Abstract
Memory CD8+ T cells in the immune system are responsible for the removal of external Ags for a long period of time to protect against re-infection. Naïve to memory CD8+ T cell differentiation and memory CD8+ T cell maintenance require many different factors including local environmental factors. Thus, it has been suggested that the migration of memory CD8+ T cells into specific microenvironments alters their longevity and functions. In this review, we have summarized the subsets of memory CD8+ T cells based on their migratory capacities and described the niche hypothesis for their survival. In addition, the basic roles of CCR7 in conjunction with the migration of memory CD8+ T cells and recent understandings of their survival niches have been introduced. Finally, the applications of altering CCR7 signaling have been discussed.
Keywords: CD8-positive T-lymphocytes; Immunologic memory; Receptors, CCR7; Cell movement; Chemotaxis; Immunotherapy
INTRODUCTION
The defense against pathogens is composed of innate and adaptive immunities. While innate response mounts in a few hours by the recognition of molecular patterns, adaptive immune system takes a few days to initiate Ag-dependent specific response. The adaptive immune system employs B cells, which produce antibodies, and T cells to mediate cellular immunity. One cardinal feature of the adaptive immune system is “memory” response to the same Ag. Once the adaptive immune system has been stimulated with vaccines or pathogens, challenges with the same Ag induce a faster and stronger response than the primary response. These memory responses are mainly mediated by Ab-producing plasma cells and memory T (TM) cells. Through these memory responses, vaccines can protect our body against fatal or serious infectious agents. Although vaccination is the most efficient way to prevent infectious disease, few vaccines have been developed to induce functional TM cells for certain infectious diseases, which is probably because of the lack of knowledge about TM cell development and homeostasis.
Migratory capacity is another important feature of the adaptive immunity, which enables adaptive immune cells to circulate constantly among tissues. Depending on their activation status, these cells generally circulate throughout the body to search for their cognate Ags. For example, naïve T cells are activated with Ags in the secondary lymphoid organs (SLOs) where adaptive immune responses initiate. When T cells are properly activated, they develop into effector T (TE) cells and migrate into infected sites. While effector CD4+ T cells provide “help” to other immune cells, effector CD8+ T cells, also called CTLs, kill infected or damaged cells. Hence, T cell homing is crucial to fight against invading pathogens so that their migration is tightly regulated. It has been well documented that stimulation of chemokine receptors and adhesion molecules on T cell surface allows for an orderly access to a specific microenvironment. Therefore, it is known that various chemokine receptors are sequentially expressed in the course of immune response to pathogens. Recently, the migratory capacities of TM cells have been highlighted because their recall response and survival are affected by their localization.
In this review, we discussed memory CD8+ T cells and their migratory capacities, particularly the C-C chemokine receptor 7 (CCR7)-dependent pathway, for their survival and longevity. We also highlighted recent studies describing the expression of CCR7 and its ligands, CCL19 and CCL21.
MEMORY CD8+ T CELLS
Development of memory CD8+ T cells
Naïve CD8+ T cells constantly circulate throughout the body through SLOs including the lymph nodes (LNs), spleen, and Peyer's patches (PPs) (1,2,3,4). The activation of T cells was first reported in 1970s using a mouse model of lymphocytic choriomeningitis virus (LCMV) infection (5,6). During viral infections, antigen presenting cells (APCs) such as dendritic cells (DCs) obtain viruses and migrate into the SLOs where naïve T cells search for their cognate Ags. Activated CD8+ T cells by these APCs undergo several pathways to proliferate and gain effector functions important to fight against infectious agents. In addition, CD8+ TE cells modulate the expression of their homing receptors to egress from the SLOs and move into the infected area to induce protective immunity (7,8). After the pathogens are cleared, majority (up to 90%) of the activated CD8+ T cells dies, while only a fraction (up to 10%) survives and are maintained as TM (9) cells. These TM cells survive for an extended period of time and provide rapid and robust secondary response to the same Ag. In addition, they undergo self-renewal without Ag exposure and subsets of TM cells undergo a series of steps to re-circulate the whole body searching for re-infections. These prominent characteristics enable TM cells to fight against secondary infections with the same pathogens efficiently (10).
The subsets of memory CD8+ T cells
CD8+ TM cells with these cardinal characteristics can be categorized into diverse subsets including effector memory T (TEM) (11), central memory T (TCM), and tissue-resident memory T (TRM) cells (Fig. 1) (12). These subsets were initially identified based on their differential localizations by the expressions of homing receptors, but they also differ from each other in terms of their cytokine production and proliferation capacities. These abilities are regulated by a series of expression of their receptors, signaling molecules, transcription factors, and other important factors. Of note, different subsets of TM cells form depending on the tropism and characteristics of infectious agents and these subsets cooperate to eradicate infections.
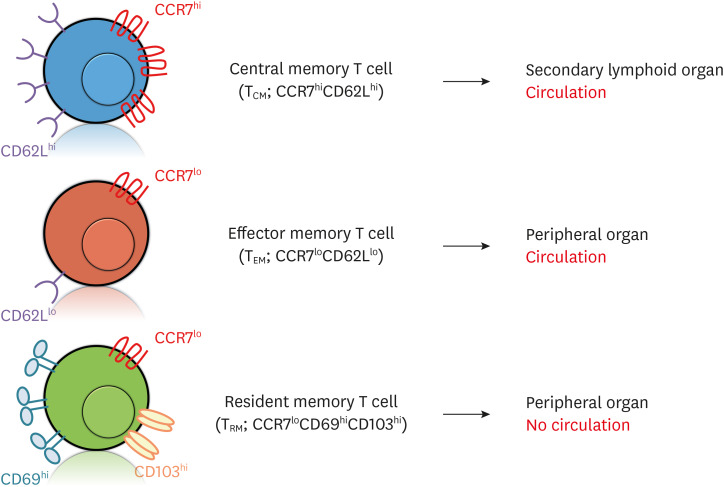
Figure 1. Subsets of TM cells. There are 3 major subsets of TM cells: TCM, TEM and TRM. These subsets are identified on the basis of differential expressions in several chemokine receptors and adhesion molecules. While TCM cells express high levels of CCR7 and CD62L (L-selectin) (CCR7hiCD62Lhi), TEM cells express low levels of CCR7 and CD62L (CCR7loCD62Llo). These phenotypes indicate that TCM cells circulate to SLOs, whereas TEM cells circulate to peripheral organs. TRM cells express low levels of CCR7 and high levels of CD69 and CD103 (CCR7loCD69hiCD103hi) to remain in peripheral organs.
TCM cells monitor infection in the body by circulating in the SLOs. In contrast, TEM and TRM cells do not survey SLOs and are located in peripheral tissues (Fig. 1). However, TEM cells can return to the SLOs if the Ags exist in the SLOs even after Ag is cleared in the infected target area (13,14,15,16). In 2009, Gebhardt et al. (17) reported that TRM cells stay in the non-lymphoid tissues (NLTs) without returning to the SLOs. They prepare to fight against local infections and are found in many different organs including the brain, lung, gut, skin, and other peripheral organs (18,19,20).
The generation of specific subsets of CD8+ TM cells has been attempted using different animal models. It is generally assumed that systemic infections or vaccinations can induce TCM and TRM cell formation, while local infections induce TRM cell development (21). Thus, Ag tropism or location determines the fate of TM cells. In addition, the generation of certain subsets may be crucial to protect against specific pathogens (22). For example, the presence of TRM cells in the liver substantially enhances protection against Plasmodium (23). Altogether, the modulation of TM cell mobility would benefit our body to fight properly against pathogens by placing TM cells in proper positions.
