Abstract
Matrigel is a commercially available substrate that is derived from the extracellular matrix. Matrigel is widely used in cell culture experiments such as the transdifferentiation of primary pancreatic acini to ductal epithelial-like cells. Difficulty arises during gene expression analysis for cells cultured on Matrigel because residual RNA in the Matrigel will not only contribute to the poor integrity of RNA isolated from Matrigel cultures, but also will impact the gene expression data. We report here a simple method of removing Matrigel from primary cultures of human or mouse pancreatic acini. Following the experiment, the cultures are placed on wet ice to liquefy the Matrigel. The cell and Matrigel mixture is then centrifuged at low speed to separate the pancreatic cells from the Matrigel solution that resides in the supernatant. RNA isolated from the pelleted cells has high integrity and may be readily used for gene expression analysis such as quantitative reverse transcription PCR.
Keywords: RNA integrity, Pancreas, Matrigel, Organoids, Extracellular matrix, Acinar ductal metaplasia
Graphical abstract
Specifications Table
| Subject Area |
|
| More specific subject area | Pancreas, organoid culture, RNA biology |
| Protocol name | Method for Improved Integrity of RNA Isolated from Matrigel Cultures |
| Reagents/tools |
Reagents:
|
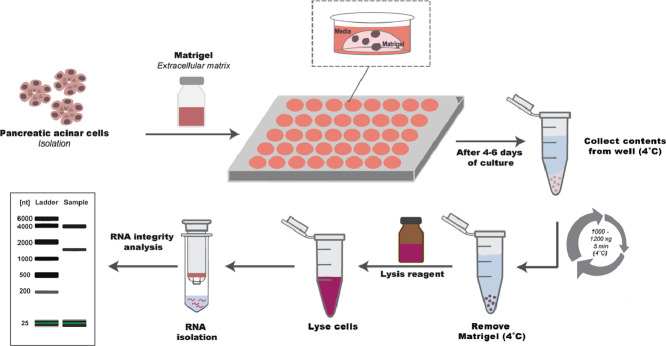
| Experimental design | After 4–6 days of culture, the culturing media is removed from each well and the cells in Matrigel are collected in two washing steps with cold PBS. After three centrifugation steps, the Matrigel is completely removed. The cell pellet is resuspended in lysis buffer and RNA isolation is performed. |
| Trial registration | If applicable, include clinical trial registry and number |
| Ethics | If applicable, include ethical details e.g. Patient informed consent, Ethics Review Board-competent authority approval, animal experimentation guidelines followed etc. |
| Value of the Protocol |
|
Description of protocol:
Method details
Reconstituted extracellular matrix (ECM) such as Matrigel are basement membrane substrates that are widely used in cell culture experiments. ECMs provide a solid support for three-dimensional culture as well as supplementing extracellular matrix proteins, growth factors and other components that facilitate cellular growth or transdifferentiation [1,2]. It is not widely appreciated, but ECMs also contain RNA, likely derived from extracellular vesicles found within the ECM [3]. Therefore, procedures to isolate RNA by direct lysis of the cells embedded in the ECM, will contain RNA from both the cells and the ECM. Methods have been reported to disrupt the cells embedded within the ECM using commercially-available enzymatic solutions such as Cell Recovery Solution [4] (Corning) or dispase [5]. We report here a simple method of removing ECM from primary cultures of pancreatic acini that involves first solving the gel at 4˚ C followed by low speed centrifugation to remove the cells from the solvated ECM. This modification results in an efficient and cost-effective method to isolate RNA from cells embedded in ECM that substantially limits the contaminating, largely degraded RNA that is found within commercially available ECMs.
Cell seeding in ECM
Primary human acinar cells obtained from a commercial source (Prodo Laboratories, AlisoViejo, CA) or freshly isolated mouse pancreas acini were cultured in Matrigel for up to 6 days prior to harvesting for RNA isolation. As we are interested in studying the changes in gene expression that occur during the culture period, we compare the expression at various time points to the day 0 culture. In order for the processing to be consistent with the cultures, the day 0 sample is prepared by placing the cells on the Matrigel and then immediately processed for RNA extraction as described below. Briefly, cells suspended in culture media were mixed with Matrigel in 1:1 ratio and 200 µL of the mix was added to each well of the 48 well-plate. After a 30 min incubation at 37° C, 5% CO2, for solidification of the Matrigel, 400 µL of culture media was carefully added to each well.
