Graphical abstract
Keywords: Subtractive genome analysis, Molecular dynamics simulation, Target analysis, Infectious and epidemic diseases, Bacteria
Abstract
Infectious and epidemic diseases induced by bacteria have historically caused great distress to people, and have even resulted in a large number of deaths worldwide. At present, many researchers are working on the discovery of viable drug and vaccine targets for bacteria through multiple methods, including the analyses of comparative subtractive genome, core genome, replication-related proteins, transcriptomics and riboswitches, which plays a significant part in the treatment of infectious and pandemic diseases. The 3D structures of the desired target proteins, drugs and epitopes can be predicted and modeled through target analysis. Meanwhile, molecular dynamics (MD) analysis of the constructed drug/epitope-protein complexes is an important standard for testing the suitability of these screened drugs and vaccines. Currently, target discovery, target analysis and MD analysis are integrated into a systematic set of drug and vaccine analysis strategy for bacteria. We hope that this comprehensive strategy will help in the design of high-performance vaccines and drugs.
1. Introduction
Human health is continuously threatened by various infectious diseases and large-scale epidemics caused by bacteria, medicines and vaccines are important means for the treatment of human diseases. In the past, owing to the fact that technology and resources still had not matured sufficiently, effective drugs or vaccines could not be developed promptly to cure the diseases under pressing epidemic situations and therefore epidemics or infectious diseases always cause a panic. With developments in medicine and technology, several vaccines and drugs were gradually developed. However, a few of them failed to achieve the desired effect, or potentially interfered with other normal functions and produced certain adverse effects [1]. Even presently, the development of innovative drugs still poses great challenges, such as extreme complexities, high risk, long development cycle and huge investment [2], [3], [4]. Thus, ensuring rapid, safe and effective development of drugs and vaccines has always been an urgent problem. The development of vaccines and drugs can be roughly divided into preclinical and clinical development, in which preclinical development plays a dominant role in the whole process [5]. If a candidate vaccine/drug is not proven to be safe and effective in preclinical studies, no further clinical studies are required. In preclinical studies, drug discovery is the first step in the drug development, which aims to achieve breakthrough progress. Therefore, we pay extra attention to the discovery of new drugs and vaccines in this review.
Investigation of new drugs and vaccines has continued throughout the history of human development. Initially, researchers isolated and identified the effective components to treat various diseases mainly from natural products [6]. However, employing natural products has certain challenges in practical applications, such as their low solubility and stability. Therefore, it is necessary to structurally modify the effective natural components. In 1796, Edward Jenner was successful in preventing a smallpox virus infection using a vaccinia vaccine. This achievement was the first victory in the history of vaccine development, and the beginning of vaccinology and immunology. Unfortunately, no new vaccine has emerged in the more than 100 years since the discovery of the first. At the end of the 19th century, Louis Pasteur et al. developed the anthrax vaccine and proposed the principle of vaccinology [7], which was a big step in the study of vaccines, and led to the development of a variety of vaccines to resist the corresponding pathogens [8], [9], [10]. Until 1932, the structural modification of drug molecules was first guided by a theory proposed by Erlenmeyer, which opened the way for further development in drug theories [11]. Subsequently, a quantitative structure-activity relationship (QSAR) was developed by Hansch et al. in 1964 [12]. QSAR can improve the success rate of candidate drugs in clinical experience, and lays a theoretical and practical foundation for quantitative drug design. Simultaneously, the development of bacterial vaccines had also progressed further before the mid-20th century. Since the late 20th century, bioinformatics, molecular biology, pharmacy, immunology, microbiology, and other related disciplines have developed rapidly, which has allowed new opportunities in the development of bacterial vaccines and drugs. Techniques for proteomic and genomic analyses have been further developed, and a large number of proteins and their coding genes have been discovered. At present, the designing of proteome- or genome-based bacterial drugs and vaccines has emerged as the new direction [13].
According to the published literature [14], [15], [16], the genome/proteome-based drug and vaccine design mainly involves four steps: selection and identification of drug target, optimization of the target molecules, discovery of compounds and peptide epitopes, and optimization of the compounds and peptide epitopes. The generation and availability of a large amount of genomic data have enabled the identification of effective targets through computational genomics methods, and completely changed the threat of pathogens to humans [17]. Among these genomics methods, the comparative subtractive genome approach has laid the foundation for target discovery and become an extensive tool for mining promising therapeutic targets [18], [19]. Other methods, including core genome [20], replication-related proteins [21], transcriptomics [22] and riboswitches analyses [23] have also garnered increasing attention for exploration of drug targets. Furthermore, target prioritization is an indispensable step in the design of drugs and vaccines. A three-dimensional (3D) model [24] for the target proteins, epitopes and drugs can be successfully predicted and constructed based on an in-depth analysis of the drug/vaccine targets. In addition, MD analysis of these modeled drug/epitope-protein complexes is a necessary standard for testing the effectiveness of drugs and vaccines. By MD simulation, the binding ability of inhibitors/peptides to proteins and the conformational changes of target proteins will be well reflected [25], [26].
Therefore, this review focuses on a combination of three important sections (target discovery, target analysis and MD analysis) to discover the preclinical inhibitors and vaccines that target bacteria-related diseases. First, we introduce five universal methods for exploring the targets: comparative subtractive genome, core genome, replication-related proteins, transcriptomics, and riboswitches analyses. Then, we summarize the basic process of the drug and vaccine design, which mainly includes target optimization, screening of drugs and vaccines, and optimization of drugs and vaccines. Finally, MD simulation and some advanced methods based on MD trajectory are described in detail.
2. Target discovery
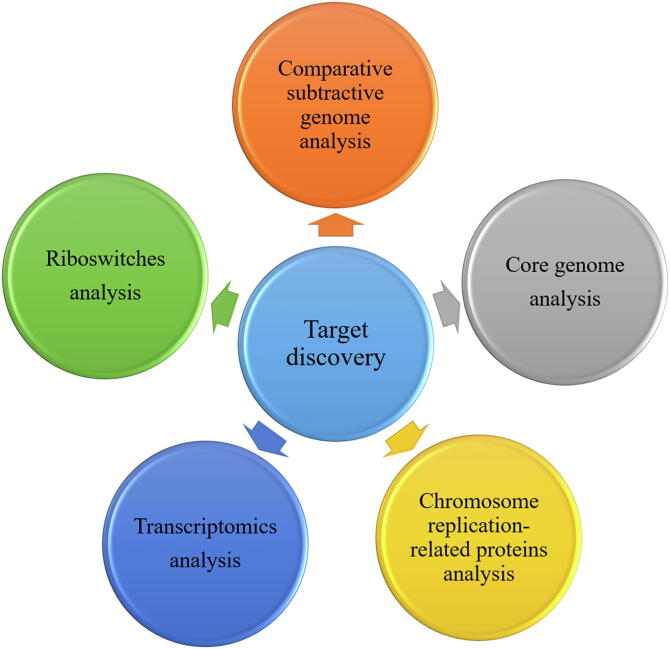
Exploring the therapeutic targets in bacteria is the first and crucial step in developing efficient vaccines and drugs. Certain essential proteins and proteins involved in basic cellular processes can serve as potential targets for novel antimicrobial agents. In this section, we summarize five analytical methods for exploration of drug targets (Fig. 1).
Fig. 1.
Multiple ways for drug discovery.
2.1. Comparative subtractive genome analysis
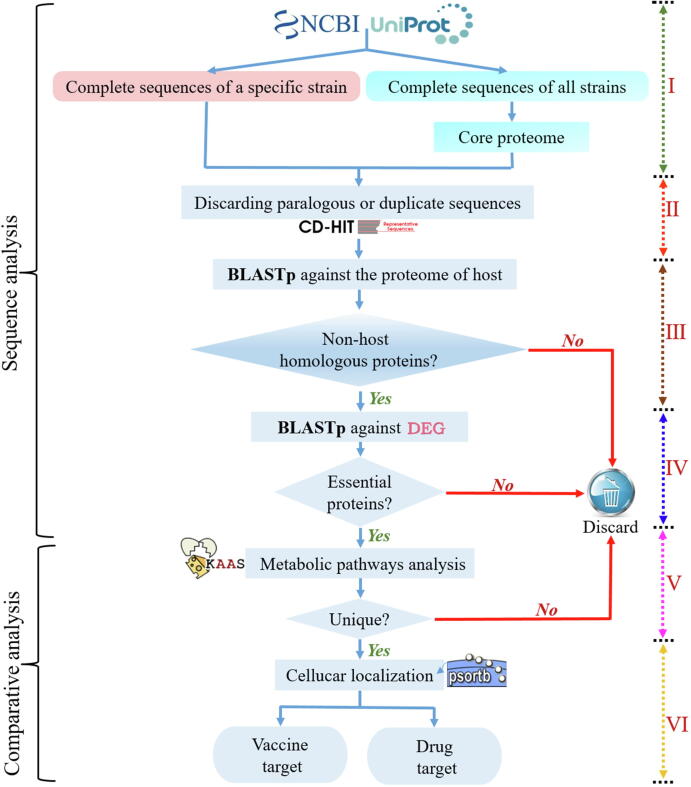
For the actual target selection, the potential candidate targets should be necessary for bacterial growth and reproduction, non-homologous to the host proteins, and have a unique metabolic pathway different from the host. With the aim of finding essential and non-homologous targets with unique metabolic pathways, subtractive genome analysis is selected to analyze the bacterial proteome through layers of screening. Since Sakharkar et al. first proposed the subtractive genome approach [1], many researchers have used this method to analyze drug and vaccine targets, which has immense potential for future experimental design of novel drugs and vaccines. For example, Sharma et al. revealed the target candidates for Lymphatic filariasisin in 2016 [18], and Sudha et al. investigated the drug targets and vaccine candidates for Clostridium botulinum in 2019 [19] using this method. In the following sections, we summarize the target screening process using the subtractive genome method. The detailed and complete workflow is shown in Fig. 2.
Fig. 2.
Workflow for screening potential drug and vaccine targets from complete sequences using comparative subtractive genome analysis and core genome analysis, respectively.
2.1.1. Getting the complete sequences of bacteria
According to published studies [19], [27], the complete sequences of bacteria for subtractive genome analysis are mainly retrieved as files in FASTA format from the National Center for Biotechnology Information (NCBI) [28] and Universal Protein (UniProt) [29] databases (Fig. 2I), which are the most informative and extensive protein databases.
2.1.2. Removing paralogous or duplicate sequences
The rapid emergence of next generation sequencing (NGS) technology has led to an explosive growth in biological sequence data, and the removal of redundant or duplicate sequence data has become one of the significant challenges to subsequent bioinformatics analyses [30]. Luckily, Li et al. created a fast online program CD-HIT [31] to search representative protein sequences based on the possible correlation and homology of certain sequences (Fig. 2II), alleviating the problem of calculation and analysis to some extent. To date, CD-HIT has been widely used to discard redundant or duplicate sequences by comparing the similarities between two sequences with expected threshold values.
2.1.3. Eliminating host-homologous sequences
Eliminating sequences that are homologous to the host, is a crucial operation in this process. If the target protein is homologous to the one in the host, the designed drug may produce nonspecific interactions with the host protein, resulting in certain negative effects [32]. Therefore, selecting proteins that are non-homologous to those in the host is necessary. Basic local alignment search tool (BLAST) [33] is the best choice for this requirement. In this section, BLASTp is applied by numerous researchers to perform a similarity search by comparing non-paralogous proteins with the entire host proteome (Fig. 2III), with the expectation value (e-value) set to widely used threshold 0.0001 [14], [34], [35]. Finally, the sequences that are homologous to those in the host are deleted.
