Abstract
Replication of hepatitis B virus (HBV) originates from covalently closed circular DNA (cccDNA) and involves reverse transcription of pregenomic RNA (pgRNA), which is also called core RNA and encodes the capsid protein. The RNA coding for hepatitis B surface antigen (HBsAg) in the envelope of viral or subviral particles is produced from cccDNA or from HBV DNA integrated into the host genome. Because only cccDNA can generate the core and the 3′ redundancy regions of HBV RNA, we aimed to clarify to what extent such HBV integrations are expressed by quantifying the different HBV RNA species in liver tissue. Digital droplet polymerase chain reaction (ddPCR) was employed to quantify six HBV RNA targets in 76 liver biopsies from patients with chronic infection, comprising 14 who were hepatitis B e antigen (HBeAg) positive and 62 who were HBeAg negative. In patients who were HBeAg negative, HBV RNA from the S RNA region was >1.6 log10 units higher than in the core and 3′ redundancy regions (P < 0.0001), indicating that >90% of S RNA was integration derived. HBeAg‐negative samples showed 10 times lower levels of pgRNA (5′ core) compared with core RNA (3′ part of core; P < 0.0001), suggesting that a large proportion of core RNA might have a downstream shift of the transcription starting point. In multiple regression analysis, HBV DNA levels in serum were most strongly dependent on pgRNA. Conclusion: In patients who were HBeAg negative, integration‐derived S RNA seemed to predominate and a large proportion of the core RNA lacked the 5′ part. Because this part comprises the down‐regulator of transcription 1 sequences, which are necessary for virus production (plus strand translocation), the finding might help to explain the low level of HBV DNA in serum that frequently is observed in patients with chronic HBV infection who are HBeAg negative.
Abbreviations
- cccDNA
covalently closed circular DNA
- ddPCR
digital droplet polymerase chain reaction
- DNase
deoxyribonuclease
- HBeAg
hepatitis B e antigen
- HBsAg
hepatitis B viral surface antigen
- HBV
hepatitis B virus
- nt
nucleotide
- ORF
open reading frame
- PCR
polymerase chain reaction
- pgRNA
pregenomic RNA
- RT‐PCR
reverse‐transcription polymerase chain reaction
Chronic hepatitis B virus (HBV) infection is a major risk factor for hepatocellular carcinoma.( 1 ) HBV is a small DNA virus (≈3,200 nucleotides) that replicates through an RNA intermediate (pregenomic RNA [pgRNA]) that is reverse transcribed inside nascent virus capsids. An episomal HBV genome, called covalently closed circular DNA (cccDNA), resides in the nucleus of infected hepatocytes and is transcribed to pgRNA as well as to four other RNA species that have a common polyadenylated 3′ end.( 2 )
HBV DNA may integrate into the chromosomal DNA in hepatocytes.( 3 , 4 ) These integrations are believed to contribute to oncogenesis by, for example, changing the function of host genes or by inducing chromosomal instability.( 5 , 6 ) In addition, integrations have been proposed to encode the hepatitis B viral surface antigen (HBsAg), which is embedded in the virus envelope as well as in so‐called subviral particles; these particles are usually present in the blood at levels more than 10,000 times higher than viral particles.
By 1980, HBsAg synthesis from integrated DNA had been discovered in the hepatoma cell line PLC/PRF/5.( 3 ) It has since been proposed to maintain high serum levels of HBsAg, in particular in subjects with low‐grade viremia.( 7 ) This hypothesis is supported by the fact that HBsAg declines much less than HBV DNA when viral replication is suppressed during immune‐mediated clearance.( 7 , 8 ) Further support for this idea was obtained by an experimental study in chimpanzees that analyzed the HBV transcriptome in liver tissue and found an absence of RNA reads representing pgRNA compared with S RNA.( 9 ) Recent human data support this view.( 10 )
There are two full‐length transcripts of HBV. The precore RNA starts at nucleotide (nt) 1795 and contains the open reading frame (ORF) for hepatitis B e antigen (HBeAg), a nonstructural protein that is secreted into the blood. The pgRNA (or core RNA) that starts at nt 1820, contains the ORF for the core antigen and is also used for reverse transcription to minus strand DNA as well as for translation to the viral polymerase. It has been proposed that a reduced ratio of the precore to core RNA might explain the reduced expression of HBeAg.( 11 ) There are also three shorter HBV RNA species: the 2.4‐kb preS1 RNA, the 2.1‐kb preS2 RNA, and the 0.8‐kb X RNA. When transcribed from cccDNA, these HBV RNA species are typically polyadenylated near nt 1930, as guided by a TATAAA signal at 1916‐1921.( 12 , 13 )
By contrast, because integrated HBV DNA is typically fused to chromosomal DNA at or slightly upstream of nt 1830, transcripts derived from integrations carry a human part downstream of this point along with variable polyadenylation sites, guided by the human sequence. Thus, by analyzing if a 3′ part of HBV RNA downstream of nt 1830 (the “redundancy”) is present, it is possible to distinguish transcripts from cccDNA from those derived from integrations. Notably, however, transcripts from both cccDNA and integrations may be shorter if either of two alternative polyadenylation sequences (CATAAA) located at nt 1653‐1658 or at 1788‐1793 are employed.( 12 , 13 )
Quantification of different HBV RNA species may be done by northern blot, but this technique is insufficiently sensitive for analysis of small tissue samples. Alternatively, quantitative polymerase chain reaction (PCR) assays that target specific parts of the genome may be used. However, a limitation of this strategy is related to overlapping HBV transcripts; for example, an assay that targets the X region unavoidably measures all the different RNA species. Another limitation is that amplification efficiencies may bias the comparison if the RNA levels are assessed by assays, such as real‐time PCR, that rely on threshold cycle (Ct) values for quantification. By using an absolute quantification technique, such as digital droplet PCR (ddPCR), this problem can be overcome.( 14 )
In the present study, we used ddPCR to quantify HBV RNA in liver biopsies from 76 patients with defined stages of chronic HBV infection. The aims were (i) to determine the profile of HBV RNA species in liver tissue and (ii) to compare levels of HBV transcripts that are only derived from cccDNA with those that are likely derived also from integrations in order to clarify if integrated HBV DNA is expressed.
