Abstract
Acute myelogenous leukemia (AML) is a complex blood malignancy leading to immature leukemic stem cells (LSCs) proliferation. T cell immunoglobulin mucin-3 (TIM-3) is known as a biomarker of AML LSCs. Several microRNAs (miRNAs) can affect gene expression in AML. In this study, the silencing effect of miR-133a-5p on TIM-3 expression in AML cell lineage (HL-60) was investigated. It’s been hypothesized that miR-133a-5p may suppress the TIM-3 expression in AML cell line. Initially, miRNA-TIM-3 prediction, enrichment, and network analysis were done. Then, miR-133a-5p mimic was transfected into HL-60 cells. The TIM-3 protein and gene expression were measured by flow cytometry analysis and real-time PCR, respectively. MTT assay was also carried out. Based on the Bioinformatics predictions, miR-133a-5p was able to silence TIM-3 expression. Also, significant pathways pertained to miR-133a-5p were obtained using enrichment analysis. According to this, miR-133a-5p was mainly engaged in the MAPK signaling pathway and Nicotine addiction pathway using the KEGG database. The TIM-3 protein expression of the transfected cells was measured as 17.15 ± 8.87% (p = 0.001). A 52.48% significant gene silencing in mRNA level was obtained in comparison to the negative control. Despite of down regulation of TIM-3, HL-60 cell viability has not been significantly changed. It has been finally confirmed that miR-133a-5p could strongly suppress TIM-3 expression in AML cell line. Presumably, down regulation of TIM-3 could affect MAPK and Nicotine addiction signaling pathways.
Electronic supplementary material
The online version of this article (10.1007/s12291-019-00834-z) contains supplementary material, which is available to authorized users.
Keywords: miR-133a-5p, TIM-3, AML, HL-60, Silencing
Introduction
Acute myelogenous leukemia (AML) is a complex blood malignancy that is prevalent in adults [1, 2]. In AML patients, both of the leukemic stem cells (LSCs) and the hematopoietic stem cells (HSCs) are observed in bone marrow. Hence, to facilitate the diagnostic and therapeutic purposes, finding a biomarker to discriminate between them is essential. Among all investigated proteins presented on LSCs surfaces, TIM-3 showed a specific expression [3, 4]; while normal HSCs do not express it. Thus, TIM-3 could be used as a target for confronting LSCs via inhibitory methods like monoclonal antibodies and microRNAs [5]. As mentioned above, TIM-3 blocking with the aim of AML remission causes autoimmunity. Therefore, a provident fight against AML with the minimum harmful consequences is needed. MicroRNAs, as the non-coding RNAs, can interact with their mRNA targets and are able to regulate the gene expression at the post-transcriptional mechanism. These non-coding RNAs can bind to their mRNA target leading to mRNA degradation or translational inhibition and also, activate the gene expression. Using the inhibitory role of microRNAs, the scientists design and synthesize the microRNA mimic molecules to suppress the mRNA targets. It is demonstrated that the critical region of microRNAs for silencing their respective mRNAs is limited to the seed region, including nucleotides 2–8 at the 5′ end of microRNA [6]. Also, microRNAs are classified as tumor suppressors or oncogenes, related to the gene they regulate. Implications of microRNAs in various cellular functions like proliferation, differentiation, and development, justify their association with cancers [7, 8]. In fact, genetic aberration leading to abnormalities in the regulation of microRNAs indicates their involvement in cancers [9]. Due to the TIM-3 specific expression on AML LSCs and also according to our bioinformatic studies, we assumed that targeting TIM-3 via miR-133a-5p mimic can possibly suppress TIM-3 expression on HL-60 cells (AML cell line) and presumably open a new window to cure the AML in the future.
