Abstract
For hundreds of thousands of years, the human genome has extensively evolved, resulting in genetic variations in almost every gene. Immunological reflections of these genetic variations become clearly visible after an allogeneic stem cell transplantation (allo-SCT) as minor Histocompatibility (H) antigens. Minor H antigens are peptides cleaved from genetically encoded variable protein regions after which they are presented at the cell surface by HLA molecules. After allo-SCT with minor H antigen mismatches between donor and recipient, donor T cells recognize the minor H antigens of the recipient as foreign, evoking strong alloreactive immune responses. Studies in the late eighties have discovered that a subset of minor H antigens are encoded by hematopoietic system-specific genes. After allo-SCT, this subset is strictly expressed on the hematopoietic malignant cells and was therefore the first well-defined highly immunogenic group of tumor-specific antigens. In the last decade, neoantigens derived from genetic mutations in tumors have been identified as another group of immunogenic tumor-specific antigens. Therefore, hematopoietic minor H antigens and neoantigens are therapeutic equivalents. This review will connect our current knowledge about the immune biology and identification of minor H antigens and neoantigens leading to novel conclusions on their prediction.
Keywords: minor histocompatibility antigen, neoantigen, antigen prediction, antigen identification, reverse antigen identification strategy
Introduction
Minor H Antigens: From Enigmatic to Well-Defined Transplantation-Antigens
Today, more than six decades after the first application of allogeneic stem cell transplantation (allo-SCT), scientists and clinicians are still impressed by the therapeutic Graft-versus-Tumor (GvT) effect established by donor T cells administered along with stem cells into the recipient (1). This therapeutic effect can be so powerful that patients can remain in long term remissions, even may be cured, after transplantation (2, 3). Therefore, allo-SCT is still being widely applied for several recurrent hematological malignancies, even though the therapeutic effects of allo-SCT are strongly associated with the development of life-threatening Graft-versus-Host-Disease (GvHD). The main mediators of GvHD as well as GvT are the alloreactive donor T cells directed at recipient antigens that are absent in the donor, responding to Major Histocompatibility Complex (MHC; HLA in humans) molecules at the cell surface (4). However, GvT and especially GvHD still occur in about 40% of patients whose stem cell donors are completely HLA-identical, indicating the existence of an additional transplantation antigen system (5). These transplantation antigens were originally designated as minor Histocompatibility (H) antigens (6).
The nature of minor H antigens recognized by donor T cells remained an enigma for more than two decades. In the mid nineties, almost a decade after the identification of MHC-bound peptides as T cell epitopes (7) and the demonstration of structure and the peptide binding groove of MHC class I molecules (8, 9), pioneering studies conducted in mice and humans demonstrated that minor H antigens are polymorphic peptides presented by MHC molecules (10, 11). A subgroup of minor H antigens, the male-specific HY antigens, were derived from “male-specific” proteins encoded by genes located on the Y-chromosome (12). All other non-gender related minor H antigens identified to date are encoded by autosomal genes that have gained allelic polymorphism through evolution over thousands of years [reviewed in Oostvogels et al. (13)]. Although some analyses suggested that minor H antigens are mainly derived from oncological relevant genes (14), this idea was not embraced by all investigators. Any non-synonymous coding variation can give rise to an immunogenic minor H antigen after allo-SCT. Of these variations, single nucleotide polymorphisms (SNPs) leading to single amino acid substitutions are currently the most common for the generation of minor H antigens (10, 15–19). But also base-pair insertions, deletions (indels) or copy number variations (CNVs) contribute to the generation of polymorphic peptides that are recognized as minor H antigens at the cell surface (20).
The Concept of the Minor H Antigen-Targeted Immunotherapy
As soon as the molecular identity of minor H antigens was unraveled, it became clear why several minor H antigen-specific T cells isolated from transplanted patients lysed only hematopoietic cells, including hematopoietic tumor cells but not the cells derived from other tissues such as fibroblasts or keratinocytes (21). In all those cases the target minor H antigen was encoded by genes, which are solely expressed in the hematopoietic system (22). This discovery underlies the concept of minor H antigen-targeted immunotherapy, which aims at targeting hematopoiesis-specific minor H antigens, which would induce GvT without GvHD after allo-SCT. Also the newly developing minor H antigen negative donor-derived hematopoietic system would remain unharmed (23). The development of this concept fueled the efforts to identify new hematopoiesis-specific minor H antigens. To be broadly therapeutically applicable, such minor H antigens are ideally presented by common HLA-alleles and have a balanced population prevalence in order to get frequent minor H antigen disparities between donor and patient (24). Now, almost 25 years later, the research resulted in the identification of about 10 genuinely hematopoiesis-specific minor H antigens (18, 25–33), some of which have been or are being tested in early phase I/II clinical trials. The approaches used in these trials include treatment of allo-transplanted patients with ex vivo generated minor H antigen-specific T cells (34, 35), with T cell receptor (TCR)-gene transferred T cells (NCT03326921, ongoing) or vaccination of allo-transplanted patients with recipient- or donor-derived dendritic cells loaded with minor H antigen peptides (13, 36) or with minor H antigen encoding mRNA (NCT02528682, ongoing) (37). Nevertheless, except the HA-1, UTA2-1, and CD19 minor H antigens (25, 27, 29), all hematopoiesis-specific minor H antigens identified till now are either presented by infrequent HLA-alleles or display an unbalanced population frequency, which makes it highly challenging to enroll sufficient minor H antigen mismatched donor-patient pairs in clinical trials. Due to this issue, all current clinical translation attempts are either progressing very slowly (38) or even terminated due to poor accrual (NCT00943293). Thus, the efficient clinical translation of this highly personalized immunotherapy approach is still largely dependent on the development of solid strategies to identify clinically relevant hematopoiesis-specific minor H antigens. These efforts are relevant not only for the application of minor H antigen-targeted immunotherapy but also for immunotherapy aiming at targeting the so-called neoantigens, because the genetic, immunogenic, and therapeutic properties of hematopoietic minor H antigens and tumor-specific neoantigens display extreme similarities.
Similarities and Differences Between Hematopoietic Minor H Antigens and Tumor-Specific Neoantigens
Minor H antigens are the immunological reflections of evolutionary established genetic polymorphisms, while neoantigens are immunological reflections of tumor-specific genetic mutations (38, 39). Thus, from a genetic point of view, the only difference between these antigens is that minor H antigens are inherited, while neoantigens are not. For subsequent gene expression, antigen processing and HLA-mediated presentation, minor H antigens and neoantigens follow identical rules. These include that HLA class I (HLA-I) antigens are liberated from the polymorphic or mutated regions of intracellular proteins by (immuno)proteasomes in the cytosol, followed by ER-translocation via transporters associated with antigen processing (TAP) in order to be loaded into HLA-I molecules (40). Next to HLA-I restricted antigens that induce CD8± cytotoxic T cells, HLA class II (HLA-II) restricted antigens that induce CD4± T cells can also play important roles in anti-tumor responses. The proteolytic processing of HLA-II restricted minor H antigens and neoantigens is generally regulated by lysosomal enzymes, followed by HLA-DM assisted loading into the HLA-II peptide binding groove (41).
From the immunological point of view, the existence of minor H antigen- and neoantigen-specific T cells in the naïve T cell repertoire is likely similar. Both antigens are foreign to the immune system and therefore there is no negative selection for the high-affinity T cells reactive with minor H antigens and neoantigens in the thymus (42). Consequently, both antigens can induce very potent T cell immune responses. The basic difference is that minor H antigens are solely immunogenic in a minor H antigen mismatched allo-SCT setting, while neoantigens can be readily immunogenic both in the allogeneic and autologous settings (38, 39).
Finally, from a clinical viewpoint, minor H antigens can be encoded by any polymorphic gene and are thus not tumor-specific antigens per se, as opposed to neoantigens. In fact, many minor H antigens are expressed by normal tissues and associate with the occurrence of detrimental GvHD as explained above (43). Nonetheless, this latter distinction does not apply for hematopoietic minor H antigens, which are tumor-specific antigens after an allo-SCT, similar to neoantigens (23, 24). This is because after allo-SCT the originally minor H antigen-positive normal hematopoietic system of the recipient is replaced by the minor H antigen-negative donor hematopoietic system. The only cells expressing the hematopoietic minor H antigens are the residual tumor cells. Therefore, it would not be wrong to state that hematopoietic minor H antigens in an allo-SCT setting are the equivalents of tumor-specific neoantigens. It should be noted that the replacement of residual minor H antigen positive host dendritic cells (DCs) after allo-SCT can take longer periods. These residual host DCs can therefore present endogenous hematopoietic minor H antigens to prime hematopoietic minor H antigen-specific T cells without the need for cross-presentation (44). In the case of neoantigens however, cross-presentation of the target antigen by DCs is an absolute requirement, because tumor cells are generally not able to prime T cells. Furthermore, specific targeting of either of these types of antigen is expected to exclusively generate a powerful anti-tumor effect without inducing direct damage to non-malignant cells.