Patients and Methods
Patients, Samples, and HBV Testing by Diagnostic Assays
Frozen biopsies from 76 patients that were included in a cross‐sectional study of 160 patients( 15 ) were available for analysis of intrahepatic HBV RNA by ddPCR. The patients represent HBeAg‐positive (n = 14) and HBeAg‐negative (n = 62) stages of infection, genotypes A‐D, and showed mild to severe liver damage (Table 1). None of the patients were coinfected with human immunodeficiency virus, hepatitis C virus, or hepatitis D virus. The serum samples used for quantification of HBV DNA and HBsAg were collected at the time of biopsy. Quantification of HBsAg and detection of HBeAg in serum were performed by Architect assays (Abbott Laboratories, Chicago, IL). Quantification of HBV DNA in serum was performed using the Cobas Amplicor HBV Monitor (Roche Diagnostics, Branchburg, NJ).
Table 1.
Characteristics of 76 Patients With Chronic HBV Infection
| Characteristic | HBeAg+ | HBeAg− |
|---|---|---|
| (n = 14) | (n = 62) | |
| Age, mean years (range) | 33 (16‐60) | 35 (18‐50) |
| Sex (female/male) | 5/9 | 24/38 |
| HBV DNA, median log10 copies/mL (IQR) | 8.51 (7.61‐9.52) | 4.05 (3.34‐4.96) |
| HBsAg, median log10 IU/mL (IQR) | 4.54 (3.93‐4.88) | 3.49 (2.96‐4.06) |
| Genotype (A/B/C/D) | 3/2/2/7 | 14/6/3/39 |
| Inflammation score* (0‐2/3‐5/7‐10) | 4/6/4 | 43/16/3 |
| Fibrosis score* (0‐1/3/4) | 10/2/2 | 52/9/1 |
| ALT/upper limit of normal, median (IQR) | 1.55 (0.81‐2.79) | 0.72 (0.55‐1.11) |
Abbreviations: ALT, alanine aminotransferase; IQR, interquartile range.
Histological index by Knodell scoring.( 23 )
Nucleic Acid Extraction
Approximately 5 mg of liver biopsies stored at −70°C was homogenized in 200 μL of Magnapure LC lysis buffer on a Magnalyser instrument (Roche Diagnostics). Nucleic acids were extracted from 80 μL of the lysate using the Magnapure LC DNA Isolation Kit II (Roche Diagnostics) on the Magnapure (Roche Diagnostics) robot according to the instructions from the manufacturer. A portion of the nucleic acid extraction was treated with the TURBO DNA‐free kit (Thermo Fisher Scientific) to remove contaminating DNA before RNA quantification. The Deoxyribonuclease (DNase) treatment was carried out according to the manufacturer's instructions. In our hands, it did not digest RNA (Ct values in real‐time PCR for hepatitis D virus RNA without DNase and after DNase treatment being 25.75 and 25.81, respectively).
PCR Target Regions
Six PCR targets were used to quantify different HBV RNA species by ddPCR (Table 2; Fig. 1). In some analyses, these RNA levels were compared with cccDNA levels quantified by real‐time PCR, as described.( 16 ) All primers and probes were located in genomic segments with no or very few variable positions compared with genotypes A‐D. Of note, the reverse primer in the core assay (nt 2454R) had mismatches compared with genotypes E‐H, but no such samples were included in this study.
Table 2.
Primers and Probes Used in Digital PCR
| Target* | Oligo | Sequence | Position in HBV Genome |
|---|---|---|---|
| 5′ precore RNA | F | GCACCAGCACCATGCAACTT | 1803‐1822 |
| R | TCAGAAGGCAAAAACGAGAGTAACT | 1941‐1965 | |
| Probe | ACTGTTCAAGCCTCCAAGCTGTGCCTT | 1859‐1885 | |
| pgRNA (5′ core) | F | TCACCTCTGCCTAATCATCTCTTG | 1825‐1848 |
| R | TCAGAAGGCAAAAACGAGAGTAACT | 1941‐1965 | |
| Probe | ACTGTTCAAGCCTCCAAGCTGTGCCTT | 1859‐1885 | |
| Core | F | GGTCCCCTAGAAGAAGAACTCCCT | 2361‐2384 |
| R | CATTGAGATTCCCGAGATTGAGAT | 2425‐2448 | |
| Probe | CGCCGCGTCGCA | 2409‐2420 | |
| S | F | TCCTCCAAYTTGTCCTGGTYATC | 349‐371 |
| R | AGATGAGGCATAGCAGCAGGAT | 410‐431 | |
| Probe | CCGCAGACACATCC | 375‐388 | |
| X | F | CGTCTGTGCCTTCTCATCTG | 1550‐1569 |
| R | GCGTTCACGGTGGTCTCCA | 1609‐1627 | |
| Probe | CACTTCGCTTCACCTC | 1582‐1597 | |
| 3′ precore RNA | F | GGAGGCTGTAGGCATAAATTGGTC | 1776‐1799 |
| R | TTCTTTATAAGGGTCAATGTCCATG | 1900‐1924 | |
| Probe | CCTCCAAGCTGTGCC | 1869‐1883 | |
| cccDNA | F | CCGTGTGCACTTCGCTTCA | 1575‐1593 |
| R | GCACAGCTTGGAGGCTTGA | 1864‐1882 | |
| Probe | CATGGAGACCACCGTGAACGCCC | 1607‐1629 | |
| Beta‐globin DNA | F | GCTCATGGCAAGAAAGTGCTC | |
| R | GCAAAGGTGCCCTTGAGGT | ||
| Probe | AGTGATGGCCTGGCTCACCTGGAC |
Minor groove‐binding probes were used in core RNA, S RNA, X RNA, and 3′ precore RNA assays.
