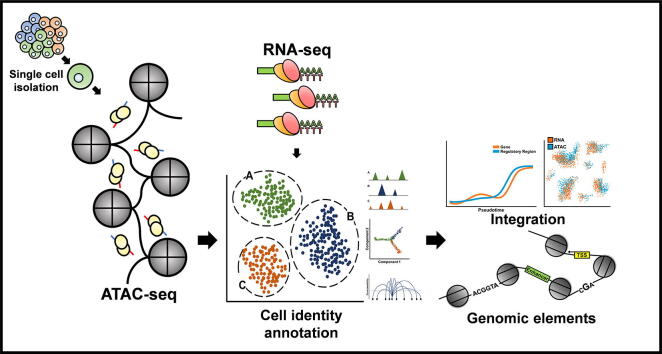
Graphical abstract
Keywords: ATAC sequencing, Chromatin accessibility, Single-cell biology, Single-cell ATAC sequencing, Single-cell RNA sequencing
Abstract
Most genetic variations associated with human complex traits are located in non-coding genomic regions. Therefore, understanding the genotype-to-phenotype axis requires a comprehensive catalog of functional non-coding genomic elements, most of which are involved in epigenetic regulation of gene expression. Genome-wide maps of open chromatin regions can facilitate functional analysis of cis- and trans-regulatory elements via their connections with trait-associated sequence variants. Currently, Assay for Transposase Accessible Chromatin with high-throughput sequencing (ATAC-seq) is considered the most accessible and cost-effective strategy for genome-wide profiling of chromatin accessibility. Single-cell ATAC-seq (scATAC-seq) technology has also been developed to study cell type-specific chromatin accessibility in tissue samples containing a heterogeneous cellular population. However, due to the intrinsic nature of scATAC-seq data, which are highly noisy and sparse, accurate extraction of biological signals and devising effective biological hypothesis are difficult. To overcome such limitations in scATAC-seq data analysis, new methods and software tools have been developed over the past few years. Nevertheless, there is no consensus for the best practice of scATAC-seq data analysis yet. In this review, we discuss scATAC-seq technology and data analysis methods, ranging from preprocessing to downstream analysis, along with an up-to-date list of published studies that involved the application of this method. We expect this review will provide a guideline for successful data generation and analysis methods using appropriate software tools and databases for the study of chromatin accessibility at single-cell resolution.
1. Introduction
Assay for Transposase Accessible Chromatin with high-throughput sequencing (ATAC-seq) was designed to identify open chromatin regions in the genome [1]. Due to the use of hyperactive Tn5 transposase, which simultaneously tags and fragments DNA sequences in open chromatin regions, ATAC-seq requires shorter sample preparation times and fewer number of cells for high quality profiling of chromatin accessibility compared to other existing methods [1]. With emergence of single-cell biology and adaptation of various sequencing-based omics technologies, the study of chromatin accessibility at single cell resolution became possible owing to the development of single-cell ATAC sequencing (scATAC-seq). However, computational analysis of scATAC-seq data remains challenging. Moreover, a wide range of potentially functional elements within the accessible genomic regions add more complexity for interpretation of scATAC-seq data, if they are not well understood. Recently, computational algorithms and software tools for scATAC-seq data analysis have been developed. However, algorithmic approaches and parameters for each step of the data analysis pipeline must be carefully selected for reliable translation of chromatin accessibility information into novel biological hypotheses.
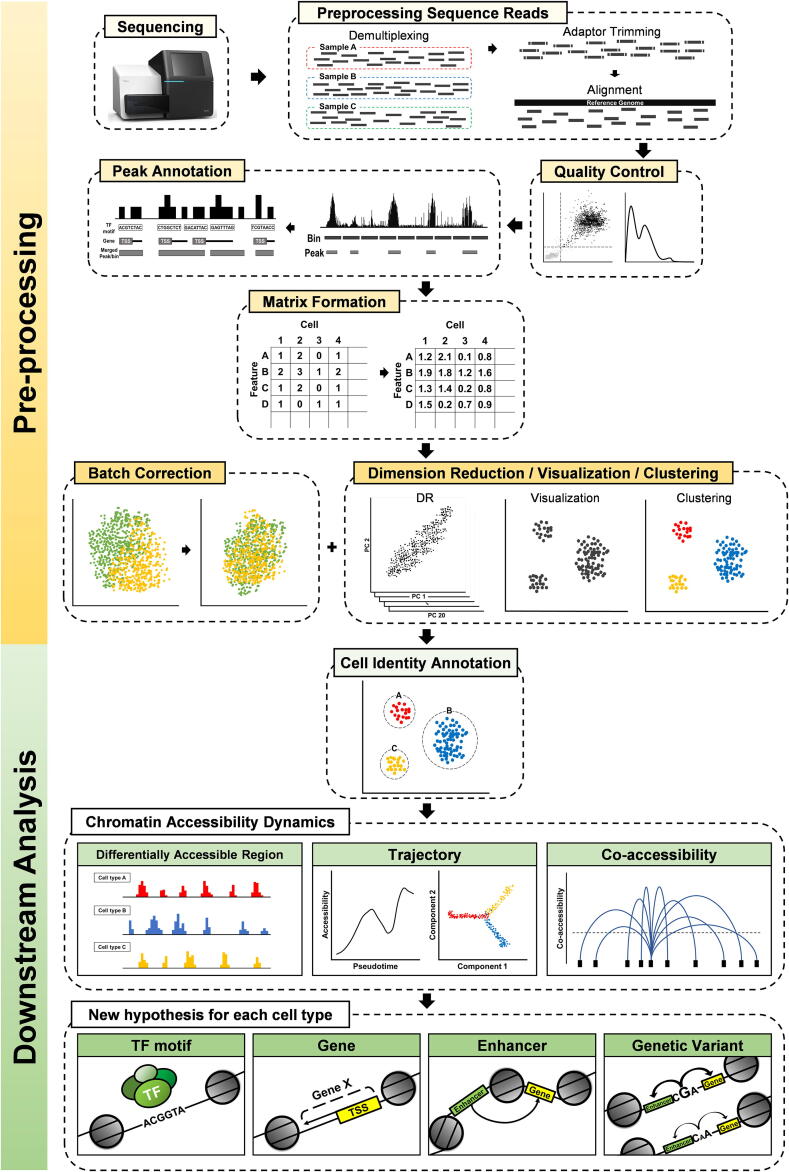
In this review, we aim to elaborate the overall workflow of scATAC-seq data analysis (Fig. 1) from data preprocessing to various downstream analyses, including integration with other types of genetics and genomics data. The analyses of sequencing read data from scATAC-seq are subject to initial data preprocessing, which is similar to those of other next-generation sequencing data [2]. Sequence files are processed with software tools widely used for quality control of sequence information, read mapping to the reference genome, and identification of read peaks that may indicate open chromatin regions [3], [4]. The generation of cell-by-feature matrix is critical for scATAC-seq data analysis and this is facilitated by the various options available for defining genomic features [5]. The preprocessed data are then used for downstream analysis to elucidate networks among cis-regulatory elements, such as promoters and enhancers, and trans-regulatory elements, such as transcription factors (TFs). Gene activity and accessibility to genetic variants can also be analyzed using scATAC-seq data [6]. Moreover, scATAC-seq can be integrated with single-cell RNA sequencing (scRNA-seq) data [7] and other omics data for multi-omics studies.
Fig. 1.
Schematic overview of a typical single-cell ATAC sequencing analysis workflow.
