Abstract
Colistin is the last resort for the treatment of infections with carbapenem-resistant (CR) Gram-negative bacteria particularly Acinetobacter baumannii (CRAB). Currently, both colistin-resistant and -heteroresistant A. baumannii isolates have been reported globally. We therefore investigated the colistin heteroresistance rate in 75 non-duplicate colistin-susceptible CRAB clinical isolates from a Thai university collected in 2016. Minimum inhibitory concentrations (MICs) of colistin for all isolates were determined by broth microdilution method and carbapenemase genes were detected by PCR methods. All isolates were genotyped by ERIC-PCR method and screened for colistin heteroresistance by modified population analysis profile (PAP) method. The colistin MIC range for the 75 isolates was 0.5–2 µg/mL, with MIC50 and MIC90 of 1 and 2 µg/mL, respectively. Thirty-three isolates (44%) were considered colistin-heteroresistant with subpopulations growing at 3–8 μg/mL of colistin. After three daily passages of the subpopulations on antibiotic-free medium, their colistin MICs ranged from 4 to > 32 µg/mL, with MIC50 and MIC90 of 32 and > 32 µg/mL, respectively. Eight different ERIC-PCR profiles were obtained among the 33 isolates and all carried blaOXA-23-like. The high rate of colistin heteroresistance in the CRAB isolates highlights the possibility of treatment failure of CRAB infections by colistin due to the selection of colistin-resistant subpopulations.
Keywords: Acinetobacter baumannii, Carbapenemase genes, Colistin heteroresistance, ERIC-PCR, Population analysis profile
Introduction
Acinetobacter baumannii is frequently associated with life-threatening nosocomial infections and also causes outbreaks particularly in intensive care units (Lee et al. 2017). It is included in the six highest-urgency risky microorganisms, the ESKAPE group, by the Infectious Diseases Society of America (IDSA) (Boucher et al. 2017). The increasing worldwide prevalence of multi-drug resistant (MDR) A. baumannii, particularly carbapenem-resistant (CRAB) strains, is of great concern since the treatment options become limited. The “old” antibiotics including polymyxin B and colistin (polymyxin E) have now been reused as the last resort antibiotics for the treatment of serious infections with carbapenem-resistant Gram-negative bacteria. However, the increased use of colistin and improper understanding of its pharmacokinetics and pharmacodynamics have led to the emergence of colistin-resistant Gram-negative bacilli (Poirel et al. 2017). A systematic review conducted on the studies across 41 countries showed that the global prevalence rates of colistin resistance in A. baumannii were 0.2–17.5% (Pormohammad et al. 2019) with the levels as high as 30.6% in Korea (Ko et al. 2007). Colistin heteroresistance, which is defined as the presence of resistant subpopulations within susceptible isolates, was also reported in this organism (Li et al. 2006) with the rates ranging from 18.7 to 100% (Cai et al. 2012). Treatment of infections caused by colistin-heteroresistant isolates may result in the selection of colistin-resistant subpopulations, thus leading to therapeutic failures (Hawley et al. 2008).
In 2011, 10% of A. baumannii clinical isolates from a university hospital in central Thailand were colistin-resistant (Naksena et al. 2012). In our hospital, this organism was the most common causative agent (approximately 20–30%) of hospital-acquired pneumonia and ventilator-associated pneumonia during 2008 and 2009 (Reechaipichitkul et al. 2013). In this period, carbapenem resistance rate was more than 90% in A. baumannii isolates, thus leading to an increased use of colistin as a therapeutic option. In the same study, the colistin resistance rate in A. baumannii was 0.6%. However, there has been little information about colistin heteroresistance in Thai clinical isolates. The recommended colistin susceptibility testing, broth microdilution (BMD) method, is laborious and unable to detect colistin-heteroresistant strains in routine laboratory. A current standard method for detection of colistin heteroresistance is a population analysis profile (PAP) method, which is also laborious and time-consuming (Li et al. 2006; Sherman et al. 2019). Therefore, we investigated colistin heteroresistance rate in our colistin-susceptible CRAB clinical isolates by the PAP method. This would be baseline data and useful information for clinicians to be aware of the interpretation of colistin susceptibility results and use with caution.