Abbreviations: F, forward; R, reverse.
Fig. 1.
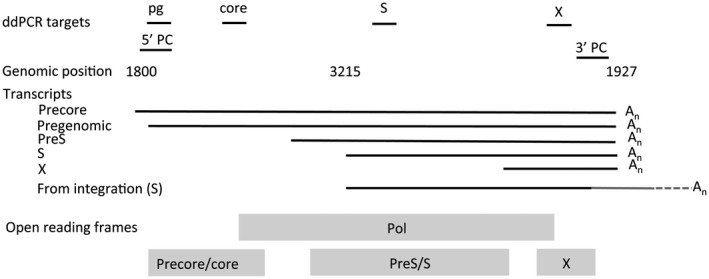
The six target regions of the ddPCR assays for quantification of HBV RNA. The five transcripts derived from cccDNA are shown with a common polyadenylation at nt 1927, although truncated forms may occur. The gray 3′ part of the S transcript from integrations indicates the part encoded by chromosomal DNA, which has a polyadenylation further downstream. Abbreviations: An, polyadenylation; Pol, polyadenylation.
ddPCR
Digital PCR was used to quantify the absolute copy numbers of HBV RNA in liver biopsies. The analysis was run as a one‐step reverse‐transcription (RT)‐PCR after DNase treatment to degrade HBV DNA. The DNase‐treated samples were also run in parallel with a DNA master mix (without reverse transcriptase) to confirm that DNA degradation had been effective, i.e., that the RT‐PCR did not amplify undigested HBV DNA. Analyses were carried out in duplicate reactions, each in a 20‐μL RT‐PCR reaction mix containing 1× One‐Step RT‐PCR mix (Bio‐Rad), primers (750 nM), probe (500 nM), and 5 μL extracted RNA. After rigorous mixing (30 times with a 10‐μL pipette), droplets were formed in the AutoDG Automated Droplet Generator (Bio‐Rad) with oil for probes (Bio‐Rad).
Amplification was performed on a Veriti Thermocycler (Applied Biosystems) with an initial reverse transcription step at 48°C for 60 minutes, followed by thermal profiling with 40 cycles of 95°C for 30 seconds and 58°C for 1 minute, 1 cycle of 98°C for 10 minutes to deactivate the enzymes, and ending at 4°C. After overnight incubation at 4°C, the plate was analyzed in the QX200 Droplet Reader (Bio‐Rad), and results were analyzed with QuantaSoft (Bio‐Rad) software. A negative template control (distilled water) and a positive control (plasmid carrying the whole HBV genome [genotype A], linearized at a BamH1 site at nt 31) were included in each run. Reactions with less than five positive droplets were considered negative and were not included in the analysis.
Ethics
All participants gave written informed consent, and the principles of the Declaration of Helsinki were followed. The Regional Ethical Review Board in Gothenburg approved this study.
Statistics
HBV RNA levels between HBeAg‐positive and HBeAg‐negative groups were compared by the Mann‐Whitney U test. Levels obtained analyzing the same sample by ddPCR amplifying different targets were compared by paired t test. Correlations between HBV markers were analyzed by Pearson's correlation analysis. Multiple regression analysis was performed to evaluate how HBV DNA was influenced by pgRNA and core RNA. Statistical analyses were performed using JMP software (SAS Institute Inc.).
Results
Patient Characteristics
The characteristics along with markers of HBV replication and liver damage for the 76 patients are shown in Table 1. The mean age of all patients was 34 years, and 40% were women. The mean HBV DNA level in serum was 8.51 log10 copies/mL in the 14 patients who were HBeAg positive and 4.05 log10 copies/mL in the 62 patients who were HBeAg negative. The clinical stage( 1 ) was HBeAg‐positive infection (previously called immune tolerance) in 9 patients, HBeAg‐positive hepatitis in 5, HBeAg‐negative hepatitis in 21, and (low‐active) HBeAg‐negative infection in 41.
Levels of HBV RNA Species
Levels of ddPCR targets, not the levels of the RNA species, are shown in Fig. 2. Because all transcripts share a common 3′ end, the ddPCR that targets X measures the level of all HBV RNA species. Therefore, to obtain the level of X RNA, the levels (as linear values) obtained by ddPCR in the S region should be subtracted from levels by X ddPCR; to obtain S RNA levels, the observed levels by core ddPCR should be subtracted from S ddPCR. In most figures and analyses, the original results were used; however, in some analyses, such subtracted values were used (as specified).
Fig. 2.