Materials and Methods
In Silico Analysis
For predicting the engagement of microRNA and TIM-3, the online server Mirwalk 2.0 (http://zmf.umm.uni-heidelberg.de/apps/zmf/mirwalk2/) was applied by TIM3 RefSeq ID (NM_032782.4). Then, TargetScan 7.2 (http://www.targetscan.org/) was used for validation of the Mirwalk predicted interaction between microRNA and TIM-3. miRTargetLink Human (https://ccb-web.cs.uni-saarland.de/mirtargetlink/) and mirDIP (http://ophid.utoronto.ca/mirDIP/index.jsp) were applied to gain the weak to strong predicted targets. The targets of miR-133a-5p were achieved using the options including weaker evidence and predicted interactions from miRTargetLink Human. Also, the targets of this microRNA were obtained according to the score class (very high, high and medium) from mirDIP. Also, Kyoto encyclopedia of genes and genomes (KEGG) biological pathway (http://www.genome.jp/) and ShinyGO v0.41 (Gene Ontology Enrichment Analysis + more) (http://bioinformatics.sdstate.edu/go/) by p value cutoff = 0.05 for false discovery rate (FDR) were used to identify the gene networks and gene ontology enrichment analysis.
Cell Culture
HL-60 cell line was purchased from the Pasture Institute, Iran. These cells were cultured in RPMI-1640 (BIO-IDEA, Iran) containing 10% Fetal bovine serum (FBS) and 1% Pen-Strep (BIO-IDEA, Iran).
Transfection
24 h prior to transfection, 105 HL-60 cells were seeded in each well of 24-well plate. For TIM-3 expression induction, the cells were treated with 50 ng PMA (Sigma Aldrich, Germany). Then, miR-133a-5p mimic (Qiagene, Germany) with an ultimate concentration of 50 nM was applied for transfection to HL-60 cells by using X-tremeGENE transfection reagent (Roche, Germany). As an indicator for transfection and also scramble siRNA, LableIT siRNA delivery control labeled with FITC (Mirus, USA) was used. Moreover, for comparison, mock (cells treated with only X-tremeGENE transfection reagent) and negative control (cells without treatment) were applied in this study. Cells were refilled with fresh medium after overnight incubation.
Cell Viability Test
HL-60 cells viability were assessed by Methylthiazole tetrazolium (MTT) assay. To do so, in each well of 96-well plate (Orang, Belgium) containing 90 μl medium, 10 μl MTT solution (5 mg/ml) in PBS buffer (Sigma-Aldrich, Germany) was added that was followed by incubation at 37 °C and 5% CO2 for 1 h. After removing the medium, the cells were frozen at − 80 °C for 1 h. Then, 100 μl dimethyl sulfoxide (DMSO) (Sigma-Aldrich, Germany) was added to each well while it was shaking for 30 min. The optical density of each well of the plate was measured using microplate reader at 590 nm and results were reported as a percentage in comparison to the negative control (untransfected cells).
Reverse Transcriptase and Quantitative Real-Time PCR
HL-60 cells whole RNA was isolated by the RNX Plus kit (Cinnagen, Iran) according to manufacturer recommended protocol. For cDNA synthesis, the Revert Identify Transcription Kit (Fermentase, Germany) was used following the manufacturer’s instruction. TIM-3 expression level was measured through quantitative real-time PCR (Applied Biosystem, USA) and maxims SYBER Green qPCR master mix kit (Fermentase, Germany). ACTB was used as a housekeeping gene in this study and specific primers for ACTB and TIM-3 were designed by Allele ID 7.0 software (Table 1). The following conditions was performed for real-time PCR, 40 cycles containing denaturation at 95 °C for 15 s, annealing at 60 °C for 1 min, and extension at 72 °C for 15 s. Finally, 2−ΔΔCt method was used for TIM-3 relative gene expression.
Table 1.
Primer sequences used for real-time PCR
| Specific primers | Direction | Sequence |
|---|---|---|
| β-Actin | Forward | 5′-TTCGAGCAAGAGATGGCCA-3′ |
| Reverse | 5′-CACAGGACTCCATGCCCAG-3′ | |
| TIM-3 | Forward | 5′-CCATCAGAATAGGCATCTACATC-3′ |
| Reverse | 5′-CCATCAGAATAGGCATCTACATC-3′ |
Flow Cytometry Analysis
TIM-3 protein expression on HL-60 cells was evaluated using FACS Callibuor device (BD Bioscience, San Jose, USA) and anti-human TIM-3-PE antibody (eBiosciences, USA) according to the manufacturer protocol. Also, the Cell Quest Pro software (BD Bioscience, San Jose, USA) was accomplished for data analysis.
