Abstract
With the evolution of biotechnology and the introduction of the high throughput sequencing, researchers have the ability to produce and analyze vast amounts of genomics data. Since genomics produce big data, most of the bioinformatics algorithms are based on machine learning methodologies, and lately deep learning, to identify patterns, make predictions and model the progression or treatment of a disease. Advances in deep learning created an unprecedented momentum in biomedical informatics and have given rise to new bioinformatics and computational biology research areas. It is evident that deep learning models can provide higher accuracies in specific tasks of genomics than the state of the art methodologies. Given the growing trend on the application of deep learning architectures in genomics research, in this mini review we outline the most prominent models, we highlight possible pitfalls and discuss future directions. We foresee deep learning accelerating changes in the area of genomics, especially for multi-scale and multimodal data analysis for precision medicine.
Keywords: Deep learning, Genomics, Computational biology, Bioinformatics, Gene expression and regulation, Precision medicine
1. Introduction
Bioinformatics has been successful to a significant extent, due to the radical influence from machine learning (ML) methodologies. Most of the well-known computational tools used by biologists have been addressed by the ML community. Nevertheless, current advances in the -omics era pose new insights for high impact collaboration and new challenges in the research community of ML. Methodologies and problems falling in the ML categories of classification, clustering and regression [1] have been proven useful for solving biological research questions such as gene signatures, functional genomics, gene-phenotype associations and gene interactions [2], [3], [4].
With the massive generation of data, the era known as ‘big’ data, deep learning (DL) approaches appeared as a discipline of ML that are considered to be more efficient and effective when we deal with big amounts of data [5]. These models have proven to achieve prediction accuracies at higher level than ever. The main limitation of ML compared to DL is that these methods cannot handle efficiently natural data in their raw form [6]. DL has also proven to provide models with higher accuracy that are efficient at discovering patterns in high-dimensional data making them applicable to a variety of domains. Like ML, DL models require training data and in the case of DL, the amount of training data is more demanding and drastically affect the predicting value of the trained model. The minimums vary with the complexity of the problem, but tens to hundreds of thousands of instances is a good place to start.
DL models are considered the state of the art predictive models for big datasets only the last decade, even though they first theorized in the 1980 s [7], a concept based on the perceptron model and the notion of neurons [8]. The hard requirements of DL models for large amounts of training data and substantial computing power, placed them unrealistic or limited until the introduction of special hardware such as the high-performance GPUs with parallel architecture. Nowadays deep learning architectures, also known as deep neural networks (DNNs), have been applied to many fields including speech recognition, natural language processing, vision and social networks analysis. The term “deep” in DL refers to the number of layers through which the data is transformed. Traditional neural networks only contain two to three hidden layers, while DL networks can have as many as two hundred layers. Nevertheless, DL networks require special hardware and massive parallelism to be effective [9]. In order to overcome resources demand and hardware limitations, the DL models use pipeline parallelism that can scale up the training phase. In the following we introduce the main DL architectures.
1.1. Deep learning architectures
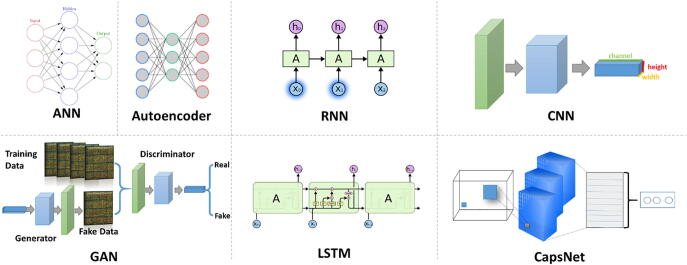
Artificial Neural Networks (ANN) were inspired by the neurons and their network that constitute human brains [10]. The ANN constitutes of a set of fully-connected nodes (neurons) modelling the stimuli propagation of brain synapses -fire or not- across the neural network. Such DL architectures are used for feature selection, classification, dimensionality reduction or as a submodule of a deeper architecture such as the convolutional neural networks.
Convolutional Neural Network (CNN) is an architecture of deep neural networks, most commonly applied to analyze visual imagery and was originally designed as a fully-automated image analysis network for classifying handcrafted characters [11]. CNNs are based on the multilayer perceptrons method and represent fully connected networks where each node/neuron in one layer is (fully) connected to all nodes of the following layer. ANN is a collection of connected and tunable units which can pass a signal from a unit to another. Contrary, CNNs have layers of convolution units that receive input from units of the previous layer and altogether produce a proximity. The fundamental principle of this deep architecture is to massively compute and combine feature maps inferring non-linear relationships between the input signal and the targeted output [12]. CNN is popular for feature extraction, selection, reduction mainly for the classification of image datasets.
Recurrent Neural Networks (RNN) exhibit similar functionality with the regular feedforward Neural Networks (FNN) [13] where connections between nodes form a directed graph along a temporal sequence [14]. This allows RNNs to exhibit temporal dynamic behavior and in addition, integrate internal memory. This short-term memory allows recurrent networks to remember information from the previously analyzed states, a perfect fit for sequential signal analysis and predictive models. One of the strengths of RNNs is the idea that models are able to connect information from a previous task to the present task.
Long short-term memory (LSTM) is a variation of the RNNs [15] capable of learning long-term dependencies and actually are designed to avoid the long-term dependency problem. In its core, a LSTM unit has a cell/node, a gate for the input, a gate for the output and a forget gate. The node takes into account values over specific time intervals while the input/output gates regulate the flow of information.
