Abstract
Up until recently, Australia was considered free of Leishmania due to the absence of phlebotomine sandfly species (Diptera: Phlebotominae) known to transmit Leishmania parasites in other parts of the world. The discovery of Leishmania (Mundinia) macropodum (Kinetoplastida: Trypanosomatidae) in Northern Australia sparked questions as to the existence of alternative vectors of Leishmania. This has added to the complexity of fully understanding the parasite's interaction with its vector, which is known to be very specific. Previous findings demonstrated L. macropodum infection beyond the blood meal stage in the day-biting midges Forcipomyia (Lasiohelea) Kieffer (Diptera: Ceratopogonidae) implicating them in the parasite's life cycle. Currently, there is no conclusive evidence demonstrating this suspected vector to transmit L. macropodum to a naïve host. Therefore, this research aimed to investigate the vector competency of day-biting midge F. (Lasiohelea) to transmit L. macropodum utilising a novel technology that preserves nucleic acids. Honey-soaked Flinders Technology Associates (FTA®) filter-paper cards were used to obtain saliva expectorated from biting midges while sugar-feeding. F. (Lasiohelea) were aspirated directly off macropods from a known Leishmania-transmission site and were kept in a waxed-paper container holding a honey-coated FTA® card for feeding. Insect identification and Taqman quantitative real-time PCR (qPCR) screening assays revealed L. macropodum DNA in F. (Lasiohelea) up to 7 days post field-collection, and in an unidentified biting midge, previously known as F. (Lasiohelea) sp.1. Moreover, 7/145 (4.83%) of FTA® cards were confirmed positive with L. macropodum DNA after exposure to field-collected F. (Lasiohelea). Additionally, FTA® cards were found to be a valuable surveillance tool, given the ease of use in the field and laboratory. Overall, our findings support previous reports on L. macropodum transmission by an alternative vector to phlebotomine sandflies. Further studies identifying and isolating infective L. macropodum promastigotes is necessary to resolve questions on the L. macropodum vector.
Keywords: Leishmania, Day-biting midge, FTA® cards, Transmission, Saliva, Australia, Macropod
Graphical abstract
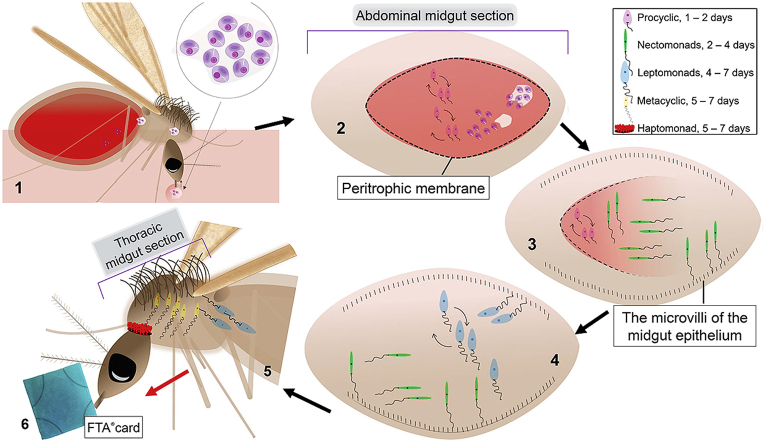
The graphical abstract illustrates the life cycle of Leishmania* within a phlebotomine sandfly vector. Infected macrophages containing Leishmania amastigotes are ingested when a sand fly feeds on an infected host (1). Within the abdominal midgut the amastigotes transform into replicative procyclic promastigotes, which are protected from digestive enzymes by the peritrophic membrane (PM) (2). The procyclic parasites differentiates into highly motile nectomonads to escape from a broken PM (3). Migrating towards the anterior midgut, nectomonads will attach to the microvilli of the midgut epithelium until reaching the stomodeal valve. To resume parasite replication, the nectomonad promastigotes are transformed into leptomonad promastigotes (4), which can either attach to the cuticle lining of the stomodeal valve as haptomonad promastigotes (5) or differentiate into metacyclic promastigotes (infective form) (5). The infective Leishmania form will colonise the stomodeal valve and are injected into the host during feeding such as on a blood meal or in this case a honey-coated FTA® card (6). *Note, the life cycle is described for suprapylarian Leishmania species where development is restricted to the sandfly midgut.
Highlights
-
•
4.83% of FTA® cards were positive for Leishmania macropodum DNA after exposure to field-collected Forcipomyia (Lasiohelea).
-
•
L. macropodum DNA was detectable on cards after 10 weeks demonstrating their utility in Leishmania surveillance programs.
-
•
Real-time qPCR confirmed L. macropodum DNA in the previously implicated vector, F. (Lasiohelea) sp.1.
-
•
Our results support previous findings suggesting L. macropodum is transmitted by an alternative vector to the sandfly.
1. Introduction
The leishmaniases are a highly complex group of vector-borne diseases caused by parasites from the genus Leishmania (Kinetoplastida: Trypanosomatidae). These parasites cause a wide spectrum of clinical manifestations and are listed by the World Health Organization as one of the most important tropical diseases (Pigott et al., 2014). The leishmaniases are endemic in more than 90 countries with approximately 1 million new infections and more than 50,000 deaths reported annually (Burza et al., 2018). Leishmania species known to be important human pathogens belong to the subgenera Leishmania (Leishmania) and Leishmania (Viannia). Infection with Leishmania is acquired through the bite of an infected phlebotomine sandfly, which are currently the only confirmed group of vectors of Leishmania. Less than 10% of phlebotomine sandfly species have been incriminated as vectors and their distribution is known to be a critical factor in acquiring leishmaniasis (Burza et al., 2018; Ready, 2013).
Australia was considered a Leishmania-free region largely due to the absence of the vector species. However, Leishmania (Mundinia) macropodum (previous nomenclature Leishmania sp. AM-2004 and Leishmania australiensis) was discovered and identified to cause cutaneous leishmaniasis in macropod species in the Darwin region of Northern Australia (Dougall et al., 2009, 2011; Rose et al., 2004). The absence of a Leishmania vector species in Australia has convoluted the understanding of the parasite's life cycle and raised new questions regarding alternative vectors.
