Abstract
Background
To develop a deep learning (DL) method based on multiphase, contrast-enhanced (CE) magnetic resonance imaging (MRI) to distinguish Liver Imaging Reporting and Data System (LI-RADS) grade 3 (LR-3) liver tumors from combined higher-grades 4 and 5 (LR-4/LR-5) tumors for hepatocellular carcinoma (HCC) diagnosis.
Methods
A total of 89 untreated LI-RADS-graded liver tumors (35 LR-3, 14 LR-4, and 40 LR-5) were identified based on the radiology MRI interpretation reports. Multiphase 3D T1-weighted gradient echo imaging was acquired at six time points: pre-contrast, four phases immediately post-contrast, and one hepatobiliary phase after intravenous injection of gadoxetate disodium. Image co-registration was performed across all phases on the center tumor slice to correct motion. A rectangular tumor box centered on the tumor area was drawn to extract subset tumor images for each imaging phase, which were used as the inputs to a convolutional neural network (CNN). The pre-trained AlexNet CNN model underwent transfer learning using liver MRI data for LI-RADS tumor grade classification. The output probability number closer to 1 or 0 indicated a higher possibility of being combined LR-4/LR-5 tumor or LR-3 tumor, respectively. Five-fold cross validation was used for training (60% dataset), validation (20%) and testing processes (20%).
Results
The DL CNN model for LI-RADS grading using inputs of multiphase liver MRI data acquired at three time points (pre-contrast, arterial, and washout phase) achieved a high accuracy of 0.90, sensitivity of 1.0, precision of 0.835, and AUC of 0.95 with reference to the expert human radiologist report. The CNN output of probability provided radiologists a confidence level of the model’s grading for each liver lesion.
Conclusions
An AlexNet CNN model for LI-RADS grading of liver lesions provided diagnostic performance comparable to radiologists and offered valuable clinical guidance for differentiating intermediate LR-3 liver lesions from more-likely malignant LR-4/LR-5 lesions in HCC diagnosis.
Keywords: Deep learning (DL), convolutional neural network (CNN), MRI, LI-RADS, hepatocellular carcinoma (HCC)
Introduction
Imaging plays a critical role in hepatocellular carcinoma (HCC) diagnosis. The American Association for the Study of Liver Diseases guidance statement (1) recommends diagnostic evaluation for HCC with either multiphase computed tomography (CT) or multiphase magnetic resonance imaging (MRI) and the use of the Liver Imaging Reporting and Data System (LI-RADS®) (2). LI-RADS provides standardized criteria for performing, interpreting, and reporting multiphase CT and MRI exams for HCC diagnosis. It is consistent with the guidelines of the National Comprehensive Cancer Network (3) and is easily convertible to the Organ Procurement and Transplantation Network classes (4) utilized by liver transplant centers in the United States. According to the LI-RADS diagnostic algorithm, a liver lesion in a high-risk patient (history of cirrhosis, chronic HBV infection, and current or prior HCC) is assigned a LI-RADS (LR) category reflecting the likelihood of being HCC: LR-1 to LR-5. Grades of LR-1 and LR-2 are definitely benign and probably benign, respectively. LR-3 indicates intermediate probability of HCC, LR-4 indicates high probability of HCC without certainty, and LR-5 indicates definite certainty of HCC.
Differentiation between LR-3 and combined LR-4/5 lesions is important because management options for LR-3 lesions are less invasive than those for LR-4 and LR-5. Many LR-3 lesions are benign hyperenhancing pseudolesions which can be followed for stability with imaging (5), whereas 80% of biopsied LR-4 lesions are HCC, and 68% of untreated LR-4 lesions become LR-5 lesions within two years. LR-4 lesions may be biopsied, while an LR-5 score indicates HCC diagnostic certainty and biopsy is usually not needed before treatment (1). Percutaneous liver lesion biopsies are not always performed in LR-4/LR-5 patients because of the risk of accidental tumor cell seeding along the biopsy needle tract and other complications. Thus, in clinical practice, the radiologist-determined LI-RADS score is often considered as the ground truth or gold standard for liver lesion characterization in high-risk patients, leading to HCC treatment intervention without biopsy confirmation. This approach emphasizes the importance of accurate imaging characterization.
Compared with CT, MRI diagnosis of HCC is more accurate and does not expose patients to harmful radiation (1,6-8). Multiphase contrast-enhanced (CE) MRI with intravenous gadoxetate disodium injection can achieve a 92% staging accuracy (6). An expert consensus statement and meta-analysis studies recommended state-of-the-art MRI as the most accurate imaging method for HCC diagnosis (9-11). Accurate LI-RADS liver lesion scoring performed on CE MRI requires comprehensive evaluation of liver lesion imaging features, notably: tumor size, arterial phase non-rim hyper-enhancement, washout, threshold growth, and presence of a capsule. Given the technical complexity in performing and interpreting multiparametric CE MRI for liver tumor LI-RADS scoring, this advanced imaging is usually conducted at expert academic centers rather than smaller community hospitals or outpatient imaging centers.
Deep learning (DL) methods empowered with convolutional neural network (CNN) algorithms have shown promise in medical imaging for disease detection (12-14), tissue segmentation (15,16), and lesion classification (17,18). These methods are able to excavate hidden or high-dimensional quantitative imaging features that currently elude identification by human experts such as radiologist physicians. DL-extracted imaging features are found to be more robust and generalizable than manually-extracted analytical features (19). DL methods have been used for differentiation of liver masses on dynamic CE CT images (20) and for classification between HCC and intrahepatic cholangiocarcinoma (21). However, to our knowledge, a DL method has not been reported for LI-RADS grading based on MRI data.