Statistical Analysis
Statistical analysis was performed by SPSS16.0 software (SPSS Inc, Chicago, USA) through One-way ANOVA test. Results were showed as mean percentage ± relative standard deviation, and p values < 0.05 were considered as meaningful.
Results
Prediction of miR-133a-5p as a TIM-3 Silencer microRNA
MiR-133a-5p was predicted as a TIM-3 expression suppresser according to the miRWalk output by 9/12 of chosen web servers (miRWalk, miRanda, Microt4, miRDB, miRMap, RNA22, PITA, Targetscan, and RNAhybrid). It should be noted that there are no experimental researches to prove this prediction. Moreover, the context score percentile of miR-133a-5p for TIM-3 expression suppression predicted as 92%, based on the Targetscan output.
Enrichment Analysis Outcomes
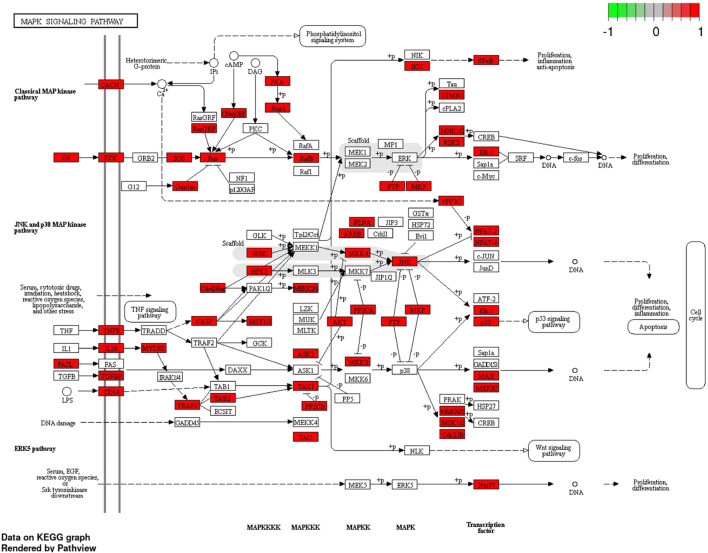
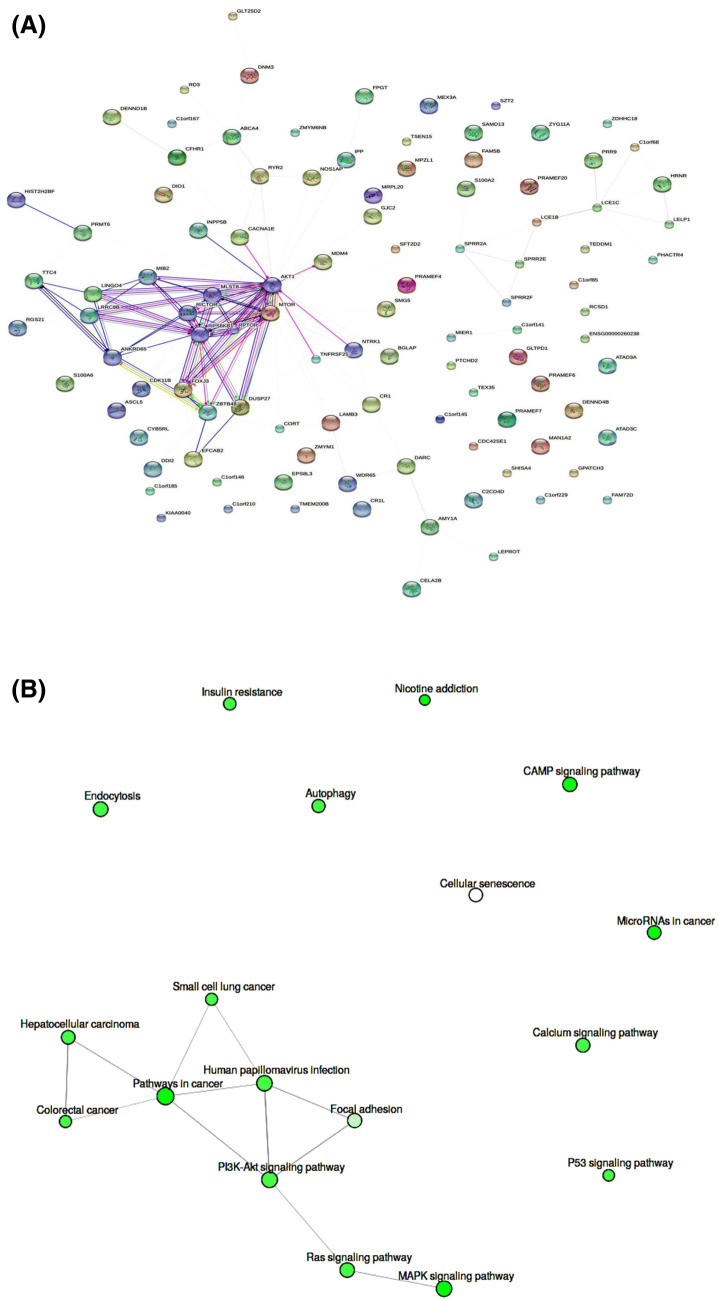
The mirDIP predicted target genes of miR-133a-5p had integrated scores ranging from 0.2 to 0.014. Also, 63 genes were resulted as the predicted targets of miR-133a-5p by miRTargetLink Human. Using KEGG server, 5131 predicted target genes were totally pertained to miR-133a-5p (Table S1) and located mainly on the chromosomes 1, 2, 3, 11, 12, and 17 (data not shown). Also, MAPK signaling pathway and Nicotine addiction pathway for miR-133a-5p were involved as the top predicted pathways by p = 2.2E−02 (Table 2). MAPK signaling pathway and its engaged components in enrichment analysis of miR-133a-5p were depicted in Fig. 1. Other significant pathways with highest Enrichment FDR raging from 2.7E−02 to 4.0E−02 for this microRNA were included: pathways in cancer, microRNAs in cancer, CAMP signaling pathway, Ras signaling pathway, Calcium