Alternative vectors of the leishmaniases have been highly debated in scientific literature but currently there is no conclusive evidence demonstrating Leishmania transmission by arthropods other than phlebotomine sandflies (Seblova et al., 2014). Various studies have investigated suspected arthropods’ susceptibility to transmit Leishmania parasites, through both experimental infections (Almeida et al., 2016; Sádlová et al., 2013; Seblova et al., 2015, 2012) and screening wild-caught arthropods using molecular techniques (Berdjane-Brouk et al., 2012; Dantas-Torres et al., 2010; Dougall et al., 2011; Jaouadi et al., 2015; Manuel et al., 2016; Mukherjee et al., 1997; Solano-Gallego et al., 2012). However, the interaction between Leishmania species and its vector is highly complex and specific, and current molecular and field methodologies have not been able to confirm new Leishmania-vectors. The parasite undergoes various developmental stages within the vector prior to reaching the infective stage that is transmitted during host blood feeding (Bates, 2007; Kamhawi, 2006). In the early stage of infection, the promastigote phase of the parasite is non-host specific and has been proven to thrive in many insects. If unable to bind to the midgut epithelium in the later stage of infection (when the blood meal is being digested), the promastigotes will be excreted with blood meal remnants. The lack of midgut attachment is the major refractory barrier for Leishmania and this phase of attachment marks a true vector (Bates, 2007; Dostálová and Volf, 2012; Sacks and Kamhawi, 2002; Seblova et al., 2012).
In 1999, Robert Killick-Kendrick established criteria to assess vector competency for the transmission of the leishmaniases. These criteria included: i) Naturally infected vectors must be collected on more than one occasion containing identical Leishmania isolates as found in human or reservoir hosts, ii) The vector must feed on humans (anthroponotic) and if zoonotic, also on the animal reservoir host, iii) There needs to be evidence for a strong ecological association between the vector and the host, iv) Full development of the parasite has to occur within the vector after digesting the blood meal, and v) It is imperative that the vector is able to transmit the parasite, via a blood meal, to a susceptible host (Killick-Kendrick, 1999). Dougall et al. (2011) implicated the Ceratopogonid subgenus Forcipomyia (Lasiohelea) Kieffer as the alternative vector in Northern Australia. However, based on the above criteria, it has yet to be proven if the parasites are transmitted by F. (Lasiohelea) during host blood feeding. The majority of Leishmania - vector competency studies have not been able to fulfil the fifth Killick-Kendrick criterion with acceptable evidence of parasite transmission by bite (Seblova et al., 2012). Therefore, novel strategies are essential to investigate transmission and incriminate these suspected vectors.
Over the last decade, an arbovirus surveillance system based on the preservation of nucleic acids has become widely applied in disease surveillance (Hall-Mendelin et al., 2017, 2010; Kurucz et al., 2019; van den Hurk et al., 2014; Wipf et al., 2019). Soaked in honey, Flinders Technology Associates (FTA®; Whatman – GE Healthcare Life technologies) cards have been used to detect pathogens during insect sugar feeding (Hall-Mendelin et al., 2017, 2010). This technique has been shown to be inexpensive and efficient when screening for arboviruses, and the implementation of this system for detecting parasites could be of international importance.
By taking advantage of this technique, this research sought to i) assess FTA® card's potential in Leishmania surveillance programs, and ii) investigate if the suspected day-biting midges, F. (Lasiohelea), were able to transmit L. macropodum onto the honey-coated FTA® cards, and thereby show evidence on a Leishmania-transmission during feeding (criterion v).
2. Materials and methods
2.1. Cultivating L. macropodum
Leishmania macropodum parasites had previously been isolated from clinical infected macropod species at the Territory Wildlife Park (TWP). Skin lesions had been grown in biphasic Novy-MacNeal-Nicolle (NNN) medium and incubated at 26 °C for promastigote growth (Dougall et al., 2009). To obtain an on-going in vitro culture, promastigotes were cultivated in Grace's Insect medium (Invitrogen, Australia), containing 20% heat-inactivated foetal bovine serum, 2 mM L-glutamine, 100 U/mL penicillin, and 100 μg/mL streptomycin (Sigma-Aldrich) at 26 °C.
2.2. FTA® card preparation
FTA® cards (2.5 cm × 2.5 cm) were coated with honey 24-48 h prior to use. In order to identify which insects had fed on the coated FTA® cards, blue food dye was added to the honey (Hall-Mendelin et al., 2010). Waxed-paper containers were converted to contain field-collected biting insects and to hold a FTA® card (Fig. 1). A total of 145 waxed-paper containers were set up, each holding one honey-coated FTA® card.
Fig. 1.
Wax-paper cups were used to contain and maintain field-collected biting midges. (A) Honey-coated FTA® cards were left at room temperature for 48 h allowing even absorption of honey into the cards. (B) A 2.5 cm slit was carved into the bottom of disposable cup and sealed with adhesive tape. (C) Insects were aspirated directly into the bottom of the containers through a small perforation created before field collection. Once biting midges were collected from the macropods, the small perforation was sealed with a rubber plug. Gauze was used as a lid to seal the top of the containers and fastened securely with a rubber band. The honey-coated FTA® card was inserted through the bottom slit after insect collection and once again sealed with adhesive tape.
2.3. Stability of L. macropodum DNA on FTA® cards
To ensure that honey-coated FTA® cards could be used to preserve and identify Leishmania DNA, a pilot experiment was designed to investigate the cards’ ability to detect cultured L. macropodum promastigotes over a 10-week time course. FTA® cards were inoculated with a parasite load between 0 to 106 parasites/card in 10-fold increments in triplicates and stability of DNA was tested at five time points (week 0, 2, 5, 8, and 10). Furthermore, cards had either been coated with honey or without (plain, control group) in order to ensure the use of honey had no effect on parasite detection.
