Abstract
Background
Glutathione peroxidases (GPXs) are an enzyme family with peroxidase activity. Abnormal GPX expression is associated with carcinogenesis. However, the potential role of the GPX gene family in acute myeloid leukemia (AML) remains to be comprehensively examined.
Methods
We analyzed GPX mRNA expression levels and determined the correlation between gene expression and the prognostic value via multiple universally acknowledged databases including the Oncomine, Gene Expression Profiling Interactive Analysis (GEPIA), PROGgeneV2, UALCAN, Cancer Cell Line Encyclopedia (CCLE), and The European Bioinformatics Institute (EMBL-EBI) databases. The functional network of differentially expressed GPXs was investigated via the NetworkAnalyst platform. Correlated genes as well as kinase, microRNA (miRNA), and transcription factor (TF) targets were identified using LinkedOmics.
Results
We observed that the transcriptional expression levels of GPX-1, -2, -4, -7, and -8 had significant difference between AML patients samples and normal samples, and that AML patients with high expression of GPX-1, -3, -4, and -7 were associated with poorer prognosis of overall survival (OS). Functional enrichment analysis showed that the differentially expressed GPXs were mainly enriched in response to oxidative stress, regulation of immune response, and inflammatory response, along with glutathione metabolism and ferroptosis. Overexpression of correlated genes, PSMB10, VPS13D, NDUFS8, ATP5D, POLR2E, and HADH were linked to adverse OS in AML. Regulatory network analysis indicated that differentially expressed GPXs regulated cell proliferation, cancer progression, apoptosis, and cell cycle signaling via pathways involving cancer-related kinases (such as DAPK1 and SRC), miRNAs (such as miR-202 and miR-181), and TFs (such as SRF and E2F1).
Conclusions
Our findings offer novel insights into the differential expression and prognostic potential of the GPX family in AML, and lay a foundation for subsequent research of GPX’s role in the carcinogenesis and regulatory network of AML.
Keywords: Acute myeloid leukemia (AML), bioinformatics analysis, functional network, glutathione peroxidases (GPXs), prognosis
Introduction
Acute myeloid leukemia (AML) is a heterogeneous disease of the blood system which is characterized by the clonal expansion and differentiation arrest of myeloid progenitor cells. The incidence of AML increases with age and mortality exceeds 90% when diagnosed after age 65 (1). In 2020, an estimated 19,940 new patients will be diagnosed with AML, and 11,180 AML deaths are projected to occur in the United States (2). AML is a highly heterogeneous malignancy, and prognosis of this disease varies widely. To our knowledge, genetic, epigenetic, and proteomic alterations are often related to the molecular mechanism underlying disease development and clinical prognosis of AML (3). Therefore, there is an urgent need to identify potential biomarkers that can be contribute to significantly improve the diagnosis, treatment, and prognosis of AML patients, and reduce the health cost and monitoring of this disease.
Glutathione peroxidases (GPXs) belong to a peroxidase enzyme family whose main biological functions are to protect organisms from oxidative damage by reducing lipid hydroperoxides and free hydrogen peroxide (4). Currently, 8 GPX sub-members have been identified in mammals (5), and studies have reported that they play significant roles in restoring reactive oxygen species (ROS)-induced damage, and protecting DNA, protein, and lipids from oxidative damage (6) and carcinogenesis (7). Among the 8 GPXs, GPX-1, GPX-3, GPX-4, and GPX-7 have important roles in different types of cancers. Reports have shown that GPX-1 is crucially involved in the development and progression of colorectal cancer (8), thyroid cancer (9), and kidney cancer (10); meanwhile GPX-2 has been found to regulate invasion, metastasis, apoptosis, and prognosis in cancers including pancreatic cancer (11), bladder cancer (12), cervical cancer (13), and nasopharyngeal carcinoma (14), suggesting that GPX2 may be an oncogene. Furthermore, GPX-3 methylation has been linked to multiple cancers, has potential value in predicting prognosis (15-17), and is also considered to be a tumor suppressor (18-21). GPX-4 regulates ferroptotic cancer cell death, and is related to the therapy resistance of cancer cells (22-24). Moreover, GPX7 functions as an epigenetic silenced tumor suppressor in gastric cancer and esophageal adenocarcinoma (25,26). A large number of publications have demonstrated the significant roles that GPX family members (expect for GPX-5, GPX-6, and GPX-8) play in carcinogenesis. Overall, identifying the role of GPXs in AML can contribute to the development and advancement of improved cancer treatment options.
Oxidative stress exists in primary human AML cells whose intrinsic property is aberrant glutathione metabolism (27). The imbalance of ROS level contributes to abnormal cell growth and differentiation, thus resulting in carcinogenesis. GPXs function as antioxidant enzymes and participate in the homeostasis balance that is associated with cancer cell renewal and differentiation. GPX-3 was reported to be associated with the frequency of leukemia stem cells in induced leukemia (28), while the promoter methylation of GPX-3 has been shown to predict adverse clinical outcome in non-M3 AML patients (29). FMS-like tyrosine kinase 3 (FLT3) mutations were the most important and common genetics aberration of AML patients. There are approximately 90% of AML patients express FLT3 (30), and the rates of FLT3 mutations in AML patients are approximately 30% (31). FLT3 was recognized as an adverse prognostic marker, regardless of cytogenetics (32). Recently, FLT3 inhibitors as single agents or combined with chemotherapy have been introduced into treating AML patients (33,34). Subsequently, the resistance for FLT3 inhibitors was observed in AML patients. As we know that GPXs and FLT3 may affect chemotherapy sensitivity. Based on these, we will explore whether have a relation between GPXs expression with FLT3 mutations status, insight whether the GPXs expression related to resistance for FLT3 inhibitors. The role of FLT3 in AML is well elaborated but the role of other individual GPXs in the prognosis and progression of AML still remains unclear. Learning about the expression, prognostic values, and regulation networks of GPXs may contribute to the development of therapeutic strategies for patients with AML. Therefore, in the study presented, we identified the potential role of the GPX family using in-depth and comprehensive bioinformatics analysis to explore the relationship between the GPX genes and AML.
Methods
Ethics statement
This study was approved by the Academic Committee of Guangxi Medical University and conducted according to the principles of the Helsinki Declaration. All the data analyzed in our study were retrieved from published literature, and we confirmed that written informed consent was obtained .
Oncomine database analysis
Oncomine, a powerful bioinformatics platform which includes 715 datasets and 86,733 samples, aims at collecting, standardizing, analyzing, and delivering cancer transcriptome data to the biomedical research community (35). The expression level of the GPX gene family in different types of cancer was identified using the Oncomine database (http://www.oncomine.org). The threshold was determined according to a P value of 0.05, a fold change of 2, and a gene rank that included the top 10%. Student’s t-test was used to analyze the difference in the GPX expression in AML.
The Gene Expression Profiling Interactive Analysis (GEPIA) analysis
GEPIA, a web-based tool to deliver fast and customizable functionalities based on The Cancer Genome Atlas (TCGA) and Genotype-Tissue Expression (GTEx) project provide key interactive functions including differential expression analysis, profiling plotting, correlation analysis, and patient survival analysis (36). We investigated the differential expression and prognostic values of GPX genes in the “Single Gene Analysis” module of the GEPIA database (http://gepia.cancer-pku.cn/). Multiple gene comparison analysis of GPXs was conducted with the “Multiple Gene Comparison” module, using the “LAML” dataset. Student’s t-test was used to perform expression analysis. The survival results were displayed with hazard ratios (HRs) and P values from a log-rank test. The P value cutoff was 0.05.
The Cancer Cell Line Encyclopedia (CCLE) dataset
CCLE (http://www.broadinstitute.org/ccle) is a compilation of gene expression, chromosomal copy number, and massively parallel sequencing data from 947 human cancer cell lines (37). Recently, CCLE has expanded the characterizations of cancer cell lines to include genetic, RNA splicing, DNA methylation, and microRNA (miRNA) expression data for 1,072 cell lines from individuals of various lineages and ethnicities (38). The expression of the GPX gene family in cancer cell lines was validated by the CCLE dataset.
The European Bioinformatics Institute (EMBL-EBI) dataset
EMBL-EBI (https://www.ebi.ac.uk) provides free and unrestricted access to data across all major areas of biology and biomedicine. The interconnectivity that exists between data resources at EMBL-EBI provides a better understanding of the relationship between different data types including sequences, genes, proteins, enzymes, and macromolecular structures (39). The expression of GPX gene family in AML cell lines was verified by the EMBL-EBI dataset.
PROGgeneV2 survival analysis
PROGgeneV2 (http://www.compbio.iupui.edu/proggene) is a comprehensive tool that can be used to study the prognostic implications of genes in various cancers. Survival analysis can be performed on a single gene, multiple genes, ratio of expression of 2 genes, and gene signatures in a new version. One unique function of this tool is that users can upload their own datasets to perform survival analysis on genes of interest (40). As such, we validated the prognostic values for GPX gene family using the PROGgeneV2 AML dataset. The P value cutoff was 0.05, and the cohort was divided by the median of gene expression. Results are displayed with HRs and P values from a log-rank test.
UALCAN database analysis
UALCAN (http://ualcan.path.uab.edu) is an easy-to-use, interactive web-portal that can perform in-depth analyses of TCGA gene expression data (41). In our study, UALCAN was used to validate the prognostic values of GPXs via the “Survival Analysis” module and the “AML” dataset. Expression data of GPXs was examined via the “Expression Analysis” module and the “AML” dataset. Student’s t-test was used for prognostic analysis. The P value cutoff was 0.05.
