Abstract
TLRs are pattern recognition receptors (PRRs) whose cytoplasmic signalling domain is similar to that of IL-1. The extracellular domain of TLRs serve as the binding site of pathogen associated molecular patterns. TLRs are found on both plasma and endosomal membranes and they mainly exert their function by activating genes which lead to production of inflammatory factors. The latest TLR to be discovered, TLR10 is a unique TLR which exhibit anti-inflammatory properties. TLR10 is found on the plasma membrane with other TLRs namely TLR1, TLR2, TLR4, TLR5 and TLR6. Studies have revealed that TLR10 is found on the same gene cluster with TLR1 and TLR6 and is also a coreceptor of TLR2. Up to date, TLR10 is the only TLR which exhibit anti-inflammatory property. Previously, TLR10 was thought to be an “orphan receptor” but much recent studies have identified ligands for TLR10. Currently there is no review article on TLR10 that has been published. In this narrative review, we are going to give an account of TLR10, its functions mainly as an anti-inflammatory PRR and its possible applications as a target in therapeutics.
Keywords: Toll-like receptor 10, Anti-inflammatory, TLR10 ligands, Polymorphism
INTRODUCTION
TLRs are the most studied pattern recognition receptors (PRRs), which are characterized by at least 3 domains namely the cytoplasmic (for signalling purpose), the transmembrane and the extracellular domain (microbial pattern recognition) (1,2,3). TLRs are called toll/IL-1R domain because they have similar cytoplasmic domain even though their extracellular domains are different. The extracellular domain of TLRs is made up of leucine rich repeats (LRRs) whilst that of IL-IR is like an immunoglobin (3,4). The main function of TLRs in innate immunity is to recognize pathogen associated molecular patterns (PAMPs) and danger associated molecular patterns (DAMPs) (5). Once PAMPs or DAMPs have been recognized by TLRs, TLRs respond by recruiting phagocytes to the site of infection, expression of mainly inflammatory mediators like chemokines and cytokines which will subsequently result in microbial killing (6,7).
Up to date, ten members of TLRs have been identified in humans and they are TLR1 through TLR10, with TLR10 being the latest to be discovered (1,8,9). There is differential distribution of TLRs within a cell and they are further grouped based on their cellular location. TLRs on plasma membranes are TLR1, TLR2, TLR4, TLR5, TLR6 and TLR10 whereas those on endosomal membranes are TLR3, TLR7, TLR8 and TLR9 (9,10). Each TLR recognizes specific PAMPs from microbes/pathogens. The PAMPs recognized by TLRs can be from viruses, bacteria, fungi and protozoa (1,7,10). Endosomal TLRs mainly recognize nucleic components of pathogens with TLR3 sensing double-strand RNA, TLR7 and TLR8 sensing single-stranded RNA, and TLR9 sensing CpG DNA (1,7,9,10,11). Extracellular TLRs sense different types of PAMPs with TLR4 sensing quite a number of them, including lipopolysaccharides and taxol, and TLR5 recognizes flagellin (7,8). TLRs play vital roles in both inflammatory and infectious diseases (12).
When a PAMP has been sensed by a TLR this result in homodimerization or heterodimerization of the TLR. TLRs can be either myeloid differentiation primary response 88 (MyD88) dependent or independent (4,5). All TLRs function by recruiting MyD88 with the exception of TLR3 which functions by recruiting TIR domain containing activator of TLR (TRIF) in a TRIF-dependent signalling pathway. Besides TLR3, TLR4 can also recruit TRIF through the use of an additional adaptor, TRIF-related adaptor molecule. The MyD88 signalling pathway is initiated by recruiting IL-1R associated kinase proteins such as TNF receptor-associated factor 6 and TGF-β activated kinase 1 which will in turn result in the activation of NF-κB which will as a result turn transcription of inflammatory cytokine genes (1,2,3,4,6,7,8,13).
TLR10
Unlike other TLRs with well-defined function and mode of action, TLR10 is considered to be an orphan receptor with ligands and functions which are not well understood (14). The biology and ligand specificity of TLR10 remains an area that need to be demystified (15).