Materials and methods
Clinical isolates
A total of 75 non-duplicate colistin-susceptible CRAB isolates obtained from patients in Srinagarind Hospital, Khon Kaen University, Thailand between January and November in 2016 were included. They were from sputum or tracheal aspirates (46 isolates), pus (10 isolates), blood (3 isolates), drain fluid (2 isolates), others (urine, pleural fluid, tip cut down and stump, 1 isolate for each) and unknown sources (10 isolates). They were identified by conventional biochemical tests including glucose, citrate and malonate utilization, growth at 41 and 44 °C and the presence of hemolysis (Bouvet and Grimont 1986), and then confirmed by the presence of an intrinsic blaOXA-51-like (Turton et al. 2006). All isolates were kept in skimmed milk with 15% glycerol at − 20 °C for further analysis.
MIC determination
Antimicrobial agents tested were amikacin and meropenem (Siam Bheasach, Bangkok, Thailand), gentamicin, ciprofloxacin, cefotaxime, ceftazidime and colistin (Sigma-Aldrich, St. Louis, MO, USA), imipenem (MSD, Whitehouse Station, NJ, USA), fosfomycin (Meiji Seika Pharma, Tokyo, Japan) and tigecycline (Pfizer Inc., Philadelphia, PA, USA). MICs of these antimicrobials were determined by agar dilution method except for that of colistin using the BMD method (CLSI 2019). The MICs were interpreted according to the criteria of CLSI (2019) except for tigecycline using those of the European Committee on Antimicrobial Susceptibility Testing (EUCAST) breakpoint (EUCAST 2019). Escherichia coli ATCC 25922 was used as an antimicrobial-susceptible control strain.
Detection of carbapenemase genes
All isolates were screened for an intrinsic blaOXA-51-like, OXA carbapenemase (blaOXA-23, blaOXA-24, blaOXA-58 and blaOXA-235) and metallo-β-lactamase (blaNDM, blaIMP and blaVIM) genes by PCR methods (Higgins et al. 2013; Poirel et al. 2011).
Modified population analysis profile (PAP) method
All isolates were screened for colistin heteroresistance by the modified PAP method described by Li et al. (2006). A volume of 50 µL of each tenfold serial dilution (final inoculum of 102 to 108 CFU/mL) of bacterial suspension was spread onto Mueller Hinton agar (MHA) plates (Oxoid, Basingstoke, Hampshire, England) containing various concentrations of colistin (0, 0.5, 1, 2, 3, 4, 5, 6 and 8 µg/mL). Bacterial colonies were counted after 48-h incubation at 37 °C. Colistin heteroresistance was defined as the presence of a colistin-susceptible isolate with MIC of ≤ 2 µg/mL in which detectable colistin-resistant subpopulations were able to grow in the presence of > 2 µg/mL of colistin (Yau et al. 2009). Colistin MICs for the heteroresistant subpopulations were determined by the BMD method after 3 daily subculturing on antibiotic-free MHA plates. Pseudomonas aeruginosa ATCC 27853 was used as a colistin-susceptible control strain.
Strain typing
The genetic relatedness of all isolates was investigated by enterobacterial repetitive intergenic consensus-PCR (ERIC-PCR) (Versalovic et al. 1991). The fingerprints were verified by BioNumerics software (version 7.6, Applied Maths, Belgium) using the Dice coefficient and the unweighted pair group method of averages (UPGMA) with 1% optimization and 1% position tolerance. Isolates showing ≥ 80% similarity were considered clonally related (Ezadi et al. 2019).
Results
Antimicrobial susceptibility
All 75 CRAB isolates were susceptible to colistin with MICs ranging from 0.5 to 2 µg/mL (MIC50 and MIC90 of 1 and 2 µg/mL, respectively). Apart from colistin, amikacin was the second agent mostly active against the CRAB isolates with 25.3% susceptibility, whereas susceptibility to tigecycline was 2.7% only (Table 1).
Table 1.