NetworkAnalyst analysis
NetworkAnalyst (https://www.networkanalyst.ca) was first released in 2014 to address the key need for interpreting gene expression data within the context of protein-protein interaction (PPI) networks (42). In addition, users can now perform gene expression profiling for 17 different species and create gene regulatory networks, gene co-expression networks, along with toxicogeneomics and pharmacogenomics studies (43). The gene ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis, and PPI network was visualized using NetworkAnalyst3.0.
The LinkedOmics dataset
The LinkedOmics database (http://www.linkedomics.org) contains multi-omics data and clinical data for 32 cancer types comprising 11,158 patients from TCGA project. It provides a unique platform for scholars to access, analyze, and compare cancer multi-omics data within and across tumor types (44). The “LinkFinder” module was used to investigate differentially expressed genes within the TCGA LAML cohort. The “LinkInterpreter” module was used to perform analysis of kinase target s, miRNA target, and transcription factor (TF) target for GPXs. Results were analyzed for significance using the Pearson’s correlation test. The P value cutoff was 0.05.
Results
Transcriptional expression levels of GPX genes in AML patients
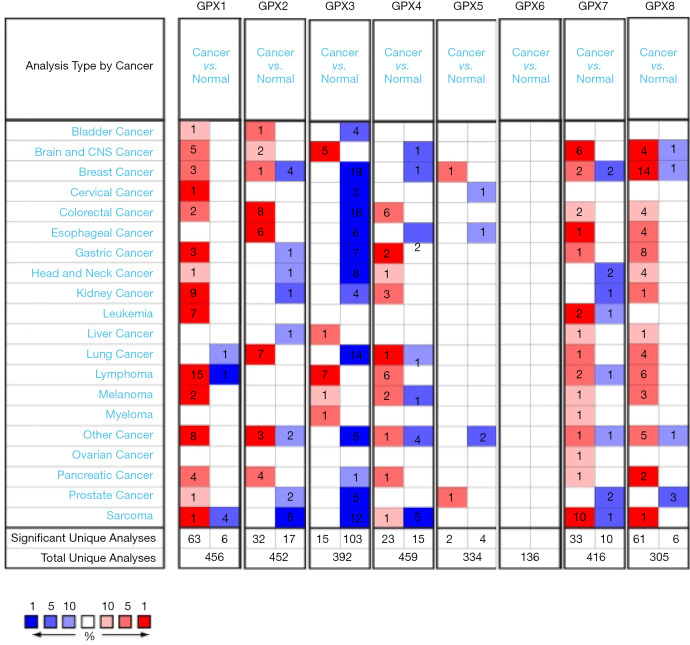
To determine the differences of GPX gene expression in leukemia samples and normal controls, the GPX mRNA levels in tumorigenic and healthy control samples were analyzed using the Oncomine database (Figure 1). Results revealed that the transcriptional expression of GPX-1 and GPX-7 was significantly upregulated in patients with leukemia. The database included a total of 456, 452, 392, 459, 334, 136, 416, and 305 unique analyses for GPX-1, GPX-2, GPX-3, GPX-4, GPX-5, GPX-6, GPX-7, and GPX-8, respectively. In total, 7 studies indicated that the mRNA expression level of GPX1 was higher in leukemia, upregulated expression of GPX7 was found in 2 datasets compared to normal samples, while 1 study suggested an opposite finding. As for GPX-2, GPX-3, GPX-4, GPX-5, GPX-6, and GPX8, no significant differences were observed in the messenger RNA (mRNA) expression levels between leukemic and healthy control samples according to the Oncomine database. In sum, the transcriptional expression levels of GPX-1 and GPX-7 were significantly higher in leukemia than in normal samples.
Figure 1.
The transcription levels of GPX gene family in different types of cancer (Oncomine database). GPX, glutathione peroxidase.
To further evaluate the GPX family expression in AML, we examined the GPX expression levels in the GEPIA database (Figures 2A,B,C). According to the results, the expression levels of GPX-1, GPX-2, and GPX-7 were overexpressed in AML samples in comparison to those of normal samples, while the expression levels of GPX-4 and GPX-8 were found to be lower in AML samples than in normal samples. However, the expression levels of GPX-3, GPX-5, and GPX-6 were not significantly different. Subsequently, together with the results obtained from the Oncomine database, we evaluated all the significantly differentially expressed GPXs (GPX-1, GPX-2, GPX-4, GPX-7, and GPX-8) in AML samples compared to normal healthy controls for further analysis.
Figure 2.
The expression levels of GPXs in AML compared with healthy normal controls (GEPIA). GPX, glutathione peroxidase; AML, acute myeloid leukemia; GEPIA, Gene Expression Profiling Interactive Analysis.
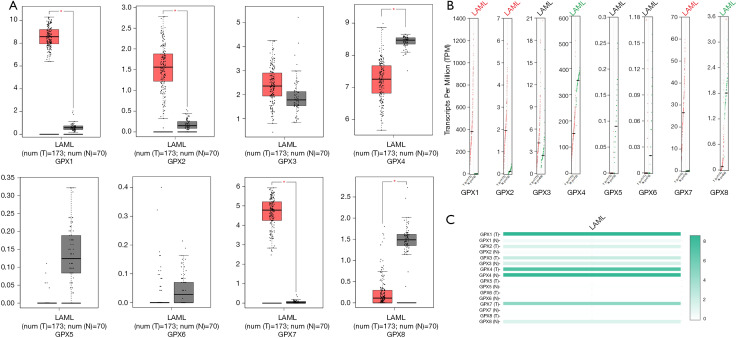
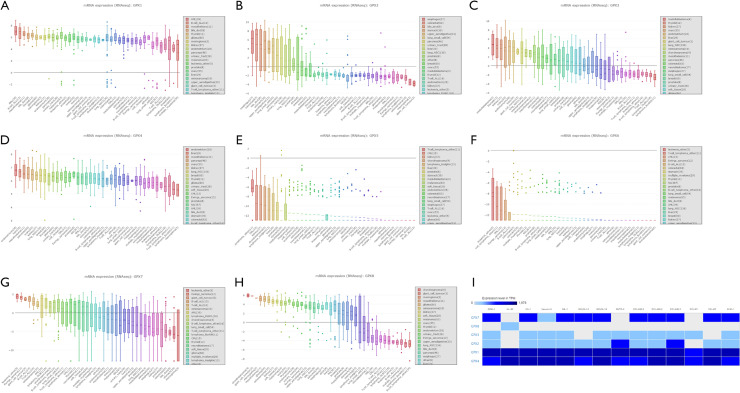
The GPX transcriptional levels in leukemia cell lines
The large number of cancer cell lines in the CCLE provided strong clues about the expression of genes on many more cancer subtypes of various tissues of origin. With the use of the CCLE database, we systematically elucidated the expression levels of the GPX family in different cancer cells (Figures 3A,B,C,D,E,F,G,H). We discovered that GPX-1, GPX-4, and GPX-7 were positively expressed in AML cell lines. Subsequently, we analyzed GPX family expression levels in EMBL-EBI panel of leukemia cell lines, and revealed that GPX-1, GPX-4, and GPX-7 were also expressed in most AML cell lines (Figure 3I). We eventually identified the differentially expression levels among the GPX gene family in AML sample s, and then evaluated the differentially expressed GPXs (GPX-1, GPX-2, GPX-4, GPX-7, and GPX-8) to identify their biology processes and gene regulation networks in AML.
Figure 3.
Expression of GPX factors in AML cell lines (CCLE and EMBL-EBI). (A,B,C,D,E,F,G,H) The expression levels of GPX family in leukemia cell lines by CCLE. (I) The expression levels of GPXs in AML cell lines by EMBL-EBI. GPX, glutathione peroxidase; AML, acute myeloid leukemia; CCLE, Cancer Cell Line Encyclopedia; EMBL-EBI, The European Bioinformatics Institute.
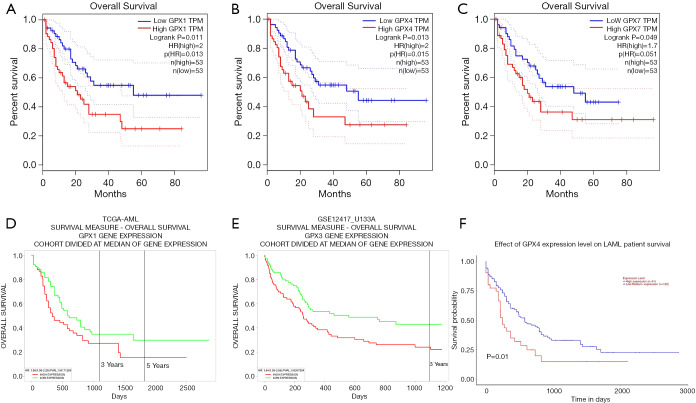
Relationship between GPX expression and prognostic values of patients with AML
To investigate whether the GPX gene family expression would impact the survival in AML patients, we used the GEPIA database clarify the relationships between GPX expression and prognostic values of AML. As shown in the results, the expression of GPX-1 and GPX-4 significantly impacted the prognosis of overall survival (OS) in AML. Notably, a high GPX1 expression was associated with poorer prognosis in patients with AML (P=0.011), and overexpression of GPX4 indicated a worse outcome in AML patients (P=0.013). In particular, high GPX7 expression was marginally associated with adverse OS (P=0.049) (Figures 4A,B,C). No significance between the expression of GPX-2, GPX-3, GPX-5, GPX-6, and GPX-8 or the prognosis in AML was found in the GEPIA database. The value of the GPX gene family in the disease-free survival of AML patients was also evaluated, but no significant differences were found. These results thus reveal that high expressions of GPX-1, GPX-4, and GPX-7 have a significant impact on the worse OS in AML.