For parasite DNA elution from FTA® cards, the cards were cut into strips and added to a 5 mL tube containing 1 mL molecular grade water kept on ice. To release the DNA from the matrix of the FTA® cards, the tubes were vortexed every 5 min for 10 s for a total of 20 min. The strips and suspension were separated with a syringe, and strips were discarded (Hall-Mendelin et al., 2010). The suspension, containing the DNA, was aliquoted and 200 μL (1/5 of the card) was used for DNA extraction using DNeasy® Blood & Tissue Kit (Qiagen) following the manufacturer's protocol for purification of total DNA from cells. DNA was eluted in 200 μL AE buffer.
2.4. Study site and insect collection
The collection site was set up at the TWP where identified cases of cutaneous leishmaniasis in macropods had previously been reported (Dougall et al., 2009; Rose et al., 2004). Additionally, F. (Lasiohelea) species have been identified with L. macropodum DNA at this site (Dougall et al., 2011). Day-biting midges were collected between January-April 2017 during the regional wet season, at the park's macropod enclosure containing agile wallabies (Macropus agilis) and antilopine wallaroos (Macropus antilopinus). During day-hours, biting insects were aspirated directly off macropods and transferred into waxed-paper containers holding honey-coated FTA® cards and maintained at 30 °C (±5 °C) with relative humidity at 85% (±5%). Insects were kept alive between 1 and 8 days post field-collection to confirm a potential promastigote infection beyond blood meal stage and to identify Leishmania-transmission onto FTA® cards during feeding. At the end of each experiment, insects were kept in 70% ethanol) and FTA® cards were stored individually in plastic sealable bags containing silica beads. Cards were kept at room temperature for up to 60 days prior to L. macropodum qPCR screening.
2.5. Insect identification and L. macropodum DNA extraction
For the purpose of this study only field-collected F. (Lasiohelea), the suspected vectors of L. macropodum, were identified and examined for parasite DNA. Forcipomyia (Lasiohelea) species were distinguished from other Ceratopogonid biting insects by their plain wing pattern covered with suberect macrotrichia, a well-developed empodium and claws. To identify F. (Lasiohelea) to species level, a representative 10% subset of F. (Lasiohelea) were mounted on slides in Hoyer's medium, consisting of acacia gum, chloral hydrate, glycerol, and distilled water. Slides were stored at 37 °C for six weeks followed by morphological identification using a taxonomic key for Australasian F. (Lasiohelea) (Debenham, 1983).
Field-collected F. (Lasiohelea) kept in containers holding an FTA® card, were randomly selected for parasite examination. In order to assess whether F. (Lasiohelea) could support L. macropodum development beyond blood meal stage, insects were selected between 1 and 8 days from day of collection. Forcipomyia (Lasiohelea) were either examined for L. macropodum DNA individually or in pools (n = 2–15) and were grouped into five categories according to the number of insects in that respective pool (1, ≤ 2, 3–4, 5–6, or ≥ 7). Leishmania macropodum DNA extractions were performed using the Qiagen DNeasy® Blood & Tissue Kit according to manufacturer's instructions. Insects were homogenised in the manufacturer's lysis buffer containing 20 mg/mL proteinase K, followed by Qiagen's protocol for purification of total DNA from insects, with a final elution volume of 200 μL AE buffer.
Moreover, to determine species of F. (Lasiohelea) carrying L. macropodum, a subset of field-collected specimens (n = 50) was processed using a non-destructive DNA extraction method (Dougall et al., 2011). Individually F. (Lasiohelea) were digested in the Qiagen's lysis buffer/Proteinase K buffer without homogenisation. After a 3-h incubation at 56 °C, insects were removed from the lysis solution for morphological identification. Purification of total DNA was hereafter performed as describe above.
2.6. Detecting and quantifying L. macropodum by real-time qPCR
Leishmania macropodum screening was performed with specific Taqman qPCR assays. Published primers rooME-F2 5′-AAACTTCCGGAACCTGTCGT-3′, rooME-R2 5′-GTAGGCACCCGAAGAGACC-3′, and the Taqman probe LeishME 5′d FAM-CCGGCAAGATTTTGGGAGCG-BHQ-1 3′ were used to amplify L. macropodum (Dougall et al., 2009). PCR reactions were made in a 10 μL reaction of 1 × SsoAdvanced™ universal probe supermix, 6 mM MgCl2 (Bio-Rad Laboratories, Australia), 0.3 μM of primers, 0.05 μM Taqman probe (Sigma-Aldrich, Australia) and 2 μL of DNA template extracted from either insect or FTA® card sample. PCR cycling conditions were as follows: 2 min at 95 °C followed by 35 cycles of 15 s at 95 °C and 40 s at 66 °C with the CFX96 Real Time System (C1000 Thermal Cycler; Bio-Rad Laboratories). Genomic L. macropodum DNA standards from cultivated promastigotes were included in every PCR run to quantify positive insect samples and FTA® cards. Standards were purified from cultured L. macropodum promastigotes and made up of serial dilutions 10−1 – 10−7 in Tris-EDTA buffer (Sigma-Aldrich, Australia). Moreover, L. macropodum DNA standards were used to determine the qPCR assay's limit of detection. Parasite detection threshold was identified at ≥ 50 cultured parasites (data not shown here).
3. Results
3.1. Insect collection and identification
Approximately 3000 female F. (Lasiohelea) were aspirated directly off macropods at the TWP macropod enclosure. All insects were kept alive in containers (approximately 20 insects/container) with a honey-coated FTA® card. From a subset of 260 biting midges, three species were identified, however only two could be identified with Debenham's taxonomic key, namely F. (L.) townsvillensis (n = 49/260) and F. (L.) peregrinator (n = 3/260) (Debenham, 1983; Taylor, 1918). The third species, which was found to be the dominant species (n = 208/260), was the undescribed species previously referred to as Forcipomyia (Lasiohelea) sp. 1 and implicated as a vector of L. macropodum by Dougall et al. (2011).
