Abstract
Knowledge graphs can support many biomedical applications. These graphs represent biomedical concepts and relationships in the form of nodes and edges. In this review, we discuss how these graphs are constructed and applied with a particular focus on how machine learning approaches are changing these processes. Biomedical knowledge graphs have often been constructed by integrating databases that were populated by experts via manual curation, but we are now seeing a more robust use of automated systems. A number of techniques are used to represent knowledge graphs, but often machine learning methods are used to construct a low-dimensional representation that can support many different applications. This representation is designed to preserve a knowledge graph’s local and/or global structure. Additional machine learning methods can be applied to this representation to make predictions within genomic, pharmaceutical, and clinical domains. We frame our discussion first around knowledge graph construction and then around unifying representational learning techniques and unifying applications. Advances in machine learning for biomedicine are creating new opportunities across many domains, and we note potential avenues for future work with knowledge graphs that appear particularly promising.
Keywords: knowledge graphs, Network embeddings, Text mining, Natural language processing, Machine learning, Lterature review
1. Introduction
Graphs are practical resources for many real-world applications. They have been used in social network mining to classify nodes [1] and create recommendation systems [2]. They have also been used in natural language processing to interpret simple questions and use relational information to provide answers [3], [4]. In a biomedical setting, graphs have been used to prioritize genes relevant to disease [5], [6], [7], [8], perform drug repurposing [9] and identify drug-target interactions [10].
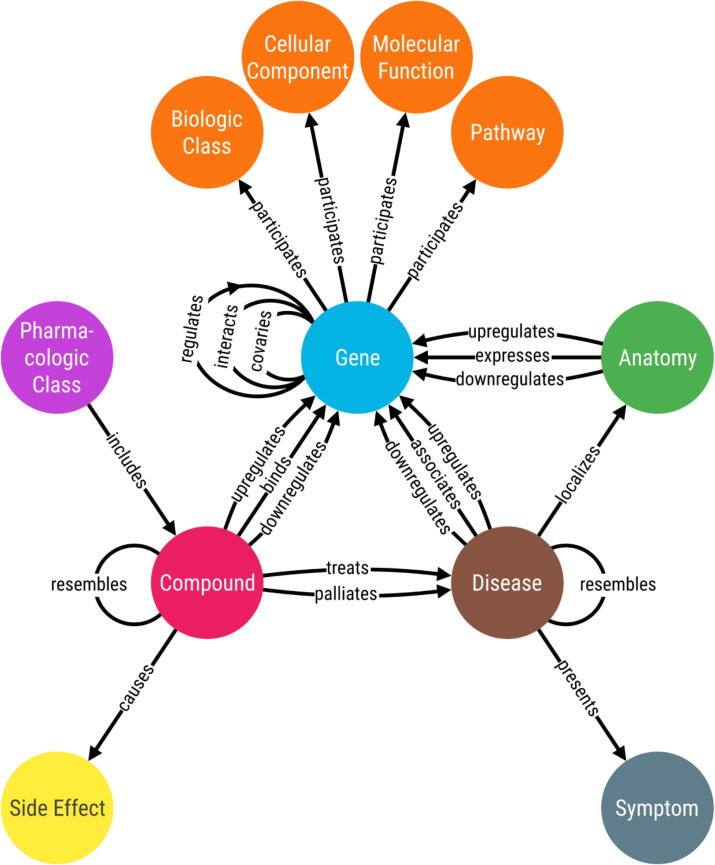
Within a biomedical setting, some graphs can be considered knowledge graphs; although, precisely defining a knowledge graph is difficult because there are multiple conflicting definitions [11]. For this review, we define a biomedical knowledge graph as the following: a resource that integrates one or more expert-derived sources of information into a graph where nodes represent biomedical entities and edges represent relationships between two entities. This definition is consistent with other definitions found in the literature [12], [13], [14], [15], [16], [17], [18]. Often relationships are considered unidirectional (e.g., a compound treats a disease, but a disease cannot treat a compound); however, there are cases where relationships can be considered bidirectional (e.g., a compound resembles another compound, or a gene interacts with another gene). A subset of graphs that meet our definition of a knowledge graph would be unsuitable for applications such as symbolic reasoning [19]; however, we chose a more liberal definition because it has been demonstrated that these broadly defined graphs have numerous uses throughout the literature. For example, Hetionet (Fig. 1) [9] would be considered a biomedical knowledge graph by this definition, and it has been used to identify drug repurposing opportunities [9]. We do not consider databases like DISEASES [20] and DrugBank [21] to be knowledge graphs. Although these resources contain essential information, they do not represent their data in the form of a graph.
Fig. 1.
The metagraph (i.e., schema) of the knowledge graph used in the Rephetio project [9]. The authors of this project refer to their resource as a heterogenous network (i.e., hetnet), and this network meets our definition of a knowledge graph. This resource depicts pharmacological and biomedical information in the form of nodes and edges. The nodes (circles) represent entities and edges (lines) represent relationships that are shared between two entities. The majority of edges in this metagraph are depicted as unidirectional, but some relationships can be considered bidirectional.
Biomedical knowledge graphs are often constructed from manually curated databases [10], [22], [23], [24], [9]. These databases provide previously established information that can be incorporated into a graph. For example, a graph using DISEASES [20] as a resource would have genes and diseases as nodes, while edges added between nodes would represent an association between a gene and a disease. This example shows a single type of relationship; however, there are graphs that use databases with multiple relationships [25], [9]. In addition to manual curation, other approaches have used natural language processing techniques to construct knowledge graphs [26], [27]. One example used a text mining system to extract sentences that illustrate a protein’s interaction with another protein [28]. Once identified, these sentences can be incorporated as evidence to establish an edge in a knowledge graph.
In this review we describe various approaches for constructing and applying knowledge graphs in a biomedical setting. We discuss the pros and cons of constructing a knowledge graph via manually curated databases and via text mining systems. We also compare assorted approaches for applying knowledge graphs to solve biomedical problems. Lastly, we conclude on the practicality of knowledge graphs and point out future applications that have yet to be explored.
2. Building biomedical knowledge graphs
Knowledge graphs can be constructed in many ways using resources such as pre-existing databases or text. Usually, knowledge graphs are constructed using pre-existing databases. These databases are constructed by domain experts using approaches ranging from manual curation to automated techniques, such as text mining. Manual curation is a time-consuming process that requires domain experts to read papers and annotate sentences that assert a relationship. Automated approaches rely on machine learning or natural language processing techniques to rapidly detect sentences of interest. We categorize these automated approaches into the following groups: rule-based extraction, unsupervised machine learning, and supervised machine learning and discuss examples of each type of approach while synthesizing their strengths and weaknesses.
2.1. Constructing databases and manual curation
Database construction dates back all the way to 1956 when the first database contained a protein sequence of the insulin molecule [29]. The process of database construction involves gathering relevant text such as journal articles, abstracts, or web-based text and having curators read the gathered text to detect sentences that implicate a relationship (i.e., relationship extraction). Notable databases constructed by this process can be in found in Table 1. An example database, COSMIC [30] was constructed by a group of domain experts scanning the literature for key cancer related genes. This database contained approximately 35 M entries in 2016 [30] and by 2018 had grown to 45 M entries [31]. Studies have shown that databases constructed in this fashion contain relatively precise data but the recall is low [32], [33], [34], [35], [36], [37], [38]. Low recall happens because the publication rate is too high for curators to keep up [39]. This bottleneck highlights a critical need for future approaches to scale fast enough to compete with the increasing publication rate.
Table 1.
A table of databases that used a form of manual curation to populate entries. Reported number of entities and relationships are relative to the time of publication.
| Database [Reference] | Short Description | Number of Entries | Entity Types | Relationship Types | Method of Population |
|---|---|---|---|---|---|
| BioGrid [52] | A database for major model organisms. It contains genetic and proteomic information. | 572,084 | Genes, Proteins | Protein-Protein interactions | Semi-automatic methods |
| Comparative Toxicogenomics Database [53] | A database that contains manually curated chemical-gene-disease interactions and relationships. | 2,429,689 | Chemicals (Drugs), Genes, Diseases | Drug-Genes, Drug-Disease, Disease-Gene mappings | Manual curation and Automated systems |
| Comprehensive Antibiotic Resistance Database [54] | Manually curated database that contains information about the molecular basis of antimicrobial resistance. | 174,443 | Drugs, Genes, Variants | Drug-Gene, Drug-Variant mappings | Manual curation |
| COSMIC [30] | A database that contains high resolution human cancer genetic information. | 35,946,704 | Genes, Variants, Tumor Types | Gene-Variant Mappings | Manual Curation |
| Entrez-Gene [55] | NCBI’s Gene annotation database that contains information pertaining to genes, gene’s organism source, phenotypes etc. | 7,883,114 | Genes, Species and Phenotypes | Gene-Phenotypes and Genes-Species mappings | Semi-automated curation |
| OMIM [56] | A database that contains phenotype and genotype information | 25,153 | Genes, Phenotypes | Gene-Phenotype mappings | Manual Curation |
| PharmGKB [57] | A database that contains genetic, phenotypic, and clinical information related to pharmacogenomic studies. | 43,112 | Drugs, Genes, Phenotypes, Variants, Pathways | Gene-Phenotypes, Pathway-Drugs, Gene-Variants, Gene-Pathways | Manual Curation and Automated Methods |
| UniProt [58] | A protein–protein interaction database that contains proteomic information. | 560,823 | Proteins, Protein sequences | Protein-Protein interactions | Manual and Automated Curation |
Semi-automatic methods are a way to accelerate the curation process [36], [40], [41], [42], [43], [44], [45]. The first step of these methods is to use an automated system to initially extract sentences from text. This process removes irrelevant sentences, which dramatically decreases the amount of text that curators must sift through. Following the pre-filtering step, curators then approve or reject the remaining sentences. This approach saved curators an average of 2–2.8 h compared to manual efforts [40], [46]. Despite automated systems excelling in identifying sentences for commonly occurring relationships, they tend to miss lesser-known relationships [40]. These systems also have a hard time parsing ambiguous sentences that naturally occur in text, which makes correcting them a challenging task [40]. Given these caveats, future approaches should look into using techniques that simplify sentences to solve the ambiguity issue [47], [48].
Despite the negatives of manual curation, it is still an essential process for extracting relationships from text. This process can be used to generate gold standard datasets that automated systems use for validation [49], [50] and can be used during the training process of these systems (i.e., active learning) [51]. It is important to remember that manual curation alone is precise but results in low recall rates [38]. Future databases should consider initially relying on automated methods to obtain sentences at an acceptable recall level, then incorporate manual curation as a way to fix or remove irrelevant results.
2.2. Text mining for relationship extraction
2.2.1. Rule-based relationship extraction
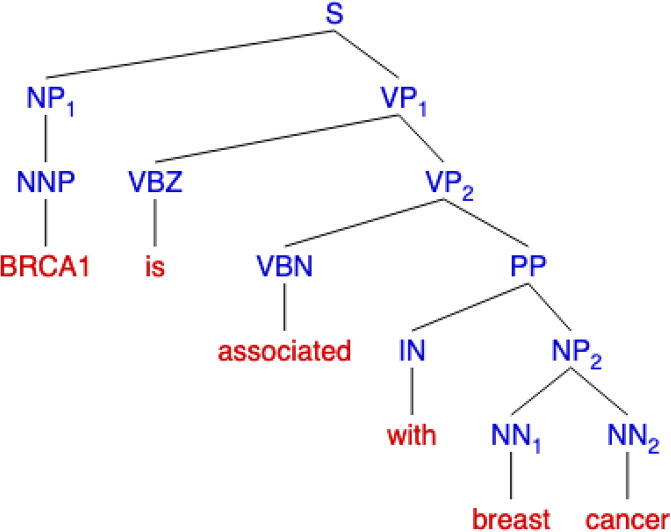
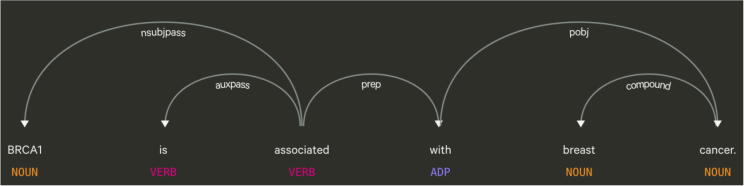
Rule-based extraction consists of identifying essential keywords and grammatical patterns to detect relationships of interest. Keywords are established via expert knowledge or through the use of pre-existing ontologies, while grammatical patterns are constructed via experts curating parse trees. Parse trees are tree data structures that depict a sentence’s grammatical structure and come in two forms: a constituency parse tree (Fig. 2) and a dependency parse tree (Fig. 3). Both trees use part of speech tags, labels that dictate the grammatical role of a word such as noun, verb, adjective, etc., for construction, but represent the information in two different forms. Constituency parse trees break a sentence into subphrases (Fig. 2) while dependency path trees analyze the grammatical structure of a sentence (Fig. 3). Many text mining approaches [59], [60], [61] use such trees to generate features for machine learning algorithms and these approaches are discussed in later sections. In this section we focus on approaches that use rule-based extraction as a primary strategy to detect sentences that allude to a relationship.
Fig. 2.
A visualization of a constituency parse tree using the following sentence: “BRCA1 is associated with breast cancer” [73]. This type of tree has the root start at the beginning of the sentence. Each word is grouped into subphrases depending on its correlating part of speech tag. For example, the word “associated” is a past participle verb (VBN) that belongs to the verb phrase (VP) subgroup.