2.1.4. Screening the essential proteins in bacteria
Choosing the essential proteins in bacteria, is another crucial step in this process. The essential proteins in the bacterial proteome are crucial for maintaining their life activities under specific conditions and vital importance for their survival, and any blocking of their functions will lead to cell death [36]. Hence, inhibiting the activity of such essential proteins can greatly improve the therapeutic effect in bacterial diseases. To select the essential proteins in bacterial proteome, an essentiality analysis is conducted on the non-homologous proteins. In subtractive genome analysis, it is common for users to perform a BLAST search against the Database of Essential Genes (DEG) [37] to remove non-essential proteins (Fig. 2IV) [38], [39], [40]. Since the DEG database was developed by Zhang et al. in 2004 [37], the content of this database has been updated continually and a large number of essential genes in prokaryotes and eukaryotes have been included [41]. Collection of a larger amount of essential gene data and availability of flexible BLAST tools [42] would contribute even more to the prediction of essential genes or proteins.
2.1.5. Metabolic pathway analysis
A metabolic pathway analysis [43] is performed on the non-homologous essential proteins by utilizing the Kyoto Encyclopedia of Genes and Genomes (KEGG) [44] Automatic Annotation Server (KAAS) [45] to identify the metabolic pathway of the targets, and similarity searches with BLASTp are conducted for all existing proteins against the latest KEGG database (Fig. 2V). Meanwhile, the metabolic pathways of the bacteria and their hosts also need to be compared. If the protein is involved in a unique metabolic pathway, it is marked for subsequent analyses; otherwise, the protein is removed from the proteome under consideration. Through this comparative pathway method, the non-homologous essential proteins following unique metabolic pathways can be mapped, and these proteins can be key targets for the treatment of diseases.
2.1.6. Subcellular localization analysis
Predicting the subcellular localization of bacterial proteins is critical to the identification of target proteins, and can quickly provide information about the protein function [46], [47]. An ideal candidate protein for a vaccine should interact with the extracellular environment and trigger the immune system of the host effectively; therefore, proteins distributed on the extracellular and outer membranes are considered effective vaccine candidates [48]. Meanwhile, it has been demonstrated that cytoplasm-related proteins can be effective drug targets [49]. At this stage, the remaining therapeutic targets are subjected to subcellular localization analysis to identify potential drug and vaccine candidates by using the most accurate and user-friendly PSORTb server [50] (Fig. 2VI). Besides, some other verified methods (CELLO [51], PA-SUB [52], SignalIP [53], Phobius [54] and ngLOC [55]) can be combined with PSORTb to achieve a more precise prediction of subcellular localization for predicted targets.
After subtractive genome analysis, the putative drug and vaccine targets have been identified separately, which is the cornerstone of future drug and vaccine design.
2.2. Core genome analysis
Studies have confirmed that the bacterial core genome plays an important role in their growth, and is also related to the essence of the species [56], [57]. The core genome dataset comprises the common genes in all the available strains of species, and the genes that belong to the core genome are closely related to the nature of the species [58], which makes core genome analysis a reasonable method to address the difficulty in obtaining therapeutic targets. Therefore, comparative subtractive genome analysis based on the core genome of bacteria is another method used to detect targets. In contrast to the subtractive genome method based on essential genes, the first step in core genome analysis is obtaining the complete sequences of all strains for a particular species (Fig. 2). According to recently published works [59], [60], [61], the core genome can be probed by Pan-Genome Analysis Pipeline (PGAP) [62], EDGAR tool version 2.0 [63], etc.
2.3. Chromosome replication-related proteins analysis
Chromosome replication-related proteins can also be used as potential targets for exploring novel and effective antimicrobials. It is well known that all bacterial cells undergo chromosome replication before they can be split into two identical daughter cells. Chromosome replication-related proteins are essential for maintaining cellular activity and the replication process of chromosomes, and represent a promising target class [21], [64]. Unfortunately, other than nonsteroidal anti-inflammatory drugs [65] and aminocoumarin [66], there are few available antimicrobials for targeting the bacterial chromosome replication. Therefore, identifying potential proteins that can interfere with or block bacterial chromosome replication through drug inhibition can be of great help in designing efficient drugs targeting a range of diseases caused by bacteria. In almost all bacterial species, chromosomal replication is triggered by the binding of the primary initiator protein (DnaA) to chromosomal replication origin (oriC), thus, DnaA and oriC are the main forces behind the formation of multimeric complexes required for the initiation of DNA replication [67]. The control of DnaA, which has multifunctional proteins required for chromosome replication, is the most prominent goal for inhibiting chromosome replication [68]. The four domains of DnaA have already been well summarized in literature, especially the N-terminal domain [69], [70]. To date, researchers have made considerable efforts to understand bacterial replication-related proteins, and the replication initiation of many bacterial species, including Escherichia coli, Bacillus subtilis, and Caulobacter crescentus, has been well studied [21], [68], [70], [71]. Meanwhile, the replication-related proteins are always present as a cluster aroud oriC. We have massively updated the information about the oriCs of bacteria in the online database DoriC 10.0 [72] based on the predicted results of Ori-Finder [73], [74], which can provide excellent opportunities to better explore the replication-related proteins of more bacteria.
2.4. Transcriptomics analysis
According to the genetic “central dogma”, transcription plays an important role in controlling the transmission of genetic information, which is the first key step in gene expression [75]. At present, transcriptomics has emerged as the leading and exciting topic in the life science field [76]. Transcriptomics is the study of cellular gene transcription and transcriptional regulation at the RNA level, and can provide a comprehensive and rapid understanding of the molecular mechanism of diseases and drug action at the transcriptome level [77]. Therefore, transcriptomics analysis has developed into a useful tool for acquiring novel antimicrobial targets [22], [78]. To better assist in the discovery of drug targets and drug design in different ways, a few technologies for transcriptomics studies, such as RNA-sequencing (RNA-seq) method for gene expression [79] and gene microarray or chip technology [80] have been developed and widely used to quickly search the transcriptomics. Practical applications of NGS-based RNA-seq method and microarray analysis in predicting genetic targets have been well reviewed [22], [75]. Recently, detailed target analyses of Escherichia coli, Clostridium difficile, Mycobacterium tuberculosis, Mycobacterium smegmatis and other pathogenic bacteria [81], [82], [83], [84], [85], [86], [87], [88] have been performed using bacterial transcriptomics, relevant techniques, and transcriptomics experiments. These successful cases of transcriptomics analyses again demonstrate that transcriptomics is a promising approach for predicting bacterial drug targets.
2.5. Riboswitches analysis
Riboswitches can mediate the expression of crucial and essential genes that are critical to the survival and virulence of bacterial pathogens [89], [90], and inhibiting the synthesis of bacterial ribosomal proteins can achieve antibacterial purposes [91]. Hence, bacterial riboswitches are considered as promising and capable antimicrobial targets for new drugs. In fact, riboswitches are widely found in the bacteria genomes and absent in human genome, which will reduce the probability of potentially harmful effects in humans and is one of the advantages of riboswitches as a useful tool for exploring bacterial drug targets [92]. In addition, riboswitches can bind to small molecules with high selectivity and are controlled by simple metabolism [93]. Given these advantages, the use of riboswitches as drug targets has attracted increasing attention, and some riboswitches-related work can provide valuable clues for future research [23], [94], [95]. It has been proven that few of the most widespread riboswitches, including lysine, cobalamin, SAM, and SAH, are useful antibacterial drug targets. In addition, certain methods for exploring potential riboswitches, such as Riboswitch Scanner [96] and drug design including high-throughput screening method have been well summarized [97]. Based on a powerful covariance model (CM), a comprehensive online database (RiboD) has recently been developed as a useful resource for predicting ribosomes in bacteria [98]. Using existing methods or developing new ones to dig deeper into riboswitches-related targets in bacteria will greatly help in the treatment of diseases associated with bacteria.
3. Target analysis
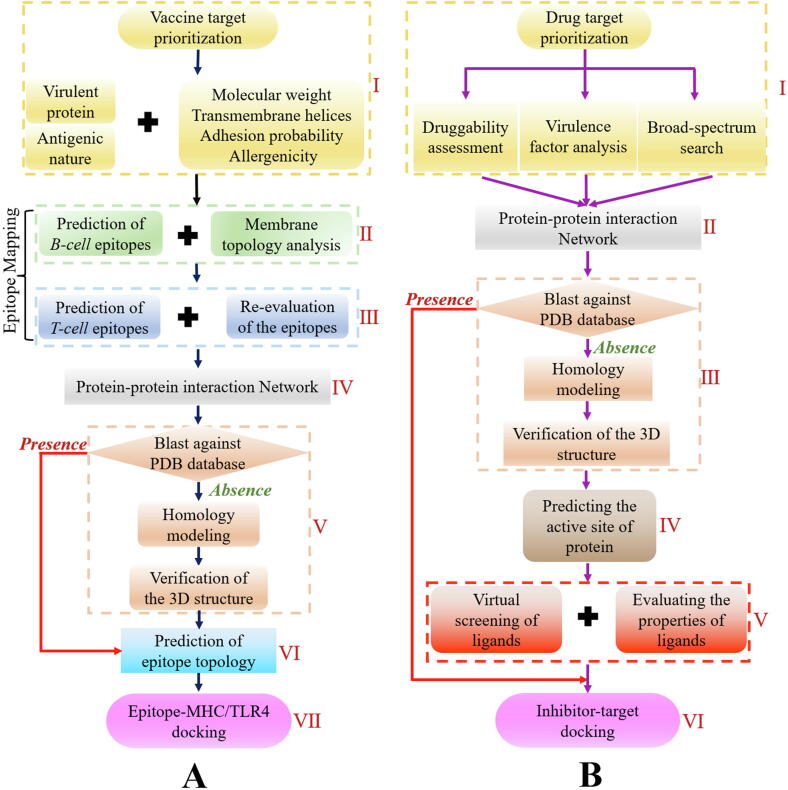
Once we have identified the vaccine and drug targets, the next important thing is to search for novel inhibitors and vaccines based on these possible targets. Notably, there are still considerable differences in the design of vaccines and drugs because of their unique properties, and the screening of vaccine targets is more complicated than that of drug targets. In this section, we present a detailed and systematic summary of the fundamental processes of target analysis, which are presented as flowcharts in Fig. 3.
Fig. 3.
Target analysis flowchart for target optimization and construction of 3D epitopes structures, target proteins and drugs: (A) Analysis of vaccine target, (B) Analysis of drug target.
3.1. Prediction of vaccine candidates
3.1.1. Vaccine target prioritization
Virulence is an important factor in the study of pathogenesis. Compared with non-virulent proteins, virulent proteins are more likely to cause serious infections and promote the survival of pathogens in the host, making it an attractive target for vaccine design [99], [100]. Therefore, virulence analysis has been incorporated into the flowchart as a necessary step (Fig. 3A-I). Currently, a few free databases, such as the Virulence Factor Database (VFDB) of pathogenic bacteria [101] and the Microbial Virulence Database (MvirDB) [102] are available that can be used to gain information on the virulence of proteins. In addition to these two databases, Garg et al. also performed protein virulence prediction using the Virulentpred server [103] with a threshold of ≥1. These selected virulent proteins are then subjected to antigenicity evaluation using the online VaxiJen server [104], where proteins with antigenicity scores ≥0.4 are marked as potential antigens that can effectively stimulate the human immune system.