MICs of various antimicrobials for the 75 colistin-susceptible CRAB isolates
| Antimicrobial agents (concentrations tested, µg/mL) |
MICs (µg/mL)a | % | ||||
|---|---|---|---|---|---|---|
| Ranges | MIC50 | MIC90 | S | I | R | |
| Amikacin (0.5–64) | ≤ 0.5 to 64 | > 64 | > 64 | 25.3 | 1.3 | 73.4 |
| Gentamicin (0.25–32) | ≤ 0.25 to 32 | > 32 | > 32 | 10.7 | 2.7 | 86.6 |
| Ciprofloxacin (0.004–32) | 2–32 | > 32 | > 32 | 0 | 1.3 | 98.7 |
| Ceftazidime (0.125–256) | 8–256 | > 256 | > 256 | 1.3 | 1.3 | 97.4 |
| Imipenem (0.125–256) | 16–256 | 64 | 64 | 0 | 0 | 100 |
| Meropenem (0.125–256) | 16–256 | 64 | 128 | 0 | 0 | 100 |
| Fosfomycin (0.25–256) | 64–256 | 128 | 256 | 10.7 | 53.3 | 36 |
| Tigecycline (0.031–32) | 0.5–16 | 4 | 8 | 2.7 | – | 97.3 |
| Colistin (0.125–32) | 0.5–2 | 1 | 2 | 100 | – | 0 |
S susceptible, I intermediate, R resistant
aMICs of all antimicrobials tested were determined by the agar dilution method except for those of colistin using the BMD method
Carbapenemase genes
All isolates contained the intrinsic blaOXA-51-like. Carbapenemase genes were found in 74 isolates (98.7%): 67 isolates with blaOXA-23-like, 2 isolates with blaNDM-like, 1 isolate with blaOXA-58-like, 3 isolates with both blaOXA-23-like and blaOXA-58-like, and 1 isolate with both blaOXA-23-like and blaNDM-like.
Colistin heteroresistance by modified PAP
Modified PAP analysis showed the growth of subpopulations in the presence of > 2 µg/mL of colistin (Fig. 1). Of the 75 colistin-susceptible isolates, 35 isolates (46.7%) with colistin MICs of 1 (11 isolates) and 2 µg/mL (24 isolates) were identified as colistin-heteroresistant. After 3 daily passages through an antibiotic-free medium, colistin heteroresistance phenotype was maintained in 33 isolates (44%). They were from sputum or tracheal aspirates (25 isolates), pus (4 isolates), blood, drain fluid, tip cut down and tissue (1 isolate for each). The colistin MIC range for the 33 subpopulation isolates was 4- > 32 µg/mL, with MIC50 and MIC90 of 32 and > 32 µg/mL, respectively (Table 2).
Fig. 1.
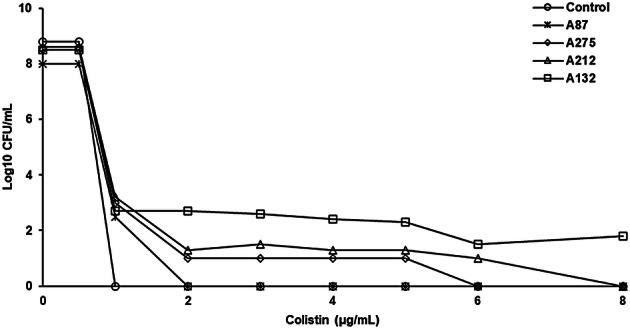
Population analysis profiles of the CRAB clinical isolates with heteroresistant subpopulations (isolates A132, A212 and A275) and colistin susceptibility (isolate A87), and colistin-susceptible control strain (P. aeruginosa ATCC 27853)
Table 2.
Characteristics of the 33 colistin-heteroresistant CRAB clinical isolates
| No. of isolates | Carbapenemase genes | ERIC patterns |
MICs (µg/mL) | Resistant subpopulations | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AK | G | CIP | CZ | IP | ME | FS | TG | CT | Growth at colistin concentration (µg/mL)a |
Colistin MICs (µg/mL)b |
|||
| 7 | blaOXA-23-like | Cluster C | > 64 | > 32 | > 32 | > 256 | 32–64 | 32–64 | 128–256 | 4–8 | 2 | 4–8 | 8- > 32 |