Fig. 3.
A visualization of a dependency parse tree using the following sentence: “BRCA1 is associated with breast cancer” [74]. For these types of trees, the root begins with the main verb of the sentence. Each arrow represents the dependency shared between two words. For example, the dependency between BRCA1 and associated is nsubjpass, which stands for passive nominal subject. This means that “BRCA1” is the subject of the sentence and it is being referred to by the word “associated”.
Grammatical patterns can simplify sentences for easy extraction [48], [62]. Jonnalagadda et al. used a set of grammar rules inspired by constituency trees to reshape complex sentences with simpler versions [48] and these simplified versions were manually curated to determine the presence of a relationship. By simplifying sentences, this approach achieved high recall, but had low precision [48]. Other approaches used simplification techniques to make extraction easier [63], [64], [65], [66]. Tudor et al. simplified sentences to detect protein phosphorylation events [65]. Their sentence simplifier broke complex sentences that contain multiple protein events into smaller sentences that contain only one distinct event. By breaking these sentences down the authors were able to increase their recall; however, sentences that contained ambiguous directionality or multiple phosphorylation events were too complex for the simplifier. As a consequence, the simplifier missed some relevant sentences [65]. These errors highlight a crucial need for future algorithms to be generalizable enough to handle various forms of complex sentences.
Pattern matching is a fundamental approach used to detect relationship asserting sentences. These patterns can consist of phrases from constituency trees, a set of keywords or some combination of both [36], [67], [68], [69], [70], [71]. Xu et al. designed a pattern matcher system to detect sentences in PubMed abstracts that indicate drug-disease treatments [70]. This system matched drug-disease pairs from ClinicalTrials.gov to drug-disease pairs mentioned in abstracts. This matching process aided the authors in identifying sentences that can be used to create simple patterns, such as “Drug in the treatment of Disease” [70], to match other sentences in a wide variety of abstracts. The authors hand curated two datasets for evaluation and achieved a high precision score of 0.904 and a low recall score of 0.131 [70]. This low recall score was based on constructed patterns being too specific to detect infrequent drug pairs. Besides constituency trees, some approaches used dependency trees to construct patterns [59], [72]. Depending upon the nature of the algorithm and text, dependency trees could be more appropriate than constituency trees and vice versa. The performance difference between the two trees remains as an open question for future exploration.
Rule-based methods provide a basis for many relationship extraction systems. Approaches in this category range from simplifying sentences for easy extraction to identifying sentences based on matched key phrases or grammatical patterns. Both require a significant amount of manual effort and expert knowledge to perform well. A future direction is to develop ways to automate the construction of these hand-crafted patterns, which would accelerate the process of creating these rule-based systems.
2.2.2. Extracting relationships without labels
Unsupervised extractors draw inferences from textual data without the use of annotated labels. These methods involve some form of clustering or statistical calculations. In this section we focus on methods that use unsupervised learning to extract relationships from text.
An unsupervised extractor can exploit the fact that two entities may appear together in text. This event is referred to as co-occurrence and studies that use this phenomenon can be found in Table 2. Two databases DISEASES [20] and STRING [75] were populated using a co-occurrence scoring method on PubMed abstracts, which measured the frequency of co-mention pairs within individual sentences as well as the abstracts themselves. This technique assumes that each individual co-occurring pair is independent from one another. Under this assumption mention pairs that occur more than expected were presumed to implicate the presence of an association or interaction. This approach identified 543,405 disease gene associations [20] and 792,730 high confidence protein–protein interactions [75] but is limited to only PubMed abstracts.
Table 2.
Table of approaches that mainly use a form of co-occurrence.
| Study | Relationship of Interest |
|---|---|
| CoCoScore [79] | Protein-Protein Interactions, Disease-Gene and Tissue-Gene Associations |
| Rastegar-Mojarad et al. [80] | Drug Disease Treatments |
| CoPub Discovery [81] | Drug, Gene and Disease interactions |
| Westergaard et al. [76] | Protein-Protein Interactions |
| DISEASES [20] | Disease-Gene associations |
| STRING [82] | Protein-Protein Interactions |
| Singhal et al. [83] | Genotype-Phenotype Relationships |
Full text articles are able to dramatically enhance relationship detection [76], [77]. Westergaard et al. used a co-occurrence approach, similar to DISEASES [20] and STRING [75], to mine full articles for protein–protein interactions and other protein related information [76]. The authors discovered that full text provided better prediction power than using abstracts alone, which suggests that future text mining approaches should consider using full text to increase detection power.
Unsupervised extractors often treat different biomedical relationships as multiple isolated problems. An alternative to this perspective is to capture all different types at once. Clustering is an approach that performs simultaneous extraction. Percha et al. used a biclustering algorithm on generated dependency parse trees to group sentences within PubMed abstracts [78]. Each cluster was manually curated to determine which relationship each group represented. This approach captured 4,451,661 dependency paths for 36 different groups [78]. Despite the success, this approach suffered from technical issues such as dependency tree parsing errors. These errors resulted in some sentences not being captured by the clustering algorithm [78]. Future clustering approaches should consider simplifying sentences to prevent this type of issue.
Overall unsupervised methods provide a means to rapidly extract relationship asserting sentences without the need of annotated text. Approaches in this category range from calculating co-occurrence scores to clustering sentences and provide a generalizable framework that can be used on large repositories of text. Full text has already been shown to meaningfully improve the performance of methods that aim to infer relationships using cooccurrences [76], and we should expect similar benefits for machine learning approaches. Furthermore, we expect that simplifying sentences would improve unsupervised methods and should be considered as an initial preprocessing step.
2.2.3. Supervised relationship extraction
Supervised extractors use labeled sentences to construct generalized patterns that bisect positive examples (sentences that allude to a relationship) from negative ones (sentences that do not allude to a relationship). Most of these approaches have flourished due to pre-labelled publicly available datasets (Table 3). These datasets were constructed by curators for shared open tasks [84], [85] or as a means to provide the scientific community with a gold standard [85], [86], [87]. Approaches that use these available datasets range from using linear classifiers such as support vector machines (SVMs) to non-linear classifiers such as deep learning techniques. The rest of this section discusses approaches that use supervised extractors to detect relationship asserting sentences.
Table 3.
A set of publicly available datasets for supervised text mining.
| Dataset | Type of Sentences |
|---|---|
| AIMed [50] | Protein-Protein Interactions |
| BioInfer [123] | Protein-Protein Interactions |
| LLL [124] | Protein-Protein Interactions |
| IEPA [125] | Protein-Protein Interactions |
| HPRD5 [86] | Protein-Protein Interactions |
| EU-ADR [49] | Disease-Gene Associations |
| BeFree [90] | Disease-Gene Associations |
| CoMAGC [87] | Disease-Gene Associations |
| CRAFT [126] | Disease-Gene Associations |
| Biocreative V CDR [85] | Compound induces Disease |
| Biocreative IV ChemProt [84] | Compound-Gene Bindings |
Some supervised extractors involve the mapping of textual input into a high dimensional space. SVMs are a type of classifier that can accomplish this task with a mapping function called a kernel [61], [88]. These kernels take information such as a sentence’s dependency tree [59], [60], part of speech tags [61] or even word counts [88] and map them onto a dense feature space. Within this space, these methods construct a hyperplane that separates sentences in the positive class (illustrates a relationship) from the negative class (does not illustrate a relationship). Kernels can be manually constructed or selected to cater to the relationship of interest [60], [61], [88], [88]. Determining the correct kernel is a nontrivial task that requires expert knowledge to be successful. In addition to single kernel methods, a recent study used an ensemble of SVMs to extract disease-gene associations [89]. This ensemble outperformed notable disease-gene association extractors [72], [90] in terms of precision, recall and F1 score. Overall, SVMs have been shown to be beneficial in terms of relationship mining; however, major focus has shifted to utilizing deep learning techniques which can perform non-linear mappings of high dimensional data.
Deep learning is an increasingly popular class of techniques that can construct their own features within a high dimensional space [91], [92]. These methods use different forms of neural networks, such as recurrent or convolutional neural networks, to perform classification.
Recurrent neural networks (RNN) are designed for sequential analysis and use a repeatedly updating hidden state to make predictions. An example of a recurrent neural network is a long short-term memory (LSTM) network [93]. Cocos et al. [94] used a LSTM to extract drug side effects from de-identified twitter posts, while Yadav et al. [95] used an LSTM to extract protein–protein interactions. Others have also embraced LSTMs to perform relationship extraction [94], [96], [97], [98], [99]. Despite the success of these networks, training can be difficult as these networks are highly susceptible to vanishing and exploding gradients [100], [101]. One proposed solution to this problem is to clip the gradients while the neural network trains [102]. Besides the gradient problem, these approaches only peak in performance when the datasets reach at least tens of thousands of data points [103].
Convolutional neural networks (CNNs), which are widely applied for image analysis, use multiple kernel filters to capture small subsets of an overall image [92]. In the context of text mining an image is replaced with words within a sentence mapped to dense vectors (i.e., word embeddings) [104], [105]. Peng et al. used a CNN to extract sentences that mentioned protein–protein interactions [106] and Zhou et al. used a CNN to extract chemical-disease relations [107]. Others have used CNNs and variants of CNNs to extract relationships from text [108], [109], [110]. Just like RNNs, these networks perform well when millions of labeled examples are present [103]; however, obtaining these large datasets is a non-trivial task. Future approaches that use CNNs or RNNs should consider solutions to obtaining these large quantities of data through means such as weak supervision [111], semi-supervised learning [112] or using pre-trained networks via transfer learning [113], [114].
Semi-supervised learning [112] and weak supervision [111] are techniques that can rapidly construct large datasets for machine learning classifiers. Semi-supervised learning trains classifiers by combining labeled data with unlabeled data. For example, one study used a variational auto encoder with a LSTM network to extract protein–protein interactions from PubMed abstracts and full text [115]. This is an elegant solution for the small dataset problem but requires labeled data to start. This dependency makes finding under-studied relationships difficult as one would need to find or construct examples of the missing relationships at the start.
Weak or distant supervision takes a different approach by using noisy or even erroneous labels to train classifiers [111], [116], [117], [118]. Under this paradigm, sentences are labeled based on their mention pair being present (positive) or absent (negative) in a database and, once labeled, a machine learning classifier can be trained to extract relationships from text [111]. For example, Thomas et al. [119] used distant supervision to train a SVM to extract sentences mentioning protein–protein interactions (PPI). Their SVM model achieved comparable performance against a baseline model; however, the noise generated via distant supervision was difficult to eradicate [119]. A number of efforts have focused on combining distant supervision with other types of labeling strategies to mitigate the negative impacts of noisy knowledge bases [120], [121], [122]. Nicholson et al. [110] found that, in some circumstances, these strategies can be reused across different types of biomedical relationships to learn a heterogeneous knowledge graph in cases where those relationships describe similar physical concepts. Combining distant supervision with other types of labeling strategies remains an active area of investigation with numerous associated challenges and opportunities. Overall, semi-supervised learning and weak supervision provide promising results in terms of relationship extraction and future approaches should consider using these paradigms to train machine learning classifiers.
3. Applying knowledge graphs to biomedical challenges
Knowledge graphs can help researchers tackle many biomedical problems such as finding new treatments for existing drugs [9], aiding efforts to diagnose patients [127] and identifying associations between diseases and biomolecules [128]. In many cases, solutions rely on representing knowledge graphs in a low dimensional space, which is a process called representational learning. The goal of this process is to retain and encode the local and/or global structure of a knowledge graph that is relevant to the problem while transforming the graph into a representation that can be readily used with machine learning methods to build predictors. In the following sections we review methods that construct a low dimensional space (Unifying Representational Learning Techniques) and discuss applications that use this space to solve biomedical problems (Unifying Applications).
3.1. Unifying representational learning techniques
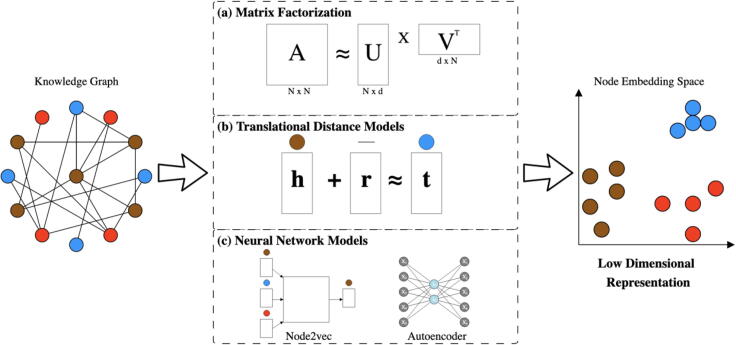
Mapping high dimensional data into a low dimensional space greatly improves modeling performance in fields such as natural language processing [104], [105] and image analysis [129]. The success of these approaches served as rationale for a sharper focus on representing knowledge graphs in a low dimensional space [130]. Methods of this class are designed to capture the essence of a knowledge graph in the form of dense vectors [131], [132]. These vectors are often assigned to nodes in a graph [133], but edges can be assigned as well [134]. Techniques that construct a low dimensional space often require information on how nodes are connected with one another [135], [136], [137], [138], while other approaches can work directly with the edges themselves [139]. Once this space has been constructed, machine learning techniques can utilize the space for downstream analyses such as classification or clustering. We group techniques that construct this space into the following three categories: matrix factorization, translational distance models, and neural network models (Fig. 4).
