Abstract
Despite effective hepatitis B virus (HBV)‐DNA suppression, HBV RNA can circulate in patients receiving nucleoside/nucleotide analogues (NAs). Current assays quantify HBV DNA by either real‐time polymerase chain reaction (PCR), which uses DNA polymerase, or transcription‐mediated amplification, which uses reverse‐transcriptase (RT) and RNA polymerase. We assessed the effect of RT capability on HBV‐DNA quantification in samples from three cohorts, including patients with quantified HBV RNA. We compared the HBV‐DNA levels by real‐time PCR (cobas HBV, Roche 6800/8800; Xpert HBV, Cepheid), transcription‐mediated amplification (Aptima HBV, Hologic), and real‐time PCR with added RT capability (cobas HBV+RT). In the first cohort (n = 45) followed over 192 weeks of NA therapy, on‐treatment HBV‐DNA levels were higher with cobas HBV+RT than cobas HBV (mean difference: 0.14 log10 IU/mL). In a second cohort (n = 50) followed over 96 weeks of NA therapy, HBV‐DNA viral load was significantly higher with the cobas HBV+RT and Aptima HBV compared with the cobas HBV test at all time points after initiation of NA therapy (mean difference: 0.65‐1.16 log10 IU/mL). A clinically significant difference was not detected between the assays at baseline. In a third cohort (n = 53), after a median of 2.2 years of NA therapy, we detected HBV RNA (median 5.6 log10 copies/mL) in 23 patients (43.4%). Median HBV‐DNA levels by Aptima HBV were 2.4 versus less than 1 log10 IU/mL in samples with HBV RNA and without HBV RNA, respectively (P = 0.0006). In treated patients with HBV RNA, Aptima HBV measured higher HBV‐DNA levels than Xpert HBV and cobas HBV. Conclusion: Tests including an RT step may overestimate HBV DNA, particularly in samples with low viral loads as a result of NA therapy. This overestimation is likely due to amplification of HBV RNA and may have an impact on clinical decisions.

Abbreviations
- BL
baseline
- cccDNA
covalently closed circular DNA
- CI
confidence interval
- HBeAG
hepatitis B e surface antigen
- HBsAG
hepatitis B surface antigen
- HBV
hepatitis B virus
- HPS
High Pure System
- IQR
interquartile range
- LL
lower limit
- LLOQ
lower limit of quantification
- NA
nucleos(t)ide analogue
- OLS
ordinary least squares
- PCR
polymerase chain reaction
- RT
reverse transcription
- TMA
transcription‐mediated amplification
- UL
upper limit
Current guidelines for the management of chronic hepatitis B virus (HBV) infection recommend quantitative measurement of circulating HBV DNA to guide therapeutic decisions and monitor response to antiviral therapy.( 1 , 2 ) The main endpoint of current treatment strategies is the induction of long‐term and complete suppression of HBV replication.( 1 ) Detection of low, residual HBV‐DNA levels during nucleos(t)ide analogue (NA) therapy may have significant clinical consequences, including a higher estimated risk of hepatocellular carcinoma.( 3 ) Repeatedly confirmed undetectable (or at least not quantifiable) levels of HBV DNA may identify patients in whom NA treatment can be discontinued under specific circumstances.( 1 , 4 ) Inaccurate or inconsistent measurement of viral load could adversely affect patient care, with some patients potentially not receiving the medication they need, and others getting antiviral treatment that might no longer be required.( 1 , 4 ) Overestimation of viral load may erroneously suggest adherence issues or virologic failure, resulting in further inconclusive resistance testing and potential changes to treatment that may be associated with risk of toxicity, additional cost, and emotional distress for the patient.
To date, general agreement has been reported when comparing commercial HBV tests across different platforms.( 5 , 6 , 7 , 8 , 9 ) However, in studies comparing HBV viral load quantification using analytical performance panels, underestimation of values at high viral loads and a lack of linearity in performance across the viral load range has been reported.( 10 ) Available assays for HBV‐DNA quantification operate according to one of two main designs. Historically, most tests used real‐time polymerase chain reaction (PCR) formats, which use different types of DNA polymerases to amplify the target DNA. Real‐time PCR can also be used to quantify RNA; however, this requires the inclusion of an extra step of reverse transcription (RT) before DNA amplification and RT activity of the polymerase. Alternative nucleic acid amplification technologies recently introduced for HBV include real‐time transcription‐mediated amplification (TMA).( 11 ) Due to its excellent sensitivity, TMA has an established track record in the context of blood safety( 12 ) and is applied to the quantification of human immunodeficiency virus RNA( 13 ) and hepatitis C virus RNA.( 14 ) The TMA assay incorporates two enzymes for nucleic acid amplification: Moloney murine leukemia virus reverse transcriptase and T7 RNA polymerase. This makes RT of RNA into DNA potentially part of any amplification process.( 11 ) As a result, it appears likely that a TMA assay may be prone to amplifying not only HBV DNA, but potentially also HBV RNA, particularly in those receiving NA therapy.
Following infection, the HBV genome forms a covalently closed circular DNA (cccDNA) episome in the nucleus of hepatocytes, which functions as the viral transcriptional template. The pregenomic HBV RNA transcribed from cccDNA must be converted into DNA by the viral polymerase to allow production of new virus particles for export. By targeting the viral polymerase, NAs effectively block production of infectious viruses. In patients with high levels of cccDNA transcription, excess pregenomic HBV RNA can bypass RT (and inhibition by NAs) and be exported in HBV‐RNA‐containing particles, which are thought to be noninfectious.( 15 ) Thus, patients receiving antiviral therapy can have circulating HBV RNA despite the NA effectively blocking HBV DNA synthesis and virus replication.
To date, the magnitude of any possible overestimation of HBV DNA as a result of amplification of HBV RNA, and whether clinically relevant discrepancies need to be taken into consideration, has not been investigated. In this study we used samples from three independent cohorts to compare HBV‐DNA measurements obtained by TMA and real‐time PCR, to explore the impact of adding an RT step to real‐time PCR, and to relate the findings to the duration of NA therapy and, in a subset of patients, the direct quantification of circulating HBV RNA.
Materials and Methods
Study Populations
All studies were performed in accordance with the International Conference on Harmonization Good Clinical Practice Guidelines, the Declaration of Helsinki, and relevant local legislation. Ethical approval was obtained from the relevant Institutional Review Board/Independent Ethics Committee. All study cohorts and evaluated HBV‐DNA tests are summarized in Tables 1 and 2.
TABLE 1.
Evaluated HBV DNA Tests
| HBV‐DNA Test (Abbreviation) | Cobas HPS | Cobas HBV | Cobas HBV+RT | Aptima HBV | Xpert HBV |
|---|---|---|---|---|---|
| Full description | Real‐time PCR COBAS TaqMan HBV test for use with the HPS | Real‐time PCR cobas HBV DNA assay for use on the cobas 6800/8800 systems | Real‐time PCR cobas HBV DNA assay for use on the cobas 6800/8800 systems, including a nonstandard total nucleic acid software | TMA Aptima HBV Quant assay for use with the Hologic Panther system | Xpert HBV Viral load |
| Method of amplification | Real‐time PCR | Real‐time PCR | Real‐time PCR | TMA | Real‐time PCR |
| Significant RT capability | No | No | Yes | Yes | No |
| Limit of detection* (IU/mL) | 5.9 | 2.7 | n.d. | 5.58 | 10 |
| LLOQ* (IU/mL) | 29 | 10 | 10 | 5.58 | 10 |
In plasma.
Abbreviation: n.d., not determined.
TABLE 2.