Meanwhile, the physiochemical properties of all potential targets, including molecular weight, transmembrane helices, adhesion probability and allergenicity, are analyzed to assist in experimental validation. These factors may improve the vaccine prediction accuracy and reduce any negative effects. For ensuring purification during the experiment, the focus in a majority of studies has been concentrated on only selecting proteins with molecular weight ≤ 110 kDa as effective drug targets [105], [106], and these shortlisted proteins measured by freely available ExPASy server [107] will simplify the purification and development process. Furthermore, the number of transmembrane helices in the proteins can affect the cloning and expression of the target, and their presence in large quantities may lead to the failure of experimental validation; thus, selecting proteins with fewer transmembrane helices is more feasible [108]. For this purpose, two popular servers, TMHMM [109] and HMMTOP [110] are widely used to evaluate the number of transmembrane helices [106], [111], [112]. In addition, it has been reported that the interaction between the bacterial surface proteins known as adhesions and the host receptors contributes greatly to the bacterial attachment, and the antibodies generated due to these adhesive proteins can prevent infections and diseases [113]. Therefore, the adhesion probability of the proteins should be taken into account, which can be effectively predicted using the data available on the Vaxign [114] or SPAAN server [115]. Finally, the allergenicity analysis of all filtered protein is performed by accessing the Allertop server [116], online AlgPred [117] or SORTALLER [118] to reduce the allergic reactions, and proteins that could cause allergic behavior are removed.
In this section, virulent and antigenic proteins with prospective physiochemical characteristics are scanned for subsequent analysis.
3.1.2. Prediction of B- and T-cell epitopes
Since Barh et al. proposed the peptide vaccine design for Neisseria gonorrhoeae in 2010 [119], epitope-based vaccine design (EBVD) has emerged as the most popular and effective strategy in vaccine design [120], [121], [122]. Epitope vaccines have several advantages over traditional vaccines, such as atoxicity, safety, stability and easy production. They can also directly stimulate the host to create a specific immune response, thus confirming the suitability of EBVD for future development directions [119]. It is known that the antigen specificity and diversity are determined by B- and T-cell epitopes. Therefore, discovering the B- and T-cell epitopes capable of stimulating B- and T-cell immune responses is an imperative step in the development of such vaccines. In the following section, we summarize the prediction process of B- and T-cell epitopes in detail.
The proteins retained in the prioritization process are ideal vaccine candidates for the preparation of epitope-based vaccines, and these proteins are used to conduct an epitope analysis to predict the B-cell epitopes by employing the software BCPreds [123] or recent BepiPred-2.0 [124]. The selected B-cell epitopes with a BCPreds threshold score > 0.8 are then subjected to membrane topology analysis to determine their exposed topology by TMHMM (Fig. 3A-II).
The T-cell epitopes are then screened from B-cell epitopes with exposed surface based on the principles put forward by Barh et al. [119]. It has been affirmed that the binding affinity of reactive peptides to both classes of major histocompatibility complex I and II (MHC-I and II) molecules plays a vital role in immune response [125]. For the selection of an efficient T-cell epitope (Fig. 3A-III), the first step is to identify the binding epitope alleles to MHC-I and MHC-II by using the Propred1 [126] and Propred [127] servers, respectively. T-cell epitopes that can bind to more than fifteen MHC molecules simultaneously, especially to HLA-DRB1*0101, are cataloged. It is worth noting that DRB*0101 is the most frequent MHC-II allele, and an antigen can produce a more effective immune recognition and immune response when bound to DRB*0101 instead of other alleles. Next, calculation of the half-maximal inhibition concentration (IC50) for all probed T-cell epitopes is performed utilizing MHCPred [128] and the epitopes with an IC50 score < 100 nM are considered. Then, the virulence, antigenicity, adhesion probability, and allergenicity of the B-cell-derived T-cell epitopes are re-confirmed using VirulentPred, VaxiJe, Vaxign and Allertop servers, respectively. Meanwhile, ProtParam, Comprehensive Antibiotic Resistance Database (CARD) [129] and CLC Sequence Viewer are separately chosen to further estimate the chemical stability, resistance sequence, and conservation of the final selected epitopes.
Finally, ideal T-cell epitopes are successfully selected from a large number of vaccine targets.
3.1.3. Interaction network
This work extends further to the selection of epitope proteins with strong cellular interactions (Fig. 3A-IV). Proteins with strong connections to neighboring proteins are regarded as hub proteins, which contribute greatly to the protein-protein interaction (PPI) network and have a direct relationship with the lethality of the pathogen [130]. If the activity of the hub proteins in the PPI network is inhibited, the entire network will be affected. Given the importance of key proteins, understanding the PPI network of the target candidates at the cellular level is also crucial, and has important implications for future vaccine and drug development [131]. The interaction analysis of all remaining epitope proteins can be achieved by searching a large number of protein relationships with the Search Tool for the Retrieval of Interacting Genes (STRING) [132], and the output results contain direct and indirect interactions from different sources. In the protein interaction network, proteins with the highest confidence score (0.9) are selected for further analysis [105], [112].
3.1.4. Homology modelling and epitope topology analysis
With the aim of visualizing the topology of the predicted epitopes, the 3D structures of the epitope proteins need to be known (Fig. 3A-Ⅴ). As the initial step, a BLASTp search against Protein Data Bank (PDB) [133] is performed to seek structural information about the epitope proteins or suitable structural templates for epitope proteins that are unidentified to date, which is important for the prediction of immunogenic domains. For protein structures that are unavailable in the PDB library, the corresponding structures can be constructed by homology modeling. Online available servers, including I-TASSER [134], Phyre2 [135], Modweb [136], RaptorX [137], Modeler [138], M4T [139] and Swiss-Model [140] can help predict the 3D structure of the vaccine candidates. Subsequently, common web servers RAMPAGE, ProSA [141] verify 3D [142], ERRAT algorithm [143], WHAT_CHECK [144] and PROCHECK program [145] can be combined to accurately validate the 3D structure. Using the Ramachandran plot and Z-score analyses, the structure with the most residues mapped in favorable regions and a few residues in disallowed regions are selected as the best structure for each protein. In addition to the tools described above, PEPFOLD [146] can also be utilized to design the 3D structures of the epitopes according to amino acid sequence.
Once we know the 3D structure of the epitope proteins, this information can help us to calculate and predict the corresponding epitope topology (Fig. 3A-VI) [147]. To ensure the epitopes that effectively trigger the host immune system have exposed surfaces, the Pepitope server [147] is used to perform an exomembrane topology analysis on the shortlisted epitopes and their respective folded proteins.
3.1.5. Molecular docking
A promising molecular docking method is subsequently performed to view the binding affinity and binding modes of epitopes to the MHC alleles [119] or Toll-like receptor 4 (TLR4) (Fig. 3A-VII) [105], [112], [148]. The precise epitope-protein docking can be achieved by ClusPro 2.0 [149], or a combination of PatchDock [150] and FireDock [151], or a combination of Autodock Vina [152] and GalaxyPepDock [153]. The detailed binding information of the peptide-protein complex can be viewed through UCSF Chimera [154] and LigPlot [155].
3.2. Prediction of drug candidates
3.2.1. Drug target prioritization
As depicted in Fig. 3B-Ⅰ, the overall prioritization of predicted drug target is mainly considered from three factors: druggability, virulence factor (VF) and broad spectrum. The ideal drug target should integrate closely with drug-like molecules to make the drug more effective, and the binding affinity of the target proteins to the drug-like molecules can be reflected by druggability [156]. To find the proteins that can develop into potential drug targets, all putative proteins undergo the BLASTp similarity analysis against the bacterial drug targets in the DrugBank database [157] to assess the druggability of each protein, and predicted proteins with a high similarity to the bacterial drug targets are regarded as druggable targets for subsequent analysis.
Virulent proteins can regulate the infection pathway and play a decisive role in the survival of the pathogens in the host [99]. Thus, VF analysis has been proven as a promising approach for identifying therapeutic drug targets. To probe the virulence-related proteins, VFDB is applied for similarity comparison using the BLAST tool with a bit score > 100, and the output data will contain multiple types of virulence factor, such as adherence and protease.
Bacterial pathogens can generate different simultaneous infections in the host, thus, screening for broad-spectrum targets is now considered preferable. In this step, a broad-spectrum search of predicted proteins is conducted to investigate the potential broad-spectrum targets by BLASTp against bacterial pathogen proteomes with an e-value of 0.005 [14], [16].
After this progressive evaluation, the non-homologous and essential proteins that pass successfully through these filtration conditions and demonstrate unique metabolic pathways to the host are listed as prospective drug targets.
3.2.2. Interaction network, homology modelling and 3D structure assessment
The PPI network analyses, homology modeling and 3D structural assessment of the drug candidates are similar to the corresponding analyses used for the prediction of vaccine targets (Fig. 3B-II and III).
3.2.3. Predicting the binding site of target proteins
Once the final model of the predicted proteins is established, the next step is to predict the binding site of the proteins, which is essential for understanding the protein function (Fig. 3B-IV). Proteins contain a large number of residues, whereas the binding site is composed of those residues that can bind specifically to the drug. Therefore, understanding the interactions between the inhibitors and the proteins is crucial in drug design. Candidate interaction-based binding sites can be forecasted with the following programs: COACH [158], Computed Atlas of Surface Topography of proteins (CASTp) [159], Active Site Finder tool, DoGSiteScorer [160], fpocket [161], MetaPocket [162], and GHECOM [163].
3.2.4. Virtual screening of ligands, evaluating the properties of ligands and molecular docking
Virtual screening (VS), also known as computer screening, is one of the latest advances in drug discovery (Fig. 3B-Ⅴ). Generally speaking, VS involves screening candidate ligands through ligand databases and investigating the possibility of these molecules binding or docking with the target proteins. ZINC is a broad platform for drug screening, and can accomplish a multi-method molecular search according to structure, properties, targets, etc. Initially, small molecules in the MOL2 format are downloaded from the ZINC database [145]. Then, the selected ligands are converted into the PDBQT format and undergo VS using the AutoDock Vina or AutoDock software tools [164], and these two software tools can realize the batch docking of molecules. The docking results are sorted in ascending order based on the binding free energy (ΔGbind) between the inhibitor and the receptor, and the first ten candidates are generally selected as the ideal inhibitors.
Molecular properties of the ligands are important for every step of the design, synthesis and clinical application of an effective drug. For the purpose of minimizing the negative effects of the selected ligands, the absorption, distribution, metabolism, excretion and toxicity (ADMET) characteristics that can influence the pharmacokinetics of the designed drugs are further evaluated by employing the SwissADME program [165] or the PreADMET server. The DrugBank database can also be used to assess the pharmaco-chemical properties of the drugs. Ultimately, the best predicted ligands with better pharmacokinetics and pharmaco-chemical features are acquired.
The docking between an inhibitor and a target protein is more complicated than the binding between two proteins, and their docking relationship is similar to that of a lock and key (Fig. 3B-VI). The inhibitor and the target protein should be paired complementarily, and the attachment of the inhibitor to the binding pocket should be as close as possible. Once the receptor proteins and inhibitors are ready, flexible molecular docking can be done through AutoDock, AutoDock Vina, GOLD [166] software, etc. Structures with the lowest binding free energy during molecular docking are considered the best and most stable initial structures for MD studies.