2. Single-cell ATAC sequencing technologies
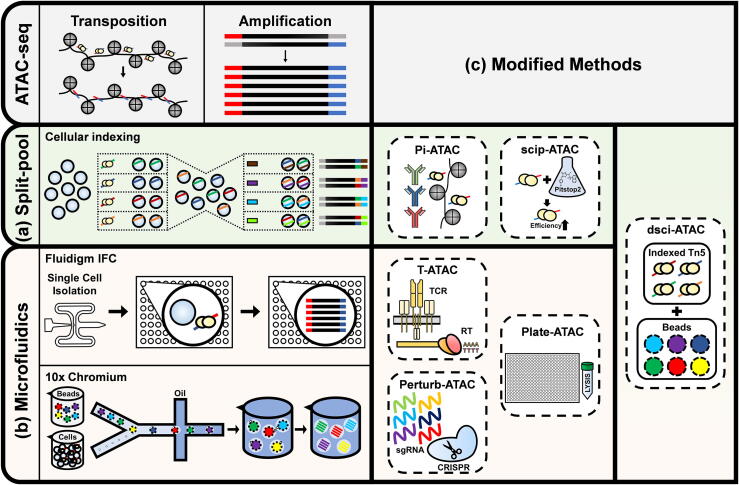
Within two years of the development of bulk ATAC-seq technology, two different strategies of single-cell adaptations were introduced: split-and-pool combinatorial cellular indexing such as sci-ATAC-seq [8] and microfluidics approach such as using integrated fluidics circuit (IFC) [9] (Fig. 2). In sci-ATAC-seq, the nuclei of lysed cells are placed in 96-well plates with uniquely barcoded transposases and pooled back together before dispensing into a second 96-well plate using fluorescence-activated cell sorter (FACS). The second barcodes are introduced during amplification. By recognizing unique combinations of two barcodes, sciATAC allows sequencing of about 1500 cells with median reads of 2500 and ~11% collision rate. In contrast, IFC scATAC-seq utilizes a Fluidigm C1 device to capture single cells and to perform transposition and PCR on IFC. While this method can obtain more than 70,000 reads per cell, only up to 96 cells can be processed in parallel. Another microfluidics-based scATAC-seq using 10x Genomics Chromium device is recently gaining popularity. Chromium system captures single transposed nucleus in a Gel bead-in EMulsion (GEM), which involves addition of unique barcodes to DNA fragments [6]. The scalability and high throughput of GEM, combined with the intuitive software called Cell Ranger ATAC, allows for scATAC-seq study on large number of cells.
Fig. 2.
Schematic summary of two major strategies for single-cell adaptation of ATAC sequencing library generation: (a) split-pool cellular indexing and (b) microfluidics-based, and (c) their modified methods.
Since the initial single-cell adaptations of ATAC-seq technology with cellular indexing and microfluidics, various modifications and improvement have been added to them. Protein-indexed scATAC-seq (Pi-ATAC) [10] profiles protein epitopes in parallel with DNA transposition to quantify protein expression and chromatin accessibility of the same individual cell. The small molecule inhibitor Pitstop2 (scip-ATAC-seq) [11] improves the efficiency of transposase entry into the nucleus and thus enhances library complexity and resolution. Transcript-indexed ATAC-seq (T-ATAC-seq) [12] using microfluidic devices allows sequencing of T cell receptor-encoding genes with ATAC-seq. Perturb-ATAC [13] adds CRISPR single guide RNA (sgRNA) after transposition and sequences both the sgRNA and ATAC DNA to study the relationship among factors regulating chromatin accessibility. Plate-based scATAC-seq facilitates high library complexity with lower amounts of mitochondrial DNA and higher fraction of reads in peaks (FRiP), along with bulk Tn5 tagging and single-nuclei sorting [14]. By combining cellular indexing with microfluidics, droplet microfluidics scATAC-seq with cellular indexing (dsciATAC-seq) [15] maintains read depth of microfluidics-based scATAC-seq while increasing cellular throughput in parallel.
Nano-well ATAC-seq (μATAC-seq) employs ICELL8 platform to offer single cell sequencing with high throughput and low library preparation costs [16]. Nevertheless, it is important to consider the availability of experimental devices, compatibility with analysis software, required read depth and cellular throughput, and overall purpose of study before selecting scATAC-seq technologies.
3. Data preprocessing
Before generating biological hypotheses with downstream analysis, scATAC-seq data must undergo preprocessing steps for accurate interpretation. Preprocessing of scATAC-seq data starts from demultiplexing of sequence files and removal of low-quality cells. Genomic regions used for cell-by-feature matrix, data transformation methods, dimension reduction (DR) approaches, and clustering methods for annotation of cell identities must be carefully selected. Additionally, batch effects must be removed if necessary. Since there is no magic bullet in data analysis, comparisons of multiple methods with complementary algorithms is necessary for obtaining the best result from a given dataset. In Table 1, we summarize 13 software packages available for scATAC-seq data analysis: ChromVAR [17], SCRAT [18], scABC [19], Cicero [20], Scasat [21], cisTopic [22], snapATAC [23], epiScanpy [24], Destin [25], SCALE [26], scATAC-pro [27], Signac [7] and ArchR [28]. Although varying in capability of downstream analysis, they all include unique preprocessing steps. Recently, many of these tools were also evaluated based on the performances in accurately identifying cell types with clustering results [5].
Table 1.
Summary of scATAC-seq analysis software packages.
| Tool | Platform | Feature Matrix | Preprocessing | Clustering | DAR | Motif/k-mer | Gene activity | Co-accessibility | Trajectory | Pathway | Enrichment analysis | scRNA integration | Reference |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ChromVAR | R | TF motifs, k-mer | O | O | X | O | X | X | X | X | X | X | [17] |
| SCRAT | R/Web | Selectable feature | O | O | O | X | X | X | X | X | X | X | [18] |
| scABC | R | Peak | O | O | X | O (ChromVAR) | X | X | X | X | X | X | [19] |
| Cicero | R | TSS | O | O | O | X | O | O | O | X | X | X | [20] |
| Scasat | Python/R | Peak | O | O | O | X | X | X | X | O (GREAT) | X | X | [21] |
| cisTopic | R | Peak | O | O | X | X | O | X | X | O | O | X | [22] |
| snapATAC | Python/R | Bin, peak | O | O | O | O (ChromVAR, Homer) | O | X | X | O (GREAT) | X | O (Seurat) | [23] |
| epiScanpy | Python | Peak | O | O | X | X | X | X | X | X | X | X | [24] |
| Destin | R | Peak | O | O | O | X | X | X | X | X | O | X | [25] |
| SCALE | Python | Peak | O | O | O | O (ChromVAR) | X | X | X | X | X | X | [26] |
| scATAC-pro | Python/R | Peak | O | O | O | O (ChromVAR) | O | O (Cicero) | X | O (GREAT) | X | X | [27] |
| Signac | R | Peak | O | O | O | O (ChromVAR) | O | X | X | X | X | O (Seurat) | [7] |
| ArchR | R | Bin, peak | O | O | O | O (ChromVAR), TF footprinting | O | O | O | X | O | O (Seurat) | [28] |
Tools used in junction are indicated in parentheses.
3.1. Preprocessing of sequencing reads
If multiple samples are indexed and sequenced in a single reaction through multiplexing, they need to be demultiplexed based on the index adapter sequence by software packages, such as Illumina’s bcl2fastq. Demultiplexed sample files are then processed by adaptor trimming, in which adaptor and primer sequences are trimmed off by Bowtie2 [3] or Trimmomatic [29]. Trimmed reads are then aligned to the genome of the same species to the prepared sample using Bowtie2 [3] or BWA [30] and sorted with Samtools [31].
3.2. Quality control
After processing sequencing read data, barcodes that are corresponding to low quality cells or doublets must be filtered out. Generally, quality control (QC) criteria for most of single-cell sequencing technologies are based on the read counts (count depth) and feature counts per barcode [32]. Barcodes with either a low count depth or too high count depth are considered to be low quality cells or doublets, respectively. The same can be applied to the feature counts. However, utilizing unique characteristics of scATAC-seq data may lead to more adequate QC. For example, fraction of reads in peaks (FRiP), ratio of reads in promoter regions, ratio of reads in blacklist sites, or enrichment of transcription start sites (TSS) are often used for barcode selection [9], [23], [28]. Barcodes that do not show nucleosomal banding patterns that are unique to high-quality ATAC-seq data are also excluded [8], [33]. In addition to barcodes, features (e.g., peaks) that are located in blacklist regions or house-keeping genes can be filtered out [23]. It is important to remember that there is no absolute QC standard fitting for all samples. Therefore, combinations of QC criteria must be carefully chosen depending on characteristics of the samples such as overall structure of data, heterogeneity, possibly exiting cell types, batches, or sequencing platforms.
3.3. Cell-by-feature matrix formation
The cells that have passed the QC are selected to generate a cell-by-feature matrix for downstream analysis. A major factor that diversifies the data matrices is defined by the genomic regions from raw peak reads and annotation of the defined regions using regulatory elements. While majority of the pipelines employ a single combination of defining and annotating genomic regions, some pipelines adapt various suited matrices for different purposes of downstream analysis. Largely, the definition of genomic regions can be classified by the use of sample-specific information and feature annotation can be varied with regulatory elements of interest.