Study Cohorts
| Study Cohorts | Hannover Cohort | DEFINE Cohort | Liverpool Cohort |
|---|---|---|---|
| Number of NA‐treated patients (n) | 45 | 50 | 53 |
| Number of retested plasma samples (n) | 191 | 346 † | 26 |
| Evaluated HBV assays | |||
| Without RT | cobas HBV (cobas HPS*) | cobas HBV (cobas HPS*) | cobas HBV |
| Xpert HBV | |||
| With RT | cobas HBV+RT | cobas HBV+RT | Aptima HBV (TMA) |
| Aptima HBV (TMA) | |||
| Specific value for the study | Longitudinal follow‐up available | Longitudinal follow‐up available | Direct HBV‐RNA quantification performed |
Original test that was used before the study.
A total of 271 samples with enough remaining volume generated valid results for HBV DNA for both cobas HBV and Aptima HBV.
Hannover Cohort: HBV DNA Quantification by Real‐Time PCR (cobas HBV) and by Real‐Time PCR Plus RT (cobas HBV+RT)
In a previous study conducted at three sites in Germany (Hannover Medical School), Switzerland and Korea, we demonstrated the concordance between the real‐time PCR cobas HBV‐DNA assay for use on the cobas 6800/8800 Systems (henceforth described as cobas HBV) and the real‐time PCR COBAS TaqMan HBV test for use with the High Pure System (HPS; Roche Molecular Diagnostics, Pleasanton, CA) (Supporting Fig. S1). The materials and methods, as well as the results for this study, have been published previously.( 8 ) In an exploratory evaluation, all available samples (n = 191) from 45 patients (Supporting Table S1) under NA therapy (principally entecavir and tenofovir disoproxil fumarate; 78%) from the Hannover Medical School (Germany) were used to explore the contribution of circulating HBV RNA to any viral load difference obtained with the standard cobas HBV software and a nonstandard total nucleic acid software (cobas HBV+RT), which includes an RT step in the PCR profile, allowing for the amplification of HBV RNA that otherwise would not be amplified at a meaningful level. The need for written informed consent was waived by the institutional review board.
DEFINE Cohort: HBV DNA Quantification by TMA (Aptima HBV), Real‐Time PCR (cobas HBV), and Real‐Time PCR Plus RT (cobas HBV+RT)
Based on the results of the first study, a second study was conducted at two sites in Germany and Spain to compare HBV‐DNA quantification by cobas HBV, cobas HBV+RT, and the TMA Aptima HBV Quant assay (henceforth described as Aptima HBV) for use with the Hologic Panther system (Hologic Inc., Marlborough, MA). A total of 346 plasma samples collected from 50 lamivudine‐resistant adult patients (≥18 years of age) starting salvage therapy with various NA combinations were collected from baseline to week 96 of treatment. Detailed information on the design of the DEFINE study and the study population has been published elsewhere.( 16 ) Viral load concentrations ranged from undetectable to greater than 9 log10 IU/mL, overlapping the medical decision points (≥20,000, ≥2,000, <2,000, and <50 IU/mL) used to direct treatment during the study. All samples with enough remaining volume that generated valid results for HBV DNA (i.e., n = 271 for both cobas HBV and Aptima HBV) were considered for further statistical analysis. All samples were anonymized by an independent ethics committee–approved procedure. All specimens were derived from archived samples that had been stored at −20°C or lower for a maximum of 10 years (DEFINE trial( 16 ); all patients had provided informed consent). Each sample was divided into sufficient aliquots to allow at least single‐replicate testing with each test. HBV‐DNA results from the original test (cobas HPS) at the time of collection were available for each sample (referred to as nominal viral load).
Liverpool Cohort: HBV‐DNA Quantification by TMA (Aptima HBV) and Real‐Time PCR (Xpert HBV and Cobas HBV) in Patients With Circulating HBV RNA
As part of an ongoing study evaluating HBV‐RNA detection during chronic HBV infection at the University of Liverpool, United Kingdom (Research Ethics Committee Approval 18/YH/0286, July 2018), plasma samples from 101 patients underwent HBV‐RNA quantification by an in‐house real‐time PCR assay that applies the method described by van Bömmel et al.( 17 ) and was performed at DDL Diagnostic Laboratory (Rijswijk, the Netherlands). The assay reported detection/quantification range that spans 2.5/4.0‐9.5 log10 copies/mL. Aptima HBV is the test used for routine care in the accredited National Health Service (NHS) diagnostic laboratory of the Royal Liverpool University Hospital. To determine whether there was any overestimation of HBV DNA by Aptima HBV, plasma samples from 26 patients who had both quantifiable HBV RNA and quantifiable HBV DNA by Aptima HBV were also tested for HBV DNA by real‐time PCR, using Xpert HBV Viral Load (henceforth described as Xpert HBV) (Cepheid, Maurens‐Scopont, France). Xpert HBV was performed in the local diagnostic laboratory, and cobas HBV was performed either at a referral NHS laboratory or at Roche Diagnostics.
HBV‐DNA Quantification
All tests were performed by trained operators in accordance with the manufacturers’ specifications.( 18 , 19 , 20 ) Runs were considered valid if both positive and negative controls were valid and no protocol deviations or incidents occurred that might affect the validity of the data. If a run was considered invalid, all samples included in that run were retested wherever possible. The cobas HBV+RT test was performed according to an in‐house protocol as described previously.
Analysis and Statistical Methods
All statistical analyses were carried out using the SAS System software version 9.4 through the SAS Enterprise Guide software version 7.12 or higher. Results were log10‐transformed and compared according to Clinical and Laboratory Standards Institute guidance EP09‐A.( 21 ) HBV viral loads were compared between assays using the Student t test, and scatter plots were overlaid with the Deming regression lines used to assess correlation; Bland−Altman plots were used to estimate bias. Longitudinal plots were used to present viral loads at different time points for individual subjects and combined means. For individual subject graphs, the viral load trajectory was measured using the slope of the regression line for each test. When comparing HBV‐DNA levels, results below the lower limit of quantification (LLOQ) were assigned a value of 0.5 × LLOQ (IU/mL) if the assay reported qualitative target detection, and a value of 0.0 log10 IU/mL if the target was not detected. The characteristics of patients with or without detectable HBV RNA were compared by chi‐square or Fisher’s exact test for categorical variables and Kruskal–Wallis test for continuous variables.
Results
Impact of an RT Step on the Measurement of HBV DNA
To investigate the potential impact of an RT step on the measurement of HBV DNA, we tested 191 clinical samples taken from 45 patients under treatment with NAs for up to 48 months with the cobas HBV and cobas HBV+RT tests (Hannover cohort). All samples (n = 191) generated valid results (Supporting Table S2). Testing these samples with the cobas HBV+RT test resulted in a proportionally biased positive linear correlation, with viral load results that were up to 4 log10 IU/mL higher than those detected with the standard cobas HBV test (Fig. 1A; mean ± SD, 0.89 log10 IU/mL ± 1.33; paired Student t test for mean difference [min, max] 0.14 log10 IU/mL [−0.70, 4.25]; P < 0.0001). Bland−Altman bias analysis confirmed these observations (Fig. 1B), with differences being particularly evident in samples with low HBV‐DNA levels. A total of 92 of 191 samples tested had HBV viral loads less than the LLOQ (detectable or undetectable) with the cobas HBV test (Supporting Table S2). Of note, 38 (41%) of these samples yielded levels at LLOQ or higher when tested with the cobas HBV+RT step, with viral loads ranging from 1.06 to 4.43 log10 IU/mL (Supporting Table S3A).
FIG. 1.