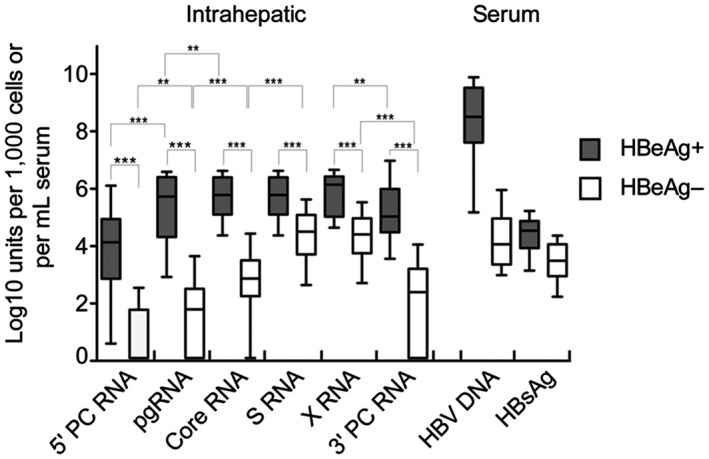
Intrahepatic HBV RNA profiles in patients positive or negative for HBeAg. The box plots shows median levels and interquartile ranges that were observed by ddPCR assays targeting different segments of the genome. The levels of HBsAg and HBV DNA in serum are shown for comparison. Whiskers show tenth and ninetieth percentile; ***P < 0.0001; **P < 0.001. Abbreviation: PC, precore.
The levels of all investigated HBV RNA targets in liver tissue as well as levels of HBV DNA and HBsAg in serum were higher in patients who were HBeAg positive than in patients who were HBeAg negative (P < 0.0001, for all comparisons) (Fig. 2). In the HBeAg‐positive group, pgRNA (targeting 5′ core), core RNA, S RNA, and X RNA levels showed similar levels, whereas 5′ precore and 3′ precore were 1‐2 log10 units lower (P values shown in Fig. 2). In patients who were HBeAg negative, lower levels were seen for pgRNA, core, and 3′ precore (the redundancy) regions than for core, S, and X regions. The largest difference (>4 log10 units) between HBeAg‐positive and HBeAg‐negative groups was observed for pgRNA in liver tissue and HBV DNA in serum. The smallest difference (1 log10 unit) was observed for HBsAg in serum.
Comparisons of Intrahepatic RNA Species
Levels of precore RNA were approximately 2 log10 units lower than those of core RNA, and this relation was similar in HBeAg‐positive and HBeAg‐negative cases (Figs. 2 and 3A). By using a forward primer at nt 1803‐1822, the precore ddPCR assay measures only levels of precore RNA, which starts at nt 1793, and excludes pgRNA starting at nt 1818‐1820.
Fig. 3.
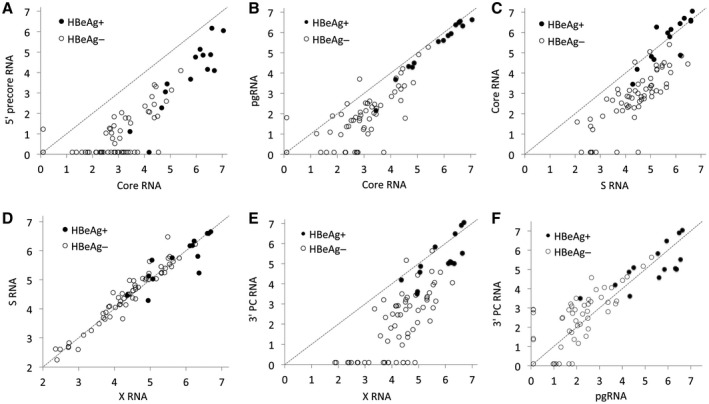
Correlations between levels of intrahepatic HBV RNA measured by ddPCR. (A) The 5′ precore RNA levels were ≈1.6 log10 units lower than core RNA. (B) pgRNA was almost 1 log10 unit lower than core RNA in patients who were HBeAg negative. (C) Core RNA levels were ≈1.6 log10 units lower than S RNA in patients who were HBeAg negative. (D) RNA levels measured in S and X regions were similar. (E) RNA levels measured in the 3′ precore (redundancy) region were much lower than RNA levels measured in the X region a short distance upstream, indicating that a large proportion of the RNA in patients who were HBeAg negative lacked the 3′ redundancy part (nt 1830‐1927), either because they were derived from integrated HBV DNA or because they were truncated by the use of an upstream polyadenylation signal. (F) Levels obtained by ddPCR targeting pgRNA and 3′ precore (redundancy) regions. The unit in all plots is log10 copies/1,000 cells. Undetected RNA is shown as 0.1 log10 units/1,000 cells. The dotted line shows y = x. Abbreviation: PC, precore.
pgRNA (nt 1825‐1966) levels correlated strongly (P < 0.0001) with but were significantly lower than core RNA (nt 2367‐2454) (Fig. 3B). This difference was statistically significant in patients who were HBeAg positive (median, 0.35 log10 units; P = 0.0005) and HBeAg negative (median, 0.95 log10 units; P < 0.0001) and with a significantly larger difference in patients who were HBeAg negative compared with those who were HBeAg positive (0.95 vs. 0.35 log10 units; P = 0.0009) (Fig. 2). Core RNA was detected in all 62 HBeAg‐negative samples, whereas pgRNA was detected in 55 (P = 0.0003). The lower levels of pgRNA than core RNA suggest that a large proportion of the core transcripts might have an atypical (later) starting point, in particular in patients who are HBeAg negative.
Levels of core RNA correlated strongly with S RNA and were similar in patients who were HBeAg positive (0.03 log10 units difference; P = 0.85) (Fig. 3C), but core RNA levels were 1.58 log10 units lower (median, P < 0.0001) than S RNA in patients who were HBeAg negative, as also shown in Fig. 2.