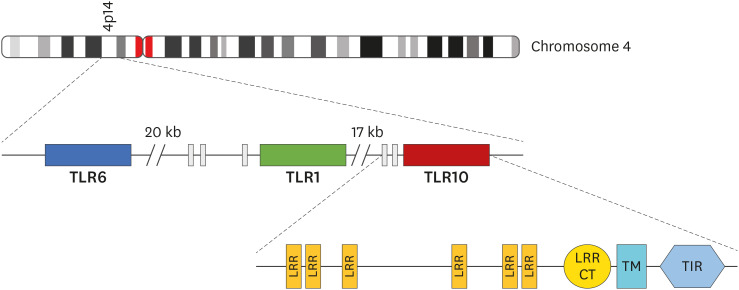
The gene that code for TLR10 is located on the same gene cluster with TLR1, and TLR6 (15,16,17,18) and its on chromosome 4p14 (16,17,19) as shown in Fig. 1. According to Chuang and Ulevitch (20), the TLR10 gene encodes 811 amino acids with a molecular weight of about 94.6 kDa. TLR10 gene can be found in other mammals like sheep, pigs, horses, rats and cattle, however mice have a pseudogene (21). Phylogenetic analysis indicated that TLR10 is more similar to TLR1 and TLR6 with an overall amino acid identity of 50% and 49% respectively (20). Even though TLR10 is homologous to TLR1 and TLR6, Shinkai et al. (22) stated that there are differences in expression patterns and signal transduction pathways in TLR10. Like other TLRs, TLR10 is highly expressed on immune cells such as eosinophils (23), dendritic cells (DCs), neutrophils, and B cells, however, its expression is not limited to cells of the immune since it can be found in non-immune cells such as trophoblasts (14,15). Unlike TLR1 and TLR6, TLR10 is expressed in a highly restricted fashion as a highly N-glycosylated protein, which was detected in B cell lines, B cells from peripheral blood, and plasmacytoid DCs from tonsils (18). Activated B cells express more TLR10 compared to resting B cells (24). Cells which express TLR10 are found in organs and tissues, lymphoid organs such as the spleen, lymph node (25) and some mucosal sites, including the small intestine, stomach, fallopian tubes and eye (14).
Figure 1. Schematic presentation of TLR10 gene. TLR10 is located on 4p14 of chromosome 4 on the same gene structure with TLR1 and TLR6. Like other TLRs, TLR10 gene also encodes LRR, TM, and TIR which are a hallmark of TLRs.
TM, transmembrane, CT, C-terminal.
When compared to other TLRs, TLR10 is not highly expressed in tissues of the lymphoid system. Surprisingly, TLR10 is also expressed on Tregs where its expression is controlled by forkhead box P3 (FOXP3), speculating that TLR10 may have a different function with other TLRs (17,26). According to Bell et al. (26), primary human T cells and a reporter assay in Jurkat T cell lines were used to dissect the regulation of TLR10, a TLR highly expressed in human Treg cells. The expression of TLR10 in human Treg cells was determined through quantitative PCR, Western blotting, and flow cytometry. DNA binding of FOXP3 to a suspected cis-regulatory region in proximity to the transcription start site of TLR10 was established through EMSA and chromatin immunoprecipitation. Transcriptional control of TLR10 by FOXP3 was determined through luciferase reporter assays in Jurkat T cell lines. Relevance of FOXP3 to TLR10 gene transcription in primary T cells was established through the transfection of primary CD4CD25FOXP3T cells with a FOXP3 expression vector, which resulted in prompt production of TLR10 mRNA. The calcium-dependent fashion was used to enhance the expression of TLR10 protein in primary Treg cells through TCR activation. The abolition of the luciferase signal upon transfection of a mutant FOXP3 devoid of NF-AT-binding activity was suspected to establish the promotional between FOXP3 and NF-AT. The findings of the study showed that human Treg cells express TLR10, and this expression is regulated through a cooperative complex of FOXP3 and NF-AT (26).
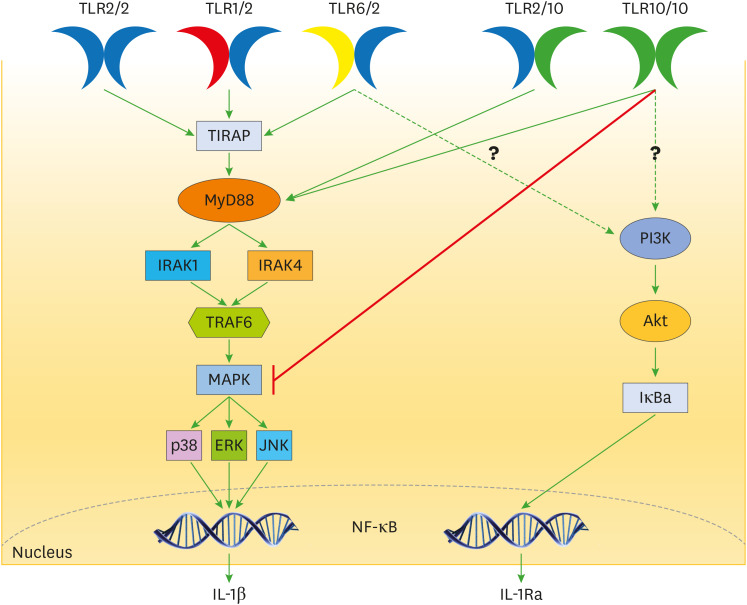
TLR10 can homodimerize as well as heterodimerize with TLR1, TLR2, and TLR6 (16,18,27,28), but the function of the individual TLR in the resultant complex remains to be elucidated (16). The homodimer (TLR10/TLR10) and heterodimer (TLR10/TLR2) can recruit MyD88 (Fig. 2), however they all failed to activate a TLR-induced signalling such as the activation of NF-κB (21).