Generative Adversarial Networks (GANs) is a more recent architecture that uses two neural networks pitting one against the other [16]. One network generates synthetic realistic data while the second evaluates the authenticity of the data (if it belongs to the real training dataset or not). GANs proved to improve the classification accuracy in many domains including genomics [17].
Autoencoders (AE) learn a representation (encoder) for the data by training the network to ignore signal “noise” [18] and are one of the well-known DL models for unsupervised learning. Neural nets typically use simple non-linearities in which a non-linear function is applied to the scalar output of a linear filter. Capsules use a much more complicated non-linearity, where a set of neurons model a part of the input by activating a small subset of its properties [19]. The CapsuleNet [20] consists of independent sets of capsules instead of kernels. This architecture is one of the newest in the DL models and have yet to be tested extensively from the research community. Fig. 1 sketches the architectures of the most common DL models.
Fig. 1.
Architecture of the main deep learning models.
Apart from the DL architectures, there are also methodologies that can combine DL or ML models to enhance the predictive accuracy. One such methodology is the multi model fusion, a meta-analysis of diverse models built on different data aiming at a single objective [21]. Decision fusion combines the outcome of multiple classifiers into a singular final prediction forming a meta-estimator by utilizing statistical methods to amplify the individual classifiers. Sequential fusion models also do exist, such as the DanQ which employs CNN and then RNN for the quantification of the function of DNA sequences [22]. Both lead to an improved accumulated predictive power and can resolve uncertainties or disagreements among singular analyses.
Another methodology that has proven to improve accuracy is transfer learning. The idea behind transfer learning is that data from a different domain can be the starting point for training a predictive model. So a model trained with widely available dataset, e.g. natural images, can be transferred to a target model that will perform similar tasks but in a different domain, e.g. medical imaging, that lack the volume of training data. In transfer learning two major methodologies can be followed namely off-the-self models and fine-tuned models. There are several available pre-trained models, especially in the domain of imaging such as the VGG-161, Inception [23], DenseNet [24], Mask R-CNN [25], employed by many authors claiming mixed results for the off-the-self method while with fine-tuning being the most promising due to its supplementary adaptation to the targeted model [26], [27], [28]. The research community supports the DL modelling with open access frameworks for DL, such as PyTorch, TensorFlow, Theano and Caffe making the implementation process easier and faster [9].
1.2. Genomics data analysis
A multiplicity of machine learning approaches [3], [29], [30], [31] have been suggested and evaluated in order to identify important data for stratification/classification of different patient groups (e.g. with respect to therapy response, probability of serious adverse events or outcome prediction). Such methodologies can select features that characterize classes, identify groups with similar feature space, classify cases or mixed data such as the Montesinos-López et al [32]. These methods have been applied in the context of genomics multi-level classification, especially for cancer research [33], [34], [35].
Furthermore, in the literature we can find precision medicine approaches that take advantage of genomic and clinical data along with the power of DL for prognostic prediction [36]. A representative paradigm for precision medicine is the precision oncology [37], founded and enabled by revolutionary post-genomics advances, that is confronted with the generation of heterogenous multi-scale genomic profiles (multi–omics) [38]. The -omics research area produces big volumes of data mainly due to the evolution that has taken place in the field of genomics and the advances of biotechnology. Indicative examples include the high-throughput platforms that measure the expression of thousands of genes or non-coding transcripts (e.g., miRNAs), the genotyping platforms and next generation sequencing (NGS) technologies and related genome-wide association studies (GWAS) that produce quantitative gene expression profiles (e.g., RNA-seq), identify large number of gene variants (SNPs, Indels) as well as other genome alterations (e.g., copy number variations CNVs) for different populations.
2. Materials & methods
The manuscript does not aim to provide a systematic literature review of deep learning methodologies for genomics but rather captures the current trends in the area. Towards this direction, studies focusing on radiogenomics where deep learning architectures used only for the image analysis [39] and then combined with genomics analysis using statistics or traditional machine learning methodologies were excluded. Also studies where deep learning used only for data augmentation and synthetic data generation paired with genomics or other analysis, were excluded as well.
In the literature we can find a few approaches of DL models applied in gene expression data. DeepTarget [40] and deepMirGene [41] use RNN and LSTM models respectively to perform miRNA and target prediction using expression data. The algorithms proved that can predict microRNA target with higher accuracy than the non-deep learning state of the art model called TargetScan [42]. Apart from the higher accuracies, the proposed methods have a major advantage over existing alternatives in that no hand-crafted feature set is needed.
Urda et al [43] provide a first approximation of how to use a multi-layer feed-forward artificial neural network to analyze RNA-Seq gene expression data. Their model outperforms LASSO in analyzing RNA-Seq gene expression profiles data. Gupta et al [44] demonstrated the empirical effectiveness of using deep networks as a pre-processing step for clustering of gene expression data. Authors employed Deep Belief Networks with AE for learning a low-dimensional representation of expression profiles, an unsupervised learning approach for gene selection. The DL model used as a pre-processing step for clustering the yeast expression microarrays into modules that simulate the cell cycle processes and the results indicate that this method outperforms the principal component analysis algorithm. Chen et al [45] used also AE on yeast cDNA microarray data in order to learn the encoding system of yeast transcriptomic machinery. Results indicate that such a methodology can be used to partially recover the organization of transcriptomic machinery.