The use of sample-specific information includes utilization of bulk ATAC-seq peaks from public data or analyzing aggregated or merged peaks from scATAC-seq data [17], [19], [20], [21], [22], [34]. Single cell aggregation can be carried out using cells from the entire sample or from every cluster (which represents a distinct cell type) obtained from initial temporary clustering results [23]. In most cases, MACS2 [4] is used for peak identification. The other definition of genomic regions is fixed-size bins or windows of the genome along with scores based on the relative abundance of sequence reads for the regions [23], [33].
After defining genomic regions by either peaks or bins/windows of fixed-size, regulatory elements, such as TF motifs and TSS, are used to generate cell-by-feature matrix. Since motifs and k-mers for TF binding are specific for cell types, cell type annotation based on the information is included in some data analysis pipelines [17], [18], [35]. The genomic regions are annotated with either the known TF motifs from public databases, such as cisBP [36], JASPAR [37], and HOMER [38], or k-mers for unsupervised annotation using motifmatchr [39]. Moreover, accessibility to TSS can be used as cell type-specific features [20]. Frequently, these genomic features are combined together to form a feature set for accurate analysis of cell heterogeneity [18]. Some tools simply merge nearby peaks or use them directly as features for matrix formation without annotation of genomic elements [22], [23].
3.4. Batch correction and data integration
When we analyze scATAC-seq data of multiple batches collectively, non-biological factors such as technical variance can lead to wrong biological hypotheses. Batch effects can occur by differences in experimenters, sample preparation protocols, sample harvest time, sequencing lanes, and sequencing technologies [32], [40]. Batch effect correction for scATAC-seq data are often indirectly carried out without specific computational tools. With careful examination, batch-specific peaks or features can be removed [21], [22], [26]. Batch effects are often corrected during other preprocessing steps such as selecting variable peaks or dimension reduction [6], [28], [33].
Batch effects of single-cell omics data can be more systematically corrected with data integration approaches based on non-linear algorithms. These methods assume that all batches share at least one cell type with another and differences between batches are smaller than those between cell types [40]. However, these methods may also remove biological variance, thus resulting in overcorrection. Therefore, both capability of batch removal and conservation of biological variation need to be considered [41]. Although there is no designated tool for integrating scATAC-seq data, those developed for scRNA-seq may be utilized. A benchmarking study of the data integration tools with atlas-level scATAC-seq data showed that most of them performed poorly, which may be attributable to the sparsity and binary nature of the data [41]. Nevertheless, Harmony [42], Seurat v3 [7] and scVI [43] showed best trade-off between batch removal and conservation of biological variation for integrating scATAC-seq data in the benchmarking study.
Data integration tools for batch correction can also be used for integrating multi-modal single-cell omics data (e.g., integration of scRNA-seq and scATAC-seq data generated from the same tissue source). They are further described in the later part of this review.
3.5. Data transformation
Though various experimental technologies are being attempted to increase the sequencing output, peak reads from a single cell have been reported to represent only about 1~10% of overall detectable peaks in scATAC-seq analysis [5]. Therefore, instead of using the initial cell-to-feature matrices directly for downstream analysis, data transformation can be applied to compensate for the limitation from data sparsity. Due to the binary nature of scATAC-seq profiles (1 for presence and 0 for absence of sequence read, respectively), classical text mining methods of topic modeling can be used for data transformation [22], [33]. Term-frequency inverse-document-frequency (TF-IDF) method transforms a cell-to-feature matrix to give more weight to rarer peaks in the cell population [33]. The transformed data matrix tends to capture peaks that are more variable (i.e., more informative) for distinct cell types. Jaccard distances can also be used to measure dissimilarity of two cells in accessibility matrix to signify unique peaks in one cell against all the other peaks [21]. Based on the assumption that higher sequencing depth attributes to the better capture of important features, some methods weigh features of each cell by its sequencing depth [19].
3.6. Dimension reduction, visualization and clustering
After transforming data to overcome inherit sparsity, the cell-by-feature matrix undergoes DR which can mitigate redundant information and potential noise of high dimensional data, and may reduce the computational time for downstream analysis [5]. Principle Component Analysis (PCA) is a widely used linear DR technique and the number of principal components to be chosen is determined based on the elbow of scree plot analysis or Jackstraw test [44]. Topic modeling methods (e.g., cisTopic) reduce dimensions of feature matrix by choosing top topics based on topic-cell distribution generated by latent Dirichlet allocation (LDA) [22]. While LDA is relatively time-consuming, it can capture cell type-specific characteristics which might improve clustering accuracy [5]. Latent Semantic indexing (LSI) is performed by using TF-IDF followed by singular value decomposition (SVD) [33]. Multidimensional scaling (MDS) is also used to reduce dimensions based on the profile similarity among cells [21]. Diffusion map is a nonlinear method of DR processing and it tends to be robust to sequencing noise [23]. While some data analysis pipelines omit a linear DR step, its application is shown to improve overall clustering results during downstream analysis [5]. Overall, the results from these DR methods are used as input for both visualization and clustering.
In order to visualize the data in a 2- or 3-dimensional space, non-linear DR techniques such as t-distributed stochastic neighborhood embedding (t-SNE) [45] and uniform manifold approximation and projection (UMAP) [46] are used. These techniques are often called as embeddings. UMAP visualization tends to preserve global structures better while t-SNE visualization preserves local neighborhoods [46]. However, there are still debates over which methods need to be used for single cell analysis, and frequently the choice of method depends on the properties of each dataset and the data preprocessing method used. Therefore, it is highly recommended to apply multiple visualization methods for given datasets and make a choice based on the results obtained.
Cells with similar accessibility profiles can be organized into clusters. For scATAC-seq data, there are several clustering methods which are widely used: hierarchical, k-means, k-medoids, and Louvain algorithm. Hierarchical clustering is useful for understanding overall relationships among clusters and the result is often visualized with a dendrogram to capture the hierarchical relationship. K-means and k-medoids clustering use parametric algorithms with predetermined number of clusters. K-medoids clustering is known to be more robust to noise but requires more computational power. Louvain clustering is a graph-based method which often takes the results of k-nearest neighbor (KNN) method as input data [7], [23]. Some analytic tools might have preferred methods for clustering but in most cases they are interchangeable. Recent benchmark results for clustering scATAC-seq data showed the most favorable results with Louvain clustering [5].
4. Downstream analysis for hypothesis generation
The main purpose of the single-cell omics study is to generate biological hypotheses about distinct subsets of complex mixtures of heterogeneous cellular population. Thus, downstream analysis generally begins with assigning cellular identity of the clusters obtained from the preprocessed scATAC-seq data. Peak calling is often repeated for each cluster to identify accessible chromatin regions for distinct cellular populations, which are then subject to a statistical test for association with various pre-defined genomic features, such as, cis- and trans-regulatory elements and genetic variants, such as, disease-associated SNPs. Downstream analysis methods mainly aim to uncover novel regulatory elements and to understand their functional roles in a cell type-specific manner. In addition, the dynamics of chromatin accessibility during cellular development can be studied during downstream analysis.
4.1. Cell identity annotation
For the analysis of single-cell omics data, cell identity annotation of clusters is preliminary yet must be carried out with care. Incorrect cell identity information can mislead to a wrong biological hypothesis during downstream analysis of scATAC-seq data. While there are a number of tools for automated cell type annotation for scRNA-seq data [47] and an extensive list of cell type-specific genes are available from various databases [32], [48], there are only a limited set of tools for scATAC-seq data and a few reference datasets for cell type specific chromatin accessibility [33]. Therefore, combined use of complementary approaches for cluster annotation is necessary for scATAC-seq data. Largely, there are two approaches to cell identity annotation; the first one is based on feature annotation of ATAC peaks, and the second one utilizes integration with reference scRNA-seq data.