The survival and homeostasis of TM cells
A remarkable aspect of TM cells is their longevity and homeostasis without further antigenic stimulation. The underlying mechanisms of their homeostasis are based on the exposure to the homeostatic cytokines such as IL-7 and -15 (24-31). IL-7 has been well documented as a survival cytokine of naïve, memory precursor (MP) and TM cells. This cytokine is provided by stromal cells including fibroblastic reticular cells (FRCs) in the spleen and LNs (32,33,34). In conjunction with IL-7, IL-15 can induce homeostatic proliferation of TM cells. IL-15 also helps for the survival of KLRG1hi terminally differentiated TE and TM cells. Therefore, it is crucial for TM cells to “see” these cytokines in order to develop and maintain homeostasis.
TM cells develop and maintain in multiple organs including the spleen, LNs, liver, lung, and bone marrow (BM) (35). After systemic infection, TM cells can survive and proliferate in these organs, particularly in the BM (36). However, different TM cell subsets are differentially localized within different organs, suggesting that these cells may be exposed to different survival factors depending on their location (37,38). Since leukocyte recruitment is tightly regulated, it is interesting to understand the homing of each subset.
CCR7—CHEMOKINE RECEPTOR FOR MEMORY CD8+ T CELLS
CCR7 is a homing receptor
CCR7 is a lymphocyte-specific G-protein-coupled receptor with 7 transmembrane spanning alpha helices for CCL19 and CCL21 as ligands. It was first named Epstein-Barr virus (EBV)-indicted gene 1, a gene induced by EBV and Burkitt's lymphoma cells in B-lymphocytes. In the same study, it was shown that it plays an important role in response to virus infection and is detected only in B- and T-lymphocytes (39,40).
In the late 1990s, a study using CCR7-deficient mice showed that CCR7 plays an important role in controlling T cell movement to SLOs, particularly LNs and PPs. In addition, the formation of T cell zone was abolished due to abnormal T cell migration. After immunization, the migration of mature skin DCs into the LNs resulted in delayed immune response to injected Ags (41,42). Based on this observation, CCR7 has been established as one of the crucial receivers responsible for lymphocyte homing (41).
CD8+ T cells and CCR7
Among the CD8+ T cells, naïve and TCM cells generally express high levels of CCR7 (3,12,43,44), hence they can migrate to the T cell zone of the LNs and spleen. These T cells can be activated in the T cell zone by the APCs and developed into TE cells. During this process, TE cells can move from the T cell zone to the red pulp and the infected area by the downregulation of CCR7 expression (45). Through this regulation of CCR7 expression, CD8+ T cells can find their cognate Ag in the SLOs to be activated and migrated into infected locus. After infections are cleared, TM cells form and circulate to different parts of the body based on the levels of CCR7 expression (45,46,47).
During TE–TM cell transition, CCR7 expression influences the fate of these cells. It was reported that the mRNA levels of CCR7 were more pronounced in memory precursor T cells (MPECs) than in short-lived effector cells (SLECs) (48). In addition, the TCM and TEM cells were found in different locations of the SLOs depending on CCR7 concentration.
CCR7 expression was inhibited in TRM cells, the recently identified TM cell subset, making it possible for TRM cells to act as the first line of defense within peripheral tissues (49). Altogether, the regulation of CCR7 expression controls the recruitment and release of CD8+ T cells from SLOs, determining the CD8+ T cell response outcome.
Transcriptional regulation of CCR7 and microRNAs (miRNAs)
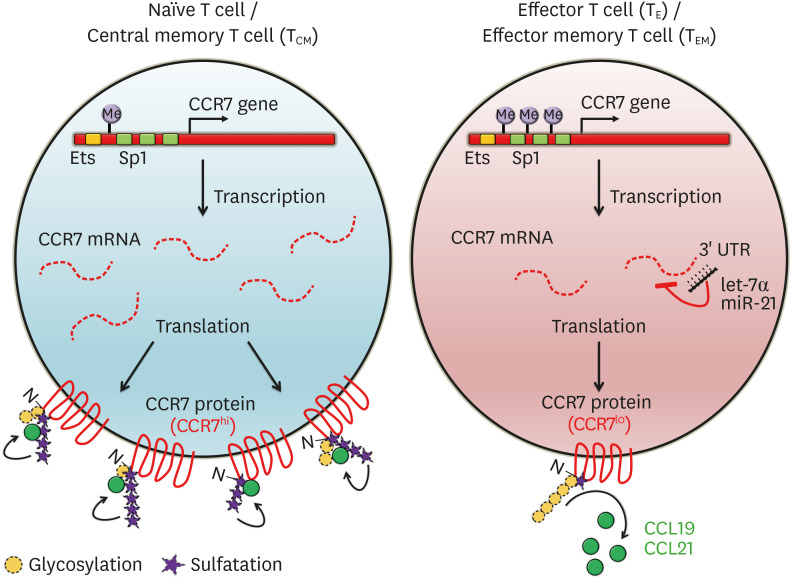
The expression of CCR7 on CD8+ T cells is regulated by several transcription factors. In the CCR7 promoter region, there are 3 binding sites specific for protein 1 (Sp1) and one Ets-1-binding site (50), which suggests that the increased expression of CCR7 is mediated at least partially by transcription factors such as Sp1 and Ets-1 (Fig. 2) (35,50). AP1 and NF-κB were also reported to upregulate CCR7 expression via binding to the CCR7 promoter locus in various cancer cell lines (51,52,53). In addition, Krüppel-like factor 2 (KLF2) and T cell factor 1 (TCF1) are significantly upregulated in naïve and TCM cells and regulate the expression of several molecules including CCR7, CD62L (L-selectin; encoded by Sell), and sphingosine-1-phosphate receptor 1 in order to modulate the migration into the SLOs (54). In contrast, CD8+ TEM and TRM cells suppress KLF2 and TCF1, while simultaneously expressing Blimp-1 and suppressing the transcription of CCR7 independently (55,56,57).
Figure 2. The regulation of CCR7 expression in T cells. The distinguishing features of naïve, TE, and TM cells reflect different programs of gene expression regulated by various factors. First, the gene encoding CCR7 protein is modulated by epigenetic mechanisms, such as methylation. The CCR7 chromatin can silenced by methylation of the promoter regions, in which transcription factor binding sites such as Sp1 and Ets-1 in effector and TEM cells are located. In contrast, this locus is demethylated, allowing the gene to be expressed in naïve and TCM cells. Additional inhibitory mechanisms of CCR7 gene expression include miRNA-dependent regulation. The indicated miRNAs such as let-7α and miR-21 bind to the 3′ UTR of CCR7 mRNA and decay the mRNA, thereby inhibiting translation efficiency. Following the translation, CCR7 protein can be post-translationally modified to modulate their affinity with their ligands, CCL19 and CCL21. The glycosylation of N-terminus of CCR7 protein suppresses CCR7 activity, while sulfation increases its affinity for the ligands.
It was also reported that Forkhead O 1 (FOXO1), another transcription factor, regulates CCR7 transcription during TE–TM cell transition, and not during naïve–CD8+ TE cell transition. A study using FOXO1 knockout (KO) mice demonstrated that FOXO1 promoted enrichment of MP cells, thus wild-type TE cells highly proliferated upon secondary infections in CD8+ T cells compared to FOXO1-deficient TE cells (58). In a related study, it was observed that FOXO1 expression is significantly higher in MPECs than in SLECs, and it increase the levels of IL-7Ra and TCF7 expression to help create and maintain TCM cells which were derived from MPECs (59).