Figure 4.
The prognostic values of GPXs expression in AML patients (GEPIA, PROGgeneV2, and UALCAN). (A,B,C) The overall survival of GPX-1, GPX-4, and GPX-7 expression in AML patients, analyzed by GEPIA. (D,E) The overall survival of GPX-1 and GPX-3 expression in AML patients, analyzed by PROGgeneV2. (F) The survival prognosis of GPX-4 expression in AML, analyzed by UALCAN. GPX, glutathione peroxidase; AML, acute myeloid leukemia; GEPIA, Gene Expression Profiling Interactive Analysis.
Subsequently, we further studied the survival significance of GPX genes in patients with AML using the PROGgeneV2 platform. The results revealed that the increased expression level of GPX1 and GPX3 were significantly associated with poor prognosis in AML patients (OS HR =1.58, P value =0.0171289; OS HR =1.64, P value =0.0307594) (Figures 4D,E). However, the expression of GPX-2, GPX-4, GPX-5, GPX-6, GPX-7, and GPX-8 showed no significant impact on the survival prognosis of AML within the PROGgeneV2 database.
In addition to the microarray analysis of GPX family in the GEPIA database and the PROGgeneV2 platform, the UALCAN database was also used to examine the prognostic potential of the GPX family in AML. High GPX4 expression was correlated with poorer prognosis in AML patients (P=0.01) (Figure 4F). However, according to the UALCAN database, there was no significant difference for the rest of GPX family except for GPX4 in survival prognosis. In summary, we found the overexpression of GPX-1, -3, -4, and -7 to be significantly associated with poorer prognosis of OS in AML patients.
Expression levels of GPXs of prognostic value based on French–American–British subtype classification and FLT3 mutation status
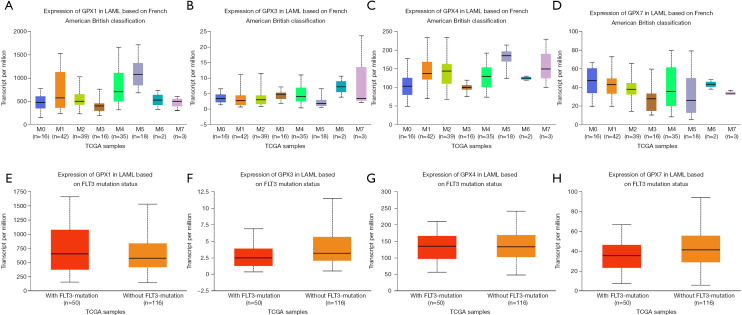
To further explore the expression information of GPXs (-1, -3, -4, -7) in relation to AML prognostic value, we analyzed the expression levels based on French-American-British (FAB) classification and FLT3 mutation status using UALCAN (Figure 5). Expression of GPX-1 in AML based on FAB classification is shown in Figure 5A; statistical significances were observed in M0 vs. M1 (P=4.016100E−03), M0 vs. M4 (P=2.594100E−04), M0 vs. M5 (P=2.773399E−08), M1 vs. M3 (P=1.277960E−03), M1 vs. M5 (P=4.970200E−04), M2 vs. M3 (P=4.062400E−02), M2 vs. M4 (P=2.770400E−02), M2 vs. M5 (P=2.388890E−07), M3 vs. M4 (P=7.704900E−05), M3 vs. M5 (P=1.043499E−08), M4 vs. M5 (P=3.619300E−03), M5 vs. M6 (P=2.216000E−02), and M5 vs. M7 (P=2.701200E−03). Expression of GPX-3 in AML based on FAB classification is shown in Figure 5B; however, there was no significant difference among FAB subtypes. Expression of GPX-4 in AML based on FAB classification is shown in Figure 5C; we found that M0 vs. M1 (P=9.841200E−04), M0 vs. M2 (P=2.379200E−03), M0 vs. M4 (P=3.055600E−02), M0 vs. M5 (P=4.942300E−02), M0 vs. M7 (P=4.942300E−02), M1 vs. M3 (P=1.028899E−08), M1 vs. M5 (P=2.595800E−04), M2 vs. M3 (P=4.578999E−06), M2 vs. M5 (P=1.209750E−02), M3 vs. M4 (P=5.242800E−04), M3 vs. M5 (P=5.819199E−08), M3 vs. M6 (P=4.318000E−02), and M4 vs. M5 (P=1.122710E−03) showed statistical significance. The GPX-7 expression profile based on FAB classification, as shown in Figure 5D; comparison of M0 vs. M3 (P=1.939960E−03), M0 vs. M5 (P=2.223800E−02), M0 vs. M7 (P=3.859900E−03), M1 vs. M3 (P=7.824500E−04), M1 vs. M5 (P=7.780200E−03), M1 vs. M7 (P=3.763000E−04), M2 vs. M3 (P=1.738430E−02), and M2 vs. M7 (P=3.124100E−02) showed significant difference. We then assessed the correlation between the expression of GPX-1, -3, -4, and -7 and FLT3 mutation status in AML, and found a significant correlation between the expression of GPX3 and FLT3 mutation status (Figure 5F). GPX3 had significantly lower expression level in AML samples with FLT3 mutation (P=4.147700E−02). Correlation in the expression of GPX-1, -4, and -7 with or without FLT3 mutation was also determined, but not significantly so (Figures 5E,G,H). These data suggest that the expression of GPX varies in different FAB subtypes and in different FLT3 mutation statuses in patients with AML.
Figure 5.
The expression levels of GPX (GPX-1, -3, -4, -7) prognostic value in AML based on FAB classification and FLT3 mutation status (UALCAN). The expression levels of GPX-1 (A), GPX-3 (B), GPX-4 (C), and GPX-7 (D) in AML based on FAB classification. (E,F,G,H) The expression levels of GPX-1 (P=2.984000E−01), GPX3 (P=4.147700E−02), GPX4 (P=7.870800E−01), and GPX7 (P=2.031200E−01) in AML based on FLT3 mutation status. GPX, glutathione peroxidase; AML, acute myeloid leukemia; FAB, French-American-British.
Functional enrichment analysis of differentially expressed GPXs in patients with AML
To explore the potential functional enrichment among differentially expressed GPXs, we subsequently compiled a list of differentially expressed GPXs (-1, -3, -4, -7, -8) for analysis of the functional network via NetworkAnalyst. Analysis of significantly enriched GO biological process (BP) terms suggested that these genes were primarily involved in regulation of immune response, inflammatory response, negative regulation of immune system process, response to oxidative stress, and regulation of defense response (Figure 6A). KEGG pathway analysis of differentially expressed GPXs showed enrichment in ferroptosis, metabolic pathway, glutathione metabolism, and arachidonic acid metabolism (Figure 6B). The PPI network was constructed based on the whole blood-specific data (Figures 6C,D,E). As shown in the results, GPX-1 was predicted to interact with ABL2, and GPX-2 was predicted to interact with MYC, TP53, and other important biological molecules. These results indicate that the differentially expressed GPXs may play important roles in the carcinogenesis and progression of AML.
Figure 6.
Functional enrichment analysis of differentially expressed GPXs (GPX-1, -2, -4, -7, -8) (NetworkAnalyst). (A) GO-BP terms of differentially expressed GPXs. (B) KEGG pathway analysis of differentially expressed GPXs. (C-E) The blood-specific PPI network of GPX-1, GPX-2, and GPX-7. GPX, glutathione peroxidase; GO-BP, gene ontology-biological process; KEGG, Kyoto Encyclopedia of Genes and Genomes.