Figure 2. Schematic signalling of TLRs via MyD88 dependent pathway. Both the homodimer of TLR10 and the heterodimer of TLR10/TLR2 can recruit the adaptor protein MyD88 and the downward signalling pathway remains an area to be elucidated. The homodimer and heterodimer of TLR10 can reduce production of IL-1β by directly inhibiting MyD88 or MAPK. TLR10 has also been shown to increase production of IL-1Ra (with anti-inflammatory) (15,29) but the mechanism still remains a mystery as shown by the question marks.
IRAK, IL-1R associated kinase, TRAF6, TNF receptor-associated factor 6.
BIOLOGICAL FUNCTIONS OF TLR10
Since the discovery of TLR10 by Chuang and Ulevitch (20,29), a lot of research in trying to find the biological significant of TLR10 has been done (30). Interestingly, discordant results were found based on TLR10 biological function. Some studies showed TLR10 to be an inflammatory PRR which can activate NF-κB, whilst others have shown TLR10 to be an anti-inflammatory PRR.
In an attempt to find the biological function of TLR10, Oosting et al. (15) used anti-TLR10 Abs, and they observed an increase in the production of inflammatory cytokines IL-1β, IL-6 and the chemoattractant, IL-8 and to a lesser significant TNF-α, when compared to the control without anti-TLR10 Abs. To find how TLR10 was exerting its suppressive effect, they cultured human embryonic kidney (HEK) in the presence TLR2, Pam3Cys (TLR2 ligand) with either TLR1 or TLR6 or TLR10. There was a reduction in the production of IL-8 in HEK cells with TLR10. To confirm that TLR10 functions by inhibiting TLR2, HEK cells were cultured with TLR5, flagellin (TLR5 ligand) and TLR10 and they found no change in the production of IL-8 (15). Another study which was carried out in a cohort of Spain rheumatoid arthritis (RA) patients was able to show the inhibitory effect of TLR10 on NF-κB transcription (16), and the details are in polymorphism section. Vitamin D active metabolite, which is a well-studied promoter of anti-inflammatory cytokines (31) was able to upregulate the expression of TLR10 in both monocytes (17) and microglial cells (32). When microglial cells were used, an upregulation of TLR10 expression was observed and this was accompanied by an increase in M2 cytokines such as IL-10 and a decrease in M1 cytokines such as TNF-α and IL-12 (32).
Another study supported the role of TLR10 as an inhibitor of MyD88 dependent and independent pathways. In their experiment Hess et al. (30) used monoclonal Abs (activator of TLR10) to TLR10 and they observed a reduction in the production of pro-inflammatory cytokines, IL-6 and TNF-α. Even though the signalling method involved is not fully elucidated, they explained that there was inhibition of both IκB degradation and phosphorylation of MAPK and PI3K/protein kinase B (Akt). They also showed the involvement of TLR10 in differentiation of DCs. TLR10 was shown to reduce the expression of costimulatory receptors on DCs and it also affected DC maturation and their ability to activate T cells (30). To investigate the downstream signalling of TLR10 as a homodimer or as a heterodimer with TLR2, Pachathundikandi and Backert (33) used HEK293 to investigate the expression of mRNA of the genes encoding pro-inflammatory cytokines and they observed that TLR2/HEK293 significantly upregulated IL-1β but not control cells or TLR10/HEK293.
The ability of the immune system to enhance a long-term immune response and resistance to heterologous infections is termed trained immunity. The role of TLR10 in trained immunity has also been studied. TLR10 can induce trained immunity in vitro as recently observed by Mourits et al. (29) who used β-glucan and BCG. In addition, these researchers also observed the role of TLR10 as an anti-inflammatory cytokine via ex vivo increased production of IL-1Ra preceded by engagement and crosslinking of TLR10 (29).
B cells are one of the immune cells that express TLR10 and the expression of TLR10 in B cells amongst RA patients in Chinese population was also studied. In this study the researchers found that both the health controls and RA subjects expressed TLR10 on CD19+ B cells, however the percentage of TLR10 expression increased with severity of disease. In this study the authors found TLR10 to be an inflammatory PRR after they observed higher levels of IL-1β in RA subjects (34). RA disease is a hyper-inflammatory one, hence it is expected that the subjects were having high levels of inflammatory cytokines, and in regard to this we strongly suggest an independent research without intermitted variables like RA, that will use the B cell subset that the authors used and do an in vitro analysis that will involve the use of inhibitors to the TLR10 and compare with one without inhibitors. Another study which used NC1-N87 gastric cells co-cultured with Helicobacter pylori concluded that, the heterodimer of TLR2 and TLR10 upregulated the activation of NF-κB to a larger extent when compared to other TLR2 heterodimers (35). In another research, silencing of TLR10 resulted in increased viability of Listeria monocytogenes, and the authors found out that the heterodimer TLR2/TLR10 was the one responsible for the activation of NF-κB (36).
The differences in the biological functions of TLR10 observed in several studies could be as a result of the complexity of TLR10 mechanism of action which include competition for the formation of TLR2/TLR10 heterodimer, competition for the ligands and induction of PI3K/Akt (16). TLR10, unlike other TLRs is also expressed on Treg cells speculating a different function. To fully understand the function of TLR10 researchers need to work on individual cells expressing TLR10. The heterodimerization of TLR10 with other TLRs such as TLR2 results in a complex where the function of each TLR in the complex is not known. Understanding the role played by each individual TLR in the complex may answer the variations observed different functions of TLR10.