After cells are assigned to distinct clusters based on profile similarities, each cluster can have Differentially Accessible Regions (DARs) which might contain various regulatory elements. The first approach to cell identity annotation can vary in genomic features to be used for identification of such cluster-specific peaks. Supervised or manual annotation of cluster identity requires databases or literature references of cell type specific genomic features, such as TF motifs, enhancers, promoters, and TSSs [6]. Promoters and TSSs are most widely used for cluster annotation due to the extensive list of cell type-specific genes. In simpler approaches, accessibility to the cell type-specific genes can be defined by the existence of ATAC peaks within certain distance from upstream of cell type-specific promoters or TSSs. More advanced analysis takes various distal and proximal regulatory elements into consideration. ‘Gene activity scores’ weigh co-accessible elements to a gene’s promoter region differently to infer gene expression from chromatin accessibility profiles more accurately [33]. As a result, gene activity scores correlate better with gene expression profiles than simple profiles of promoter accessibility [33]. A software called Garnett also employs gene activity scores and a priori profile with known cell types along with their marker genes for supervised classification of cell types [49].
The second approach takes advantage of extensively available scRNA-seq data for diverse cell types. Gene expression matrix from scRNA-seq data can be integrated with gene activity score matrix from scATAC-seq data for the same cell types. After projecting them onto the maximally correlated dimensions, mutual nearest neighbors (MNN) algorithm is used to transfer cell-labels from the scRNA-seq data to the scATAC-seq data [7], [33]. While samples with a highly dominant cell type or non-matching cell types to the other omics data show limitations in accuracy, overall results of cell identity annotations are concordant with matching datasets [33]. With semi-supervised identification of cell populations in scATAC-seq data (SSIPs), existing reference scRNA-seq and bulk ATAC-seq data are used to generate a network of scATAC-seq data for the sample of interest, with reference cells from external data sources to transfer cell-labels [50].
4.2. Study of chromatin accessibility dynamics
Annotated clusters proceed to the study of chromatin accessibility dynamics. Hypotheses about regulation of cellular development can be generated using various genomic elements associated with DARs, pseudotime-dependent changes, and co-accessibility. DAR analysis is used to identify regulatory elements specific for each cell type. In general, cell type-specific DARs are identified by comparing chromatin accessibility in cells for a particular cluster with all the other cells in the dataset. Various statistical tests have been employed for DAR analysis, including a binomial test [33], negative binomial generalized linear model [20], a Wald test [19], Fisher’s exact test [23], unequal variances t-test [17], and information gain [21] along with 1% or 5% false discovery rate (FDR) adjustment with Benjamini-Hochberg [6], [23], [33] or Bonferroni correction [21].
Single-cell trajectory analysis utilizes pseudotemporal ordering of cells to reconstruct differentiation processes or cell lineages. Trajectory analysis is useful if chromatin accessibility changes continuously within cell population. Cicero [20] is an extension of Monocle2 [51], a widely used trajectory analysis tool for scRNA-seq data, for scATAC-seq data. Nearby peaks are aggregated for dealing with sparsity and DARs are selected to define temporal states. After cells are ordered in pseudotime using DDRTree [52] method, accessibility kinetics at selected genomic regions can be depicted. STREAM [53] is a trajectory analysis tool that can handle both transcriptomic and epigenomic data. For analyzing scATAC-seq data, k-mer score matrix in accessibility variable regions is used to construct pseudotime trajectories. The strength of STREAM lies in an unbiased end-to-end pipeline starting with unprocessed raw data files. Trajectory analysis with such tools can be used for identification of cell type-specific regulatory elements associated with cellular development from one cell type to another [6], [20], [54], [55], [56]. For example, if accessibility of TF motifs changes significantly during differentiation process, the matching TFs can be studied further for their involvement in activation or initiation of differentiation [11], [53], [57].
Interactions between different genomic elements are important for understanding regulatory networks. Such interactions can be analyzed with co-accessibility of different genomic regions. Cicero groups similar cells to generate cell accessibility matrix to calculate covariance between each pair of genomic elements in overlapping genomic windows. Co-accessibility is used for analysis of interactions between TSS and enhancers [8], [11], [57], [58], [59], promoters [20], and other genomic elements.
4.3. TF motif-based hypothesis generation
TFs are major trans-acting regulators of gene expression. Analysis of scATAC-seq enables identifying specific TFs for different cell types within heterogeneous cell population [17]. Since TFs are highly involved in the developmental process, the analysis of cell-to-cell variation of TF expression will facilitate understanding of their roles during cellular differentiation [35]. Furthermore, scATAC-seq allows for simultaneous analysis of cis-regulatory elements that are associated with the activities of relevant TFs.
Study of TFs with scATAC-seq data requires both software packages and databases for TFs and their binding motifs. Initially, methods for scATAC-seq data analysis mainly utilized the known TF motifs [8], [9]. Though not invented solely for analysis of scATAC-seq data, bioinformatics tools, such as, Homer [38] and FIMO [60], are useful in identifying TF motifs within open chromatin regions. A software package chromVAR, which was developed for scATAC-seq analysis, is a widely used algorithm for calculating bias-corrected deviations and z-scores of TF motifs and k-mers [17]. TFs related to various cell types, such as immune cells [12], cardiac progenitor cells [55], and neuronal cells [61], have been analyzed with chromVAR deviations and z-scores. Furthermore, TF motif accessibility can be compared with TF expression values calculated from scRNA-seq data [62]. For identification of cell type specific TFs and prediction of cell types from those TF motifs, several models, such as convolutional neural network [33] and random forest classification [57], can be used.
4.4. Gene-based hypothesis generation
scRNA-seq has been widely used for studying gene expression profiles of heterogeneous cell populations [63]. Gene expression can be inferred from chromatin accessibility information at TSSs, gene body, and other regulatory elements. TSSs and transcription termination sites of active genes are located at open chromatin regions or nucleosome-depleted regions [64] and so, accessibility profiles at TSSs can be utilized for gene-based downstream analysis of scATAC-seq data.
UROPA [65] can assign TSSs to scATAC-seq peaks using genomic annotation databases. Peaks annotated by TSSs can be used for further analysis, such as comparison of opening and closing of chromatin at TSSs [55], calculation of TSS gene set deviation [58], and identification of chromatin accessibility at known marker genes to identify cell types and states [61], [66], [67]. However, considering only the chromatin states of TSSs might not fully indicate gene expression [20], calculation of ‘gene activity score’, which takes information from regulatory elements, can improve translation of accessibility information into gene expression [20]. Cicero gene activity scores consider accessibility at sites proximal and distal to TSS of a gene and weigh them by their co-accessibility. Gene activity scores have been used to compare TF motif accessibility to TF gene activity scores from the same scATAC-seq data [11], to annotate cells using cell type specific marker genes [6], [68], and to transfer cell-labels from scRNA-seq datasets to matched scATAC-seq datasets [69]. Lastly, for visualization of DARs at gene bodies, Deeptools [70] and MACS2 [4] generates bigwig files, which can be displayed with genomic browsers, such as Gviz [71], Integrative Genomics Viewer (IGV) [72], and UCSC Genome Browser [73].
Gene set enrichment analysis for a distinct cell population is useful for identifying pathways relevant to the cell identity. Gene Ontology (GO) [74] and KEGG [75] are the most widely used databases for pathway gene sets. Pathways associated with a cell population are analyzed based on genes associated with cell type-specific accessible (peak) regions. Peaks within upstream and downstream extension of gene body [61], [76], TSS [55], or with gene activity scores [57] are used as input data for pathway analysis. Various gene set enrichment tools, such as GREAT [77] or clusterProfiler [78], can be applied to scATAC-seq data.
4.5. Enhancer-based hypothesis generation
Enhancers are cis-regulatory elements distal from their regulatory target genes. Proximal and distal interactions of enhancers with other regulatory elements have been identified by analyzing 3D structures of chromatin [79]. Moreover, enhancer dense regions, called super-enhancers, are known to be cell type and state specific [80] and are involved in disease-associated regulatory nodes [81]. Studies on enhancers at single cell resolution are useful in predicting specific cell types, as it has higher accuracy than other cis-regulatory elements and transcriptomes [82].
Various studies have focused on identifying cell type-specific enhancers and their involvement in developmental processes. The most common types of enhancer analyses include identification of cell type-specific distal and proximal enhancers [76] and relative enrichment of enhancer activities [54], [83], [84]. Notably, various enhancer databases, such as VISTA [85], CRM Activity Database (CAD) [54], Redfly Enhancer [86], and Vienna Tiles library [87], can be utilized for such analysis. Furthermore, evaluating the interactions of enhancers to promoters or genes with co-accessibility [57], paired scRNA-seq data [88], virtual latent space [69], and Activity-by-Contact model [83] have been suggested in several data analysis pipelines.