Various CNN architectures pre-trained using a large scale of annotated images of real-world objects have been exploited in the medical imaging field through transfer learning (22,23) to improve the prediction performance when dealing with relatively scarce data. Deep CNNs such as ResNet (24) and GoogleNet (25) demonstrated promising classification performance in the ImageNet Large-Scale Visual Recognition Challenge (26) and have been used in the detection and grading of brain tumors based on multiparametric MRI data (27,28). Nonetheless, a deep and complex CNN may be unsuitable for classification tasks using insufficient and scarce datasets due to the overfitting problem (29). In contrast, CNN models with a less depth and less-complex architecture have proven to be more robust in solving medical imaging problems (30). The AlexNet model contains a traditional architecture with 8 layers, which has less depth but improved generalization performance compared with other models (31). Therefore, in this study, we modified the commonly used AlexNet CNN for tumor grade classification.
The purpose of our study was to develop a DL method based on multiphase CE MRI data to distinguish LR-3 liver lesions (intermediate chance of malignancy) from a combined group of LR-4 and LR-5 lesions (high chance of malignancy), using the expert radiologist report as the reference standard. The hypothesis was that the DL-driven LI-RADS grading system can provide diagnostic performance comparable to experienced radiologists and provide valuable guidance to radiologists.
Methods
Dataset
This retrospective study was approved by Rush University Medical Center institutional review board and written informed consent was waived. By searching the electronic health record system at Rush University Medical Center, we identified 89 untreated liver tumors in 59 patients with suspected HCC who underwent baseline MRI with corresponding LI-RADS grades. The study radiologist reviewed and annotated a total of 35 LR-3, 14 LR-4, and 40 LR-5 classified liver tumors, as reported by reading radiologists. The LI-RADS grades confirmed by the study radiologist were used as the reference standard.
Multi-phase contrast enhanced MRI
A dynamic multiphase three-dimensional (3D) T1-weighted (T1W) gradient-echo sequence was acquired before and after contrast administration (0.1 mL/kg Eovist® Gadoxetate Disodium) with an injection rate of 2 mL/sec. Images were recorded at six time points (TPs) including a pre-contrast phase (TP1) and five post-contrast phases (TP2-TP6). We collected four immediate post-contrast acquisitions which included the early arterial, late arterial, portal venous, and transitional phases. The hepatic arterial phase was typically acquired around 30 seconds after an injection started. The next three post-contrast acquisitions were acquired every 30 seconds to evaluate lesion contrast enhancement and washout dynamics. The last time point of the immediate post-contrast acquisitions represented the washout phase of the lesion, which corresponded approximately to a late portal venous phase or transitional phase. The final acquisition was during the hepatobiliary phase at 20 minutes after injection.
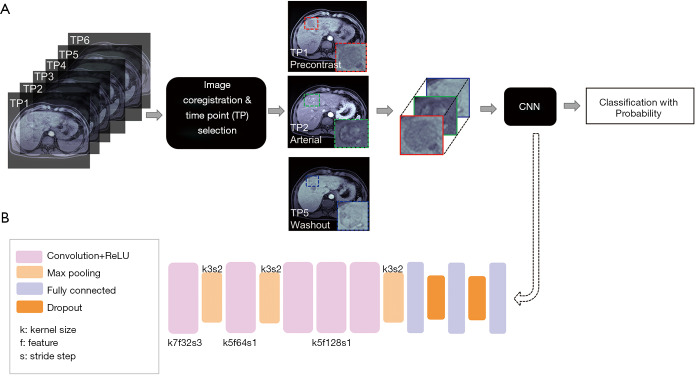
Image co-registration across acquisitions at different time points was performed through projective geometric transformation based on multiple paired control points that identified the same feature or landmark in the images using the ‘fitgeotrans’ function of the Matlab software (Image Processing Toolbox, Matlab, MathWorks, Inc., Natick, Massachusetts, United States). For each liver lesion/tumor, a center image slice with the best lesion delineation was selected by the study radiologist, on which one region-of-interest (ROI) was placed around the tumor area and the other ROI on adjacent liver tissue. The ROIs were then copied onto the same slice position of the multiphase images. Averaged signal intensities of the tumor and liver ROIs (Stumor and Sliver), and the signal ratio ∆S = (Stumor − Sliver)/Sliver were calculated. Of the multiple MRI acquisitions obtained at different TPs, the two clinically-important imaging phases demonstrating maximum liver lesion enhancement (TPme) and maximum lesion washout (TPmw) were identified as those with the largest ∆S and smallest ∆S values, respectively. Lastly, a rectangular tumor bounding box centered on the liver lesion (and including 1–2 cm of surrounding liver tissue) was drawn to extract a subset tumor images for each image phase. The workflow of the DL LI-RADS grading system is illustrated in Figure 1A.
Figure 1.
The design of the DL LI-RADS grading system using contrast enhanced multiphase liver MRI. (A) The workflow of the DL-driven LI-RADS grading system; (B) AlexNet model architecture used for CNN-based image feature extraction and classification. DL, deep learning; LI-RADS, Liver Imaging Reporting and Data System; CNN, convolutional neural network.
DL CNN model
Image augmentation
Each subset tumor image was rotated by degrees of {−60, −30, 30, 60}, and flipped both horizontally and vertically. The original subset tumor images and rotated images were zero-padded to the same matrix size of 100×100. As a result, a total of 7 images for each subset tumor image were used as inputs to the CNN. This step of image augmentation increased the sample size of the training dataset to avoid the potential overfitting problem.