From the therapeutic point of view, one final common and challenging aspect is the execution of clinical studies. As stated above, many minor H antigen-based clinical studies are facing with poor recruitment issues. Similar poor recruitment for adoptive T cell transfer trials is also expected for neoantigens due to the highly personalized character of most tumor-specific mutations. Since vaccination studies can include several antigens in one study, they are more easily applied (45) as compared to adoptive T cell transfer, but their success is still critically dependent on the development of effective (DC) vaccination strategies that can induce robust and long lasting T cell responses (46).
All these similarities between minor H antigens and neoantigens together show that it is of paramount importance to combine the knowledge of both fields toward the effective identification of both types of antigens and their application in the clinic.
Most Successful Methods for the Identification of Minor H Antigens and Neoantigens
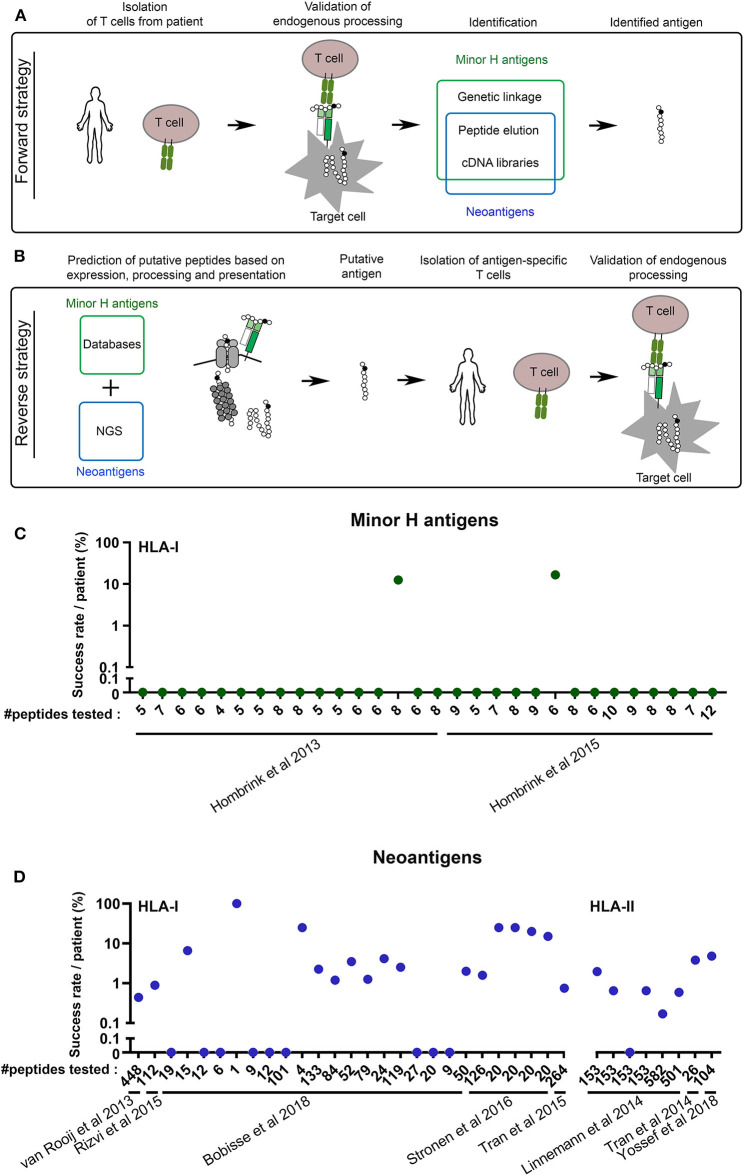
In general, methods for the identification of a peptide antigen recognized by T cells fall into two main categories. The direct (forward) strategy aims to identify the antigen of a T cell clone that has already been isolated from a patient or a healthy individual. The “reverse” strategy follows the opposite direction through the isolation of a T cell clone that recognizes an in silico predicted antigen of interest (Figures 1A,B).
Figure 1.
The reverse antigen identification strategy still requires major improvement. (A) Schematic overview of the forward antigen identification strategy. (B) Schematic overview of the reverse antigen identification strategy. The cartoons depict processing by the proteasome, TAP transporter and HLA class I. NGS, Next Generation Sequencing. (C,D) The success rate of the reverse immunology approach is low. (C) From two papers in the field of minor H antigens, data were collected about the number of potential minor H antigens that were tested (#peptides tested) vs. the percentage of peptides against which T cell reactivity was detected or raised. Only papers were selected in which reactive T cells were confirmed to recognize the endogenous (or naturally processed) antigen. (D) The same was done with eight key papers in the field of neoantigens, five on HLA-I and three on HLA-II antigens.
Forward Antigen Identification Strategies
There are several forward methods to identify minor H antigens and a fewer to identify neoantigens (Figure 1A). Initially, specialized biochemical peptide elution and fractionation techniques were used to identify peptides recognized by minor H antigen- and tumor-specific T cell clones (10, 12, 18, 25). At the same time, laborious cDNA library screening approaches were utilized to identify minor H antigen and tumor antigen encoding mRNA (47, 48). However, after the discovery that minor H antigens were encoded by inheritable genetic variations, genetic analyses were developed specifically for minor H antigens (32). Over the last 15 years, we and others have advanced these analyses from conventional pairwise linkage analysis into rapid and convenient SNP-based genome-wide association studies (15, 29, 49, 50). Moreover, we have implemented major resolution upgrades of those screens that initially used self-made databases toward publically available databases first from the HapMap Project and later from the 1,000 Genomes Project, which highly improved the success rate of genetic HLA-I and HLA-II restricted minor H antigen identification efforts (49). With these methods, a minor H antigen recognized by a T cell can be identified within 3–4 months (15). It is therefore not surprising that the vast majority of the more than 50 known minor H antigens to date has been identified by genetic linkage analyses (15, 27–29, 32, 33, 49, 51–53). Since neoantigens are not encoded in the germline and thus are not polymorphic in the population, the highly convenient forward genetic approaches are not applicable to their identification.
Despite its evident success, the forward T cell-to-antigen strategy has clear drawbacks when it comes to the identification of minor H antigens with a desired HLA-restriction, population frequency and tissue distribution. Although minor H antigen-specific T cell clones can be readily isolated from many, if not all, allo-transplanted patients, the available T cell isolation techniques cannot be adapted to isolate only those T cell clones with the required characteristics (24). All generated T cell clones need to be tested for the desired HLA restriction and minor H antigen frequency using cell line panels. Moreover, there are no convenient and reliable strategies to select T cell clones directed at minor H antigens expressed only in the hematopoietic system. A better and more convenient control of HLA restriction, population frequency and tissue distribution is key toward more efficient identification of clinically relevant minor H antigens.
The Reverse Antigen Identification Strategies
While forward methods hamper at the identification of clinically relevant minor H antigens and neoantigens, opportunities are offered by the “reverse immunology” approach. The reverse method first predicts potential T cell antigens based on in silico analyses of polymorphic or mutated genomic sites (Figure 1B). The 1,000 Genomes Project has cataloged most of human polymorphism and is therefore the database of choice for selection of putative minor H antigen encoding variations, preferably with a balanced allele frequency to allow for optimal donor-recipient disparity (54, 55). Because of the personalized character of neoantigens, the current state-of-the-art for mapping individual mutations is through tumor exome sequencing (56). Next tumor transcriptome analyses based on RNAseq or online databases are usually utilized to filter for tumor expression. Candidate minor H antigens should go through an additional selection for selective hematopoietic restricted expression. Finally, algorithms are applied to determine HLA-binding and sometimes the antigen processing efficiency for each possible peptide covering the polymorphism or mutation (56). The combined predictions for HLA-I presented peptides generally provide a score that accounts for the C-terminal cleavage of the protein by the proteasome, the TAP-mediated translocation of the peptide into the ER and the binding of the peptide to HLA with high “on-” and low “off-” rates. The multistep predictions are then validated by isolating the antigen-specific T cells from relevant patients or individuals. The last, but very essential steps are the confirmation that the targeted antigens are naturally processed and that these endogenous antigens are effectively recognized by the isolated T cells or their TCRs (Figure 1B).