LIGANDS FOR TLR10
Given the potential role of TLR10 as a receptor which induce anti-inflammatory activity, there is need to look for ligands specific for this TLR. Ligands which interact with TLR2 are also thought to be ligands for TLR10 and these include Pam3Cys and FSL-1. Borrelia burgdorferi, the causative agent of Lyme disease which is mainly sensed by TLR/TLR2 heterodimer is also likely to be a source of PAMPs for TLR10 (15). Using computational modelling, it has been suggested that, the homodimer of TLR10 senses diacylated lipopeptides (17,21). An artificially created chimeric receptor which was made by combining the extracellular domain of TLR10 and the intracellular/cytoplasmic domain of TLR1 showed that TLR2/TLR10 heterodimer recognizes several PAMPs which are recognized by TLR2/TLR1 heterodimer (17). Lipopolysaccharides were also identified as potential ligands for the TLR2/TLR10 heterodimer in H. pylori infection. This finding is of interest, because lipopolysaccharides of H. pylori do not vigorously cause a TLR4 response as expected (35). Regan et al. (36) pointed out that L. monocytogenes is a source of a TLR10 PAMP since it was able to cause the activation NF-κB through the TLR2/TLR10 heterodimer. Recently, HIV-gp41 has been identified as a ligand for TLR10 (14).
POLYMORPHISM IN TLR10 AND RELATED DISEASES
Genetic variations found in the TLR10 gene may cause a shift in the levels of pro- and anti-inflammatory responses and hence, enhance the susceptibility to autoimmune diseases, cancers and infections. In a cohort that involved Spain RA patients, polymorphism was observed at position 437 within the β-strand of TLR10 which is part of the LRR domain. This polymorphism resulted in the change of the hydrophobic amino acid isoleucine to the polar amino acid threonine. The other members of TLR10 family have hydrophobic amino acids at position 437 as well, and these are isoleucine and valine for TLR2 and TLR1 respectively (16). The change from a non-polar amino acid to a polar amino acid may cause the resultant protein not to perform its function properly. The functional consequences of the I437T variant was tested by generating the I437T variant via site-directed mutagenesis. K562 cell lines were co-transfected with either the wild type or the mutant TLR10 and their results showed that the wild type TLR10 inhibited the transcription of NF-κB after K562 cells were stimulated with TNF-α (16).
The non-synonymous single nucleotide polymorphism (SNP), rs11096957, which was found in a Han Chinese population of hip osteoarthritis, was found to be significantly correlated to increased risk of hip osteoarthritis (37). In regards to Crohn's disease, 4 SNPs were found to be associated with susceptibility to the disease and these are, rs7653908 (38), rs7658893 (12,38), rs6941698, rs10024216, and rs4274855 (12). In these 2 studies discordant results were observed, Morgan et al. (12) was able to find an association in 2 SNPs which were considered to have no association by Abad et al., (38) and one SNP, where Abad et al. (38) was able to find an association, Morgan et al. (12) did not find an association. Despite their discordant results which might need another research in a different population, it is relatively clear that a variant in TLR10 gene may increase susceptibility to Crohn's disease. No TLR10 SNPs were found to be associated with aspergillosis in a study that was carried out by Smith et al. (39) in a selected Caucasian population, however their results showed that TLR10 was lowly expressed in monocyte-derived-macrophages of aspergillosis subjects as compared to the healthy ones. Their findings do not rule out the effect of TLR10 variant in aspergillosis, but mark the beginning of an area that need to be studied. The TLR10 rs10004195 SNP was found to increase susceptibility to H. pylori (40). In addition, abnormalities in TLR10 (SNP rs10004195) were also found to be positively correlated with Hashimoto's disease (41).
A study which was carried out in Korea found a SNP which was more common in papillary-thyroid-carcinoma (PTC) patients as compared to those without the disease. The rs11466653 SNP which was found at position 326 resulted in a change of amino acid from methionine to threonine hence Met326Thr. The frequency of this SNP in the diseased subjects was 87.2% indicating that this SNP might increase the risk of developing PTC (42). Purdue et al. (43) also linked variations in the TLR10 gene to increased risk of cancer but in this case to Non-Hodgkin lymphoma (NHL). A reduced risk to prostate cancer was observed as a result of TLR10 rs11096955 (Ile369Leu) and TLR10 rs11096957 (Asn241His) (44). Another disease which has been associated with TLR10 polymorphism is IgA Nephropathy. This was carried out in Korean population and they concluded that the rs1004195 SNP may have increased susceptibility to IgA Nephropathy (25). Other TLR10 polymorphisms which have been associated with disease severity and or increased risk include 720A/C and 992/TA in Crimean-Congo haemorrhagic fever (CCHF) (45), rs10004195 in H. pylori (46), variants increasing susceptibility to tuberculosis (47) and Niemann type C (48).