Model architecture
AlexNet is a well-known CNN architecture pre-trained with the quality-controlled and annotated real-world images from ImageNet Large Scale Visual Recognition Challenge database (26). It consists of 8 trainable layers: 5 convolutional layers plus 3 fully connected layers. The activation function Rectified Linear Unit (ReLU) is applied in each layer to accelerate the convergence of gradient descent. The model also consists of non-trainable layers including pooling layers to reduce the dimension of the parameters and dropout layers to alleviate the overfitting problem. The last fully connected layer outputs a probability number ranging from 0 to 1 for each liver lesion, with a number closer to 1 indicating a higher probability that the lesion represents an LR-4/LR-5 tumor, whereas a number closer to 0 indicates a greater probability of being an LR-3 tumor. Thus, a liver lesion with a probability number closer to 1 was more likely malignant and a lesion with number closer to 0 was less likely malignant. The architecture of the CNN model is illustrated in Figure 1B.
Training, validation, and testing dataset
Five-fold cross validation was used for training, validation, and testing processes. Each fold contained 60% of the total number of liver tumors as a training dataset, 20% as a validation dataset, and the remaining 20% as a testing dataset. The batch size was set to be 8 images in each iteration of the training process. The number of epochs was originally set to be 200 and then reduced to 53 with an early stop, where the training process stopped if the performance of a validation dataset started to degrade. The training and testing processes were conducted using the Python programming language (version 3.6) on a single GPU (Nvidia GeForce 2080Ti) workstation with training time per fold averaging 18 minutes.
Transfer learning
Transfer learning was used to fine-tune the weights of the AlexNet model parameters with liver MRI images as the inputs. The initial weights of AlexNet pre-trained with the ImageNet dataset were kept fixed in the five convolutional layers, whereas the weights of the three fully connected layers were fine-tuned using the liver MRI training dataset. Next, all eight of the trainable (convolutional and fully connected) layers were fine-tuned together using the liver MRI validation dataset. AlexNet model performance was evaluated using the liver MRI testing dataset. The model’s performance both with and without transfer learning was also evaluated.
Cyclical learning rate
To increase the speed of convergence with fewer epochs and to avoid local minima, the optimal learning rate was determined by the approach of a cyclical learning rate algorithm (32). First, a random learning rate was assigned for one epoch of training and the loss function was calculated on a validation dataset. The loss values were plotted as a function of the corresponding learning rates that were randomly assigned for each epoch of training. The minimal and maximal boundaries of the learning rate were determined from the loss value curve, where the minimal boundary represented the point where the loss started to decrease, and the maximal boundary represented the point where the loss started to increase (Figure 2A). After the learning rate boundaries were chosen, the optimal learning rate was determined dynamically during the full training process along with multiple epochs, where the learning rate oscillated cyclically within the minimal and maximal boundaries (Figure 2B). The rising course of the oscillation helped the training loss jump out of the local minimum, while the declining course helped alleviate the overfitting problem.
Figure 2.
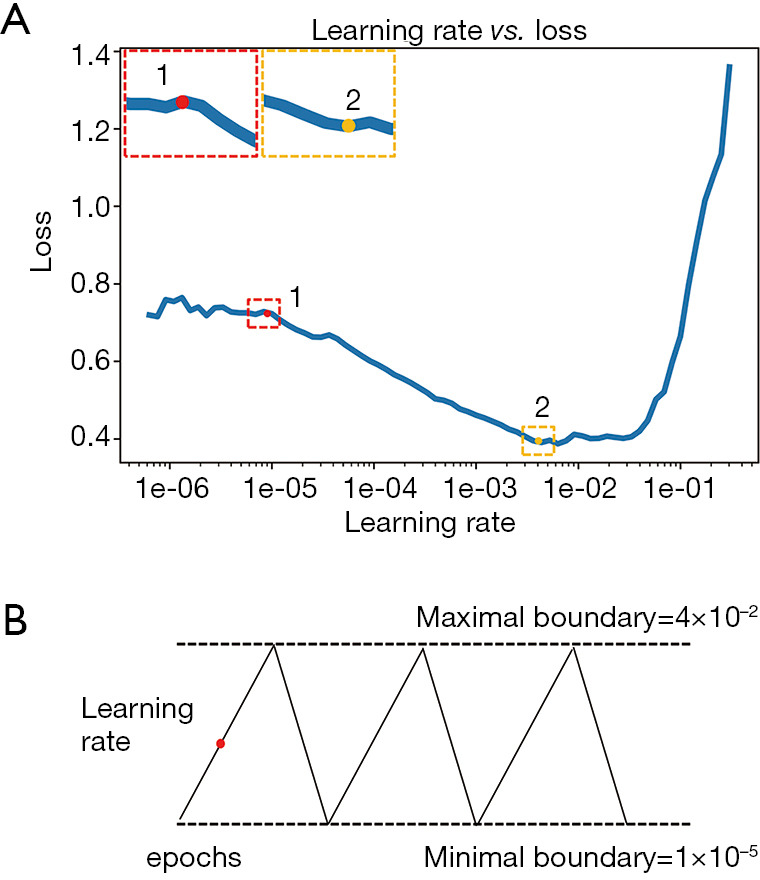
The cyclical learning rate algorithm for determining the optimal learning rate of the CNN model. (A) Loss value curve as a function of randomly selected learning rate. The red dot is the minimal boundary where the loss begins to drop. The yellow dot is the maximal boundary where the loss begins to increase. (B) The process of determining the optimal learning rate during the full training process. The learning rate oscillates cyclically between the minimal and maximal boundaries along a triangular waveform.
Performance evaluation
The optimal acquisition scheme was determined by separately using four different combinations of image phases as inputs to the CNN and comparing the model performances. Combination 1 (C1) dataset: all phases acquired at six time points (TPs); Combination 2 (C2) dataset: pre-contrast (TP1), arterial phase (TP2), and washout phase (TP5); Combination 3 (C3) dataset: C2 plus the hepatobiliary phase (TP6); and Combination 4 (C4) dataset: pre-contrast (TP1), maximum enhancement phase (TPme), and maximum washout phase (TPmw). For each combination dataset, images acquired at the chosen TPs were concatenated into multiple channels as the inputs to the CNN classifier.