The reverse strategy contained several highly challenging aspects until the last decade, which underlie the limited success of minor H antigen or neoantigen identification attempts in that period. Thanks to the recent advances in human genomics [e.g., RNAseq, exome sequencing, 1,000 Genomes Project (54, 56)], tissue expression profiling [e.g., Single Cell Expression Atlas, The Human Protein Atlas, BioGPS (57–59)], antigen processing and binding algorithms [e.g., NetCTL/IEDB (60, 61)] and large-scale peptide-specific T cell detection tools [e.g., UV-exchangeable HLA-I multimers, multimer barcoding (56)], the reverse methodology has majorly improved especially for HLA-I antigens.
This has led to the identification of a vast number of neoantigens, the antigen category for which the forward identification strategies offered only limited options. At the same time, the in silico predictions have even led to the generation of multiple libraries of thousands of putative new minor H antigens (55, 62). Nonetheless, these thousands of putative minor H antigens identified by such strategies have only resulted in the actual identification of a handful of minor H antigens (17, 26, 63), because to date none of these reverse strategies account for all minor H antigen-specific features (see below). The success of this gene-to-T cell approach highly depends on the strategy of antigen selection and intensity of T cell isolation efforts. In order to quantify the current efficiency of the reverse strategy, we have analyzed seven recent studies that applied a reverse strategy to identify novel minor H antigens or neoantigens (63–69). We only included studies from which we could extract the number of predicted antigens actually tested, as compared to the number of antigens that were endogenously expressed and against which a true T cell response could be raised. These analyses show that the efficiency of the reverse identification approach is between 0 and 20% (Figures 1C,D). These data argue that the reverse identification pipeline is currently far from optimal, with a dominant pool of false positive predicted candidates against which no T cell reactivity can be detected or raised (Figures 1C,D). One of the causes of suboptimal prediction is the incompleteness of the human reference proteome, despite huge progress in the last two decades. This is illustrated by the fact that many newly identified antigens were derived from supposedly non-coded regions (70–73). Furthermore, different efforts have shown that more stringent selection on expression and HLA binding score improves the success rate (66). Because not all neoantigens or minor H antigens behave according to these stringent criteria, we expect the number of false negative antigens to increase, which could effect the amount of therapeutic opportunities for individual patients in the long term. Thus, there is still much room for improvement of the reverse approach, which in theory is the best directed and straight forward strategy to identify HLA-I restricted hematopoiesis-specific minor H antigens as well as neoantigens.
For HLA-II restricted antigens, the development of a reverse method is even more complicated, because the rules for antigen processing and HLA-binding are more promiscuous and less defined. Nevertheless, some pioneering studies have combined minimalistic in silico analyses, without including antigen processing or HLA-binding predictions, but with large plasmid- or peptide-library screening strategies to identify HLA-II restricted neoantigen-specific T cell responses in cancer patients (67, 74, 75). Similar to HLA-I restricted antigens, these studies resulted in a low discovery rate (0–6%, Figure 1D). Recently, predictions for HLA-II binding have been incorporated in these analyses, but endogenous processing of immunogenic peptides was not confirmed (45, 76). So far, no HLA-II restricted minor H antigens have been identified following a reverse strategy. For a more successful identification of HLA-II restricted minor H antigens or neoantigens through reverse strategies, prediction algorithms for peptide processing and HLA-binding, but also cognate T cell detection tools (such as HLA-II multimers) still require revolutionary improvements.
As an additional layer of confirmation before isolating T cells, recent studies applied selection of candidate minor H antigen peptides from a large pool of HLA-I derived peptides as detected by mass spectrometry (Figure 1C) (63, 69, 77). Nonetheless, these studies also generated many false positive candidates, indicating that starting analyses from HLA-derived peptide repertoire may not necessarily compensate the current drawbacks of T cell epitope prediction algorithms.
Differential Peptide Processing and Presentation Is a Major Opportunity in Minor H Antigen and Neoantigen Reverse Identification
When studying the immunogenicity of genetic variations, it is not sufficient to consider only the antigen processing steps such as peptide cleavage, TAP translocation, HLA binding. The proper execution of these processing steps is definitely required, but not sufficient for the majority of minor H antigens and neoantigens to become immunogenic. This is because the immunogenicity of a polymorphic or mutated peptide depends on the existence of peptide-specific T cells in the (donor) T cell repertoire. The extent of the T cell repertoire against a specific antigen can be negatively affected by the presence of similar antigens in the HLA-presented peptidome during thymic development (78). Since in most cases minor H antigens and neoantigens differ only in a single amino acid from their respective allelic or wildtype counterpart peptides, it is crucial to consider potential effects on the shaping of the T cell repertoire.
If both mutated and wild type peptides are equally well-presented at the cell surface, then T cells may discriminate between these two peptides depending on the position of the amino acid substitution. This is for instance the case for two minor H antigens that both have a single amino acid substitution due to a SNP, HB-1 and ACC1 (Table 1) (26, 32). Separate T cell clones have been isolated that specifically recognize either one or the other allelic peptide at the cell surface (26, 32).
Table 1.
The majority of minor H antigens (30/50) identified by an unbiased forward strategy are (predicted to be) differentially processed.