Fig. 4.
Pipeline for representing knowledge graphs in a low dimensional space. Starting with a knowledge graph, this space can be generated using one of the following options: Matrix Factorization (a), Translational Models (b) or Neural Network Models (c). The output of this pipeline is an embedding space that clusters similar node types together.
3.1.1. Matrix factorization
Matrix factorization is a class of techniques that use linear algebra to map high dimensional data into a low dimensional space. This projection is accomplished by decomposing a matrix into a set of small rectangular matrices (Fig. 4 (a)). Notable methods for matrix decomposition include Isomap [140], Laplacian eigenmaps [132] and Principal Component Analysis (PCA) [141]/Singular Vector Decomposition (SVD) [131]. These methods were designed to be used on many different types of data; however, we discuss their use in the context of representing knowledge graphs in a low dimensional space and focus particularly on SVD and laplacian eigenmaps.
SVD [131] is an algorithm that uses matrix factorization to portray knowledge graphs in a low dimensional space. The input for this algorithm is an adjacency matrix (), which is a square matrix where rows and columns represent nodes and each entry is a binary representation of the presence of an edge between two nodes. is constructed based on the knowledge graph’s structure itself and collapses all edges between two nodes into one unique entity. Following construction, is decomposed into the following three parts: a square matrix and a set of two small rectangular matrices and . Values within are called singular values, which are akin to eigenvalues [131]. Each row in and each column in represents nodes within a low dimensional space [131], [141]. In practice, is usually used to represent nodes in a knowledge graph and can be used as input for machine learning classifiers to perform tasks such as link prediction or node classification [142]; however, has also been used [131], [143]. Typically, matrix factorization algorithms such as SVD are used for recommendation systems via collaborative filtering [144]; however, this technique can also provide a standalone baseline for other relational learning approaches [142].
Laplacian eigenmaps assume there is low dimensional structure in a high dimensional space and preserves this structure when projecting data into a low dimensional space [132]. The first step of this technique is to preserve the low dimensional structure by representing data in the form of a graph where nodes are datapoints and edges are the distance between two points. Knowledge graphs already provide this representation, so no additional processing is necessary at this stage. The second step of this technique is to obtain both an adjacency matrix () and a degree matrix () from the graph representation. A degree matrix is a diagonal matrix where each entry represents the number of edges connected to a node. The adjacency and degree matrices are converted into a laplacian matrix (), which is a matrix that shares the same properties as the adjacency matrix. The laplacian matrix is generated by subtracting the adjacency matrix from the degree matrix () and, once constructed, the algorithm uses linear algebra to calculate the laplacian’s eigenvalues and eigenvectors (). The generated eigenvectors represent the knowledge graph’s nodes represented in a low dimensional space [132]. Other efforts have used variants of this algorithm to construct low dimensional representations of knowledge graphs [135], [136], [145]. Typically, eigenmaps work well when knowledge graphs have a sparse number of edges between nodes but struggle when presented with denser networks [142], [145], [146]. An open area of exploration is to adapt these methods to accommodate knowledge graphs that have a large number of edges.
Matrix factorization is a powerful technique that represents high dimensional data in a low dimensional space. The representation of a knowledge graph in this reduced space does not meet our definition of a knowledge graph; however, this representation supports many use cases including similarity-based (e.g., cosine similarity [147]) and machine learning applications. Common matrix factorization approaches involve using SVD, Laplacian eigenmaps or variants of the two to decompose matrices into smaller rectangular forms. Regarding knowledge graphs, the adjacency matrix () is the typical matrix that gets decomposed, but the laplacian matrix () can be used as well. Despite reported success, the dependence on matrices creates an issue of scalability as matrices of large networks may reach memory limitations. Furthermore, the approaches we discussed consider all edge types as equivalent. These limitations could be mitigated by new approaches designed to accommodate multiple node and edge types separately.
3.1.2. Translational distance models
Translational distance models treat edges in a knowledge graph as linear transformations. For example, one such algorithm, TransE [134], treats every node-edge pair as a triplet with head nodes represented as , edges represented as , and tail nodes represented as . These representations are combined into an equation that mimics the iconic word vectors translations () from the word2vec model [105]. The described equation is shown as follows: . Starting at the head node (), one adds the edge vector () and the result should be the tail node (). TransE optimizes vectors for , , , while guaranteeing the global equation () is satisfied [134]. A caveat to the TransE approach is that it forces relationships to have a one to one mapping, which may not be appropriate for all relationship types.
Wang et al. attempted to resolve the one to one mapping issue by developing the TransH model [148]. TransH treats relations as hyperplanes rather than a regular vector and projects the head () and tail () nodes onto a hyperplane. Following this projection, a distance vector () is calculated between the projected head and tail nodes. Finally, each vector is optimized while preserving the global equation: [148]. Other efforts have built off of the TransE and TransH models [149], [150]. In the future, it may be beneficial for these models to incorporate other types of information such as edge confidence scores, textual information, or edge type information when optimizing these distance models.
3.1.3. Neural networks
Neural networks are a class of machine learning models inspired by the concept of biological neural networks [151]. These networks are reputable for making non-linear transformations of high dimensional data to solve classification and regression problems [151]. In the context of knowledge graphs, the most commonly used structures are based on word2vec [104], [105]. The word2vec term applies to a set of conceptually related approaches that are widely used in the natural language processing field. The goal of word2vec is to project words onto a low dimensional space that preserves their semantic meaning. Strategies for training word2vec models use one of two neural network architectures: skip-gram and continuous bag of words (CBOW). Both models are feed-forward neural networks, but CBOW models are trained to predict a word given its context while skip-gram models are trained to predict the context given a word [104], [105]. Once training is completed, words will be associated with dense vectors that downstream models, such as feed forward networks or recurrent networks, can use for input.
Deepwalk is an early method that represents knowledge graphs in a low dimensional space [152]. The first step of this method is to perform a random walk along a knowledge graph. During the random walk, every generated sequence of nodes is recorded and treated as a sentence in word2vec [104], [105]. After every node has been processed, a skip-gram model is trained to predict the context of each node thereby constructing a low dimensional representation of a knowledge graph [152]. A limitation for deepwalk is that the random walk cannot be controlled, so every node has an equal chance to be reached. Grover and Leskovec demonstrated that this limitation can hurt performance when classifying edges between nodes and developed node2vec as a result [133]. Node2vec operates in the same fashion as deepwalk; however, this algorithm specifies a parameter that lets the random walk be biased when traversing nodes [133]. A caveat to both deepwalk and node2vec is that they ignore information such as edge type and node type. Various approaches have evolved to fix this limitation by incorporating node, edge and even path types when representing knowledge graphs in a low dimensional space [153], [154], [155], [156]. An emerging area of work is to develop approaches that capture both the local and global structure of a graph when constructing this low dimensional space.
Though word2vec is the most common framework used to represent graphs, neural networks are sometimes designed to use the adjacency matrix as input [104], [105]. These approaches use models called autoencoders [157], [158], [159]. Autoencoders are designed to map input into a low dimensional space and then back to a reconstruction of the same input [160], [161]. It is possible to layer on additional objectives by modifying the loss function to take into account criteria above and beyond reconstruction loss [162], [163]. In the context of knowledge graphs, the generated space correlates nodes with dense vectors that capture a graph’s connectivity structure [157], [158], [159]. Despite the high potential of autoencoders, this method relies on an adjacency matrix for input which can run into scalability issues as a knowledge graph asymptotically increases in size [164]. Plus, Khosla et al. discovered that approaches akin to node2vec outperformed algorithms using autoencoders when undergoing link prediction and node classification [164].
Overall, the performance of neural network models largely depends upon the structure of nodes and edges within a knowledge graph [164]. Furthermore, when these approaches are used only nodes are explicitly represented by these vectors. This means a represented knowledge graph no longer meets our definition of a knowledge graph; however, this representation can make it more suitable for many biomedical applications. Future areas of exploration should include hybrid models that use both node2vec and autoencoders to construct complementary low dimensional representations of knowledge graphs.
3.2. Unifying applications
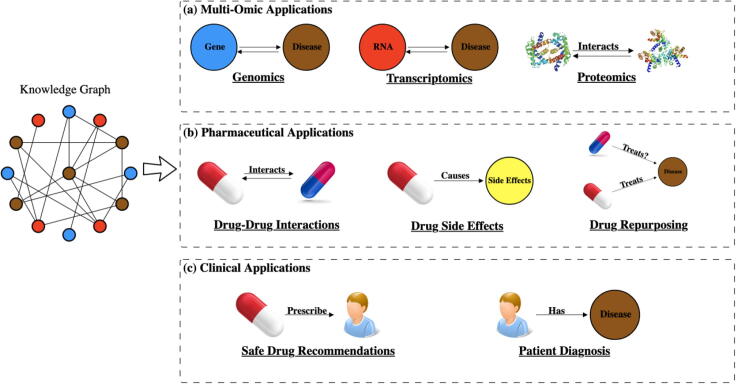
Knowledge graphs have been applied to many biomedical challenges ranging from identifying proteins’ functions [165] to prioritizing cancer genes [166] to recommending safer drugs for patients [167], [168] (Fig. 5). In this section we review how knowledge graphs are applied in biomedical settings and put particular emphasis on an emerging set of techniques that represent knowledge graphs in a low dimensional space.
Fig. 5.
Overview of various biomedical applications that make use of knowledge graphs. Categories consist of: (a) Multi-Omic applications, (b) Pharmaceutical Applications and (c) Clinical Applications.
3.2.1. Multi-omic applications
Multi-omic applications employ knowledge graphs to study the genome, how genes are expressed in the transcriptome, and how the products of those transcripts interact in the proteome. These graphs are used to establish connections between -omic entities as well as diseases. Tasks in this context include gene-symptom prioritization [169], protein–protein interaction prediction [170], [171] and detecting miRNA-disease associations [128]. We focus specifically on multi-omic applications that represent knowledge graphs in a low dimensional space to make connections.
Recommendation systems make use of knowledge graphs to establish links between RNA with disease and proteins with other proteins. Shen et al. used an algorithm called collaborative filtering to establish an association between miRNA and diseases [128]. The authors constructed a miRNA-Disease network using the Human MicroRNA Disease database (HMDD) [172] and generated an adjacency matrix with the rows representing miRNA and the columns representing diseases. This matrix was decomposed into small rectangular matrices using SVD, then these small matrices were used to calculate similarity scores between miRNAs and diseases. High scores implied a high likelihood that a given miRNA had an association with a given disease [128]. Other approaches built off of Shen et al.’s work by incorporating novel ways to perform matrix factorization [173], [174], [175] or by integrating machine learning models in conjunction with matrix factorization [176]. These approaches achieved high area under the receiver operating curve (AUROC), but new discoveries have been hard to validate as experiments in this space are costly and time consuming at best [128]. Apart from miRNA, collaborative filtering has been used to predict protein–protein interactions [170], [171], [177]. Although extensive validation of newly generated candidates may be impractical, it would be helpful to see future efforts in this space include a blinded literature search for prioritized and randomly selected candidates as part of the standard evaluation pipeline.
Applications of neural network models have mainly used the node2vec model [133] or variants of it. Yang et al. used node2vec to create a recommendation system to infer associations between genes and disease symptoms [169]. The authors constructed a gene-disease symptom knowledge graph by combining two bipartite graphs: genes with diseases and diseases with disease symptoms. The generated graph was embedded via node2vec and similarity scores were calculated for every gene-symptom pair in the graph. High scores implied a high likelihood of an association [169]. This approach outperformed methods that didn’t use a knowledge graph; however, validation was difficult as it involved manual curation of the literature [169]. Similar approaches used variants of node2vec to predict gene-disease associations [8], [178], [179] analyze RNA-seq data [180] and infer novel protein information [165], [181], [182], [183].
Knowledge graphs benefited the multi-omics field as a resource for generating novel discoveries. Most approaches to date use matrix factorization and node2vec to project knowledge graph into a low dimensional space, while translational models (Fig. 4 (b)) may be an untapped resource that could aid future efforts. Another area of exploration could be incorporating multiple sources of information such as compounds, anatomic locations or genetic pathways to improve the specificity of findings (i.e., to predict that a protein–protein interaction happens in a specific cell type or tissue).
3.2.2. Pharmaceutical applications
There are a multitude of examples where knowledge graphs have been applied to identify new properties of drugs. Tasks in this field involve predicting drugs interacting with other drugs [184], identifying molecular targets a drug might interact with [185] and identifying new disease treatments for previously established drugs [186]. In this section we concentrate on applications that apply these graphs to discover new properties of drugs and focus on approaches that use these graphs in a low-dimensional space.
Similar to multi-omic applications, recommendation systems have utilized knowledge graphs to infer novel links between drugs and diseases. Dai et al. used collaborative filtering to infer drug-disease associations [185]. The authors constructed a drug-disease network by integrating two bipartite networks: a drug-gene interaction network and a disease-gene interaction network. They integrated both networks under the assumption that drugs associated with a disease interact with the same gene of interest. Following construction, the authors generated an adjacency matrix where rows represent drugs and columns represent diseases. This matrix was decomposed into two small rectangular matrices and these matrices were used to calculate similarity scores between all drugs and all diseases. High values implied a high chance of an association [185]. Related approaches used this technique to infer drug-target interactions [187], [188], [189] and drug-disease treatments [190], [191], [192], [193], [194]. In spite of reported success, these approaches are limited to the drugs and diseases contained in the graph. Combining these approaches with representations of chemical structures might make it possible to one day make predictions about novel compounds.