Correlated genes of differentially expressed GPXs and their survival analysis in AML
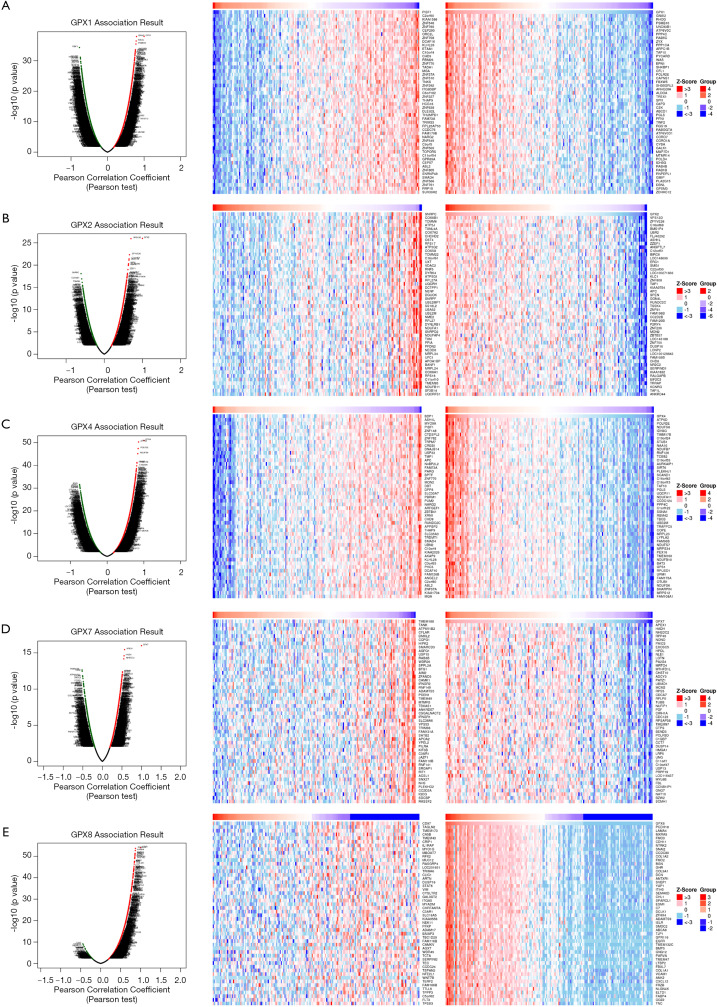
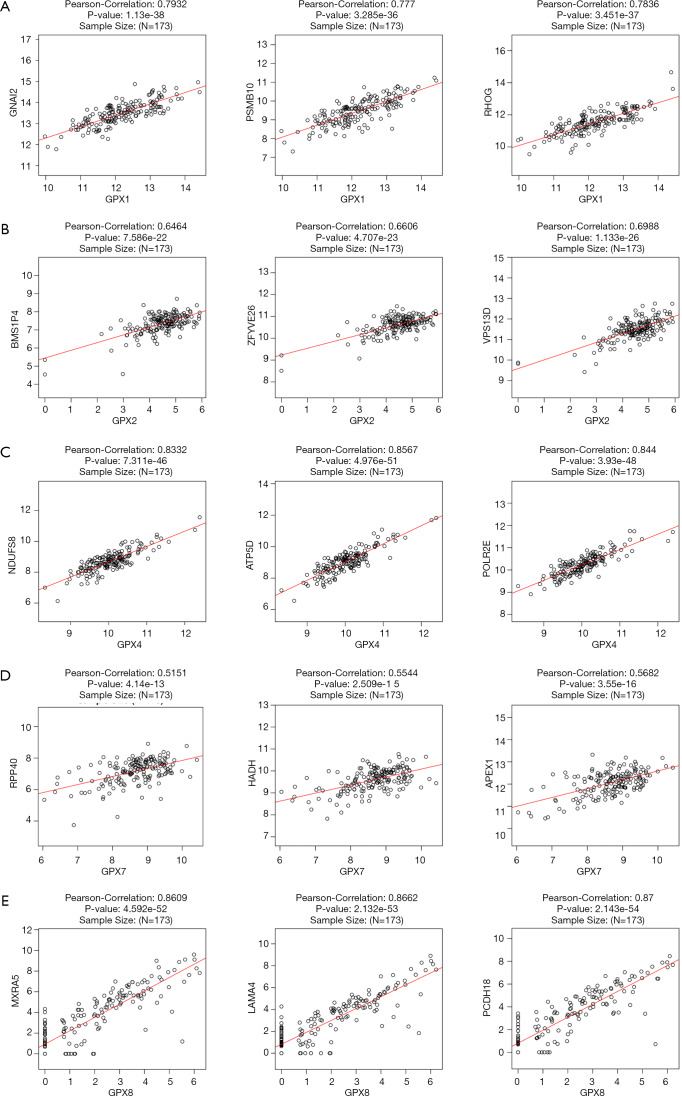
To further investigate the potential role of differentially expressed GPXs in AML, the LinkFinder module of LinkedOmics was used to analyze mRNA sequencing data from 173 patients in TCGA. In the volcano plot, the dark red dots show a significantly positive correlation with GPX-1, -2, -4, -7, and -8, whereas dark green dots show a significantly negative correlation (false discovery rate, FDR <0.01). The top 50 significant gene sets positively and negatively associated with GPX-1, -2, -4, -7, and -8, as shown in the heat maps (Figures 7A,B,C,D,E). Meanwhile, the statistical scatter plots show that the GPX-1 expression had a strong positive association with the expression of GNAI2 (Pearson’s correlation =0.7932, P value =1.13E−38), PSMB10 (Pearson’s correlation =0.777, P value =3.285E−36), and RHOG (Pearson’s correlation =0.7836, P value =3.451E−37) (Figure 8A). GPX2 expression showed a strong positive correlation with the expression of NMS1P4 (Pearson’s correlation =0.6464, P value =7.586E−22), ZFYVE26 (Pearson’s correlation =0.6606, P value =4.707E−23), and VPS13D (Pearson’s correlation =0.6988, P value =1.133E−26) (Figure 8B). GPX4 expression showed a strong positive correlation with the expression of NDUFS8 (Pearson’s correlation =0.8332, P value =7.311E−46), ATP5D (Pearson’s correlation =0.8567, P value =4.976E−51), and POLR2E (Pearson’s correlation =0.844, P value =3.93E−48) (Figure 8C). GPX7 expression showed a strong positive association with the expression of RPP40 (Pearson’s correlation =0.5151, P value =4.14E−13), HADH (Pearson’s correlation =0.5544, P value =2.509E−15), and APEX1 (Pearson’s correlation =0.5682, P value =3.55E−16) (Figure 8D). GPX8 expression showed a strong positive association with the expression of MXRA5 (Pearson’s correlation =0.8609, P value =4.592E−52), LAMA4 (Pearson’s correlation =0.8662, P value =2.132E−53), and PCDH18 (Pearson’s correlation =0.87, P value =2.143E−54) (Figure 8E).
Figure 7.
Differentially expressed genes correlated with differentially expressed GPXs in AML (LinkedOmics). A Pearson test was used to identify the correlations between GPX-1, -2, -4, -7, and -8 genes differentially expressed in AML cohort (n=173). (A,B,C,D,E) Volcano plots and heat maps indicating genes positively and negatively genes correlated with GPX-1, -2, -4, -7, and -8 in AML, respectively (top 50). Red suggests positively correlated genes, and green suggests negatively correlated genes. GPX, glutathione peroxidase; AML, acute myeloid leukemia.
Figure 8.
Gene correlation expression analysis for differentially expressed GPXs (LinkedOmics). (A) The scatter plots show Pearson’s correlation of GPX-1 expression with expression of GNAI2, PSMB10, and RHOG. (B) The scatter plots show Pearson’s correlation of GPX-2 expression with expression of BMS1P4, ZFYVE26, and VPS13D. (C) The scatter plots show Pearson’s correlation of GPX-4 expression with expression of NDUFS8, ATP5D, and POLR2E. (D) The scatter plots show Pearson’s correlation of GPX-7 expression with expression of RPP40, HADH, and APEX1. (E) The scatter plots show Pearson’s correlation of GPX-8 expression with the expression of MXRA5, LAMA4, and PCDH18. GPX, glutathione peroxidase.
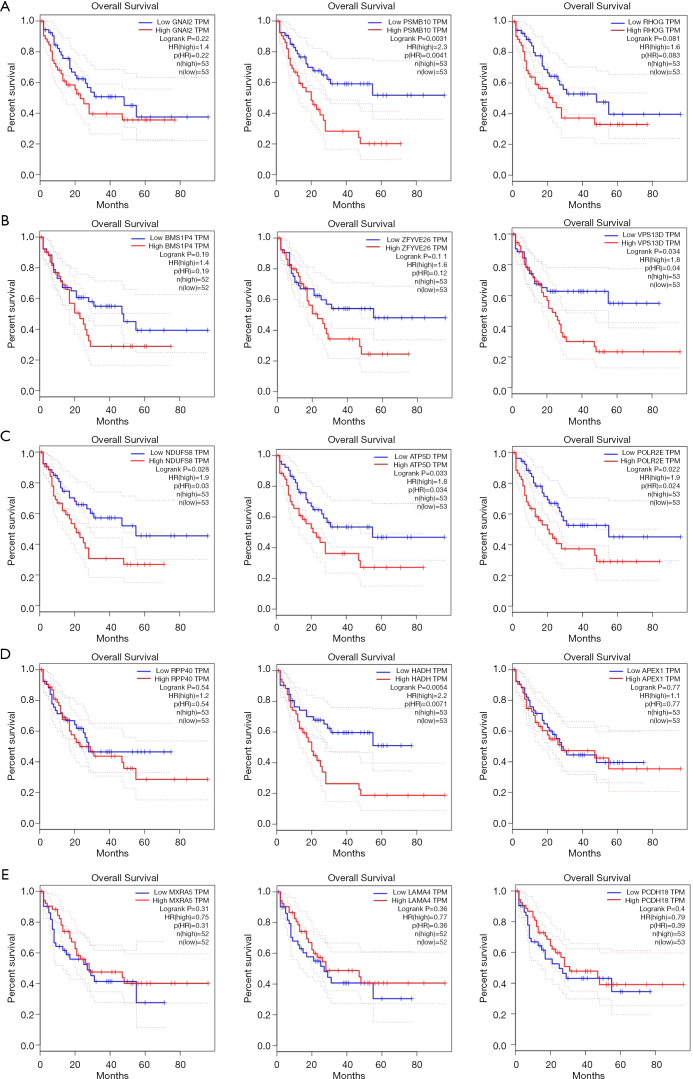
To evaluate the prognostic value of the most correlated genes of the differentially expressed GPXs in AML, we assessed the value of the GPX-1, -2, -4, -7, and -8 correlated genes in the OS of AML patients using GEPIA. We found that AML patients with high transcriptional levels of PSMB10 (P=0.0031), VPS13D (P=0.034), NDUFS8 (P=0.0.028), ATP5D (P=0.033), POLR2E (P=0.022), and HADH (P=0.0054) were significantly associated with poorer OS (Figure 9). Disease-free survival curves were also performed, but there was no significant difference.