Harvesting the cells from ECM
-
1.
Completely aspirate the media from each well and place the 48 well-plate on ice for 5 min.
-
2.
Wash each well with 1 mL of cold PBS removing all contents by pipetting up/down three times.
-
3.
Transfer all of the contents to a 2 mL microtube placed on ice.
-
4.
Wash each well again using 800 µL of cold PBS. Transfer the remaining contents to the 2 mL microtube on ice.
-
5.
Mix the contents in the 2 mL microtube (pipetting up/down three times) and centrifuge for 5 min, at 4º C, 1000 x g.
-
6.
A small cell pellet may be visually observed alongside of the tube's wall. Carefully aspirate the supernatant in two steps, leaving ~200 µL in the tube.
-
7.
Resuspend the pellet in 1.8 mL of PBS and repeat step 5.
-
8.
Repeat step 6 and centrifuge for 5 min, at 4º C 1200 x g.
-
9.
Repeat step 6 and using a 200 µL pipette tip, aspirate as much as possible of the supernatant without aspirating the cell pellet.
-
10.
Resuspend the pellet in 700 µL of QIazol lysis buffer (Qiagen) and pipette up/down five times until completely homogenized.
-
11.
Proceed with RNA extraction and purification (Qiagen RNeasy Mini kit), or store the lysed samples at −80° C until processing.
Method validation
RNA integrity and concentration
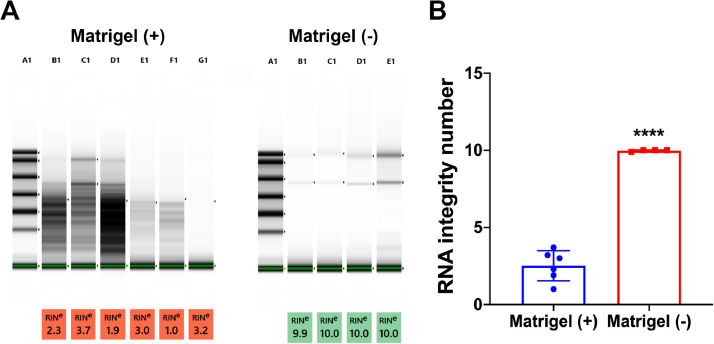
The integrity of the RNA extracted from the cell samples was determined by the ratio of the 18S to 28S ribosomal RNA and the calculated RNA integrity number (RIN). The application of the method to remove the Matrigel resulted in an improvement in the quality of cellular RNA as represented by increased RIN values (Fig. 1).
Fig. 1.
Integrity Analysis of RNA isolated from human pancreatic acinar cultures containing Matrigel or from those in which the Matrigel was removed. RNA from pancreatic acinar cells cultured on Matrigel was isolated either by first removing the Matrigel from the cultures (Matrigel -) or by direct lysis of both the cells and Matrigel (Matrigel +). (A) results of Agilent 2100 Bioanalyzer for Matrigel (+) and Matrigel (–) RNA Integrity Number (RIN). (B) Mean ± SD from the RINs shown in (A). **** p < 0.0001, Student's t-test.
Comparison of method to dispase treated cultures
The method of solvating the Matrigel at 4˚ C was compared to enzymatic digestion of the Matrigel using dispase [5]. Remove the media and add 75 µL of dispase solution in Waymouth's media (10 mg/mL). Incubate for 30 min at 37° C. Collect the cells plus Matrigel in Waymouth's media and centrifuge at 1200 xg, room temperature. Wash pellet three times with 2 mL of Waymouth's media with the centrifugation step in between washes. Lyse the pellet in 700 µL of QIazol and extract RNA. Our method of solvating the Matrigel at 4˚ C greatly improved the RIN compared to enzymatic digestion of the Matrigel with dispase (Fig. 2).
Fig. 2.