CCR7 expression is also controlled by miRNAs. MiRNAs consisting of small RNA fragments under 23 nucleotides generally bind to 3' UTRs of complementary target sequences on target mRNAs to decay or stabilize the mRNA (60). Using big data analysis, several T-cell-associated genes including CCR7 were discovered to be targeted by miR-21, an anti-apoptotic factor (61,62,63). MiR-21, a miRNA inhibiting post-transcription levels of CCR7, led to a significant reduction of CCR7 protein expression when naïve and CD4+ TM cells are activated. Indeed, it is activated in inverse correlation to the amount of CCR7 protein (Fig. 2) (64). Further, it was reported that the expression of CCR7 was adjusted by miRNAs in several cancers, such as let-7α (in breast cancer cells and patients) (65), miR-320d (in oral squamous cell carcinoma) (66), miR-532-3p (in tongue squamous cell carcinoma) (67) and head and neck squamous cell carcinoma (68), and miR-199a (in human mantle cell lymphoma) (69).
Epigenetic and post-translational modification (PTM) of CCR7
Although there is little report of epigenetic regulators for CCR7, it has been documented based on CCR7 gene methylation using big data analysis. CCR7 methylation in human CD8+ TM cells increased in the order of CD8+ TM cell differentiation state: naïve<TSCM (stem cell memory)<TCM<TEM, when this locus was analyzed using whole-genome bisulfite sequencing, a next-generation sequencing method (Fig. 2) (4). As the DNA methylation state is known as silent chromatin (called as heterochromatin), the expression of the gene was inhibited (70). Another report showed that monocyte-derived DCs in the mouse lung homing into SLOs are significantly lower in number than conventional DCs. In correlation with this observation, monocyte-derived DCs was highly methylated in H3K27 (H3K27me3), which forms heterochromatin, thus blocking their homing into the SLOs (71).
CCR7 can also be regulated by PTM (Fig. 2). Glycosylation in the N-terminus of CCR7 protein in TE cells not only blocks, but also decreases receptor sensitivity, resulting in chemokine-induced downstream signaling modulation. It was also reported that de-glycosylation with enzymes produced by DCs changed the folding of CCR7 protein to increase the activity of this receptor (9). Conversely, the induction of sulfation in tyrosine residue of the CCR7 protein N-terminus can increase the affinity between CCR7 and its ligands (72).
Altogether the expression of CCR7 can be altered by many different factors in several steps, possibly indicating that the regulation of this receptor is critical for the immune response to pathogen. In addition, it has been suggested that this receptor can be modulated using multiple methods in order to enhance adaptive immunity.
CCL19 and CCL21, ligands of CCR7
CCR7hi CD8+ naïve and TM cells have chemotaxis features toward CCL19 and CCL21 in a concentration-dependent manner (73,74,75,76). These chemokines were reported to be produced by high endothelial venules (63) in the T cell zone of SLOs (77,78). When naïve T cells enter the SLOs, they are located in the T and B cell zones. Particularly, naïve CD8+ T cells with CCR7 expression are enriched by FRCs in the T cell zones within the spleen and LNs, where CCL19 and CCL21 are highly enriched (33,79). Correlated with the role of these chemokines, the rate of movement of naïve T cells in the SLOs decreases in plt/plt mouse (paucity of LN; naturally weak CCL19 and CCL21 mutant mice) regardless of T cells lacking CCR7 (80,81). In the CCL21-deficient-special niche, CCR7-positive naïve and TM cells circulate themselves within the interstitial lymphoid organs after being ejected from SLOs.
The quality and quantity of these microenvironmental niches were dynamically changed by several factors during the state of immune responses. Among these factors, IFN-γ, an antiviral-related cytokine, can transiently but substantially downregulate the expressions of CCL19 and CCL21 in the spleen and LNs. During viral infections, IFN-γ is released by effector immune cells including CD8+ TE cells, which then reduces the concentration of CCL19 and CCL21 in the T cell zone of the SLOs. In turn, TE cells escape SLOs possibly due to the low levels of CCL19 and CCL21 (82).
CCL21 was also reported to participate in cancer metastasis. Certain types of cancer cells express CCR7, so that they have the ability to migrate towards the SLOs, particularly LNs (83). This migration facilitates cancer metastasis by pushing cancer cells into lymphatic vessels through LNs. Indeed, the patients with CCR7-positive tumors have significantly poorer prognosis than the patients with CCR7-negative tumors (84,85,86). These studies showed that the regulation of these chemokines is also important for the movement and localization of T and cancer cells.
CCR7 PROVIDES IMPORTANT HOMING SIGNALS FOR MEMORY CD8+ T CELLS
Niche for the survival of TM cells
Survival cytokines are required for the longevity of TM cells; however, it is incompletely defined how these cells “see” these cytokines. There are at least 2 possible explanations for these mechanisms. One hypothesis is systemic exposure, where one organ provides these cytokines and TM cells recognize them anywhere in the body. Another possibility is that these cytokines are present only in specialized microenvironments called niches and TM cells need to home these anatomical locations to receive the required signals. Although systemic exposure cannot be ruled out, evidence have been accumulated to explain the niche hypothesis. First, BM is the preferred site for the homeostatic proliferation, suggesting that survival cytokines are provided locally. Second, differential localization of TE cell subsets within the organ may indicate that they receive different signals depending on their location (37,38). Third, IL-7 has been suggested to be bound to the extracellular matrix near the IL-7 producers (87). Finally, IL-15 has been shown to be trans-presented so that cell-to-cell contacts with IL-15 presenting cells are required for optimal simulation (88). Altogether, it is strongly suggested that TM cells migrate to specialized microenvironments within the organ in order to survive.
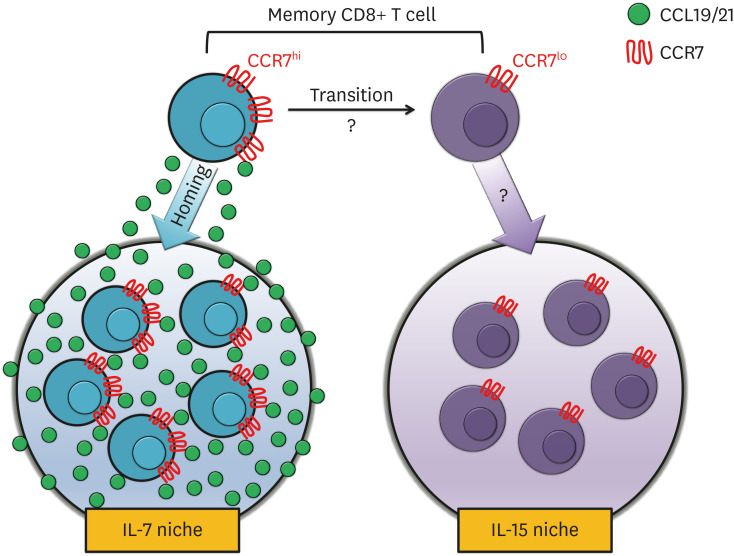
Among the cytokines that are critical for TM cells, it is known that CCL19 and CCL21 maintain a collaborative relationship with IL-7 and IL-15 (Fig. 3) (89). A rich CCL19 within the LNs works in conjunction with IL-7 to improve the survival of the T cells, but no exact mechanism has been identified (33). Whereas, the adjustment of CD8+ TM cells by IL-15 was found within the NLTs, such as the BM and lung, where distinct from SLOs with large amount of CCL19 and CCL21 (36,90). This implies that the importance of the physical microenvironment is formed by CCL19 and CCL21, as well as sufficient CCR7 expression within the T cells.