Applications that use neural network models have used node2vec [195], [196] and autoencoders [197], [198] approaches to represent knowledge graphs in a low dimensional space. Zong et al. used a node2vec-like model to predict drug-target associations [195]. The authors constructed a disease-target-disease network using drug centered databases: Drugbank [199] and Diseasome [200]. Next, the authors applied a random walk to the graph and trained a skip-gram model to generate a low dimensional representation of the graph. Lastly, the authors constructed a similarity metric that used this space to rank how similar drugs are to their targets [195]. A limitation to this approach is that their graph is missing information such as pharmacological class or drug chemical structure that could improve prediction performance. Overall, neural networks provide a robust set of techniques that have been shown to outperform most linear approaches in this context [201], [202].
Applications that discover new properties of drugs have benefited from using knowledge graphs as a resource. Most methods to date use matrix factorization and neural network models to produce a low-dimensional representation. Due to the success of neural networks [201], [202] much of the field’s focus has shifted to these techniques; however, a possible improvement is to use an ensemble of neural network models and linear methods to improve performance. Another potential avenue for future work would be to incorporate entity-specific hierarchical information or similarity information to improve detection power. For drugs, this could include pharmaceutical classes or chemical structure similarities.
3.2.3. Clinical applications
Clinical applications that use knowledge graphs are in early stages of development, but the long-term goal is to use analyses of these graphs to aid patient care. Typically, graphs for these applications are constructed from electronic health records (EHR): nodes represent patients, drugs and diseases while edges represent relationships such as a patient being prescribed a treatment or a patient being diagnosed with a disease [203], [204], [205], [26]. Tasks within this field range from improving patient diagnoses [206], [207] to recommending safer drugs for patients [167], [207]. We briefly discuss efforts that use knowledge graphs to accomplish such tasks.
Early work in this field applied translational models (Fig. 4 (b)) to knowledge graphs with the goal of recommending safe drugs. Wang et al. used a variant of the TransH [148] model to create such a system for patients [167]. They constructed a disease-patient-drug network by integrating a patient-disease bipartite network with a patient-drug bipartite network. Every node in the newly constructed graph was embedded while satisfying the following equation: . Following the embedding step, the authors formulated their own similarity metric that selected drug combinations with a low number of interactions [167]. Researchers in [150] applied a similar variant of the TransH model to a medical knowledge graph and evaluated their model for link prediction rather than patient recommendation.
In contrast with most applications where node2vec and autoencoder models have become established, this field have focused on using graph attention models [208]. These models mimic machine translation models [209] and aim to simultaneously represent knowledge graphs in a low dimensional space and perform the task at hand. Choi et al. used a graph attention model to predict patient diagnoses [127]. The authors constructed a directed graph using medical concepts from patient EHR data. This directed graph was fed into a graph attention network and then used to predict a patient’s likelihood of heart failure [127]. Other approaches have used graph attention models to perform clinical tasks such as drug safety recommendations [168] and patient diagnoses [210].
Knowledge graphs have shown promising results when used for clinical applications; however, there is still room for improvement. Most approaches have run into the common problem of missing data within EHR [127], [167], [168]. Future directions for the field consist of designing algorithms that can fill in this missing data gap or construct models that can take missing data into account.
4. Conclusion
Knowledge graphs are becoming widely used in biomedicine, and we expect their use to continue to grow. At the moment, most are constructed from databases derived from manual curation or from co-occurrences in text. We expect that machine learning approaches will play a key role in quickly deriving new findings from these graphs. Representing these knowledge graphs in a low dimensional space that captures a graph’s local and global structure can enable many downstream machine learning analyses, and methods to capture this structure are an active area of research.
As with any field, rigorous evaluation that can identify key factors that drive success is critical to moving the field forward. In regard to knowledge graphs, evaluation remains difficult. Experiments in this context require a significant amount of time and consequently resources [128], [169]. Moving from open ended and uncontrolled evaluations that consist of describing findings that are consistent with the literature to blinded evaluations of the literature that corroborate predictions and non-predictions would be a valuable first step. There are also well-documented biases related to node degree and degree distribution that must be considered for accurate evaluation [211]. Furthermore, the diversity of applications hinders the development of a standardized set of expected evaluations.
We anticipate that a fruitful avenue of research will be techniques that can produce low dimensional representations of knowledge graphs which distinguish between multiple node and edge types. There are many different sources of bias that lead to spurious edges or incompleteness, and modeling these biases may support better representations of knowledge graphs. It is a promising time for research into the construction and application of knowledge graphs. The peer reviewed literature is growing at an increasing rate and maintaining a complete understanding is becoming increasingly challenging for scientists. One path that scientists can take to maintain awareness is to become hyper-focused on specific areas of knowledge graph literature. If advances in how these graphs are constructed, represented and applied can enable the linking of fields, we may be able to savor the benefits of this detailed knowledge without losing the broader contextual links.
Funding
David Nicholson Funded by The Gordon and Betty Moore Foundation (GBMF4552) and the National Institutes of Health (T32 HG000046)
Casey S. Greene Funded by The Gordon and Betty Moore Foundation (GBMF4552) and the National Institutes of Health (R01 HG010067).
CRediT authorship contribution statement
David N. Nicholson: Conceptualization, Funding acquisition, Investigation, Writing - original draft, Visualization. Casey S. Greene: Conceptualization, Funding acquisition, Supervision, Writing - review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2020.05.017.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Node Classification in Social Networks Smriti Bhagat, Graham Cormode, S. Muthukrishnan Social Network Data Analytics (2011) https://doi.org/fjj48w DOI: 10.1007/978-1-4419-8462-3_5
- 2.Network Embedding Based Recommendation Method in Social Networks Yufei Wen, Lei Guo, Zhumin Chen, Jun Ma Companion of the The Web Conference 2018 on The Web Conference 2018 - WWW ’18 (2018) https://doi.org/gf6rtt DOI: 10.1145/3184558.3186904
- 3.Open Question Answering with Weakly Supervised Embedding Models Antoine Bordes, Jason Weston, Nicolas Usunier arXiv (2014-04-16) https://arxiv.org/abs/1404.4326v1
- 4.Neural Network-based Question Answering over Knowledge Graphs on Word and Character Level Denis Lukovnikov, Asja Fischer, Jens Lehmann, Sören Auer Proceedings of the 26th International Conference on World Wide Web (2017-04-03) https://doi.org/gfv8hp DOI: 10.1145/3038912.3052675
- 5.Towards integrative gene prioritization in Alzheimer’s disease. Jang H Lee, Graciela H Gonzalez Pacific Symposium on Biocomputing. Pacific Symposium on Biocomputing (2011) https://www.ncbi.nlm.nih.gov/pubmed/21121028 DOI: 10.1142/9789814335058_0002 · PMID: 21121028 [DOI] [PubMed]
- 6.PhenoGeneRanker: A Tool for Gene Prioritization Using Complete Multiplex Heterogeneous Networks Cagatay Dursun, Naoki Shimoyama, Mary Shimoyama, Michael Schläppi, Serdar Bozdag Cold Spring Harbor Laboratory (2019-05-27) https://doi.org/gf6rtr DOI: 10.1101/651000
- 7.Biological Random Walks: Integrating heterogeneous data in disease gene prioritization Michele Gentili, Leonardo Martini, Manuela Petti, Lorenzo Farina, Luca Becchetti 2019 IEEE Conference on Computational Intelligence in Bioinformatics and Computational Biology (CIBCB) (2019-07) https://doi.org/gf6rts DOI: 10.1109/cibcb.2019.8791472
- 8.Semantic Disease Gene Embeddings (SmuDGE): phenotype-based disease gene prioritization without phenotypes Mona Alshahrani, Robert Hoehndorf Bioinformatics (2018-09-01) https://doi.org/gd9k8n DOI: 10.1093/bioinformatics/bty559 · PMID: 30423077 · PMCID: PMC6129260 [DOI] [PMC free article] [PubMed]
- 9.Systematic integration of biomedical knowledge prioritizes drugs for repurposing Daniel Scott Himmelstein, Antoine Lizee, Christine Hessler, Leo Brueggeman, Sabrina L Chen, Dexter Hadley, Ari Green, Pouya Khankhanian, Sergio E BaranzinieLife (2017-09-22) https://doi.org/cdfk DOI: 10.7554/elife.26726 · PMID: 28936969 · PMCID: PMC5640425 [DOI] [PMC free article] [PubMed]
- 10.Assessing Drug Target Association Using Semantic Linked Data Bin Chen, Ying Ding, David J. Wild PLoS Computational Biology (2012-07-05) https://doi.org/rn6 DOI: 10.1371/journal.pcbi.1002574 · PMID: 22859915 · PMCID: PMC3390390 [DOI] [PMC free article] [PubMed]
- 11.Towards a definition of knowledge graphs Lisa Ehrlinger, Wolfram Wöß SEMANTiCS (2016).
- 12.Knowledge graph refinement: A survey of approaches and evaluation methods Heiko Paulheim Semantic Web (2016-12-06) https://doi.org/gc9zzx DOI: 10.3233/sw-160218
- 13.Knowledge Graphs and Knowledge Networks: The Story in Brief Amit Sheth, Swati Padhee, Amelie Gyrard, Amit Sheth IEEE Internet Computing (2019-07-01) https://doi.org/ggtmq6 DOI: 10.1109/mic.2019.2928449
- 14.A review: Knowledge reasoning over knowledge graph Xiaojun Chen, Shengbin Jia, Yang Xiang Expert Systems with Applications (2020-03) https://doi.org/ggdq8x DOI: 10.1016/j.eswa.2019.112948
- 15.Privacy Inference on Knowledge Graphs: Hardness and Approximation Jianwei Qian, Shaojie Tang, Huiqi Liu, Taeho Jung, Xiang-Yang Li 2016 12th International Conference on Mobile Ad-Hoc and Sensor Networks (MSN) (2016-12) https://doi.org/ggtjgz DOI: 10.1109/msn.2016.030
- 16.A Review of Relational Machine Learning for Knowledge Graphs Maximilian Nickel, Kevin Murphy, Volker Tresp, Evgeniy Gabrilovich Proceedings of the IEEE (2016-01) https://doi.org/f75f5k DOI: 10.1109/jproc.2015.2483592