TLR10 AS A TARGET IN THERAPEUTICS AND DIAGNOSTICS
Several studies have proven the relationship between genetic variation in TLR10 with the risk of developing autoimmune diseases such as RA, infectious diseases such as tuberculosis, inflammatory diseases such as asthma as well as cancers amongst others (34). Other diseases which have been associated with polymorphism to TLR10 include Crohn's disease, influenza, HIV (14,28) and H. pylori infections (33), aspergillosis, allogenic stem cell transplantation, thyroid disease, NHL, bladder and nasopharyngeal carcinomas (39,42,43), and IgA nephropathy (25).
In brain pathology, M1 macrophages are considered as neurotoxic whereas M2 macrophages are neuroprotective. Microglial cells express TLR10 and this receptor was shown to inhibit MI cytokines but promote M2 cytokines indicating that TLR10 may have a protective role in the brain. Compounds or metabolites that promote the differentiation of M2 macrophages in the brain can be used in brain therapeutics (32). RA is an autoimmune disease which is characterised by hyper-inflammation. TLR10 may be a good immunotherapy to patients with diseases such as RA due to its function in inhibiting inflammatory cytokines. Inhibition of NF-B by TLR10 will limit the activation of inflammatory genes which when properly regulated will result in autoimmune disorders (16). In a study carried out by Zhang et al., (34) they observed that the percentage of TLR10 expressed on CD19+B cell subsets increased with disease severity, suggesting TLR10 to be a possible marker for disease severity in RA. In a Turkish population, the TLR10 720/C and 992TA polymorphism was found to be positively correlated with disease severity and the authors concluded that TLR 10 polymorphisms may also be an important biomarker for CCHF susceptibility and fatality rate (45). TLR10 gene was found to have an impact on adipose tissue morphology in obesity. The authors recommended further studies in humans to assess its potential value as therapeutic target in metabolic syndrome and type 2 diabetes (49).
CONCLUSION
TLR10 is an endosomal TLRs whose biological functions in humans is yet to be fully elucidated. To exert its function, TLR10 can homodimerize or heterodimerize with TLR2. Unlike other TLRs which are highly expressed in immune cells of the innate immune system, TLR10 is also expressed on Treg cells indicating a different biological function to other TLRs. When TLR10 heterodimerize with TLR2, most ligands that are recognized by TLR1/TLR2 heterodimer are also recognized by TLR10 heterodimer. HIV-gp41 has been newly identified as a ligand for TLR10. Both the homodimer and heterodimer can recruit MyD88, however they all fail to activate the downstream signalling pathway. Quite a number of studies have proven the role of TLR10 and as an inhibitor of inflammatory cytokines even though others have shown it exert inflammatory properties. The differences in the biological functions of TLR10 observed in several studies could be as a result of the complexity of TLR1o mechanism of action which include competition for the formation of TLR2/TLR10 heterodimer, competition for the ligands and induction of PI3K/Akt. SNP in TLR10 gene has been associated with increased susceptibility to diseases such as RA, hip osteoarthritis, thyroid carcinoma and increased resistance to prostate cancer.
Understanding the complex mechanism employed by TLR10 to exert its function will be beneficial to the field of health as this will give a starting point in therapeutics and diagnostics of diseases which can be directly affected by polymorphisms in TLR10. There is a lot of research that should be done to understand the signalling of TLR10. We recommend the use of rat as an animal model because a complete TLR10 gene was sequenced in rats.
Abbreviations
- Akt
protein kinase B
- CCHF
Crimean-Congo haemorrhagic fever
- DAMP
danger associated molecular pattern
- DC
dendritic cell
- FOXP3
forkhead box P3
- HEK
human embryonic kidney
- LRR
leucine rich repeat
- MyD88
myeloid differentiation primary response 88
- NHL
Non-Hodgkin lymphoma
- PAMP
pathogen associated molecular pattern
- PRR
pattern recognition receptor
- PTC
papillary-thyroid-carcinoma
- RA
rheumatoid arthritis
- SNP
single nucleotide polymorphism
- TIR
toll-interleukin 1 receptor
- TRIF
TIR domain containing activator of TLR
Footnotes
Conflict of Interest: The authors declare no potential conflicts of interest.
- Conceptualization: Fore F.
- Formal analysis: Fore F.
- Methodology: Fore F.
- Supervision: Nugraha J.
- Writing - original draft: Fore F, Indriputri C, Mamutse J.
- Writing - review & editing: Fore F, Indriputri C, Nugraha J.