| 3 | blaOXA-23-like | Cluster C | > 64 | > 32 | > 32 | > 256 | 64 | 64–128 | 128–256 | 2 | 1–2 | 4–8 | 32- > 32 |
| 2 | blaOXA-23-like | Cluster C | > 64 | > 32 | > 32 | > 256 | 64 | 128 | 128–256 | 8 | 1–2 | 5–6 | 32- > 32 |
| 2 | blaOXA-23-like | Cluster C | > 64 | > 32 | > 32 | > 256 | 64–128 | 64–128 | > 256 | 2 | 1–2 | 5 | 8–16 |
| 1 | blaOXA-23-like | Cluster C | > 64 | > 32 | > 32 | > 256 | 16 | 64 | 128 | 4 | 1 | 3 | > 32 |
| 1 | blaOXA-23-like | Cluster C | > 64 | > 32 | > 32 | > 256 | 256 | 256 | 128 | 4 | 2 | 5 | > 32 |
| 1 | blaOXA-23-like | Cluster C | > 64 | > 32 | > 32 | > 256 | 128 | 128 | > 256 | 4 | 1 | 5 | > 32 |
| 1 | blaOXA-23-like | Cluster C | 4 | 8 | > 32 | > 256 | 32 | 64 | 128 | 4 | 2 | 4 | > 32 |
| 1 | blaOXA-23-like & blaNDM-like | Cluster C | > 64 | > 32 | > 32 | > 256 | 128 | 128 | 128 | 4 | 2 | 3 | 32 |
| 1 | blaOXA-23-like | Cluster E | 4 | > 32 | > 32 | > 256 | 32 | 32 | 256 | 4 | 1 | 5 | > 32 |
| 1 | blaOXA-23-like | Cluster E | 2 | 0.5 | > 32 | > 256 | 32 | 32 | 128 | 2 | 2 | 3 | 4 |
| 1 | blaOXA-23-like | Cluster E | < 0.5 | ≤ 0.25 | 32 | > 256 | 64 | 64 | 256 | 2 | 2 | 6 | 16 |
| 2 | blaOXA-23-like | Cluster B | > 64 | > 32 | > 32 | > 256 | 32–64 | 32–64 | 256 | 2–4 | 2 | 8 | 32- > 32 |
| 1 | blaOXA-23-like | Cluster A | 4 | 8 | > 32 | 16 | 64 | 64 | 256 | 8 | 2 | 5 | > 32 |
| 1 | blaOXA-23 -like & blaOXA-58-like | Cluster A | 8 | > 32 | > 32 | > 256 | 64 | 32 | 256 | 4 | 2 | 8 | 4 |
| 2 | blaOXA-23-like | Cluster D | 1 | 16–32 | > 32 | 64 | 32 | 16–32 | 128–256 | 2 | 2 | 8 | 32- > 32 |
| 1 | blaOXA-23-like | Cluster D | < 0.5 | 4 | 8 | 128 | 32 | 16 | 128 | 0.5 | 1 | 6 | > 32 |
| 1 | blaOXA-23-like & blaOXA-58-like | Singleton | 1 | > 32 | > 32 | > 256 | 32 | 32 | 128 | 4 | 1 | 8 | 16 |
| 1 | blaOXA-23-like | Cluster F | 4 | 4 | > 32 | > 256 | 32 | 32 | 128 | 16 | 2 | 8 | 32 |
| 1 | blaOXA-23 -like & blaOXA-58-like | Cluster F | 8 | > 32 | > 32 | > 256 | 16 | 32 | 128 | 16 | 2 | 6 | 8 |
| 1 | blaOXA-23-like | Singleton | > 64 | > 32 | > 32 | > 256 | 64 | 64 | 64 | 4 | 1 | 4 | > 32 |
AK amikacin, CIP ciprofloxacin, CT colistin, CZ ceftazidime, G gentamicin, FS fosfomycin, IP imipenem, ME meropenem, TG tigecycline
aHighest colistin concentration that the isolate grew in the PAP method
bMICs of heteroresistant subpopulations after 3 daily passages onto the colistin-free medium
Strain typing
Cluster analysis using 80% similarity cut-off for clonal relatedness of the 75 CRAB isolates revealed 12 different ERIC-PCR patterns, 8 clusters and 4 singletons (data not shown). Thirty-eight isolates (50.7%) belonged to the same cluster. Among the 33 colistin-heteroresistant isolates, 6 clusters and 2 singletons were obtained (Fig. 2).
Fig. 2.