The confidence level of the classification outcomes generated by the CNN model was defined as the probability of being LR-3 or combined LR-4/LR-5 for any given tumor. The probability threshold used for classification between LR-3 and combined LR-4/LR-5 tumors was set to be 0.5. The tumor with a probability score <0.5 was classified as LR-3, whereas a probability score >0.5 was classified as LR-4/LR-5. Probability scores between 0.4 and 0.6 indicated less confidence in the classification outcomes and suggested further evaluation by study radiologists based on other clinical and imaging information.
Statistical analysis
The mean accuracy, precision, sensitivity, and F1 score for differentiation between LR-3 and combined LR-4/LR-5 liver lesions were calculated for each of the five cross-validation testing datasets. The receiver operating characteristic (ROC) analysis and the area under the receiver operating characteristic curve (AUC) were calculated to evaluate the performance of the CNN classifier. McNemar’s test was performed to compare the accuracy and sensitivity of the C1-C4 models, and between C2 models with and without transfer learning. The P values were adjusted for multiple comparisons using false discovery rate. A P value <0.05 was considered statistically significant. In addition, a confusion matrix was generated for each cross-validation process, which presented the number ratio of lesions that were correctly or incorrectly classified by CNN in either the LR-3 or LR-4/LR-5 category.
Results
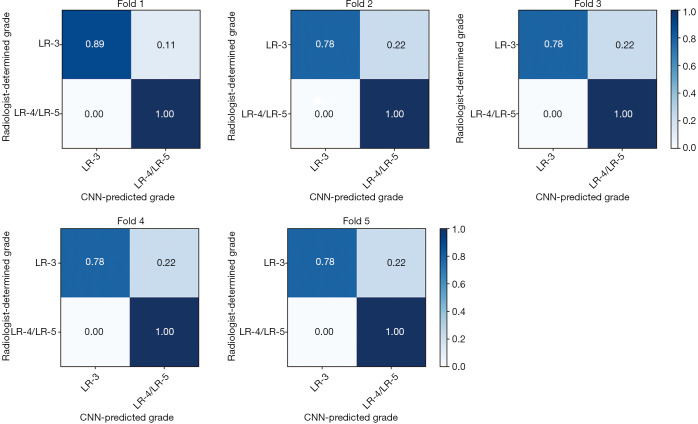
The classification performance using datasets acquired from different combinations of MRI imaging phases are shown in Table 1. Among the four combinations (C1–C4), the C2 scheme (with the three time points of pre-contrast, arterial phase, and washout phase) provided the best performance with the highest accuracy (0.900), precision (0.835), sensitivity (1.0), F1 score (0.909), and AUC (0.95). McNemar’s test showed that the accuracy of C2 was significantly higher than C1, C3, and C4 with all P values =0.02. The sensitivity of C2 was significantly higher than C4 with P value of 0.01. However, there was no significant difference in sensitivity between C2 and C1 or C3 with P value of 0.16 and 0.21, respectively. Neither the use of all six time points (C1) nor addition of hepatobiliary phase (C3) to the C2 dataset improved the performance of the CNN classifier. The transfer learning approach greatly improved the classification outcomes when comparing the performances of the C2 dataset with and without transfer learning (P value =0.02 for both accuracy and sensitivity comparisons). The sensitivity of CNN using the C2 dataset with transfer learning reached 1.0, indicating that all liver lesions in the combined LR-4/LR-5 group were correctly classified by the CNN as compared to a human radiologist gold standard. Similarly, the confusion matrix, which provided the number ratio of correctly-classified and misclassified CNN-predicted LI-RADS grades with reference to radiologist-determined LI-RADS grades, demonstrated that none of the LR-4/LR-5 lesions were classified as LR-3 in all five folds of validation (Figure 3). The AUC of ROC curve using the C2 dataset was 0.95±0.2 (mean ± standard deviation) across all five folds of validation (Figure 4).
Table 1. The classification performance of the CNN model using datasets acquired at different combinations of image phases.
| Dataset | Accuracy | Precision | Sensitivity | F1 score | AUC |
|---|---|---|---|---|---|
| C1 (all 6 phases) | 0.833 | 0.800 | 0.889 | 0.842 | 0.92 |
| C2 (pre, arterial, washout) | 0.900 | 0.835 | 1.0 | 0.909 | 0.95 |
| C3 (C2 + hepatobiliary phase) | 0.833 | 0.803 | 0.889 | 0.843 | 0.92 |
| C4 (pre, me, mw) | 0.789 | 0.814 | 0.756 | 0.780 | 0.91 |
| C2 without transfer learning | 0.767 | 0.777 | 0.778 | 0.767 | 0.90 |
pre, pre-contrast; arterial, arterial phase; washout, washout phase; me, maximum enhancement phase; mw, maximum washout phase; CNN, convolutional neural network; AUC, area under the receiver operating characteristic curve.
Figure 3.
The confusion matrix describes CNN classification model performance using C2 dataset in each fold of cross-validation. The number ratio of correctly classified and misclassified CNN-predicted grades is listed with reference to each radiologist-determined LI-RADS grade. LI-RADS, Liver Imaging Reporting and Data System; CNN, convolutional neural network.
Figure 4.