Figure 9.
Prognostic analysis of genes correlated with differentially expressed GPXs in patients with AML (GEPIA). (A) The overall survival curves of GNAI2, PSMB10, and RHOG in AML. (B) The overall survival curves of BMS1P4, ZFYVE26, and VPS13D in AML. (C) The overall survival curves of NDUFS8, ATP5D, and POLR2E in AML. (D) The overall survival curves of RPP40, HADH, and APEX1 in AML. (E) The overall survival curves of MXRA5, LAMA4, and PCDH18 in AML. GPX, glutathione peroxidase; AML, acute myeloid leukemia; GEPIA, Gene Expression Profiling Interactive Analysis.
Kinase targets, miRNA targets, and TF targets of differentially expressed GPXs
To deepen the understanding of the differentially expressed GPXs regulators in AML, we further analyzed the significant kinase, miRNA, and TF targets of GPX-1, -2, -4, -7, and -8 via the LinkedOmics database. The most significant kinase targets are shown in Table 1. DAPK1, PAK1, SRC, PRKCD, and CHUK were the top 5 kinase targets of both GPX-1 and GPX-4. GPX2 kinase targets were mainly associated with DAPK1, SGK1, CDK5, MAPK1, and MYLK. TTK, CDK2, ATR, EGFR, while PRKDC was found to be the kinase target of GPX-7. GPX-8 kinase targets were primary related to DAPK1 and PRKCZ, along with NTRK2, TTK, and BUB1. Therefore, DAPK1, PAK1, SRC, PRKCD, TTK, and CHUK were the most associated kinase targets for differentially expressed GPXs.
Table 1. The kinase targets of differentially expressed glutathione peroxidases in AML (LinkedOmics).
| GPXs | Gene set | Description | Leading edge number | P value |
|---|---|---|---|---|
| GPX-1 | Kinase_DAPK1 | Death associated protein kinase 1 | 11 | 1.08E−02 |
| Kinase_PAK1 | p21 (RAC1) activated kinase 1 | 15 | 0.00E+00 | |
| Kinase_SRC | SRC proto-oncogene, non-receptor tyrosine kinase | 48 | 0.00E+00 | |
| Kinase_PRKCD | Protein kinase C delta | 23 | 0.00E+00 | |
| Kinase_CHUK | Conserved helix-loop-helix ubiquitous kinase | 11 | 0.00E+00 | |
| GPX-2 | Kinase_DAPK1 | Death associated protein kinase 1 | 8 | 3.76E−02 |
| Kinase_SGK1 | Serum/glucocorticoid regulated kinase 1 | 9 | 0.00E+00 | |
| Kinase_CDK5 | Cyclin dependent kinase 5 | 18 | 8.15E−03 | |
| Kinase_MAPK1 | Mitogen-activated protein kinase 1 | 62 | 4.91E−02 | |
| Kinase_MYLK | Myosin light chain kinase | 5 | 0.00E+00 | |
| GPX-4 | Kinase_PAK1 | P21 (RAC1) activated kinase 1 | 15 | 3.44E−03 |
| Kinase_DAPK1 | Death associated protein kinase 1 | 8 | 3.83E−03 | |
| Kinase_SRC | SRC proto-oncogene, non-receptor tyrosine kinase | 37 | 9.93E−03 | |
| Kinase_PRKCD | Protein kinase C delta | 25 | 9.97E−03 | |
| Kinase_CHUK | Conserved helix-loop-helix ubiquitous kinase | 10 | 1.06E−02 | |
| GPX-7 | Kinase_TTK | TTK protein kinase | 9 | 7.22E−03 |
| Kinase_CDK2 | Cyclin dependent kinase 2 | 108 | 0.00E+00 | |
| Kinase_ATR | ATR serine/threonine kinase | 28 | 0.00E+00 | |
| Kinase_EGFR | Epidermal growth factor receptor | 15 | 0.00E+00 | |
| Kinase_PRKDC | Protein kinase, DNA-activated, catalytic polypeptide | 16 | 9.23E−03 | |
| GPX-8 | Kinase_DAPK1 | Death associated protein kinase 1 | 2 | 3.70E−02 |
| Kinase_PRKCZ | Protein kinase C zeta | 7 | 6.00E−03 | |
| Kinase_NTRK2 | Neurotrophic receptor tyrosine kinase 2 | 4 | 1.75E−02 | |
| Kinase_TTK | TTK protein kinase | 6 | 3.02E−02 | |
| Kinase_BUB1 | BUB1 mitotic checkpoint serine/threonine kinase | 3 | 2.18E−02 |
AML, acute myeloid leukemia; GPX, glutathione peroxidase; LeadingEdgeNum, the number of leading edge genes.
In addition, the potential miRNA targets of differentially expressed GPXs were also explored (Table 2). We identified (ATAGGAA) MIR-202, (CTTGTAT) MIR-381, (ATGCAGT) MIR-217, (ATGTTTC) MIR-494, and (ACACTAC) MIR-142-3p as the top 5 miRNA targets associated with differentially expressed GPXs.
Table 2. The miRNA targets of differentially expressed glutathione peroxidases in AML (LinkedOmics).
| GPXs | Gene set | Leading edge number | P value |
|---|---|---|---|
| GPX-1 | ATAGGAA, MIR-202 | 41 | 0.00E+00 |
| CTTGTAT, MIR-381 | 90 | 0.00E+00 | |
| ATGCAGT, MIR-217 | 50 | 0.00E+00 | |
| CATTTCA, MIR-203 | 109 | 0.00E+00 | |
| ATGTTTC, MIR-494 | 51 | 0.00E+00 | |
| GPX-2 | ATAGGAA, MIR-202 | 32 | 0.00E+00 |
| CTTGTAT, MIR-381 | 61 | 0.00E+00 | |
| ATGCAGT, MIR-217 | 41 | 0.00E+00 | |
| CTACTGT, MIR-199A | 67 | 0.00E+00 | |
| ACACTAC, MIR-142-3p | 42 | 0.00E+00 | |
| GPX-4 | CATTTCA, MIR-203 | 112 | 0.00E+00 |
| ATGTTTC, MIR-494 | 66 | 0.00E+00 | |
| TGAATGT, MIR-181A, MIR-181B, MIR-181C, MIR-181D | 161 | 0.00E+00 | |
| ATAGGAA, MIR-202 | 46 | 0.00E+00 | |
| ATACTGT, MIR-144 | 70 | 0.00E+00 | |
| GPX-7 | TGAATGT, MIR-181A, MIR-181B, MIR-181C, MIR-181D | 134 | 0.00E+00 |
| ATCATGA, MIR-433 | 34 | 0.00E+00 | |
| ACACTAC, MIR-142-3p | 51 | 0.00E+00 | |
| GTATTAT, MIR-369-3p | 62 | 0.00E+00 | |
| TGTTTAC, MIR-30A-5p, MIR-30C, MIR-30D, MIR-30B, MIR-30E−5P | 138 | 0.00E+00 | |
| GPX-8 | TAGCTTT, MIR-9 | 60 | 6.00E−03 |
| TAATGTG, MIR-323 | 50 | 2.40E−02 | |
| ATGTACA, MIR-493 | 82 | 4.00E−03 | |
| AATGTGA, MIR-23A, MIR-23B | 99 | 2.00E−03 | |
| CATGTAA, MIR-496 | 37 | 1.40E−02 |
AML, acute myeloid leukemia; GPX, glutathione peroxidase; LeadingEdgeNum, the number of leading edge genes.
Finally, the enrichment of TFs showed that several key TFs were associated with the regulation of differentially expressed GPXs (Table 3). Notably, AP1, SRF, and E2F1 were suggested as the key targets for the GPX-1, -4, and -7 TF-target network. Meanwhile, V$FOX_Q2, V$USF_C, V$ELK1_02, V$NRF2_01, and V$YY1_01 were primarily related to GPX2. V$NFY_Q6, V$IRF1_01, V$HNF1_01, V$AP3_Q6, and V$SMAD3_Q6 were suggested as targets for GPX8. Thus, these essential cancer-related regulators warrant further investigation. Overall, these results contribute to a greater understanding of the associations between differentially expressed GPXs and AML pathogenesis and prognosis.
Table 3. The transcription factor targets of differentially expressed glutathione peroxidases in AML (LinkedOmics).
| GPXs | Gene set | Leading edge number | P value |
|---|---|---|---|
| GPX-1 | V$AP1_Q4_01 | 58 | 0.00E+00 |
| V$AP1_Q6 | 59 | 0.00E+00 | |
| V$AP1_Q6_01 | 50 | 0.00E+00 | |
| V$LFA1_Q6 | 58 | 0.00E+00 | |
| V$SRF_01 | 19 | 0.00E+00 | |
| GPX-2 | V$FOX_Q2 | 50 | 0.00E+00 |
| V$USF_C | 48 | 0.00E+00 | |
| V$ELK1_02 | 65 | 0.00E+00 | |
| V$NRF2_01 | 60 | 0.00E+00 | |
| V$YY1_01 | 53 | 0.00E+00 | |
| GPX-4 | V$SRF_01 | 15 | 3.51E−03 |
| V$SP1_Q6_01 | 66 | 0.00E+00 | |
| V$AP1_Q4_01 | 57 | 0.00E+00 | |
| V$LFA1_Q6 | 68 | 0.00E+00 | |
| V$SOX5_01 | 61 | 0.00E+00 | |
| GPX-7 | V$E2F1DP1_01 | 69 | 0.00E+00 |
| V$E2F1_Q4_01 | 65 | 0.00E+00 | |
| V$E2F1_Q6 | 70 | 0.00E+00 | |
| V$E2F1DP1RB_01 | 70 | 0.00E+00 | |
| V$E2F1DP2_01 | 69 | 0.00E+00 | |
| GPX-8 | V$NFY_Q6 | 41 | 0.00E+00 |
| V$IRF1_01 | 43 | 0.00E+00 | |
| V$HNF1_01 | 36 | 0.00E+00 | |
| V$AP3_Q6 | 56 | 0.00E+00 | |
| V$SMAD3_Q6 | 71 | 0.00E+00 |
AML, acute myeloid leukemia; GPX, glutathione peroxidase; LeadingEdgeNum, the number of leading edge genes; V$, the annotation found in Molecular Signatures Database (MSigDB) for transcription factor (TF).