4. MD analysis
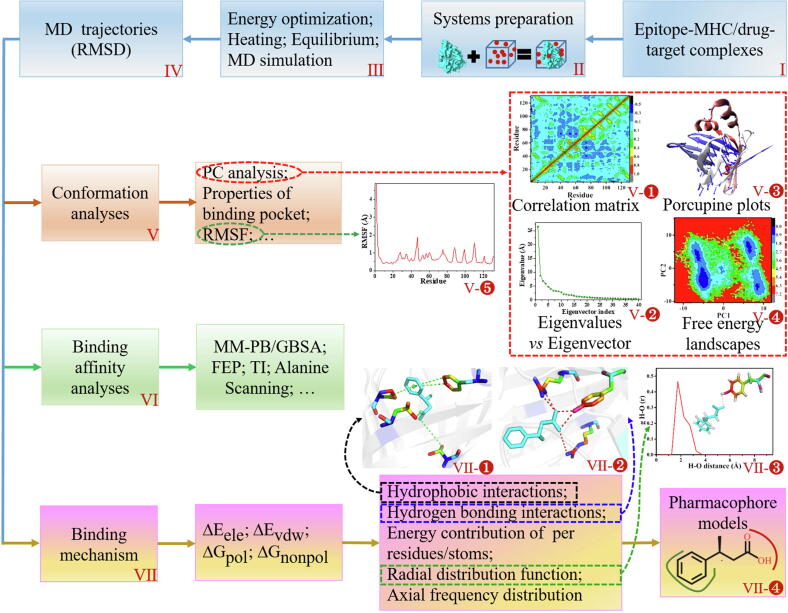
MD analysis is a computer-based simulation method used widely in several fields, such as physics, chemistry, and biology. With the development of computer simulation technology, MD simulation [167], [168], [169], [170] has become a common tool for studying the binding mechanism of the inhibitors/peptides to the proteins and conformational changes of target proteins based on equilibrium MD trajectories. MD simulation can respond well to the dynamic characteristics of biomolecules, which helps provide a better theoretical basis for efficient vaccine and drug design. At present, existing mature software packages such as AMBER [171] and GROMACS [172] can provide strong technical support for MD simulations. To understand MD simulation better, we now summarize the basic MD simulation process and the advanced methods used for analyzing the conformation of target proteins and the binding affinity between the protein and the drug/vaccine (Fig. 4).
Fig. 4.
Flow diagram of MD analysis for studying the binding mechanism of inhibitors/epitopes to proteins and conformational changes in proteins.
4.1. System preparation
Prior to the MD simulation, the selected systems should be properly prepared by applying the following four steps: adding missing hydrogen atoms to their respective heavy atoms; setting force fields for the proteins, inhibitors, or peptide epitopes; adding a certain amount of Na and Cl ions to neutralize the system; and immersing all the systems into a water box (Fig. 4II).
After the systems are well prepared, three critical operations (energy optimization, heating, equilibrium) are executed stepwise to ensure that the MD simulations are performed in an ideal experimental environment (Fig. 4III). First, the energy of all the studied models is optimized to eliminate any possible adverse effect on the structural deformation and simulation stability by combining the steepest descent and conjugate gradient methods. Subsequently, the system is gradually heated until the expected temperature of 300 K is reached. In the next step, the simulation is continued at the same temperature and characteristics including pressure, energy, and structure are evaluated. The simulation continues until these characteristics stop showing any changes over time. Finally, long-time MD simulation is performed at room temperature (300 K) and atmospheric pressure (1 atm). To better understand the universality of MD simulations and the various analytical methods based on the MD trajectories, we arbitrarily selected an example (PDB ID: 3P6F) to simulate the MD of 150 ns and the calculated results are presented in Fig. 4.
4.2. Root-mean-square deviation
The root mean square deviation (RMSD) value represents the deviation of backbone atoms in the proteins relative to their respective initial optimized structures, and is a commonly used method to evaluate the stability of the system. Smaller the RMSD value, the more stable the system is during the simulation. Generally, the equilibrium MD trajectories are selected for later analysis (Fig. 4IV).
4.3. Conformation analysis of proteins, and assessment of binding affinity and binding mechanism between inhibitors/peptides and proteins
To be effective, a drug must reach the binding site of the target protein and generate a strong interaction with the residues at the active site to form a stable complex. Meanwhile, proteins binding with the inhibitors/peptides will cause changes in their conformation. Therefore, a deeper understanding of the binding affinity and binding mechanism of inhibitors/peptides to the proteins and the conformation changes in the proteins induced by the binding will be of great help in the design of effective drugs and vaccines.
4.3.1. Conformation analysis
The most popular tool for performing conformation analysis is the principal component analysis (PCA) [173]. Fundamentally, this method constructs a covariance matrix based on the coordinates of atoms using a dimensional reduction method, which can then reflect the deviations of the atoms from their respective average positions. Thus, cross-correlation matrices (Fig. 4Ⅴ-1) corresponding to the correlated motion between residues can be constructed. In a diagonalized covariance matrix, the eigenvalues and the eigenvector plot (Fig. 4Ⅴ-2) representing the motion intensity and direction of the residues, respectively, can be obtained, and then a porcupine plot (Fig. 4Ⅴ-3) can be established to characterize the movement of the residues. By projecting MD trajectories onto the first and second principal components (PC1 and PC2), the binding free energy landscapes (Fig. 4Ⅴ-4) of the proteins can also be constructed to better reflect their conformational distribution. Existing work has shown that a combination of RMSD and gyration radiuses (GR) can also be used to construct the free energy landscapes [174].
In addition to PCA, a few other methods are also used to analyze the protein structure based on the equilibrium MD trajectories. For example, the root mean square fluctuation (RMSF) of Cα atoms can be used to indicate the flexibility of the protein during the MD simulation (Fig. 4Ⅴ-5). The stability, continuity, correlation and volume of the binding pocket for each protein can also be separately evaluated by employing the D3Pockets server [175] and the POVME procedure [176] to characterize the structural changes in the proteins.
4.3.2. Binding affinity analysis
The binding ability of inhibitors/peptides to proteins can be confirmed by calculating the ΔGbind between the inhibitors/peptides and the proteins (Fig. 4VI). Numerous methods have been developed to predict the ΔGbind, including molecular mechanics Poisson Boltzmann/generalized Born surface area (MM-PB/GBSA) [177], thermodynamics integration (TI) [178], and free energy perturbation (FEP) [179]. Considering the computational resources and time, MM-PBSA and MM-GBSA have been the most widely used methods in recent years. In this method, the ΔGbind between inhibitor and protein can be determined by the following formula.
| (1) |
The items on the right side of the equation represent the contributions of the electrostatic interaction (Δ), van der Waals interaction (), polar interaction (), nonpolar interaction () and entropy change (ΔS) to , respectively. Notably, the calculation of entropy is time-consuming; therefore, only 50–100 conformations are generally calculated by normal mode method [180]. In addition, the new interaction entropy (IE) method proposed by Duan et al. can also help in the calculation of entropy [181].
To further understand the influence of key residues on binding affinity, a computational alanine scanning method [182] based on MM-PBSA and MM-GBSA methods can also be applied to estimate the change in and binding mechanism caused by the mutation of residues. Alanine mutant structures are generated by altering the coordinates of the wild-type (WT) residues, and the alanine residue parameters then replace all the parameters of the WT residue in the topology file. Subsequently, computational alanine scanning is performed based on the same snapshots as implemented in the MM-PBSA method. The difference in ΔGbind can be determined by the following equation.
| (2) |
where the first two terms (, ) are the binding free energies of WT and mutant complexes. The measurement unit for terms , , , , , , , and is kcal/mol.
4.3.3. Binding mechanism analysis
Through continuous efforts of a large number of researchers, the binding mechanism between drugs/peptides and proteins has been extensively studied (Fig. 4VII). With the calculation of ΔGbind, analyses of , , and interactions between inhibitors/peptides and proteins have been performed [168], [169], [183]. In these analyses, as the electrostatic and van der Waals interactions play a major role in the binding of drugs/peptides with proteins, further research on these two interactions has also been performed. Presently, the energy contributions of individual residues in proteins and individual atoms on residues to electrostatic and van der Waals interactions have also been calculated [184]. Simultaneously, detailed analyses of the hydrogen bonding (Fig. 4VII-1) and hydrophobic interactions (Fig. 4VII-2) between residues and inhibitors/peptides have also been performed to reveal the source of these two interactions. Furthermore, the radial distribution function (RDF) (Fig. 4VII-3) can partially contribute to the analysis and identification of hydrogen bonds [185]. Recently, a comprehensive method of axial frequency distribution (AFD) has also been proposed. This method can not only reflect the conformational characteristics, such as structural stability and flexibility, but also be used to analyze bi-molecular interactions including hydrogen bonds, van der Waals, and polar or ionic interactions [186]. We believe that due to long-term efforts, the prediction of binding affinity and binding mechanisms between inhibitors/peptides and proteins is no longer a puzzle.
After an in-depth analysis of the interaction mechanism, the optimal pharmacophore model [169], [187] is generated, as shown in Fig. 4VII-4. Generally speaking, the red-labeled region indicates that this region is easy to produce hydrogen bonding interactions with drug, while the green-labeled region make hydrophobic interactions with drug. Once the theoretical pharmacophore models of the relevant drugs are identified, pharmacophore-based VS can be performed to explore additional drugs, as proved by the great success of this method [188], [189].
5. Summary and outlook
Developing drugs or vaccines for highly contagious bacterial diseases in a short period of time can be challenging, and some drugs/vaccines can also show adverse effects during clinical treatment, which poses a great challenge to the clinical treatment, and necessitates a strict and careful monitoring of each step of the drug/vaccine design.
In this review, the methods for target discovery, target analysis, and MD analysis are summarized to present a complete and systematic scheme for the design of effective drugs and vaccines. In the first step, five common analytical methods, including comparative subtractive genome, core genome, replication-related proteins, transcriptomics, and riboswitches analyses are used to obtain promising drug and vaccine targets. Then, an in-depth analysis of selected targets is performed to minimize the negative effects of drugs and vaccines. Finally, each model is analyzed and verified by MD simulations to facilitate a deeper understanding of the binding mechanism of inhibitors/peptides to proteins and the structural changes in the proteins caused by the binding of inhibitors/peptides. We have also summarized the online software/database and corresponding websites used in each step to facilitate the readers to use and consult them, and the results are listed in Table 1.
Table 1.
Online software and corresponding websites used in each step.
The development and application of effective drugs still need to undergo long-term and numerous clinical trials, and researchers have performed numerous clinical investigations on vaccines and drugs [190], [191], [192].We expect that this review will provide useful ideas and guidance for the clinical development of effective drugs or vaccines to cure potential infectious diseases or epidemics caused by bacteria.
CRediT authorship contribution statement
Fangfang Yan: Conceptualization, Methodology, Investigation, Writing - original draft. Feng Gao: Supervision, Project administration, Funding acquisition, Writing - review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by the National Key Research and Development Program of China (grant no 2018YFA0903700) and the National Natural Science Foundation of China (grant nos. 31571358, 21621004 and 9174611). The authors would like to thank Prof. Chun-Ting Zhang for the invaluable assistance and inspiring discussions.