3.2. Detection of L. macropodum in F. (Lasiohelea)
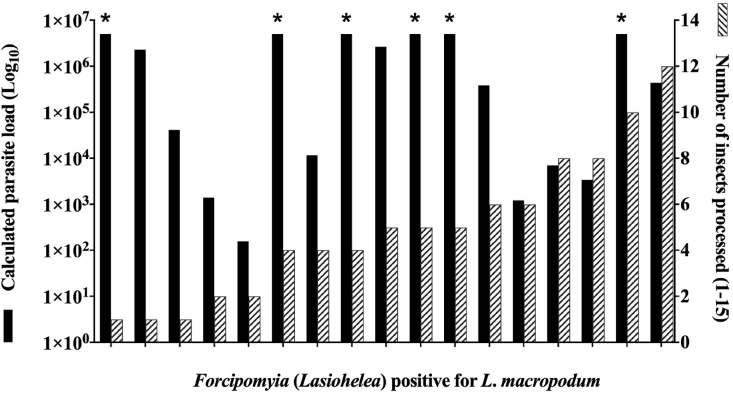
To confirm ongoing Leishmania circulation at the TWP site, 123 pools (a total of 536 individuals) and 47 individual F. (Lasiohelea) were screened for L. macropodum by real-time qPCR. 14/123 pools and 1/47 individual F. (Lasiohelea) were positive for L. macropodum DNA. High parasitemia was detected in several pools and individual F. (Lasiohelea) with ≥ 5 × 106 parasites (Fig. 2). Moreover, non-destructive DNA extractions from field-collected F. (Lasiohelea) (n = 50) species confirmed L. macropodum DNA in specimens identified as F. (Lasiohelea) sp.1 (n = 2/39; Table 1).
Fig. 2.
Leishmania macropodum DNA detection by qPCR. Individual or pools of F. (Lasiohelea) species were assessed for the presence of L. macropodum DNA. Only positive samples are shown, with each pair of columns representing results from one sample. Black columns depict the parasitic load detected and the dashed columns show the number of insects processed in that sample. Asterisks represent groups that contained ≥ 5 × 106 F. (Lasiohelea) parasites.
Table 1.
Summary of F. (Lasiohelea) identified to species-level and those identified as positive for L. macropodum by qPCR from non-destructive insect processing.
| Species | Identification | qPCR positive |
|---|---|---|
| Forcipomyia (Lasiohelea) sp.1 | 39/50 | 2/39 |
| Forcipomyia (Lasiohelea) townsvillensis | 10/50 | 0/10 |
| Forcipomyia (Lasiohelea) peregrinator | 1/50 | 0/1 |
In addition to the steady-state detection of L. macropodum in randomly selected F. (Lasiohelea) species, we further examined the possibility of biting midges to sustain infection and support a successful development of L. macropodum. Towards this, the presence of L. macropodum DNA by qPCR was performed over an 8-day period post insect field-collection. Our data shows that parasite DNA was detected up to 7 days (Table 2) suggesting possible parasite development beyond the blood meal stage (day 2–3).
Table 2.
Summary of F. (Lasiohelea) species detected positive for L. macropodum DNA over 1–8 days after collection. Insects were screened either individually (1) or in pools (2 – ≥7).
| Number of insects present in the tested sample |
|||||
|---|---|---|---|---|---|
| 1 | ≤2 | 3–4 | 5–6 | ≥7 | |
| Day 1 | 2/46 | 1/15 | 3/19 | 4/22 | 5/19 |
| Day 2 | 0/8 | 0/1 | 0/2 | 1/2 | 0/1 |
| Day 3 | 0/12 | 0/4 | 0/7 | 0/3 | 0/1 |
| Day 4 | 0/8 | 0/6 | 0/1 | 0/1 | 0/2 |
| Day 5 | 1/6 | 0/6 | 0/1 | – | – |
| Day 6 | 0/5 | 0/3 | 0/1 | – | – |
| Day 7 | 0/3 | 1/2 | 0/1 | – | – |
| Day 8 | 0/3 | 0/1 | – | – | – |
– Pools of F. (Lasiohelea) were not screened on that day.
3.3. Detection of L. macropodum on FTA® cards after exposure to field-collected F. (Lasiohelea)
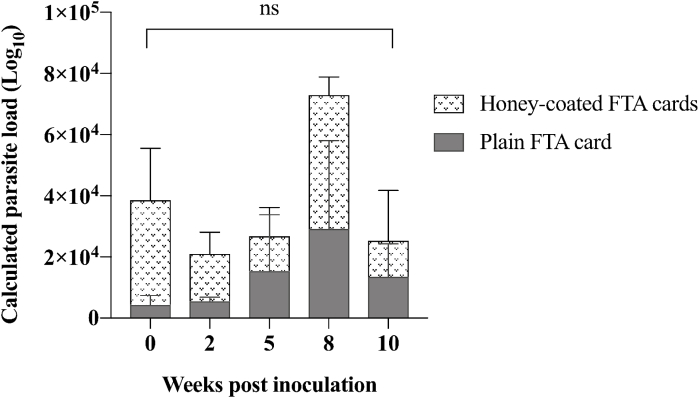
Stability of L. macropodum DNA on FTA® cards was assessed over a 10-week time course by qPCR. Results are shown for cards inoculated with 106 parasites per card over the 10-week period (Fig. 3). A two-way ANOVA with a post Tukey's comparison test confirmed the detected parasite load over the 10 weeks were not significantly different. Furthermore, the use of honey was not found to be associated with any interference of parasite load detection or the assay's sensitivity.
Fig. 3.
Evaluation of L. macropodum viability on FTA® cards and the effect of honey. To determine L. macropodum detection on honey-coated and plain FTA® cards, an experiment was designed over a 10-week time course. FTA® cards (2.5 × 2.5 cm) were inoculated with 106L. macropodum per card in triplicates and DNA was tested at five time points (week 0, 2, 5, 8 and 10).