References
- 1.Kumar H, Kawai T, Akira S. Pathogen recognition by the innate immune system. Int Rev Immunol. 2011;30:16–34. doi: 10.3109/08830185.2010.529976. [DOI] [PubMed] [Google Scholar]
- 2.Moresco EM, LaVine D, Beutler B. Toll-like receptors. Curr Biol. 2011;21:R488–R493. doi: 10.1016/j.cub.2011.05.039. [DOI] [PubMed] [Google Scholar]
- 3.Akira S, Takeda K. Toll-like receptor signalling. Nat Rev Immunol. 2004;4:499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- 4.Akira S. Toll-like receptor signaling. J Biol Chem. 2003;278:38105–38108. doi: 10.1074/jbc.R300028200. [DOI] [PubMed] [Google Scholar]
- 5.Nie L, Cai SY, Shao JZ, Chen J, Chen J. Toll-like receptors, associated biological roles, and signaling networks in non-mammals. Front Immunol. 2018;9:1523. doi: 10.3389/fimmu.2018.01523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vidya MK, Kumar VG, Sejian V, Bagath M, Krishnan G, Bhatta R. Toll-like receptors: significance, ligands, signaling pathways, and functions in mammals. Int Rev Immunol. 2018;37:20–36. doi: 10.1080/08830185.2017.1380200. [DOI] [PubMed] [Google Scholar]
- 7.Kawasaki T, Kawai T. Toll-like receptor signaling pathways. Front Immunol. 2014;5:461. doi: 10.3389/fimmu.2014.00461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiménez-Dalmaroni MJ, Gerswhin ME, Adamopoulos IE. The critical role of toll-like receptors--from microbial recognition to autoimmunity: a comprehensive review. Autoimmun Rev. 2016;15:1–8. doi: 10.1016/j.autrev.2015.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kang JY, Lee JO. Structural biology of the toll-like receptor family. Annu Rev Biochem. 2011;80:917–941. doi: 10.1146/annurev-biochem-052909-141507. [DOI] [PubMed] [Google Scholar]
- 10.Takeda K, Akira S. Toll-like receptors in innate immunity. Int Immunol. 2005;17:1–14. doi: 10.1093/intimm/dxh186. [DOI] [PubMed] [Google Scholar]
- 11.Blasius AL, Beutler B. Intracellular toll-like receptors. Immunity. 2010;32:305–315. doi: 10.1016/j.immuni.2010.03.012. [DOI] [PubMed] [Google Scholar]
- 12.Morgan AR, Lam WJ, Han DY, Fraser AG, Ferguson LR. Genetic variation within TLR10 is associated with Crohn's disease in a New Zealand population. Hum Immunol. 2012;73:416–420. doi: 10.1016/j.humimm.2012.01.015. [DOI] [PubMed] [Google Scholar]
- 13.Lim KH, Staudt LM. Toll-like receptor signaling. Cold Spring Harb Perspect Biol. 2013;5:a011247. doi: 10.1101/cshperspect.a011247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Henrick BM, Yao XD, Zahoor MA, Abimiku A, Osawe S, Rosenthal KL. TLR10 senses HIV-1 proteins and significantly enhances HIV-1 infection. Front Immunol. 2019;10:482. doi: 10.3389/fimmu.2019.00482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oosting M, Cheng SC, Bolscher JM, Vestering-Stenger R, Plantinga TS, Verschueren IC, Arts P, Garritsen A, van Eenennaam H, Sturm P, et al. Human TLR10 is an anti-inflammatory pattern-recognition receptor. Proc Natl Acad Sci U S A. 2014;111:E4478–E4484. doi: 10.1073/pnas.1410293111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Torices S, Julia A, Muñoz P, Varela I, Balsa A, Marsal S, Fernández-Nebro A, Blanco F, López-Hoyos M, Martinez-Taboada V, et al. A functional variant of TLR10 modifies the activity of NFkB and may help predict a worse prognosis in patients with rheumatoid arthritis. Arthritis Res Ther. 2016;18:221. doi: 10.1186/s13075-016-1113-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Verma R, Jung JH, Kim JY. 1,25-Dihydroxyvitamin D3 up-regulates TLR10 while down-regulating TLR2, 4, and 5 in human monocyte THP-1. J Steroid Biochem Mol Biol. 2014;141:1–6. doi: 10.1016/j.jsbmb.2013.12.012. [DOI] [PubMed] [Google Scholar]
- 18.Hasan U, Chaffois C, Gaillard C, Saulnier V, Merck E, Tancredi S, Guiet C, Brière F, Vlach J, Lebecque S, et al. Human TLR10 is a functional receptor, expressed by B cells and plasmacytoid dendritic cells, which activates gene transcription through MyD88. J Immunol. 2005;174:2942–2950. doi: 10.4049/jimmunol.174.5.2942. [DOI] [PubMed] [Google Scholar]
- 19.Xiao W, Liu Z, Lin J, Li J, Wu K, Ma Y, Gong Y, Liu Z. Polymorphisms in TLR1, TLR6 and TLR10 genes and the risk of Graves' disease. Autoimmunity. 2015;48:13–18. doi: 10.3109/08916934.2014.939269. [DOI] [PubMed] [Google Scholar]