Shallow denoising AE, a special case of AE where the model feeds the input data with noise, have been evaluated for their usefulness in the domain of genomics. Tan et al [46] applied analysis using denoising autoencoders of gene expression (ADAGE) on a publicly available gene expression data compendium for pseudomonas aeruginosa in order to identify differences between strains and predict the involvement of biological processes based on low-level gene expression differences. The same research group generated an ensemble ADAGE that integrates stable biological patterns, enables cross-experiment comparisons and can highlight measured but undiscovered relationships [47].
Authors of D-GEX provide a deep learning architecture to infer the expression of target genes from the expression of landmark genes [48]. D-GEX trained a multi-layer feedforward deep neural network with three hidden layers using 111,000 public expression profiles from Gene expression Omnibus2. The DL models provide better accuracy than linear regression in inferring the expression of the human genes (about 21000) based on a set of landmark genes (about 1000). Even though the DL model provided better accuracy than existing ML models it still displayed poor performance indicating that there is room for improvement in the architecture of the model.
The DeepChrome CNN method [49] automatically learns combinatorial interactions among histone modification marks in order to predict the gene expression. DeepChrome proved the improvement of the prediction accuracy over existing methods such as Support Vector Machines (SVM) and Random Forests (FR) for Boolean (high/low) gene expression prediction using histone modifications as input. The same research team also provided the AttentiveChrome [50], an LSTM DL model to further enhance DeepChrome using a unified architecture to interpret dependencies among chromatin factors for controlling gene regulation.
DeepVariant [51] is a CNN variant caller that proved to outperform all the non-DL state-of-the-art variant callers. Furthermore, authors proved that DeepVariant generalizes beyond its training data using different versions of the human genome built as train and test datasets. Also when DeepVariant was trained using human reads and tested against a mouse dataset achieved accuracy that outperforms training on the mouse data itself. DeepFIGV [52] is a DL model able to predict locus-specific signals from epigenetic assays using DNA sequence. DeepFIGV models quantitative variation in the epigenome using many experiments from the same cell type and assay and integrates whole genome sequencing to create a personalized genome sequence for each individual.
Sakellaropoulos et al [53] implemented a DL model for predicting response therapy in cancer. The authors used a pharmacogenomics database of 1001 cancer cell lines to train the model in order to predict drug response and proved that DL outperforms the current state in machine learning frameworks for the specific task. Liang et al. [54] provided a multimodal deep belief network able to integrate DNA methylation, gene and miRNA expression data for the identification of cancer subtypes. The proposed method exploits both deep intrinsic statistical properties of each input modality and complex cross-modality correlations among multi-platform input data. Another multi-modal DL model in genomics is the DeePathology [55], a DL method that is capable of simultaneously infer various properties of biological samples, through multi-task and transfer learning. The model encodes the whole transcription profile and can accurately predict tissue and disease type.
Yuan et al [56] introduced a convolutional neural network for coexpression (CNNC) that improves upon prior methods in inferring gene relationships from single-cell expression data tasks. The method can be used for a wide range of -omics research questions ranging from predicting transcription factor targets to identifying disease-related genes to causality inference. DeepCpG [57] is another computational approach for low-coverage single-cell methylation data using a CNN model. DeepCpG predicts missing methylation states and detects sequence motifs that are associated with changes in methylation levels and cell-to-cell variability better than the state of the art ML methods.
It is evident that DL architectures have also been applied to genomics lately with promising results. Table 1 summarizes the discussed DL models in the genomics domain, highlighting the omics data used, the research question, the DL model and the evaluation results.
Table 1.
. List of deep learning methodologies in genomics. From left to right the columns represent the DL model acronym (if any), the respective publication, DL model, omics data used as input, prediction/research question, evaluation metrics and the comparison with other classic ML methods (if any).