Figure 3. CCR7 introduces microenvironmental niche for memory CD8+ T cells. The schematic model indicates the trafficking patterns of CCR7hi or CCR7lo CD8+ TM cells to find individual niches for their survival. CCR7hi CD8+ TM cells migrate toward high concentration of CCL19 or CCL21, where IL-7 is also expressed. In the contrary, CCR7lo TM cells migrate toward IL-15-rich microenvironment, even though the homing receptors for this migration have not been identified yet. Altogether, CCR7hi and CCR7lo CD8+ TM cells do not share their survival cytokines, and CCR7 biases the survival of CD8+ TM cells toward IL-7 niche.
CCR7 expression alters the fate decision of CD8+ T cells
To understand the roles of CCR7, Junt et al. (91) first reported normal T cell response to viral infection in plt/plt mice with spontaneous mutations in CCL19 and CCL21 locus. Thus, the viral clearance and secondary response to the same pathogen were comparable to those in littermate controls. However, they also observed that the formation of TM cells were significantly reduced, when CCR7-deficient mice were infected with the same virus (92). It was also observed that viral clearance in CCR7 KO mice was slower than that in wild-type mice, if completely cleared. Probably due to the reduced number of TM cells, CCR7-deficient mice contained reduced number of IFN-γ+ CD8+ T cells in response to secondary infections. Since 2 different results were reported, it was suggested that plt/plt mice express CCL21b, a leucine isoform of CCL21, in lymphatic vessels. In addition, CCL21b was also found in peripheral tissue; thus, the recruitment of CCR7hi cells into the peripheral tissues was compensated (78,93). In agreement with this report, mice with ubiquitous overexpression of CCL21 showed abnormal viral clearance (94). These altered immune responses are not restricted to viral infections, because CCR7 KO mice mounted reduced inflammation in response to Toxoplasma gondii infection, an intracellular protozoan parasite (95).
In order to exclude the possibility of delayed viral clearance and emphasize the role of CCR7 in the fate decision of CD8+ TM cells, P14 transgenic mice, which recognize the DbGP33–41 epitope of LCMV, were employed (90). By transferring small number of CCR7 wild-type or KO P14 CD8+ T cells into wild-type C57/Bl6 mice, these mice were able to clear the viral infection regardless of the genotypes of P14 cells. Using this model, increased number of CCR7 KO TM cells was observed with normal recall abilities. Enhanced survival of CCR7 KO TM cells by IL-15 signaling were found in the lung and BM, because the removal of IL-15, but not IL-7, maintained the comparable number of CCR7-deficient and -sufficient TM cells. In addition, these CCR7-deficient TM cells showed increased ratio of homeostatic turnover compared to CCR7-sufficient cells. These results suggest that CCR7 signaling directs the migration toward IL-7-niches, while CCR7lo TM cells migrate toward IL-15 niches for their survival. In addition, inhibition of CCR7 signaling may increase the number of TM cells in the lung, where it has been suggested to be difficult for TM cells to survive (Fig. 3).
When CCR7 was overexpressed in all T cells, the numbers of TM cells in the spleen and LNs of CCR7-tg mice were increased compared to that in wild-type mice, while CCR7-tg TM cell number reduced in the liver and lung. These mice showed difficulty in clearance of skin infection, suggesting that CCR7 forces the migration of TM cells into the SLOs. It would be interesting to see if increased survival of CCR7-tg TM cells in the SLOs is IL-7-dependent. Taken together, the CCR7 expression of CD8+ TM cells determines the fates of CCR7hi and CCR7lo CD8+ T cells by properly guiding them into survival niches.
CONCLUSION AND FUTURE PERSPECTIVES
The importance of TM cell subsets has been well documented, and the roles of each subset shed light on individual infectious disease in the last decade. However, it is still not evident how each subset receives crucial signals for their longevity. Accumulating data suggest that specialized niches are present for the survival of TM cells and the migration of MEPCs and TM cells into these niches is crucial for their maintenance. CCR7 may determine the niche suitable for their survival, depending on the subtypes or functions of CD8+ TM cells. Therefore, it can be inferred that CCR7 plays a crucial role in the activation of naïve T cells as well as the development and maintenance of CD8+ TM cell subsets according to these studies.
Currently, the therapies using T cells, such as chimeric Ag receptor T cells, have shown remarkably effective progresses into clinical phases (96,97,98). Despite these successes, the cancer types where these therapies can be applied are limited till date. One of the reasons for this limitation is the type of T cells used in these therapies. Most therapies use naïve or in vitro activated T cells, which can quickly be inactivated in tumor microenvironments. Thus, TM cells have been suggested to be utilized for these applications, particularly since they are seldom exhausted even in various types of immune diseases including cancer, allergic disease, autoimmune disease, and inflammatory bowel disease.
In addition to this suggestion, controlling the migration of TM cells by CCR7 modulation may determine the destiny of CD8+ TM cells by changing their location as discussed in this review. Depending on the diseases or cancers, the localization of TM cells is crucial to promote optimal immune response. Taken together, TM cells may be present and respond effectively at the target sites regardless of whether these sites are present in the SLOs or peripheral organs by inducing or inhibiting the CCR7 signaling in TM cells.
In the past few decades, new respiratory diseases have been caused by virus infections such as influenza virus, severe acute respiratory syndrome coronavirus (SARS-CoV), middle east respiratory syndrome coronavirus, and severe acute respiratory syndrome coronavirus 2 repeatedly. All of these pathogenic viruses were derived from coronaviruses that commonly infect humans, but new mutations have made it difficult for our immune system to get rid of these viral infections. However, the survival of memory CD8+ T cell responding to SARS-CoV in peripheral blood of the patients possibly suggest the importance of TM cells for the recovery from these diseases (99,100). Particularly, the Ab response to SARS-CoV was short-lived, indicating the biased immune responses toward T cell immunity. However, it is still not clear whether virus-specific TM cells are also present in the lungs of these survived patients. Since the lungs were proposed to provide unfavorable microenvironments for TM cell homeostasis, it would be particularly challenging to develop proper vaccines or therapies. However, Jung et al. (90) showed that CCR7-deficient TM cells can heavily populate the lung, possibly indicating that the regulation of CCR7 signaling may give us a chance to prevent these diseases. These approaches may illuminate the future of our novel vaccines or therapies.
ACKNOWLEDGEMENTS
We thank Sang-Hoon Kim and Aryeong Choi for their useful and constructive suggestions on this review. This work was supported by the National Research Foundation of Korea (NRF) funded by Ministry of Education, Science and Technology (NRF-2019R1A6A1A03031807).
Abbreviations
- APC
antigen presenting cell
- BM
bone marrow
- CCR7
C-C chemokine receptor 7
- DC
dendritic cell
- FOXO1
Forkhead O 1
- FRC
fibroblastic reticular cell
- KLF2
Krüppel-like factor 2
- KO
knockout
- LCMV
lymphocytic choriomeningitis virus
- LN
lymph node
- miRNA
microRNA
- MP
memory precursor
- MPEC
memory precursor T cell
- NLT
non-lymphoid tissue
- PP
Peyer's patch
- PTM
post-translational modification
- SARS-CoV
severe acute respiratory syndrome coronavirus
- SLEC
short-lived effector cell
- SLO
secondary lymphoid organ
- Sp1
specific for protein 1
- TCF1
T cell factor 1
- TCM
central memory T
- TE
effector T
- TEM
effector memory T
- TM
memory T
- TRM
tissue-resident memory T
Footnotes
Conflict of Interest: The authors declare no potential conflicts of interest.