signaling pathway, P53 signaling pathway, Autophagy, Endocytosis, PI3K–Akt signaling pathway, Insulin resistance, Human papillomavirus infection, Colorectal cancer, Small cell lung cancer, Hepatocellular carcinoma, Focal adhesion, and Cellular senescence (Table 2). Also, the STRING protein interaction in the total predicted network and pathway interactions were depicted (Fig. 2a, b). Most of the pathway interactions were occurred among MAPK signaling pathway, Ras signaling pathway, PI3K–Akt signaling pathway, Focal adhesion, Human papillomavirus infection, pathways in cancer, Colorectal cancer, Hepatocellular carcinoma, and Small cell lung cancer (Fig. 2b).
Table 2.
The enrichment analysis results of miR-133a-5p by ShinyGO v0.41 web server
| Enrichment FDR | Genes in list | Total genes | Functional category |
|---|---|---|---|
| 2.20E−02 | 95 | 295 | MAPK signaling pathway |
| 2.20E−02 | 20 | 40 | Nicotine addiction |
| 2.70E−02 | 152 | 523 | Pathways in cancer |
| 2.70E−02 | 53 | 150 | MicroRNAs in cancer |
| 3.00E−02 | 65 | 196 | CAMP signaling pathway |
| 3.30E−02 | 72 | 230 | Ras signaling pathway |
| 3.30E−02 | 59 | 180 | Calcium signaling pathway |
| 3.30E−02 | 28 | 72 | P53 signaling pathway |
| 3.30E−02 | 45 | 128 | Autophagy |
| 3.30E−02 | 76 | 243 | Endocytosis |
| 3.30E−02 | 106 | 353 | PI3K–Akt signaling pathway |
| 3.30E−02 | 38 | 107 | Insulin resistance |
| 3.30E−02 | 95 | 318 | Human papillomavirus infection |
| 3.30E−02 | 33 | 86 | Colorectal cancer |
| 3.30E−02 | 34 | 93 | Small cell lung cancer |
| 3.30E−02 | 55 | 167 | Hepatocellular carcinoma |
| 3.90E−02 | 63 | 199 | Focal adhesion |
| 4.00E−02 | 52 | 159 | Cellular senescence |
Fig. 1.