| Name | Publication | DL model | omics data | Purpose / Prediction | accuracy | performance gap over other methods |
|---|---|---|---|---|---|---|
| DeepTarget | [40] | RNN | miRNA-mRNA pairing | target prediction | 0,96 | +25% f-measure |
| DeepMirGene | [41] | LSTM | positive pre-miRNA and non-miRNA | miRNA target | 0.89 sensitivity | +4% f-measure |
| DeepNet | [43] | ANN | RNA-Seq | control-cases | ~0.7 | same or worst AUC from LASSO |
| [44] | AE | time-series gene expression | pre-processing step for clustering | Better than PCA | ||
| [45] | AE | cDNA microarrays | Predict the organization of transcriptomic machinery | – | significant overlap with previous studies | |
| ADAGE | [47] | AE | gene expression | identification/reconstruction of biological signals | – | significant overlap with post-hoc analysis KEGG |
| eADAGE | [47] | AE | gene expression | identification of biological patterns | – | significant overlap with post-hoc analysis KEGG |
| D-GEX | [48] | RNN | expression of landmark genes | Gene expression inference | overall error 0.3204 ± 0.0879 | Outperforms Linear Regression(LR) (+15.33%) and KNN-GE in most of the target genes |
| DeepChrome | [49] | CNN | histone modifications | classify gene expression | Average area under the curve (AUC) = 0.80 | (+5%) from support vector machines (SVM), (+21% from random forest (RF) |
| AttentiveChrome | [50] | LSTM | histone modifications | classify gene expression | Average AUC = 0.81 | Marginally better than DeepChrome |
| Multimodal deep belief network | [54] | DBN | gene expression, DNA methylation and miRNA expression | Identification of Key Genes and miRNAs | average correlations 0.91, 0.73 and 0.69 for the GE, DM and ME | – |
| DeepVariant | [51] | CNN | whole-genome sequence | variant caller | 99,45% F1 | produced more accurate results with greater consistency across a variety of quality metrics |
| [53] | ANN | cell-line with drug response | predict drug response | 0.65 AUC | Outperformed FR 0.54 AUC and elastic nets 0.51 AUC | |
| DeepFIGV | [52] | CNN | whole-genome sequence | predict quantitative epigenetic variation | z-scores DNase rho = 0.0802, P = 5.32e–16 | |
| DeePathology | [55] | Multiple AEs | mRNA and miRNA | predict tissue-of-origin, normal or disease state and cancer type | 99.4% accuracy for cancer subtype | 95.1% for SVM |
| DeepCpG | [57] | CNN | Single cell methylation | predicts missing methylation states and detects sequence motifs | 89% AUC | 86% AUC for Random Forest |
| CNNC | [56] | CNN | scRNA-seq | predicting transcription factor target | ~70% accuracy for multiple experiments | Outperformed GBA (guilt by association) and DNN (fully connected DL) across a variety of experiments |
| DanQ | [22] | CNN and RNN | DNA-seq | predicting the function of DNA directly from sequence alone | AUC score ~ 70% | Outperformed LR and DeepSEA (CNN DL), with over 10% improvement in AUC |
| FBGAN | [17] | GANs | DNA-seq | optimize the synthetic gene sequences | Train accuracy 0.94 test accuracy 0.84 | Outperformed kmer and Wasserstein GAN trained directly on AMPs |
3. Results and discussion
Deep learning models are considered the state of the art for classification and clustering when we deal with big data such as the -omics area. Nevertheless, we are still far from providing DL models for -omics data that can be used in the precision medicine since the proposed methodologies have not been validated yet in the clinical practice. The success of DL depends on finding an architecture to fit the research question and be capable to handle the respective data. Over the years, various DL methods introduced making the selection of the most appropriate method a non-trivial path [68]. For example, LSTM networks are an advanced version of RNN, capsNets try to overcome limitations of CNNs such as the viewpoints of the data and GANs provide promising results for automatically training a generative model by treating the unsupervised problem as supervised.
3.1. Limitations of DL in genomics
DL models are in its infancy in the genomics area and still far from complete. In the following, we provide five major limitations of the DL models in the genomics area:
-
1.
Model interpretation (the black box): One of the major issues for DL architectures in general, is the interpretation of the model [58]. Due to the structure of the DL models it is difficult to understand the rational and the learned patterns if one would like to extract the causality relationship between the data and the outcome. This is more evident in the bioinformatics domain, given that researchers prefer ‘white-box’ approaches to ‘black-box’ approaches [59]. The use of explainable AI techniques [60] has start to gain momentum in the genomics area [61].
-
2.
The curse of dimensionality: The most pronounced limitation of artificial intelligence in the genomics is the so-called “curse of dimensionality” of the -omics data [62]. Even though genomics is considered a big data domain in terms of volume, the genomic datasets usually represent a very large number of variables and a small number of samples. This is a known problem in genomics not only for DL but also for less demanding (in terms of samples) ML algorithms [63]. Fortunately, in the genomics area there are repositories that provide access to public data and one can combine datasets from multiple sources. Nevertheless, in order to collect a representative cohort for DL training, a lot of preprocessing and harmonization is needed.
-
3.
Imbalanced classes: Most of the DL and ML models for genomics deal with classification problems e.g. discrimination between disease and healthy samples. It is well-known that genomics trials and data gathered from various sources are usually inherently class imbalanced and ML/DL models cannot be effective until a sufficient number of instances per class has been fitted. Fortunately, transfer learning can provide a solution to tackle the class imbalanced problem since the model can be initially trained to a general dataset [64].
-
4.
Heterogeneity of data: The data in most of the genomic applications is heterogeneous since we deal with subgroups of the population. Even in the individual level, genomic data include: (i) sequencing of genes or non-coding transcripts, (ii) quantitative gene expression profiles (iii) gene variants (iv) genome alternations and (v) gene interactions in the system’s biology level. One of the obstacles in integrating different data is the covariates between the underling interdependencies among these heterogeneous data. Bioinformatics community, taking advantage of the plethora of data sources, have provide many analysis tools but in most of the cases this combination is troubling researchers to use the available resources effectively [65].
-
5.
Parameters and hyper-parameters tuning: One of the most difficult steps for DL is the tuning of the model. Careful analysis of initial results may prove really helpful during tuning since the tuning is correlated with the dataset and the research question. The main tuning hyper-parameters for every DL architecture are the learning rate, the batch size, the momentum, and the weight decay. Learning rate is a tuning parameter that determines the step size at each iteration while moving toward a minimum of a loss function, batch size is the number of training samples used in each iteration, momentum tries to find the optimal training path and weight decay is a process where after each update, the weights are multiplied by a factor. These hyper-parameters act as knobs which can be tweaked during the training of the model. A wrong setting in any of these parameters may result to under-fitting or over-fitting [66].