- Conceptualization: Choi H, Jung YW.
- Data curation: Choi H, Jung YW.
- Funding acquisition: Jung YW.
- Supervision: Jung YW.
- Visualization: Choi H, Song H.
- Writing - original draft: Choi H, Song H, Jung YW.
- Writing - review & editing: Jung YW.
References
- 1.Weninger W, Crowley MA, Manjunath N, von Andrian UH. Migratory properties of naive, effector, and memory CD8+ T cells. J Exp Med. 2001;194:953–966. doi: 10.1084/jem.194.7.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Butcher EC, Williams M, Youngman K, Rott L, Briskin M. Lymphocyte trafficking and regional immunity. Adv Immunol. 1999;72:209–253. doi: 10.1016/s0065-2776(08)60022-x. [DOI] [PubMed] [Google Scholar]
- 3.Campbell JJ, Murphy KE, Kunkel EJ, Brightling CE, Soler D, Shen Z, Boisvert J, Greenberg HB, Vierra MA, Goodman SB, et al. CCR7 expression and memory T cell diversity in humans. J Immunol. 2001;166:877–884. doi: 10.4049/jimmunol.166.2.877. [DOI] [PubMed] [Google Scholar]
- 4.Abdelsamed HA, Moustaki A, Fan Y, Dogra P, Ghoneim HE, Zebley CC, Triplett BM, Sekaly RP, Youngblood B. Human memory CD8 T cell effector potential is epigenetically preserved during in vivo homeostasis. J Exp Med. 2017;214:1593–1606. doi: 10.1084/jem.20161760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Volkert M, Marker O, Bro-Jørgensen K. Twp populations of T lymphocytes immune to the lymphocytic choriomeningitis virus. J Exp Med. 1974;139:1329–1343. doi: 10.1084/jem.139.5.1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnson ED, Cole GA. Functional heterogeneity of lymphocytic choriomeningitis virus-specific T lymphocytes. I. Identification of effector and memory subsets. J Exp Med. 1975;141:866–881. [PMC free article] [PubMed] [Google Scholar]
- 7.Huster KM, Busch V, Schiemann M, Linkemann K, Kerksiek KM, Wagner H, Busch DH. Selective expression of IL-7 receptor on memory T cells identifies early CD40L-dependent generation of distinct CD8+ memory T cell subsets. Proc Natl Acad Sci U S A. 2004;101:5610–5615. doi: 10.1073/pnas.0308054101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Joshi NS, Cui W, Chandele A, Lee HK, Urso DR, Hagman J, Gapin L, Kaech SM. Inflammation directs memory precursor and short-lived effector CD8+ T cell fates via the graded expression of T-bet transcription factor. Immunity. 2007;27:281–295. doi: 10.1016/j.immuni.2007.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Badovinac VP, Harty JT. Programming, demarcating, and manipulating CD8+ T-cell memory. Immunol Rev. 2006;211:67–80. doi: 10.1111/j.0105-2896.2006.00384.x. [DOI] [PubMed] [Google Scholar]
- 10.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 11.Sallusto F, Kremmer E, Palermo B, Hoy A, Ponath P, Qin S, Förster R, Lipp M, Lanzavecchia A. Switch in chemokine receptor expression upon TCR stimulation reveals novel homing potential for recently activated T cells. Eur J Immunol. 1999;29:2037–2045. doi: 10.1002/(SICI)1521-4141(199906)29:06<2037::AID-IMMU2037>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 12.Debes GF, Arnold CN, Young AJ, Krautwald S, Lipp M, Hay JB, Butcher EC. Chemokine receptor CCR7 required for T lymphocyte exit from peripheral tissues. Nat Immunol. 2005;6:889–894. doi: 10.1038/ni1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bromley SK, Thomas SY, Luster AD. Chemokine receptor CCR7 guides T cell exit from peripheral tissues and entry into afferent lymphatics. Nat Immunol. 2005;6:895–901. doi: 10.1038/ni1240. [DOI] [PubMed] [Google Scholar]
- 14.Mackay CR, Marston WL, Dudler L, Spertini O, Tedder TF, Hein WR. Tissue-specific migration pathways by phenotypically distinct subpopulations of memory T cells. Eur J Immunol. 1992;22:887–895. doi: 10.1002/eji.1830220402. [DOI] [PubMed] [Google Scholar]
- 15.Mackay CR, Marston WL, Dudler L. Naive and memory T cells show distinct pathways of lymphocyte recirculation. J Exp Med. 1990;171:801–817. doi: 10.1084/jem.171.3.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim SK, Reed DS, Heath WR, Carbone F, Lefrançois L. Activation and migration of CD8 T cells in the intestinal mucosa. J Immunol. 1997;159:4295–4306. [PubMed] [Google Scholar]
- 17.Gebhardt T, Wakim LM, Eidsmo L, Reading PC, Heath WR, Carbone FR. Memory T cells in nonlymphoid tissue that provide enhanced local immunity during infection with herpes simplex virus. Nat Immunol. 2009;10:524–530. doi: 10.1038/ni.1718. [DOI] [PubMed] [Google Scholar]
- 18.Wakim LM, Woodward-Davis A, Bevan MJ. Memory T cells persisting within the brain after local infection show functional adaptations to their tissue of residence. Proc Natl Acad Sci U S A. 2010;107:17872–17879. doi: 10.1073/pnas.1010201107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mueller SN, Zaid A, Carbone FR. Tissue-resident T cells: dynamic players in skin immunity. Front Immunol. 2014;5:332. doi: 10.3389/fimmu.2014.00332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shin H, Iwasaki A. A vaccine strategy that protects against genital herpes by establishing local memory T cells. Nature. 2012;491:463–467. doi: 10.1038/nature11522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rosato PC, Wijeyesinghe S, Stolley JM, Masopust D. Integrating resident memory into T cell differentiation models. Curr Opin Immunol. 2020;63:35–42. doi: 10.1016/j.coi.2020.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fernandez-Ruiz D, Ng WY, Holz LE, Ma JZ, Zaid A, Wong YC, Lau LS, Mollard V, Cozijnsen A, Collins N, et al. Liver-resident memory CD8+ T cells form a front-line defense against malaria liver-stage infection. Immunity. 2016;45:889–902. doi: 10.1016/j.immuni.2016.08.011. [DOI] [PubMed] [Google Scholar]
- 23.Welch PA, Namen AE, Goodwin RG, Armitage R, Cooper MD. Human IL-7: a novel T cell growth factor. J Immunol. 1989;143:3562–3567. [PubMed] [Google Scholar]
- 24.Schluns KS, Kieper WC, Jameson SC, Lefrançois L. Interleukin-7 mediates the homeostasis of naïve and memory CD8 T cells in vivo . Nat Immunol. 2000;1:426–432. doi: 10.1038/80868. [DOI] [PubMed] [Google Scholar]
- 25.Zhang X, Sun S, Hwang I, Tough DF, Sprent J. Potent and selective stimulation of memory-phenotype CD8+ T cells in vivo by IL-15. Immunity. 1998;8:591–599. doi: 10.1016/s1074-7613(00)80564-6. [DOI] [PubMed] [Google Scholar]
- 26.Kanegane H, Tosato G. Activation of naive and memory T cells by interleukin-15. Blood. 1996;88:230–235. [PubMed] [Google Scholar]