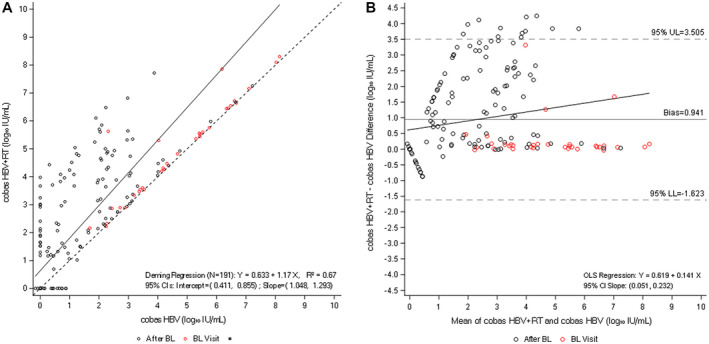
Comparison of cobas HBV and cobas HBV+RT (Hannover cohort). (A) Deming regression analysis of viral load quantification for cobas HBV versus cobas HBV+RT. (B) Bland−Altman bias plot for measurement of HBV with both tests. Abbreviations: BL: Baseline; CI, confidence interval; LL, lower limit; OLS, ordinary least squares; UL, upper limit.
To confirm these results, we tested samples from a second, independent patient cohort. This unique cohort consisted of lamivudine‐resistant patients starting salvage therapy with entecavir, entecavir + adefovir, or adefovir + lamivudine (DEFINE cohort).( 18 ) A total of 346 samples from seven selected time points (baseline, and weeks 12, 24, 48, 72, 84, and 96) were tested with the cobas HBV and cobas HBV+RT tests. Data from all samples that had enough volume and generated valid complete paired observations (i.e., 271 for both cobas HBV and Aptima HBV) were included in the analysis. As was the case in the previous analysis, HBV‐DNA levels were consistently higher with the cobas HBV+RT test, particularly at lower HBV‐DNA levels (Fig. 2A,B). Fourteen samples had viral levels less than the LLOQ with the cobas HBV test. Of these, five (36%) yielded levels at the LLOQ or higher with the cobas HBV+RT test (range: 1.12‐3.07 log10 IU/mL) (Supporting Table S3B).
FIG. 2.
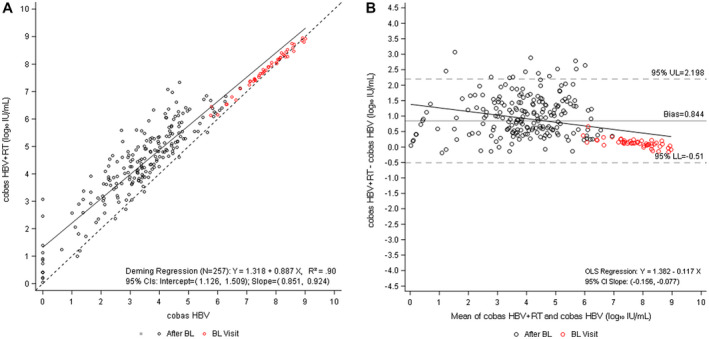
Comparison of cobas HBV and cobas HBV+RT (DEFINE cohort). (A) Deming regression analysis of viral load quantification for cobas HBV versus cobas HBV+RT. (B) Bland−Altman bias plot for measurement of HBV with both tests. Of the 346 longitudinal observations available from seven selected time points (day 1, weeks 12, 24, 48, 72, 84, and 96), 257 samples with enough remaining volume generated valid results for both tests and are included in these plots.
To further evaluate the impact of RT in tests developed to quantify HBV‐DNA levels, samples from the DEFINE cohort were additionally tested with the Aptima HBV test, a TMA‐based assay that includes an RT step. Of the samples available, 253 were within the overlapping linear range (1‐9 log10 IU/mL) for quantitative HBV DNA for both the cobas HBV and Aptima HBV tests. Six samples that generated results less than the LLOQ with the cobas HBV test (five undetectable and one detectable but less than the LLOQ) generated results at the LLOQ or higher with the Aptima HBV test, with levels ranging from 1.11‐2.92 log10 IU/mL (Supporting Table S3C). Comparison of these results similarly indicated a proportionally biased positive linear correlation among the viral loads quantified with each test (Fig. 3A). In line with previous findings, Bland−Altman bias analysis showed that the Aptima HBV quantified higher HBV‐DNA levels than the cobas HBV test (relative to the unity line) at the lower end of the test range, where HBV DNA is being suppressed by treatment, progressing closer to the unity line toward the upper end of the test range (Fig. 3B).
FIG. 3.
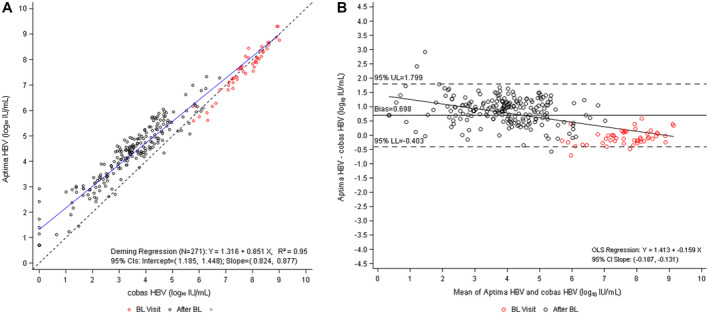
Comparison of Aptima HBV versus cobas HBV (DEFINE cohort). (A) Deming regression analysis of viral load quantification for Aptima HBV versus cobas HBV. (B) Bland−Altman bias plot for measurement of HBV with Aptima and cobas HBV tests. Of the 346 longitudinal observations available from seven selected time points (day 1, weeks 12, 24, 48, 72, 84, and 96), 271 samples with enough remaining volume generated valid results for both tests and are included in these plots.
Finally, the two tests incorporating the RT step, cobas HBV+RT and Aptima HBV, were compared with each other (Fig. 4 and Supporting Table S4) and each was compared versus cobas HBV (without RT). Across each of the study time points, the differences between the cobas HBV test versus the cobas HBV+RT, and the cobas HBV versus Aptima HBV, were similar (Table 3). Comparison of samples showing quantification of HBV DNA with both the cobas HBV+RT and Aptima HBV indicated a near 1:1 positive linear correlation in HBV‐DNA viral load (Fig. 4A). Bland−Altman bias analysis further revealed minimal alterations between the two different tests that include the RT step (Fig. 4B).
FIG. 4.
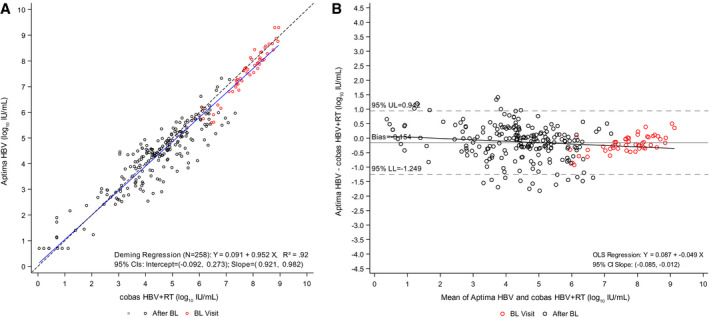
Comparison of Aptima HBV versus cobas HBV+RT (DEFINE cohort). (A) Deming regression analysis of viral load quantification for Aptima HBV versus cobas HBV+RT. (B) Bland−Altman bias plot for measurement of HBV with Aptima and cobas HBV+RT tests. Of the 346 longitudinal observations available from seven selected time points (day 1, weeks 12, 24, 48, 72, 84, and 96), 258 samples with enough remaining volume generated valid results for both tests and are included in these plots.
TABLE 3.