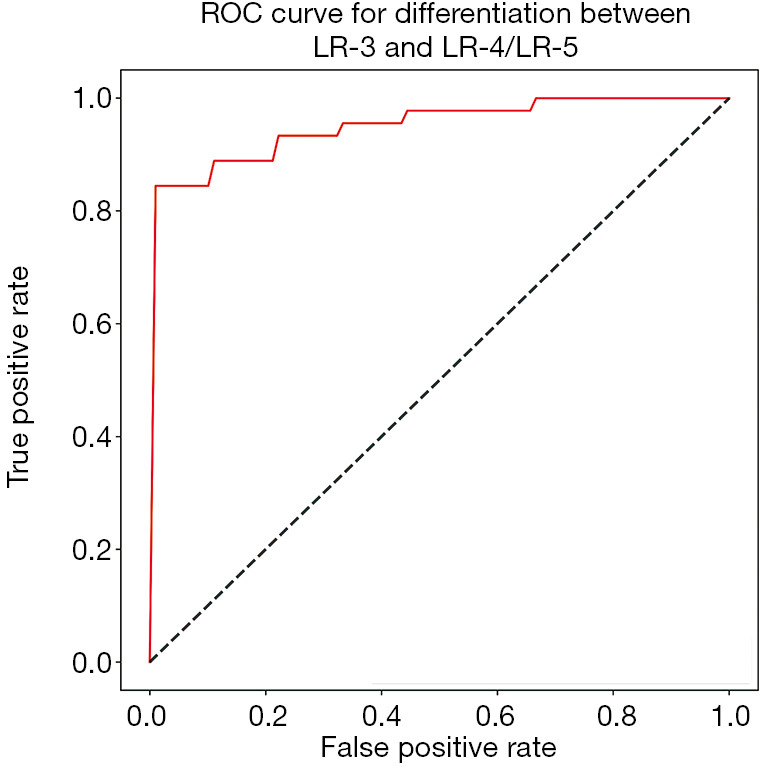
ROC curve of CNN classification performance using the C2 dataset. The AUC was 0.95±0.2 (mean ± standard deviation) across five-fold validations. CNN, convolutional neural network; AUC, area under the receiver operating characteristic curve.
The certainty score (i.e., probability) of the classification result for any given tumor in each cross-validation testing dataset was calculated (Figure 5). A probability higher than 0.6 or lower than 0.4 indicated more certainty in the CNN classification, whereas a probability falling between 0.4 and 0.6 indicated a less reliable classification. Overall, none of the LR-4/LR-5 lesions were misclassified as LR-3 lesions, in concordance with the high certainty scores and suggested the great sensitivity of this model. A total of four LR-3 lesions were misclassified as LR-4/LR-5, two of which showed a probability number (0.52 and 0.56) falling in the uncertain area, prompting further evaluation by the radiologist. The other two misclassified LR-3 lesions had a relatively higher probability number (0.86 and 0.66).
Figure 5.
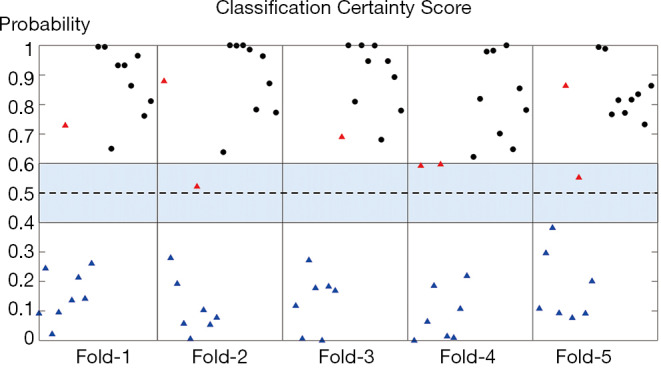
The probability of classification result for any given tumor in each cross-validation testing dataset. Black dots represent correctly-classified LR-4/LR-5 lesions. Blue triangles represent correctly-classified LR-3 lesions. Red triangles represent misclassified LR-3 lesions. Probability numbers shown in the vertical axis closer to 1 or 0 indicate higher possibilities of being LR-4/LR-5 or LR-3, respectively. The shaded area between 0.4 and 0.6 represents the uncertain area where the classification result is considered as not reliable if the probability number falls in this range.
Discussion
This study is the first to implement a CNN DL model based on multiphase CE MRI images to facilitate LI-RADS HCC grading of clinically-relevant lesions (LR-3 and LR-4/L4-5), which achieved a high accuracy of 0.90, sensitivity of 1.0, and AUC of 0.95 with reference to the expert radiologist report. Images acquired at three time points (pre-contrast, arterial phase, and washout phase) as CNN inputs provided the best performance with significantly higher accuracy, whereas including the other three time points did not improve performance. This result suggests that fewer dynamic acquisitions during contrast injections are needed than typically performed in current clinical practice. Reducing the number of CE MRI acquisitions needed can beneficially shorten MRI exam time, leading to a decrease in patient motion artifacts associated with uncomfortable, mandatory patient breath-holding during imaging. The hepatobiliary phase acquired at 20 minutes after the contrast injection did not contribute to the LI-RADS classification performance of the CNN model, indicating that acquisition of this delayed phase may not be necessary for the purpose of LI-RADS grading.