- 27.Hasan MS, Kallas EG, Thomas EK, Looney J, Campbell M, Evans TG. Effects of interleukin-15 on in vitro human T cell proliferation and activation. J Interferon Cytokine Res. 2000;20:119–124. doi: 10.1089/107999000312513. [DOI] [PubMed] [Google Scholar]
- 28.Ku CC, Murakami M, Sakamoto A, Kappler J, Marrack P. Control of homeostasis of CD8+ memory T cells by opposing cytokines. Science. 2000;288:675–678. doi: 10.1126/science.288.5466.675. [DOI] [PubMed] [Google Scholar]
- 29.Kieper WC, Tan JT, Bondi-Boyd B, Gapin L, Sprent J, Ceredig R, Surh CD. Overexpression of interleukin (IL)-7 leads to IL-15-independent generation of memory phenotype CD8+ T cells. J Exp Med. 2002;195:1533–1539. doi: 10.1084/jem.20020067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tan JT, Ernst B, Kieper WC, LeRoy E, Sprent J, Surh CD. Interleukin (IL)-15 and IL-7 jointly regulate homeostatic proliferation of memory phenotype CD8+ cells but are not required for memory phenotype CD4+ cells. J Exp Med. 2002;195:1523–1532. doi: 10.1084/jem.20020066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hara T, Shitara S, Imai K, Miyachi H, Kitano S, Yao H, Tani-ichi S, Ikuta K. Identification of IL-7-producing cells in primary and secondary lymphoid organs using IL-7-GFP knock-in mice. J Immunol. 2012;189:1577–1584. doi: 10.4049/jimmunol.1200586. [DOI] [PubMed] [Google Scholar]
- 32.Link A, Vogt TK, Favre S, Britschgi MR, Acha-Orbea H, Hinz B, Cyster JG, Luther SA. Fibroblastic reticular cells in lymph nodes regulate the homeostasis of naive T cells. Nat Immunol. 2007;8:1255–1265. doi: 10.1038/ni1513. [DOI] [PubMed] [Google Scholar]
- 33.Onder L, Narang P, Scandella E, Chai Q, Iolyeva M, Hoorweg K, Halin C, Richie E, Kaye P, Westermann J, et al. IL-7-producing stromal cells are critical for lymph node remodeling. Blood. 2012;120:4675–4683. doi: 10.1182/blood-2012-03-416859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fang LW, Kao YH, Chuang YT, Huang HL, Tai TS. Ets-1 enhances tumor migration through regulation of CCR7 expression. BMB Rep. 2019;52:548–553. doi: 10.5483/BMBRep.2019.52.9.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Becker TC, Coley SM, Wherry EJ, Ahmed R. Bone marrow is a preferred site for homeostatic proliferation of memory CD8 T cells. J Immunol. 2005;174:1269–1273. doi: 10.4049/jimmunol.174.3.1269. [DOI] [PubMed] [Google Scholar]
- 36.Guilliams M, Scott CL. Does niche competition determine the origin of tissue-resident macrophages? Nat Rev Immunol. 2017;17:451–460. doi: 10.1038/nri.2017.42. [DOI] [PubMed] [Google Scholar]
- 37.T'Jonck W, Guilliams M, Bonnardel J. Niche signals and transcription factors involved in tissue-resident macrophage development. Cell Immunol. 2018;330:43–53. doi: 10.1016/j.cellimm.2018.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Birkenbach M, Josefsen K, Yalamanchili R, Lenoir G, Kieff E. Epstein-Barr virus-induced genes: first lymphocyte-specific G protein-coupled peptide receptors. J Virol. 1993;67:2209–2220. doi: 10.1128/jvi.67.4.2209-2220.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Burgstahler R, Kempkes B, Steube K, Lipp M. Expression of the chemokine receptor BLR2/EBI1 is specifically transactivated by Epstein-Barr virus nuclear antigen 2. Biochem Biophys Res Commun. 1995;215:737–743. doi: 10.1006/bbrc.1995.2525. [DOI] [PubMed] [Google Scholar]
- 40.Förster R, Schubel A, Breitfeld D, Kremmer E, Renner-Müller I, Wolf E, Lipp M. CCR7 coordinates the primary immune response by establishing functional microenvironments in secondary lymphoid organs. Cell. 1999;99:23–33. doi: 10.1016/s0092-8674(00)80059-8. [DOI] [PubMed] [Google Scholar]
- 41.Yoshida R, Imai T, Hieshima K, Kusuda J, Baba M, Kitaura M, Nishimura M, Kakizaki M, Nomiyama H, Yoshie O. Molecular cloning of a novel human CC chemokine EBI1-ligand chemokine that is a specific functional ligand for EBI1, CCR7. J Biol Chem. 1997;272:13803–13809. doi: 10.1074/jbc.272.21.13803. [DOI] [PubMed] [Google Scholar]
- 42.Sallusto F, Lenig D, Förster R, Lipp M, Lanzavecchia A. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature. 1999;401:708–712. doi: 10.1038/44385. [DOI] [PubMed] [Google Scholar]
- 43.Sallusto F, Lenig D, Mackay CR, Lanzavecchia A. Flexible programs of chemokine receptor expression on human polarized T helper 1 and 2 lymphocytes. J Exp Med. 1998;187:875–883. doi: 10.1084/jem.187.6.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Potsch C, Vöhringer D, Pircher H. Distinct migration patterns of naive and effector CD8 T cells in the spleen: correlation with CCR7 receptor expression and chemokine reactivity. Eur J Immunol. 1999;29:3562–3570. doi: 10.1002/(SICI)1521-4141(199911)29:11<3562::AID-IMMU3562>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 45.Khanna KM, McNamara JT, Lefrançois L. In situ imaging of the endogenous CD8 T cell response to infection. Science. 2007;318:116–120. doi: 10.1126/science.1146291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dauner JG, Williams IR, Jacob J. Differential microenvironment localization of effector and memory CD8 T cells. J Immunol. 2008;180:291–299. doi: 10.4049/jimmunol.180.1.291. [DOI] [PubMed] [Google Scholar]
- 47.Jung YW, Rutishauser RL, Joshi NS, Haberman AM, Kaech SM. Differential localization of effector and memory CD8 T cell subsets in lymphoid organs during acute viral infection. J Immunol. 2010;185:5315–5325. doi: 10.4049/jimmunol.1001948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schenkel JM, Masopust D. Tissue-resident memory T cells. Immunity. 2014;41:886–897. doi: 10.1016/j.immuni.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chuang CW, Pan MR, Hou MF, Hung WC. Cyclooxygenase-2 up-regulates CCR7 expression via AKT-mediated phosphorylation and activation of Sp1 in breast cancer cells. J Cell Physiol. 2013;228:341–348. doi: 10.1002/jcp.24136. [DOI] [PubMed] [Google Scholar]
- 50.Mathas S, Hinz M, Anagnostopoulos I, Krappmann D, Lietz A, Jundt F, Bommert K, Mechta-Grigoriou F, Stein H, Dörken B, et al. Aberrantly expressed c-Jun and JunB are a hallmark of Hodgkin lymphoma cells, stimulate proliferation and synergize with NF-κ B. EMBO J. 2002;21:4104–4113. doi: 10.1093/emboj/cdf389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Höpken UE, Foss HD, Meyer D, Hinz M, Leder K, Stein H, Lipp M. Up-regulation of the chemokine receptor CCR7 in classical but not in lymphocyte-predominant Hodgkin disease correlates with distinct dissemination of neoplastic cells in lymphoid organs. Blood. 2002;99:1109–1116. doi: 10.1182/blood.v99.4.1109. [DOI] [PubMed] [Google Scholar]