- 20.Chuang T, Ulevitch RJ. Identification of hTLR10: a novel human toll-like receptor preferentially expressed in immune cells. Biochim Biophys Acta. 2001;1518:157–161. doi: 10.1016/s0167-4781(00)00289-x. [DOI] [PubMed] [Google Scholar]
- 21.Tarlinton RE, Alder L, Moreton J, Maboni G, Emes RD, Tötemeyer S. RNA expression of TLR10 in normal equine tissues. BMC Res Notes. 2016;9:353. doi: 10.1186/s13104-016-2161-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shinkai H, Muneta Y, Suzuki K, Eguchi-Ogawa T, Awata T, Uenishi H. Porcine toll-like receptor 1, 6, and 10 genes: complete sequencing of genomic region and expression analysis. Mol Immunol. 2006;43:1474–1480. doi: 10.1016/j.molimm.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 23.Nagase H, Okugawa S, Ota Y, Yamaguchi M, Tomizawa H, Matsushima K, Ohta K, Yamamoto K, Hirai K. Expression and function of toll-like receptors in eosinophils: activation by toll-like receptor 7 ligand. J Immunol. 2003;171:3977–3982. doi: 10.4049/jimmunol.171.8.3977. [DOI] [PubMed] [Google Scholar]
- 24.Bourke E, Bosisio D, Golay J, Polentarutti N, Mantovani A. The toll-like receptor repertoire of human B lymphocytes: inducible and selective expression of TLR9 and TLR10 in normal and transformed cells. Blood. 2003;102:956–963. doi: 10.1182/blood-2002-11-3355. [DOI] [PubMed] [Google Scholar]
- 25.Park HJ, Hahn WH, Suh JS, Kim MJ, Kang SW, Lee JS, Kim JW, Chung JH, Cho BS. Association between toll-like receptor 10 (TLR10) gene polymorphisms and childhood IgA nephropathy. Eur J Pediatr. 2011;170:503–509. doi: 10.1007/s00431-010-1325-1. [DOI] [PubMed] [Google Scholar]
- 26.Bell MP, Svingen PA, Rahman MK, Xiong Y, Faubion WA., Jr FOXP3 regulates TLR10 expression in human T regulatory cells. J Immunol. 2007;179:1893–1900. doi: 10.4049/jimmunol.179.3.1893. [DOI] [PubMed] [Google Scholar]
- 27.Kim D, Kim YJ, Koh HS, Jang TY, Park HE, Kim JY. Reactive oxygen species enhance TLR10 expression in the human monocytic cell line THP-1. Int J Mol Sci. 2010;11:3769–3782. doi: 10.3390/ijms11103769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sindhu S, Akhter N, Kochumon S, Thomas R, Wilson A, Shenouda S, Tuomilehto J, Ahmad R. Increased expression of the innate immune receptor TLR10 in obesity and type-2 diabetes: association with ROS-mediated oxidative stress. Cell Physiol Biochem. 2018;45:572–590. doi: 10.1159/000487034. [DOI] [PubMed] [Google Scholar]
- 29.Mourits VP, Arts RJW, Novakovic B, Matzaraki V, de Bree LCJ, Koeken VACM, Moorlag SJCFM, van Puffelen JH, Groh L, van der Heijden CDCC, et al. The role of Toll-like receptor 10 in modulation of trained immunity. Immunology. 2020;159:289–297. doi: 10.1111/imm.13145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hess NJ, Felicelli C, Grage J, Tapping RI. TLR10 suppresses the activation and differentiation of monocytes with effects on DC-mediated adaptive immune responses. J Leukoc Biol. 2017;101:1245–1252. doi: 10.1189/jlb.3A1116-492R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiménez-Sousa MÁ, Martínez I, Medrano LM, Fernández-Rodríguez A, Resino S. Vitamin D in human immunodeficiency virus infection: influence on immunity and disease. Front Immunol. 2018;9:458. doi: 10.3389/fimmu.2018.00458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Verma R, Kim JY. 1,25-Dihydroxyvitamin D3 facilitates M2 polarization and upregulates TLR10 expression on human microglial cells. Neuroimmunomodulation. 2016;23:75–80. doi: 10.1159/000444300. [DOI] [PubMed] [Google Scholar]
- 33.Pachathundikandi SK, Backert S. Differential expression of interleukin 1β during Helicobacter pylori infection of toll-like receptor 2 (TLR2)- and TLR10-expressing HEK293 cell lines. J Infect Dis. 2016;214:166–167. doi: 10.1093/infdis/jiw154. [DOI] [PubMed] [Google Scholar]
- 34.Zhang Y, Cao R, Ying H, Du J, Chen S, Wang N, Shen B. Increased expression of TLR10 in B cell subsets correlates with disease activity in rheumatoid arthritis. Mediators Inflamm. 2018;2018:9372436. doi: 10.1155/2018/9372436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nagashima H, Iwatani S, Cruz M, Jiménez Abreu JA, Uchida T, Mahachai V, Vilaichone RK, Graham DY, Yamaoka Y. Toll-like receptor 10 in Helicobacter pylori infection. J Infect Dis. 2015;212:1666–1676. doi: 10.1093/infdis/jiv270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Regan T, Nally K, Carmody R, Houston A, Shanahan F, Macsharry J, Brint E. Identification of TLR10 as a key mediator of the inflammatory response to Listeria monocytogenes in intestinal epithelial cells and macrophages. J Immunol. 2013;191:6084–6092. doi: 10.4049/jimmunol.1203245. [DOI] [PubMed] [Google Scholar]