Tumor size is an important factor in the diagnosis of HCC. In this study, the average tumor diameter was 14.7±4.7 mm for LR-3 lesions and 22.6±11.8 mm for LR-4/5 lesions. Although tumor size was not used as a manually extracted feature for the CNN model, DL networks are capable of extracting discriminant features such as size and shape from the inner layers of the network by learning the abstract and useful relationships between the input data and features (33). This CNN model demonstrated a sensitivity of 1.0, indicating that not a single high-grade (LR-4 or LR-5) liver lesion was misdiagnosed as the lower LR-3 grade, which can help minimize delayed diagnosis and treatment of the higher-grade HCC lesions. However, misclassification occurred in four radiologist-determined low-grade lesions in this study. The probability score proposed in this study provided radiologists and clinicians a confidence level for each lesion classification that facilitates clinical decision making and flags uncertain cases requiring further human radiologist review. The probability of two of the misclassified lesions (0.53 and 0.59) suggested uncertainty (i.e., low confidence level) in the CNN classification and therefore required re-evaluation by radiologist. These two lesions were originally graded by the reading radiologists as LR-3 because of their small size and hyper-enhancement on arterial phase while lacking tumor washout compared to liver tissue. CNN classification of a lesion as a higher grade than indicated in the radiology report prompted additional radiologist review to reduce misdiagnosis. The other two misclassified LR-3 tumors had a relatively high probability number (0.86 and 0.66). Interestingly, the tumor with a very high probability (0.86) was re-classified as a LR-5 tumor later in a 6-month follow-up MRI exam. The study radiologist reviewed this case carefully based on the LI-RADS guideline and suspected that suboptimal timing of arterial phase acquisition and severe motion artifact in the venous phase images may have misled the interpreting radiologist’s judgement of this lesion as LR-3. Or, the CNN identified some as-yet-unknown feature of the lesion resulting in a higher-grade LI-RADS score. It is noteworthy that the CNN model successfully predicted the ‘true’ grade of LR-5 for this lesion with a high confidence level under the limited conditions of suboptimal image quality. The last misclassified LR-3 lesion by the CNN with the probability of 0.66 was confirmed by the study radiologist as an LR-3 lesion because the lesion was small, arterial-phase hyper-enhancing, and demonstrated no washout in later phases. The over-estimation of higher-grade tumor by the CNN model was hypothesized to be secondary to the presence of heterogenous, nodular, cirrhotic liver parenchyma in this patient, which also presents a diagnostic challenge for human radiologists. The radiologist should be aware of this confounding factor and re-evaluate the CNN outcomes.
One general problem with DL networks is the “black box” problem. Unlike analytical imaging features used in traditional machine learning, DL-extracted imaging features are difficult for humans to interpret, and precisely how predicted outcomes and conclusions are drawn by DL networks can evade our understanding. Data inputs and probability outputs are processed inside the “black box” of the DL networks without complete human understanding of which features are used for CNN predicted outcomes, making it difficult to identify failures of the DL tool. For future study, a graphical Class Activation Map (CAM) (34) will be generated and overlaid upon actual MRI images to highlight potential ‘hotspot’ liver lesions for the radiologist to review at the time of MRI interpretation. This additional context can improve physician performance, minimize human “missed diagnosis” of liver lesions, and improve patient care. Further, the CAM can yield insights into which features the “black box” CNN DL network model uses to creat its prediction outcomes. This CAM ‘evidence map’ may also provide information for tvalidating the reliability of such CNN DL network methods.
Another promising future study will be to add non-imaging data to the CNN model inputs to improve model performance, such as relevant patient clinical and laboratory information. For example, blood testing for known liver tumor marker serum alpha fetoprotein (AFP) may improve model performance, as higher AFP levels can predict the presence of HCC. Liver function tests (LFTs) are a suite of blood tests which may be helpful CNN inputs, as LFTs often help diagnose hepatocellular diseases. The Child-Pugh score based on clinical and laboratory data assesses the prognosis of chronic liver diseases or cirrhosis and helps guide treatment planning. Evidence of a patient’s liver morphology obtained from imaging, including liver nodularity, heterogeneity, and size, is also clinically important and may help improve the CNN model performance. Patient clinical information, such as a history of known liver cirrhosis and other risk factors of hepatitis virus infection and non-alcoholic steatohepatitis (NASH), can be utilized as inputs to the CNN model to further improve performance.
Compared with AlexNet, both ResNet and GoogleNet are larger networks with special and deep architectures of more than one hundred layers. With a limited number of training datasets, large networks are usually prone to the problem of overfitting (35). AlexNet performs well in image classification with fewer layers and alleviates the overfitting problem by adding dropout layers (36) and data augmentation (37). In addition, transfer learning played an important role in a small size dataset by refining the AlexNet model through learning discriminant features from the MRI dataset in fully connected layers, which helped reduce overfitting and further improve the model performance as shown in Table 1.
Due to the greater technical complexity of multiparametric liver MRI acquisition and interpretation, imaging studies for HCC diagnosis are usually conducted at academic centers with liver MRI expertise and are not easily confirmed in community hospitals with fewer MRI technical, radiologic, and clinical expertise. MRI datasets in this study were acquired from different MRI scanners and platforms with slightly different protocols; as a result, this DL model is robust and generalizable for larger, more-varied imaging datasets which should help the validation of the model through multi-institutional studies. The DL methods should be validated with rigorous, continuous clinical studies using independent and multi-institutional patient cohorts prior to implementation in clinical practice. The authors anticipate that this DL-driven, automated LI-RADS grading system can provide valuable guidance to radiologists, and reduce intra- and inter-reader variability with the ultimate goal of improving cancer diagnosis, treatment, and patient care.
There are several limitations of this study. Two-dimensional (2D) image co-registration for pre-selected tumor image slices acquired at multiple image phases was performed by manually identifying the landmarks on each image slice. After co-registration, a rectangular tumor box was drawn manually. Researchers are currently working on developing volumetric tumor segmentation and ROI extraction methods using specific CNN models to automate and accelerate these processes. Lastly, larger and more diverse imaging datasets are needed to enable validation and deployment of these tools in multi-institutional studies.
In conclusion, the DL-driven LI-RADS grading system developed in this study provided diagnostic performance comparable to experienced radiologists and provided valuable guidance to radiologists by differentiating LR-3 liver tumors from a group of higher-grade, combined LR-4/LR-5 liver lesions. Accurate LI-RADS scoring is essential for accurate diagnosis and treatment of patients with HCC.
Supplementary
The article’s supplementary files as
Acknowledgments
This research is partially supported by the Swim Across America Grant from Rush Translational Sciences Consortium. The authors would also like to thank Dr. Sharon E. Byrd for her administrative support in establishing the medical imaging artificial intelligence research program.
Funding: None.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This retrospective study was approved by Rush University Medical Center institutional review board and written informed consent was waived.