- 52.Mburu YK, Egloff AM, Walker WH, Wang L, Seethala RR, van Waes C, Ferris RL. Chemokine receptor 7 (CCR7) gene expression is regulated by NF-κB and activator protein 1 (AP1) in metastatic squamous cell carcinoma of head and neck (SCCHN) J Biol Chem. 2012;287:3581–3590. doi: 10.1074/jbc.M111.294876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Choi HS, Kim KH, Jin S, Kim J, Yoo I, Pack SP, Ha UH, Park TW, Choi SA, Yuk SH, et al. Decreased expression of sphingosine-1-phosphate receptor 1 in the blood leukocyte of rheumatoid arthritis patients. Immune Netw. 2018;18:e39. doi: 10.4110/in.2018.18.e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kallies A, Xin A, Belz GT, Nutt SL. Blimp-1 transcription factor is required for the differentiation of effector CD8+ T cells and memory responses. Immunity. 2009;31:283–295. doi: 10.1016/j.immuni.2009.06.021. [DOI] [PubMed] [Google Scholar]
- 55.Rutishauser RL, Martins GA, Kalachikov S, Chandele A, Parish IA, Meffre E, Jacob J, Calame K, Kaech SM. Transcriptional repressor Blimp-1 promotes CD8+ T cell terminal differentiation and represses the acquisition of central memory T cell properties. Immunity. 2009;31:296–308. doi: 10.1016/j.immuni.2009.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mackay LK, Minnich M, Kragten NA, Liao Y, Nota B, Seillet C, Zaid A, Man K, Preston S, Freestone D, et al. Hobit and Blimp1 instruct a universal transcriptional program of tissue residency in lymphocytes. Science. 2016;352:459–463. doi: 10.1126/science.aad2035. [DOI] [PubMed] [Google Scholar]
- 57.Kim MV, Ouyang W, Liao W, Zhang MQ, Li MO. The transcription factor FOXO1 controls central-memory CD8+ T cell responses to infection. Immunity. 2013;39:286–297. doi: 10.1016/j.immuni.2013.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hess Michelini R, Doedens AL, Goldrath AW, Hedrick SM. Differentiation of CD8 memory T cells depends on FOXO1. J Exp Med. 2013;210:1189–1200. doi: 10.1084/jem.20130392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Croce O, Chevenet F, Christen R. OligoHeatMap (OHM): an online tool to estimate and display hybridizations of oligonucleotides onto DNA sequences. Nucleic Acids Res. 2008;36:W154–W156. doi: 10.1093/nar/gkn221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029–6033. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- 63.Smigielska-Czepiel K, van den Berg A, Jellema P, Slezak-Prochazka I, Maat H, van den Bos H, van der Lei RJ, Kluiver J, Brouwer E, Boots AM, et al. Dual role of miR-21 in CD4+ T-cells: activation-induced miR-21 supports survival of memory T-cells and regulates CCR7 expression in naive T-cells. PLoS One. 2013;8:e76217. doi: 10.1371/journal.pone.0076217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kim SJ, Shin JY, Lee KD, Bae YK, Sung KW, Nam SJ, Chun KH. MicroRNA let-7a suppresses breast cancer cell migration and invasion through downregulation of C-C chemokine receptor type 7. Breast Cancer Res. 2012;14:R14. doi: 10.1186/bcr3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xu Z, Han X, Tang Z, Tian G, Gao J, Xu X. Interaction between MALAT-1, CCR7 and correlated genes in oral squamous cell carcinoma. Int J Clin Exp Pathol. 2017;10:10730–10739. [PMC free article] [PubMed] [Google Scholar]
- 66.Feng C, So HI, Yin S, Su X, Xu Q, Wang S, Duan W, Zhang E, Sun C, Xu Z. MicroRNA-532-3p suppresses malignant behaviors of tongue squamous cell carcinoma via regulating CCR7. Front Pharmacol. 2019;10:940. doi: 10.3389/fphar.2019.00940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kim C, Hu B, Jadhav RR, Jin J, Zhang H, Cavanagh MM, Akondy RS, Ahmed R, Weyand CM, Goronzy JJ. Activation of miR-21-regulated pathways in immune aging selects against signatures characteristic of memory T cells. Cell Rep. 2018;25:2148–2162.e5. doi: 10.1016/j.celrep.2018.10.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li W, Xue W, Wang X, Fu X, Sun Z, Li Z, Chang Y, Zhang X, Zhou Z, Chen C, et al. MiR-199a mediated the dissemination of human mantle cell lymphoma by interacting with the CCR7/CCL21 pair. Anticancer Drugs. 2018;29:861–870. doi: 10.1097/CAD.0000000000000656. [DOI] [PubMed] [Google Scholar]
- 69.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 70.Moran TP, Nakano H, Kondilis-Mangum HD, Wade PA, Cook DN. Epigenetic control of Ccr7 expression in distinct lineages of lung dendritic cells. J Immunol. 2014;193:4904–4913. doi: 10.4049/jimmunol.1401104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hauser MA, Kindinger I, Laufer JM, Späte AK, Bucher D, Vanes SL, Krueger WA, Wittmann V, Legler DF. Distinct CCR7 glycosylation pattern shapes receptor signaling and endocytosis to modulate chemotactic responses. J Leukoc Biol. 2016;99:993–1007. doi: 10.1189/jlb.2VMA0915-432RR. [DOI] [PubMed] [Google Scholar]
- 72.Phillips AJ, Taleski D, Koplinski CA, Getschman AE, Moussouras NA, Richard AM, Peterson FC, Dwinell MB, Volkman BF, Payne RJ, et al. CCR7 sulfotyrosine enhances CCL21 binding. Int J Mol Sci. 2017;18:E1857. doi: 10.3390/ijms18091857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lo JC, Chin RK, Lee Y, Kang HS, Wang Y, Weinstock JV, Banks T, Ware CF, Franzoso G, Fu YX. Differential regulation of CCL21 in lymphoid/nonlymphoid tissues for effectively attracting T cells to peripheral tissues. J Clin Invest. 2003;112:1495–1505. doi: 10.1172/JCI19188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kim CH, Pelus LM, White JR, Applebaum E, Johanson K, Broxmeyer HE. CK β-11/macrophage inflammatory protein-3 β/EBI1-ligand chemokine is an efficacious chemoattractant for T and B cells. J Immunol. 1998;160:2418–2424. [PubMed] [Google Scholar]
- 75.Ngo VN, Tang HL, Cyster JG. Epstein-Barr virus-induced molecule 1 ligand chemokine is expressed by dendritic cells in lymphoid tissues and strongly attracts naive T cells and activated B cells. J Exp Med. 1998;188:181–191. doi: 10.1084/jem.188.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yoshida R, Nagira M, Kitaura M, Imagawa N, Imai T, Yoshie O. Secondary lymphoid-tissue chemokine is a functional ligand for the CC chemokine receptor CCR7. J Biol Chem. 1998;273:7118–7122. doi: 10.1074/jbc.273.12.7118. [DOI] [PubMed] [Google Scholar]