- 37.Tang H, Cheng Z, Ma W, Liu Y, Tong Z, Sun R, Liu H. TLR10 and NFKBIA contributed to the risk of hip osteoarthritis: systematic evaluation based on Han Chinese population. Sci Rep. 2018;8:10243. doi: 10.1038/s41598-018-28597-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abad C, González-Escribano MF, Diaz-Gallo LM, Lucena-Soto JM, Márquez JL, Leo E, Crivell C, Gómez-García M, Martín J, Núñez-Roldán A, et al. Association of toll-like receptor 10 and susceptibility to Crohn's disease independent of NOD2. Genes Immun. 2011;12:635–642. doi: 10.1038/gene.2011.41. [DOI] [PubMed] [Google Scholar]
- 39.Smith NL, Hankinson J, Simpson A, Denning DW, Bowyer P. Reduced expression of TLR3, TLR10 and TREM1 by human macrophages in chronic cavitary pulmonary aspergillosis, and novel associations of VEGFA, DENND1B and PLAT. Clin Microbiol Infect. 2014;20:O960–O968. doi: 10.1111/1469-0691.12643. [DOI] [PubMed] [Google Scholar]
- 40.Ravishankar Ram M, Goh KL, Leow AH, Poh BH, Loke MF, Harrison R, Shankar EM, Vadivelu J. Polymorphisms at locus 4p14 of toll-like receptors TLR-1 and TLR-10 confer susceptibility to gastric carcinoma in Helicobacter pylori infection. PLoS One. 2015;10:e0141865. doi: 10.1371/journal.pone.0141865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li M, Han W, Zhu L, Jiang J, Qu W, Zhang L, Jia L, Zhou Q. IRAK2 and TLR10 confer risk of Hashimoto's disease: a genetic association study based on the Han Chinese population. J Hum Genet. 2019;64:617–623. doi: 10.1038/s10038-019-0613-5. [DOI] [PubMed] [Google Scholar]
- 42.Kim SK, Park HJ, Hong IK, Chung JH, Eun YG. A missense polymorphism (rs11466653, Met326Thr) of toll-like receptor 10 (TLR10) is associated with tumor size of papillary thyroid carcinoma in the Korean population. Endocrine. 2013;43:161–169. doi: 10.1007/s12020-012-9783-z. [DOI] [PubMed] [Google Scholar]
- 43.Purdue MP, Lan Q, Wang SS, Kricker A, Menashe I, Zheng TZ, Hartge P, Grulich AE, Zhang Y, Morton LM, et al. A pooled investigation of toll-like receptor gene variants and risk of non-Hodgkin lymphoma. Carcinogenesis. 2009;30:275–281. doi: 10.1093/carcin/bgn262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stevens VL, Hsing AW, Talbot JT, Zheng SL, Sun J, Chen J, Thun MJ, Xu J, Calle EE, Rodriguez C. Genetic variation in the toll-like receptor gene cluster (TLR10-TLR1-TLR6) and prostate cancer risk. Int J Cancer. 2008;123:2644–2650. doi: 10.1002/ijc.23826. [DOI] [PubMed] [Google Scholar]
- 45.Kızıldağ S, Arslan S, Özbilüm N, Engin A, Bakır M. Effect of TLR10 (2322A/G, 720A/C, and 992T/A) polymorphisms on the pathogenesis of Crimean Congo hemorrhagic fever disease. J Med Virol. 2018;90:19–25. doi: 10.1002/jmv.24924. [DOI] [PubMed] [Google Scholar]
- 46.Tongtawee T, Bartpho T, Kaewpitoon S, Kaewpitoon N, Dechsukhum C, Leeanansaksiri W, Loyd RA, Talabnin K, Matrakool L, Panpimanmas S. Genetic polymorphisms in TLR1, TLR2, TLR4, and TLR10 of Helicobacter pylori-associated gastritis: a prospective cross-sectional study in Thailand. Eur J Cancer Prev. 2018;27:118–123. doi: 10.1097/CEJ.0000000000000347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ma X, Liu Y, Gowen BB, Graviss EA, Clark AG, Musser JM. Full-exon resequencing reveals toll-like receptor variants contribute to human susceptibility to tuberculosis disease. PLoS One. 2007;2:e1318. doi: 10.1371/journal.pone.0001318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou XX, Jia WH, Shen GP, Qin HD, Yu XJ, Chen LZ, Feng QS, Shugart YY, Zeng YX. Sequence variants in toll-like receptor 10 are associated with nasopharyngeal carcinoma risk. Cancer Epidemiol Biomarkers Prev. 2006;15:862–866. doi: 10.1158/1055-9965.EPI-05-0874. [DOI] [PubMed] [Google Scholar]
- 49.Boutens L, Mirea AM, van den Munckhof I, Doppenberg-Oosting M, Jaeger M, Hijmans A, Netea MG, Joosten LAB, Stienstra R. A role for TLR10 in obesity and adipose tissue morphology. Cytokine. 2018;108:205–212. doi: 10.1016/j.cyto.2018.03.021. [DOI] [PubMed] [Google Scholar]