MAPK signaling pathway and its engaged components in enrichment analysis of miR-133a-5p
Fig. 2.
a The STRING protein interaction in the total predicted network. In this image, 105 nodes and 124 edges were considered. b pathway interactions resulted from the analysis of miR-133a-5p
TIM-3 Expression Induction on HL-60 Cells by PMA
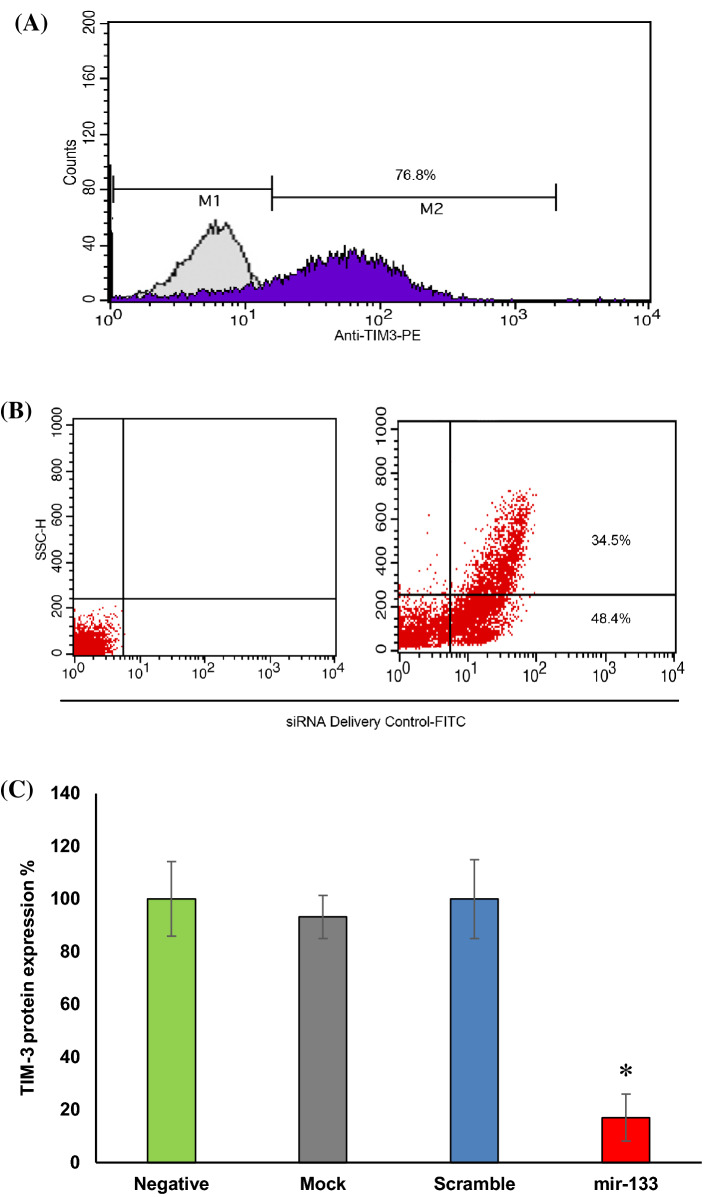
After 72 h of HL-60 cells treatment by PMA, TIM-3 protein expression on the cells was measured 76.8% while it was 0.08% without PMA treatment (Fig. 3a). Also, 82.9% of the cells were successfully transfected after 24 h of using LableIT siRNA Delivery Control-FITC (Fig. 3b).
Fig. 3.
Flow cytometry analysis. a The histogram represents that 76.8% of the HL-60 cells were expressed TIM-3 protein after PMA treatment. b The histogram demonstrates the 82.9% successful transfection of the cells with FITC labeled siRNA. c The graph indicates the significant suppression of TIM-3 protein expression (p < 0.05) after cells transfection with miR133a-5p mimic in comparison to the negative control. Among the negative, mock, and scrambled groups no significant difference was observed. The asterisk (*) represents statistical significance
TIM-3 Expression Suppression on HL-60 Cell Line by miR-133a-5p Mimic
The TIM-3 protein expression suppression on the cells was confirmed by anti-TIM-3-PE antibody after 48 h of transfection with miR-133a-5p mimic (72 h after TIM-3 induction by PMA) using flow cytometry method. The TIM-3 protein expression on the transfected cells was measured as 17.15 ± 8.87% (p = 0.001).