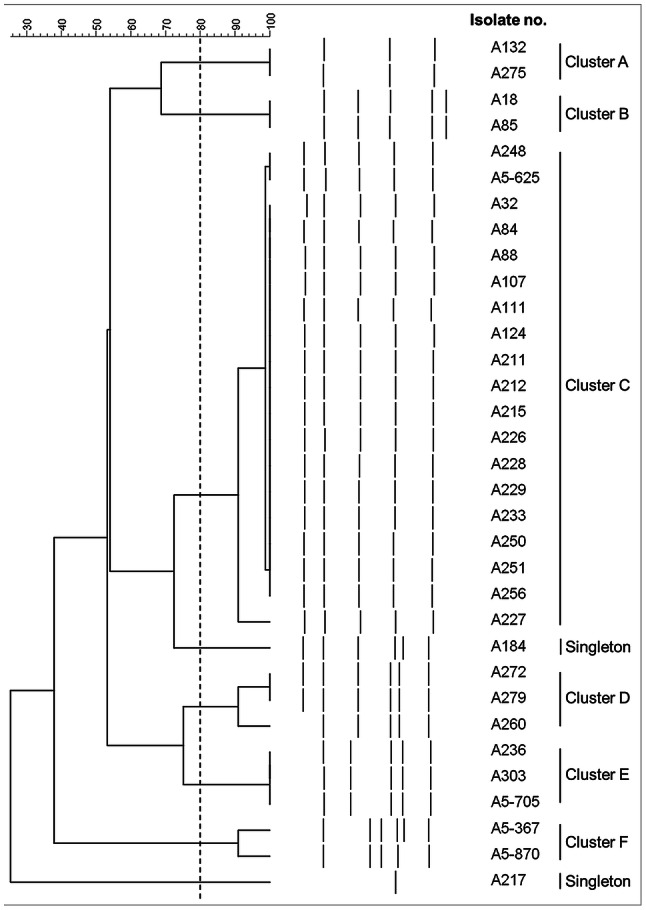
Dendrogram of ERIC-PCR from the 33 colistin-heteroresistant CRAB clinical isolates generated by BioNumerics using the UPGMA and the Dice coefficient. Dashed line indicates 80% similarity cut-off
Discussion
Multidrug resistance (MDR) to currently available antibiotics in Gram-negative bacteria is the major problem of public health worldwide. A critical problem is the increasing prevalence of carbapenem resistance, particularly by the mechanism of carbapenemase production (Codjoe and Donkor 2017). In this study, most of the CRAB isolates harbored one or two OXA carbapenemase genes, blaOXA-23-like and/or blaOXA-58-like, similar to previous studies from Thailand (Lertsrisatit et al. 2017; Leungtongkam et al. 2018). In agreement with other reports, blaOXA-23-like was the major resistance determinant (40–100%) among the CRAB isolates, indicating that carbapenem monotherapy is now no longer used as a therapeutic option against the MDR A. baumannii infections (Ezadi et al. 2019; Salehi et al. 2018).
Colistin has been used as the last resort for either MDR or extremely drug-resistant (XDR) A. baumannii infections (Cai et al. 2012). However, colistin resistance in this organism has been reported globally. Furthermore, colistin heteroresistance was also discovered in 15 of 16 (93.8%) colistin-susceptible MDR A. baumannii isolates from Australia (Li et al. 2006). Hawley et al. (2008) described colistin heteroresistance in 19 MDR A. baumannii clinical isolates from Texas between 2003 and 2005 as defined by their growth on plates containing 8 μg/mL of colistin and giving colistin MICs of > 8 μg/mL, whereas Yau et al. (2009) reported the heteroresistance rate of 23% from Thailand in 2008. Colistin heteroresistance was also observed in 20 of 24 (83%) A. baumannii isolates obtained between 2013 and 2015 from the USA (Srinivas et al. 2018). These isolates were from blood and respiratory tract, and susceptible to colistin with MICs of ≤ 0.25 to 0.5 µg/mL but grew on MHA containing 4 µg/mL of colistin. Recently, 9 of 44 (20.5%) colistin-susceptible CRAB isolates from northern Iran have been identified as colistin-heteroresistant with subpopulations growing in the presence of 6–8 µg/mL of colistin (Ezadi et al. 2019). The colistin heteroresistance rates may be different due to inoculum size applied (10–100 μL) and cut-off for identifying heteroresistant subpopulations (4 or ≥ 8 μg/mL) (Hawley et al. 2008; Li et al. 2006; Srinivas et al. 2018). Although colistin was the most active agent against the CRAB isolates in this study, colistin heteroresistance was observed in 44% of them as detected by the modified PAP method. Unfortunately, colistin-heteroresistant A. baumannii strains cannot be discriminated from colistin-susceptible strains by the standard BMD susceptibility testing (Ezadi et al. 2019).