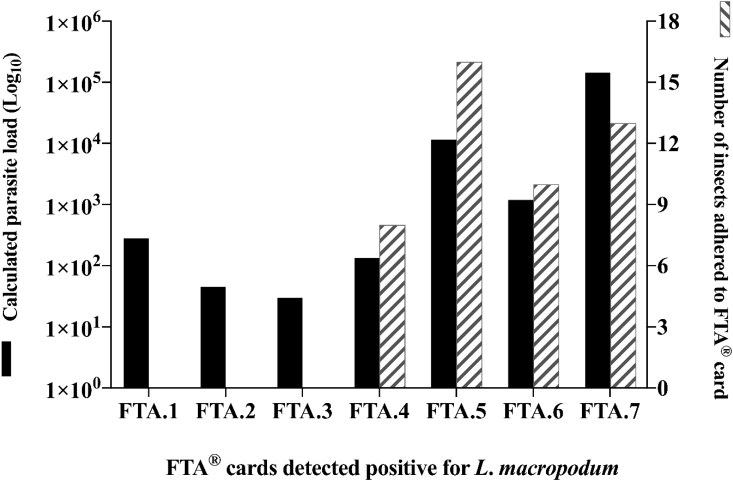
From the field experiments, 145 FTA® cards were screened for L. macropodum DNA by qPCR within 60 days' post-exposure to field collected F. (Lasiohelea). Due to the insect's small size, a high proportion of F. (Lasiohelea) became stuck in the coated honey, and prior to screening with qPCR 44/145 cards (30%) had insects removed. Overall, 7/145 (4.83%) of FTA® cards were detected positive for L. macropodum DNA when screened with (n = 4/44) and without (3/101) adhered F. (Lasiohelea) (Fig. 4).
Fig. 4.
Assessment of L. macropodum DNA using FTA® card technology. FTA® cards were exposed to field-collected F. (Lasiohelea). Cards with and without insects adhered were processed and parasite load was determined with qPCR. 4.83% (7/145) FTA® cards were positive for L. macropodum DNA. Black columns show parasite load detected on each positive FTA® card (numbered FTA.1 – FTA.7). Dashed columns show the number of insects adhered to each card. When dashed columns are absent, this signifies the absence of insects on the positive cards.
4. Discussion
This study had two overall aims. First to investigate the potential use of FTA® cards in Leishmania surveillance programs, and second whether this novel technique could provide evidence in support of the outstanding fifth Killick-Kendrick criterion demonstrating that F. (Lasiohelea) can transmit L. macropodum.
Firstly, this study found that FTA® cards are a valuable surveillance tool, given the ease of use compared to today's insect screening protocols. These protocols can be time consuming and costly due to the large number of field-collected samples that may need to be processed to confirm disease transmission by a vector. Converting catch containers of CO2-bated light traps to hold honey-coated FTA® cards has previously shown to be an efficient method for arboviral surveillance in Australia (Hall-Mendelin et al., 2010; Kurucz et al., 2019). Given that Leishmania parasites are highly prevalent in developing countries, FTA® cards may offer an alternative inexpensive tool to enhance field surveillance activities for leishmaniasis. Not only will the simple approach of applying the cards in elimination programs substitute the necessary extensive training of personnel, it can preclude the need to screen large samples and analysing insect population to provide evidence of disease transmission. This may benefit programs in remote areas where accessibility to laboratory facilities are limited and samples need to be stored for long-term. Leishmania macropodum DNA was shown to be stable on FTA® cards for the entire 10-week time course at room temperature, supporting their suitability for projects where long-term storage is unavoidable. Although not found to be statistically significant, our results suggest that the addition of honey may aid the survival of the promastigotes. However, more work needs to be performed to assess this. Overall, our data does demonstrate that the addition of honey does not have a detrimental effect. One limitation was the ability to detect low parasite load in our long-term experiment when L. macropodum promastigotes were inoculated onto FTA® cards (threshold identified at 102 parasites). Although quantification of parasite load might not be important in surveillance programs, insects harbouring a low parasite load, might lead to false-negative results if PCR cannot detect less than 102 parasites on FTA® cards. However, the average number of Leishmania promastigotes harboured by phlebotomine sandflies is 5 × 104/sandfly thereby minimising the risk of false-negative PCR results (Rogers et al., 2004).
Overall, these findings suggest that FTA® cards could become a valuable public health surveillance tool to survey the emergence and re-emergence of the leishmaniases.
Several studies have suggested the existence of alternative vectors that can transmit Leishmania parasites, and these have been highly debated in literature (Seblova et al., 2014). Fulfilling all five Killick-Kendrick criteria with convincing evidence is a difficult task. Most often, studies have been unable to confirm a successful parasite infection within the suspected vector or unable to demonstrate successful transmission to a naïve host during blood feeding. For this study's second aim, blood fed F. (Lasiohelea) were aspirated directly off the macropod host with the purpose of i) assessing a successful Leishmania infection in this suspected vector over an 8 day period and ii) allow F. (Lasiohelea) to feed on a honey-coated FTA® card thereby potentially depositing infective promastigotes with saliva.
We showed that after exposure to field-collected biting midges, L. macropodum DNA was detected on 7/145 (4.83%) of FTA® cards (with and without adhered insects) when screened with qPCR. The limitations with modern molecular screening techniques used in Leishmania-vector studies is the identification of the infectious parasite. In the Leishmania life cycle, parasites take two distinct forms depending on if they are found in an insect vector (promastigote form) or a mammalian host (amastigote form). These stages are immediately triggered by the change in pH and temperature within the respective hosts (Bates, 2007). Within its vector, the Leishmania promastigotes undergo various obligated development stages before becoming infectious promastigotes that are in turn transmitted during blood feeding. The challenge is to confirm the developmental change of amastigote to promastigote forms as well as identifying the various stages of the promastigotes with these molecular screening techniques. Standard PCR is widely used today, however a reverse transcription PCR is required to detect metacyclic-specific transcripts (Giraud et al., 2019). The detected Leishmania DNA in this study, can therefore not verify that DNA originated from the infective promastigote stage expectorated with saliva, which limits the cards’ potential. Interestingly, we detected L. macropodum DNA from FTA® cards without insects adhered. This could suggest a successful development of L. macropodum in F. (Lasiohelea) indicating parasites were expectorated during sugar feeding. However, it is important to emphasise the complexity currently faced in vector studies. Various groups of hematophagous insects are known to defecate during blood feeding, and it is possible that the positive Leishmania detection on cards had originated from parasites excreted with faeces, suggesting parasites were not able to develop beyond the blood meal stage (Bates, 2007). Moreover, phlebotomine sandflies are known to undergo pre-diuresis (excrete urine) during blood feeding and one study frequently detected free-swimming forms of infective Leishmania promastigotes in urine droplets from phlebotomine sandflies while blood feeding (Sádlová and Volf, 1999). For these reasons, the purpose of adapting this technique in vector competence studies, FTA® cards currently have limitations and thus cannot alone conclusively incriminate a suspected Leishmania vector.