Difference in HBV‐DNA Viral Loads (log10 IU/mL): Cobas HBV Versus Cobas HBV+RT and Cobas HBV Versus Aptima HBV, Cobas HPS Versus Cobas HBV and Aptima HBV Versus HBV+RT, and Aptima HBV Versus Cobas HPS
| Week | Test Comparison | Number of Samples | Mean ± SD (log10 IU/mL) | Adjusted* P Value for Difference |
|---|---|---|---|---|
| Baseline | cobas HBV+RT vs. cobas HBV | 49 | 0.12 ± 0.15 | 0.0000 |
| Aptima HBV vs. cobas HBV | 49 | −0.08 ± 0.26 | 1.0000 | |
| cobas HBV vs. cobas HPS | 49 | 0.12 ± 0.24 | 0.0205 | |
| Aptima HBV vs. cobas HBV+RT | 49 | −0.20 ± 0.29 | 0.0005 | |
| Aptima HBV vs. cobas HPS | 49 | 0.04 ± 0.29 | 1.0000 | |
| 12 | cobas HBV+RT vs. cobas HBV | 49 | 0.93 ± 0.53 | 0.0000 |
| Aptima HBV vs. cobas HBV | 49 | 0.65 ± 0.4 | 0.0000 | |
| cobas HBV vs. cobas HPS | 49 | −0.17 ± 0.36 | 0.0519 | |
| Aptima HBV vs. cobas HBV+RT | 49 | −0.29 ± 0.53 | 0.0151 | |
| Aptima HBV vs. cobas HPS | 49 | 0.48 ± 0.38 | 0.0000 | |
| 24 | cobas HBV+RT vs. cobas HBV | 50 | 1.16 ± 0.64 | 0.0000 |
| Aptima HBV vs. cobas HBV | 50 | 0.89 ± 0.43 | 0.0000 | |
| cobas HBV vs. cobas HPS | 50 | −0.22 ± 0.36 | 0.0027 | |
| Aptima HBV vs. cobas HBV+RT | 50 | −0.27 ± 0.55 | 0.0426 | |
| Aptima HBV vs. cobas HPS | 50 | 0.67 ± 0.39 | 0.0000 | |
| 48 | cobas HBV+RT vs. cobas HBV | 48 | 1.16 ± 0.72 | 0.0000 |
| Aptima HBV vs. cobas HBV | 48 | 0.96 ± 0.38 | 0.0000 | |
| cobas HBV vs. cobas HPS | 48 | −0.28 ± 0.39 | 0.0003 | |
| Aptima HBV vs. cobas HBV+RT | 48 | −0.20 ± 0.62 | 0.9520 | |
| Aptima HBV vs. cobas HPS | 48 | 0.68 ± 0.4 | 0.0000 | |
| 72 | cobas HBV+RT vs. cobas HBV | 25 | 0.97 ± 0.68 | 0.0000 |
| Aptima HBV vs. cobas HBV | 25 | 1.06 ± 0.65 | 0.0000 | |
| cobas HBV vs. cobas HPS | 25 | −0.26 ± 0.59 | 1.0000 | |
| Aptima HBV vs. cobas HBV+RT | 25 | 0.09 ± 0.63 | 1.0000 | |
| Aptima HBV vs. cobas HPS | 25 | 0.80 ± 0.55 | 0.0000 | |
| 84 | cobas HBV+RT vs. cobas HBV | 15 | 0.80 ± 0.68 | 0.0144 |
| Aptima HBV vs. cobas HBV | 15 | 1.11 ± 0.45 | 0.0000 | |
| cobas HBV vs. cobas HPS | 15 | −0.37 ± 0.59 | 0.9959 | |
| Aptima HBV vs. cobas HBV+RT | 15 | 0.31 ± 0.73 | 1.0000 | |
| Aptima HBV vs. cobas HPS | 15 | 0.74 ± 0.43 | 0.0004 | |
| 96 | cobas HBV+RT vs. cobas HBV | 20 | 0.85 ± 0.75 | 0.0026 |
| Aptima HBV vs. cobas HBV | 20 | 0.89 ± 0.64 | 0.0002 | |
| cobas HBV vs. cobas HPS | 20 | −0.37 ± 0.63 | 0.5458 | |
| Aptima HBV vs. cobas HBV+RT | 20 | 0.04 ± 0.55 | 1.0000 | |
| Aptima HBV vs. cobas HPS | 20 | 0.52 ± 0.65 | 0.0771 |
All 256 valid observations with complete paired results are included. Target not detected observations are imputed with a zero value. Values below the LLOQ are imputed as 0.5 × LLOQ = 5 IU/mL for cobas HBV, cobas HBV+RT and Aptima HBV, and imputed as 0.5 × LLOQ = 14.5 IU/mL for the cobas HPS assay. Only complete data within each week across tests are included.
Adjusted for multiplicity using the Bonferroni method.
Longitudinal Comparison of Tests With and Without an RT Step
As the documented differences between the HBV tests with and without the RT step were most prominent in samples with suppressed HBV‐DNA levels to less than the LLOQ, we decided to investigate the impact of NA treatment in more detail. For this purpose, we first compared HBV‐DNA results at baseline and after initiation of salvage therapy in the DEFINE cohort (weeks 12, 24, 48, 72, 84, and 96).
Mean viral loads detected by the cobas HBV, cobas HBV+RT, and Aptima HBV tests at each time point are shown in Fig. 5, along with nominal HBV‐DNA levels measured by cobas HPS. At baseline, discrepancies between the cobas HBV and the two tests that include RT were low (mean differences 0.12 and ‐0.08 log10 IU/mL for cobas HBV+RT and Aptima HBV, respectively). In contrast, there were significant differences in the HBV viral load determined using the assays with RT versus no RT after only 12 weeks of NA therapy, ranging from 0.65‐0.93 log10 IU/mL (P < 0.0001). The difference between Aptima HBV and cobas HBV increased from 0.65 log10 IU/mL (P > 0.0001) at week 12 to a maximum of 1.11 log10 IU/mL (P < 0.0001) at week 84 of therapy. The difference between cobas HBV and cobas HBV+RT increased from 0.93 log10 IU/mL (P < 0.001) at week 12 to a maximum of 1.16 log10 IU/mL (P < 0.0001) at treatment week 48. Differences between the assays slightly decreased at week 96, yielding 0.89 log10 IU/mL (P = 0.0002) and 0.85 log10 IU/mL (P = 0.003) with the cobas HBV and the Aptima HBV, and the cobas HBV and the cobas HBV+RT, respectively. Of note, no statistically significant difference was documented between the cobas HBV+RT and the Aptima HBV (RT assays) during treatment, with the exception of week 12 and 24 (P = 0.02 and P = 0.0426, respectively). Similarly, no significant differences were observed between the cobas HBV and the cobas HPS (no RT assays) at weeks 12, 72, 84, and 96 of therapy (Table 3).
FIG. 5.
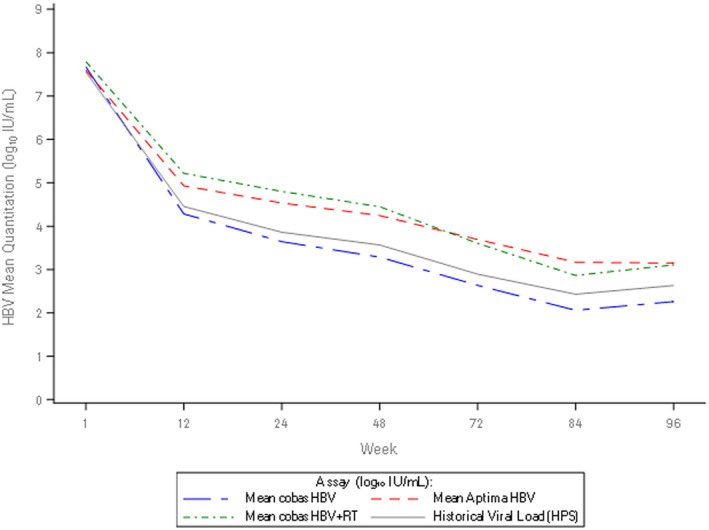
Longitudinal assessment of mean on‐treatment viral load. Longitudinal plot of mean viral load (across patients) from baseline to week 96 of treatment with NAs for cobas HBV, Aptima HBV, cobas HBV+RT, and historical viral load.