- 17.Yago Fabian M. Suchanek, Gjergji Kasneci, Gerhard Weikum Proceedings of the 16th international conference on World Wide Web - WWW ’07 (2007) https://doi.org/c427cr DOI: 10.1145/1242572.1242667
- 18.Knowledge Graph Embedding: A Survey of Approaches and Applications Quan Wang, Zhendong Mao, Bin Wang, Li Guo IEEE Transactions on Knowledge and Data Engineering (2017-12-01) https://doi.org/gcj4mp DOI: 10.1109/tkde.2017.2754499
- 19.Symbolic Artificial Intelligence and Numeric Artificial Neural Networks: Towards a Resolution of the Dichotomy Vasant Honavar The Springer International Series In Engineering and Computer Science https://doi.org/c6ndzz DOI: 10.1007/978-0-585-29599-2_11
- 20.DISEASES: Text mining and data integration of disease–gene associations Sune Pletscher-Frankild, Albert Pallejà, Kalliopi Tsafou, Janos X. Binder, Lars Juhl Jensen Methods (2015-03) https://doi.org/f3mn6s DOI: 10.1016/j.ymeth.2014.11.020 · PMID: 25484339 [DOI] [PubMed]
- 21.DrugBank 5.0: a major update to the DrugBank database for 2018 David S Wishart, Yannick D Feunang, An C Guo, Elvis J Lo, Ana Marcu, Jason R Grant, Tanvir Sajed, Daniel Johnson, Carin Li, Zinat Sayeeda, … Michael Wilson Nucleic Acids Research (2017-11-08) https://doi.org/gcwtzk DOI: 10.1093/nar/gkx1037 · PMID: 29126136 · PMCID: PMC5753335 [DOI] [PMC free article] [PubMed]
- 22.A network integration approach for drug-target interaction prediction and computational drug repositioning from heterogeneous information Yunan Luo, Xinbin Zhao, Jingtian Zhou, Jinglin Yang, Yanqing Zhang, Wenhua Kuang, Jian Peng, Ligong Chen, Jianyang Zeng Nature Communications (2017-09-18) https://doi.org/gbxwrc DOI: 10.1038/s41467-017-00680-8 · PMID: 28924171 · PMCID: PMC5603535 [DOI] [PMC free article] [PubMed]
- 23.Inferring new indications for approved drugs via random walk on drug-disease heterogenous networks Hui Liu, Yinglong Song, Jihong Guan, Libo Luo, Ziheng Zhuang BMC Bioinformatics (2016-12) https://doi.org/gf6v27 DOI: 10.1186/s12859-016-1336-7 · PMID: 28155639 · PMCID: PMC5259862 [DOI] [PMC free article] [PubMed]
- 24.Finding disease similarity based on implicit semantic similarity Sachin Mathur, Deendayal Dinakarpandian Journal of Biomedical Informatics (2012-04) https://doi.org/b7b3tw DOI: 10.1016/j.jbi.2011.11.017 · PMID: 22166490 [DOI] [PubMed]
- 25.Bio2RDF: Towards a mashup to build bioinformatics knowledge systems François Belleau, Marc-Alexandre Nolin, Nicole Tourigny, Philippe Rigault, Jean Morissette Journal of Biomedical Informatics (2008-10) https://doi.org/frqkq5 DOI: 10.1016/j.jbi.2008.03.004 · PMID: 18472304 [DOI] [PubMed]
- 26.KnowLife: a versatile approach for constructing a large knowledge graph for biomedical sciences Patrick Ernst, Amy Siu, Gerhard Weikum BMC Bioinformatics (2015-05-14) https://doi.org/gb8w8d DOI: 10.1186/s12859-015-0549-5 · PMID: 25971816 · PMCID: PMC4448285 [DOI] [PMC free article] [PubMed]
- 27.Constructing biomedical domain-specific knowledge graph with minimum supervision Jianbo Yuan, Zhiwei Jin, Han Guo, Hongxia Jin, Xianchao Zhang, Tristram Smith, Jiebo Luo Knowledge and Information Systems (2019-03-23) https://doi.org/gf6v26 DOI: 10.1007/s10115-019-01351-4
- 28.Feature assisted stacked attentive shortest dependency path based Bi-LSTM model for protein–protein interaction Shweta Yadav, Asif Ekbal, Sriparna Saha, Ankit Kumar, Pushpak Bhattacharyya Knowledge-Based Systems (2019-02) https://doi.org/gf4788 DOI: 10.1016/j.knosys.2018.11.020
- 29.Biological Databases- Integration of Life Science Data Nishant Toomula, Arun Kumar, Sathish Kumar D, Vijaya Shanti Bheemidi Journal of Computer Science & Systems Biology (2012) https://doi.org/gf8qcb DOI: 10.4172/jcsb.1000081
- 30.COSMIC: somatic cancer genetics at high-resolution Simon A. Forbes, David Beare, Harry Boutselakis, Sally Bamford, Nidhi Bindal, John Tate, Charlotte G. Cole, Sari Ward, Elisabeth Dawson, Laura Ponting, … Peter J. Campbell Nucleic Acids Research (2016-11-28) https://doi.org/f9v865 DOI: 10.1093/nar/gkw1121 · PMID: 27899578 · PMCID: PMC5210583 [DOI] [PMC free article] [PubMed]
- 31.COSMIC: the Catalogue Of Somatic Mutations In Cancer John G Tate, Sally Bamford, Harry C Jubb, Zbyslaw Sondka, David M Beare, Nidhi Bindal, Harry Boutselakis, Charlotte G Cole, Celestino Creatore, Elisabeth Dawson, … Simon A Forbes Nucleic Acids Research (2018-10-29) https://doi.org/gf9hxg DOI: 10.1093/nar/gky1015 · PMID: 30371878 · PMCID: PMC6323903 [DOI] [PMC free article] [PubMed]
- 32.Recurated protein interaction datasets Lukasz Salwinski, Luana Licata, Andrew Winter, David Thorneycroft, Jyoti Khadake, Arnaud Ceol, Andrew Chatr Aryamontri, Rose Oughtred, Michael Livstone, Lorrie Boucher, … Henning Hermjakob Nature Methods (2009-12) https://doi.org/fgvkmf DOI: 10.1038/nmeth1209-860 · PMID: 19935838 [DOI] [PubMed]
- 33.Literature-curated protein interaction datasets Michael E Cusick, Haiyuan Yu, Alex Smolyar, Kavitha Venkatesan, Anne-Ruxandra Carvunis, Nicolas Simonis, Jean-François Rual, Heather Borick, Pascal Braun, Matija Dreze, … Marc Vidal Nature Methods (2008-12-30) https://doi.org/d4j62p DOI: 10.1038/nmeth.1284 · PMID: 19116613 · PMCID: PMC2683745 [DOI] [PMC free article] [PubMed]
- 34.Curation accuracy of model organism databases I. M. Keseler, M. Skrzypek, D. Weerasinghe, A. Y. Chen, C. Fulcher, G.-W. Li, K. C. Lemmer, K. M. Mladinich, E. D. Chow, G. Sherlock, P. D. Karp Database (2014-06-12) https://doi.org/gf63jz DOI: 10.1093/database/bau058 · PMID: 24923819 · PMCID: PMC4207230 [DOI] [PMC free article] [PubMed]
- 35.OMIM.org: Online Mendelian Inheritance in Man (OMIM®), an online catalog of human genes and genetic disorders Joanna S. Amberger, Carol A. Bocchini, François Schiettecatte, Alan F. Scott, Ada Hamosh Nucleic Acids Research (2014-11-26) https://doi.org/gf8qb6 DOI: 10.1093/nar/gku1205 · PMID: 25428349 · PMCID: PMC4383985 [DOI] [PMC free article] [PubMed]
- 36.Textpresso Central: a customizable platform for searching, text mining, viewing, and curating biomedical literature H.-M. Müller, K. M. Van Auken, Y. Li, P. W. Sternberg BMC Bioinformatics (2018-03-09) https://doi.org/gf7rbz DOI: 10.1186/s12859-018-2103-8 · PMID: 29523070 · PMCID: PMC5845379 [DOI] [PMC free article] [PubMed]
- 37.Text mining and expert curation to develop a database on psychiatric diseases and their genes Alba Gutiérrez-Sacristán, Àlex Bravo, Marta Portero-Tresserra, Olga Valverde, Antonio Armario, M. C. Blanco-Gandía, Adriana Farré, Lierni Fernández-Ibarrondo, Francina Fonseca, Jesús Giraldo, … Laura I. Furlong Database (2017-01-01) https://doi.org/gf8qb5 DOI: 10.1093/database/bax043 · PMID: 29220439 · PMCID: PMC5502359 [DOI] [PMC free article] [PubMed]
- 38.Manual curation is not sufficient for annotation of genomic databases William A. Baumgartner Jr, K. Bretonnel Cohen, Lynne M. Fox, George Acquaah-Mensah, Lawrence Hunter Bioinformatics (2007-07-01) https://doi.org/dtck86 DOI: 10.1093/bioinformatics/btm229 · PMID: 17646325 · PMCID: PMC2516305 [DOI] [PMC free article] [PubMed]
- 39.The rate of growth in scientific publication and the decline in coverage provided by Science Citation Index Peder Olesen Larsen, Markus von Ins Scientometrics (2010-03-10) https://doi.org/c4hb8r DOI: 10.1007/s11192-010-0202-z · PMID: 20700371 · PMCID: PMC2909426 [DOI] [PMC free article] [PubMed]
- 40.Semi-automatic semantic annotation of PubMed queries: A study on quality, efficiency, satisfaction Aurélie Névéol, Rezarta Islamaj Doğan, Zhiyong Lu Journal of Biomedical Informatics (2011-04) https://doi.org/bq34sj DOI: 10.1016/j.jbi.2010.11.001 · PMID: 21094696 · PMCID: PMC3063330 [DOI] [PMC free article] [PubMed]
- 41.Assisting manual literature curation for protein-protein interactions using BioQRator D. Kwon, S. Kim, S.-Y. Shin, A. Chatr-aryamontri, W. J. Wilbur Database (2014-07-22) https://doi.org/gf7hm3 DOI: 10.1093/database/bau067 · PMID: 25052701 · PMCID: PMC4105708 [DOI] [PMC free article] [PubMed]
- 42.R. Rak A. Rowley W. Black S. Ananiadou Argo: an integrative, interactive, text mining-based workbench supporting curation Database 2012 0 2012 bas010 bas10 10.1093/database/bas010 https://academic.oup.com/database/article-lookup/doi/10.1093/database/bas010 [DOI] [PMC free article] [PubMed]
- 43.CurEx Michael Loster, Felix Naumann, Jan Ehmueller, Benjamin Feldmann Proceedings of the 27th ACM International Conference on Information and Knowledge Management (2018-10-17) https://doi.org/gf8qb8 DOI: 10.1145/3269206.3269229
- 44.Charles Tapley Hoyt Daniel Domingo-Fernández Rana Aldisi Lingling Xu Kristian Kolpeja Sandra Spalek Esther Wollert John Bachman Benjamin M Gyori Patrick Greene Martin Hofmann-Apitius Re-curation and rational enrichment of knowledge graphs in Biological Expression Language 2019 2019 2019 10.1093/database/baz068 https://academic.oup.com/database/article/doi/10.1093/database/baz068/5521414 [DOI] [PMC free article] [PubMed]
- 45.Juan Miguel Cejuela Shrikant Vinchurkar Tatyana Goldberg Madhukar Sollepura Prabhu Shankar Ashish Baghudana Aleksandar Bojchevski Carsten Uhlig André Ofner Pandu Raharja-Liu Lars Juhl Jensen Burkhard Rost LocText: relation extraction of protein localizations to assist database curation BMC Bioinformatics 19 1 2018 10.1186/s12859-018-2021-9 https://bmcbioinformatics.biomedcentral.com/articles/10.1186/s12859-018-2021-9 [DOI] [PMC free article] [PubMed]
- 46.Todd Lingren Louise Deleger Katalin Molnar Haijun Zhai Jareen Meinzen-Derr Megan Kaiser Laura Stoutenborough Qi Li Imre Solti Evaluating the impact of pre-annotation on annotation speed and potential bias: natural language processing gold standard development for clinical named entity recognition in clinical trial announcements J Am Med Inform Assoc 21 3 2014 406 413 10.1136/amiajnl-2013-001837 https://academic.oup.com/jamia/article-lookup/doi/10.1136/amiajnl-2013-001837 [DOI] [PMC free article] [PubMed]
- 47.iSimp in BioC standard format: enhancing the interoperability of a sentence simplification system Y. Peng, C. O. Tudor, M. Torii, C. H. Wu, K. Vijay-Shanker Database (2014-05-21) https://doi.org/gf9hxf DOI: 10.1093/database/bau038 · PMID: 24850848 · PMCID: PMC4028706 [DOI] [PMC free article] [PubMed]
- 48.BioSimplify: an open source sentence simplification engine to improve recall in automatic biomedical information extraction. Siddhartha Jonnalagadda, Graciela Gonzalez AMIA … Annual Symposium proceedings. AMIA Symposium (2010-11-13) https://www.ncbi.nlm.nih.gov/pubmed/21346999 PMID: 21346999 · PMCID: PMC3041388 [PMC free article] [PubMed]
- 49.E.U.-A.D.R. The corpus: Annotated drugs, diseases, targets, and their relationships Erik M. van Mulligen, Annie Fourrier-Reglat, David Gurwitz, Mariam Molokhia, Ainhoa Nieto, Gianluca Trifiro, Jan A. Kors, Laura I. Furlong, Journal of Biomedical Informatics 2012–10 https://doi.org/f36vn6 10.1016/j.jbi.2012.04.004 · PMID: 22554700 [DOI] [PubMed]
- 50.Razvan Bunescu Ruifang Ge Rohit J. Kate Edward M. Marcotte Raymond J. Mooney Arun K. Ramani Yuk Wah Wong Comparative experiments on learning information extractors for proteins and their interactions Artificial Intelligence in Medicine 33 2 2005 139 155 10.1016/j.artmed.2004.07.016 https://linkinghub.elsevier.com/retrieve/pii/S0933365704001319 [DOI] [PubMed]