| Levels of differential peptide processing | Minor H antigen | HLA restrictiona | Peptide sequenceb,c | C-terminal cleavage scorec,d | TAP scorec,d | HLA affinity score (nM)c,d | References |
|---|---|---|---|---|---|---|---|
| Transcription | UGT2B17/A29 | HLA-A*29:02 | AELLNIPFLY | (79) | |||
| – | |||||||
| UGT2B17/B44 | HLA-B*44:03 | AELLNIPFLY | (79) | ||||
| – | |||||||
| UGT2B17/A2 | HLA-A*02:06 | CVATMIFMI | (50) | ||||
| – | |||||||
| ACC-6 | HLA-B*44:02/03 | MEIFIEVFSHF | (31) | ||||
| – | |||||||
| ZAPHIR | HLA-B*07:02 | IPRDSWWVEL | (80) | ||||
| – | |||||||
| Translation | LRH-1 | HLA-B*07:02 | TPNQRQNVC | (28) | |||
| – | |||||||
| PANE1 | HLA-A*03:01 | RVWDLPGVLK | (81) | ||||
| – | |||||||
| C-terminal or internal proteasome cleavage | ACC-4 | HLA-A*31:01 | ATLPLLCAR | 0.26 | 0.68 | 17 | (82) |
| ATLPLLCAG | 0.03 | −0.57 | 11313 | ||||
| ACC-5 | HLA-A*33:03 | WATLPLLCAR | 0.26 | 0.65 | 210 | (82) | |
| WATLPLLCAG | 0.03 | −0.59 | 29173 | ||||
| HA-3e | HLA-A*01:01 | VTEPGTAQY | 0.97 | 1.25 | 13 | (16) | |
| VMEPGTAQY | 0.97 | 1.31 | 134 | ||||
| SP110e | HLA-A*03:01 | SLPRGTSTPK | 54 | (83) | |||
| SLPGGTSTPK | 155 | ||||||
| LB-FUCA2-1V | HLA-B*0702 | RLRQVGSWL | 0.90 | 0.48 | 38 | (84) | |
| RLRQMGSWL | 0.53 | 0.48 | 24 | ||||
| LB-GEMIN4-1V | HLA-B*07:02 | FPALRFVEV | 0.97 | 0.04 | 65 | (53) | |
| FPALRFVEE | 0.24 | −0.78 | 3208 | ||||
| LB-GEMIN4-2V | HLA-B*08:01 | FPALRFVEV | 0.97 | 0.04 | 23 | (71) | |
| FPALRFVEE | 0.24 | −0.78 | 405 | ||||
| TAP transport | HA-8 | HLA-A*02:01 | RTLDKVLEV | 0.96 | 0.23 | 35 | (30) |
| PTLDKVLEV | 0.96 | −0.08 | 3665 | ||||
| HLA-binding | HA-1 | HLA-A*02:01 | VLHDDLLEA | 0.95 | −0.19 | 29 | (25) |
| VLRDDLLEA | 0.93 | −0.18 | 321 | ||||
| HA-2 | HLA-A*02:01 | YIGEVLVSV | 0.96 | 0.12 | 7 | (10) | |
| YIGEVLVSM | 0.96 | 0.11 | 58 | ||||
| TRIM22 | HLA-A*02:01 | MAVPPCCIGV | 0.95 | 0.17 | 620 | (85) | |
| MAVPPCRIGV | 0.89 | 0.17 | 3046 | ||||
| LB-APOBEC3B-1K | HLA-B*07:02 | KPQYHAEMCF | 0.26 | 0.95 | 278 | (53) | |
| EPQYHAEMCF | 0.26 | 0.82 | 9507 | ||||
| LB-BCAT2-1R | HLA-B*07:02 | QPRRALLFVIL | 0.94 | 0.33 | 253 | (53) | |
| QPTRALLFVIL | 0.92 | 0.30 | 2753 | ||||
| DPH1 | HLA-B*57:01 | SVLPEVDVW | 0.45 | 0.50 | 217 | (34) | |
| SLLPEVDVW | 0.59 | 0.44 | 1582 | ||||
| LB-TRIP10-1EPC | HLA-B*40:01 | GEPQDLCTL | 0.96 | 0.26 | 176 | (52) | |
| GGSQDLGTL | 0.87 | 0.21 | 15676 | ||||
| LB-C16ORF-1R | HLA-B*07:02 | RPCPSVGLSFL | 0.9 | 0.38 | 643 | (71) | |
| WPCPSVGLSFL | 0.9 | 0.26 | 3010 | ||||
| LB-NCAPD3-1Q | HLA-A*02:01 | WLQGVVPVV | 0.91 | 0.18 | 13 | (71) | |
| WLRGVVPVV | 0.91 | 0.22 | 108 | ||||
| UTA2-1 | HLA-A*02:01 | QLLNSVLTL | 0.97 | 0.46 | 39 | (27) | |
| QLPNSVLTL | 0.97 | 0.45 | 222 | ||||
| LB-TMEM8A-1I | HLA-B*07:02 | RPRSVTIQPLL | 0.97 | 0.41 | 11 | (71) | |
| RPRSVTVQPLL | 0.97 | 0.41 | 28826 | ||||
| LB-ERAP1-1R | HLA-B*07:02 | HPRQEQIALLA | 0.96 | −0.45 | 692 | (53) | |
| HPPQEQIALLA | 0.97 | −0.48 | 12460 | ||||
| LB-ADIR-1F | HLA-A*02:01 | SVAPALALSPA | 0.89 | −0.08 | 490 | (86) | |
| SVAPALALFPA | 0.91 | −0.08 | 1555 | ||||
| TCR affinity | HB-1 | HLA-B*44:03 | EEKRGSLHVW | 0.9 | 0.29 | 184 | (48) |
| EEKRGSLYVW | 0.89 | 0.29 | 188 | ||||
| ACC1 | HLA-A*24:02 | DYLQYVLQI | 0.9 | 0.20 | 115 | (31) | |
| DYLQCVLQI | 0.77 | 0.20 | 197 | ||||
| Yet unknown | LB-ECGF-1 | HLA-B*07:02 | RPHAIRRPLAL | 0.91 | 0.42 | 9 | (73) |
| RPRAIRRPLAL | 0.91 | 0.43 | 5 | ||||
| SLC5A1 | HLA-B*40:02 | AEATANGGLAL | 0.96 | 0.49 | 48 | (50) | |
| AEPTANGGLAL | 0.96 | 0.48 | 50 | ||||
| LB-WNK1-1I | HLA-A*02:01 | RTLSPEIITV | 0.97 | 0.30 | 58 | (53) | |
| RTLSPEMITV | 0.95 | 0.30 | 78 | ||||
| LB-NDC80-1P | HLA-A*02:01 | HLEEQIPKV | 0.97 | 0.09 | 82 | (71) | |
| HLEEQIAKV | 0.97 | 0.09 | 184 | ||||
| LB-ZDHHC6-1Y | HLA-B*07:02 | RPRYWILLVKI | 0.97 | 0.23 | 338 | (71) | |
| RPRHWILLVKI | 0.95 | 0.18 | 334 | ||||
| LB-SON-1R | HLA-B*40:01 | SETKQRTVL | 0.92 | 0.37 | 58 | (52) | |
| SETKQCTVL | 0.95 | 0.37 | 28 | ||||
| LB-SWAP70-1Q | HLA-B*40:01 | MEQLEQLEL | 0.94 | 0.43 | 178 | (52) | |
| MEQLEELEL | 0.92 | 0.43 | 141 | ||||
| LB-NUP133-1R | HLA-B*40:01 | SEDLILCRL | 0.90 | 0.28 | 194 | (52) | |
| SEDLILCQL | 0.90 | 0.28 | 80 | ||||
| P2RX7 | HLA-A*29:02 | WFHHCHPKY | 0.95 | 1.40 | 6 | (34) | |
| WFHHCRPKY | 0.85 | 1.40 | 15 | ||||
| LB-TTK-1D | HLA-A*02:01 | RLHDGRVFV | 0.89 | 0.26 | 30 | (51) | |
| RLHEGRVFV | 0.84 | 0.26 | 33 | ||||
| LB-EBI3-1I | HLA-B*07:02 | RPRARYYIQV | 0.96 | 0.15 | 26 | (53) | |
| RPRARYYVQV | 0.96 | 0.15 | 19 | ||||
| LB-ARHGDIB-1R | HLA-B*07:02 | LPRACWREA | 0.42 | −0.07 | 10 | (53) | |
| LPRACWPEA | 0.42 | −0.07 | 35 | ||||
| LB-SSR1-1S | HLA-A*02:01 | VLFRGGPRGSLAVA | 0.89 | −0.14 | 1403 | (87) | |
| VLFRGGPRGLLAVA | 0.86 | −0.14 | 649 | ||||
| LB-PRCP-1D | HLA-A*02:01 | FMWDVAEDL | 0.92 | 0.49 | 8 | (53) | |
| FMWDVAEEL | 0.95 | 0.49 | 3 | ||||
| LB-MOB3A-1C | HLA-B*07:02 | CPRPGTWTC | NAf | −0.13 | 442 | (71) | |
| SPRPGTWTC | −0.09 | 69 | |||||
| LB-PNP-1S | HLA-B*13:01 | TQAQIFDYSEI | 0.57 | 0.28 | NAg | (71) | |
| TQAQIFDYGEI | 0.4 | 0.28 | |||||
| LB-GSTP1-1V | HLA-B*08:01 | DLRCKYVSL | 0.77 | 0.24 | 8 | (71) | |
| DLRCKYISL | 0.71 | 0.24 | 13 | ||||
| C19ORF48 | HLA-A*02:01 | TAWPGAPEV | 0.97 | 0.38 | 163 | (72) | |
| TAWPGAPGV | 0.96 | 0.38 | 268 | ||||
| Predicted not to bind HLAh | LB-C19ORF48-2E | HLA-B*51:01 | TAWPGAPEV | 0.97 | 0.38 | 24822 | (71) |
| TAWPGAPGV | 0.96 | 0.38 | 25920 | ||||
| LB-PDCD11-1F | HLA-B*07:02 | GPDSSKTFLCL | 0.97 | 0.15 | 7364 | (53) | |
| GPDSSKTLLCL | 0.96 | 0.15 | 7408 | ||||
| LB-ZNFX1-1Q | HLA-B*40:01 | NEIEDVWQLDL | 0.94 | 0.47 | 5834 | (71) | |
| NEIEDVWHLDL | 0.96 | 0.47 | 4026 | ||||
| LB-APOBEC3B-1K | HLA-B*08:01 | KPQYHAEMCF | 0.26 | 0.95 | 9287 | (71) | |
| EPQYHAEMCF | 0.26 | 0.82 | 4709 | ||||
| LB-CCL4-1T | HLA-A*02:01 | CADPSETWV | 0.15 | 0.08 | 8014 | (71) | |
| CADPSESWV | 0.29 | 0.08 | 9995 |
For B
44:02/03 binding mHags, only scores for B*44:03 are depicted.
The upper peptide sequence corresponds to minor H antigen and the bottom to allelic counterpart.
Bold values indicate the level of (predicted) differential peptide processing.
Predictions made by NetChop3.1, IEDB and NetMHC4.0.
HA-3 and SP110 are generated through differential internal proteasome cleavage and proteosome-catalyzed peptide splicing.
The C-terminus of the MOB3A antigen is the C-terminus of the MOB3A protein.