References
- 1.Sakharkar K.R., Sakharkar M.K., Chow V.T. A novel genomics approach for the identification of drug targets in pathogens, with special reference to Pseudomonas aeruginosa. Silico Biol. 2004;4:355–360. [PubMed] [Google Scholar]
- 2.Pammolli F., Magazzini L., Riccaboni M. The productivity crisis in pharmaceutical R&D. Nat Rev Drug Discov. 2011;10:428–438. doi: 10.1038/nrd3405. [DOI] [PubMed] [Google Scholar]
- 3.Dimasi J.A., Grabowski H.G., Hansen R.W. Innovation in the pharmaceutical industry: new estimates of R&D costs. J Health Econ. 2016;47:20–33. doi: 10.1016/j.jhealeco.2016.01.012. [DOI] [PubMed] [Google Scholar]
- 4.Moffat J.G., Vincent F., Lee J.A., Eder J.R., Prunotto M. Opportunities and challenges in phenotypic drug discovery: an industry perspective. Nat Rev Drug Discov. 2017;16:531–543. doi: 10.1038/nrd.2017.111. [DOI] [PubMed] [Google Scholar]
- 5.Plotkin S., André F., Poolman J., Robbins J., Salisbury D., Wood D. Preclinical and clinical development of new vaccines. Biologicals. 1998;26:247–251. doi: 10.1006/biol.1998.9998. [DOI] [PubMed] [Google Scholar]
- 6.Macht D.I. The history of opium and some of its preparations and alkaloids. J Am Med Assoc. 1915;LXIV:477–481. [Google Scholar]
- 7.Burke D.S. Joseph-alexandre auzias-turenne, louis pasteur, and early concepts of virulence, attenuation, and vaccination. Perspect Biol Med. 1996;39:171–186. doi: 10.1353/pbm.1996.0037. [DOI] [PubMed] [Google Scholar]
- 8.Mcaleer W.J., Buynak E.B., Maigetter R.Z., Wampler D.E., Miller W.J., Hilleman M.R. Human hepatitis B vaccine from recombinant yeast. Nature. 1984;307:178–180. doi: 10.1038/307178a0. [DOI] [PubMed] [Google Scholar]
- 9.Hilleman M.R., Warfield M.S., Anderson S.A., Werner J.H. Adenovirus (RI-APC-ARD) vaccine for prevention of acute respiratory Illness. 1. Vaccine development. J Am Chem Soc. 1957;163:4–9. doi: 10.1001/jama.1957.02970360006002. [DOI] [PubMed] [Google Scholar]
- 10.Buynak E.B., Weibel R.E., Whitman J.E., Stokes J., Hilleman M.R. Combined live measles, mumps, and rubella virus vaccines. J Am Med Assoc. 1969;207:2259–2262. [PubMed] [Google Scholar]
- 11.Erlenmeyer H., Leo M. über Pseudoatome und isostere Verbindungen. Vergleichende Studien mit Benzol, Thiophen und Furan. Helv Chim Acta. 1933;16:1381–1389. [Google Scholar]
- 12.Hansch C., Fujita T. A method for the correlation of biological activity and chemical structure. J Am Chem Soc. 1964;86:1616–1626. [Google Scholar]
- 13.Doolan D.L., Apte S.H., Proietti C. Genome-based vaccine design: the promise for malaria and other infectious diseases. Int J Parasitol. 2014;44:901–913. doi: 10.1016/j.ijpara.2014.07.010. [DOI] [PubMed] [Google Scholar]
- 14.Hossain M.U., Khan M.A., Hashem A., Islam M.M., Morshed M.N., Keya C.A. Finding potential therapeutic targets against shigella flexneri through proteome exploration. Front Microbiol. 2016;7:1817. doi: 10.3389/fmicb.2016.01817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hassan A., Naz A., Obaid A., Paracha R.Z., Naz K., Awan F.M. Pangenome and immuno-proteomics analysis of Acinetobacter baumannii strains revealed the core peptide vaccine targets. BMC Genomics. 2016;17:732. doi: 10.1186/s12864-016-2951-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Omeershffudin U., Kumar S. In silico approach for mining of potential drug targets from hypothetical proteins of bacterial proteome. Int J Mol Biol Open Access. 2019;4:145–152. [Google Scholar]
- 17.Miesel L., Greene J., Black T.A. Microbial genetics: genetic strategies for antibacterial drug discovery. Nat Rev Genet. 2003;4:442–456. doi: 10.1038/nrg1086. [DOI] [PubMed] [Google Scholar]
- 18.Sharma O.P., Kumar M.S. Essential proteins and possible therapeutic targets of Wolbachia endosymbiont and development of FiloBase-a comprehensive drug target database for Lymphatic filariasis. Sci Rep. 2016;6:19842. doi: 10.1038/srep19842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sudha R., Katiyar A., Katiyar P., Singh H., Prasad P. Identification of potential drug targets and vaccine candidates in Clostridium botulinum using subtractive genomics approach. Bioinformation. 2019;15:18–25. doi: 10.6026/97320630015018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tettelin H., Riley D., Cattuto C., Medini D. Comparative genomics: The bacterial pan-genome. Curr Opin Microbiol. 2008;11:472–477. doi: 10.1016/j.mib.2008.09.006. [DOI] [PubMed] [Google Scholar]
- 21.Van Eijk E., Wittekoek B., Kuijper E.J., Smits W.K. DNA replication proteins as potential targets for antimicrobials in drug-resistant bacterial pathogens. J Antimicrob Chemoth. 2017;72:1275–1284. doi: 10.1093/jac/dkw548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Domínguez Á., Muñoz E., López M.C., Cordero M., Martínez J.P., Viñas M. Transcriptomics as a tool to discover new antibacterial targets. Biotechnol Lett. 2017;39:819–828. doi: 10.1007/s10529-017-2319-0. [DOI] [PubMed] [Google Scholar]
- 23.A Reyes-Darias J, Krell T. Riboswitches as potential targets for the development of anti-biofilm drugs. Curr Top Medicinal Chem 2017;17:1945-1953. [DOI] [PubMed]
- 24.Kauzmann W. The three dimensional structures of protein.s. Biophys J. 1964;4:43–54. doi: 10.1016/s0006-3495(64)86925-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zafar S., Nguyen M.E., Muthyala R., Jabeen I., Sham Y.Y. Modeling and simulation of hGAT1: A mechanistic investigation of the GABA transport process. Comput Struct Biotec. 2019;17:61–69. doi: 10.1016/j.csbj.2018.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yan F.F., Liu X.G., Zhang S.L., Su J., Zhang Q.G., Chen J.Z. Effect of double mutations T790M/L858R on conformation and drug-resistant mechanism of epidermal growth factor receptor explored by molecular dynamics simulations. RSC Adv. 2018;8:39797–39810. doi: 10.1039/c8ra06844e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khalid Z., Ahmad S., Raza S., Azam S.S. Subtractive proteomics revealed plausible drug candidates in the proteome of multi-drug resistant Corynebacterium diphtheriae. Meta Gene. 2018;17:34–42. [Google Scholar]
- 28.Sayers E.W., Barrett T., Benson D.A., Bolton E., Bryant S.H., Canese K. Database resources of the National Center for Biotechnology Information. Nucl Acids Res. 2010;39:D38–D51. doi: 10.1093/nar/gkq1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Consortium U Activities at the universal protein resource (UniProt) Nucl Acids Res. 2013;42:D191–D198. doi: 10.1093/nar/gkt1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yooseph S., Sutton G., Rusch D.B., Halpern A.L., Williamson S.J., Remington K. The sorcerer II global ocean sampling expedition: expanding the universe of protein families. PLoS Biol. 2007;5:432–466. doi: 10.1371/journal.pbio.0050016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang Y., Niu B.F., Gao Y., Fu L.M., Li W.Z. CD-HIT Suite: a web server for clustering and comparing biological sequences. Bioinformatics. 2010;26:680–682. doi: 10.1093/bioinformatics/btq003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shiragannavar S.S., Shettar A.K., Madagi S.B., Sarawad S. Subtractive genomics approach in identifying polysacharide biosynthesis protein as novel drug target against Eubacterium nodatum. Asian J Pharm Pharmacol. 2019;5:382–392. [Google Scholar]
- 33.Johnson M., Zaretskaya I., Raytselis Y., Merezhuk Y., McGinnis S., Madden T.L. NCBI BLAST: a better web interface. Nucl Acids Res. 2008;36:W5–W9. doi: 10.1093/nar/gkn201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sivashanmugam M., Nagarajan H., Vetrivel U., Ramasubban G., Therese K.L., Hajib Narahari M. In silico analysis and prioritization of drug targets in Fusarium solani. Med Hypotheses. 2015;84:81–84. doi: 10.1016/j.mehy.2014.12.015. [DOI] [PubMed] [Google Scholar]
- 35.Habib A.M., Islam M.S., Sohel M., Mazumder M.H.H., Sikder M.O.F., Shahik S.M. Mining the proteome of Fusobacterium nucleatum subsp. nucleatum ATCC 25586 for potential therapeutics discovery: an in silico approach. Genomics Inform. 2016;14:255–264. doi: 10.5808/GI.2016.14.4.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kamath R.S., Fraser A.G., Dong Y., Poulin G., Durbin R., Gotta M. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- 37.Zhang R., Ou H.Y., Zhang C.T. DEG: a database of essential genes. Nucl Acids Res. 2004;32:D271–D272. doi: 10.1093/nar/gkh024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Uddin R., Siddiqui Q.N., Azam S.S., Saima B., Wadood A. Identification and characterization of potential druggable targets among hypothetical proteins of extensively drug resistant Mycobacterium tuberculosis (XDR KZN 605) through subtractive genomics approach. Eur J Pharm Sci. 2018;114:13–23. doi: 10.1016/j.ejps.2017.11.014. [DOI] [PubMed] [Google Scholar]
- 39.Ul Ain Q., Ahmad S., Azam S.S. Subtractive proteomics and immunoinformatics revealed novel B-cell derived T-cell epitopes against Yersinia enterocolitica: an etiological agent of Yersiniosis. Microb Pathogenesis. 2018;125:336–348. doi: 10.1016/j.micpath.2018.09.042. [DOI] [PubMed] [Google Scholar]
- 40.Peng C., Lin Y., Luo H., Gao F. A comprehensive overview of online resources to identify and predict bacterial essential genes. Front Microbiol. 2017;8:2331. doi: 10.3389/fmicb.2017.02331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang R., Lin Y. DEG 5.0, a database of essential genes in both prokaryotes and eukaryotes. Nucl Acids Res. 2008;37:D455–D458. doi: 10.1093/nar/gkn858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luo H., Lin Y., Gao F., Zhang C.T., Zhang R. DEG 10, an update of the database of essential genes that includes both protein-coding genes and noncoding genomic elements. Nucl Acids Res. 2013;42:D574–D580. doi: 10.1093/nar/gkt1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schilling C.H., Schuster S., Palsson B.O., Heinrich R. Metabolic pathway analysis: basic concepts and scientific applications in the post-genomic era. Biotechnol Progr. 1999;15:296–303. doi: 10.1021/bp990048k. [DOI] [PubMed] [Google Scholar]
- 44.Kanehisa M., Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucl Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moriya Y., Itoh M., Okuda S., Yoshizawa A.C., Kanehisa M. KAAS: an automatic genome annotation and pathway reconstruction server. Nucl Acids Res. 2007;35:W182–W185. doi: 10.1093/nar/gkm321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nevo-Dinur K., Govindarajan S., Amster-Choder O. Subcellular localization of RNA and proteins in prokaryotes. Trends Genet. 2012;28:314–322. doi: 10.1016/j.tig.2012.03.008. [DOI] [PubMed] [Google Scholar]
- 47.Peng C., Gao F. Protein localization analysis of essential genes in prokaryotes. Sci Rep. 2015;4:6001. doi: 10.1038/srep06001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zagursky R.J., Olmsted S.B., Russell D.P., Wooters J.L. Bioinformatics: how it is being used to identify bacterial vaccine candidates. Expert Rev Vaccines. 2003;2:417–436. doi: 10.1586/14760584.2.3.417. [DOI] [PubMed] [Google Scholar]
