G. L. (2012). Carbapenemases in Klebsiella pneumoniae and other Enterobacteriaceae: an evolving crisis of global dimensions. Clin. Microbiol. Rev. 25:682. 10.1128/cmr.05035-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urvashi B., Lukacs N. W., Osterholzer J. J., Newstead M. W., Xianying Z., Moore T. A., et al. (2007). TLR9 is required for protective innate immunity in Gram-negative bacterial pneumonia: role of dendritic cells. J. Immunol. 179 3937–3946. 10.4049/jimmunol.179.6.3937 [DOI] [PubMed] [Google Scholar]
- Uzoechi S. C., Abu-Lail N. I. (2019). Changes in cellular elasticities and conformational properties of bacterial surface biopolymers of multidrug-resistant escherichia coli (mdr-e. coli) strains in response to ampicillin. Cell Surface 5:100019. 10.1016/j.tcsw.2019.100019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Duan Y.-J., Wang S.-P., Wang L.-T., Hou Z.-L., Cui Y.-X., et al. (2020). Occurrence and distribution of clinical and veterinary antibiotics in the faeces of a Chinese population. J. Hazardous Mater. 383:121129. 10.1016/j.jhazmat.2019.121129 [DOI] [PubMed] [Google Scholar]
- Zhang Z., Wu X., Cao S., Wang L., Wang D., Yang H., et al. (2016). Caffeic acid ameliorates colitis in association with increased Akkermansia population in the gut microbiota of mice. Oncotarget 7:31790. 10.18632/oncotarget.9306 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This Whole Genome Shotgun project has been deposited at NCBI SRA SRR9891530–SRR9891532 and SRR9891535–SRR9891537.