The HLA-B*13:01 binding prediction is not available at NetMHC4.0, but both peptides harbor the HLA-B*13 binding motif.
Because the NetMHC4.0 affinity scores indicated no binding to HLA (>> 1,000 nM), it is currently complicated to assess potential differential HLA-binding, so we excluded these five minor H antigens from the analyses.
However, if the amino acid alteration is not at a position in the peptide that is exposed to the TCR, and if it is not affecting peptide conformation, then T cells can not discriminate between two of such slightly different peptides. In this case, the allelic or wildtype peptide variants already induce the cognate T cells to be deleted from the T cell repertoire by negative selection in the thymus, similar to self-peptide-reactive T cells (42). Consequently, individuals (donors or patients) lack the majority of the minor H antigen or neoantigen-specific T cells before even being exposed to these antigens. Furthermore, the few T cells in the repertoire that are antigen-specific will have a low affinity TCR with cross-reactivity against the allelic or wildtype peptide. Therapeutic use of such T cells may put patients at risk of detrimental self-recognition on healthy cells, similar to what has been observed for tumor antigens MART-1 and MAGE-A3 (88–91).
Such restrictions do not apply for peptide pairs that are differentially expressed on the cell surface. In fact, for several minor H antigens this is the case. For instance HA-1-, HA-2- and HA-3-specific T cells make no distinction between the allelic peptides (16–18, 25). The strong immunogenicity of such minor H antigens occurs due to the fact that the non-immunogenic peptide has impaired HLA-binding (HA-1) (92), proteasome cleavage (HA-3) (16), TAP translocation (HA-8) (30) or even a yet unknown processing event (HA-2) (Table 1). Similar one-sided lack of peptide presentation occurs if the genetic variation causes loss of conventional gene transcription such as through alternative splicing (31) or loss of genomic DNA (Table 1) (79). Finally, alternative translation events may also cause differential surface expression, for example due to a frame shift (28) or the introduction of a stop-codon (81) (Table 1). Moreover, in silico analysis of all currently known HLA-I restricted minor H antigens, which have been identified using unbiased forward strategies, revealed that at least 28/48 of the non-immunogenic counterpart peptides are likely not expressed on the cell surface. This is probably an underestimation because various parameters that affect intracellular peptide processing were (largely) disregarded in this overview, such as proteosome-catalyzed peptide splicing, internal proteosomal cleavage, ERAP1 trimming (16, 83, 93). In addition, the prediction algorithms have been trained on positive datasets and therefore currently have limited power to predict the absence of processing. Taken together, this dataset suggests that more than 58% of genetically variable antigens are immunogenic because the non-immunogenic peptide is simply not present on the cell surface (Table 1). Mono-allelic (by some investigators called as “dominant”) presentation was also seen in mass spectrometry data of HLA-I derived peptides (77, 94).
Recently, SNP-induced differential processing was also found for an HLA-II restricted minor H antigen (95), indicating that development of HLA-II epitope processing prediction algorithms may be valuable for future identification of immunogenic HLA-II presented antigens.
Strikingly, in the exploding field of neoantigen prediction models, differential surface expression of mutated vs. wild type peptide is largely neglected, which might be a reason of the large number of false positive neoantigen predictions (Figure 1D). Thus, the addition of a specific differential surface presentation module, based on the molecular features of minor H antigens and their allelic counterparts, to the current prediction models may improve both minor H antigen and neoantigen reverse identification strategies.
Conclusions and Future Directions
There is no doubt that the genetic alterations encoding hematopoietic minor H antigens in the allo-SCT setting and neoantigens in the autologous setting can induce potent T cell responses in patients. For clinical application of both antigen types, the most important challenge is to include sufficient patients in the clinical trials. It is currently unclear what the best strategy will be to provide the personalized immunotherapy necessary to target either of these antigens. Furthermore, there are also major challenges for their identification. The current T cell epitope prediction algorithms need significant improvement. We here postulate based on the published data of the last 25 years that also predictions for the non-immunogenic allelic or wild type peptide should be included in the algorithms. Selecting only those candidate peptides that will be differentially expressed at the cell surface may increase the success rate to detect or raise antigen-specific T cells from the naïve repertoire.
Author Contributions
TM and RS developed the concept, designed, and wrote the manuscript. AX performed literature analyses and generated the figures.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This work was financially supported by ZonMw (TM), KWF (TM), Janssen Research and Development (TM), Genmab (TM), Takeda (TM), Gadeta (TM), Onkimmune (TM), the Netherlands organization for scientific research (VIDI 91719369; RS), KWF Alpe d'HuZes (Bas Mulder Award 2015-7982; RS), the Landsteiner Foundation for Blood Transfusion Research (1842F; RS).
References
- 1.Kolb HJ. Graft-versus-leukemia effects of transplantation and donor lymphocytes. Blood. (2008) 112:4371–83. 10.1182/blood-2008-03-077974 [DOI] [PubMed] [Google Scholar]
- 2.Gratwohl A, Pfirrmann M, Zander A, Kroger N, Beelen D, Novotny J, et al. Long-term outcome of patients with newly diagnosed chronic myeloid leukemia: a randomized comparison of stem cell transplantation with drug treatment. Leukemia. (2016) 30:562–9. 10.1038/leu.2015.281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heinzelmann F, Bethge W, Beelen DW, Engelhard M, Kroger N, Dreger P, et al. Allogeneic hematopoietic cell transplantation as curative therapy for non-transformed follicular lymphomas. Bone Marrow Transplant. (2016) 51:654–62. 10.1038/bmt.2015.348 [DOI] [PubMed] [Google Scholar]
- 4.van Rood JJ, van Leeuwen A, Schippers A, Balner H. Human histocompatibility antigens in normal and neoplastic tissues. Cancer Res. (1968) 28:1415–22. [PubMed] [Google Scholar]
- 5.Gratwohl A, Hermans J, Apperley J, Arcese W, Bacigalupo A, Bandini G, et al. Acute graft-versus-host disease: grade and outcome in patients with chronic myelogenous leukemia. Working party chronic leukemia of the European group for blood and marrow transplantation. Blood. (1995) 86:813–8. 10.1182/blood.V86.2.813.bloodjournal862813 [DOI] [PubMed] [Google Scholar]
- 6.Goulmy E. Minor histocompatibility antigens: allo target molecules for tumor-specific immunotherapy. Cancer J. (2004) 10:1–7. 10.1097/00130404-200401000-00001 [DOI] [PubMed] [Google Scholar]
- 7.Guillet JG, Lai MZ, Briner TJ, Smith JA, Gefter ML. Interaction of peptide antigens and class II major histocompatibility complex antigens. Nature. (1986) 324:260–2. 10.1038/324260a0 [DOI] [PubMed] [Google Scholar]
- 8.Bjorkman PJ, Saper MA, Samraoui B, Bennett WS, Strominger JL, Wiley DC. Structure of the human class I histocompatibility antigen, HLA-A2. Nature. (1987) 329:506–12. 10.1038/329506a0 [DOI] [PubMed] [Google Scholar]
- 9.Bjorkman PJ, Saper MA, Samraoui B, Bennett WS, Strominger JL, Wiley DC. The foreign antigen binding site and T cell recognition regions of class I histocompatibility antigens. Nature. (1987) 329:512–8. 10.1038/329512a0 [DOI] [PubMed] [Google Scholar]