A subset of 8 patients had samples available at all seven selected time points that were tested across all platforms. Descriptive statistics of HBV‐DNA levels across these patients for all tests are presented in Supporting Table S5, along with the mean and median log differences between tests (Supporting Table S6). Analyses demonstrated that results were comparable for all four tests at baseline and at week 96 (Table 4). Results for cobas HBV+RT versus Aptima HBV were comparable throughout all time points, and the cobas HBV and HPS test results were comparable at all time points (apart from week 12; Table 4). However, from week 12 until week 84, the Aptima HBV test results were significantly different from the cobas HPS (mean difference: 0.58‐0.99 log10 IU/mL) and cobas HBV results (mean difference: 0.94‐1.2 log10 IU/mL). Similarly, results with cobas HBV were significantly different from those with cobas HBV+RT (mean difference: 1.17‐1.42 log10 IU/mL). Longitudinal assessment of the clinical samples throughout the treatment of these 8 patients generally corroborated this picture (Supporting Fig. S2A‐H). Longitudinal assessment of samples from all other patients with cobas HBV and Aptima HBV is provided in Supporting Fig. S3.
TABLE 4.
Difference in HBV‐DNA Viral Loads (log10 IU/mL): Cobas HBV Versus Cobas HBV+RT and Cobas HBV Versus Aptima HBV, Cobas HPS Versus Cobas HBV and Aptima HBV Versus HBV+RT, and Aptima HBV Versus Cobas HPS in 8 Patients With Samples Available at All Seven Time Points Across All HBV Tests
| Week | Test Comparison | Number of Samples | Mean ± SD (log10 IU/mL) | Adjusted* P Value for Difference |
|---|---|---|---|---|
| Baseline | cobas HBV+RT vs. cobas HBV | 8 | 0.15 ± 0.11 | 0.1928 |
| Aptima HBV vs. cobas HBV | 8 | 0.06 ± 0.22 | 1.0000 | |
| cobas HBV vs. cobas HPS | 8 | 0.02 ± 0.16 | 1.0000 | |
| Aptima HBV vs. cobas HBV+RT | 8 | −0.09 ± 0.21 | 1.0000 | |
| Aptima HBV vs. cobas HPS | 8 | 0.09 ± 0.12 | 1.0000 | |
| 12 | cobas HBV+RT vs. cobas HBV | 8 | 1.17 ± 0.44 | 0.0050 |
| Aptima HBV vs. cobas HBV | 8 | 0.94 ± 0.33 | 0.0033 | |
| cobas HBV vs. cobas HPS | 8 | −0.36 ± 0.19 | 0.0413 | |
| Aptima HBV vs. cobas HBV+RT | 8 | −0.24 ± 0.51 | 1.0000 | |
| Aptima HBV vs. cobas HPS | 8 | 0.58 ± 0.26 | 0.0153 | |
| 24 | cobas HBV+RT vs. cobas HBV | 8 | 1.41 ± 0.46 | 0.0019 |
| Aptima HBV vs. cobas HBV | 8 | 1.19 ± 0.3 | 0.0003 | |
| cobas HBV vs. cobas HPS | 8 | −0.37 ± 0.25 | 0.1444 | |
| Aptima HBV vs. cobas HBV+RT | 8 | −0.21 ± 0.58 | 1.0000 | |
| Aptima HBV vs. cobas HPS | 8 | 0.82 ± 0.19 | 0.0002 | |
| 48 | cobas HBV+RT vs. cobas HBV | 8 | 1.42 ± 0.57 | 0.0075 |
| Aptima HBV vs. cobas HBV | 8 | 1.2 ± 0.31 | 0.0004 | |
| cobas HBV vs. cobas HPS | 8 | −0.38 ± 0.25 | 0.1329 | |
| Aptima HBV vs. cobas HBV+RT | 8 | −0.21 ± 0.65 | 1.0000 | |
| Aptima HBV vs. cobas HPS | 8 | 0.82 ± 0.25 | 0.0011 | |
| 72 | cobas HBV+RT vs. cobas HBV | 8 | 1.23 ± 0.52 | 0.0099 |
| Aptima HBV vs. cobas HBV | 8 | 1.08 ± 0.63 | 0.0638 | |
| cobas HBV vs. cobas HPS | 8 | −0.1 ± 0.61 | 1.0000 | |
| Aptima HBV vs. cobas HBV+RT | 8 | −0.16 ± 0.75 | 1.0000 | |
| Aptima HBV vs. cobas HPS | 8 | 0.97 ± 0.35 | 0.0038 | |
| 84 | cobas HBV+RT vs. cobas HBV | 8 | 1.17 ± 0.59 | 0.0289 |
| Aptima HBV vs. cobas HBV | 8 | 1.07 ± 0.51 | 0.0194 | |
| cobas HBV vs. cobas HPS | 8 | −0.08 ± 0.51 | 1.0000 | |
| Aptima HBV vs. cobas HBV+RT | 8 | −0.1 ± 0.68 | 1.0000 | |
| Aptima HBV vs. cobas HPS | 8 | 0.99 ± 0.2 | 0.0001 | |
| 96 | cobas HBV+RT vs. cobas HBV | 8 | 1.14 ± 0.83 | 0.2133 |
| Aptima HBV vs. cobas HBV | 8 | 0.98 ± 0.8 | 0.3558 | |
| cobas HBV vs. cobas HPS | 8 | −0.17 ± 0.71 | 1.0000 | |
| Aptima HBV vs. cobas HBV+RT | 8 | −0.16 ± 0.67 | 1.0000 | |
| Aptima HBV vs. cobas HPS | 8 | 0.81 ± 0.53 | 0.1188 |
All 256 valid observations with complete paired results across all seven selected time points, across all assays, are included. Target not detected observations are imputed with a zero value. Values below the LLOQ are imputed as 0.5 × LLOQ = 5 IU/mL for cobas HBV, cobas HBV+RT and Aptima HBV, and imputed as 0.5 × LLOQ = 14.5 IU/mL for the cobas HPS assay. Only complete data within each week across tests are included.
Adjusted for multiplicity using the Bonferroni method.
Similar results were documented when comparing the cobas HBV and the cobas HBV+RT tests in the Hannover cohort. In the overall analyses, differences were evident from week 12 until week 192 (end of follow‐up) (Supporting Fig. S4). Longitudinal data for individual patients are presented in Supporting Fig. S5.