4.6. Hypothesis generation with disease-associated genetic variants
Disease-associated SNPs detected via genome-wide association study (GWAS) and expression quantitative trait loci (eQTL) analysis are useful resources for understanding genomic regulation in diseases. Since most SNPs are located in non-coding regions [68], it is anticipated that many GWAS SNPs and eQTLs are associated with cis-regulatory elements; therefore, the study of open chromatin regions are useful in identifying their functional effects [89], [90]. In addition, the identification of cell types relevant to disease-associated variants is crucial for in-depth understanding of these variants [91]. Using scATAC-seq, genetic variants can be linked to their cellular and functional targets through the identification of both DNA sequences and chromatin accessibility of regulatory elements at single cell resolution. While relating various epigenetic features to GWAS signals through various bulk sequencing methods have provided useful results, single cell resolution analysis additionally enables us to overcome the limitations imposed by cell-type heterogeneity [33]. Indeed, several studies have demonstrated the importance of providing enrichment profiles of GWAS SNPs in cell-type specific peaks [25], [33], [67], [92]. The modified version of chromVAR, called gchromVAR, scores each single cell for GWAS enrichment to identify causal variants in genomic regions and putative target genes of those variants in a cell type specific manner [92]. By utilizing co-accessibility measurements, interconnected peaks overlapping GWAS SNPs and GTEx eQTL to other peaks containing regulatory elements can be analyzed [6]. GREGOR [93] is also used to annotate enrichment of disease-associated SNPs from various databases [67]. More complex models using deep learning and machine learning framework to identify cell type-specific functional SNPs and associated novel functional genes were also implemented in some recent studies [67], [68].
5. Integrative analysis with single-cell transcriptome data
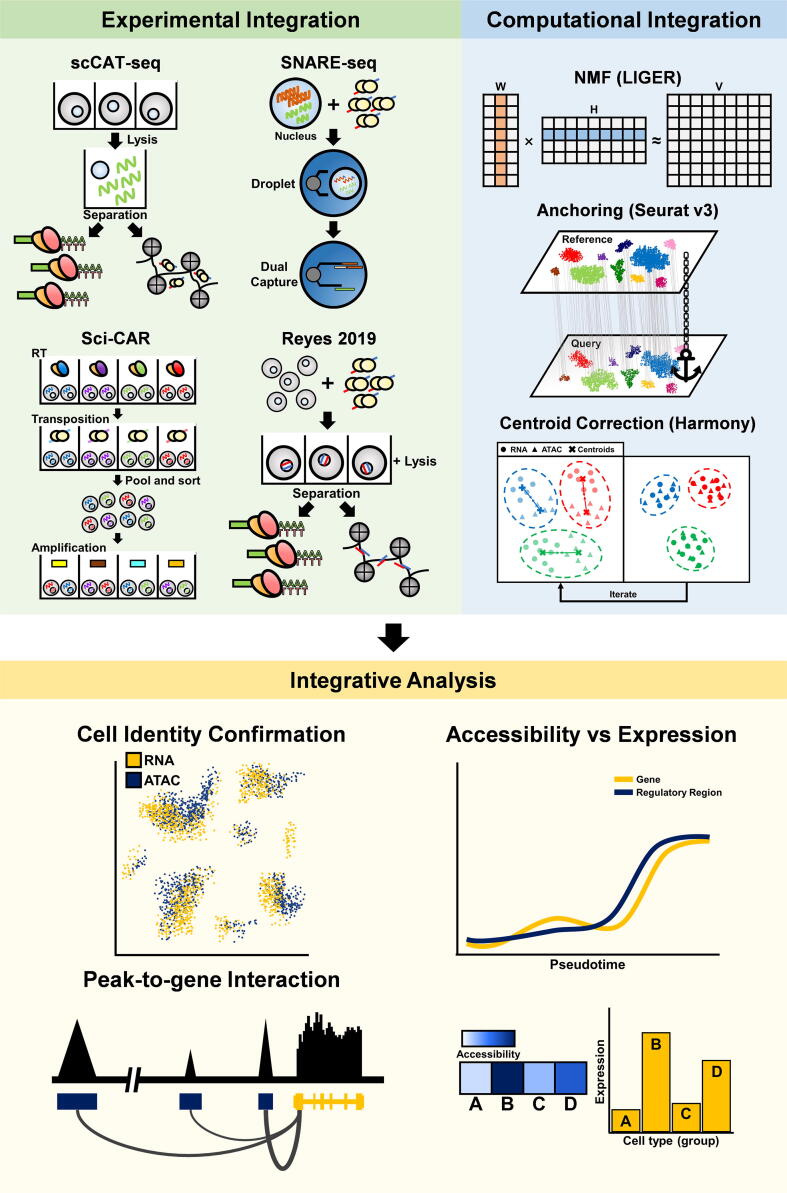
Integration of single-cell gene expression and chromatin accessibility data may improve cell identity annotation. More importantly, joint analysis of multimodal data will facilitate detecting correlations between trans- and cis-regulatory elements underlying the cellular state of interest. Integrative analysis of single-cell transcriptome and chromatin accessibility can be achieved by both experimental and computational approaches (Fig. 3).
Fig. 3.
Integration of single-cell ATAC sequencing data with single-cell RNA sequencing data via experimental approaches and computational approaches. Integrative analysis of gene expression and chromatin accessibility for the same cell types can be used for confirming cell identity annotation and for facilitating generating new hypotheses for regulatory elements. For example, identification of peak-to-gene interactions can infer enhancer-promoter interactions; comparison between expression of a gene and accessibility of its TF-enriched regions across pseudotime can reveal kinetic relationship between transcription and regulatory regions; comparison between expression of a gene and accessibility of its TF-enriched regions across cell types or sample groups can reveal expression and accessibility signature associated with a cell type or subpopulation.
Experimental approaches to integrative analysis focus on obtaining transcriptome and epigenome data from the same cells simultaneously. The multimodal single-cell analysis method sci-CAR employs combinatorial indexing method for both scRNA-seq and scATAC-seq to increase throughput [94]. Another method, single-cell chromatin accessibility and transcriptome sequencing (scCAT-seq), separates cytoplasm components and nuclei for scRNA-seq and scATAC-seq, respectively [95]. Single-Nucleus chromatin Accessibility and mRNA Expression sequencing (SNARE-seq) method utilizes linked barcodes for capturing both gDNA from transposed DNA fragments and mRNA from a nucleus in a single droplet for parallel sequencing using the same barcodes for each cell [96]. There is a method that involves cell fixation with chemical reagents, followed by bulk transposition for single cell sorting to reduce the cost and simplify overall procedures [97]. Using multimodal single-cell technologies, chromatin accessibility can be directly compared to gene expression for understanding the functional relationships between cis/trans-regulatory elements and associated gene expressions.
At present, there are algorithmic approaches for computational integration of single-cell genomics data derived from different sample groups, experiments, or even technologies. Methods based on non-negative matrix factorization (NMF), such as CoupledNMF [62] and LIGER [98], have been proven useful in multimodal single-cell data integration. Seurat v3 is a widely used method for scRNA-seq and scATAC-seq integration [7]. Seurat v3 integrates multimodal single-cell data by projecting two different datasets into a sub-space defined by correlated variables and then identifying anchors between datasets. Harmony is a fast and scalable algorithm of single-cell data integration based on iterative adjustment of data-specific clusters [42]. Recently, more approaches for data integration were reported, including the maximum mean discrepancy manifold alignment (MMD-MA) algorithm [99] and De-Convolution and Coupled-Clustering (DC3) [100]. The single-cell multi-omics integration has been used for validation of cell identity assignments [57], [58], [69], linking differentially expressed genes (DEGs) to DARs for inference of enhancer-promoter (E-P) interactions [88], observation of a trend for accessibility of enhancers predicted by TF-motif to precede changes in gene expression [83] and identification of conserved chromatin accessibility and transcription across cell types or sample groups [101].