- 51.A Unified Active Learning Framework for Biomedical Relation Extraction Hong-Tao Zhang, Min-Lie Huang, Xiao-Yan Zhu Journal of Computer Science and Technology (2012-11) https://doi.org/gf8qb4 DOI: 10.1007/s11390-012-1306-0
- 52.The BioGRID interaction database: 2013 update Andrew Chatr-aryamontri, Bobby-Joe Breitkreutz, Sven Heinicke, Lorrie Boucher, Andrew Winter, Chris Stark, Julie Nixon, Lindsay Ramage, Nadine Kolas, Lara O’Donnell, … Mike Tyers Nucleic Acids Research (2012-11-30) https://doi.org/f4jmz4 DOI: 10.1093/nar/gks1158 · PMID: 23203989 · PMCID: PMC3531226 [DOI] [PMC free article] [PubMed]
- 53.The Comparative Toxicogenomics Database: update 2019 Allan Peter Davis, Cynthia J Grondin, Robin J Johnson, Daniela Sciaky, Roy McMorran, Jolene Wiegers, Thomas C Wiegers, Carolyn J Mattingly Nucleic Acids Research (2018-09-24) https://doi.org/gf8qb7 DOI: 10.1093/nar/gky868 · PMID: 30247620 · PMCID: PMC6323936 [DOI] [PMC free article] [PubMed]
- 54.CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database Baofeng Jia, Amogelang R. Raphenya, Brian Alcock, Nicholas Waglechner, Peiyao Guo, Kara K. Tsang, Briony A. Lago, Biren M. Dave, Sheldon Pereira, Arjun N. Sharma, … Andrew G. McArthur Nucleic Acids Research (2016-10-26) https://doi.org/f9wbjs DOI: 10.1093/nar/gkw1004 · PMID: 27789705 · PMCID: PMC5210516 [DOI] [PMC free article] [PubMed]
- 55.Entrez Gene: gene-centered information at NCBI D. Maglott, J. Ostell, K. D. Pruitt, T. Tatusova Nucleic Acids Research (2010-11-28) https://doi.org/fsjcqz DOI: 10.1093/nar/gkq1237 · PMID: 21115458 · PMCID: PMC3013746 [DOI] [PMC free article] [PubMed]
- 56.OMIM.org: leveraging knowledge across phenotype-gene relationships. Jo7anna S Amberger, Carol A Bocchini, Alan F Scott, Ada Hamosh Nucleic acids research (2019-01-08) https://www.ncbi.nlm.nih.gov/pubmed/30445645 DOI: 10.1093/nar/gky1151 · PMID: 30445645 · PMCID: PMC6323937 [DOI] [PMC free article] [PubMed]
- 57.Pharmacogenomics Knowledge for Personalized Medicine M Whirl-Carrillo, EM McDonagh, JM Hebert, L Gong, K Sangkuhl, CF Thorn, RB Altman, TE Klein Clinical Pharmacology & Therapeutics (2012-10) https://doi.org/gdnfzr DOI: 10.1038/clpt.2012.96 · PMID: 22992668 · PMCID: PMC3660037 [DOI] [PMC free article] [PubMed]
- 58.UniProt: a worldwide hub of protein knowledgeNucleic Acids Research (2018-11-05) https://doi.org/gfwqck DOI: 10.1093/nar/gky1049 · PMID: 30395287 · PMCID: PMC6323992
- 59.LPTK: a linguistic pattern-aware dependency tree kernel approach for the BioCreative VI CHEMPROT task Neha Warikoo, Yung-Chun Chang, Wen-Lian Hsu Database (2018-01-01) https://doi.org/gfhjr6 DOI: 10.1093/database/bay108 · PMID: 30346607 · PMCID: PMC6196310 [DOI] [PMC free article] [PubMed]
- 60.DTMiner: identification of potential disease targets through biomedical literature mining Dong Xu, Meizhuo Zhang, Yanping Xie, Fan Wang, Ming Chen, Kenny Q. Zhu, Jia Wei Bioinformatics (2016-08-09) https://doi.org/f9nw36 DOI: 10.1093/bioinformatics/btw503 · PMID: 27506226 · PMCID: PMC5181534 [DOI] [PMC free article] [PubMed]
- 61.Exploiting graph kernels for high performance biomedical relation extraction Nagesh C. Panyam, Karin Verspoor, Trevor Cohn, Kotagiri Ramamohanarao Journal of Biomedical Semantics (2018-01-30) https://doi.org/gf49nn DOI: 10.1186/s13326-017-0168-3 · PMID: 29382397 · PMCID: PMC5791373 [DOI] [PMC free article] [PubMed]
- 62.iSimp in BioC standard format: enhancing the interoperability of a sentence simplification system. Yifan Peng, Catalina O Tudor, Manabu Torii, Cathy H Wu, K Vijay-Shanker Database : the journal of biological databases and curation (2014-05-21) https://www.ncbi.nlm.nih.gov/pubmed/24850848 DOI: 10.1093/database/bau038 · PMID: 24850848 · PMCID: PMC4028706 [DOI] [PMC free article] [PubMed]
- 63.BELMiner: adapting a rule-based relation extraction system to extract biological expression language statements from bio-medical literature evidence sentences K. E. Ravikumar, Majid Rastegar-Mojarad, Hongfang Liu Database (2017-01-01) https://doi.org/gf7rbx DOI: 10.1093/database/baw156 · PMID: 28365720 · PMCID: PMC5467463 [DOI] [PMC free article] [PubMed]
- 64.A generalizable NLP framework for fast development of pattern-based biomedical relation extraction systems Yifan Peng, Manabu Torii, Cathy H Wu, K Vijay-Shanker BMC Bioinformatics (2014-08-23) https://doi.org/f6rndz DOI: 10.1186/1471-2105-15-285 · PMID: 25149151 · PMCID: PMC4262219 [DOI] [PMC free article] [PubMed]
- 65.Construction of phosphorylation interaction networks by text mining of full-length articles using the eFIP system Catalina O. Tudor, Karen E. Ross, Gang Li, K. Vijay-Shanker, Cathy H. Wu, Cecilia N. Arighi Database (2015-01-01) https://doi.org/gf8fpt DOI: 10.1093/database/bav020 · PMID: 25833953 · PMCID: PMC4381107 [DOI] [PMC free article] [PubMed]
- 66.miRTex: A Text Mining System for miRNA-Gene Relation Extraction Gang Li, Karen E. Ross, Cecilia N. Arighi, Yifan Peng, Cathy H. Wu, K. Vijay-Shanker PLOS Computational Biology (2015-09-25) https://doi.org/f75mwb DOI: 10.1371/journal.pcbi.1004391 · PMID: 26407127 · PMCID: PMC4583433 [DOI] [PMC free article] [PubMed]
- 67.LimTox: a web tool for applied text mining of adverse event and toxicity associations of compounds, drugs and genes Andres Cañada, Salvador Capella-Gutierrez, Obdulia Rabal, Julen Oyarzabal, Alfonso Valencia, Martin Krallinger Nucleic Acids Research (2017-05-22) https://doi.org/gf479h DOI: 10.1093/nar/gkx462 · PMID: 28531339 · PMCID: PMC5570141 [DOI] [PMC free article] [PubMed]
- 68.DiMeX: A Text Mining System for Mutation-Disease Association Extraction A. S. M. Ashique Mahmood, Tsung-Jung Wu, Raja Mazumder, K. Vijay-Shanker PLOS ONE (2016-04-13) https://doi.org/f8xktj DOI: 10.1371/journal.pone.0152725 · PMID: 27073839 · PMCID: PMC4830514 [DOI] [PMC free article] [PubMed]
- 69.Automated extraction of mutation data from the literature: application of MuteXt to G protein-coupled receptors and nuclear hormone receptors F. Horn, A. L. Lau, F. E. Cohen Bioinformatics (2004-01-22) https://doi.org/d7cjgj DOI: 10.1093/bioinformatics/btg449 · PMID: 14990452 [DOI] [PubMed]
- 70.Large-scale extraction of accurate drug-disease treatment pairs from biomedical literature for drug repurposing Rong Xu, QuanQiu Wang BMC Bioinformatics (2013-06-06) https://doi.org/gb8v3k DOI: 10.1186/1471-2105-14-181 · PMID: 23742147 · PMCID: PMC3702428 [DOI] [PMC free article] [PubMed]
- 71.RLIMS-P 2.0: A Generalizable Rule-Based Information Extraction System for Literature Mining of Protein Phosphorylation Information Manabu Torii, Cecilia N. Arighi, Gang Li, Qinghua Wang, Cathy H. Wu, K. Vijay-Shanker IEEE/ACM Transactions on Computational Biology and Bioinformatics (2015-01-01) https://doi.org/gf8fpv DOI: 10.1109/tcbb.2014.2372765 · PMID: 26357075 · PMCID: PMC4568560 [DOI] [PMC free article] [PubMed]
- 72.PKDE4J: Entity and relation extraction for public knowledge discovery Min Song, Won Chul Kim, Dahee Lee, Go Eun Heo, Keun Young Kang Journal of Biomedical Informatics (2015-10) https://doi.org/f7v7jj DOI: 10.1016/j.jbi.2015.08.008 · PMID: 26277115 [DOI] [PubMed]
- 73.PhpSyntaxTree tool A Eisenbach, M Eisenbach (2006)
- 74.Spacy 2: Natural language understanding with bloom embeddings, convolutional neural networks and incremental parsing Matthew Honnibal, Ines Montani To appear (2017)
- 75.STRING v9.1: protein-protein interaction networks, with increased coverage and integration Andrea Franceschini, Damian Szklarczyk, Sune Frankild, Michael Kuhn, Milan Simonovic, Alexander Roth, Jianyi Lin, Pablo Minguez, Peer Bork, Christian von Mering, Lars J. Jensen Nucleic Acids Research (2012-11-29) https://doi.org/gf5kcd DOI: 10.1093/nar/gks1094 · PMID: 23203871 · PMCID: PMC3531103 [DOI] [PMC free article] [PubMed]
- 76.A comprehensive and quantitative comparison of text-mining in 15 million full-text articles versus their corresponding abstracts David Westergaard, Hans-Henrik Stærfeldt, Christian Tønsberg, Lars Juhl Jensen, Søren Brunak PLOS Computational Biology (2018-02-15) https://doi.org/gcx747 DOI: 10.1371/journal.pcbi.1005962 · PMID: 29447159 · PMCID: PMC5831415 [DOI] [PMC free article] [PubMed]
- 77.STITCH 4: integration of protein–chemical interactions with user data Michael Kuhn, Damian Szklarczyk, Sune Pletscher-Frankild, Thomas H. Blicher, Christian von Mering, Lars J. Jensen, Peer Bork Nucleic Acids Research (2013-11-28) https://doi.org/f5shb4 DOI: 10.1093/nar/gkt1207 · PMID: 24293645 · PMCID: PMC3964996 [DOI] [PMC free article] [PubMed]
- 78.A global network of biomedical relationships derived from text Bethany Percha, Russ B Altman Bioinformatics (2018-02-27) https://doi.org/gc3ndk DOI: 10.1093/bioinformatics/bty114 · PMID: 29490008 · PMCID: PMC6061699 [DOI] [PMC free article] [PubMed]
- 79.CoCoScore: context-aware co-occurrence scoring for text mining applications using distant supervision Alexander Junge, Lars Juhl Jensen Bioinformatics (2019-06-14) https://doi.org/gf4789 DOI: 10.1093/bioinformatics/btz490 · PMID: 31199464 · PMCID: PMC6956794 [DOI] [PMC free article] [PubMed]
- 80.A new method for prioritizing drug repositioning candidates extracted by literature-based discovery Majid Rastegar-Mojarad, Ravikumar Komandur Elayavilli, Dingcheng Li, Rashmi Prasad, Hongfang Liu 2015 IEEE International Conference on Bioinformatics and Biomedicine (BIBM) (2015-11) https://doi.org/gf479j DOI: 10.1109/bibm.2015.7359766
- 81.Literature Mining for the Discovery of Hidden Connections between Drugs, Genes and Diseases Raoul Frijters, Marianne van Vugt, Ruben Smeets, René van Schaik, Jacob de Vlieg, Wynand Alkema PLoS Computational Biology (2010-09-23) https://doi.org/bhrw7x DOI: 10.1371/journal.pcbi.1000943 · PMID: 20885778 · PMCID: PMC2944780 [DOI] [PMC free article] [PubMed]
- 82.STRING v10: protein–protein interaction networks, integrated over the tree of life Damian Szklarczyk, Andrea Franceschini, Stefan Wyder, Kristoffer Forslund, Davide Heller, Jaime Huerta-Cepas, Milan Simonovic, Alexander Roth, Alberto Santos, Kalliopi P. Tsafou, … Christian von Mering Nucleic Acids Research (2014-10-28) https://doi.org/f64rfn DOI: 10.1093/nar/gku1003 · PMID: 25352553 · PMCID: PMC4383874 [DOI] [PMC free article] [PubMed]
- 83.Text Mining Genotype-Phenotype Relationships from Biomedical Literature for Database Curation and Precision Medicine Ayush Singhal, Michael Simmons, Zhiyong Lu PLOS Computational Biology (2016-11-30) https://doi.org/f9gz4b DOI: 10.1371/journal.pcbi.1005017 · PMID: 27902695 · PMCID: PMC5130168 [DOI] [PMC free article] [PubMed]
- 84.Overview of the biocreative vi chemical-protein interaction track Martin Krallinger, Obdulia Rabal, Saber A Akhondi, others Proceedings of the sixth biocreative challenge evaluation workshop (2017) https://www.semanticscholar.org/paper/Overview-of-the-BioCreative-VI-chemical-protein-Krallinger-Rabal/eed781f498b563df5a9e8a241c67d63dd1d92ad5