Discussion
GPXs belong to an enzyme family whose main biological functions are to protect organisms from oxidative stress damage. Oxidative stress is involved in various BPs including proliferation, DNA damage, and survival. Functioning as anti-oxidants, GPXs were reported to be involved in carcinogenesis (45,46). To gain more insight into the potential role of GPXs in AML and its regulation networks, we carried out a bioinformatics analysis based on several large public databases to comprehensively guide future research in AML.
We discovered that the expression levels of GPX-1, -2, -4, -7, and -8 were differentially expressed in AML compared with normal tissues. Analysis of the Oncomine and GEPIA datasets revealed that the mRNA expression levels of GPX-1, -2, and -7 were significantly higher in AML than in normal samples. Meanwhile, the transcriptional expression levels of GPX-4 and -8 were decreased in AML patients compared with healthy controls according to GEPIA. Survival analysis showed that AML patients with high expression of GPX-1, -3, -4, and -7 were significantly related to poorer prognosis of OS. Based on this, our results suggested that the upregulation of GPX-1, -3, -4, and -7 is related to adverse clinical outcome in AML patients, and these GPXs merit further large-scale clinical validation as potential therapeutic targets and prognostic biomarkers.
Previous research has shown that GPX-1 was highly expressed in variety of human malignancies, and high GPX-1 expression was positively correlated with shorter OS time in different cancer types (10,47), which is line with the results of the present study. High mRNA expression of GPX1 was correlated with worse OS in AML patients, which indicates that GPX-1 may serve as an oncogene.
Several studies have examined the prognostic values of GPX-3 and GPX-4 in malignant blood diseases. For example, GPX-4-positive expression contributed to poor OS and progression-free survival in diffuse large B-cell lymphoma, and overexpression of GPX-4 served as an independent prognostic predictor (48). This was consistent with the result observed in our study, and GPX-4 may have an impact on cancer progression and carcinogenesis. On the other hand, another study reported that GPX-3 methylation predicted adverse prognosis and served as an independent prognostic biomarker in non-M3 AML (29), which suggests GPX3 could be an epidemic silenced tumor suppressor gene, which conflicts with our study’s results. However, it is worth noting that our study could not find a significant correlation between GPX-3 expression and promoter methylation in AML patient samples. Since there is heterogeneity in the molecular biological properties and complex epigenetic mechanisms of leukemia, we consider the survival role of GPX-3 in AML patients to be controversial, necessitating further examination. Taken together, our results ultimately show that some members of the GPX family (GPX-1, GPX-3, GPX-4, and GPX-7) may have prognostic potential for adverse OS in patients with AML. Further research is needed to verify these findings and to identify whether GPX genes interact among themselves to affect the survival prognosis of AML patients.
The deregulation of multiple GPX genes in AML further interested us. We identified the specific roles and gene regulation network among significantly differentially expressed GPXs (-1, -2, -4, -7, -8) in AML, and explored their GO enrichment and KEGG pathway analysis. As expected, the functions of these genes were primarily related to glutathione metabolism, ferroptosis, and metabolic pathway, in addition to regulation of immune response, inflammatory response, negative response of immune system process, response to oxidative stress, and regulation of defense response. Organism metabolic abnormalities and homeostasis imbalance often lead to malignant transformation and tumor progression. These data suggest that the GPX gene family, which was found to be differentially expressed in AML, is potentially involved in carcinogenesis, cell death, immune regulation, and inflammatory response.
The blood-specific PPI network indicated that ABL2 is one of the key proteins that interacts with GPX-1. ABL2, also known as ARG or the ABL-related gene, belongs to the mammalian Abelson (ABL) family of non-receptor tyrosine kinases. Notably, the breakpoint cluster region (BCR)-ABL fusion gene is the major oncogene in chronic myelogenous leukemia (CML). Recent study suggests that the overexpression of ABL2 inhibits FLT3-ITD-dependent cell proliferation and colony formation, revealing that ABL2 acts as a negative regulator of signaling downstream of FLT3 (49). Therefore, targeting ABL2 may be an alternative and promising approach to treating FLT3-dependent AML.
The correlation of the expression genes associated with differentially expressed GPXs was carried out by the LinkedOmics database. Overexpression of GPX-1 showed a strong positive correlation with the expression of GNAI2, PSMB10, and RHOG, which reflect regulations in oncogenesis, inflammation, and angiogenesis (50-53). The current study was the first to identify the overexpression of PSMB10 as being associated with adverse OS in AML, and we await further large-scale validation of this discovery. Furthermore, GPX-2 expression was significantly positively correlated with the expression of MBS1P4, ZFYVE26, and VPS13D. Prognostic analysis revealed high VPS13D expression to be associated with the adverse OS in AML. VPS13D was reported to play an important role in autophagy and mitochondria functions (54). However, there is yet little research concerning its prognostic role in AML.
Meanwhile, GPX-4 showed a strong positive correlation with the expression of NDUFS8, ATP5D, and POLR2E, and their increased expression levels were all predicted to associate with poorer OS in patients with AML. NDUFS8 and ATP5D have been shown to be involved in tumor metabolism and disease progression (55,56). Meanwhile, high NDUFS8 IHC and RNA expression level was reported to have poor OS in lung cancer (56). However, a meta-analysis demonstrated that POLR2E rs3787016 polymorphism was associated with overall risk and prostate cancer risk (57). However, how these genes relate to the susceptibility and pathogenesis of AML has not been extensively examined.
Overexpression of GP-7 showed a strong positive correlation with the expression of RPP40, HADH, and APEX1, among which high HADH expression indicated a poorer OS in AML. One report showed that HADH was significantly associated with worse survival for kidney renal clear cell carcinoma (58), but our results showed no correlation between this gene and AML progression and prognosis.
The expression of GPX-8 has been positively correlated with the expression of MXRA5, LAMA4, and PCDH18, while High MXRA5 and LAMA4 expression has been correlated with tumor progression and poor survival in non-small cell lung cancer and gastric cancer, respectively (59,60). However, PCDH18 was considered to be an epigenetic silenced tumor suppressor in colorectal cancer (61). Our analysis of these genes showed no significant difference in AML prognosis.
To further explore the regulators that could be potentially responsible for GPXs dysregulation in AML, we explored the target networks of kinase, miRNA, and TF targets of differentially expressed GPXs. We found that differentially expressed GPXs in AML were associated with a network of kinases including death-associated protein kinase 1 (DAPK1), p21 (RAC1) activated kinase 1 (PAK1), and SRC proto-oncogene (SRC). These kinases are involved in the cell cycle, immune response, autophagy, and DNA damage. In fact, aberrant DNA methylation and transcriptional silence of DAPK1 participate in the pathogenesis, malignant transformation, and apoptosis in hematologic malignancies. Demethylating drugs such as 5-Azacytidine (AZA) and 5-aza-2'deoxycytidine (DAC), have been applied for the treatment of neoplastic cells in myelodysplastic syndromes (MDS) and AML (62). PAK1 was shown to be relevant for AML pathogenesis and patient survival in MDS, and inhibition of PAK1 inhibited primary human leukemic cells including immature leukemic stem cell-enriched populations (63). As key driver genes, high expression of SRC kinases was found to predict poor survival in AML patients (64). In AML, differentially expressed GPXs may be involved in genomic stability, pathogenesis, and apoptosis by interacting with these kinases.
MicroRNAs are essential for post-transcriptional regulation of gene expression and play an important role in human carcinogenesis and leukemogenesis. Our results found several miRNAs (miR-202, miR-381, and miR-181) to be associated with differentially expressed GPXs. A causal link has been established between miR-202 and cancer cell proliferation, migration, and invasion (65,66). In particular, miR-381 has been linked to multi-drug resistance in leukemic cells (67). Moreover, it was found that, miR-181, being functionally relevant for myeloid differentiation, could be responsible for AML development and disease activity and thus a potential therapeutic target (68,69). Our analyses then suggest that the differentially expressed GPXs may act through these miRNAs to regulate cell proliferation, disease progression, and drug resistance of AML.