3.2. Future directions
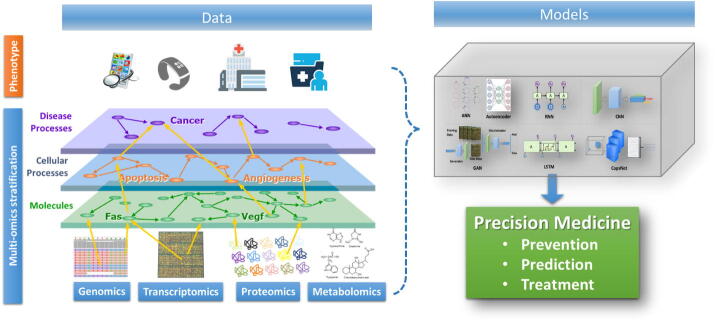
In bioinformatics and computational biology, the methods for heterogeneous data and sources integration are in rapid evolution. The ability of describing and representing biomedical findings on different data layers is already a state-of-art. A multi-layer model approach has been motivated by the very nature of systems biology [67], and it is an accepted basis for system approaches towards precision medicine. Multi-scale dynamic modelling approaches have been recently explored to model the human body as a single complex dynamical system. Although exciting, this global approach has proven challenging with statistical methods [68]. Deep learning has the ability to deal with multimodal data effectively and genomics offers extremely heterogeneous data. The notion of precision medicine is based on the multimodal data analysis and a typical example of multi-level and multi scale genomics data is depicted in Fig. 2.
Fig. 2.
Multi level and multi scale -omics models.
DL models have an advantage over other genomics algorithms in the preprocessing steps that traditionally are manually curated, error prone and time consuming. DL is fed with all the data and the abstraction ability of the model can select or define features that often increase predictive power. Zou et al [69] provides a guide for the design of deep learning systems for genomics that best augment and complement human experience in making medical decisions, while Eraslan et al [70] proposes DL models based on the research question in the domain of genomics. Nevertheless, in many cases genomics data do not conform to the requirements posed by most of the DL architectures [71]. An example comes from the text mining DL architectures for chatbots or text auto-completion that at first sight one could imagine to be a solution for single-nucleotide polymorphism (SNP) analysis and prediction using each SNP as one word. Unfortunately, such DL architectures currently cannot handle ‘dictionaries’ larger than a few hundred of thousand ‘words’ and the known SNPs for the human genome are about 90 million.
Irrespective of the genomics modeling methodologies, the process of translating the knowledge acquired in genomics research into clinically useful tools has been extremely slow. This is in part due to the requirements for validation and standardization, which are sometimes slow to fulfill due to the fragmentation of genomics research and inadequacies of analysis set-ups and platforms. Fortunately, FDA is considering a regulatory framework for computational technologies that would allow modifications to be made from real-world learning and adaptation, while still ensuring that the safety and effectiveness of the software as a medical device is maintained3.
Availability of patient data for precision medicine, especially the small information-rich data sets, are often not representative for the overall population whilst most models in DL need a lot of data to be able to generalize findings and predict on future classes of patients. Genomics data often comprise features of heterogeneous data types (numerical, categorical, and possibly other data types like functions), which are only handled adequately when using correspondingly different dissimilarities [72] Models that are capable of such an integration are however often not easy to be interpreted by human experts; and whilst being sometime successful in classifying patients to groups they are often complex and difficult to grasp for medical and biological experts. The notion of explainable-AI is still far from complete in the bioinformatics domain [73].
4. Conclusions
In this mini review we discuss the concepts of DL models in genomics while we outline the most prominent DL architectures in the area of genomics and as DL is practically a new methodology, the studies discussed, have been proposed the last years. Based on our research, it is evident that DL models can provide higher accuracies in specific tasks of genomics than the state of the art methodologies. In addition, deep learning has the ability to deal with multimodal data effectively and genomics offers extremely heterogeneous data making them an excellent candidate for the realization of precision medicine. Nevertheless, the process of translating the knowledge acquired in genomics research into clinically useful tools has been extremely slow. More efforts should be made to analyze and combine datasets (private and public) in order to enhance the role of DL genomics in prediction and prognosis. Furthermore, explainable DL models can pave the way for identifying not only novel biomarkers but also regulatory interactions in different pathology conditions such as tissues and disease states.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We acknowledge support of this work by the project “ELIXIR-GR: Hellenic Research Infrastructure for the Management and Analysis of Data from the Biological Sciences” (MIS 5002780) which is implemented under the Action “Reinforcement of the Research and Innovation Infrastructure”, funded by the Operational Programme “Competiveness, Entrepreneurship and Innovation” (NSRF 2014-2020) and co-financed by Greece and the European Union (European Regional Development Fund).
Footnotes
References
- 1.Zhang Y.Q., Rajapakse J.C. Mach Learn Bioinform. 2008 doi: 10.1002/9780470397428. [DOI] [Google Scholar]
- 2.Potamias G, Koumakis L, Moustakis V. Gene Selection via Discretized Gene-Expression Profiles and Greedy Feature-Elimination. Methods Appl. Artif. Intell. Third Helenic Conf. AI, {SETN} 2004, Samos, Greece, May 5-8, 2004, Proc., 2004, p. 256–66. https://doi.org/10.1007/978-3-540-24674-9_27.