RNA levels obtained by ddPCR targeting X and S were similar (Fig. 3D), whereas RNA levels in the 3′ precore (redundancy) part, which is anticipated to be present in all transcripts from cccDNA (but not from integrated DNA), were much lower (Fig. 3E). This difference was small in patients who were HBeAg positive but large (median, 2.2 log10 units) in patients who were HBeAg negative, with a difference that increased with decreasing levels of RNA. Levels obtained by the 5′ core and the 3′ precore ddPCR showed a strong correlation (P < 0.0001; Fig. 3F).
cccDNA, Intrahepatic HBV RNA, and Serum Levels of HBV DNA and HBsAg
Correlations between cccDNA and HBV DNA or HBsAg in serum, as well as between cccDNA and pgRNA or S RNA in liver tissue are shown in Fig. 4. A dotted line indicates the levels that would be expected if the values on the y axis had been proportional to cccDNA levels with a 1:1 slope (starting at the highest values). By comparing observed values with this line, it is apparent that in patients who were HBeAg negative, both HBV DNA in serum and pgRNA levels in liver tissue were much lower than explained by cccDNA levels and conversely that both HBsAg in serum and S RNA levels in liver tissue were much higher than explained by cccDNA levels. Multiple regression analysis showed that the association with HBV DNA in serum was stronger for pgRNA (P < 0.0001) than for core RNA (P = 0.92).
Fig. 4.
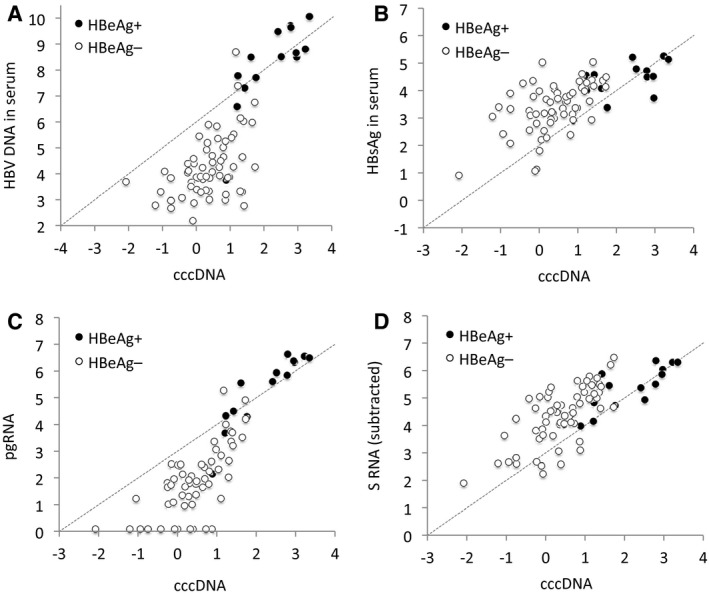
HBV DNA levels in serum and pgRNA and S RNA levels in liver tissue as a function of cccDNA in liver tissue. (A‐D) In patients who were HBeAg negative, the (A) HBV DNA levels in serum and (C) pgRNA levels in liver tissue were 1‐2 log10 units lower than expected from the cccDNA levels. (B) HBsAg levels in serum and (D) S RNA levels in liver tissue were 0.5‐1 log10 higher than expected from the cccDNA levels. The unit is log IU/mL serum or log10 copies/1,000 cells. Undetected RNA is shown as 0.1 log10 units/1,000 cells. S RNA levels in D were obtained by subtraction of observed core RNA from observed S RNA in patients who were HBeAg negative and by dividing the observed S RNA level by 2 in patients who were HBeAg positive. The dotted line shows the expected level if the y axis values had changed similarly (1:1) as x axis values (starting as the highest values).
Intrahepatic HBV Ratios and the HBV DNA/HBsAg Ratio in Serum
Further explorations of the associations between intrahepatic RNA and intrahepatic HBV DNA as well as serum markers are shown in Fig. 5. The cccDNA fraction of total intrahepatic HBV DNA correlated significantly with the ratios between pgRNA (which can only be derived from cccDNA) and X HBV RNA (which can be derived from both cccDNA and integrated HBV DNA) (Fig. 5A). Similarly, the ratio between pgRNA and X RNA correlated with the ratio between serum levels of HBV DNA (which is derived from pgRNA) and HBsAg (which can be derived from both cccDNA and integrations) (Fig. 5B).
Fig. 5.