- 49.Bakheet T.M., Doig A.J. Properties and identification of antibiotic drug targets. BMC Bioinf. 2010;11:195. doi: 10.1186/1471-2105-11-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yu N.Y., Wagner J.R., Laird M.R., Melli G., Rey S., Lo R. PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics. 2010;26:1608–1615. doi: 10.1093/bioinformatics/btq249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yu C.S., Chen Y.C., Lu C.H., Hwang J.K. Prediction of protein subcellular localization. Proteins Struct Funct Bioinforma. 2006;64:643–651. doi: 10.1002/prot.21018. [DOI] [PubMed] [Google Scholar]
- 52.Lu Z.Y., Szafron D., Greiner R., Lu P., Wishart D.S., Poulin B. Predicting subcellular localization of proteins using machine-learned classifiers. Bioinformatics. 2004;20:547–556. doi: 10.1093/bioinformatics/btg447. [DOI] [PubMed] [Google Scholar]
- 53.Petersen T.N., Brunak S., Von Heijne G., Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 54.Käll L., Krogh A., Sonnhammer E.L. Advantages of combined transmembrane topology and signal peptide prediction—the Phobius web server. Nucl Acids Res. 2007;35:W429–W432. doi: 10.1093/nar/gkm256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.King B.R., Guda C. ngLOC: an n-gram-based Bayesian method for estimating the subcellular proteomes of eukaryotes. Genome Biol. 2007;8:R68. doi: 10.1186/gb-2007-8-5-r68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tettelin H., Masignani V., Cieslewicz M.J., Donati C., Medini D., Ward N.L. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome”. Proc Natl Acad Sci USA. 2005;102:13950–13955. doi: 10.1073/pnas.0506758102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang Z.K., Luo H., Zhang Y.M., Wang B.J., Gao F. Pan-genomic analysis provides novel insights into the association of E. coli with human host and its minimal genome. Bioinformatics. 2018;35:1987–1991. doi: 10.1093/bioinformatics/bty938. [DOI] [PubMed] [Google Scholar]
- 58.Vernikos G., Medini D., Riley D.R., Tettelin H. Ten years of pan-genome analyses. Curr Opin Microbiol. 2014;23:148–154. doi: 10.1016/j.mib.2014.11.016. [DOI] [PubMed] [Google Scholar]
- 59.Yang X.W., Li Y.J., Zang J., Li Y.X., Bie P.F., Lu Y.L. Analysis of pan-genome to identify the core genes and essential genes of Brucella spp. Mol Genet Genomics. 2016;291:905–912. doi: 10.1007/s00438-015-1154-z. [DOI] [PubMed] [Google Scholar]
- 60.Aslam M., Shehroz M., Shah M., Khan M.A., Afridi S.G., Khan A. Potential druggable proteins and chimeric vaccine construct prioritization against Brucella melitensis from species core genome data. Genomics. 2019;112:1734–1745. doi: 10.1016/j.ygeno.2019.10.009. [DOI] [PubMed] [Google Scholar]
- 61.Wu H., Wang D., Gao F. Toward a high-quality pan-genome landscape of Bacillus subtilis by removal of confounding strains. Brief Bioinform. 2020 doi: 10.1093/bib/bbaa1013. [DOI] [PubMed] [Google Scholar]
- 62.Zhao Y.B., Sun C., Zhao D.Y., Zhang Y.D., You Y., Jia X.M. PGAP-X: extension on pan-genome analysis pipeline. BMC Genomics. 2018;19:115–124. doi: 10.1186/s12864-017-4337-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Blom J., Kreis J., Spänig S., Juhre T., Bertelli C., Ernst C. 2.0: an enhanced software platform for comparative gene content analyses. Nucl Acids Res. 2016;44:W22–W28. doi: 10.1093/nar/gkw255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Grimwade J.E., Leonard A.C. Targeting the bacterial orisome in the search for new antibiotics. Front Microbiol. 2017;8:2352. doi: 10.3389/fmicb.2017.02352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yin Z., Wang Y., Whittell L.R., Jergic S., Liu M., Harry E. DNA replication is the target for the antibacterial effects of nonsteroidal anti-inflammatory drugs. Chem Biol. 2014;21:481–487. doi: 10.1016/j.chembiol.2014.02.009. [DOI] [PubMed] [Google Scholar]
- 66.Heide L. New aminocoumarin antibiotics as gyrase inhibitors. Int J Med Microbiol. 2014;304:31–36. doi: 10.1016/j.ijmm.2013.08.013. [DOI] [PubMed] [Google Scholar]
- 67.Katayama T., Ozaki S., Keyamura K., Fujimitsu K. Regulation of the replication cycle: conserved and diverse regulatory systems for DnaA and oriC. Nat Rev Microbiol. 2010;8:163–170. doi: 10.1038/nrmicro2314. [DOI] [PubMed] [Google Scholar]
- 68.Mott M.L., Berger J.M. DNA replication initiation: mechanisms and regulation in bacteria. Nat Rev Microbiol. 2007;5:343–354. doi: 10.1038/nrmicro1640. [DOI] [PubMed] [Google Scholar]
- 69.Zawilak-Pawlik A., Nowaczyk M., Zakrzewska-Czerwińska J. The role of the N-terminal domains of bacterial initiator DnaA in the assembly and regulation of the bacterial replication initiation complex. Genes. 2017;8:136. doi: 10.3390/genes8050136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Grimwade J.E., Leonard A.C. Blocking the trigger: inhibition of the initiation of bacterial chromosome replication as an antimicrobial strategy. Antibiotics. 2019;8:111. doi: 10.3390/antibiotics8030111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jameson K.H., Wilkinson A.J. Control of initiation of DNA replication in Bacillus subtilis and Escherichia coli. Genes. 2017;8:22. doi: 10.3390/genes8010022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Luo H., Gao F. DoriC 10.0: an updated database of replication origins in prokaryotic genomes including chromosomes and plasmids. Nucl Acids Res. 2019;47:D74–D77. doi: 10.1093/nar/gky1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gao F., Zhang C.T. Ori-Finder: a web-based system for finding oriCs in unannotated bacterial genomes. BMC Bioinf. 2008;9:79. doi: 10.1186/1471-2105-9-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Luo H., Quan C.L., Peng C., Gao F. Recent development of Ori-Finder system and DoriC database for microbial replication origins. Brief Bioinforma. 2018;20:1114–1124. doi: 10.1093/bib/bbx174. [DOI] [PubMed] [Google Scholar]
- 75.Dong Z.C., Chen Y. Transcriptomics: advances and approaches. Sci China Life Sci. 2013;56:960–967. doi: 10.1007/s11427-013-4557-2. [DOI] [PubMed] [Google Scholar]
- 76.Lockhart D.J., Winzeler E.A. Genomics, gene expression and DNA arrays. Nature. 2000;405:827–836. doi: 10.1038/35015701. [DOI] [PubMed] [Google Scholar]
- 77.Wang Z., Gerstein M., Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pabon N.A., Xia Y., Estabrooks S.K., Ye Z., Herbrand A.K., Süß E. Predicting protein targets for drug-like compounds using transcriptomics. PLoS Comput Biol. 2018;14:e1006651. doi: 10.1371/journal.pcbi.1006651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nagalakshmi U., Wang Z., Waern K., Shou C., Raha D., Gerstein M. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 2008;320:1344–1349. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Schena M., Shalon D., Davis R.W., Brown P.O. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 81.Chan J.P., Wright J.R., Wong H.T., Ardasheva A., Brumbaugh J., McLimans C. Using bacterial transcriptomics to investigate targets of host-bacterial interactions in caenorhabditis elegans. Sci Rep. 2019;9:5545. doi: 10.1038/s41598-019-41452-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Klitgaard R.N., Jana B., Guardabassi L., Nielsen K.L., Løbner-Olesen A. DNA damage repair and drug efflux as potential targets for reversing low or intermediate ciprofloxacin resistance in E. coli K-12. Front Microbiol. 2018;9:1438. doi: 10.3389/fmicb.2018.01438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Beydokhti S.S., Stork C., Dobrindt U., Hensel A. Orthosipon stamineus extract exerts inhibition of bacterial adhesion and chaperon-usher system of uropathogenic Escherichia coli—a transcriptomic study. Appl Microbiol Biot. 2019;103:8571–8584. doi: 10.1007/s00253-019-10120-w. [DOI] [PubMed] [Google Scholar]
- 84.Kashaf S.S., Angione C., Lió P. Making life difficult for Clostridium difficile: augmenting the pathogen’s metabolic model with transcriptomic and codon usage data for better therapeutic target characterization. BMC Syst Biol. 2017;11:25. doi: 10.1186/s12918-017-0395-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Płociński P., Macios M., Houghton J., Niemiec E., Płocińska R., Brzostek A. Proteomic and transcriptomic experiments reveal an essential role of RNA degradosome complexes in shaping the transcriptome of Mycobacterium tuberculosis. Nucl Acids Res. 2019;47:5892–5905. doi: 10.1093/nar/gkz251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Maarsingh J.D., Yang S., Park J.G., Haydel S.E. Comparative transcriptomics reveals PrrAB-mediated control of metabolic, respiration, energy-generating, and dormancy pathways in Mycobacterium smegmatis. BMC Genomics. 2019;20:942. doi: 10.1186/s12864-019-6105-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chung M., Teigen L.E., Libro S., Bromley R.E., Olley D., Kumar N. Drug repurposing of bromodomain inhibitors as potential novel therapeutic leads for lymphatic filariasis guided by multispecies transcriptomics. mSystems. 2019;4:e00596–19. doi: 10.1128/mSystems.00596-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Somani D., Adhav R., Prashant R., Kadoo N.Y. Transcriptomics analysis of propiconazole-treated Cochliobolus sativus reveals new putative azole targets in the plant pathogen. Funct Integr Genomic. 2019;19:453–465. doi: 10.1007/s10142-019-00660-9. [DOI] [PubMed] [Google Scholar]
- 89.Cheah M.T., Wachter A., Sudarsan N., Breaker R.R. Control of alternative RNA splicing and gene expression by eukaryotic riboswitches. Nature. 2007;447:497–500. doi: 10.1038/nature05769. [DOI] [PubMed] [Google Scholar]
- 90.Loh E., Dussurget O., Gripenland J., Vaitkevicius K., Tiensuu T., Mandin P. A trans-acting riboswitch controls expression of the virulence regulator PrfA in Listeria monocytogenes. Cell. 2009;139:770–779. doi: 10.1016/j.cell.2009.08.046. [DOI] [PubMed] [Google Scholar]
- 91.Poehlsgaard J., Douthwaite S. The bacterial ribosome as a target for antibiotics. Nat Rev Microbiol. 2005;3:870–881. doi: 10.1038/nrmicro1265. [DOI] [PubMed] [Google Scholar]
- 92.Pavlova N., Kaloudas D., Penchovsky R. Riboswitch distribution, structure, and function in bacteria. Gene. 2019;708:38–48. doi: 10.1016/j.gene.2019.05.036. [DOI] [PubMed] [Google Scholar]
- 93.Blount K.F., Breaker R.R. Riboswitches as antibacterial drug targets. Nat Biotechnol. 2006;24:1558–1564. doi: 10.1038/nbt1268. [DOI] [PubMed] [Google Scholar]
- 94.Yan L.H., Le Roux A., Boyapelly K., Lamontagne A.M., Archambault M.A., Frédéric P.J. Purine analogs targeting the guanine riboswitch as potential antibiotics against Clostridioides difficile. Eur J Med Chem. 2018;143:755–768. doi: 10.1016/j.ejmech.2017.11.079. [DOI] [PubMed] [Google Scholar]
- 95.Pavlova N., Penchovsky R. Genome-wide bioinformatics analysis of FMN, SAM-I, glmS, TPP, lysine, purine, cobalamin, and SAH riboswitches for their applications as allosteric antibacterial drug targets in human pathogenic bacteria. Expert Opin Ther Tar. 2019;23:631–643. doi: 10.1080/14728222.2019.1618274. [DOI] [PubMed] [Google Scholar]