Confirmation of Alteration in TIM-3 Expression on HL-60 Cell Line by Real-Time PCR
Our results represented a significant 82.85% gene silencing in translation level compared with the negative control. Moreover, no meaningful difference was observed among the negative, mock, and scrambled controls which demonstrated the silencing process was done specifically (Fig. 3c, Table 3). Also, the silencing effect of miR-133a-5p on TIM-3 expression in transcription level using real-time PCR was determined as 47.52 (p = 0.0001). According to our analysis, 52.48% significant gene silencing in mRNA level was obtained in comparison to the negative control. Again there was no meaningful difference between the negative, mock, and scrambled groups (Fig. 4a, Table 3).
Table 3.
The fold change of gene expression of TIM-3 in this study and their statistical results
| Negative | Mock | Scramble | miR-133a | |
|---|---|---|---|---|
| 1.11 | 1.48 | 0.98 | 0.48 | |
| 0.91 | 0.52 | 1.04 | 0.48 | |
| 1.01 | 1.00 | 1.01 | 0.48 | AVE |
| 0.14 | 0.68 | 0.04 | 0.00 | SD |
| 100.00 | 99.01 | 100.00 | 47.52 | % |
| 1.00 | 4.80 | 0.30 | 0.00 | Rel. SD |
| p = 0.0001 |
Fig. 4.
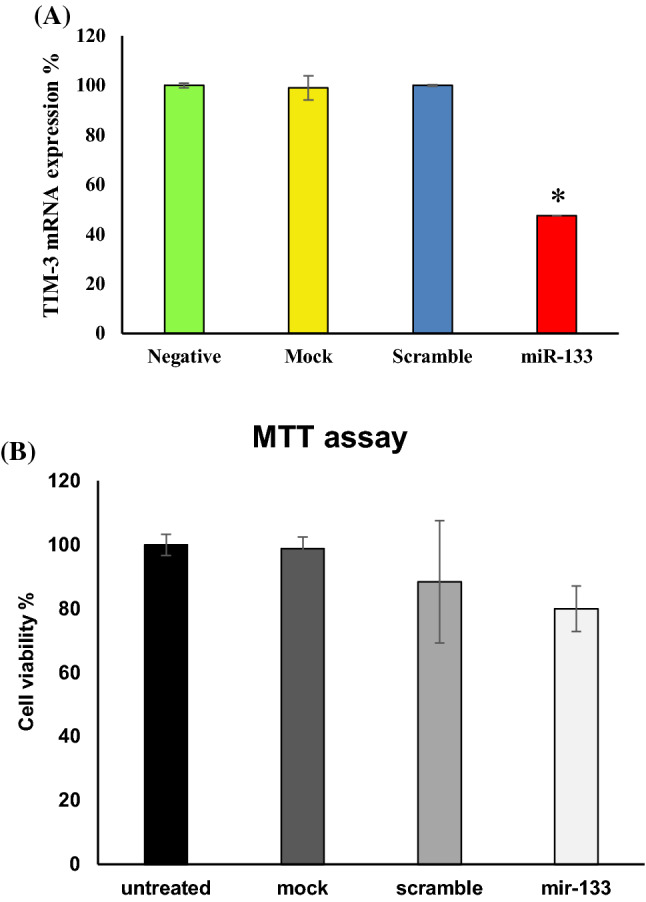
a Measurment of the TIM-3 expression by real-time PCR analysis. Transfected HL-60 cells with miR133a-5p mimic showed a significant down regulation of TIM-3 (p < 0.05) compared with the negative control. There was no meaningful difference between the negative, mock, and scrambled controls. The asterisk (*) expresses statistical significance. b Evaluation of cell viability by MTT assay. Cell viability in transfected cells was measured as 80.037% compared with the negative control which was not statistically meaningful
HL-60 Cells Viability Evaluation After Transfection with miR-133a-5p
Using MTT assay, the transfected cells viability was evaluated as 80.037 ± 7.031% which had no meaningful difference with the negative, mock, and scrambled controls (p > 0.05). This indicated that in spite of reducing HL-60 cell line viability by silencing TIM-3 gene expression utilizing has-miR-133a-5p mimic, this decrease was not statistically significant (Fig. 4b).