- 10.den Haan JM, Sherman NE, Blokland E, Huczko E, Koning F, Drijfhout JW, et al. Identification of a graft versus host disease-associated human minor histocompatibility antigen. Science. (1995) 268:1476–80. 10.1126/science.7539551 [DOI] [PubMed] [Google Scholar]
- 11.Malarkannan S, Shih PP, Eden PA, Horng T, Zuberi AR, Christianson G, et al. The molecular and functional characterization of a dominant minor H antigen, H60. J Immunol. (1998) 161:3501–9. [PubMed] [Google Scholar]
- 12.Wang W, Meadows LR, den Haan JM, Sherman NE, Chen Y, Blokland E, et al. Human H-Y: a male-specific histocompatibility antigen derived from the SMCY protein. Science. (1995) 269:1588–90. 10.1126/science.7667640 [DOI] [PubMed] [Google Scholar]
- 13.Oostvogels R, Kneppers E, Minnema MC, Doorn RC, Franssen LE, Aarts T, et al. Efficacy of host-dendritic cell vaccinations with or without minor histocompatibility antigen loading, combined with donor lymphocyte infusion in multiple myeloma patients. Bone Marrow Transplant. (2017) 52:228–37. 10.1038/bmt.2016.250 [DOI] [PubMed] [Google Scholar]
- 14.Spierings E, Wieles B, Goulmy E. Minor histocompatibility antigens–big in tumour therapy. Trends Immunol. (2004) 25:56–60. 10.1016/j.it.2003.12.002 [DOI] [PubMed] [Google Scholar]
- 15.Spaapen RM, de Kort RA, van den Oudenalder K, van Elk M, Bloem AC, Lokhorst HM, et al. Rapid identification of clinical relevant minor histocompatibility antigens via genome-wide zygosity-genotype correlation analysis. Clin Cancer Res. (2009) 15:7137–43. 10.1158/1078-0432.CCR-09-1914 [DOI] [PubMed] [Google Scholar]
- 16.Spierings E, Brickner AG, Caldwell JA, Zegveld S, Tatsis N, Blokland E, et al. The minor histocompatibility antigen HA-3 arises from differential proteasome-mediated cleavage of the lymphoid blast crisis (Lbc) oncoprotein. Blood. (2003) 102:621–9. 10.1182/blood-2003-01-0260 [DOI] [PubMed] [Google Scholar]
- 17.Mommaas B, Kamp J, Drijfhout JW, Beekman N, Ossendorp F, Van Veelen P, et al. Identification of a novel HLA-B60-restricted T cell epitope of the minor histocompatibility antigen HA-1 locus. J Immunol. (2002) 169:3131–6. 10.4049/jimmunol.169.6.3131 [DOI] [PubMed] [Google Scholar]
- 18.Pierce RA, Field ED, Mutis T, Golovina TN, Von Kap-Herr C, Wilke M, et al. The HA-2 minor histocompatibility antigen is derived from a diallelic gene encoding a novel human class I myosin protein. J Immunol. (2001) 167:3223–30. 10.4049/jimmunol.167.6.3223 [DOI] [PubMed] [Google Scholar]
- 19.Warren EH, Gavin MA, Simpson E, Chandler P, Page DC, Disteche C, et al. The human UTY gene encodes a novel HLA-B8-restricted H-Y antigen. J Immunol. (2000) 164:2807–14. 10.4049/jimmunol.164.5.2807 [DOI] [PubMed] [Google Scholar]
- 20.Griffioen M, van Bergen CA, Falkenburg JH. Autosomal minor histocompatibility antigens. How genetic variants create diversity in immune targets. Front Immunol. (2016) 7:100. 10.3389/fimmu.2016.00100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Bueger M, Bakker A, Van Rood JJ, Van der Woude F, Goulmy E. Tissue distribution of human minor histocompatibility antigens. Ubiquitous versus restricted tissue distribution indicates heterogeneity among human cytotoxic T lymphocyte-defined non-MHC antigens. J Immunol. (1992) 149:1788–94. [PubMed] [Google Scholar]
- 22.Mutis T, Goulmy E. Hematopoietic system-specific antigens as targets for cellular immunotherapy of hematological malignancies. Semin Hematol. (2002) 39:23–31. 10.1053/shem.2002.29248 [DOI] [PubMed] [Google Scholar]
- 23.Goulmy E. Human minor histocompatibility antigens: new concepts for marrow transplantation and adoptive immunotherapy. Immunol Rev. (1997) 157:125–40. 10.1111/j.1600-065X.1997.tb00978.x [DOI] [PubMed] [Google Scholar]
- 24.Spaapen R, Mutis T. Targeting haematopoietic-specific minor histocompatibility antigens to distinguish graft-versus-tumour effects from graft-versus-host disease. Best Pract Res Clin Haematol. (2008) 21:543–57. 10.1016/j.beha.2008.06.001 [DOI] [PubMed] [Google Scholar]
- 25.den Haan JM, Meadows LM, Wang W, Pool J, Blokland E, Bishop TL, et al. The minor histocompatibility antigen HA-1: a diallelic gene with a single amino acid polymorphism. Science. (1998) 279:1054–7. 10.1126/science.279.5353.1054 [DOI] [PubMed] [Google Scholar]
- 26.Dolstra H, de Rijke B, Fredrix H, Balas A, Maas F, Scherpen F, et al. Bi-directional allelic recognition of the human minor histocompatibility antigen HB-1 by cytotoxic T lymphocytes. Eur J Immunol. (2002) 32:2748–58. [DOI] [PubMed] [Google Scholar]
- 27.Oostvogels R, Minnema MC, van Elk M, Spaapen RM, te Raa GD, Giovannone B, et al. Towards effective and safe immunotherapy after allogeneic stem cell transplantation: identification of hematopoietic-specific minor histocompatibility antigen UTA2-1. Leukemia. (2013) 27:642–9. 10.1038/leu.2012.277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Rijke B, van Horssen-Zoetbrood A, Beekman JM, Otterud B, Maas F, Woestenenk R, et al. A frameshift polymorphism in P2X5 elicits an allogeneic cytotoxic T lymphocyte response associated with remission of chronic myeloid leukemia. J Clin Invest. (2005) 115:3506–16. 10.1172/JCI24832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Spaapen RM, Lokhorst HM, van den Oudenalder K, Otterud BE, Dolstra H, Leppert MF, et al. Toward targeting B cell cancers with CD4+ CTLs: identification of a CD19-encoded minor histocompatibility antigen using a novel genome-wide analysis. J Exp Med. (2008) 205:2863–72. 10.1084/jem.20080713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brickner AG, Warren EH, Caldwell JA, Akatsuka Y, Golovina TN, Zarling AL, et al. The immunogenicity of a new human minor histocompatibility antigen results from differential antigen processing. J Exp Med. (2001) 193:195–206. 10.1084/jem.193.2.195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kawase T, Akatsuka Y, Torikai H, Morishima S, Oka A, Tsujimura A, et al. Alternative splicing due to an intronic SNP in HMSD generates a novel minor histocompatibility antigen. Blood. (2007) 110:1055–63. 10.1182/blood-2007-02-075911 [DOI] [PubMed] [Google Scholar]
- 32.Akatsuka Y, Nishida T, Kondo E, Miyazaki M, Taji H, Iida H, et al. Identification of a polymorphic gene, BCL2A1, encoding two novel hematopoietic lineage-specific minor histocompatibility antigens. J Exp Med. (2003) 197:1489–500. 10.1084/jem.20021925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pont MJ, Hobo W, Honders MW, van Luxemburg-Heijs SA, Kester MG, van Oeveren-Rietdijk AM, et al. LB-ARHGDIB-1R as a novel minor histocompatibility antigen for therapeutic application. Haematologica. (2015) 100:e419–22. 10.3324/haematol.2015.125021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Warren EH, Fujii N, Akatsuka Y, Chaney CN, Mito JK, Loeb KR, et al. Therapy of relapsed leukemia after allogeneic hematopoietic cell transplantation with T cells specific for minor histocompatibility antigens. Blood. (2010) 115:3869–78. 10.1182/blood-2009-10-248997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meij P, Jedema I, van der Hoorn MA, Bongaerts R, Cox L, Wafelman AR, et al. Generation and administration of HA-1-specific T-cell lines for the treatment of patients with relapsed leukemia after allogeneic stem cell transplantation: a pilot study. Haematologica. (2012) 97:1205–8. 10.3324/haematol.2011.053371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Franssen LE, Roeven MWH, Hobo W, Doorn R, Oostvogels R, Falkenburg JHF, et al. A phase I/II minor histocompatibility antigen-loaded dendritic cell vaccination trial to safely improve the efficacy of donor lymphocyte infusions in myeloma. Bone Marrow Transplant. (2017) 52:1378–83. 10.1038/bmt.2017.118 [DOI] [PubMed] [Google Scholar]