6. Conclusion and outlook
Despite potentially wide applications in the study of cellular systems, a relatively high cost of single-cell sequencing technologies and high complexity of the data might limit accessibility to single-cell biology for many researchers. However, there have been many community-wide efforts for improving both experimental and computational methods of single-cell omics, including scATAC-seq data analysis. While a reasonable consensus in data analysis pipelines has not been achieved yet, the number of publications on data generation technology and data analysis methods for scATAC-seq are growing exponentially during recent times. Benchmarking studies utilizing different methods for data generation and analysis would provide useful information to the community for establishing the best practices of scATAC-seq data analysis [27]. Moreover, integration with other types of single-cell and bulk omics data, as well as genomic variation data, will greatly potentiate scATAC-seq studies aimed at elucidating complex circuits of gene regulation involved in disease progression. Especially, integration of scATAC-seq with other epigenetic technologies, such as ChIP-seq and Hi-C, will unravel 3D chromatin structures [68], [102]. Such integrative multimodal analysis will facilitate identification of key regulators involved in disease progression, which are often potential therapeutic targets and biomarkers for diagnosis. Conclusively, we anticipate that scATAC-seq will promote a holistic view of epigenetic regulation and regulatory networks involved in the development of normal cells and disease progression in human and other multi-cellular organisms.
CRediT authorship contribution statement
Seungbyn Baek: Conceptualization, Writing - original draft, Writing - review & editing. Insuk Lee: Conceptualization, Writing - review & editing, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This study was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (2018M3C9A5064709, 2018R1A5A2025079, 2019M3A9B6065192) and Brain Korea 21 (BK21) PLUS program.
References
- 1.Buenrostro J.D., Giresi P.G., Zaba L.C., Chang H.Y., Greenleaf W.J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Valouev A., Johnson D.S., Sundquist A., Medina C., Anton E., Batzoglou S. Genome-wide analysis of transcription factor binding sites based on ChIP-Seq data. Nat Methods. 2008;5:829–834. doi: 10.1038/nmeth.1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen H., Lareau C., Andreani T., Vinyard M.E., Garcia S.P., Clement K. Assessment of computational methods for the analysis of single-cell ATAC-seq data. Genome Biol. 2019;20:241. doi: 10.1186/s13059-019-1854-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Satpathy A.T., Granja J.M., Yost K.E., Qi Y., Meschi F., McDermott G.P. Massively parallel single-cell chromatin landscapes of human immune cell development and intratumoral T cell exhaustion. Nat Biotechnol. 2019;37:925–936. doi: 10.1038/s41587-019-0206-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM, 3rd, Hao Y, Stoeckius M, Smibert P, Satija R. Comprehensive Integration of Single-Cell Data. Cell 2019;177:1888–02 e21. [DOI] [PMC free article] [PubMed]
- 8.Cusanovich D.A., Daza R., Adey A., Pliner H.A., Christiansen L., Gunderson K.L. Multiplex single cell profiling of chromatin accessibility by combinatorial cellular indexing. Science. 2015;348:910–914. doi: 10.1126/science.aab1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buenrostro J.D., Wu B., Litzenburger U.M., Ruff D., Gonzales M.L., Snyder M.P. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature. 2015;523:486–490. doi: 10.1038/nature14590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen X., Litzenburger U.M., Wei Y., Schep A.N., LaGory E.L., Choudhry H. Joint single-cell DNA accessibility and protein epitope profiling reveals environmental regulation of epigenomic heterogeneity. Nat Commun. 2018;9:4590. doi: 10.1038/s41467-018-07115-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mulqueen RM, DeRosa BA, Thornton CA, Sayar Z, Torkenczy KA, Fields AJ, Wright KM, Nan X, Ramji R, Steemers FJ, et al. Improved single-cell ATAC-seq reveals chromatin dynamics of in vitro corticogenesis. bioRxiv 2019:637256.
- 12.Satpathy A.T., Saligrama N., Buenrostro J.D., Wei Y., Wu B., Rubin A.J. Transcript-indexed ATAC-seq for precision immune profiling. Nat Med. 2018;24:580–590. doi: 10.1038/s41591-018-0008-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rubin AJ, Parker KR, Satpathy AT, Qi Y, Wu B, Ong AJ, Mumbach MR, Ji AL, Kim DS, Cho SW, et al. Coupled Single-Cell CRISPR Screening and Epigenomic Profiling Reveals Causal Gene Regulatory Networks. Cell 2019;176:361–76 e317. [DOI] [PMC free article] [PubMed]
- 14.Chen X., Miragaia R.J., Natarajan K.N., Teichmann S.A. A rapid and robust method for single cell chromatin accessibility profiling. Nat Commun. 2018;9:5345. doi: 10.1038/s41467-018-07771-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lareau C.A., Duarte F.M., Chew J.G., Kartha V.K., Burkett Z.D., Kohlway A.S. Droplet-based combinatorial indexing for massive-scale single-cell chromatin accessibility. Nat Biotechnol. 2019;37:916–924. doi: 10.1038/s41587-019-0147-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mezger A., Klemm S., Mann I., Brower K., Mir A., Bostick M. High-throughput chromatin accessibility profiling at single-cell resolution. Nat Commun. 2018;9:3647. doi: 10.1038/s41467-018-05887-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schep A.N., Wu B., Buenrostro J.D., Greenleaf W.J. chromVAR: inferring transcription-factor-associated accessibility from single-cell epigenomic data. Nat Methods. 2017;14:975–978. doi: 10.1038/nmeth.4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ji Z., Zhou W., Ji H. Single-cell regulome data analysis by SCRAT. Bioinformatics. 2017;33:2930–2932. doi: 10.1093/bioinformatics/btx315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zamanighomi M., Lin Z., Daley T., Chen X., Duren Z., Schep A. Unsupervised clustering and epigenetic classification of single cells. Nat Commun. 2018;9:2410. doi: 10.1038/s41467-018-04629-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pliner HA, Packer JS, McFaline-Figueroa JL, Cusanovich DA, Daza RM, Aghamirzaie D, Srivatsan S, Qiu X, Jackson D, Minkina A, et al. Cicero Predicts cis-Regulatory DNA Interactions from Single-Cell Chromatin Accessibility Data. Mol Cell 2018;71:858–71 e8. [DOI] [PMC free article] [PubMed]
- 21.Baker S.M., Rogerson C., Hayes A., Sharrocks A.D., Rattray M. Classifying cells with Scasat, a single-cell ATAC-seq analysis tool. Nucleic Acids Res. 2019;47 doi: 10.1093/nar/gky950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bravo Gonzalez-Blas C., Minnoye L., Papasokrati D., Aibar S., Hulselmans G., Christiaens V. cisTopic: cis-regulatory topic modeling on single-cell ATAC-seq data. Nat Methods. 2019;16:397–400. doi: 10.1038/s41592-019-0367-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fang R, Preissl S, Hou X, Lucero J, Wang X, Motamedi A, Shiau AK, Mukamel EA, Zhang Y, Behrens MM, et al. Fast and Accurate Clustering of Single Cell Epigenomes Reveals Cis-Regulatory Elements in Rare Cell Types. bioRxiv 2019:615179.
- 24.Danese A, Richter ML, Fischer DS, Theis FJ, Colomé-Tatché M. EpiScanpy: integrated single-cell epigenomic analysis. bioRxiv 2019:648097. [DOI] [PMC free article] [PubMed]
- 25.Urrutia E., Chen L., Zhou H., Jiang Y. Destin: toolkit for single-cell analysis of chromatin accessibility. Bioinformatics. 2019;35:3818–3820. doi: 10.1093/bioinformatics/btz141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xiong L., Xu K., Tian K., Shao Y., Tang L., Gao G. SCALE method for single-cell ATAC-seq analysis via latent feature extraction. Nat Commun. 2019;10:4576. doi: 10.1038/s41467-019-12630-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu W., Uzun Y., Zhu Q., Chen C., Tan K. scATAC-pro: a comprehensive workbench for single-cell chromatin accessibility sequencing data. Genome Biol. 2020;21:94. doi: 10.1186/s13059-020-02008-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Granja JM, Corces MR, Pierce SE, Bagdatli ST, Choudhry H, Chang H, Greenleaf W. ArchR: An integrative and scalable software package for single-cell chromatin accessibility analysis. bioRxiv 2020:04.28.066498.