Footnotes
Provenance and Peer Review: This article was commissioned by the Guest Editors (Haotian Lin and Limin Yu) for the series “Medical Artificial Intelligent Research” published in Annals of Translational Medicine. The article was sent for external peer review organized by the Guest Editors and the editorial office.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm.2019.12.151). The series “Medical Artificial Intelligent Research” was commissioned by the editorial office without any funding or sponsorship. JD reports grants from Swim Across American, during the conduct of the study. The other authors have no other conflicts of interest to declare.
References
- 1.Marrero JA, Kulik LM, Sirlin CB, et al. Diagnosis, Staging, and Management of Hepatocellular Carcinoma: 2018 Practice Guidance by the American Association for the Study of Liver Diseases. Hepatology 2018;68:723-50. 10.1002/hep.29913 [DOI] [PubMed] [Google Scholar]
- 2.CT/MRI LI-RADS v2018 [Internet]. [cited 2019 Sep 6]. Available online: https://www.acr.org/Clinical-Resources/Reporting-and-Data-Systems/LI-RADS/CT-MRI-LI-RADS-v2018
- 3.NCCN - Evidence-Based Cancer Guidelines, Oncology Drug Compendium, Oncology Continuing Medical Education [Internet]. [cited 2019 Sep 6]. Available online: https://www.nccn.org/
- 4.optn_committee_initiatives_2014-15.pdf [Internet]. [cited 2019 Sep 6]. Available online: https://optn.transplant.hrsa.gov/media/1524/optn_committee_initiatives_2014-15.pdf
- 5.Choi JY, Cho HC, Sun M, et al. Indeterminate observations (liver imaging reporting and data system category 3) on MRI in the cirrhotic liver: fate and clinical implications. AJR Am J Roentgenol 2013;201:993-1001. 10.2214/AJR.12.10007 [DOI] [PubMed] [Google Scholar]
- 6.Choi SH, Byun JH, Kim SY, et al. Liver Imaging Reporting and Data System v2014 With Gadoxetate Disodium-Enhanced Magnetic Resonance Imaging: Validation of LI-RADS Category 4 and 5 Criteria. Invest Radiol 2016;51:483. 10.1097/RLI.0000000000000258 [DOI] [PubMed] [Google Scholar]
- 7.Abd Alkhalik Basha M, Abd El Aziz El Sammak D, El Sammak AA. Diagnostic efficacy of the Liver Imaging-Reporting and Data System (LI-RADS) with CT imaging in categorising small nodules (10-20 mm) detected in the cirrhotic liver at screening ultrasound. Clin Radiol 2017;72:901.e1-901.e11. 10.1016/j.crad.2017.05.019 [DOI] [PubMed] [Google Scholar]
- 8.Kim YY, An C, Kim S, et al. Diagnostic accuracy of prospective application of the Liver Imaging Reporting and Data System (LI-RADS) in gadoxetate-enhanced MRI. Eur Radiol 2018;28:2038-46. 10.1007/s00330-017-5188-y [DOI] [PubMed] [Google Scholar]
- 9.Vauthey JN, Dixon E, Abdalla EK, et al. Pretreatment assessment of hepatocellular carcinoma: expert consensus statement. HPB 2010;12:289-99. 10.1111/j.1477-2574.2010.00181.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee YJ, Lee JM, Lee JS, et al. Hepatocellular Carcinoma: Diagnostic Performance of Multidetector CT and MR Imaging—A Systematic Review and Meta-Analysis. Radiology 2015;275:97-109. 10.1148/radiol.14140690 [DOI] [PubMed] [Google Scholar]
- 11.Consensus report from the 6th International forum for liver MRI using gadoxetic acid - Sirlin - 2014 - Journal of Magnetic Resonance Imaging - Wiley Online Library [Internet]. [cited 2019 Aug 30]. Available online: https://onlinelibrary.wiley.com/doi/full/10.1002/jmri.24419 [DOI] [PubMed]
- 12.Shin HC, Roth HR, Gao M, et al. Deep Convolutional Neural Networks for Computer-Aided Detection: CNN Architectures, Dataset Characteristics and Transfer Learning. IEEE Trans Med Imaging 2016;35:1285-98. 10.1109/TMI.2016.2528162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rajpurkar P, Irvin J, Zhu K, et al. CheXNet: Radiologist-Level Pneumonia Detection on Chest X-Rays with Deep Learning 2017 Nov 14 [cited 2019 Aug 30]; Available online: https://arxiv.org/abs/1711.05225v3
- 14.Li Q, Cai W, Wang X, et al. Medical image classification with convolutional neural network. In: 2014 13th International Conference on Control Automation Robotics Vision (ICARCV) 2014:844-8. [Google Scholar]
- 15.Havaei M, Davy A, Warde-Farley D, et al. Brain tumor segmentation with Deep Neural Networks. Med Image Anal 2017;35:18-31. 10.1016/j.media.2016.05.004 [DOI] [PubMed] [Google Scholar]
- 16.Akkus Z, Galimzianova A, Hoogi A, et al. Deep Learning for Brain MRI Segmentation: State of the Art and Future Directions. J Digit Imaging 2017;30:449-59. 10.1007/s10278-017-9983-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheng JZ, Ni D, Chou Y-H, et al. Computer-Aided Diagnosis with Deep Learning Architecture: Applications to Breast Lesions in US Images and Pulmonary Nodules in CT Scans. Sci Rep 2016;6:24454. 10.1038/srep24454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lopez AR, Giro-i-Nieto X, Burdick J, et al. Skin lesion classification from dermoscopic images using deep learning techniques. In: 2017 13th IASTED International Conference on Biomedical Engineering (BioMed), 2017:49-54. [Google Scholar]