Discussion
AML is a disease of blood malignancy in which LSCs are abnormally proliferated [1, 2]. LSCs and HSCs could be distinguished by the molecules expressed on their surfaces. Among them, TIM-3 has been characterized for its particular expression on LSCs [3, 4]. So it could be considered as a suitable candidate for confronting AML LSCs [4]. Combining these findings with the role of microRNA mimics in the experimental procedures, the silencing effect of miR-133-5p mimic on TIM-3 expression in AML cell line (HL-60) was investigated in the present study. According to our bioinformatics studies, miR-133a-5p with 92% suppressing effect on TIM-3 expression has not been yet validated in vitro. Here, we found that miR-133a-5p functioned as a negative regulator of TIM-3 expression on HL-60 cells in both transcription and translation levels, according to real-time PCR and flow cytometry analysis, respectively. The significant difference of silencing effect between TIM-3’s mRNA and protein levels represents that this microRNA might play its suppressing role on TIM-3 expression at the mRNA translational level by steric hindrance instead of mRNA degradation, but this probability should be proved in the future in vitro and in vivo researches.
In a study, Fooladinezhad et al. [10] found that TIM-3 expression was suppressed in AML cell line using miR-330-5p and in another research by Emamdoost et al. [11] the inhibitory effect of miR-125a-3p on TIM-3 expression in HL-60 cells was proved. Also, several studies have demonstrated that miR-133a-5p had tumor suppressor properties in various human cancers, including gastric cancer [12], colorectal cancer [13], esophageal squamous cell carcinoma [14], non-small cell lung cancer [15], osteosarcoma [16], ovarian cancer [17] and breast cancer [18]. It seemed that the pathogenesis and the molecular basis of these cancers are different, and thus the roles and the gene targets and engaged signaling pathways of miR-133a-5p in cancers can be possibly varied. According to our enrichment analysis, miR-133a-5p was mainly engaged in MAPK signaling pathway and Nicotine addiction pathway using KEGG database. Presumably, down regulation of TIM-3 by miR-133a-5p mimic could influence the yield of these signaling pathways and their interaction, but these in silico results have to evaluate by further in vitro and in vivo experiments. In the current study, in spite of miR-133a-5p silencing effect, MTT assay showed the decreased viability and apoptosis of HL-60 cells which was not statistically significant. However, further experimental investigations can assess these results.
Conclusion
Altogether, the predicted bioinformatic results showed that transfected miR-133a-5p mimic could suppress TIM-3 gene expression. Also, the enrichment and network analysis indicated that miR-133a-5p was mainly engaged in MAPK signaling pathway and Nicotine addiction pathway using KEGG database. The prediction outcomes were proved at the mRNA and protein levels by quantitative RT-PCR and flow cytometry, respectively. However, MTT assay represented the reduced viability and apoptosis of HL-60 cells which was not statistically significant. Probably, the down regulation of TIM-3 can influence the yield of the obtained signaling pathways.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We greatly acknowledge Department of Research at University of Isfahan, Isfahan, Iran for their collaborations.
Funding
This study was performed at the University of Isfahan and Isfahan University of Medical Sciences and financially supported by the Graduate office of University of Isfahan, Isfahan, Iran.