- 37.Levenga H, Schaap N, Maas F, Esendam B, Fredrix H, Greupink-Draaisma A, et al. Partial T cell-depleted allogeneic stem cell transplantation following reduced-intensity conditioning creates a platform for immunotherapy with donor lymphocyte infusion and recipient dendritic cell vaccination in multiple myeloma. Biol Blood Marrow Transplant. (2010) 16:320–32. 10.1016/j.bbmt.2009.10.006 [DOI] [PubMed] [Google Scholar]
- 38.Oostvogels R, Lokhorst HM, Mutis T. Minor histocompatibility Ags: identification strategies, clinical results and translational perspectives. Bone Marrow Transplant. (2016) 51:163–71. 10.1038/bmt.2015.256 [DOI] [PubMed] [Google Scholar]
- 39.Schumacher TN, Schreiber RD. Neoantigens in cancer immunotherapy. Science. (2015) 348:69–74. 10.1126/science.aaa4971 [DOI] [PubMed] [Google Scholar]
- 40.Jongsma MLM, Guarda G, Spaapen RM. The regulatory network behind MHC class I expression. Mol Immunol. (2019) 113:16–21. 10.1016/j.molimm.2017.12.005 [DOI] [PubMed] [Google Scholar]
- 41.van den Hoorn T, Paul P, Jongsma ML, Neefjes J. Routes to manipulate MHC class II antigen presentation. Curr Opin Immunol. (2011) 23:88–95. 10.1016/j.coi.2010.11.002 [DOI] [PubMed] [Google Scholar]
- 42.Starr TK, Jameson SC, Hogquist KA. Positive and negative selection of T cells. Annu Rev Immunol. (2003) 21:139–76. 10.1146/annurev.immunol.21.120601.141107 [DOI] [PubMed] [Google Scholar]
- 43.Spierings E, Kim YH, Hendriks M, Borst E, Sergeant R, Canossi A, et al. Multicenter analyses demonstrate significant clinical effects of minor histocompatibility antigens on GvHD and GvL after HLA-matched related and unrelated hematopoietic stem cell transplantation. Biol Blood Marrow Transplant. (2013) 19:1244–53. 10.1016/j.bbmt.2013.06.001 [DOI] [PubMed] [Google Scholar]
- 44.Toubai T, Mathewson N, Reddy P. The role of dendritic cells in graft-versus-tumor effect. Front Immunol. (2014) 5:66. 10.3389/fimmu.2014.00066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen F, Zou Z, Du J, Su S, Shao J, Meng F, et al. Neoantigen identification strategies enable personalized immunotherapy in refractory solid tumors. J Clin Invest. (2019) 129:2056–70. 10.1172/JCI99538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wculek SK, Cueto FJ, Mujal AM, Melero I, Krummel MF, Sancho D. Dendritic cells in cancer immunology and immunotherapy. Nat Rev Immunol. (2020) 20:7–24. 10.1038/s41577-019-0210-z [DOI] [PubMed] [Google Scholar]
- 47.Cerottini JC, von V, Boon T. Recognition of tumor-associated antigens by T lymphocytes: from basic concepts to new approaches. Ann Oncol. (1992) 3:11–6. 10.1093/oxfordjournals.annonc.a058055 [DOI] [PubMed] [Google Scholar]
- 48.Dolstra H, Fredrix H, Maas F, Coulie PG, Brasseur F, Mensink E, et al. A human minor histocompatibility antigen specific for B cell acute lymphoblastic leukemia. J Exp Med. (1999) 189:301–8. 10.1084/jem.189.2.301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Oostvogels R, Lokhorst HM, Minnema MC, van Elk M, van den Oudenalder K, Spierings E, et al. Identification of minor histocompatibility antigens based on the 1000 genomes project. Haematologica. (2014) 99:1854–9. 10.3324/haematol.2014.109801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kamei M, Nannya Y, Torikai H, Kawase T, Taura K, Inamoto Y, et al. HapMap scanning of novel human minor histocompatibility antigens. Blood. (2009) 113:5041–8. 10.1182/blood-2008-07-171678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pont MJ, Oostvogels R, van Bergen CAM, van der Meijden ED, Honders MW, Bliss S, et al. T cells specific for an unconventional natural antigen fail to recognize leukemic cells. Cancer Immunol Res. (2019) 7:797–804. 10.1158/2326-6066.CIR-18-0137 [DOI] [PubMed] [Google Scholar]
- 52.Griffioen M, Honders MW, van der Meijden ED, van Luxemburg-Heijs SA, Lurvink EG, Kester MG, et al. Identification of 4 novel HLA-B*40:01 restricted minor histocompatibility antigens and their potential as targets for graft-versus-leukemia reactivity. Haematologica. (2012) 97:1196–204. 10.3324/haematol.2011.049478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Van Bergen CA, Rutten CE, Van Der Meijden ED, Van Luxemburg-Heijs SA, Lurvink EG, Houwing-Duistermaat JJ, et al. High-throughput characterization of 10 new minor histocompatibility antigens by whole genome association scanning. Cancer Res. (2010) 70:9073–83. 10.1158/0008-5472.CAN-10-1832 [DOI] [PubMed] [Google Scholar]
- 54.The 1000 Genomes Project Consortium, Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. (2012) 491:56–65. 10.1038/nature11632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bykova NA, Malko DB, Efimov GA. In silico analysis of the minor histocompatibility antigen landscape based on the 1000 genomes project. Front Immunol. (2018) 9:1819. 10.3389/fimmu.2018.01819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.van Buuren MM, Calis JJ, Schumacher TN. High sensitivity of cancer exome-based CD8 T cell neo-antigen identification. Oncoimmunology. (2014) 3:e28836. 10.4161/onci.28836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu C, Orozco C, Boyer J, Leglise M, Goodale J, Batalov S, et al. BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol. (2009) 10:R130. 10.1186/gb-2009-10-11-r130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, et al. Proteomics. Tissue-based map of the human proteome. Science. (2015) 347:1260419. 10.1126/science.1260419 [DOI] [PubMed] [Google Scholar]
- 59.Regev A, Teichmann SA, Lander ES, Amit I, Benoist C, Birney E, et al. Human cell atlas meeting: the human cell atlas. Elife. (2017) 6:e27041. 10.7554/eLife.27041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fleri W, Paul S, Dhanda SK, Mahajan S, Xu X, Peters B, et al. The immune epitope database and analysis resource in epitope discovery and synthetic vaccine design. Front Immunol. (2017) 8:278. 10.3389/fimmu.2017.00278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Larsen MV, Lundegaard C, Lamberth K, Buus S, Lund O, Nielsen M. Large-scale validation of methods for cytotoxic T-lymphocyte epitope prediction. BMC Bioinform. (2007) 8:424. 10.1186/1471-2105-8-424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.DeLuca DS, Eiz-Vesper B, Ladas N, Khattab BA, Blasczyk R. High-throughput minor histocompatibility antigen prediction. Bioinformatics. (2009) 25:2411–7. 10.1093/bioinformatics/btp404 [DOI] [PubMed] [Google Scholar]
- 63.Hombrink P, Hassan C, Kester MG, Jahn L, Pont MJ, de Ru AH, et al. Identification of biological relevant minor histocompatibility antigens within the B-lymphocyte-derived HLA-ligandome using a reverse immunology approach. Clin Cancer Res. (2015) 21:2177–86. 10.1158/1078-0432.CCR-14-2188 [DOI] [PubMed] [Google Scholar]
- 64.Bobisse S, Genolet R, Roberti A, Tanyi JL, Racle J, Stevenson BJ, et al. Sensitive and frequent identification of high avidity neo-epitope specific CD8 (+) T cells in immunotherapy-naive ovarian cancer. Nat Commun. (2018) 9:1092. 10.1038/s41467-018-03301-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. (2015) 348:124–8. 10.1126/science.aaa1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stronen E, Toebes M, Kelderman S, van Buuren MM, Yang W, van Rooij N, et al. Targeting of cancer neoantigens with donor-derived T cell receptor repertoires. Science. (2016) 352:1337–41. 10.1126/science.aaf2288 [DOI] [PubMed] [Google Scholar]
- 67.Tran E, Ahmadzadeh M, Lu YC, Gros A, Turcotte S, Robbins PF, et al. Immunogenicity of somatic mutations in human gastrointestinal cancers. Science. (2015) 350:1387–90. 10.1126/science.aad1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.van Rooij N, van Buuren MM, Philips D, Velds A, Toebes M, Heemskerk B, et al. Tumor exome analysis reveals neoantigen-specific T-cell reactivity in an ipilimumab-responsive melanoma. J Clin Oncol. (2013) 31:e439–42. 10.1200/JCO.2012.47.7521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hombrink P, Hassan C, Kester MG, de Ru AH, van Bergen CA, Nijveen H, et al. Discovery of T cell epitopes implementing HLA-peptidomics into a reverse immunology approach. J Immunol. (2013) 190:3869–77. 10.4049/jimmunol.1202351 [DOI] [PubMed] [Google Scholar]