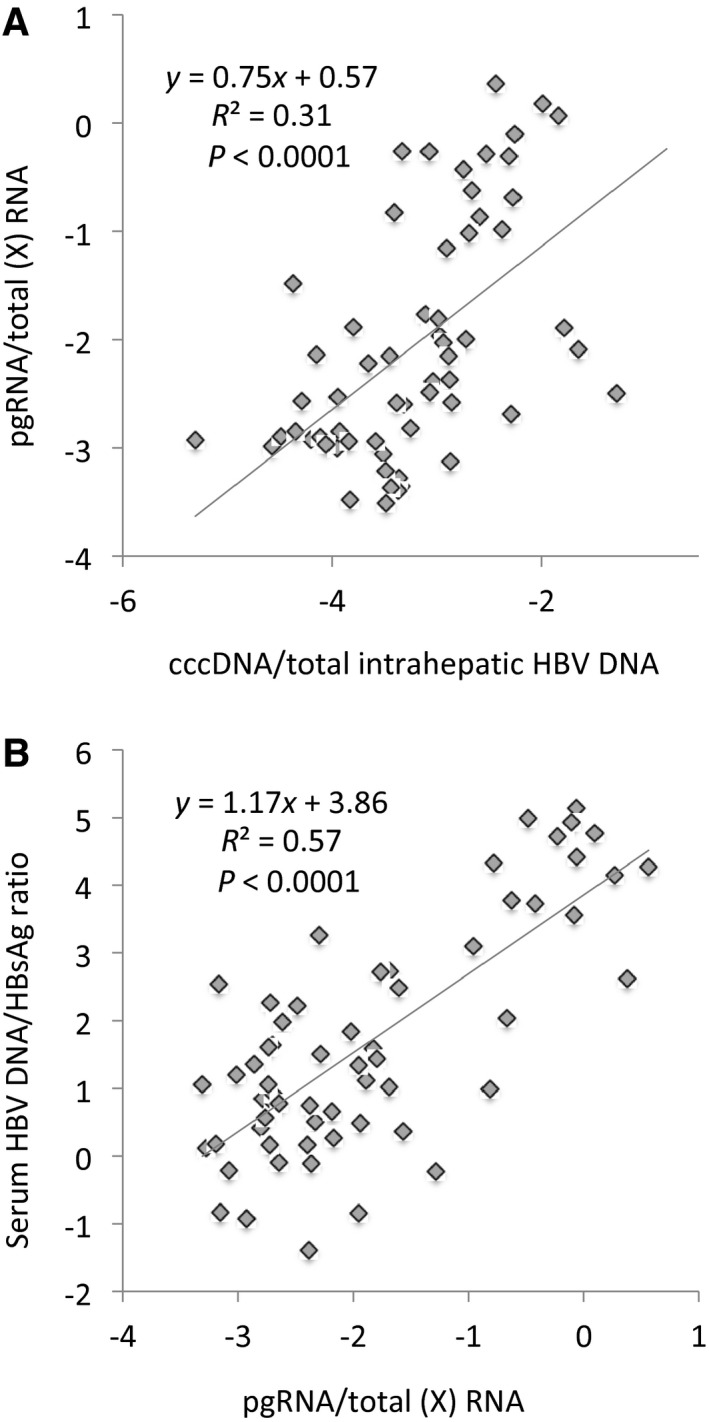
Correlations between liver and serum marker ratios. Correlations between and regression lines for (A) the cccDNA/total intrahepatic HBV DNA ratio and the pgRNA/total HBV RNA ratio and (B) the pgRNA/total HBV RNA ratio and the serum HBV DNA/serum HBsAg ratio.
Discussion
To our knowledge, this is the first study that quantifies HBV RNA in liver tissue by using ddPCR targeting several segments of the genome. This approach provides an RNA profile similar to that obtained by RNA sequencing (massive parallel sequencing) and allows analysis of correlations between the levels of different RNA species. The results suggest that most of the very large reduction in HBV DNA levels between the HBeAg‐positive and HBeAg‐negative stages of infection is a combined effect of eradication of infected cells (cccDNA) and a reduced or altered transcription of pgRNA from cccDNA. The smaller reduction in HBsAg seems to be explained by expression from integrated HBV DNA. The findings agree with transcriptome data from chimpanzees( 9 ) and humans( 10 ) but extend these findings and provide quantitative data of the levels of different RNA species.
On average, S RNA levels were 40 times higher (1.6 log10 units higher) than core RNA levels in patients who were HBeAg negative. This reflects that S RNA was enhanced whereas core RNA was reduced compared with cccDNA levels. We propose that the high levels of S RNA in patients who were HBeAg negative are due to expression from HBV DNA integrated in the human genome.( 7 ) Noteworthy, the proportion of S RNA that is expressed from integrated HBV DNA is much higher in patients who were HBeAg negative than in those who were HBeAg positive because the cccDNA levels are around 50 times higher in the latter patients compared to patients who were HBeAg negative. An alternative explanation to the high levels of S RNA might be that its expression from cccDNA is selectively enhanced. This seems less likely because to our knowledge no such mechanism has been described. Also, S RNA derived from cccDNA should carry a 3′ redundancy that would be detected by 3′ precore ddPCR. Hence, the RNA levels measured by the S or X should be similar to those measured by the 3′ precore assays if S RNA were derived from cccDNA. However, the levels were similar only in patients who were HBeAg positive, as shown in Fig. 3E, whereas the levels differed markedly (on average by 2.2 log10) in patients who were HBeAg negative. This observation suggests that the fraction of S RNA derived from integrated HBV DNA becomes greater as transcription from cccDNA declines. The finding that among the patients with the lowest total HBV RNA levels (and the lowest degree of replication) the levels of S RNA were more than 2 log10 units higher than 3′ precore RNA suggests that more than 99% of the S RNA in these cases might be derived from integrated DNA.
The relations between the different intrahepatic RNA species are also reflected in serum where the ratio between levels of HBV DNA and HBsAg strongly correlated with the ratios between RNA species that can only be derived from cccDNA (core RNA and 3′ precore RNA) and total intrahepatic HBV RNA (Fig. 5). These findings agree with the idea that a large proportion of HBsAg in patients who are HBeAg negative originates from integrated DNA. The contribution from integrated HBV DNA to HBsAg levels in serum should be significant only if the number of integrations per cell is greater than the number of cccDNA copies. Previous studies suggest that cccDNA is present in ≈1‐5 copies per cell and that the level is on average 100‐fold lower in the HBeAg‐negative stage.( 17 ) If integrations were present in 10% of the hepatocytes, they would be rarer than cccDNA in the HBeAg‐positive stage but more frequent than cccDNA in the HBeAg‐negative stage. We propose that such relative copy numbers explain why integrations contribute to HBsAg in serum only in the HBeAg‐negative stage.