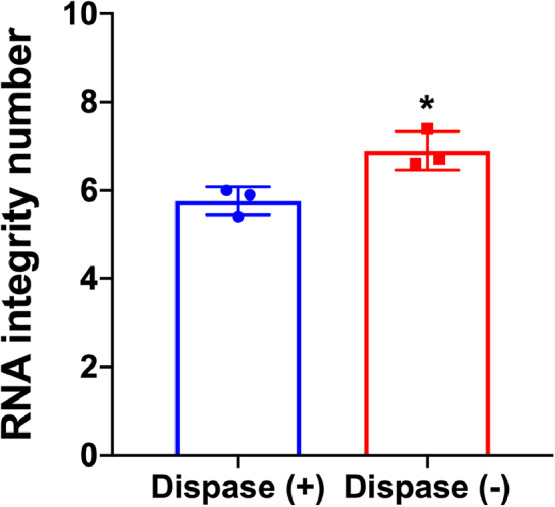
Effect of dispase on the integrity of RNA isolated from human pancreatic acinar cultures containing Matrigel. Pancreatic acinar cells plated on Matrigel were treated with Dispase as described in the method's validation section. The integrity number of the RNA isolated from the dispase (+) cultures are compared to that obtained using the method described herein, i.e. dispase (-).
qPCR analysis of isolated RNA
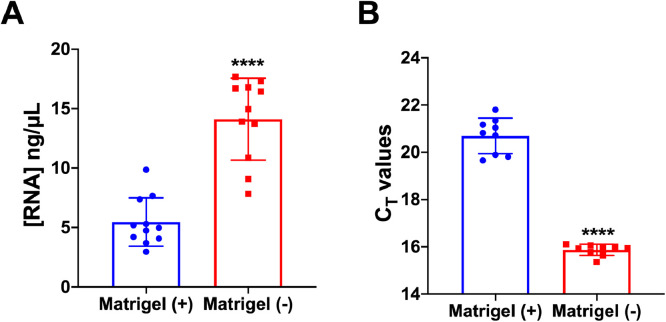
qPCR was used to determine the 18S rRNA threshold cycle (CT). The improvement in the amount and quality of RNA extracted from the cultures with the ECM removed was reflected in the higher RNA yield (Fig. 3A) and lower 18S rRNA CT values (Fig. 3B).
Fig. 3.
Quantitative assessment of RNA isolated along with and without Matrigel. (A) The concentration of RNA isolated from mouse pancreatic acinar cells from the Matrigel (+) and Matrigel (–) cultures. (B) CT values for 18S rRNA as determined by qPCR from the Matrigel (+) and Matrigel (–) cultures (n = 9) presented as the mean ± SD, **** p < 0.0001, Student's t-test.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
Supported by grant U54CA233441. The technical assistance of Alyssa Gosling is greatly appreciated. JKB was supported by NCI fellowship F31CA220937.
References
- 1.Benton G., Arnaoutova I., George J., Kleinman H.K., Koblinski J. Matrigel: from discovery and ECM mimicry to assays and models for cancer research. Adv. Drug Deliv. Rev. 2014;79-80:3–18. doi: 10.1016/j.addr.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 2.Collins M.A., Yan W., Sebolt-Leopold J.S., Pasca di Magliano M. MAPK signaling is required for dedifferentiation of acinar cells and development of pancreatic intraepithelial neoplasia in mice. Gastroenterology. 2014;146(3):822–834. doi: 10.1053/j.gastro.2013.11.052. e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.An M., Kwon K., Park J., Ryu D.R., Shin J.A., Lee Kang J., Choi J.H., Park E.M., Lee K.E., Woo M., Kim M. Extracellular matrix-derived extracellular vesicles promote cardiomyocyte growth and electrical activity in engineered cardiac atria. Biomaterials. 2017;146:49–59. doi: 10.1016/j.biomaterials.2017.09.001. [DOI] [PubMed] [Google Scholar]
- 4.Jogalekar M.P., Serrano E.E. Total RNA isolation from separately established monolayer and hydrogel cultures of human glioblastoma cell line. Bio Protoc. 2019;9(14) doi: 10.21769/BioProtoc.3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mroue R., Bissell M.J. Three-dimensional cultures of mouse mammary epithelial cells. Methods Mol. Biol. 2013;945:221–250. doi: 10.1007/978-1-62703-125-7_14. [DOI] [PMC free article] [PubMed] [Google Scholar]