By mining the TF targets of the differentially expressed GPXs, we found that SRF and E2F1 may be key TFs in the regulation of GPXs. Serum response factor SRF, an essential TF in hematopoiesis and mature myeloid cell function, has been reported to participate in cellular processes including those of cell proliferation, differentiation, apoptosis, and immune system activity (70). TF E2F1 has been shown to be involved in cell cycle progression, DNA damage response, apoptosis, and cancer metabolism (71). Furthermore, deregulation of E2F1 has been linked to the perturbation of cell cycle and apoptosis in AML. Importantly, a feedback regulatory loop for E2F1, C/EBPα, and Trib2 in AML cell proliferation and survival has been identified. E2F1 was observed playing a key role in AML cell proliferation and leukemic cell survival through targeting Trib2 (72). Therefore, our analyses suggest that E2F1 is a promising regulator target of differentially expressed GPXs. Our data provide additional useful information about the importance of E2F1 in the molecular pathogenesis of AML. Differentially expressed GPXs may regulate myeloid proliferation, cell-cycle progression, and apoptosis via interacting with TFs in AML.
Conclusions
In this study, we collectively analyzed the expression and prognostic value of GPXs and preliminary explored the biological events associated with AML progression. Our results suggest that GPX-1, -2, -4, -7, and -8 were differentially expressed in AML, and overexpression of GPX-1, -3, -4, and -7 was associated with poorer OS. Furthermore, our study indicated the importance of tumor metabolism status in AML prognosis and its potential as a novel prognostic predictor. Regulatory network analysis suggested that differentially expressed GPXs regulate cell proliferation, cell-cycle progression, apoptosis, and survival via pathways involving several tumor-associated kinases (such as DAPK1 and SRC), miRNAs (such as miR-202 and miR-181), and TFs (such as SRF and E2F1). There are some limitations in present study such as further large-scale clinical sample research and subsequent functional verification do not be performed and only analyse miRNA in correlated genes.
Supplementary
The article’s supplementary files as
Acknowledgments
We would like to thank the Oncomine and LinkedOmics databases for open access to the AML sequencing datasets.
Funding: This study was supported by the funds of National Natural Science Foundation of China (grant No. 81560028 and 81160072).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was approved by the Academic Committee of Guangxi Medical University and conducted according to the principles of the Helsinki Declaration. All the data analyzed in our study were retrieved from published literature, and we confirmed that written informed consent was obtained.
Footnotes
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (http://dx.doi.org/10.21037/atm-20-3296). The authors have no conflicts of interest to declare.
References
- 1.Aldoss I, Marcucci G. More options for older patients with acute myeloid leukemia: venetoclax in combination with low dose cytarabine. Chin Clin Oncol 2019;8:S25. 10.21037/cco.2019.09.03 [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin 2020;70:7-30. 10.3322/caac.21590 [DOI] [PubMed] [Google Scholar]
- 3.Hu L, Gao Y, Shi Z, et al. DNA methylation-based prognostic biomarkers of acute myeloid leukemia patients. Ann Transl Med 2019;7:737. 10.21037/atm.2019.11.122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takebe G, Yarimizu J, Saito Y, et al. A comparative study on the hydroperoxide and thiol specificity of the glutathione peroxidase family and selenoprotein P. J Biol Chem 2002;277:41254-8. 10.1074/jbc.M202773200 [DOI] [PubMed] [Google Scholar]
- 5.Margis R, Dunand C, Teixeira FK, et al. Glutathione peroxidase family–an evolutionary overview. FEBS J 2008;275:3959-70. 10.1111/j.1742-4658.2008.06542.x [DOI] [PubMed] [Google Scholar]
- 6.Brigelius-Flohé R, Maiorino M. Glutathione peroxidases. Biochim Biophys Acta 2013;1830:3289-303. [DOI] [PubMed]
- 7.Jiao Y, Wang Y, Guo S, et al. Glutathione peroxidases as oncotargets. Oncotarget 2017;8:80093. 10.18632/oncotarget.20278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nalkiran I, Turan S, Arikan S, et al. Determination of gene expression and serum levels of MnSOD and GPX1 in colorectal cancer. Anticancer Res 2015;35:255-9. [PubMed] [Google Scholar]
- 9.Metere A, Frezzotti F, Graves CE, et al. A possible role for selenoprotein glutathione peroxidase (GPx1) and thioredoxin reductases (TrxR1) in thyroid cancer: our experience in thyroid surgery. Cancer Cell Int 2018;18:7. 10.1186/s12935-018-0504-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng Y, Xu T, Li S, et al. GPX1, a biomarker for the diagnosis and prognosis of kidney cancer, promotes the progression of kidney cancer. Aging (Albany NY) 2019;11:12165. 10.18632/aging.102555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li F, Dai L, Niu J. GPX2 silencing relieves epithelial-mesenchymal transition, invasion, and metastasis in pancreatic cancer by downregulating Wnt pathway. J Cell Physiol 2019. [Epub ahead of print]. 10.1002/jcp.29391 [DOI] [PubMed] [Google Scholar]
- 12.Naiki T, Naiki-Ito A, Iida K, et al. GPX2 promotes development of bladder cancer with squamous cell differentiation through the control of apoptosis. Oncotarget 2018;9:15847. 10.18632/oncotarget.24627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Y, Cao P, Alshwmi M, et al. GPX2 suppression of H2O2 stress regulates cervical cancer metastasis and apoptosis via activation of the β-catenin-WNT pathway. Onco Targets Ther 2019;12:6639. 10.2147/OTT.S208781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu C, He X, Wu X, et al. Clinicopathological and prognostic significance of GPx2 protein expression in nasopharyngeal carcinoma. Cancer Biomarkers 2017;19:335-40. 10.3233/CBM-160542 [DOI] [PubMed] [Google Scholar]
- 15.Lin Y, Zhang Y, Chen Y, et al. Promoter methylation and clinical significance of GPX3 in esophageal squamous cell carcinoma. Pathol Res Pract 2019;215:152676. 10.1016/j.prp.2019.152676 [DOI] [PubMed] [Google Scholar]
- 16.Zhou C, Pan R, Li B, et al. GPX3 hypermethylation in gastric cancer and its prognostic value in patients aged over 60. Future Oncol 2019;15:1279-89. 10.2217/fon-2018-0674 [DOI] [PubMed] [Google Scholar]
- 17.Zhou C, Hu H, Zheng Z, et al. Association between GPX3 promoter methylation and malignant tumors: A meta-analysis. Pathol Res Pract 2019;215:152443. 10.1016/j.prp.2019.152443 [DOI] [PubMed] [Google Scholar]
- 18.An BC, Choi YD, Oh IJ, et al. GPx3-mediated redox signaling arrests the cell cycle and acts as a tumor suppressor in lung cancer cell lines. PLoS One 2018;13(9). 10.1371/journal.pone.0204170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cai M, Sikong Y, Wang Q, et al. Gpx3 prevents migration and invasion in gastric cancer by targeting NFкB/Wnt5a/JNK signaling. Int J Clin Exp Pathol 2019;12:1194. [PMC free article] [PubMed] [Google Scholar]
- 20.Yi Z, Jiang L, Zhao L, et al. Glutathione peroxidase 3 (GPX3) suppresses the growth of melanoma cells through reactive oxygen species (ROS)-dependent stabilization of hypoxia-inducible factor 1-α and 2-α. J Cell Biochem 2019;120:19124-36. 10.1002/jcb.29240 [DOI] [PubMed] [Google Scholar]
- 21.Zhu X, Wang J, Li L, et al. GPX3 suppresses tumor migration and invasion via the FAK/AKT pathway in esophageal squamous cell carcinoma. Am J Transl Res 2018;10:1908. [PMC free article] [PubMed] [Google Scholar]
- 22.Yang WS, SriRamaratnam R, Welsch ME, et al. Regulation of ferroptotic cancer cell death by GPX4. Cell 2014;156:317-31. 10.1016/j.cell.2013.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Viswanathan VS, Ryan MJ, Dhruv HD, et al. Dependency of a therapy-resistant state of cancer cells on a lipid peroxidase pathway. Nature 2017;547:453-7. 10.1038/nature23007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hangauer MJ, Viswanathan VS, Ryan MJ, et al. Drug-tolerant persister cancer cells are vulnerable to GPX4 inhibition. Nature 2017;551:247-50. 10.1038/nature24297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen Z, Hu T, Zhu S, et al. Glutathione peroxidase 7 suppresses cancer cell growth and is hypermethylated in gastric cancer. Oncotarget 2017;8:54345. 10.18632/oncotarget.17527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peng D, Hu T, Soutto M, et al. Glutathione peroxidase 7 has potential tumour suppressor functions that are silenced by location-specific methylation in oesophageal adenocarcinoma. Gut 2014;63:540-51. 10.1136/gutjnl-2013-304612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pei S, Minhajuddin M, Callahan KP, et al. Targeting aberrant glutathione metabolism to eradicate human acute myelogenous leukemia cells. J Biol Chem 2013;288:33542-58. 10.1074/jbc.M113.511170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Herault O, Hope KJ, Deneault E, et al. A role for GPx3 in activity of normal and leukemia stem cells. J Exp Med 2012;209:895-901. 10.1084/jem.20102386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou JD, Yao DM, Zhang YY, et al. GPX3 hypermethylation serves as an independent prognostic biomarker in non-M3 acute myeloid leukemia. Am J Cancer Res 2015;5:2047. [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 30.Carow CE, Levenstein M, Kaufmann SH, et al. Expression of the Hematopoietic Growth Factor Receptor FLT3 (STK-UFIk2) in Human Leukemias. Blood 1996;87:1089-96. 10.1182/blood.V87.3.1089.bloodjournal8731089 [DOI] [PubMed] [Google Scholar]