The higher RNA levels obtained by the core ddPCR (nt 2361‐2448) compared with those detected using the assay that targets pgRNA (the 5′ part of the core, nt 1825‐1965) has to our knowledge not been reported. However, a study that used a 5′ target (nt 1826‐1974)( 18 ) to quantify pgRNA reported lower levels relative to intrahepatic HBV DNA, whereas a study that used a target in the mid core (nt 2267‐2440) reported the opposite, i.e., that core RNA (called pgRNA in that study) levels were higher than intrahepatic HBV DNA levels.( 17 ) These previous data along with our finding that the lower pgRNA levels were mainly observed in patients who were HBeAg negative support that the observation is correct and not an artifact caused by degradation of the 5′ region of core RNA. We cannot offer a well‐founded explanation to this unexpected finding, which may reflect that a large fraction (≈90%) of core RNA in patients who are HBeAg negative starts downstream of the established 5′ starting point at nt 1818‐1820. As long as the transcription starts upstream of the start codon for the core ORF at nt 1901, the RNA may be used for core antigen synthesis. A later start point might have two critical effects. A lack of direct repeat 1 (nt 1825‐1834) would prevent the formation of a relaxed circular genome (by precluding translocation of the plus strand), whereas a start later than nt 1845 would likely break up the epsilon loop and prevent encapsidation and reverse transcription.( 19 ) Such a putative loss of function in a large proportion of the core RNA molecules might help explain the lower levels of HBV DNA in patients who are HBeAg negative. If correct, this could be a novel mechanism that reduces serum HBV DNA levels through pgRNA reduction in addition to reported epigenetic suppression.( 20 ) We have no explanation for a downstream shift of the starting point for pgRNA/core RNA transcription. Based on the link between HBeAg negativity and total number of mutations in the HBV genome,( 21 ) one might speculate that mutations in the core promoter or elsewhere influence the fidelity of the transcriptional machinery, resulting in a shift in the starting point. Additional studies are required to confirm our finding and to explain the mechanisms and consequences of this putative change in transcription initiation.
We observed lower RNA levels with the ddPCR targeting 3′ precore RNA redundancy (nt 1776‐1924), not only for S and X RNA but also compared with the core assay (nt 2361‐2448). Both these regions can only be expressed from cccDNA and not from integrated HBV DNA. One would, however, anticipate that RNA levels by the 3′ precore assay would be higher than those recorded using the core assay because the 3′ redundancy should be present not only in core RNA but also in the S (actually preS1 and preS2) and X transcripts that originate from cccDNA (Fig. 1). The lower RNA levels by the 3′ precore assay compared to the core may to some extent be due to degradation but would also be explained if a significant fraction of the cccDNA‐derived RNA was not polyadenylated at nt 1927 but at either of the upstream signals.( 12 , 13 ) Interestingly, we still observed a strong correlation between the 5′ core RNA and the 3′ precore RNA levels. This suggests that transcripts that start at nt 1820 (and are detected by the 5′ core assay) are likely to also contain the 3′ redundancy and may serve as pgRNA (be reverse transcribed), whereas core transcripts that are truncated in either the 5′ or 3′ ends can only serve as messenger RNA for core antigen (or another viral protein). Further investigations using single molecule strategies are needed to clarify this.
Previous work has suggested that suppressed transcription of precore RNA caused by mutations in the core promoter may be a mechanism to reduce the expression of HBeAg.( 11 ) In our patients, the levels obtained by the ddPCR that only amplifies precore RNA were ≈2 log10 units lower than the core RNA levels, but this difference was similar in HBeAg‐positive and HBeAg‐negative samples. We have no explanation for the difference between our results and previous findings. Additional studies, including sequencing of the core promoter, are warranted in order to determine the effect of core promoter mutations on the RNA expression and synthesis of HBeAg.
This study quantifying different HBV RNA species using ddPCR has several advantages. It provides absolute quantification and is less sensitive to bias from differences in amplification efficiency real‐time PCR, which was used for quantification of different HBV RNA species in cell culture in a recent report.( 22 ) A potential problem with cccDNA PCR assays is cross‐reaction with relaxed circular DNA (rcDNA). Such amplification can result in (false) cccDNA results at levels that are ≈2‐3 log10 units below the rcDNA level.( 16 ) The cccDNA levels in the present study were much higher than 2 log10 units below rcDNA (not shown), indicating that unspecificity had a small effect on the observed cccDNA levels. Moreover, cccDNA data were used mainly as reference to relate to RNA levels, showing that S RNA expression was enhanced and pgRNA suppressed in patients who were HBeAg negative.
In summary, by analyzing HBV RNA in a large number of liver biopsies from patients with chronic HBV infection, we found support for the concept that high serum levels of HBsAg in patients who are HBeAg negative are explained by expression from integrated HBV DNA. Our results also suggest that a large fraction of core RNA in patients who are HBeAg negative may have a starting point that precludes their function as pgRNA, which might contribute to explain the much lower levels of HBV DNA in these patients.
See Editorial on Page 949
Supported by the Swedish Cancer Society (CAN 2017/731 to M.L.) and by governmental funds to the Sahlgrenska University Hospital (ALFGBG‐146611).
Potential conflict of interest: Nothing to report.