- 3.Koumakis L., Kanterakis A., Kartsaki E., Chatzimina M., Zervakis M., Tsiknakis M. MinePath: Mining for Phenotype Differential Sub-paths in Molecular Pathways. PLoS Comput Biol. 2016;12 doi: 10.1371/journal.pcbi.1005187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang B.E., Mulyasasmita W., Rajagopal G. The path from big data to precision medicine. Expert Rev Precis Med Drug Dev. 2016;1:129–143. doi: 10.1080/23808993.2016.1157686. [DOI] [Google Scholar]
- 5.Zhang L., Tan J., Han D., Zhu H. From machine learning to deep learning: progress in machine intelligence for rational drug discovery. Drug Discov Today. 2017 doi: 10.1016/j.drudis.2017.08.010. [DOI] [PubMed] [Google Scholar]
- 6.Lecun Y., Bengio Y., Hinton G. Deep learning. Nature. 2015 doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 7.Dechter Rina. Learning While Searching In Constraint-Satisfaction-Problems. Ann Math. 1986 [Google Scholar]
- 8.Rosenblatt F. The perceptron: A probabilistic model for information storage and organization in the brain. Psychol Rev. 1958 doi: 10.1037/h0042519. [DOI] [PubMed] [Google Scholar]
- 9.Lecun Y. 1.1 Deep Learning Hardware: Past, Present, and Future. Dig. Tech. Pap. - IEEE Int. Solid-State Circuits Conf. 2019 doi: 10.1109/ISSCC.2019.8662396. [DOI] [Google Scholar]
- 10.Nelson D., Wang J. Introduction to artificial neural systems. Neurocomputing. 1992 doi: 10.1016/0925-2312(92)90018-k. [DOI] [Google Scholar]
- 11.Lecun Y, Bottou L, Bengio Y, Haffner P. Gradient-Based Learning Applied to Document Recognition. Proc. IEEE, 1998.
- 12.Gu J, Wang Z, Kuen J, Ma L, Shahroudy A, Shuai B, et al. Recent Advances in Convolutional Neural Networks. n.d.
- 13.Montana DJ, Davis L. Training Feedforward Neural Networks Using Genetic Algorithms. Proc 11th Int Jt Conf Artif Intell - Vol 1 1989.
- 14.Williams R.J., Zipser D. A Learning Algorithm for Continually Running Fully Recurrent Neural Networks. Neural Comput. 1989 doi: 10.1162/neco.1989.1.2.270. [DOI] [Google Scholar]
- 15.Hochreiter S., Schmidhuber J. Long Short-Term Memory. Neural Comput. 1997 doi: 10.1162/neco.1997.9.8.1735. [DOI] [PubMed] [Google Scholar]
- 16.Radford A, Metz L, Chintala S. Unsupervised representation learning with deep convolutional generative adversarial networks. 4th Int. Conf. Learn. Represent. ICLR 2016 - Conf. Track Proc., 2016.
- 17.Gupta A., Zou J. Feedback GAN for DNA optimizes protein functions. Nat Mach Intell. 2019 doi: 10.1038/s42256-019-0017-4. [DOI] [Google Scholar]
- 18.Baldi P. Autoencoders, Unsupervised Learning, and Deep Architectures. ICML Unsupervised Transf Learn 2012. https://doi.org/10.1561/2200000006.
- 19.Hinton G, Sabour S, Frosst N. Matrix capsules with EM routing. 6th Int. Conf. Learn. Represent. ICLR 2018 - Conf. Track Proc., 2018.
- 20.Sabour S., Frosst N., Hinton G.E. Dynamic Routing Between Capsules. Adv Neural Inf Process Syst. 2017:3856–3866. doi: 10.1177/1535676017742133. [DOI] [Google Scholar]
- 21.Shi Y., Tranchevent L.-C., De Moor B., Moreau Y. Kernel-based Data Fusion for Machine Learning Studies in Computational. Intelligence. 2011;Volume:345. [Google Scholar]
- 22.Quang D., Xie X. DanQ: A hybrid convolutional and recurrent deep neural network for quantifying the function of DNA sequences. Nucl Acids Res. 2016 doi: 10.1093/nar/gkw226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Szegedy C, Vanhoucke V, Ioffe S, Shlens J, Wojna Z. Rethinking the Inception Architecture for Computer Vision. Proc. IEEE Comput. Soc. Conf. Comput. Vis. Pattern Recognit., vol. 2016- Decem, IEEE Computer Society; 2016, p. 2818–26. https://doi.org/10.1109/CVPR.2016.308.
- 24.Huang G, Liu Z, van der Maaten L, Weinberger KQ. Densely Connected Convolutional Networks 2016.
- 25.He K, Gkioxari G, Dollar P, Girshick R. Mask R-CNN. Proc. IEEE Int. Conf. Comput. Vis., vol. 2017- Octob, Institute of Electrical and Electronics Engineers Inc.; 2017, p. 2980–8. https://doi.org/10.1109/ICCV.2017.322.
- 26.Gopalakrishnan K., Khaitan S.K., Choudhary A., Agrawal A. Deep Convolutional Neural Networks with transfer learning for computer vision-based data-driven pavement distress detection. Constr Build Mater. 2017 doi: 10.1016/j.conbuildmat.2017.09.110. [DOI] [Google Scholar]
- 27.Kolar Z., Chen H., Luo X. Transfer learning and deep convolutional neural networks for safety guardrail detection in 2D images. Autom Constr. 2018 doi: 10.1016/j.autcon.2018.01.003. [DOI] [Google Scholar]
- 28.Soudani A., Barhoumi W. An image-based segmentation recommender using crowdsourcing and transfer learning for skin lesion extraction. Expert Syst Appl. 2019 doi: 10.1016/j.eswa.2018.10.029. [DOI] [Google Scholar]
- 29.Bishop CM. Pattern Recognition and Machine Learning. vol. 4. 2006. https://doi.org/10.1117/1.2819119.