Impact of colistin heteroresistance on the clinical outcomes is of concern. The pharmacokinetic study has demonstrated that plasma colistin methanesulphonate concentrations usually achieve in the range of 1 to 4 µg/mL after intravenous administration (Li et al. 2005). However, the treatment with low dose of colistin may not be effective and can cause the selection of heteroresistance. Increased colistin dose also leads to its nephrotoxicity. Hawley et al. (2008) demonstrated a statistically higher rate of colistin heteroresistance (7 from 19 isolates, 36.8%) among MDR A. baumannii isolates from patients with previous colistin exposure. Rodriguez et al. (2009) described the selection of colistin-resistant subpopulations from a colistin-heteroresistant A. baumannii isolate during the treatment with intrathecal colistin in a case of postneurosurgical meningitis. Gazel and Otkun (2017) also demonstrated that after sub-inhibitory exposure to colistin, heteroresistance or resistance had been developed in all CRAB isolates. In the present study, patients’ clinical data and treatment outcomes, which may provide important information for the treatment of patients infected with colistin-heteroresistant CRAB isolates, were not available. However, we found the high rate of colistin heteroresistance among our CRAB isolates. It is possible that the colistin-resistant subpopulations from these isolates may be selected after exposure to colistin, leading to ineffective treatment. Therefore, combination therapies of colistin with other agents were suggested for patients infected with MDR A. baumannii isolates (Gazel and Otkun 2017; Kengkla et al. 2018; Rodriguez et al. 2010).
The main mechanism of colistin resistance in A. baumannii is the modification of lipopolysaccharide (LPS) by mutations in PmrA/PmrB two-component system (Ko et al. 2017). These mutations cause upregulation of the pmrCAB operon, which results in the synthesis and addition of positively charged phosphoethanolamine to the LPS. An increase in positive charge of the LPS leads to a decrease in the binding between colistin (positive charge) and lipid A (negative charge) of the LPS, thus resulting in colistin resistance. Mutations in PmrB such as S144KLAGS, P170L and M308R, and that in PmrA such as M12I were responsible to colistin heteroresistance (Charretier et al. 2018). Unfortunately, molecular resistance mechanisms of the colistin-heteroresistant CRAB isolates were not investigated in the present study.
In conclusion, the rate of colistin heteroresistance was high (44%) among the CRAB clinical isolates from our hospital. They were of different strains and their subpopulations exhibited high-level colistin resistance. This highlights that clinical use of colistin for the treatment of the CRAB infections may be failure because of the selection of heteroresistant subpopulations.
Acknowledgements
This work was supported by Khon Kaen University Research Grant (Project Numbers 600011 and 6100017). We thank Faculty of Associated Medical Sciences, Khon Kaen University and the CMDL, Khon Kaen University for providing a scholarship for Khin Thet Thet. We are grateful to the staff of Clinical Microbiology Unit, Srinagarind Hospital for collecting the clinical isolates.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
The present study was conducted under the protocol approved by the Khon Kaen University Ethics Committee for Human Research based on the Declaration of Helsinki and the ICH Good Clinical Practice Guidelines (Project ID: HE581434).
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Bouvet PJM, Grimont PAD. Taxonomy of the genus Acinetobacter with the recognition of Acinetobacter baumannii sp. nov. Acinetobacter haemolyticus sp. nov. Acinetobacter johnsonii sp. nov. and Acinetobacter junii sp. nov. and emended descriptions of Acinetobacter calcoaceticus and Acinetobacter lwoffii. Int J Sys Bacteriol. 1986;36:228–240. doi: 10.1099/00207713-36-2-228. [DOI] [Google Scholar]
- Boucher HW, Talbot GH, Bradley JS, Edwards JE, Gilbert D, Rice LB, Scheld M, Spellberg B, Bartlett J. Bad bugs, no drugs: No ESKAPE ! An update from the Infectious Diseases Society of America. Clin Infect Dis. 2017;48:1–12. doi: 10.1086/595011. [DOI] [PubMed] [Google Scholar]
- Cai Y, Chai D, Wang R, Liang B, Bai N. Colistin resistance of Acinetobacter baumannii: Clinical reports, mechanisms and antimicrobial strategies. J Antimicrob Chemother. 2012;67:1607–1615. doi: 10.1093/jac/dks084. [DOI] [PubMed] [Google Scholar]
- Charretier Y, Diene SM, Baud D, Chatellier S, Santiago-Allexant E, van Belkum A, Guigon G, Schrenzel J. Colistin heteroresistance and involvement of the PmrAB regulatory system in Acinetobacter baumannii. Antimicrob Agents Chemother. 2018;62:e00788–e818. doi: 10.1128/AAC.00788-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CLSI (2019) Performance Standards for Antimicrobial Susceptibility Testing: 29th edn. CLSI document M100-S29.