- 70.Laumont CM, Vincent K, Hesnard L, Audemard E, Bonneil E, Laverdure JP, et al. Noncoding regions are the main source of targetable tumor-specific antigens. Sci Transl Med. (2018) 10:eaau5516. 10.1126/scitranslmed.aau5516 [DOI] [PubMed] [Google Scholar]
- 71.van Bergen CA, van Luxemburg-Heijs SA, de Wreede LC, Eefting M, von PA, van Balen P, et al. Selective graft-versus-leukemia depends on magnitude and diversity of the alloreactive T cell response. J Clin Invest. (2017) 127:517–29. 10.1172/JCI86175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tykodi SS, Fujii N, Vigneron N, Lu SM, Mito JK, Miranda MX, et al. C19orf48 encodes a minor histocompatibility antigen recognized by CD8+ cytotoxic T cells from renal cell carcinoma patients. Clin Cancer Res. (2008) 14:5260–9. 10.1158/1078-0432.CCR-08-0028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Slager EH, Honders MW, van der Meijden ED, van Luxemburg-Heijs SA, Kloosterboer FM, Kester MG, et al. Identification of the angiogenic endothelial-cell growth factor-1/thymidine phosphorylase as a potential target for immunotherapy of cancer. Blood. (2006) 107:4954–60. 10.1182/blood-2005-09-3883 [DOI] [PubMed] [Google Scholar]
- 74.Linnemann C, van Buuren MM, Bies L, Verdegaal EM, Schotte R, Calis JJ, et al. High-throughput epitope discovery reveals frequent recognition of neo-antigens by CD4+ T cells in human melanoma. Nat Med. (2015) 21:81–5. 10.1038/nm.3773 [DOI] [PubMed] [Google Scholar]
- 75.Tran E, Turcotte S, Gros A, Robbins PF, Lu YC, Dudley ME, et al. Cancer immunotherapy based on mutation-specific CD4+ T cells in a patient with epithelial cancer. Science. (2014) 344:641–5. 10.1126/science.1251102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Malekzadeh P, Pasetto A, Robbins PF, Parkhurst MR, Paria BC, Jia L, et al. Neoantigen screening identifies broad TP53 mutant immunogenicity in patients with epithelial cancers. J Clin Invest. (2019) 129:1109–114. 10.1172/JCI123791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Granados DP, Sriranganadane D, Daouda T, Zieger A, Laumont CM, Caron-Lizotte O, et al. Impact of genomic polymorphisms on the repertoire of human MHC class I-associated peptides. Nat Commun. (2014) 5:3600. 10.1038/ncomms4600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kessels HW, de Visser KE, Tirion FH, Coccoris M, Kruisbeek AM, Schumacher TN. The impact of self-tolerance on the polyclonal CD8+ T cell repertoire. J Immunol. (2004) 172:2324–31. 10.4049/jimmunol.172.4.2324 [DOI] [PubMed] [Google Scholar]
- 79.Murata M, Warren EH, Riddell SR. A human minor histocompatibility antigen resulting from differential expression due to a gene deletion. J Exp Med. (2003) 197:1279–89. 10.1084/jem.20030044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Broen K, Levenga H, Vos J, van Bergen K, Fredrix H, Greupink-Draaisma A, et al. A polymorphism in the splice donor site of ZNF419 results in the novel renal cell carcinoma-associated minor histocompatibility antigen ZAPHIR. PLoS One. (2011) 6:e21699. 10.1371/journal.pone.0021699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Brickner AG, Evans AM, Mito JK, Xuereb SM, Feng X, Nishida T, et al. The PANE1 gene encodes a novel human minor histocompatibility antigen that is selectively expressed in B-lymphoid cells and B-CLL. Blood. (2006) 107:3779–86. 10.1182/blood-2005-08-3501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Torikai H, Akatsuka Y, Miyazaki M, Tsujimura A, Yatabe Y, Kawase T, et al. The human cathepsin H gene encodes two novel minor histocompatibility antigen epitopes restricted by HLA-A*3101 and -A*3303. Br J Haematol. (2006) 134:406–16. 10.1111/j.1365-2141.2006.06205.x [DOI] [PubMed] [Google Scholar]
- 83.Warren EH, Vigneron NJ, Gavin MA, Coulie PG, Stroobant V, Dalet A, et al. An antigen produced by splicing of noncontiguous peptides in the reverse order. Science. (2006) 313:1444–7. 10.1126/science.1130660 [DOI] [PubMed] [Google Scholar]
- 84.van Bergen CAM, Verdegaal EME, Honders MW, Hoogstraten C, Steijn-van Tol AQM, de Quartel L, et al. Durable remission of renal cell carcinoma in conjuncture with graft versus host disease following allogeneic stem cell transplantation and donor lymphocyte infusion: rule or exception? PLoS One. (2014) 9:e85198 10.1371/journal.pone.0085198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wölfel C, Lennerz V, Lindemann E, Hess G, Derigs HG, Huber C, et al. Dissection and molecular analysis of alloreactive CD8+ T cell responses in allogeneic haematopoietic stem cell transplantation. Cancer Immunol Immunother. (2008) 57:849–57. 10.1007/s00262-007-0421-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.van Bergen CAM, Kester MGD, Jedema I, Heemskerk MHM, van Luxemburg-Heijs SAP, Kloosterboer FM, et al. Multiple myeloma-reactive T cells recognize an activation-induced minor histocompatibility antigen encoded by the ATP-dependent interferon-responsive (ADIR) gene. Blood. (2007) 109:4089–96. 10.1182/blood-2006-08-043935 [DOI] [PubMed] [Google Scholar]
- 87.Bijen HM, Hassan C, Kester MGD, Janssen GMC, Hombrink P, de Ru AH, et al. Specific T Cell responses against minor histocompatibility antigens cannot generally be explained by absence of their allelic counterparts on the cell surface. Proteomics. (2018)18:e1700250. 10.1002/pmic.201700250 [DOI] [PubMed] [Google Scholar]
- 88.Yee C, Thompson JA, Roche P, Byrd DR, Lee PP, Piepkorn M, et al. Melanocyte destruction after antigen-specific immunotherapy of melanoma: direct evidence of t cell-mediated vitiligo. J Exp Med. (2000) 192:1637–44. 10.1084/jem.192.11.1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Linette GP, Stadtmauer EA, Maus MV, Rapoport AP, Levine BL, Emery L, et al. Cardiovascular toxicity and titin cross-reactivity of affinity-enhanced T cells in myeloma and melanoma. Blood. (2013) 122:863–71. 10.1182/blood-2013-03-490565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Morgan RA, Chinnasamy N, Abate-Daga D, Gros A, Robbins PF, Zheng Z, et al. Cancer regression and neurological toxicity following anti-MAGE-A3 TCR gene therapy. J Immunother. (2013) 36:133–51. 10.1097/CJI.0b013e3182829903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.van den Berg JH, Gomez-Eerland R, van de Wiel B, Hulshoff L, van den Broek D, Bins A, et al. Case report of a fatal serious adverse event upon administration of T cells transduced with a MART-1-specific T-cell receptor. Mol Ther. (2015) 23:1541–50. 10.1038/mt.2015.60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Spierings E, Gras S, Reiser JB, Mommaas B, Almekinders M, Kester MG, et al. Steric hindrance and fast dissociation explain the lack of immunogenicity of the minor histocompatibility HA-1Arg null allele. J Immunol. (2009) 182:4809–16. 10.4049/jimmunol.0803911 [DOI] [PubMed] [Google Scholar]
- 93.Alvarez-Navarro C, Martin-Esteban A, Barnea E, Admon A, de Castro LJA. Endoplasmic Reticulum Aminopeptidase 1 (ERAP1) polymorphism relevant to inflammatory disease shapes the peptidome of the birdshot chorioretinopathy-associated HLA-A*29:02 antigen. Mol Cell Proteomics. (2015) 14:1770–80. 10.1074/mcp.M115.048959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Granados DP, Rodenbrock A, Laverdure JP, Cote C, Caron-Lizotte O, Carli C, et al. Proteogenomic-based discovery of minor histocompatibility antigens with suitable features for immunotherapy of hematologic cancers. Leukemia. (2016) 30:1344–54. 10.1038/leu.2016.22 [DOI] [PubMed] [Google Scholar]
- 95.Kremer AN, Bausenwein J, Lurvink E, Kremer AE, Rutten CE, van Bergen CAM, et al. Discovery and differential processing of HLA class II-restricted minor histocompatibility antigen LB-PIP4K2A-1S and its allelic variant by asparagine endopeptidase. Front Immunol. (2020) 11:381. 10.3389/fimmu.2020.00381 [DOI] [PMC free article] [PubMed] [Google Scholar]