- 96.Mukherjee S., Sengupta S. Riboswitch Scanner: an efficient pHMM-based web-server to detect riboswitches in genomic sequences. Bioinformatics. 2016;32:776–778. doi: 10.1093/bioinformatics/btv640. [DOI] [PubMed] [Google Scholar]
- 97.Aghdam E.M., Hejazi M.S., Barzegar A. Riboswitches: from living biosensors to novel targets of antibiotics. Gene. 2016;592:244–259. doi: 10.1016/j.gene.2016.07.035. [DOI] [PubMed] [Google Scholar]
- 98.Mukherjee S., Das Mandal S., Gupta N., Drory-Retwitzer M., Barash D., Sengupta S. RiboD: a comprehensive database for prokaryotic riboswitches. Bioinformatics. 2019;35:3541–3543. doi: 10.1093/bioinformatics/btz093. [DOI] [PubMed] [Google Scholar]
- 99.Eisenreich W., Heesemann J., Rudel T., Goebel W. Metabolic host responses to infection by intracellular bacterial pathogens. Front Cell Infect Mi. 2013;3:24. doi: 10.3389/fcimb.2013.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.La M.V., Crapoulet N., Barbry P., Raoult D., Renesto P. Comparative genomic analysis of Tropheryma whipplei strains reveals that diversity among clinical isolates is mainly related to the WiSP proteins. BMC Genomics. 2007;8:349. doi: 10.1186/1471-2164-8-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chen L.H., Xiong Z.H., Sun L.L., Yang J., Jin Q. VFDB 2012 update: toward the genetic diversity and molecular evolution of bacterial virulence factors. Nucl Acids Res. 2011;40:D641–D645. doi: 10.1093/nar/gkr989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhou C., Smith J., Lam M., Zemla A., Dyer M.D., Slezak T. MvirDB—a microbial database of protein toxins, virulence factors and antibiotic resistance genes for bio-defence applications. Nucl Acids Res. 2006;35:D391–D394. doi: 10.1093/nar/gkl791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Garg A., Gupta D. VirulentPred: a SVM based prediction method for virulent proteins in bacterial pathogens. BMC Bioinf. 2008;9:62. doi: 10.1186/1471-2105-9-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Doytchinova I.A., Flower D.R. VaxiJen: a server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinf. 2007;8:4. doi: 10.1186/1471-2105-8-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ahmad S., Ranaghan K.E., Azam S.S. Combating tigecycline resistant Acinetobacter baumannii: a leap forward towards multi-epitope based vaccine discovery. Eur J Pharm Sci. 2019;132:1–17. doi: 10.1016/j.ejps.2019.02.023. [DOI] [PubMed] [Google Scholar]
- 106.Naz A., Awan F.M., Obaid A., Muhammad S.A., Paracha R.Z., Ahmad J. Identification of putative vaccine candidates against Helicobacter pylori exploiting exoproteome and secretome: a reverse vaccinology based approach. Infect Genet Evol. 2015;32:280–291. doi: 10.1016/j.meegid.2015.03.027. [DOI] [PubMed] [Google Scholar]
- 107.Artimo P., Jonnalagedda M., Arnold K., Baratin D., Csardi G., De Castro E. ExPASy: SIB bioinformatics resource portal. Nucl Acids Res. 2012;40:W597–W603. doi: 10.1093/nar/gks400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Korepanova A., Gao F.P., Hua Y.Z., Qin H.J., Nakamoto R.K., Cross T.A. Cloning and expression of multiple integral membrane proteins from Mycobacterium tuberculosis in Escherichia coli. Protein Sci. 2005;14:148–158. doi: 10.1110/ps.041022305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Krogh A., Larsson B., Von Heijne G., Sonnhammer E.L. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 110.Tusnady G.E., Simon I. The HMMTOP transmembrane topology prediction server. Bioinformatics. 2001;17:849–850. doi: 10.1093/bioinformatics/17.9.849. [DOI] [PubMed] [Google Scholar]
- 111.Baseer S., Ahmad S., Ranaghan K.E., Azam S.S. Towards a peptide-based vaccine against Shigella sonnei: a subtractive reverse vaccinology based approach. Biologicals. 2017;50:87–99. doi: 10.1016/j.biologicals.2017.08.004. [DOI] [PubMed] [Google Scholar]
- 112.Sajjad R., Ahmad S., Azam S.S. In silico screening of antigenic B-cell derived T-cell epitopes and designing of a multi-epitope peptide vaccine for Acinetobacter nosocomialis. J Mol Graph Model. 2020;94:107477. doi: 10.1016/j.jmgm.2019.107477. [DOI] [PubMed] [Google Scholar]
- 113.Wizemann T.M., Adamou J.E., Langermann S. Adhesins as targets for vaccine development. Emerg Infect Dis. 1999;5:395–403. doi: 10.3201/eid0503.990310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.He Y.Q., Xiang Z.S., Mobley H.L. Vaxign: the first web-based vaccine design program for reverse vaccinology and applications for vaccine development. BioMed Res Int. 2010;2010:297505. doi: 10.1155/2010/297505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sachdeva G., Kumar K., Jain P., Ramachandran S. SPAAN: a software program for prediction of adhesins and adhesin-like proteins using neural networks. Bioinformatics. 2004;21:483–491. doi: 10.1093/bioinformatics/bti028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Dimitrov I., Bangov I., Flower D.R., Doytchinova I. AllerTOP v. 2—a server for in silico prediction of allergens. J Mol Model. 2014;20:2278. doi: 10.1007/s00894-014-2278-5. [DOI] [PubMed] [Google Scholar]
- 117.Saha S., Raghava G. AlgPred: prediction of allergenic proteins and mapping of IgE epitopes. Nucl Acids Res. 2006;34:W202–W209. doi: 10.1093/nar/gkl343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zhang L.D., Huang Y.Y., Zou Z.H., He Y., Chen X.M., Tao A.L. SORTALLER: predicting allergens using substantially optimized algorithm on allergen family featured peptides. Bioinformatics. 2012;28:2178–2179. doi: 10.1093/bioinformatics/bts326. [DOI] [PubMed] [Google Scholar]
- 119.Barh D., Misra A.N., Kumar A., Vasco A. A novel strategy of epitope design in Neisseria gonorrhoeae. Bioinformation. 2010;5:77–85. doi: 10.6026/97320630005077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.González-Díaz H., Pérez-Montoto L.G., Ubeira F.M. Model for vaccine design by prediction of B-epitopes of IEDB given perturbations in peptide sequence, in vivo process, experimental techniques, and source or host organisms. J Immunol Res. 2014;2014:768515. doi: 10.1155/2014/768515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Nazir Z., Afridi S.G., Shah M., Shams S., Khan A. Reverse vaccinology and subtractive genomics-based putative vaccine targets identification for Burkholderia pseudomallei Bp1651. Microb Pathogenesis. 2018;125:219–229. doi: 10.1016/j.micpath.2018.09.033. [DOI] [PubMed] [Google Scholar]
- 122.Ojha R., Nandani R., Prajapati V.K. Contriving multiepitope subunit vaccine by exploiting structural and nonstructural viral proteins to prevent Epstein-Barr virus-associated malignancy. J Cell Physiol. 2019;234:6437–6448. doi: 10.1002/jcp.27380. [DOI] [PubMed] [Google Scholar]
- 123.EL‐Manzalawy Y, Dobbs D, Honavar V. Predicting linear B‐cell epitopes using string kernels. J Mol Recognit 2008;21:243-255. [DOI] [PMC free article] [PubMed]
- 124.Jespersen MC, Peters B, Nielsen M, Marcatili P. BepiPred-2.0: improving sequence-based B-cell epitope prediction using conformational epitopes. Nucleic Acids Res 2017;45:W24-W29. [DOI] [PMC free article] [PubMed]
- 125.Kuhns J.J., Batalia M.A., Yan S., Collins E.J. Poor binding of a HER-2/neu epitope (GP2) to HLA-A2. 1 is due to a lack of interactions with the center of the peptide. J Biol Chem. 1999;274:36422–36427. doi: 10.1074/jbc.274.51.36422. [DOI] [PubMed] [Google Scholar]
- 126.Singh H., Raghava G. ProPred1: prediction of promiscuous MHC Class-I binding sites. Bioinformatics. 2003;19:1009–1014. doi: 10.1093/bioinformatics/btg108. [DOI] [PubMed] [Google Scholar]
- 127.Singh H., Raghava G. ProPred: prediction of HLA-DR binding sites. Bioinformatics. 2001;17:1236–1237. doi: 10.1093/bioinformatics/17.12.1236. [DOI] [PubMed] [Google Scholar]
- 128.Guan P.P., Doytchinova I.A., Zygouri C., Flower D.R. MHCPred: a server for quantitative prediction of peptide–MHC binding. Nucl Acids Res. 2003;31:3621–3624. doi: 10.1093/nar/gkg510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Jia B.F., Raphenya A.R., Alcock B., Waglechner N., Guo P., Tsang K.K. expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucl Acids Res. 2017;2016:D566–D573. doi: 10.1093/nar/gkw1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Asensio N.C., Giner E.M., De Groot N.S., Burgas M.T. Centrality in the host–pathogen interactome is associated with pathogen fitness during infection. Nat Commun. 2017;8:14092. doi: 10.1038/ncomms14092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kitano H. Systems biology: a brief overview. Science. 2002;295:1662–1664. doi: 10.1126/science.1069492. [DOI] [PubMed] [Google Scholar]
- 132.Szklarczyk D., Franceschini A., Kuhn M., Simonovic M., Roth A., Minguez P. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucl Acids Res. 2010;39:D561–D568. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Sussman J.L., Lin D., Jiang J., Manning N.O., Prilusky J., Ritter O. Protein Data Bank (PDB): database of three-dimensional structural information of biological macromolecules. Acta Crystallogr D Biol Crystallogr. 1998;54:1078–1084. doi: 10.1107/s0907444998009378. [DOI] [PubMed] [Google Scholar]
- 134.Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinf. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kelley L.A., Mezulis S., Yates C.M., Wass M.N., Sternberg M.J. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Pieper U., Eswar N., Davis F.P., Braberg H., Madhusudhan M.S., Rossi A. MODBASE: a database of annotated comparative protein structure models and associated resources. Nucl Acids Res. 2006;34:D291–D295. doi: 10.1093/nar/gkj059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Källberg M., Wang H., Wang S., Peng J., Wang Z.Y., Lu H. Template-based protein structure modeling using the RaptorX web server. Nat Protoc. 2012;7:1511–1522. doi: 10.1038/nprot.2012.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Šali A., Blundell T.L. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 139.Fernandez-Fuentes N., Madrid-Aliste C.J., Rai B.K., Fajardo J.E., Fiser A. M4T: a comparative protein structure modeling server. Nucl Acids Res. 2007;35:W363–W368. doi: 10.1093/nar/gkm341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Schwede T., Kopp J., Guex N., Peitsch M.C. SWISS-MODEL: an automated protein homology-modeling server. Nucl Acids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Wiederstein M., Sippl M.J. ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucl Acids Res. 2007;35:W407–W410. doi: 10.1093/nar/gkm290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Eisenberg D., Lüthy R., Bowie J.U. VERIFY3D: assessment of protein models with three-dimensional profiles. Nature. 1992;356:83–85. doi: 10.1038/356083a0. [DOI] [PubMed] [Google Scholar]
- 143.Colovos C., Yeates T. ERRAT: an empirical atom-based method for validating protein structures. Protein Sci. 1993;2:1511–1519. doi: 10.1002/pro.5560020916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Hooft R.W., Vriend G., Sander C., Abola E.E. Errors in protein structures. Nature. 1996;381:272. doi: 10.1038/381272a0. [DOI] [PubMed] [Google Scholar]