- Codjoe FS, Donkor ES. Carbapenem resistance: a review. Med Sci (Basel) 2017;6:1. doi: 10.3390/medsci6010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EUCAST (2019) Breakpoint tables for interpretation of MICs and zone diameters. Version 9.0, 2019. https://www.eucast.org
- Ezadi F, Jamali A, Heidari A, Javid N, Ardebili A. Heteroresistance to colistin in oxacillinases-producing carbapenem-resistant Acinetobacter baumannii clinical isolates from Gorgan. J Glob Antimicrob Resist. 2019;21:380–385. doi: 10.1016/j.jgar.2019.11.010. [DOI] [PubMed] [Google Scholar]
- Gazel D, Otkun MT. Investigation of Colistin Heteroresistance and some factors affecting heteroresistance in carbapenem-resistant A baumannii strains. Mediterr J Infect Microb Antimicrob. 2017;6:1. [Google Scholar]
- Hawley JS, Murray CK, Jorgensen JH. Colistin heteroresistance in Acinetobacter and its association with previous colistin therapy. Antimicrob Agents Chemother. 2008;52:351–352. doi: 10.1128/AAC.00766-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins PG, Perez-Llarena FJ, Zander E, Fernandez A, Bou G, Seifert H. OXA-235, a novel class D β-lactamase involved in resistance to carbapenems in Acinetobacter baumannii. Antimicrob Agents Chemother. 2013;57:2121–2126. doi: 10.1128/AAC.02413-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kengkla K, Kongpakwattana K, Saokaew S, Apisarnthanarak A, Chaiyakunapruk N. Comparative efficacy and safety of treatment options for MDR and XDR Acinetobacter baumannii infections: a systematic review and network meta-analysis. J Antimicrob Chemother. 2018;73:22–32. doi: 10.1093/jac/dkx368. [DOI] [PubMed] [Google Scholar]
- Ko KS, Suh JY, Kwon KT, Jung SI, Park KH, Kang CI, Chung DR, Peck KR, Song JH. High rates of resistance to colistin and polymyxin B in subgroups of Acinetobacter baumannii isolates from Korea. J Antimicrob Chemother. 2007;60:1163–1167. doi: 10.1093/jac/dkm305. [DOI] [PubMed] [Google Scholar]
- Ko KS, Choi Y, Lee JY. Old drug, new findings: colistin resistance and dependence of Acinetobacter baumannii. Precis Future Med. 2017;1:159–167. doi: 10.23838/pfm.2017.00184. [DOI] [Google Scholar]
- Li J, Nation RL, Milne RW, Turnidge JD, Coulthard K. Evaluation of colistin as an agent against multi-resistant Gram-negative bacteria. Int J Antimicrob Agents. 2005;25:11–25. doi: 10.1016/j.ijantimicag.2004.10.001. [DOI] [PubMed] [Google Scholar]
- Li J, Rayner CR, Nation RL, Owen RJ, Spelman D, Tan KE, Liolios L. Heteroresistance to colistin in multidrug-resistant Acinetobacter baumannii. Antimicrob Agents Chemother. 2006;50:2946–2950. doi: 10.1128/AAC.00103-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CR, Lee JH, Park M, Park KS, Bae IK, Kim YB, Cha CJ, Jeong BC, Lee SH. Biology of Acinetobacter baumannii: Pathogenesis, antibiotic resistance mechanisms, and prospective treatment options. Front Cell Infect Microbiol. 2017;7:55. doi: 10.3389/fcimb.2017.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lertsrisatit Y, Santimaleeworagun W, Thunyaharn S, Traipattanakul J. In vitro activity of colistin mono- and combination therapy against colistin-resistant Acinetobacter baumannii, mechanism of resistance, and clinical outcomes of patients infected with colistin-resistant A. baumannii at a Thai university hospital. Infect Drug Resist. 2017;10:437–443. doi: 10.2147/IDR.S148185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leungtongkam U, Thummeepak R, Tasanapak K, Sitthisak S. Acquisition and transfer of antibiotic resistance genes in association with conjugative plasmid or class 1 integrons of Acinetobacter baumannii. PLoS ONE. 2018;13:e0208468. doi: 10.1371/journal.pone.0208468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naksena P, Prombhul S, Pelletier M, Ernst R, Tribuddharat C, Thaipisuttikul I. Determination of colistin resistance mechanism of Acinetobacter baumannii isolated in Siriraj Hospital, Thailand. Int J Infect Dis. 2012;16:e433–e434. doi: 10.1016/j.ijid.2012.05.609. [DOI] [Google Scholar]