Difference in HBV‐RNA Levels Between Samples With and Without Overestimation of Circulating HBV DNA by TMA‐Based Assays
To further evaluate whether the observed differences could be attributed to the amplification of HBV RNA, we accessed a third independent patient population (Liverpool cohort). In this particular cohort, HBV RNA was amplified by directly applying a specific PCR‐based protocol. The cross‐sectional Liverpool cohort consisted of 101 patients, of whom 41 (40.6%) had detectable HBV RNA at a median level of 5.6 log10 copies/mL (interquartile range (IQR) 2.9‐6.8 log10 copies/mL). Patients with detectable HBV RNA were more frequently females, of Asian ethnicity, hepatitis B e antigen (HBeAg)–positive, and had higher HBV‐DNA levels by Aptima HBV than patients without HBV RNA (Table 5). Among the 101 patients, there were 53 on antiviral treatment and, at the time of sampling, had received NA therapy for a median of 2.2 years (IQR 1.1‐4.5). Of the treated patients, 23 (43.4%) had detectable HBV RNA at a median level of 5.6 log10 copies/mL (IQR 3.4‐6.3 log10 copies/mL), and treatment duration was slightly shorter in patients with detectable HBV RNA relative to those without (Table 5). Among treated patients, median HBV‐DNA levels were 2.4 log10 IU/mL (ranging from <1 to 9.4 log10 IU/mL) in patients with HBV RNA versus less than 1 log10 IU/mL (ranging from <1 to 3.5 log10 IU/mL) in those without HBV RNA (P = 0.0006). A total of 26 samples from patients with both detectable HBV RNA and quantifiable HBV DNA underwent repeat HBV‐DNA testing using real‐time PCR (Fig. 6). Overall, HBV‐DNA levels were higher with Aptima HBV than Xpert HBV, and the greatest difference was seen in patients on antiviral therapy (up to 1.5 log10 IU/mL). Details of 10 treated patients who showed both detectable HBV RNA and quantifiable HBV DNA are given in Table 6. After a median of 1.7 years of antiviral therapy, median HBV‐RNA levels were 5.9 log10 copies/mL, and median HBV‐DNA levels by Aptima HBV were 3.5 log10 IU/mL. HBV‐DNA levels were a median of 1.0 log10 IU/mL higher by Aptima HBV relative to Xpert HBV, with similar results obtained with cobas HBV.
TABLE 5.
Characteristics of Patients Who Underwent HBV‐RNA Quantification (Liverpool Cohort)
| Characteristics | Total | HBV RNA Detected | P Value | ||
|---|---|---|---|---|---|
| Yes (n = 41) | No (n = 60) | ||||
| Age, median years (IQR) | 36 (29, 43) | 37 (30, 42) | 36 (29, 44) | 0.9948 | |
| Female, n (%) | 40 (39.6) | 22 (55.0) | 18 (45.0) | 0.0228* | |
| Male, n (%) | 61 (60.4) | 19 (31.1) | 42 (68.9) | — | |
| Ethnicity, n (%) | |||||
| Asian | 53 (52.5) | 29 (54.7) | 24 (45.3) | 0.0042 † | |
| Black African | 32 (31.7) | 6 (18.8) | 26 (81.2) | — | |
| White | 16 (15.8) | 6 (37.5) | 10 (62.5) | — | |
| HBeAg, n (%) | |||||
| Negative | 76 (75.2) | 22 (28.9) | 54 (71.1) | <0.0001 ‡ | |
| Positive | 25 (24.8) | 19 (76.0) | 6 (24.0) | — | |
| On antiviral therapy, n (%) | 53 (52.5) | 23 (43.4) | 30 (56.6) | 0.1713 § | |
| Duration of therapy, median years (IQR) || | 2.2 (1.1, 4.5) | 1.3 (0.8, 3.5) | 3.9 (1.6, 6.8) | 0.0494 | |
| HBV DNA (Aptima HBV), median log10 IU/mL | 2.4 (<1, 3.7) | 3.6 (2.4‐4.6) | <1 (<1, 2.7) | <0.0001 | |
Female versus male.
Asian versus non‐Asian.
HBeAg‐positive versus HBeAg‐negative.
On antiviral therapy versus untreated.
Among treated patients.
FIG. 6.
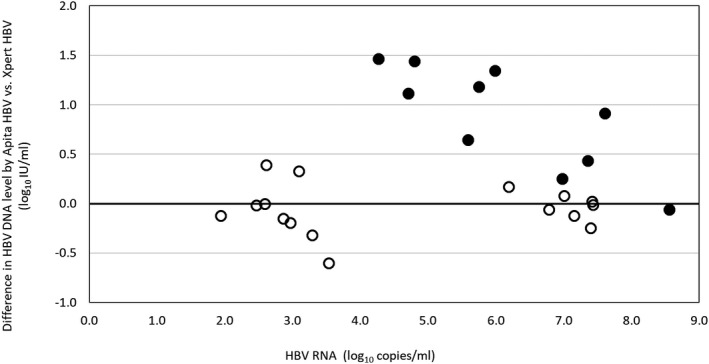
Difference in HBV‐DNA levels quantified by TMA (Aptima HBV) and real‐time PCR (Xpert HBV). Each dot indicates a single patient (n = 26). The solid dots indicate patients who were receiving antiviral therapy at the time of sampling (n = 10), whose characteristics are detailed in Table 6.
TABLE 6.
Characteristics of Patients on Antiviral Therapy and Both Detectable HBV RNA and Quantifiable HBV DNA by Aptima HBV, Ranked by Duration of Treatment
| Subject | Country of Origin | Age | HBeAg | NA | Duration of Therapy (Years) | HBV RNA log10 Copies/mL | HBV DNA (log10 IU/mL) | ||
|---|---|---|---|---|---|---|---|---|---|
| Aptima | Xpert | Cobas | |||||||
| M1 | Vietnam | 24 | + | TDF | 0.1 | 4.3 | 5.0 | 3.5 | 3.5 |
| M2 | China | 56 | + | TDF | 0.2 | 4.7 | 3.9 | 2.8 | 2.7 |
| M3 | Zambia | 26 | + | TDF | 0.3 | 8.6 | 9.4 | 9.5 | 9.6 |
| F1 | United Kingdom | 22 | + | ETV | 0.9 | 7.6 | 4.3 | 3.4 | 3.5 |
| M4 | United Kingdom | 24 | + | TDF | 1.1 | 7.4 | 3.4 | 3.0 | Not done |
| F2 | China | 29 | + | ETV | 2.3 | 7.0 | 3.5 | 3.2 | 3.3 |
| M5 | China | 37 | + | TDF | 2.8 | 6.0 | 2.3 | <LLOQ | <LLOQ |
| M6 | China | 38 | + | TDF | 3.1 | 5.6 | 2.6 | 2.0 | 2.1 |
| F3 | China | 35 | + | TDF | 4.1 | 5.8 | 1.9 | <LLOQ | <LLOQ |
| F4 | China | 33 | + | TDF | 4.1 | 4.8 | 2.4 | <LLOQ | <LLOQ |
Abbreviations: ETV, entecavir; F, female; M, male; TDF, tenofovir disoproxil fumarate.
A review of medical records indicated that in 3 of these patients, after continuous NA therapy for between 2.8 and 4.1 years, persistent quantification of HBV DNA by Aptima HBV (the local routine test) led to a diagnosis of suboptimal treatment responses, increased frequency of follow‐up, failed attempts at drug‐resistance testing by HBV‐DNA sequencing, unsuccessful treatment intensification to dual tenofovir/entecavir therapy, and distress for the patients when their adherence to treatment was repeatedly questioned. These three cases were resolved once testing by real‐time PCR demonstrated effective HBV‐DNA suppression; details of one of the cases, with results of retrospective HBV‐DNA testing by cobas HBV and cobas HBV+RT, are shown in Supporting Fig. S6.
Discussion
Accurate quantification of HBV‐DNA levels guides the clinical management of chronic HBV infection.( 1 , 2 ) We investigated HBV‐DNA quantification in three independent cohorts of patients with the specific aim of assessing how inclusion of RT capability in a viral load assay influenced the results. The key finding was that tests with an RT step (cobas HBV+RT and Aptima HBV) differed in their quantification of HBV DNA for patients undergoing treatment with NAs compared to tests without an RT step (cobas HBV, cobas HPS, and Xpert HBV). Although the cobas HBV+RT and Aptima HBV tests demonstrated good agreement with tests without the RT step at baseline, they appeared to consistently overestimate HBV‐DNA viral load in samples collected during NA therapy, specifically when HBV DNA is declining or even completely suppressed. Our data indicate that the difference is dependent on both the persistence of circulating HBV RNA during treatment and the decreasing HBV‐DNA to HBV‐RNA ratio. Subsequently, HBV‐DNA levels were fully or almost fully suppressed by real‐time PCR but quantifiable by Aptima HBV in some patients.