- 31.Kayser S, Schlenk RF, Londono MC, et al. Insertion of FLT3 internal tandem duplication in the tyrosine kinase domain-1 is associated with resistance to chemotherapy and inferior outcome. Blood 2009;114:2386-92. 10.1182/blood-2009-03-209999 [DOI] [PubMed] [Google Scholar]
- 32.Döhner H, Estey E, Grimwade D, et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood 2017;129:424-47. 10.1182/blood-2016-08-733196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pratcorona M, Brunet S, Nomdedeu J, et al. Favorable outcome of patients with acute myeloid leukemia harboring a low-allelic burden FLT3-ITD mutation and concomitant NPM1 mutation: relevance to post-remission therapy. Blood 2013;121:2734-8. 10.1182/blood-2012-06-431122 [DOI] [PubMed] [Google Scholar]
- 34.Fröhling S, Schlenk RF, Breitruck J, et al. Prognostic significance of activating FLT3 mutations in younger adults (16 to 60 years) with acute myeloid leukemia and normal cytogenetics: a study of the AML Study Group Ulm. Blood 2002;100:4372-80. 10.1182/blood-2002-05-1440 [DOI] [PubMed] [Google Scholar]
- 35.Rhodes DR, Kalyana-Sundaram S, Mahavisno V, et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia 2007;9:166. 10.1593/neo.07112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 2017;45:W98-102. 10.1093/nar/gkx247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barretina J, Caponigro G, Stransky N, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 2012;483:603-7. 10.1038/nature11003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ghandi M, Huang FW, Jané-Valbuena J, et al. Next-generation characterization of the cancer cell line encyclopedia. Nature 2019;569:503-8. 10.1038/s41586-019-1186-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Squizzato S, Park YM, Buso N, et al. The EBI Search engine: providing search and retrieval functionality for biological data from EMBL-EBI. Nucleic Acids Res 2015;43:W585-8. 10.1093/nar/gkv316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goswami CP, Nakshatri H. PROGgeneV2: enhancements on the existing database. BMC Cancer 2014;14:970. 10.1186/1471-2407-14-970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia 2017;19:649-58. 10.1016/j.neo.2017.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xia J, Benner MJ, Hancock RE. NetworkAnalyst-integrative approaches for protein-protein interaction network analysis and visual exploration. Nucleic Acids Res 2014;42:W167-74. 10.1093/nar/gku443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou G, Soufan O, Ewald J, et al. NetworkAnalyst 3.0: a visual analytics platform for comprehensive gene expression profiling and meta-analysis. Nucleic Acids Res 2019;47:W234-41. 10.1093/nar/gkz240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vasaikar SV, Straub P, Wang J, et al. LinkedOmics: analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res 2018;46:D956-63. 10.1093/nar/gkx1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brigelius-Flohé R, Kipp AP. Physiological functions of GPx2 and its role in inflammation-triggered carcinogenesis. Ann N Y Acad Sci 2012;1259:19-25. 10.1111/j.1749-6632.2012.06574.x [DOI] [PubMed] [Google Scholar]
- 46.Zhuo P, Diamond AM. Molecular mechanisms by which selenoproteins affect cancer risk and progression. Biochim Biophys Acta 2009;1790:1546-54. [DOI] [PMC free article] [PubMed]
- 47.Liu K, Jin M, Xiao L, et al. Distinct prognostic values of mRNA expression of glutathione peroxidases in non-small cell lung cancer. Cancer Manag Res 2018;10:2997. 10.2147/CMAR.S163432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kinowaki Y, Kurata M, Ishibashi S, et al. Glutathione peroxidase 4 overexpression inhibits ROS-induced cell death in diffuse large B-cell lymphoma. Lab Invest 2018;98:609-19. 10.1038/s41374-017-0008-1 [DOI] [PubMed] [Google Scholar]
- 49.Kazi JU, Rupar K, Marhäll A, et al. ABL2 suppresses FLT3-ITD-induced cell proliferation through negative regulation of AKT signaling. Oncotarget 2017;8:12194. 10.18632/oncotarget.14577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li ZW, Sun B, Gong T, et al. GNAI1 and GNAI3 reduce colitis-associated tumorigenesis in mice by blocking IL6 signaling and down-regulating expression of GNAI2. Gastroenterology 2019;156:2297-312. 10.1053/j.gastro.2019.02.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Raymond JR, Appleton KM, Pierce JY, et al. Suppression of GNAI2 message in ovarian cancer. J Ovarian Res 2014;7:6. 10.1186/1757-2215-7-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li J, Wang S, Bai J, et al. Novel role for the Immunoproteasome subunit PSMB10 in angiotensin II-induced atrial fibrillation in mice. Hypertension 2018;71:866-76. 10.1161/HYPERTENSIONAHA.117.10390 [DOI] [PubMed] [Google Scholar]
- 53.El Atat O, Fakih A, El-Sibai M. RHOG Activates RAC1 through CDC42 Leading to Tube Formation in Vascular Endothelial Cells. Cells 2019;8:171. 10.3390/cells8020171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Anding AL, Wang C, Chang TK, et al. Vps13D encodes a ubiquitin-binding protein that is required for the regulation of mitochondrial size and clearance. Curr Biol 2018;28:287-295.e286. 10.1016/j.cub.2017.11.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Oláhová M, Yoon WH, Thompson K, et al. Biallelic mutations in ATP5F1D, which encodes a subunit of ATP synthase, cause a metabolic disorder. Am J Hum Genet 2018;102:494-504. 10.1016/j.ajhg.2018.01.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Su CY, Chang YC, Yang CJ, et al. The opposite prognostic effect of NDUFS1 and NDUFS8 in lung cancer reflects the oncojanus role of mitochondrial complex I. Sci Rep 2016;6:31357. 10.1038/srep31357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen B, Wang S, Ma G, et al. The association of POLR2E rs3787016 polymorphism and cancer risk: a Chinese case–control study and meta-analysis. Biosci Rep 2018;38:BSR20180853. 10.1042/BSR20180853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang B, Wu Q, Wang Z, et al. The promising novel biomarkers and candidate small molecule drugs in kidney renal clear cell carcinoma: Evidence from bioinformatics analysis of high-throughput data. Mol Genet Genomic Med 2019;7:e607. 10.1002/mgg3.607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.He Y, Chen X, Liu H, et al. Matrix-remodeling associated 5 as a novel tissue biomarker predicts poor prognosis in non-small cell lung cancers. Cancer Biomarkers 2015;15:645-51. 10.3233/CBM-150504 [DOI] [PubMed] [Google Scholar]
- 60.Peng L, Li Y, Wei S, et al. LAMA4 activated by Androgen receptor induces the cisplatin resistance in gastric cancer. Biomed Pharmacother 2020;124:109667. 10.1016/j.biopha.2019.109667 [DOI] [PubMed] [Google Scholar]
- 61.Zhou D, Tang W, Su G, et al. PCDH18 is frequently inactivated by promoter methylation in colorectal cancer. Sci Rep 2017;7:2819. 10.1038/s41598-017-03133-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Karlic H, Herrmann H, Varga F, et al. The role of epigenetics in the regulation of apoptosis in myelodysplastic syndromes and acute myeloid leukemia. Crit Rev Oncol/Hematol 2014;90:1-16. 10.1016/j.critrevonc.2013.10.003 [DOI] [PubMed] [Google Scholar]
- 63.Pandolfi A, Stanley RF, Yu Y, et al. PAK1 is a therapeutic target in acute myeloid leukemia and myelodysplastic syndrome. Blood 2015;126:1118-27. 10.1182/blood-2014-12-618801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Patel RK, Weir MC, Shen K, et al. Expression of myeloid Src-family kinases is associated with poor prognosis in AML and influences Flt3-ITD kinase inhibitor acquired resistance. PLoS One 2019;14:e0225887. 10.1371/journal.pone.0225887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang L, Xu J, Yang G, et al. miR-202 inhibits cell proliferation, migration, and invasion by targeting epidermal growth factor receptor in human bladder cancer. Oncol Res 2018;26:949-57. 10.3727/096504018X15149787144385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xu F, Li H, Hu C. MiR-202 inhibits cell proliferation, invasion, and migration in breast cancer by targeting ROCK1 gene. J Cell Biochem 2019;120:16008-18. 10.1002/jcb.28879 [DOI] [PubMed] [Google Scholar]
- 67.Xu Y, Ohms SJ, Li Z, et al. Changes in the expression of miR-381 and miR-495 are inversely associated with the expression of the MDR1 gene and development of multi-drug resistance. PLoS One 2013;8(11). 10.1371/journal.pone.0082062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lee YG, Kim I, Oh S, et al. Small RNA sequencing profiles of mir-181 and mir-221, the most relevant microRNAs in acute myeloid leukemia. Korean J Intern Med 2019;34:178. 10.3904/kjim.2017.102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Su R, Lin H, Zhang X, et al. MiR-181 family: regulators of myeloid differentiation and acute myeloid leukemia as well as potential therapeutic targets. Oncogene 2015;34:3226-39. 10.1038/onc.2014.274 [DOI] [PubMed] [Google Scholar]
- 70.Taylor A, Halene S. The regulatory role of serum response factor pathway in neutrophil inflammatory response. Curr Opin Hematol 2015;22:67. 10.1097/MOH.0000000000000099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Denechaud PD, Fajas L, Giralt A. E2F1, a novel regulator of metabolism. Front Endocrinol (Lausanne) 2017;8:311. 10.3389/fendo.2017.00311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rishi L, Hannon M, Salomè M, et al. Regulation of Trib2 by an E2F1-C/EBPα feedback loop in AML cell proliferation. Blood 2014;123:2389-400. 10.1182/blood-2013-07-511683 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as