- 29.Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N. Genome project data processing S: the sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luecken M.D., Theis F.J. Current best practices in single-cell RNA-seq analysis: a tutorial. Mol Syst Biol. 2019;15 doi: 10.15252/msb.20188746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cusanovich DA, Hill AJ, Aghamirzaie D, Daza RM, Pliner HA, Berletch JB, Filippova GN, Huang X, Christiansen L, DeWitt WS, et al. A Single-Cell Atlas of In Vivo Mammalian Chromatin Accessibility. Cell 2018;174:1309–24 e18. [DOI] [PMC free article] [PubMed]
- 34.Zhao C., Hu S., Huo X., Zhang Y. Dr.seq2: A quality control and analysis pipeline for parallel single cell transcriptome and epigenome data. PLoS ONE. 2017;12 doi: 10.1371/journal.pone.0180583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.de Boer C.G., Regev A. BROCKMAN: deciphering variance in epigenomic regulators by k-mer factorization. BMC Bioinf. 2018;19:253. doi: 10.1186/s12859-018-2255-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weirauch M.T., Yang A., Albu M., Cote A.G., Montenegro-Montero A., Drewe P. Determination and inference of eukaryotic transcription factor sequence specificity. Cell. 2014;158:1431–1443. doi: 10.1016/j.cell.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mathelier A., Fornes O., Arenillas D.J., Chen C.Y., Denay G., Lee J. JASPAR 2016: a major expansion and update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2016;44:D110–D115. doi: 10.1093/nar/gkv1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schep A. motifmatchr: Fast Motif Matching in R R package version 1.10.0. edition; 2020.
- 40.Tran H.T.N., Ang K.S., Chevrier M., Zhang X., Lee N.Y.S., Goh M. A benchmark of batch-effect correction methods for single-cell RNA sequencing data. Genome Biol. 2020;21:12. doi: 10.1186/s13059-019-1850-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luecken MD, Büttner M, Chaichoompu K, Danese A, Interlandi M, Mueller MF, Strobl DC, Zappia L, Dugas M, Colomé-Tatché M, Theis FJ. Benchmarking atlas-level data integration in single-cell genomics. bioRxiv 2020:05.22.111161. [DOI] [PMC free article] [PubMed]
- 42.Korsunsky I., Millard N., Fan J., Slowikowski K., Zhang F., Wei K. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat Methods. 2019;16:1289–1296. doi: 10.1038/s41592-019-0619-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lopez R., Regier J., Cole M.B., Jordan M.I., Yosef N. Deep generative modeling for single-cell transcriptomics. Nat Methods. 2018;15:1053–1058. doi: 10.1038/s41592-018-0229-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chung N.C., Storey J.D. Statistical significance of variables driving systematic variation in high-dimensional data. Bioinformatics. 2015;31:545–554. doi: 10.1093/bioinformatics/btu674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kobak D., Berens P. The art of using t-SNE for single-cell transcriptomics. Nat Commun. 2019;10:5416. doi: 10.1038/s41467-019-13056-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Becht E., McInnes L., Healy J., Dutertre C.A., Kwok I.W.H., Ng L.G. Dimensionality reduction for visualizing single-cell data using UMAP. Nat Biotechnol. 2018;37:38–44. doi: 10.1038/nbt.4314. [DOI] [PubMed] [Google Scholar]
- 47.Abdelaal T., Michielsen L., Cats D., Hoogduin D., Mei H., Reinders M.J.T. A comparison of automatic cell identification methods for single-cell RNA sequencing data. Genome Biol. 2019;20:194. doi: 10.1186/s13059-019-1795-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Macosko E.Z., Basu A., Satija R., Nemesh J., Shekhar K., Goldman M. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pliner H.A., Shendure J., Trapnell C. Supervised classification enables rapid annotation of cell atlases. Nat Methods. 2019;16:983–986. doi: 10.1038/s41592-019-0535-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Przytycki PF, Pollard KS. Semi-supervised identification of cell populations in single-cell ATAC-seq. bioRxiv 2019:847657.
- 51.Trapnell C., Cacchiarelli D., Grimsby J., Pokharel P., Li S., Morse M. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol. 2014;32:381–386. doi: 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mao Q, Wang L, Goodison S, Sun Y. Dimensionality Reduction Via Graph Structure Learning. In: Proceedings of the 21th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining - KDD '15;2015, p. 765–74.
- 53.Chen H., Albergante L., Hsu J.Y., Lareau C.A., Lo Bosco G., Guan J. Single-cell trajectories reconstruction, exploration and mapping of omics data with STREAM. Nat Commun. 1903;2019:10. doi: 10.1038/s41467-019-09670-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cusanovich D.A., Reddington J.P., Garfield D.A., Daza R.M., Aghamirzaie D., Marco-Ferreres R. The cis-regulatory dynamics of embryonic development at single-cell resolution. Nature. 2018;555:538–542. doi: 10.1038/nature25981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jia G., Preussner J., Chen X., Guenther S., Yuan X., Yekelchyk M. Single cell RNA-seq and ATAC-seq analysis of cardiac progenitor cell transition states and lineage settlement. Nat Commun. 2018;9:4877. doi: 10.1038/s41467-018-07307-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kanton S, Boyle MJ, He Z, Santel M, Weigert A, Calleja FS, Sidow L, Fleck J, Guijarro P, Han D, et al:. ingle-cell genomic atlas of great ape cerebral organoids uncovers humanspecific features of brain development. bioRxiv 2019:685057. [DOI] [PubMed]
- 57.Chung CY, Ma Z, Dravis C, Preissl S, Poirion O, Luna G, Hou X, Giraddi RR, Ren B, Wahl GM. Single-Cell Chromatin Analysis of Mammary Gland Development Reveals Cell-State Transcriptional Regulators and Lineage Relationships. Cell Rep 2019;29:495–510 e6. [DOI] [PMC free article] [PubMed]
- 58.Sinnamon J.R., Torkenczy K.A., Linhoff M.W., Vitak S.A., Mulqueen R.M., Pliner H.A. The accessible chromatin landscape of the murine hippocampus at single-cell resolution. Genome Res. 2019;29:857–869. doi: 10.1101/gr.243725.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xing QR, Farran CEL, Yi Y, Warrier T, Gautam P, Collins J, Xu J, Li H, Zhang L-F, Loh Y-H. Parallel Bimodal Single-cell Sequencing of Transcriptome and Chromatin Accessibility. bioRxiv 2019:829960. [DOI] [PMC free article] [PubMed]
- 60.Grant C.E., Bailey T.L., Noble W.S. FIMO: scanning for occurrences of a given motif. Bioinformatics. 2011;27:1017–1018. doi: 10.1093/bioinformatics/btr064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tangherloni A, Ricciuti F, Besozzi D, Liò P, Cvejic A. Single cell ATAC-seq identifies broad changes in neuronal abundance and chromatin accessibility in Down Syndrome. bioRxiv 2019:727867.
- 62.Duren Z., Chen X., Zamanighomi M., Zeng W., Satpathy A.T., Chang H.Y. Integrative analysis of single-cell genomics data by coupled nonnegative matrix factorizations. Proc Natl Acad Sci U S A. 2018;115:7723–7728. doi: 10.1073/pnas.1805681115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hwang B., Lee J.H., Bang D. Single-cell RNA sequencing technologies and bioinformatics pipelines. Exp Mol Med. 2018;50:96. doi: 10.1038/s12276-018-0071-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Natarajan A., Yardimci G.G., Sheffield N.C., Crawford G.E., Ohler U. Predicting cell-type-specific gene expression from regions of open chromatin. Genome Res. 2012;22:1711–1722. doi: 10.1101/gr.135129.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kondili M., Fust A., Preussner J., Kuenne C., Braun T., Looso M. UROPA: a tool for Universal RObust Peak Annotation. Sci Rep. 2017;7:2593. doi: 10.1038/s41598-017-02464-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mich JK, Graybuck LT, Hess EE, Mahoney JT, Kojima Y, Ding Y, Somasundaram S, Miller JA, Weed N, Omstead V, et al. Epigenetic landscape and AAV targeting of human neocortical cell classes. bioRxiv 2020:555318.