- 30.Mitchell T.M. The Discipline of Machine Learning. Mach Learn. 2006 doi: 10.1080/026404199365326. [DOI] [Google Scholar]
- 31.Domingos P. A few useful things to know about machine learning. Commun ACM. 2012 doi: 10.1145/2347736.2347755. [DOI] [Google Scholar]
- 32.Montesinos-López OA, Martín-Vallejo J, Crossa J, Gianola D, Hernández-Suárez CM, Montesinos-López A, et al. New deep learning genomic-based prediction model for multiple traits with binary, ordinal, and continuous phenotypes. G3 Genes, Genomes, Genet 2019. https://doi.org/10.1534/g3.119.300585. [DOI] [PMC free article] [PubMed]
- 33.Ibrahim R, Yousri NA, Ismail MA, El-Makky NM. Multi-level gene/MiRNA feature selection using deep belief nets and active learning. 2014 36th Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. EMBC 2014, 2014. https://doi.org/10.1109/EMBC.2014.6944490. [DOI] [PubMed]
- 34.Mehta S., Hughes N.P., Li S., Jubb A., Adams R., Lord S. Radiogenomics Monitoring in Breast Cancer Identifies Metabolism and Immune Checkpoints as Early Actionable Mechanisms of Resistance to Anti-angiogenic Treatment. EBioMedicine. 2016 doi: 10.1016/j.ebiom.2016.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koumakis L., Roussos P., Potamias G. Minepath.org: A free interactive pathway analysis web server. Nucl Acids Res. 2017;45:W116–W121. doi: 10.1093/nar/gkx278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhu W., Xie L., Han J., Guo X. The application of deep learning in cancer prognosis prediction. Cancers (Basel) 2020 doi: 10.3390/cancers12030603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Prasad V., Fojo T., Brada M. Precision oncology: Origins, optimism, and potential. Lancet Oncol. 2016 doi: 10.1016/S1470-2045(15)00620-8. [DOI] [PubMed] [Google Scholar]
- 38.Civelek M., Lusis A.J. Systems genetics approaches to understand complex traits. Nat Rev Genet. 2014 doi: 10.1038/nrg3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trivizakis E., Papadakis G.Z., Souglakos I., Papanikolaou N., Koumakis L., Spandidos D.A. Artificial intelligence radiogenomics for advancing precision and effectiveness in oncologic care (Review) Int J Oncol. 2020 doi: 10.3892/ijo.2020.5063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee B., Baek J., Park S., Yoon S. deepTarget: End-to-end Learning Framework for microRNA Target Prediction using Deep Recurrent Neural Networks. ACM-BCB 2016–7th ACM. Conf Bioinformatics, Comput Biol Heal Informatics. 2016:434–442. [Google Scholar]
- 41.Park S., Min S., Choi H., Yoon S. deepMiRGene: Deep Neural Network based Precursor microRNA. Prediction. 2016 [Google Scholar]
- 42.Lewis B.P., Shih I.H., Jones-Rhoades M.W., Bartel D.P., Burge C.B. Prediction of Mammalian MicroRNA Targets. Cell. 2003 doi: 10.1016/S0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 43.Urda D, Montes-Torres J, Moreno F, Franco L, Jerez JM. Deep learning to analyze RNA-Seq gene expression data. Lect. Notes Comput. Sci. (including Subser. Lect. Notes Artif. Intell. Lect. Notes Bioinformatics), 2017. https://doi.org/10.1007/978-3-319-59147-6_5.
- 44.Gupta A, Wang H, Ganapathiraju M. Learning structure in gene expression data using deep architectures, with an application to gene clustering. Proc. - 2015 IEEE Int. Conf. Bioinforma. Biomed. BIBM 2015, 2015. https://doi.org/10.1109/BIBM.2015.7359871.