- Poirel L, Walsh TR, Cuvillier V, Nordmann P. Multiplex PCR for detection of acquired carbapenemase genes. Diagn Microbiol Infect Dis. 2011;70:119–123. doi: 10.1016/j.diagmicrobio.2010.12.002. [DOI] [PubMed] [Google Scholar]
- Poirel L, Jayol A, Nordmann P. Polymyxins: Antibacterial activity, susceptibility testing, and resistance mechanisms encoded by plasmids or chromosomes. Clin Microbiol Rev. 2017;30:557–596. doi: 10.1128/CMR.00064-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pormohammad A, Mehdinejadiani K, Gholizadeh P, Nasiri MJ, Mohtavinejad N, Dadashi M, Karimaei S, Safari H, Azimi T. Global prevalence of colistin resistance in clinical isolates of Acinetobacter baumannii: a systematic review and meta-analysis. Microb Pathog. 2019;139:103887. doi: 10.1016/j.micpath.2019.103887. [DOI] [PubMed] [Google Scholar]
- Reechaipichitkul W, Phondongnok S, Bourpoern J, Chaimanee P. Causative agents and resistance among hospital-acquired and ventilator-associated pneumonia patients at Srinagarind Hospital, northeastern Thailand, Southeast Asian. J Trop Med Public Health. 2013;44:490–502. [PubMed] [Google Scholar]
- Rodriguez CH, Bombicino K, Granados G, Nastro M, Vay C, Famiglietti A. Selection of colistin-resistant Acinetobacter baumannii isolates in postneurosurgical meningitis in an intensive care unit with high presence of heteroresistance to colistin. Diagn Microbiol Infect Dis. 2009;65:188–191. doi: 10.1016/j.diagmicrobio.2009.05.019. [DOI] [PubMed] [Google Scholar]
- Rodriguez CH, De Ambrosio A, Bajuk M, Aznar J. In vitro antimicrobials activity against endemic Acinetobacter baumannii multiresistant clones. J Infect Dev Ctries. 2010;4:164–167. doi: 10.3855/jidc.604. [DOI] [PubMed] [Google Scholar]
- Salehi B, Goudarzi H, Nikmanesh B, Houri H, Alavi-Moghaddam M, Ghalavand Z. Emergence and characterization of nosocomial multidrug-resistant and extensively drug-resistant Acinetobacter baumannii isolates in Tehran. Iran J Infect Chemother. 2018;24:515–523. doi: 10.1016/j.jiac.2018.02.009. [DOI] [PubMed] [Google Scholar]
- Sherman EX, Wozniak JE, Weiss DS. Methods to evaluate colistin heteroresistance in Acinetobacter baumannii. Methods Mol Biol. 2019;1946:39–50. doi: 10.1007/978-1-4939-9118-1_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivas P, Hunt LN, Pouch SM, Thomas K, Goff DA, Pancholi P, Balada-Llasat JM, Bauer KA. Detection of colistin heteroresistance in Acinetobacter baumannii from blood and respiratory isolates. Diagn Microbiol Infect Dis. 2018;91:194–198. doi: 10.1016/j.diagmicrobio.2018.01.028. [DOI] [PubMed] [Google Scholar]
- Turton JF, Woodford N, Glover J, Yarde S, Kaufmann ME, Pitt TL. Identification of Acinetobacter baumannii by detection of the blaOXA-51-like carbapenemase gene intrinsic to this species. J Clin Microbiol. 2006;44:2974–2976. doi: 10.1128/JCM.01021-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versalovic J, Koeuth T, Lupski JR. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 1991;19:6823–6831. doi: 10.1093/nar/19.24.6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yau W, Owen RJ, Poudyal A, Bell JM, Turnidge JD, Yu HH, Nation RL, Li J. Colistin hetero-resistance in multidrug-resistant Acinetobacter baumannii clinical isolates from the Western Pacific sregion in the SENTRY Antimicrobial Surveillance Programme. J Infect. 2009;58:138–144. doi: 10.1016/j.jinf.2008.11.002. [DOI] [PubMed] [Google Scholar]