References
- 1. European Association for the Study of the Liver . EASL 2017 clinical practice guidelines on the management of hepatitis B virus infection. J Hepatol 2017;67:370‐398. [DOI] [PubMed] [Google Scholar]
- 2. Beck J, Nassal M. Hepatitis B virus replication. World J Gastroenterol 2007;13:48‐64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Edman JC, Gray P, Valenzuela P, Rall LB, Rutter WJ. Integration of hepatitis B virus sequences and their expression in a human hepatoma cell. Nature 1980;286:535‐538. [DOI] [PubMed] [Google Scholar]
- 4. Mason WS, Gill US, Litwin S, Zhou Y, Peri S, Pop O, et al. HBV DNA integration and clonal hepatocyte expansion in chronic hepatitis B patients considered immune tolerant. Gastroenterology 2016;151:986‐998.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sung WK, Zheng H, Li S, Chen R, Liu X, Li Y, et al. Genome‐wide survey of recurrent HBV integration in hepatocellular carcinoma. Nat Genet 2012;44:765‐769. [DOI] [PubMed] [Google Scholar]
- 6. Tang KW, Alaei‐Mahabadi B, Samuelsson T, Lindh M, Larsson E. The landscape of viral expression and host gene fusion and adaptation in human cancer. Nat Commun 2013;4:2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lindh M, Rydell GE, Larsson SB. Impact of integrated viral DNA on the goal to clear hepatitis B surface antigen with different therapeutic strategies. Curr Opin Virol 2018;30:24‐31. [DOI] [PubMed] [Google Scholar]
- 8. Chevaliez S, Hezode C, Bahrami S, Grare M, Pawlotsky JM. Long‐term hepatitis B surface antigen (HBsAg) kinetics during nucleoside/nucleotide analogue therapy: finite treatment duration unlikely. J Hepatol 2013;58:676‐683. [DOI] [PubMed] [Google Scholar]
- 9. Wooddell CI, Yuen MF, Chan HL, Gish RG, Locarnini SA, Chavez D, et al. RNAi‐based treatment of chronically infected patients and chimpanzees reveals that integrated hepatitis B virus DNA is a source of HBsAg. Sci Transl Med 2017;9:pii:eaan0241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Podlaha O, Wu G, Downie B, Ramamurthy R, Gaggar A, Subramanian M, et al. Genomic modeling of hepatitis B virus integration frequency in the human genome. PLoS One 2019;14:e0220376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Laras A, Koskinas J, Hadziyannis SJ. In vivo suppression of precore mRNA synthesis is associated with mutations in the hepatitis B virus core promoter. Virology 2002;295:86‐96. [DOI] [PubMed] [Google Scholar]
- 12. Freitas N, Lukash T, Gunewardena S, Chappell B, Slagle BL, Gudima SO. Relative abundance of integrant‐derived viral RNAs in infected tissues harvested from chronic hepatitis B virus carriers. J Virol 2018;92:pii:e02221‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schutz T, Kairat A, Schroder CH. DNA sequence requirements for the activation of a CATAAA polyadenylation signal within the hepatitis B virus X reading frame: rapid detection of truncated transcripts. Virology 1996;223:401‐405. [DOI] [PubMed] [Google Scholar]
- 14. Kuypers J, Jerome KR. Applications of digital PCR for clinical microbiology. J Clin Microbiol 2017;55:1621‐1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lindh M, Horal P, Dhillon AP, Norkrans G. Hepatitis B virus DNA levels, precore mutations, genotypes and histological activity in chronic hepatitis B. J Viral Hepat 2000;7:258‐267. [DOI] [PubMed] [Google Scholar]
- 16. Malmstrom S, Larsson SB, Hannoun C, Lindh M. Hepatitis B viral DNA decline at loss of HBeAg is mainly explained by reduced cccDNA load–down‐regulated transcription of PgRNA has limited impact. PLoS One 2012;7:e36349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Volz T, Lutgehetmann M, Wachtler P, Jacob A, Quaas A, Murray JM, et al. Impaired intrahepatic hepatitis B virus productivity contributes to low viremia in most HBeAg‐negative patients. Gastroenterology 2007;133:843‐852. [DOI] [PubMed] [Google Scholar]
- 18. Laras A, Koskinas J, Dimou E, Kostamena A, Hadziyannis SJ. Intrahepatic levels and replicative activity of covalently closed circular hepatitis B virus DNA in chronically infected patients. Hepatology 2006;44:694‐702. [DOI] [PubMed] [Google Scholar]
- 19. Nassal M, Rieger A. A bulged region of the hepatitis B virus RNA encapsidation signal contains the replication origin for discontinuous first‐strand DNA synthesis. J Virol 1996;70:2764‐2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Levrero M, Pollicino T, Petersen J, Belloni L, Raimondo G, Dandri M. Control of cccDNA function in hepatitis B virus infection. J Hepatol 2009;51:581‐592. [DOI] [PubMed] [Google Scholar]
- 21. Hannoun C, Horal P, Lindh M. Long‐term mutation rates in the hepatitis B virus genome. J Gen Virol 2000;81:75‐83. [DOI] [PubMed] [Google Scholar]
- 22. D'Arienzo V, Magri A, Harris JM, Wing PAC, Ko C, Rubio CO, et al. A PCR assay to quantify patterns of HBV transcription. J Gen Virol 2019; doi: 10.1099/jgv.0.001373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Knodell RG, Ishak KG, Black WC, Chen TS, Craig R, Kaplowitz N, et al. Formulation and application of a numerical scoring system for assessing histological activity in asymptomatic chronic active hepatitis. Hepatology 1981;1:431‐435. [DOI] [PubMed] [Google Scholar]