Alongside FTA® cards, F. (Lasiohelea) species were likewise screened for L. macropodum DNA. Overall, our results showed that 3/97 individual and 14/123 pools of screened F. (Lasiohelea) were positive with L. macropodum DNA. From identifying L. macropodum infection over 8-days in field-collected F. (Lasiohelea), DNA was confirmed up to day 7 in 1/5 F. (Lasiohelea), which could indicate a full development and successful infection in stomodeal valve. The Leishmania (Leishmania) development within the phlebotomine sandfly takes approximately 1 week, though this is dependent on the vector species, parasite species and climatic factors (Kamhawi, 2006). The critical stage for successful Leishmania infection within the vector is the blood meal stage as parasites needs to attach to the midgut epithelium to avoid excretion with blood meal remnants (day 2–4) (Bates, 2007; Dostálová and Volf, 2012; Sacks and Kamhawi, 2002; Seblova et al., 2012). Infection beyond the blood meal stage define a true vector. Though our results confirm Leishmania DNA at day 7 with qPCR, these assays must be interpreted with caution to avoid misleading results, as stated above. PCR has previously been confirmed to detect Leishmania DNA after 7 days post blood meal while microscopic examination failed to detect living parasites by manual gut dissection after day 3 (Seblova et al., 2012). Therefore, it is uncertain if the molecular field screening results from this study indicates L. macropodum in its infective stage without microscopic confirmation of living parasites. However, from previous microscopic examination by Dougall et al. (2011), promastigotes in the gut of F. (Lasiohelea) midges were identified. More importantly, the presence of promastigote secretory gel (PSG) plug containing leptomonad promastigotes and parasites resembling metacyclic promastigotes (both developed in the parasite's late stage of infection) were observed in the biting midge when not containing blood meal remnants (Dougall et al., 2011). Whether the entire F. (Lasiohelea) subgenus is competent to transmit Leishmania is still uncertain. From our non-destructive DNA extraction of insects, we identified L. macropodum DNA in F. (Lasiohelea) sp.1.Blue dye had been added to the honey to identify which F. (Lasiohelea) species had sugar-fed. However, when insects died, they would immediately desiccate, which affected the visibility of the blue dye in honey-fed midges limiting the verification of honey-fed and blood-fed insects.
Forcipomyia (Lasiohelea) species were intentionally collected due to their previous implication in the L. macropodum life cycle and already fulfilling the Killick-Kendrick criteria i-iv (Dougall et al., 2011). Since the incrimination of the biting midge, they have become a speculative group of vectors of Leishmania parasites, with studies investigating their vector competence under laboratory settings (Chanmol et al., 2019; Seblova et al., 2015, 2012). Particularly the biting midge, Culicoides (Diptera: Ceratopogonidae), has been used as a model to identify and confirm Leishmania infection specifically assessing the L. (Mundinia) subgenus (Chanmol et al., 2019; Seblova et al., 2015, 2012). Microscopic examinations provided evidence that C. sonorensis was highly susceptible to L. macropodum and L. (Mundinia) enrietti. Both parasites developed through to late-stage infections, migrated successfully to the thoracic midgut and colonised the stomodeal valve, as previously observed by Dougall et al. (2011) in F. (Lasiohelea) (Seblova et al., 2015). Interestingly, a recent study found further evidence that L. (Mundinia) orientalis, causing human leishmaniasis in Thailand, was likewise able to establish successful infection in C. sonorensis with the development of promastigote stages successfully identified from post-infected blood meal (Chanmol et al., 2019). In contrast, parasite species from the subgenus Leishmania were unable to develop successful infection in Culicoides similar to Mundinia species development in phlebotomine sandflies (Chanmol et al., 2019; Seblova et al., 2015, 2012). Though Culicoides and Forcipomyia are two different genera and their vector competence should not be considered analogous, these results do support the hypothesis that the Ceratopogonid subgenus Forcipomyia (Lasiohelea) Kieffer might be an alternative vector to the phlebotomine sandflies causing macropod cutaneous leishmaniasis in the Darwin region.
5. Conclusion
FTA® cards were shown to be a useful tool in Leishmania surveillance programs due to their ability for long-term storage and preservation of parasite DNA. Their use in elimination programs can be valuable as they are inexpensive and simple to use in the field. Showing acceptable evidence of Leishmania transmission to a naïve host by an alternative vector has yet been demonstrated. This is the first report to investigate the fifth Killick-Kendrick criteria by using FTA® cards. Leishmania macropodum DNA was detected on FTA® cards screened with and without F. (Lasiohelea) adhered, indicating insects could have possibly fed and expectorated parasites onto the cards. However, due to the identified limitations with FTA® cards in vector competence studies, this research was not able to conclusively confirm L. macropodum had successfully infected F. (Lasiohelea) beyond the blood meal stage or that the DNA originated from infective promastigotes and expectorated during sugar feeding. L. macropodum DNA was identified in F. (Lasiohelea) 7 days post field-collection suggesting the parasites had established infection beyond the blood meal stage. However, isolation of parasites and microscopic evidence of infection is the only reliable method to confirm established infection. Taken together, our study was not able to confirm F. (Lasiohelea) as the vectors of. L macropodum in Northern Australia, however it does support previous findings. Further evidence is required to i) confirm their competence to transmit infective L. macropodum during feeding and ii) identify the specific vector(s) of F. (Lasiohelea).