- 145.Laskowski R.A., MacArthur M.W., Moss D.S., Thornton J.M. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 146.Shen Y., Maupetit J., Derreumaux P., Tufféry P. Improved PEP-FOLD approach for peptide and miniprotein structure prediction. J Chem Theory Comput. 2014;10:4745–4758. doi: 10.1021/ct500592m. [DOI] [PubMed] [Google Scholar]
- 147.Mayrose I., Penn O., Erez E., Rubinstein N.D., Shlomi T., Freund N.T. Pepitope: epitope mapping from affinity-selected peptides. Bioinformatics. 2007;23:3244–3246. doi: 10.1093/bioinformatics/btm493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Ohto U., Yamakawa N., Akashi-Takamura S., Miyake K., Shimizu T. Structural analyses of human Toll-like receptor 4 polymorphisms D299G and T399I. J Biol Chem. 2012;287:40611–40617. doi: 10.1074/jbc.M112.404608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Kozakov D., Hall D.R., Xia B., Porter K.A., Padhorny D., Yueh C. The ClusPro web server for protein–protein docking. Nat Protoc. 2017;12:255–278. doi: 10.1038/nprot.2016.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Schneidman-Duhovny D., Inbar Y., Nussinov R., Wolfson H.J. PatchDock and SymmDock: servers for rigid and symmetric docking. Nucl Acids Res. 2005;33:W363–W367. doi: 10.1093/nar/gki481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Mashiach E., Schneidman-Duhovny D., Andrusier N., Nussinov R., Wolfson H.J. FireDock: a web server for fast interaction refinement in molecular docking. Nucl Acids Res. 2008;36:W229–W232. doi: 10.1093/nar/gkn186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Lee H., Heo L., Lee M.S., Seok C. GalaxyPepDock: a protein–peptide docking tool based on interaction similarity and energy optimization. Nucl Acids Res. 2015;43:W431–W435. doi: 10.1093/nar/gkv495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 155.Laskowski RA, Swindells MB. LigPlot+: Multiple Ligand–Protein Interaction Diagrams for Drug Discovery. J Chem Inf Model;51:2778-2786. [DOI] [PubMed]
- 156.Agüero F., Al-Lazikani B., Aslett M., Berriman M., Buckner F.S., Campbell R.K. Genomic-scale prioritization of drug targets: the TDR Targets database. Nat Rev Drug Discov. 2008;7:900–907. doi: 10.1038/nrd2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Knox C., Law V., Jewison T., Liu P., Ly S., Frolkis A. DrugBank 3.0: a comprehensive resource for ‘omics’ research on drugs. Nucl Acids Res. 2010;39:D1035–D1041. doi: 10.1093/nar/gkq1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Yang J.Y., Roy A., Zhang Y. Protein–ligand binding site recognition using complementary binding-specific substructure comparison and sequence profile alignment. Bioinformatics. 2013;29:2588–2595. doi: 10.1093/bioinformatics/btt447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Tian W., Chen C., Lei X., Zhao J.L., Liang J. CASTp 3.0: computed atlas of surface topography of proteins. Nucl Acids Res. 2018;46:W363–W367. doi: 10.1093/nar/gky473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Volkamer A., Kuhn D., Rippmann F., Rarey M. DoGSiteScorer: a web server for automatic binding site prediction, analysis and druggability assessment. Bioinformatics. 2012;28:2074–2075. doi: 10.1093/bioinformatics/bts310. [DOI] [PubMed] [Google Scholar]
- 161.Le Guilloux V., Schmidtke P., Tuffery P. Fpocket: an open source platform for ligand pocket detection. BMC Bioinf. 2009;10:168. doi: 10.1186/1471-2105-10-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Huang B.D. MetaPocket: a meta approach to improve protein ligand binding site prediction. OMICS. 2009;13:325–330. doi: 10.1089/omi.2009.0045. [DOI] [PubMed] [Google Scholar]
- 163.Kawabata T. Detection of multiscale pockets on protein surfaces using mathematical morphology. Proteins Struct Funct Bioinforma. 2010;78:1195–1211. doi: 10.1002/prot.22639. [DOI] [PubMed] [Google Scholar]
- 164.Goodsell D.S., Morris G.M., Olson A.J. Automated docking of flexible ligands: applications of AutoDock. J Mol Recognit. 1996;9:1–5. doi: 10.1002/(sici)1099-1352(199601)9:1<1::aid-jmr241>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 165.Daina A., Michielin O., Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep. 2017;7:42717. doi: 10.1038/srep42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Jones G., Willett P., Glen R.C., Leach A.R., Taylor R. Development and validation of a genetic algorithm for flexible docking. J Mol Biol. 1997;267:727–748. doi: 10.1006/jmbi.1996.0897. [DOI] [PubMed] [Google Scholar]
- 167.Geng H., Chen F.F., Ye J., Jiang F. Applications of molecular dynamics simulation in structure prediction of peptides and proteins. Comput Struct Biotec. 2019:1162–1170. doi: 10.1016/j.csbj.2019.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Chen J.Z., Wang X.Y., Pang L.X., Zhang J.Z., Zhu T. Effect of mutations on binding of ligands to guanine riboswitch probed by free energy perturbation and molecular dynamics simulations. Nucl Acids Res. 2019;47:6618–6631. doi: 10.1093/nar/gkz499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Yan F.F., Liu X.G., Zhang S.L., Su J., Zhang Q.G., Chen J.Z. Molecular dynamics exploration of selectivity of dual inhibitors 5M7, 65X, and 65Z toward fatty acid binding proteins 4 and 5. Int J Mol Sci. 2018;19:2496. doi: 10.3390/ijms19092496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Settanni G., Schäfer T., Muhl C., Barz M., Schmid F. Poly-Sarcosine and Poly (ethylene-glycol) interactions with proteins investigated using molecular dynamics simulations. Comput Struct Biotec. 2018;16:543–550. doi: 10.1016/j.csbj.2018.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Pearlman D.A., Case D.A., Caldwell J.W., Ross W.S., Cheatham T.E., III, DeBolt S. AMBER, a package of computer programs for applying molecular mechanics, normal mode analysis, molecular dynamics and free energy calculations to simulate the structural and energetic properties of molecules. Comput Phys Commun. 1995;91:1–41. [Google Scholar]
- 172.Berendsen H.J., van der Spoel D., van Drunen R. GROMACS: a message-passing parallel molecular dynamics implementation. Comput Phys Commun. 1995;91:43–56. [Google Scholar]
- 173.Ichiye T., Karplus M. Collective motions in proteins: a covariance analysis of atomic fluctuations in molecular dynamics and normal mode simulations. Proteins Struct Funct Bioinforma. 1991;11:205–217. doi: 10.1002/prot.340110305. [DOI] [PubMed] [Google Scholar]
- 174.Yan F.F., Liu X.G., Zhang S.L., Zhang Q.G., Chen J.Z. Understanding conformational diversity of heat shock protein 90 (HSP90) and binding features of inhibitors to HSP90 via molecular dynamics simulations. Chem Biol Drug Des. 2020;95:87–103. doi: 10.1111/cbdd.13623. [DOI] [PubMed] [Google Scholar]
- 175.Chen Z.Q., Zhang X.B., Peng C., Wang J.N., Xu Z.J., Chen K.X. D3Pockets: a method and web server for systematic analysis of protein pocket dynamics. J Chem Inf Model. 2019;59:3353–3358. doi: 10.1021/acs.jcim.9b00332. [DOI] [PubMed] [Google Scholar]
- 176.Durrant J.D., de Oliveira C.A.F., McCammon J.A. POVME: an algorithm for measuring binding-pocket volumes. J Mol Graph Model. 2011;29:773–776. doi: 10.1016/j.jmgm.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Gohlke H., Case D.A. Converging free energy estimates: MM-PB (GB) SA studies on the protein–protein complex Ras-Raf. J Comput Chem. 2004;25:238–250. doi: 10.1002/jcc.10379. [DOI] [PubMed] [Google Scholar]
- 178.Zacharias M., Straatsma T., McCammon J. Separation-shifted scaling, a new scaling method for Lennard-Jones interactions in thermodynamic integration. J Chem Phys. 1994;100:9025–9031. [Google Scholar]
- 179.Jorgensen W.L., Thomas L.L. Perspective on free-energy perturbation calculations for chemical equilibria. J Chem Theory Comput. 2008;4:869–876. doi: 10.1021/ct800011m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Xu B.S., Shen H.J., Zhu X., Li G.H. Fast and accurate computation schemes for evaluating vibrational entropy of proteins. J Computat Chem. 2011;32:3188–3193. doi: 10.1002/jcc.21900. [DOI] [PubMed] [Google Scholar]
- 181.Duan L.L., Liu X., Zhang J.Z.H. Interaction entropy – a new paradigm for highly efficient and reliable computation of protein-ligand binding free energy. J Am Chem Soc. 2016;138:5722–5728. doi: 10.1021/jacs.6b02682. [DOI] [PubMed] [Google Scholar]
- 182.Massova I., Kollman P.A. Computational alanine scanning to probe protein−protein Interactions: a novel approach to evaluate binding free energies. J Am Chem Soc. 1999;121:8133–8143. [Google Scholar]
- 183.Duan L.L., Feng G.Q., Wang X.W., Wang L.Z., Zhang Q.G. Effect of electrostatic polarization and bridging water on CDK2–ligand binding affinities calculated using a highly efficient interaction entropy method. Phys Chem Chem Phys. 2017;19:10140–10152. doi: 10.1039/c7cp00841d. [DOI] [PubMed] [Google Scholar]
- 184.Cong Y.L., Li Y.C., Jin K., Zhong S.S., Zhang J.Z.H., Li H. Exploring the reasons for decrease in binding affinity of HIV-2 against HIV-1 protease complex using interaction entropy under polarized force field. Front Chem. 2018;6:380. doi: 10.3389/fchem.2018.00380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Golubkov P.A., Ren P. Generalized coarse-grained model based on point multipole and Gay-Berne potentials. J Chem Phys. 2006;125:64103. doi: 10.1063/1.2244553. [DOI] [PubMed] [Google Scholar]
- 186.Raza S., Azam S.S. AFD: an application for bi-molecular interaction using axial frequency distribution. J Mol Model. 2018;24:84. doi: 10.1007/s00894-018-3601-3. [DOI] [PubMed] [Google Scholar]
- 187.Kurogi Y., Guner O.F. Pharmacophore modeling and three-dimensional database searching for drug design using catalyst. Curr Med Chem. 2001;8:1035–1055. doi: 10.2174/0929867013372481. [DOI] [PubMed] [Google Scholar]
- 188.Sun H.M. Pharmacophore-based virtual screening. Curr Med Chem. 2008;15:1018–1024. doi: 10.2174/092986708784049630. [DOI] [PubMed] [Google Scholar]
- 189.Liu C.S., Yin J.H., Yao J.Q., Xu Z.J., Tao Y., Zhang H.B. Pharmacophore-based virtual screening toward the discovery of novel anti-echinococcal vompounds. Front Cell Infect Mi. 2020;10:118. doi: 10.3389/fcimb.2020.00118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Pappalardo F., Pennisi M., Castiglione F., Motta S. Vaccine protocols optimization: In silico experiences. Biotechnol Adv. 2010;28:82–93. doi: 10.1016/j.biotechadv.2009.10.001. [DOI] [PubMed] [Google Scholar]
- 191.Azman A.S., Luquero F.J., Ciglenecki I., Grais R.F., Sack D.A., Lessler J. The impact of a one-dose versus two-dose oral cholera vaccine regimen in outbreak settings: a modeling study. Plos Med. 2015;12:e1001867. doi: 10.1371/journal.pmed.1001867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Chang H.I., Yeh M.K. Clinical development of liposome-based drugs: formulation, characterization, and therapeutic efficacy. Int J Nanomed. 2012;7:49–60. doi: 10.2147/IJN.S26766. [DOI] [PMC free article] [PubMed] [Google Scholar]