- 85.BioCreative V CDR task corpus: a resource for chemical disease relation extraction Jiao Li, Yueping Sun, Robin J. Johnson, Daniela Sciaky, Chih-Hsuan Wei, Robert Leaman, Allan Peter Davis, Carolyn J. Mattingly, Thomas C. Wiegers, Zhiyong Lu Database (2016) https://doi.org/gf5hfw DOI: 10.1093/database/baw068 · PMID: 27161011 · PMCID: PMC4860626 [DOI] [PMC free article] [PubMed]
- 86.RelEx–Relation extraction using dependency parse trees K. Fundel, R. Kuffner, R. Zimmer Bioinformatics (2006-12-01) https://doi.org/cz7q4d DOI: 10.1093/bioinformatics/btl616 · PMID: 17142812 [DOI] [PubMed]
- 87.CoMAGC: a corpus with multi-faceted annotations of gene-cancer relations Hee-Jin Lee, Sang-Hyung Shim, Mi-Ryoung Song, Hyunju Lee, Jong C Park BMC Bioinformatics (2013) https://doi.org/gb8v5s DOI: 10.1186/1471-2105-14-323 · PMID: 24225062 · PMCID: PMC3833657 [DOI] [PMC free article] [PubMed]
- 88.Text Mining for Protein Docking Varsha D. Badal, Petras J. Kundrotas, Ilya A. Vakser PLOS Computational Biology (2015-12-09) https://doi.org/gcvj3b DOI: 10.1371/journal.pcbi.1004630 · PMID: 26650466 · PMCID: PMC4674139 [DOI] [PMC free article] [PubMed]
- 89.Automatic extraction of gene-disease associations from literature using joint ensemble learning Balu Bhasuran, Jeyakumar Natarajan PLOS ONE (2018-07-26) https://doi.org/gdx63f DOI: 10.1371/journal.pone.0200699 · PMID: 30048465 · PMCID: PMC6061985 [DOI] [PMC free article] [PubMed]
- 90.Extraction of relations between genes and diseases from text and large-scale data analysis: implications for translational research Àlex Bravo, Janet Piñero, Núria Queralt-Rosinach, Michael Rautschka, Laura I Furlong BMC Bioinformatics (2015-02-21) https://doi.org/f7kn8s DOI: 10.1186/s12859-015-0472-9 · PMID: 25886734 · PMCID: PMC4466840 [DOI] [PMC free article] [PubMed]
- 91.Deep learning Ian Goodfellow, Yoshua Bengio, Aaron Courville The MIT Press (2016) ISBN: 0262035618, 9780262035613
- 92.Deep learning Yann LeCun, Yoshua Bengio, Geoffrey Hinton Nature (2015-05) https://doi.org/bmqp DOI: 10.1038/nature14539 · PMID: 26017442 [DOI] [PubMed]
- 93.Long Short-Term Memory Sepp Hochreiter, Jürgen Schmidhuber Neural Computation (1997-11) https://doi.org/bxd65w DOI: 10.1162/neco.1997.9.8.1735 · PMID: 9377276 [DOI] [PubMed]
- 94.Deep learning for pharmacovigilance: recurrent neural network architectures for labeling adverse drug reactions in Twitter posts Anne Cocos, Alexander G Fiks, Aaron J Masino Journal of the American Medical Informatics Association (2017-02-22) https://doi.org/gbp9nj DOI: 10.1093/jamia/ocw180 · PMID: 28339747 [DOI] [PMC free article] [PubMed]
- 95.Semantic Relations in Compound Nouns: Perspectives from Inter-Annotator Agreement Yadav Prabha, Jezek Elisabetta, Bouillon Pierrette, Callahan Tiffany J., Bada Michael, Hunter Lawrence E., Cohen K. Bretonnel Studies in Health Technology and Informatics (2017) https://doi.org/ggmk8t DOI: 10.3233/978-1-61499-830-3-644 [PMC free article] [PubMed]
- 96.Cross-Sentence N-ary Relation Extraction with Graph LSTMs Nanyun Peng, Hoifung Poon, Chris Quirk, Kristina Toutanova, Wen-tau Yih arXiv (2017-08-12) https://arxiv.org/abs/1708.03743v1
- 97.Drug drug interaction extraction from biomedical literature using syntax convolutional neural network Zhehuan Zhao, Zhihao Yang, Ling Luo, Hongfei Lin, Jian Wang Bioinformatics (2016-07-27) https://doi.org/f9nsq7 DOI: 10.1093/bioinformatics/btw486 · PMID: 27466626 · PMCID: PMC5181565 [DOI] [PMC free article] [PubMed]
- 98.N-ary Relation Extraction using Graph State LSTM Linfeng Song, Yue Zhang, Zhiguo Wang, Daniel Gildea arXiv (2018-08-28) https://arxiv.org/abs/1808.09101v1
- 99.A neural joint model for entity and relation extraction from biomedical text Fei Li, Meishan Zhang, Guohong Fu, Donghong Ji BMC Bioinformatics (2017-03-31) https://doi.org/gcgnx2 DOI: 10.1186/s12859-017-1609-9 · PMID: 28359255 · PMCID: PMC5374588 [DOI] [PMC free article] [PubMed]
- 100.The problem of learning long-term dependencies in recurrent networks Y. Bengio, P. Frasconi, P. Simard IEEE International Conference on Neural Networks https://doi.org/d7zs24 DOI: 10.1109/icnn.1993.298725 [DOI] [PubMed]
- 101.Fundamentals of Recurrent Neural Network (RNN) and Long Short-Term Memory (LSTM) network Alex Sherstinsky Physica D: Nonlinear Phenomena (2020-03) https://doi.org/ggmzpd DOI: 10.1016/j.physd.2019.132306
- 102.On the difficulty of training Recurrent Neural Networks Razvan Pascanu, Tomas Mikolov, Yoshua Bengio arXiv (2012-11-21) https://arxiv.org/abs/1211.5063v2
- 103.Revisiting Unreasonable Effectiveness of Data in Deep Learning Era Chen Sun, Abhinav Shrivastava, Saurabh Singh, Abhinav Gupta arXiv (2017-07-10) https://arxiv.org/abs/1707.02968v2
- 104.Efficient Estimation of Word Representations in Vector Space Tomas Mikolov, Kai Chen, Greg Corrado, Jeffrey Dean arXiv (2013-01-16) https://arxiv.org/abs/1301.3781v3
- 105.Distributed Representations of Words and Phrases and their Compositionality Tomas Mikolov, Ilya Sutskever, Kai Chen, Greg Corrado, Jeffrey Dean arXiv (2013-10-16) https://arxiv.org/abs/1310.4546v1
- 106.Deep learning for extracting protein-protein interactions from biomedical literature Yifan Peng, Zhiyong Lu arXiv (2017-06-05) https://arxiv.org/abs/1706.01556v2
- 107.Knowledge-guided convolutional networks for chemical-disease relation extraction Huiwei Zhou, Chengkun Lang, Zhuang Liu, Shixian Ning, Yingyu Lin, Lei Du BMC Bioinformatics (2019-05-21) https://doi.org/gf45zn DOI: 10.1186/s12859-019-2873-7 · PMID: 31113357 · PMCID: PMC6528333 [DOI] [PMC free article] [PubMed]
- 108.Extraction of protein–protein interactions (PPIs) from the literature by deep convolutional neural networks with various feature embeddings Sung-Pil Choi Journal of Information Science (2016-11-01) https://doi.org/gcv8bn DOI: 10.1177/0165551516673485
- 109.Extracting chemical–protein relations with ensembles of SVM and deep learning models Yifan Peng, Anthony Rios, Ramakanth Kavuluru, Zhiyong Lu Database (2018-01-01) https://doi.org/gf479f DOI: 10.1093/database/bay073 · PMID: 30020437 · PMCID: PMC6051439 [DOI] [PMC free article] [PubMed]
- 110.Expanding a Database-derived Biomedical Knowledge Graph via Multi-relation Extraction from Biomedical Abstracts David N. Nicholson, Daniel S. Himmelstein, Casey S. Greene Cold Spring Harbor Laboratory (2019-08-08) https://doi.org/gf6qxh DOI: 10.1101/730085 [DOI] [PMC free article] [PubMed]
- 111.Distant supervision for relation extraction without labeled data Mike Mintz, Steven Bills, Rion Snow, Dan Jurafsky Proceedings of the Joint Conference of the 47th Annual Meeting of the ACL and the 4th International Joint Conference on Natural Language Processing of the AFNLP: Volume 2 - ACL-IJCNLP ’09 (2009) https://doi.org/fg9q43 DOI: 10.3115/1690219.1690287
- 112.Introduction to Semi-Supervised Learning Xiaojin Zhu, Andrew B. Goldberg Synthesis Lectures on Artificial Intelligence and Machine Learning (2009-01) https://doi.org/bq7pm2 DOI: 10.2200/s00196ed1v01y200906aim006
- 113.A Survey on Transfer Learning Sinno Jialin Pan, Qiang Yang IEEE Transactions on Knowledge and Data Engineering (2010-10) https://doi.org/bc4vws DOI: 10.1109/tkde.2009.191
- 114.A survey of transfer learning Karl Weiss, Taghi M. Khoshgoftaar, DingDing Wang Journal of Big Data (2016-05-28) https://doi.org/gfkr2w DOI: 10.1186/s40537-016-0043-6
- 115.Exploring Semi-supervised Variational Autoencoders for Biomedical Relation Extraction Yijia Zhang, Zhiyong Lu arXiv (2019-01-18) https://arxiv.org/abs/1901.06103v1 [DOI] [PMC free article] [PubMed]
- 116.Large-scale extraction of gene interactions from full-text literature using DeepDive Emily K. Mallory, Ce Zhang, Christopher Ré, Russ B. Altman Bioinformatics (2015-09-03) https://doi.org/gb5g7b DOI: 10.1093/bioinformatics/btv476 · PMID: 26338771 · PMCID: PMC4681986 [DOI] [PMC free article] [PubMed]
- 117.Snorkel Alexander Ratner, Stephen H. Bach, Henry Ehrenberg, Jason Fries, Sen Wu, Christopher Ré Proceedings of the VLDB Endowment (2017-11-01) https://doi.org/ch44 DOI: 10.14778/3157794.3157797 · PMID: 29770249 · PMCID: PMC5951191 [DOI] [PMC free article] [PubMed]
- 118.Snorkel MeTaL Alex Ratner, Braden Hancock, Jared Dunnmon, Roger Goldman, Christopher Ré Proceedings of the Second Workshop on Data Management for End-To-End Machine Learning - DEEM’18 (2018) https://doi.org/gf3xk7 DOI: 10.1145/3209889.3209898 · PMID: 30931438 · PMCID: PMC6436830 [DOI] [PMC free article] [PubMed]
- 119.Learning protein protein interaction extraction using distant supervision Philippe Thomas, Illés Solt, Roman Klinger, Ulf Leser (2011-01)
- 120.Robust Distant Supervision Relation Extraction via Deep Reinforcement Learning Pengda Qin, Weiran Xu, William Yang Wang arXiv (2018-05-24) https://arxiv.org/abs/1805.09927v1
- 121.DSGAN: Generative Adversarial Training for Distant Supervision Relation Extraction Pengda Qin, Weiran Xu, William Yang Wang arXiv (2018-05-24) https://arxiv.org/abs/1805.09929v1
- 122.Noise Reduction Methods for Distantly Supervised Biomedical Relation Extraction Gang Li, Cathy Wu, K. Vijay-Shanker BioNLP 2017 (2017) https://doi.org/ggmk8s DOI: 10.18653/v1/w17-2323
- 123.BioInfer: a corpus for information extraction in the biomedical domain Sampo Pyysalo, Filip Ginter, Juho Heimonen, Jari Björne, Jorma Boberg, Jouni Järvinen, Tapio Salakoski BMC Bioinformatics (2007-02-09) https://doi.org/b7bhhc DOI: 10.1186/1471-2105-8-50 · PMID: 17291334 · PMCID: PMC1808065 [DOI] [PMC free article] [PubMed]
- 124.Learning language in logic - genic interaction extraction challenge C. Nédellec Proceedings of the learning language in logic 2005 workshop at the international conference on machine learning (2005)
- 125.Mining medline: Abstracts, sentences, or phrases? Jing Ding, Daniel Berleant, Dan Nettleton, Eve Syrkin Wurtele Pacific symposium on biocomputing (2002) http://helix-web.stanford.edu/psb02/ding.pdf [DOI] [PubMed]
- 126.Concept annotation in the CRAFT corpus Michael Bada, Miriam Eckert, Donald Evans, Kristin Garcia, Krista Shipley, Dmitry Sitnikov, William A Baumgartner Jr, K Bretonnel Cohen, Karin Verspoor, Judith A Blake, Lawrence E Hunter BMC Bioinformatics (2012-07-09) https://doi.org/gb8vdr DOI: 10.1186/1471-2105-13-161 · PMID: 22776079 · PMCID: PMC3476437 [DOI] [PMC free article] [PubMed]
- 127.GRAM: Graph-based Attention Model for Healthcare Representation Learning Edward Choi, Mohammad Taha Bahadori, Le Song, Walter F. Stewart, Jimeng Sun arXiv (2016-11-21) https://arxiv.org/abs/1611.07012v3 [DOI] [PMC free article] [PubMed]
- 128.miRNA-Disease Association Prediction with Collaborative Matrix Factorization Zhen Shen, You-Hua Zhang, Kyungsook Han, Asoke K. Nandi, Barry Honig, De-Shuang Huang Complexity (2017) https://doi.org/ggmrpm DOI: 10.1155/2017/2498957
- 129.Deep Residual Learning for Image Recognition Kaiming He, Xiangyu Zhang, Shaoqing Ren, Jian Sun arXiv (2015-12-10) https://arxiv.org/abs/1512.03385v1
- 130.Representation Learning on Graphs: Methods and Applications William L. Hamilton, Rex Ying, Jure Leskovec arXiv (2017-09-17) https://arxiv.org/abs/1709.05584v3
- 131.The approximation of one matrix by another of lower rank Carl Eckart, Gale Young Psychometrika (1936-09) https://doi.org/c2frtd DOI: 10.1007/bf02288367