- 77.Gunn MD, Tangemann K, Tam C, Cyster JG, Rosen SD, Williams LT. A chemokine expressed in lymphoid high endothelial venules promotes the adhesion and chemotaxis of naive T lymphocytes. Proc Natl Acad Sci U S A. 1998;95:258–263. doi: 10.1073/pnas.95.1.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Luther SA, Tang HL, Hyman PL, Farr AG, Cyster JG. Coexpression of the chemokines ELC and SLC by T zone stromal cells and deletion of the ELC gene in the plt/plt mouse. Proc Natl Acad Sci U S A. 2000;97:12694–12699. doi: 10.1073/pnas.97.23.12694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Malhotra D, Fletcher AL, Astarita J, Lukacs-Kornek V, Tayalia P, Gonzalez SF, Elpek KG, Chang SK, Knoblich K, Hemler ME, et al. Transcriptional profiling of stroma from inflamed and resting lymph nodes defines immunological hallmarks. Nat Immunol. 2012;13:499–510. doi: 10.1038/ni.2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Okada T, Cyster JG. CC chemokine receptor 7 contributes to Gi-dependent T cell motility in the lymph node. J Immunol. 2007;178:2973–2978. doi: 10.4049/jimmunol.178.5.2973. [DOI] [PubMed] [Google Scholar]
- 81.Worbs T, Mempel TR, Bölter J, von Andrian UH, Förster R. CCR7 ligands stimulate the intranodal motility of T lymphocytes in vivo . J Exp Med. 2007;204:489–495. doi: 10.1084/jem.20061706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mueller SN, Hosiawa-Meagher KA, Konieczny BT, Sullivan BM, Bachmann MF, Locksley RM, Ahmed R, Matloubian M. Regulation of homeostatic chemokine expression and cell trafficking during immune responses. Science. 2007;317:670–674. doi: 10.1126/science.1144830. [DOI] [PubMed] [Google Scholar]
- 83.Shields JD, Emmett MS, Dunn DB, Joory KD, Sage LM, Rigby H, Mortimer PS, Orlando A, Levick JR, Bates DO. Chemokine-mediated migration of melanoma cells towards lymphatics--a mechanism contributing to metastasis. Oncogene. 2007;26:2997–3005. doi: 10.1038/sj.onc.1210114. [DOI] [PubMed] [Google Scholar]
- 84.Mashino K, Sadanaga N, Yamaguchi H, Tanaka F, Ohta M, Shibuta K, Inoue H, Mori M. Expression of chemokine receptor CCR7 is associated with lymph node metastasis of gastric carcinoma. Cancer Res. 2002;62:2937–2941. [PubMed] [Google Scholar]
- 85.Müller A, Homey B, Soto H, Ge N, Catron D, Buchanan ME, McClanahan T, Murphy E, Yuan W, Wagner SN, et al. Involvement of chemokine receptors in breast cancer metastasis. Nature. 2001;410:50–56. doi: 10.1038/35065016. [DOI] [PubMed] [Google Scholar]
- 86.Xu B, Zhou M, Qiu W, Ye J, Feng Q. CCR7 mediates human breast cancer cell invasion, migration by inducing epithelial-mesenchymal transition and suppressing apoptosis through AKT pathway. Cancer Med. 2017;6:1062–1071. doi: 10.1002/cam4.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yeon S, Choi A, Hong MS, Jung YW. Mediators of the homeostasis and effector functions of memory Th2 cells as novel drug targets in intractable chronic allergic diseases. Arch Pharm Res. 2019;42:754–765. doi: 10.1007/s12272-019-01159-4. [DOI] [PubMed] [Google Scholar]
- 88.Burkett PR, Koka R, Chien M, Chai S, Boone DL, Ma A. Coordinate expression and trans presentation of interleukin (IL)-15Rα and IL-15 supports natural killer cell and memory CD8+ T cell homeostasis. J Exp Med. 2004;200:825–834. doi: 10.1084/jem.20041389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Schluns KS, Lefrançois L. Cytokine control of memory T-cell development and survival. Nat Rev Immunol. 2003;3:269–279. doi: 10.1038/nri1052. [DOI] [PubMed] [Google Scholar]
- 90.Jung YW, Kim HG, Perry CJ, Kaech SM. CCR7 expression alters memory CD8 T-cell homeostasis by regulating occupancy in IL-7- and IL-15-dependent niches. Proc Natl Acad Sci U S A. 2016;113:8278–8283. doi: 10.1073/pnas.1602899113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Junt T, Nakano H, Dumrese T, Kakiuchi T, Odermatt B, Zinkernagel RM, Hengartner H, Ludewig B. Antiviral immune responses in the absence of organized lymphoid T cell zones in plt/plt mice. J Immunol. 2002;168:6032–6040. doi: 10.4049/jimmunol.168.12.6032. [DOI] [PubMed] [Google Scholar]
- 92.Junt T, Scandella E, Förster R, Krebs P, Krautwald S, Lipp M, Hengartner H, Ludewig B. Impact of CCR7 on priming and distribution of antiviral effector and memory CTL. J Immunol. 2004;173:6684–6693. doi: 10.4049/jimmunol.173.11.6684. [DOI] [PubMed] [Google Scholar]
- 93.Chen SC, Vassileva G, Kinsley D, Holzmann S, Manfra D, Wiekowski MT, Romani N, Lira SA. Ectopic expression of the murine chemokines CCL21a and CCL21b induces the formation of lymph node-like structures in pancreas, but not skin, of transgenic mice. J Immunol. 2002;168:1001–1008. doi: 10.4049/jimmunol.168.3.1001. [DOI] [PubMed] [Google Scholar]
- 94.Unsoeld H, Mueller K, Schleicher U, Bogdan C, Zwirner J, Voehringer D, Pircher H. Abrogation of CCL21 chemokine function by transgenic over-expression impairs T cell immunity to local infections. Int Immunol. 2007;19:1281–1289. doi: 10.1093/intimm/dxm098. [DOI] [PubMed] [Google Scholar]
- 95.Noor S, Habashy AS, Nance JP, Clark RT, Nemati K, Carson MJ, Wilson EH. CCR7-dependent immunity during acute Toxoplasma gondii infection. Infect Immun. 2010;78:2257–2263. doi: 10.1128/IAI.01314-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Porter DL, Levine BL, Kalos M, Bagg A, June CH. Chimeric antigen receptor-modified T cells in chronic lymphoid leukemia. N Engl J Med. 2011;365:725–733. doi: 10.1056/NEJMoa1103849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Maude SL, Frey N, Shaw PA, Aplenc R, Barrett DM, Bunin NJ, Chew A, Gonzalez VE, Zheng Z, Lacey SF, et al. Chimeric antigen receptor T cells for sustained remissions in leukemia. N Engl J Med. 2014;371:1507–1517. doi: 10.1056/NEJMoa1407222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kong W, Lacey SF, Melenhorst JJ, Fraietta JA. Biomarkers in chimeric antigen receptor T-cell therapy. Biomark Med. 2018;12:415–418. doi: 10.2217/bmm-2018-0054. [DOI] [PubMed] [Google Scholar]
- 99.Channappanavar R, Fett C, Zhao J, Meyerholz DK, Perlman S. Virus-specific memory CD8 T cells provide substantial protection from lethal severe acute respiratory syndrome coronavirus infection. J Virol. 2014;88:11034–11044. doi: 10.1128/JVI.01505-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ng OW, Chia A, Tan AT, Jadi RS, Leong HN, Bertoletti A, Tan YJ. Memory T cell responses targeting the SARS coronavirus persist up to 11 years post-infection. Vaccine. 2016;34:2008–2014. doi: 10.1016/j.vaccine.2016.02.063. [DOI] [PMC free article] [PubMed] [Google Scholar]