HBV tests operate using different designs. Tests such as cobas HBV or Xpert HBV use real‐time PCR amplification,( 18 , 20 ) whereas tests such as Aptima HBV use TMA.( 19 ) TMA is not a DNA‐specific detection tool, as it includes intrinsic RT capability that can amplify HBV RNA once the target is detected, a process that creates RNA and DNA fragments.( 11 ) Most previous studies comparing labeled HBV‐DNA tests on different platforms have found overall correlation between the viral loads.( 5 , 6 , 7 , 8 , 9 ) However, an overestimation with the Aptima HBV test later in NA treatment compared with the cobas HPS test has been previously reported, but the authors suggested that further analysis into the samples would be needed to understand the effect.( 6 ) Our data indicate that presence of circulating HBV RNA in a subset of NA‐treated patients is likely to be responsible for these reported discrepancies. It should be noted that the cobas HBV assay uses a DNA polymerase also capable of RT. Although this may theoretically also allow amplification of RNA in the standard setup of the cobas assay, estimated levels of HBV DNA were far higher by cobas HBV+RT and Aptima HBV. However, HBV‐DNA levels measured by cobas HBV were not different from those measured by Xpert HBV or by cobas HPS, which was used widely to determine the currently recommended HBV‐DNA thresholds for the initiation and monitoring of NA therapy. Thus, we conclude that a theoretically low‐level quantification of HBV RNA by cobas HBV would not be of clinical significance.
In our longitudinal evaluations, the difference in HBV‐DNA levels measured by assays with and without RT capability became clearly evident after the start of NA therapy, persisting in consecutive samples collected over long‐term follow‐up. In comparison with the marked effect of NA therapy on levels of HBV DNA, its influence on HBV RNA is limited. Differences in viral kinetics between HBV DNA and HBV RNA have been reported previously, with serum HBV RNA being reported as pregenomic RNA in virus‐like particles.( 17 , 22 ) At baseline, HBV‐RNA levels have been described to be 1.2 to 2.2 log10 IU/mL lower than HBV DNA.( 17 ) This may explain why all tests evaluated quantified HBV‐DNA levels comparably at baseline. However, NA treatment inhibits the viral polymerase preventing the formation of HBV DNA from the pregenomic HBV‐RNA template already encapsidated, which can be enveloped and then secreted in plasma as virus‐like particles that are resistant to plasma RNAse.( 17 , 23 ) Consequently, the balance becomes shifted toward RNA‐containing particles, which become predominant as DNA levels decline over time. As a result, relatively higher HBV‐RNA levels are likely leading to the enhanced difference between HBV tests with and without an RT step during NA therapy, as documented in our study. This is clinically important, as medical decisions during NA therapy are based on HBV‐DNA levels.( 1 , 2 , 4 ) Changes in viral load of 1 log10 IU/mL are used to define different levels of response or virologic breakthrough, depending on the treatment guidelines. Therefore, overestimations greater than 1 log10 IU/mL may lead to the misclassification of a patient and have a meaningful impact on their treatment. Undetectable, or at least not quantifiable, HBV DNA is considered as the definition of complete response to NA treatment, and the detection of HBV‐RNA particles influencing the reported result can lead to an HBV‐DNA‐suppressed patient being classified as failing therapy, as it was the case in at least 3 patients of the Liverpool cohort.
Circulating HBV RNA has recently been suggested as a potential surrogate of transcriptionally active hepatic cccDNA.( 15 , 24 , 25 ) Although HBV‐DNA suppression through treatment with NAs is the main endpoint, undetectable HBV DNA does not indicate suppressed cccDNA activity.( 17 ) Although hepatitis B surface antigen (HBsAg) levels correlate better with cccDNA levels in this population, some of the HBsAgs present in plasma could originate from integrated HBV DNA( 24 , 25 ); this makes HBsAg an unreliable surrogate for cccDNA. A more reliable surrogate could be HBV RNA. The difference in HBV RNA decline relative to HBV DNA suggests that cccDNA activity persists and is of interest for the development of new therapies that target clinical cure in chronic hepatitis B. Monitoring of HBV RNA could be useful in the assessment of treatment response to therapies, such as core protein allosteric modulators, with the potential to predict sustained response that would be classified as clinical cure when monitoring response from therapies targeting the cccDNA or other steps in the viral life cycle.( 17 ) To achieve this, an ability to distinguish between DNA and RNA would be critical, and most recently, undetectable HBV RNA has been suggested as an indicator of safe cessation of currently available NA therapy.( 26 ) The evaluation of tests with an RT step, as well as the direct detection in a subset of patients (Liverpool cohort), afforded us the opportunity to investigate HBV‐RNA kinetics during NA therapy in more detail. It is clear from our study that HBV RNA persists in many patients and is likely to be detectable for several months during treatment. Data from the Hannover cohort support the persistence of HBV RNA in HBV‐DNA‐suppressed patients being treated with the most potent regimens (entecavir or tenofovir disoproxil fumarate) for up to 196 weeks. This is in line with recently published data reporting detectable HBV RNA in 30% and 14% of patients after 3 and 5 years of NA therapy, respectively.( 26 ) However, longitudinal data from our patients indicate two patterns of HBV‐RNA kinetics: one in which HBV RNA persists despite treatment with NAs, and another in which HBV RNA declines with HBV DNA. This observation indicates the need for further studies.
Our study has several limitations. In the current study, the results generated with the cobas 6800 and Panther platforms were compared with historical results that were used for medical decision making, and therefore considered the benchmark; this was due to there not being enough volume to retest samples in parallel with the reference method used in these original studies. Although samples across the HBV genotypes were assessed, it was beyond the scope of this analysis to determine whether any of the effects were genotype‐specific. In addition, the conduct of the exploratory analysis with the Hannover cohort samples predates the availability of the Aptima HBV test; as such, analysis of these samples with this test was not possible.
Comparison of HBV‐DNA tests in this study suggests that HBV‐DNA levels are consistently overestimated, on average, with those tests that incorporate an RT step throughout the first 96 weeks of therapy, as HBV RNA and HBV DNA can be amplified. Of note, this affects widely used tests that are based on TMA. The overestimation of viral load due to the interference of HBV RNA may lead to the misclassification of treatment responses with subsequent unnecessary clinical interventions.
Supporting information
Supplementary Material
Acknowledgment
The authors thank Julia Heagerty, Ph.D., for the medical writing; she is associated with Obsidian Healthcare Group, Westerham, United Kingdom.
See Editorial on Page 949
This work was funded partly by Roche Diagnostics, a Ph.D. fellowship awarded to H.A., and an award from Roche Pharma Research and Early Development.
Potential conflict of interest: Dr. Canchola is employed by Roche. Dr. Cornberg advises and received grants from Roche. He advises and is on the speakers’ bureau for Gilead, MSD, AbbVie, and Janssen. He advises Biogen. He is on the speakers’ bureau for Falk. He received grants from Roche. Dr. Delgado received grants from Roche. Dr. Frontzek consults for and is on the speakers’ bureau for Roche. Dr. Garcia‐Alvarez received grants from Gilead, Roche, and Hologic. Dr. Geretti is employed by Roche. Dr. Marins owns stock in and is employed by Roche. Dr. Maasoumy consults for Abbott. He is on the speakers’ bureau for Roche. He received grants from Fujirebio. Dr. Simon owns stock in and is employed by Roche. Dr. Wedemeyer consults for, advises, and received grants from Roche. He consults for and received grants from Abbott. He consults, advises, and is on the speakers’ bureau for Siemens. Dr. Maasoumy received grants from Abbott, Roche and Fujirebio.