Compliance with Ethical Standards
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical Standards
The experiments comply with the current laws of the country in which they were performed.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Shipley JL, Butera JN. Acute myelogenous leukemia. Exp Hematol. 2009;37(6):649–658. doi: 10.1016/j.exphem.2009.04.002. [DOI] [PubMed] [Google Scholar]
- 2.De Kouchkovsky I, Abdul-Hay M. Acute myeloid leukemia: a comprehensive review and 2016 update. Blood Cancer J. 2016;6(7):e441. doi: 10.1038/bcj.2016.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jan M, Chao MP, Cha AC, Alizadeh AA, Gentles AJ, Weissman IL, et al. Prospective separation of normal and leukemic stem cells based on differential expression of TIM3, a human acute myeloid leukemia stem cell marker. Proc Natl Acad Sci USA. 2011;108(12):5009–5014. doi: 10.1073/pnas.1100551108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kikushige Y, Shima T, Takayanagi S-I, Urata S, Miyamoto T, Iwasaki H, et al. TIM-3 is a promising target to selectively kill acute myeloid leukemia stem cells. Cell Stem Cell. 2010;7(6):708–717. doi: 10.1016/j.stem.2010.11.014. [DOI] [PubMed] [Google Scholar]
- 5.Kikushige Y, Miyamoto T. TIM-3 as a novel therapeutic target for eradicating acute myelogenous leukemia stem cells. Int J Hematol. 2013;98(6):627–633. doi: 10.1007/s12185-013-1433-6. [DOI] [PubMed] [Google Scholar]
- 6.O’Brien J, Hayder H, Zayed Y, Peng C. Overview of microrna biogenesis, mechanisms of actions, and circulation. Front Endocrinol. 2018;9:402. doi: 10.3389/fendo.2018.00402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cho WC. MicroRNAs in cancer-from research to therapy. BBA Rev Cancer. 2010;1805(2):209–217. doi: 10.1016/j.bbcan.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 8.Wiemer EA. The role of microRNAs in cancer: no small matter. Eur J Cancer. 2007;43(10):1529–1544. doi: 10.1016/j.ejca.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 9.Cho WC. OncomiRs: the discovery and progress of microRNAs in cancers. Mol Cancer. 2007;6(1):60. doi: 10.1186/1476-4598-6-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fooladinezhad H, Khanahmad H, Ganjalikhani-Hakemi M, Doosti A. Negative regulation of TIM-3 expression in AML cell line (HL-60) using miR-330-5p. Br J Biomed Sci. 2016;73(3):129–133. doi: 10.1080/09674845.2016.1194564. [DOI] [PubMed] [Google Scholar]
- 11.Emamdoost F, Khanahmad H, Ganjalikhani-hakemi M, Doosti A. The miR-125a-3p inhibits TIM-3 expression in AML cell line HL-60 in vitro. Indian J Hematol Blood Transfus. 2017;33(3):342–47. doi: 10.1007/s12288-016-0733-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gong Y, Ren J, Liu K, Tang L-M. Tumor suppressor role of miR-133a in gastric cancer by repressing IGF1R. World J Gastroenterol. 2015;21(10):2949. doi: 10.3748/wjg.v21.i10.2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dong Y, Zhao J, Wu C-W, Zhang L, Liu X, Kang W, et al. Tumor suppressor functions of miR-133a in colorectal cancer. Mol Cancer Res. 2013;11(9):1051–1060. doi: 10.1158/1541-7786.MCR-13-0061. [DOI] [PubMed] [Google Scholar]
- 14.Akanuma N, Hoshino I, Akutsu Y, Murakami K, Isozaki Y, Maruyama T, et al. MicroRNA-133a regulates the mRNAs of two invadopodia-related proteins, FSCN1 and MMP14, in esophageal cancer. Br J Cancer. 2014;110(1):189. doi: 10.1038/bjc.2013.676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang L-K, Hsiao T-H, Hong T-M, Chen H-Y, Kao S-H, Wang W-L, et al. MicroRNA-133a suppresses multiple oncogenic membrane receptors and cell invasion in non-small cell lung carcinoma. PLoS ONE. 2014;9(5):e96765. doi: 10.1371/journal.pone.0096765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ji F, Zhang H, Wang Y, Li M, Xu W, Kang Y, et al. MicroRNA-133a, downregulated in osteosarcoma, suppresses proliferation and promotes apoptosis by targeting Bcl-xL and Mcl-1. Bone. 2013;56(1):220–226. doi: 10.1016/j.bone.2013.05.020. [DOI] [PubMed] [Google Scholar]
- 17.Luo J, Zhou J, Cheng Q, Zhou C, Ding Z. Role of microRNA-133a in epithelial ovarian cancer pathogenesis and progression. Oncol Lett. 2014;7(4):1043–1048. doi: 10.3892/ol.2014.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cui W, Zhang S, Shan C, Zhou L, Zhou Z. microRNA-133a regulates the cell cycle and proliferation of breast cancer cells by targeting epidermal growth factor receptor through the EGFR/Akt signaling pathway. FEBS J. 2013;280(16):3962–3974. doi: 10.1111/febs.12398. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.