- 19.Poplin R, Varadarajan AV, Blumer K, et al. Prediction of cardiovascular risk factors from retinal fundus photographs via deep learning. Nat Biomed Eng 2018;2:158-64. 10.1038/s41551-018-0195-0 [DOI] [PubMed] [Google Scholar]
- 20.Yasaka K, Akai H, Abe O, et al. Deep Learning with Convolutional Neural Network for Differentiation of Liver Masses at Dynamic Contrast-enhanced CT: A Preliminary Study. Radiology 2018;286:887-96. 10.1148/radiol.2017170706 [DOI] [PubMed] [Google Scholar]
- 21.Midya A, Chakraborty J, Pak LM, et al. Deep convolutional neural network for the classification of hepatocellular carcinoma and intrahepatic cholangiocarcinoma. In: Medical Imaging 2018: Computer-Aided Diagnosis [Internet]. International Society for Optics and Photonics; 2018 [cited 2019 Aug 30]. p. 1057528. Available online: https://www.spiedigitallibrary.org/conference-proceedings-of-spie/10575/1057528/Deep-convolutional-neural-network-for-the-classification-of-hepatocellular-carcinoma/10.1117/12.2293683.short
- 22.Kermany DS, Goldbaum M, Cai W, et al. Identifying Medical Diagnoses and Treatable Diseases by Image-Based Deep Learning. Cell 2018;172:1122-1131.e9. 10.1016/j.cell.2018.02.010 [DOI] [PubMed] [Google Scholar]
- 23.Huynh BQ, Li H, Giger ML. Digital mammographic tumor classification using transfer learning from deep convolutional neural networks. J Med Imaging (Bellingham) 2016;3:034501. 10.1117/1.JMI.3.3.034501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He K, Zhang X, Ren S, et al. Deep Residual Learning for Image Recognition. ArXiv151203385 Cs [Internet] 2015 Dec 10 [cited 2019 Aug 30]; Available online: http://arxiv.org/abs/1512.03385
- 25.Szegedy C, Liu W, Jia Y, et al. Going Deeper With Convolutions. In 2015 [cited 2019 Aug 30]. p. 1-9. Available online: https://www.cv-foundation.org/openaccess/content_cvpr_2015/html/Szegedy_Going_Deeper_With_2015_CVPR_paper.html
- 26.Krizhevsky A, Sutskever I, Hinton GE. ImageNet Classification with Deep Convolutional Neural Networks. In: Pereira F, Burges CJC, Bottou L, et al. editors. Advances in Neural Information Processing Systems 25 [Internet]. Curran Associates, Inc.; 2012 [cited 2019 Aug 30]. p. 1097-1105. Available online: http://papers.nips.cc/paper/4824-imagenet-classification-with-deep-convolutional-neural-networks.pdf [Google Scholar]
- 27.Deep learning enables automatic detection and segmentation of brain metastases on multisequence MRI - Grøvik - Journal of Magnetic Resonance Imaging - Wiley Online Library [Internet]. [cited 2019 Aug 30]. Available online: https://onlinelibrary.wiley.com/doi/full/10.1002/jmri.26766 [DOI] [PMC free article] [PubMed]
- 28.Banerjee S, Mitra S, Masulli F, et al. Deep Radiomics for Brain Tumor Detection and Classification from Multi-Sequence MRI. ArXiv190309240 Cs Eess [Internet] 2019 Mar 21 [cited 2019 Aug 30]; Available online: http://arxiv.org/abs/1903.09240
- 29.Shen D, Wu G, Suk HI. Deep Learning in Medical Image Analysis. Annu Rev Biomed Eng 2017;19:221-48. 10.1146/annurev-bioeng-071516-044442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bertrand H, Perrot M, Ardon R, et al. Classification of MRI data using deep learning and Gaussian process-based model selection. In: 2017 IEEE 14th International Symposium on Biomedical Imaging (ISBI 2017) 2017:745-8. [Google Scholar]
- 31.Zhang C, Bengio S, Hardt M, et al. Understanding deep learning requires rethinking generalization. ArXiv161103530 Cs [Internet] 2017 Feb 26 [cited 2019 Nov 3]; Available online: http://arxiv.org/abs/1611.03530
- 32.Smith LN. Cyclical Learning Rates for Training Neural Networks. ArXiv150601186 Cs [Internet] 2015 Jun 3 [cited 2019 Mar 5]; Available online: http://arxiv.org/abs/1506.01186
- 33.Zhou B, Khosla A, Lapedriza A, et al. Object Detectors Emerge in Deep Scene CNNs. ArXiv14126856 Cs [Internet] 2015 Apr 15 [cited 2019 Nov 4]; Available online: http://arxiv.org/abs/1412.6856
- 34.Zhou B, Khosla A, Lapedriza A, et al. Learning Deep Features for Discriminative Localization. In: 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) [Internet]. Las Vegas, NV, USA: IEEE; 2016:921-9. Available online: http://ieeexplore.ieee.org/document/7780688/ [Google Scholar]
- 35.Caruana R, Lawrence S, Giles CL. Overfitting in Neural Nets: Backpropagation, Conjugate Gradient, and Early Stopping. In: Leen TK, Dietterich TG, Tresp V, editors. Advances in Neural Information Processing Systems 13 [Internet]. MIT Press, 2001:402-8. Available online: http://papers.nips.cc/paper/1895-overfitting-in-neural-nets-backpropagation-conjugate-gradient-and-early-stopping.pdf [Google Scholar]
- 36.Srivastava N, Hinton G, Krizhevsky A, et al. Dropout: A Simple Way to Prevent Neural Networks from Overfitting. JMLR 2014;15:1929-58. [Google Scholar]
- 37.Perez L, Wang J. The Effectiveness of Data Augmentation in Image Classification using Deep Learning. ArXiv171204621 Cs [Internet] 2017 Dec 13 [cited 2019 Oct 29]; Available online: http://arxiv.org/abs/1712.04621
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as