- 45.Chen L., Cai C., Chen V., Lu X. Learning a hierarchical representation of the yeast transcriptomic machinery using an autoencoder model. BMC Bioinf. 2016 doi: 10.1186/s12859-015-0852-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tan J., Hammond J.H., Hogan D.A., Greene C.S. ADAGE-Based Integration of Publicly Available Pseudomonas aeruginosa Gene Expression Data with Denoising Autoencoders Illuminates Microbe-Host Interactions. MSystems. 2016 doi: 10.1128/msystems.00025-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tan J., Doing G., Lewis K.A., Price C.E., Chen K.M., Cady K.C. Unsupervised Extraction of Stable Expression Signatures from Public Compendia with an Ensemble of Neural Networks. Cell Syst. 2017 doi: 10.1016/j.cels.2017.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen Y., Li Y., Narayan R., Subramanian A., Xie X. Gene expression inference with deep learning. Bioinformatics. 2016 doi: 10.1093/bioinformatics/btw074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Singh R., Lanchantin J., Robins G., Qi Y. DeepChrome: Deep-learning for predicting gene expression from histone modifications. Bioinformatics. 2016 doi: 10.1093/bioinformatics/btw427. [DOI] [PubMed] [Google Scholar]
- 50.Singh R., Lanchantin J., Sekhon A., Qi Y. Attend and predict: Understanding gene regulation by selective attention on chromatin. Adv. Neural Inf. Process. Syst. 2017 [PMC free article] [PubMed] [Google Scholar]
- 51.Poplin R., Chang P.C., Alexander D., Schwartz S., Colthurst T., Ku A. A universal snp and small-indel variant caller using deep neural networks. Nat Biotechnol. 2018 doi: 10.1038/nbt.4235. [DOI] [PubMed] [Google Scholar]
- 52.Hoffman G.E., Bendl J., Girdhar K., Schadt E.E., Roussos P. Functional interpretation of genetic variants using deep learning predicts impact on chromatin accessibility and histone modification. Nucleic Acids Res. 2019 doi: 10.1093/nar/gkz808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sakellaropoulos T., Vougas K., Narang S., Koinis F., Kotsinas A., Polyzos A. A Deep Learning Framework for Predicting Response to Therapy in Cancer. Cell Rep. 2019 doi: 10.1016/j.celrep.2019.11.017. [DOI] [PubMed] [Google Scholar]
- 54.Liang M., Li Z., Chen T., Zeng J. Integrative Data Analysis of Multi-Platform Cancer Data with a Multimodal Deep Learning Approach. IEEE/ACM Trans Comput Biol Bioinforma. 2015 doi: 10.1109/TCBB.2014.2377729. [DOI] [PubMed] [Google Scholar]
- 55.Azarkhalili B., Saberi A., Chitsaz H., Sharifi-Zarchi A. DeePathology: Deep Multi-Task Learning for Inferring Molecular Pathology from Cancer Transcriptome. Sci Rep. 2019 doi: 10.1038/s41598-019-52937-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yuan Y., Bar-Joseph Z. Deep learning for inferring gene relationships from single-cell expression data. Proc Natl Acad Sci U S A. 2019 doi: 10.1073/pnas.1911536116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Angermueller C., Lee H.J., Reik W., Stegle O. DeepCpG: Accurate prediction of single-cell DNA methylation states using deep learning. Genome Biol. 2017 doi: 10.1186/s13059-017-1189-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ghorbani A., Abid A., Zou J. Interpretation of Neural Networks Is Fragile. Proc AAAI Conf Artif Intell. 2019 doi: 10.1609/aaai.v33i01.33013681. [DOI] [Google Scholar]
- 59.Min S., Lee B., Yoon S. Deep learning in bioinformatics. Brief Bioinform. 2017 doi: 10.1093/bib/bbw068. [DOI] [PubMed] [Google Scholar]
- 60.Ribeiro M.T., Singh S., Guestrin C. “Why should i trust you?” Explaining the predictions of any classifier. Proc. ACM SIGKDD Int. Conf. Knowl. Discov. Data Min. 2016 doi: 10.1145/2939672.2939778. [DOI] [Google Scholar]
- 61.Graham G., Csicsery N., Stasiowski E., Thouvenin G., Mather W.H., Ferry M. Genome-scale transcriptional dynamics and environmental biosensing. Proc Natl Acad Sci U S A. 2020 doi: 10.1073/pnas.1913003117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Barbour D.L. Precision medicine and the cursed dimensions. Npj Digit Med. 2019 doi: 10.1038/s41746-019-0081-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang L., Wang Y., Chang Q. Feature selection methods for big data bioinformatics: A survey from the search perspective. Methods. 2016 doi: 10.1016/j.ymeth.2016.08.014. [DOI] [PubMed] [Google Scholar]
- 64.Al-Stouhi S., Reddy C.K. Transfer learning for class imbalance problems with inadequate data. Knowl Inf Syst. 2016 doi: 10.1007/s10115-015-0870-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lathe, W., Williams, J., Mangan, M. & Karolchik D. Genomic Data Resources: Challenges and Promises. Nat Educ 2008.
- 66.Smith LN. Disciplined Approach To Neural Network. 2018.
- 67.Liebermeister W, Wierling C, Kowald A, Lehrach H, Herwig R. Systems Biology: A Textbook Answers to Problems. 2009.
- 68.Deisboeck TS, Stamatakos GS. Multiscale Cancer Modeling. 2010. https://doi.org/10.1201/b10407.
- 69.Zou J., Huss M., Abid A., Mohammadi P., Torkamani A., Telenti A. A primer on deep learning in genomics. Nat Genet. 2019 doi: 10.1038/s41588-018-0295-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Eraslan G., Avsec Ž., Gagneur J., Theis F.J. Deep learning: new computational modelling techniques for genomics. Nat Rev Genet. 2019 doi: 10.1038/s41576-019-0122-6. [DOI] [PubMed] [Google Scholar]
- 71.Grapov D., Fahrmann J., Wanichthanarak K., Khoomrung S. Rise of deep learning for genomic, proteomic, and metabolomic data integration in precision medicine. Omi A J Integr Biol. 2018 doi: 10.1089/omi.2018.0097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ziegel E.R. The Elements of Statistical Learning. Technometrics. 2003 doi: 10.1198/tech.2003.s770. [DOI] [Google Scholar]
- 73.Holzinger A., Biemann C., Pattichis C.S., Kell D.B. What do we need to build explainable AI systems for the medical domain? ArXiv Prepr ArXiv171209923. 2017 [Google Scholar]