- 67.Rai V., Quang D.X., Erdos M.R., Cusanovich D.A., Daza R.M., Narisu N. Single-cell ATAC-Seq in human pancreatic islets and deep learning upscaling of rare cells reveals cell-specific type 2 diabetes regulatory signatures. Mol Metab. 2020;32:109–121. doi: 10.1016/j.molmet.2019.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Corces MR, Shcherbina A, Kundu S, Gloudemans MJ, Frésard L, Granja JM, Louie BH, Shams S, Bagdatli ST, Mumbach MR, et al. Single-cell epigenomic identification of inherited risk loci in Alzheimer’s and Parkinson’s disease. bioRxiv 2020:896159. [DOI] [PMC free article] [PubMed]
- 69.González-Blas CB, Quan X-J, Duran-Romaña R, Taskiran II, Koldere D, Davie K, Christiaens V, Makhzami S, Hulselmans G, de Waegeneer M, et al. Identification of genomic enhancers through spatial integration of single-cell transcriptomics and epigenomics. bioRxiv 2019:12.19.882381. [DOI] [PMC free article] [PubMed]
- 70.Ramirez F., Ryan D.P., Gruning B., Bhardwaj V., Kilpert F., Richter A.S. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016;44:W160–W165. doi: 10.1093/nar/gkw257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hahne F, Ivanek R. Visualizing Genomic Data Using Gviz and Bioconductor. In: Mathé E, Davis S (Eds.) Statistical Genomics: Methods and Protocols. New York, NY: Springer New York; 2016, p. 335–51. [DOI] [PubMed]
- 72.Robinson J.T., Thorvaldsdottir H., Winckler W., Guttman M., Lander E.S., Getz G. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kanehisa M., Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Preissl S., Fang R., Huang H., Zhao Y., Raviram R., Gorkin D.U. Single-nucleus analysis of accessible chromatin in developing mouse forebrain reveals cell-type-specific transcriptional regulation. Nat Neurosci. 2018;21:432–439. doi: 10.1038/s41593-018-0079-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.McLean C.Y., Bristor D., Hiller M., Clarke S.L., Schaar B.T., Lowe C.B. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yu G., Wang L.G., Han Y., He Q.Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schoenfelder S., Fraser P. Long-range enhancer-promoter contacts in gene expression control. Nat Rev Genet. 2019;20:437–455. doi: 10.1038/s41576-019-0128-0. [DOI] [PubMed] [Google Scholar]
- 80.Heinz S., Romanoski C.E., Benner C., Glass C.K. The selection and function of cell type-specific enhancers. Nat Rev Mol Cell Biol. 2015;16:144–154. doi: 10.1038/nrm3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Vahedi G., Kanno Y., Furumoto Y., Jiang K., Parker S.C., Erdos M.R. Super-enhancers delineate disease-associated regulatory nodes in T cells. Nature. 2015;520:558–562. doi: 10.1038/nature14154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Corces M.R., Buenrostro J.D., Wu B., Greenside P.G., Chan S.M., Koenig J.L. Lineage-specific and single-cell chromatin accessibility charts human hematopoiesis and leukemia evolution. Nat Genet. 2016;48:1193–1203. doi: 10.1038/ng.3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ziffra RS, Kim CN, Wilfert A, Turner TN, Haeussler M, Casella AM, Przytycki PF, Kreimer A, Pollard KS, Ament SA, et al. Single cell epigenomic atlas of the developing human brain and organoids. bioRxiv 2020:12.30.891549.
- 84.Graybuck LT, Daigle TL, Sedeño-Cortés AE, Walker M, Kalmbach B, Lenz GH, Nguyen TN, Garren E, Kim TK, Siverts LA, et al. Prospective, brain-wide labeling of neuronal subclasses with enhancer-driven AAVs. bioRxiv 2020:525014.
- 85.Visel A., Minovitsky S., Dubchak I., Pennacchio L.A. VISTA Enhancer Browser–a database of tissue-specific human enhancers. Nucleic Acids Res. 2007;35:D88–D92. doi: 10.1093/nar/gkl822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rivera J., Keranen S.V.E., Gallo S.M., Halfon M.S. REDfly: the transcriptional regulatory element database for Drosophila. Nucleic Acids Res. 2019;47:D828–D834. doi: 10.1093/nar/gky957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kvon E.Z., Kazmar T., Stampfel G., Yanez-Cuna J.O., Pagani M., Schernhuber K. Genome-scale functional characterization of Drosophila developmental enhancers in vivo. Nature. 2014;512:91–95. doi: 10.1038/nature13395. [DOI] [PubMed] [Google Scholar]
- 88.Zhu Q, Gao P, Tober J, Bennett L, Chen C, Uzun Y, Li Y, Mumau M, Yu W, He B, et al. Developmental trajectory of pre-hematopoietic stem cell formation from endothelium. bioRxiv 2019:848846. [DOI] [PMC free article] [PubMed]
- 89.Fogarty M.P., Panhuis T.M., Vadlamudi S., Buchkovich M.L., Mohlke K.L. Allele-specific transcriptional activity at type 2 diabetes-associated single nucleotide polymorphisms in regions of pancreatic islet open chromatin at the JAZF1 locus. Diabetes. 2013;62:1756–1762. doi: 10.2337/db12-0972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Groop L. Open chromatin and diabetes risk. Nat Genet. 2010;42:190–192. doi: 10.1038/ng0310-190. [DOI] [PubMed] [Google Scholar]
- 91.Boyle E.A., Li Y.I., Pritchard J.K. An expanded view of complex traits: from polygenic to omnigenic. Cell. 2017;169:1177–1186. doi: 10.1016/j.cell.2017.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ulirsch J.C., Lareau C.A., Bao E.L., Ludwig L.S., Guo M.H., Benner C. Interrogation of human hematopoiesis at single-cell and single-variant resolution. Nat Genet. 2019;51:683–693. doi: 10.1038/s41588-019-0362-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Schmidt E.M., Zhang J., Zhou W., Chen J., Mohlke K.L., Chen Y.E. GREGOR: evaluating global enrichment of trait-associated variants in epigenomic features using a systematic, data-driven approach. Bioinformatics. 2015;31:2601–2606. doi: 10.1093/bioinformatics/btv201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cao J., Cusanovich D.A., Ramani V., Aghamirzaie D., Pliner H.A., Hill A.J. Joint profiling of chromatin accessibility and gene expression in thousands of single cells. Science. 2018;361:1380–1385. doi: 10.1126/science.aau0730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Liu L., Liu C., Quintero A., Wu L., Yuan Y., Wang M. Deconvolution of single-cell multi-omics layers reveals regulatory heterogeneity. Nat Commun. 2019;10:470. doi: 10.1038/s41467-018-08205-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chen S., Lake B.B., Zhang K. High-throughput sequencing of the transcriptome and chromatin accessibility in the same cell. Nat Biotechnol. 2019;37:1452–1457. doi: 10.1038/s41587-019-0290-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Reyes M., Billman K., Hacohen N., Blainey P.C. Simultaneous profiling of gene expression and chromatin accessibility in single cells. Adv Biosyst. 2019;3:1900065. doi: 10.1002/adbi.201900065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Welch JD, Kozareva V, Ferreira A, Vanderburg C, Martin C, Macosko EZ. Single-Cell Multi-omic Integration Compares and Contrasts Features of Brain Cell Identity. Cell 2019;177:1873–87 e17. [DOI] [PMC free article] [PubMed]
- 99.Liu J, Huang Y, Singh R, Vert J-P, Noble WS. Jointly embedding multiple single-cell omics measurements. bioRxiv 2019:644310. [DOI] [PMC free article] [PubMed]
- 100.Zeng W., Chen X., Duren Z., Wang Y., Jiang R., Wong W.H. DC3 is a method for deconvolution and coupled clustering from bulk and single-cell genomics data. Nat Commun. 2019;10:4613. doi: 10.1038/s41467-019-12547-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Granja J.M., Klemm S., McGinnis L.M., Kathiria A.S., Mezger A., Corces M.R. Single-cell multiomic analysis identifies regulatory programs in mixed-phenotype acute leukemia. Nat Biotechnol. 2019;37:1458–1465. doi: 10.1038/s41587-019-0332-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Norrie J.L., Lupo M.S., Xu B., Al Diri I., Valentine M., Putnam D. Nucleome dynamics during retinal development. Neuron. 2019;104(512–528) doi: 10.1016/j.neuron.2019.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]