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors. L.J.H is the recipient of the Australian National Health and Medical Research Council Career Development Award (ID: 105760). We thank the Cai Qvesehls Funds for their continuous financial support contributed to E.P's master dissertation and Ph.D.
Declaration of competing interest
The authors declare no conflicts of interest.
Acknowledgement
We deeply thank Associate Professor Christopher Peacock from the University of Western Australia for kindly providing L. macropodum isolates and assisting with cultivating Leishmania promastigotes. We are very grateful for all the contribution and support from staff at the Territory Wildlife Park Northern Territory Government, especially thanks to Damien Stanioch, Donna Jensen, Trine Kruse, Simon Hollamby and Shael Martin for their assistance during field work. Thanks to Mr. Peter Whelan for guidance on field collection of biting insects.
Contributor Information
Elina Panahi, Email: elina.panahi@griffithuni.edu.au.
Lara J. Herrero, Email: l.herrero@griffith.edu.au.
References
- Almeida V.dos A., da Hora T.N., Leça Junior N.F., Carvalho F.S., sa Silva A.L., Wenceslau A.A., Albuquerque G.R., Silva F.L. Detection of Leishmania infantum DNA in hamsters infested with ticks collected from naturally infected dogs. Rev. Bras. Med. Veterinária. 2016;38:329–333. [Google Scholar]
- Bates P.A. Transmission of Leishmania metacyclic promastigotes by phlebotomine sand flies. Int. J. Parasitol. 2007;37:1097–1106. doi: 10.1016/j.ijpara.2007.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berdjane-Brouk Z., Koné A.K., Djimdé A.A., Charrel R.N., Ravel C., Delaunay P., del Giudice P., Diarra A.Z., Doumbo S., Goita S., Thera M.A., Depaquit J., Marty P., Doumbo O.K., Izri A. First detection of Leishmania major DNA in Sergentomyia (Spelaeomyia) darlingi from cutaneous leishmaniasis foci in Mali. PloS One. 2012;7:1–5. doi: 10.1371/journal.pone.0028266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burza S., Croft S.L., Boelaert M. Leishmaniasis. Lancet. 2018;392:951–970. doi: 10.1016/S0140-6736(18)31204-2. [DOI] [PubMed] [Google Scholar]
- Chanmol W., Jariyapan N., Somboon P., Bates M.D., Bates P.A. Development of Leishmania orientalis in the sand fly Lutzomyia longipalpis (Diptera: psychodidae) and the biting midge Culicoides soronensis (Diptera: Ceratopogonidae) Acta Trop. 2019 doi: 10.1016/j.actatropica.2019.105157. [DOI] [PubMed] [Google Scholar]
- Dantas-Torres F., Lorusso V., Testini G., De Paiva-Cavalcanti M., Figueredo L.A., Stanneck D., Mencke N., Brandão-Filho S.P., Alves L.C., Otranto D. Detection of Leishmania infantum in Rhipicephalus sanguineus ticks from Brazil and Italy. Parasitol. Res. 2010;106:857–860. doi: 10.1007/s00436-010-1722-4. [DOI] [PubMed] [Google Scholar]
- Debenham M. Australasian species of the blood-feeding Forcipomyia subgenera, lasiohelea and dacnoforcipomyia (Diptera : Ceratopogonidae) Aust. J. Zool. Suppl. Ser. 1983;31:1. doi: 10.1071/AJZS095. [DOI] [Google Scholar]
- Dostálová A., Volf P. Leishmania development in sand flies: parasite-vector interactions overview. Parasites Vectors. 2012;5:276. doi: 10.1186/1756-3305-5-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougall A., Shilton C., Low Choy J., Alexander B., Walton S. New reports of Australian cutaneous leishmaniasis in Northern Australian macropods. Epidemiol. Infect. 2009;137:1516–1520. doi: 10.1017/S0950268809002313. [DOI] [PubMed] [Google Scholar]
- Dougall A.M., Alexander B., Holt D.C.D.C., Harris T., Sultan A.H., Bates P.A., Rose K., Walton S.F. Evidence incriminating midges (Diptera: Ceratopogonidae) as potential vectors of Leishmania in Australia. Int. J. Parasitol. 2011;41:571–579. doi: 10.1016/j.ijpara.2010.12.008. [DOI] [PubMed] [Google Scholar]
- Giraud E., Martin O., Yakob L., Rogers M. Quantifying Leishmania metacyclic promastigotes from individual sandfly bites reveals the efficiency of vector transmission. Commun. Biol. 2019;2:84. doi: 10.1038/s42003-019-0323-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall-Mendelin S., Hewitson G.R., Genge D., Burtonclay P.J., De Jong A.J., Pyke A.T., van den Hurk A.F. FTA cards facilitate storage, shipment, and detection of arboviruses in infected Aedes aegypti collected in adult mosquito traps. Am. J. Trop. Med. Hyg. 2017;96:1241–1243. doi: 10.4269/ajtmh.16-0981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall-Mendelin S., Ritchie S.A., Johansen C.A., Zborowski P., Cortis G., Dandridge S., Hall R.A., van den Hurk A.F. Exploiting mosquito sugar feeding to detect mosquito-borne pathogens. Proc. Natl. Acad. Sci. Unit. States Am. 2010;107:11255–11259. doi: 10.1073/pnas.1002040107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaouadi K., Ghawar W., Salem S., Gharbi M., Bettaieb J., Yazidi R., Harrabi M., Hamarsheh O., Ben Salah A. First report of naturally infected Sergentomyia minuta with Leishmania major in Tunisia. Parasites Vectors. 2015;8:649. doi: 10.1186/s13071-015-1269-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamhawi S. Phlebotomine sand flies and Leishmania parasites: friends or foes? Trends Parasitol. 2006;22:439–445. doi: 10.1016/j.pt.2006.06.012. [DOI] [PubMed] [Google Scholar]