References
Author names in bold designate shared co‐first authorship.
- 1. European Association for the Study of the Liver . EASL 2017 Clinical Practice Guidelines on the management of hepatitis B virus infection. J Hepatol 2017;67:370‐398. [DOI] [PubMed] [Google Scholar]
- 2. Terrault NA, Lok ASF, McMahon BJ, Chang KM, Hwang JP, Jonas MM, et al. Update on prevention, diagnosis, and treatment of chronic hepatitis B: AASLD 2018 Hepatitis B Guidance. Hepatology 2016;63:261‐283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kim JH, Sinn DH, Kang W, Gwak GY, Paik YH, Choi MS, et al. Low‐level viremia and the increased risk of hepatocellular carcinoma in patients receiving entecavir treatment. Hepatology 2017;66:335‐343. [DOI] [PubMed] [Google Scholar]
- 4. Sarin SK, Kumar M, Lau GK, Abbas Z, Chan HL, Chen CJ, et al. Asian‐Pacific clinical practice guidelines on the management of hepatitis B: a 2015 update. Hepatol Int 2016;10:1‐98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Braun P, Delgado R, Drago M, Fanti D, Fleury H, Izopet J, et al. A European multicentre study on the comparison of HBV viral loads between VERIS HBV assay and Roche COBAS® TAQMAN® HBV test, Abbott RealTime HBV assay, Siemens VERSANT HBV assay, and Qiagen artus HBV RG kit. J Clin Virol 2017;95:76‐83. [DOI] [PubMed] [Google Scholar]
- 6. Schalasta G, Börner A, Speicher A, Enders M. Evaluation of the Aptima HBV Quant assay vs. the COBAS TaqMan HBV test using the High Pure System for the quantitation of HBV DNA in plasma and serum samples. Clin Chem Lab Med 2018;56:634‐641. [DOI] [PubMed] [Google Scholar]
- 7. Wirden M, Larrouy L, Mahjoub N, Todesco E, Damond F, Delagreverie H, et al. Multicenter comparison of the new Cobas 6800 system with Cobas Ampliprep/Cobas TaqMan and Abbott RealTime for the quantification of HIV, HBV and HCV viral load. J Clin Virol 2017;96:49‐53. [DOI] [PubMed] [Google Scholar]
- 8. Maasoumy B, Bremer B, Lehmann P, Marins EG, Michel‐Treil V, Simon CO, et al. Commutability and concordance of four hepatitis B virus DNA assays in an international multicenter study. Therap Adv Gastroenterol 2017;10:609‐618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Abravanel F, Lhomme S, Trémeaux P, Migueres M, Harter A, Haslé C, et al. Performance of the Xpert HBV Viral Load assay versus the Aptima Quant assay for quantifying hepatitis B virus DNA. Diagn Microbiol Infect Dis 2019;96:114946. [DOI] [PubMed] [Google Scholar]
- 10. Paxinos E, Marins E, Duncan D, Marlowe EM, Canchola JA. Analytical performance and workflow comparisons of two high throughput viral load platforms Poster presented at the 34th American Society for Microbiology Clinical Virology Symposium, West Palm Beach, FL, 2018. [Google Scholar]
- 11. Weber J, Sahoo MK, Taylor N, Uy E, Shi RZ, Pinsky BA. Comparison of transcription‐mediated amplification and real‐time PCR assays for hepatitis B virus DNA quantitation in serum. J Appl Lab Med 2019;4:383‐390. [DOI] [PubMed] [Google Scholar]
- 12. Heim A. Evaluation of the Procleix Ultrio Elite assay and the Panther system for individual NAT screening of blood, hematopoietic stem cell, tissue and organ donors. Transfus Med Hemother 2016;43:177‐182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hopkins M, Hau S, Tiernan C, Papadimitropoulos A, Chawla A, Beloukas A, et al. Comparative performance of the new Aptima HIV‐1 Quant Dx assay with three commercial PCR‐based HIV‐1 RNA quantitation assays. J Clin Virol 2015;69:56‐62. [DOI] [PubMed] [Google Scholar]
- 14. Chevaliez S, Dubernet F, Dauvillier C, Hézode C, Pawlotsky JM. The new Aptima HCV Quant Dx real‐time TMA assay accurately quantifies hepatitis C virus genotype 1‐6 RNA. J Clin Virol 2017;91:5‐11. [DOI] [PubMed] [Google Scholar]
- 15. Liu S, Zhou B, Valdes JD, Sun J, Guo H. Serum hepatitis B virus RNA: a new potential biomarker for chronic hepatitis B virus infection. Hepatology 2019;69:1816‐1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Entecavir plus adefovir combination therapy versus entecavir monotherapy vs therapy with adefovir plus lamivudine for chronic hepatitis B infected subjects with lamivudine‐resistant virus (DEFINE). https://clinicaltrials.gov/ct2/show/results/NCT00410202?term=Define+Study%2C+entecavir&cond=Hepatitis+B%2C+Chronic&rank=1. Accessed December 2019. [Google Scholar]
- 17. van Bömmel F, Bartens A, Mysickova A, Hofmann J, Krüger DH, Berg T, et al. Serum hepatitis B virus RNA levels as an early predictor of hepatitis B envelope antigen seroconversion during treatment with polymerase inhibitors. Hepatology 2015;61:66‐76. [DOI] [PubMed] [Google Scholar]
- 18. Cobas® AmpliPrep/cobas® TaqMan® HBV Test, v2.0 . https://diagnostics.roche.com/global/en/products/params/cobas‐ampliprep‐cobas‐taqman‐hbv‐test‐v2‐0.html. Accessed December 2019.
- 19. Aptima® HBV Quant Assay [package insert]. https://www.hologic.com/package‐inserts/diagnostic‐products/aptima‐hbv‐quant‐assay. Accessed December 2019.
- 20. Xpert® HBV Viral Load . https://www.cepheid.com/uk/cepheid‐solutions/clinical‐ivd‐tests/virology/xpert‐hbv‐viral‐load. Accessed December 2019.
- 21. Clinical and Laboratory Standards Institute (CLSI) . Measurement procedure comparison and bias estimation using patient samples. 3rd ed. CLSI guideline EP09c. Wayne, PA: Clinical and Laboratory Standards Institute; 2018. [Google Scholar]
- 22. Wang J, Shen T, Huang X, Kumar GR, Chen X, Zeng Z, et al. Serum hepatitis B virus RNA is encapsidated pregenome RNA that may be associated with persistence of viral infection and rebound. J Hepatol 2016;65:700‐710. [DOI] [PubMed] [Google Scholar]
- 23. Koczera P, Martin L, Marx G, Schuerholz T. The ribonuclease A superfamily in humans: canonical RNases as the buttress of innate immunity. Int J Mol Sci 2016;17:pii: E1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lok AS, Zoulim F, Dusheiko G, Ghany MG. Hepatitis B cure: from discovery to regulatory approval. J Hepatol 2017;67:847‐861. [DOI] [PubMed] [Google Scholar]
- 25. Coffin CS, Zhou K, Terrault NA. New and old biomarkers for diagnosis and management of chronic hepatitis B virus infection. Gastroenterology 2019;156:355‐368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Carey I, Gersch J, Wang B, Moigboi C, Kuhns M, Cloherty G, et al. Pre‐genomic HBV RNA and HBcrAg predict outcomes in HBeAg negative chronic hepatitis B patients suppressed on nucleos(t)ide analogue therapy. Hepatology 2019. Nov 7. 10.1002/hep.31026. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material