- Killick-Kendrick R. The biology and control of Phlebotomine sand flies. Clin. Dermatol. 1999;17:279–289. doi: 10.1016/S0738-081X(99)00046-2. [DOI] [PubMed] [Google Scholar]
- Kurucz N., Minney-Smith C.A., Johansen C.A. Arbovirus surveillance using FTATM cards in modified CO2-baited encephalitis virus surveillance traps in the Northern Territory, Australia. J. Vector Ecol. 2019;44:187–194. doi: 10.1111/jvec.12343. [DOI] [PubMed] [Google Scholar]
- Manuel J., Rebêlo M., Rodrigues B.L., Abreu C., Luiz J., Moraes P., Fonteles R.S., Regina S., Pereira F. Detection of Leishmania amazonensis and Leishmania braziliensis in Culicoides 8Deptera, Ceratopogonidae) in an endemic area of cutaneous leishmaniasis in. the Brazilian Amazonia. 2016;41:303–308. doi: 10.1111/jvec.12227. [DOI] [PubMed] [Google Scholar]
- Mukherjee S., Hassan M.Q., Ghosh A., Ghosh K.N., Bhattacharya A., Adhya S. Short report: Leishmania DNA in Phlebotomus and Sergentomyia species during a kala-azar epidemic. Am. J. Trop. Med. Hyg. 1997;57:423–425. doi: 10.4269/ajtmh.1997.57.423. [DOI] [PubMed] [Google Scholar]
- Pigott D.M., Bhatt S., Golding N., Duda K.A., Battle K.E., Brady O.J., Messina J.P., Balard Y., Bastien P., Pratlong F., Brownstein J.S., Freifeld C.C., Mekaru S.R., Gething P.W., George D.B., Myers M.F., Reithinger R., Hay S.I. Global distribution maps of the leishmaniases. Elife. 2014;3:1–21. doi: 10.7554/eLife.02851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ready P.D. Biology of phlebotomine sand flies as vectors of disease agents. Annu. Rev. Entomol. 2013;58:227–250. doi: 10.1146/annurev-ento-120811-153557. [DOI] [PubMed] [Google Scholar]
- Rogers M.E., Ilg T., Nikolaev A.V., Ferguson M.A.J., Bates P.A. Transmission of cutaneous leishmaniasis by sand flies is enhanced by regurgitation of fPPG. Nature. 2004 doi: 10.1038/nature02675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose K., Curtis J., Baldwin T., Mathis A., Kumar B., Sakthianandeswaren A., Spurck T., Low Choy J., Handman E. Cutaneous leishmaniasis in red kangaroos: isolation and characterisation of the causative organisms. Int. J. Parasitol. 2004;34:655–664. doi: 10.1016/j.ijpara.2004.03.001. [DOI] [PubMed] [Google Scholar]
- Sacks D., Kamhawi S. Molecular aspects of parasite-vector and vector-host interactions in leishmaniasis. Annu. Rev. Microbiol. 2002 doi: 10.1146/annurev.micro.55.1.453. [DOI] [PubMed] [Google Scholar]
- Sádlová J., Dvorak V., Seblova V., Warburg A., Votypka J., Volf P. Sergentomyia schwetzi is not a competent vector for Leishmania donovani and other Leishmania species pathogenic to humans. Parasites Vectors. 2013;6:186. doi: 10.1186/1756-3305-6-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sádlová J., Volf P. Occurrence of Leishmania major in sandfly urine. Parasitology. 1999;118:455–460. doi: 10.1017/S0031182099004254. [DOI] [PubMed] [Google Scholar]
- Seblova V., Sádlová J., Carpenter S., Volf P. Speculations on biting midges and other bloodsucking arthropods as alternative vectors of Leishmania. Parasites Vectors. 2014;7:222. doi: 10.1186/1756-3305-7-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seblova V., Sádlová J., Carpenter S., Volf P. Development of Leishmania parasites in Culicoides nubeculosus (Diptera: Ceratopogonidae) and implications for screening vector competence. J. Med. Entomol. 2012;49:967–970. doi: 10.1603/ME12053. [DOI] [PubMed] [Google Scholar]
- Seblova V., Sádlová J., Vojtkova B., Votypka J., Carpenter S., Bates P.A., Volf P. The biting midge Culicoides sonorensis (Diptera: Ceratopogonidae) is capable of developing late stage infections of Leishmania enriettii. PLoS Neglected Trop. Dis. 2015;9:1–15. doi: 10.1371/journal.pntd.0004060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solano-Gallego L., Rossi L., Scroccaro A.M., Montarsi F., Caldin M., Furlanello T., Trotta M. Detection of Leishmania infantum DNA mainly in Rhipicephalus sanguineus male ticks removed from dogs living in endemic areas of canine leishmaniosis. Parasites Vectors. 2012;5:98. doi: 10.1186/1756-3305-5-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor F. Studies in phlebotomic Diptera, No. 1. New species of simuliidæ and chironomidæ. Aust. Zool. 1918 [Google Scholar]
- van den Hurk A.F., Hall-Mendelin S., Townsend M., Kurucz N., Edwards J., Ehlers G., Rodwell C., Moore F.A., McMahon J.L., Northill J.A., Simmons R.J., Cortis G., Melville L., Whelan P.I., Ritchie S.A. Applications of a sugar-based surveillance system to track arboviruses in wild mosquito populations. Vector Borne Zoonotic Dis. 2014;14:66–73. doi: 10.1089/vbz.2013.1373. [DOI] [PubMed] [Google Scholar]
- Wipf N.C., Guidi V., Tonolla M., Ruinelli M., Müller P., Engler O. Evaluation of honey-baited FTA cards in combination with different mosquito traps in an area of low arbovirus prevalence. Parasites Vectors. 2019;12:554. doi: 10.1186/s13071-019-3798-8. [DOI] [PMC free article] [PubMed] [Google Scholar]