- 132.Laplacian Eigenmaps for Dimensionality Reduction and Data Representation Mikhail Belkin, Partha Niyogi Neural Computation (2003-06) https://doi.org/bbr9cw DOI: 10.1162/089976603321780317
- 133.node2vec: Scalable Feature Learning for Networks Aditya Grover, Jure Leskovec arXiv (2016-07-03) https://arxiv.org/abs/1607.00653v1 [DOI] [PMC free article] [PubMed]
- 134.Translating embeddings for modeling multi-relational data Antoine Bordes, Nicolas Usunier, Alberto García-Durán, Jason Weston, Oksana Yakhnenko NIPS (2013)
- 135.Signed laplacian embedding for supervised dimension reduction Chen Gong, Dacheng Tao, Jie Yang, Keren Fu Proceedings of the twenty-eighth aaai conference on artificial intelligence (2014) http://dl.acm.org/citation.cfm?id=2892753.2892809
- 136.A Semi-NMF-PCA Unified Framework for Data Clustering Kais Allab, Lazhar Labiod, Mohamed Nadif IEEE Transactions on Knowledge and Data Engineering (2017-01-01) https://doi.org/f9hm9g DOI: 10.1109/tkde.2016.2606098
- 137.Partially supervised graph embedding for positive unlabelled feature selection Yufei Han, Yun Shen Proceedings of the twenty-fifth international joint conference on artificial intelligence (2016) http://dl.acm.org/citation.cfm?id=3060832.3060837 ISBN: 978-1-57735-770-4
- 138.GraRep Shaosheng Cao, Wei Lu, Qiongkai Xu Proceedings of the 24th ACM International on Conference on Information and Knowledge Management - CIKM ’15 (2015) https://doi.org/gf8rgf DOI: 10.1145/2806416.2806512
- 139.Improved Knowledge Base Completion by Path-Augmented TransR Model Wenhao Huang, Ge Li, Zhi Jin arXiv (2016-10-06) https://arxiv.org/abs/1610.04073v1
- 140.A Global Geometric Framework for Nonlinear Dimensionality Reduction J. B. Tenenbaum Science (2000-12-22) https://doi.org/cz8wgk DOI: 10.1126/science.290.5500.2319 · PMID: 11125149 [DOI] [PubMed]
- 141.Principal component analysis Svante Wold, Kim Esbensen, Paul Geladi Chemometrics and Intelligent Laboratory Systems (1987-08) https://doi.org/bm8dnf DOI: 10.1016/0169-7439(87)80084-9
- 142.Graph embedding on biomedical networks: methods, applications and evaluations Xiang Yue, Zhen Wang, Jingong Huang, Srinivasan Parthasarathy, Soheil Moosavinasab, Yungui Huang, Simon M Lin, Wen Zhang, Ping Zhang, Huan Sun Bioinformatics (2019-10-04) https://doi.org/ggmzpf DOI: 10.1093/bioinformatics/btz718 · PMID: 31584634 [DOI] [PMC free article] [PubMed]
- 143.Network embedding as matrix factorization: Unifying deepwalk, line, pte, and node2vec Jiezhong Qiu, Yuxiao Dong, Hao Ma, Jian Li, Kuansan Wang, Jie Tang Proceedings of the eleventh acm international conference on web search and data mining (2018) https://doi.org/10.1145/3159652.3159706 DOI: 10.1145/3159652.3159706 · ISBN: 9781450355810
- 144.A Survey of Collaborative Filtering Techniques Xiaoyuan Su, Taghi M. Khoshgoftaar Advances in Artificial Intelligence (2009) https://doi.org/fk9jjg DOI: 10.1155/2009/421425
- 145.GLEE: Geometric Laplacian Eigenmap Embedding Leo Torres, Kevin S Chan, Tina Eliassi-Rad arXiv (2019-05-23) https://arxiv.org/abs/1905.09763v2 DOI: 10.1093/comnet/cnaa007
- 146.Vicus: Exploiting local structures to improve network-based analysis of biological data Bo Wang, Lin Huang, Yuke Zhu, Anshul Kundaje, Serafim Batzoglou, Anna Goldenberg PLOS Computational Biology (2017-10-12) https://doi.org/gb368p DOI: 10.1371/journal.pcbi.1005621 · PMID: 29023470 · PMCID: PMC5638230. [DOI] [PMC free article] [PubMed]
- 147.A Comparison of Semantic Similarity Methods for Maximum Human Interpretability Pinky Sitikhu, Kritish Pahi, Pujan Thapa, Subarna Shakya arXiv (2019-10-21) https://arxiv.org/abs/1910.09129v2
- 148.Knowledge graph embedding by translating on hyperplanes Zhen Wang, Jianwen Zhang, Jianlin Feng, Zheng Chen Proceedings of the twenty-eighth aaai conference on artificial intelligence (2014) http://dl.acm.org/citation.cfm?id=2893873.2894046
- 149.Learning entity and relation embeddings for knowledge graph completion Yankai Lin, Zhiyuan Liu, Maosong Sun, Yang Liu, Xuan Zhu Proceedings of the twenty-ninth aaai conference on artificial intelligence (2015) http://dl.acm.org/citation.cfm?id=2886521.2886624 ISBN: 0-262-51129-0
- 150.PrTransH: Embedding Probabilistic Medical Knowledge from Real World EMR Data Linfeng Li, Peng Wang, Yao Wang, Jinpeng Jiang, Buzhou Tang, Jun Yan, Shenghui Wang, Yuting Liu arXiv (2019-09-02) https://arxiv.org/abs/1909.00672v1
- 151.Artificial neural networks: fundamentals, computing, design, and application. IA Basheer, M Hajmeer Journal of microbiological methods (2000-12-01) https://www.ncbi.nlm.nih.gov/pubmed/11084225 DOI: 10.1016/s0167-7012(00)00201-3 · PMID: 11084225 [DOI] [PubMed]
- 152.DeepWalk Bryan Perozzi, Rami Al-Rfou, Steven Skiena Proceedings of the 20th ACM SIGKDD international conference on Knowledge discovery and data mining - KDD ’14 (2014) https://doi.org/gfkpqt DOI: 10.1145/2623330.2623732
- 153.struc2vec Leonardo F. R. Ribeiro, Pedro H. P. Saverese, Daniel R. Figueiredo Proceedings of the 23rd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining - KDD ’17 (2017) https://doi.org/gd874b DOI: 10.1145/3097983.3098061
- 154.metapath2vec Yuxiao Dong, Nitesh V. Chawla, Ananthram Swami Proceedings of the 23rd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining - KDD ’17 (2017) https://doi.org/gfsqzn DOI: 10.1145/3097983.3098036
- 155.edge2vec: Representation learning using edge semantics for biomedical knowledge discovery Zheng Gao, Gang Fu, Chunping Ouyang, Satoshi Tsutsui, Xiaozhong Liu, Jeremy Yang, Christopher Gessner, Brian Foote, David Wild, Qi Yu, Ying Ding arXiv (2018-09-07) https://arxiv.org/abs/1809.02269v3 [DOI] [PMC free article] [PubMed]
- 156.Learning Graph Embeddings from WordNet-based Similarity Measures Andrey Kutuzov, Mohammad Dorgham, Oleksiy Oliynyk, Chris Biemann, Alexander Panchenko arXiv (2018-08-16) https://arxiv.org/abs/1808.05611v4
- 157.Learning to Make Predictions on Graphs with Autoencoders Phi Vu Tran 2018 IEEE 5th International Conference on Data Science and Advanced Analytics (DSAA) (2018-10) https://doi.org/ggmzpg DOI: 10.1109/dsaa.2018.00034
- 158.Variational Graph Auto-Encoders Thomas N. Kipf, Max Welling arXiv (2016-11-21) https://arxiv.org/abs/1611.07308v1
- 159.Adversarially Regularized Graph Autoencoder for Graph Embedding Shirui Pan, Ruiqi Hu, Guodong Long, Jing Jiang, Lina Yao, Chengqi Zhang arXiv (2018-02-13) https://arxiv.org/abs/1802.04407v2
- 160.Deep learning in neural networks: An overview Jürgen Schmidhuber Neural Networks (2015-01) https://doi.org/f6v78n DOI: 10.1016/j.neunet.2014.09.003 · PMID: 25462637 [DOI] [PubMed]
- 161.Autoencoders, unsupervised learning and deep architectures Pierre Baldi Proceedings of the 2011 international conference on unsupervised and transfer learning workshop - volume 27 (2011)
- 162.Auto-Encoding Variational Bayes Diederik P Kingma, Max Welling arXiv (2013-12-20) https://arxiv.org/abs/1312.6114v10
- 163.GraphVAE: Towards Generation of Small Graphs Using Variational Autoencoders Martin Simonovsky, Nikos Komodakis arXiv (2018-02-09) https://arxiv.org/abs/1802.03480v1
- 164.A Comparative Study for Unsupervised Network Representation Learning Megha Khosla, Vinay Setty, Avishek Anand IEEE Transactions on Knowledge and Data Engineering (2019) https://doi.org/ggmzph DOI: 10.1109/tkde.2019.2951398
- 165.Neural networks for link prediction in realistic biomedical graphs: a multi-dimensional evaluation of graph embedding-based approaches Gamal Crichton, Yufan Guo, Sampo Pyysalo, Anna Korhonen BMC Bioinformatics (2018-05-21) https://doi.org/ggkm7q DOI: 10.1186/s12859-018-2163-9 · PMID: 29783926 · PMCID: PMC5963080 [DOI] [PMC free article] [PubMed]
- 166.Network-based integration of multi-omics data for prioritizing cancer genes Christos Dimitrakopoulos, Sravanth Kumar Hindupur, Luca Häfliger, Jonas Behr, Hesam Montazeri, Michael N Hall, Niko Beerenwinkel Bioinformatics (2018-03-14) https://doi.org/gc6953 DOI: 10.1093/bioinformatics/bty148 · PMID: 29547932 · PMCID: PMC6041755 [DOI] [PMC free article] [PubMed]
- 167.Safe Medicine Recommendation via Medical Knowledge Graph Embedding Meng Wang, Mengyue Liu, Jun Liu, Sen Wang, Guodong Long, Buyue Qian arXiv (2017-10-16) https://arxiv.org/abs/1710.05980v2
- 168.GAMENet: Graph Augmented MEmory Networks for Recommending Medication Combination Junyuan Shang, Cao Xiao, Tengfei Ma, Hongyan Li, Jimeng Sun Proceedings of the AAAI Conference on Artificial Intelligence (2019-07-17) https://doi.org/ggkm7r DOI: 10.1609/aaai.v33i01.33011126
- 169.Heterogeneous network embedding for identifying symptom candidate genes Kuo Yang, Ning Wang, Guangming Liu, Ruyu Wang, Jian Yu, Runshun Zhang, Jianxin Chen, Xuezhong Zhou Journal of the American Medical Informatics Association (2018-10-23) https://doi.org/gfg6nr DOI: 10.1093/jamia/ocy117 · PMID: 30357378 [DOI] [PMC free article] [PubMed]
- 170.Predicting Protein–Protein Interactions from Multimodal Biological Data Sources via Nonnegative Matrix Tri-Factorization Hua Wang, Heng Huang, Chris Ding, Feiping Nie Journal of Computational Biology (2013-04) https://doi.org/f4thrx DOI: 10.1089/cmb.2012.0273 · PMID: 23509857 [DOI] [PubMed]
- 171.Protein functional properties prediction in sparsely-label PPI networks through regularized non-negative matrix factorization Qingyao Wu, Zhenyu Wang, Chunshan Li, Yunming Ye, Yueping Li, Ning Sun BMC Systems Biology (2015) https://doi.org/gb5tvr DOI: 10.1186/1752-0509-9-s1-s9 · PMID: 25708164 · PMCID: PMC4331684 [DOI] [PMC free article] [PubMed]
- 172.HMDD v3.0: a database for experimentally supported human microRNA–disease associations Zhou Huang, Jiangcheng Shi, Yuanxu Gao, Chunmei Cui, Shan Zhang, Jianwei Li, Yuan Zhou, Qinghua Cui Nucleic Acids Research (2018-10-26) https://doi.org/ggmrph DOI: 10.1093/nar/gky1010 · PMID: 30364956 · PMCID: PMC6323994 [DOI] [PMC free article] [PubMed]
- 173.Predicting MiRNA-Disease Association by Latent Feature Extraction with Positive Samples Kai Che, Maozu Guo, Chunyu Wang, Xiaoyan Liu, Xi Chen Genes (2019-01-24) https://doi.org/ggmrpr DOI: 10.3390/genes10020080 · PMID: 30682853 · PMCID: PMC6410147 [DOI] [PMC free article] [PubMed]
- 174.NPCMF: Nearest Profile-based Collaborative Matrix Factorization method for predicting miRNA-disease associations Ying-Lian Gao, Zhen Cui, Jin-Xing Liu, Juan Wang, Chun-Hou Zheng BMC Bioinformatics (2019-06-24) https://doi.org/ggmrpn DOI: 10.1186/s12859-019-2956-5 · PMID: 31234797 · PMCID: PMC6591872 [DOI] [PMC free article] [PubMed]
- 175.RCMF: a robust collaborative matrix factorization method to predict miRNA-disease associations Zhen Cui, Jin-Xing Liu, Ying-Lian Gao, Chun-Hou Zheng, Juan Wang BMC Bioinformatics (2019-12) https://doi.org/ggmrpp DOI: 10.1186/s12859-019-3260-0 · PMID: 31874608 · PMCID: PMC6929455 [DOI] [PMC free article] [PubMed]
- 176.LWPCMF: Logistic Weighted Profile-based Collaborative Matrix Factorization for Predicting MiRNA-Disease Associations Meng-Meng Yin, Zhen Cui, Ming-Ming Gao, Jin-Xing Liu, Ying-Lian Gao IEEE/ACM Transactions on Computational Biology and Bioinformatics (2019) https://doi.org/ggmrpk DOI: 10.1109/tcbb.2019.2937774 · PMID: 31478868 [DOI] [PubMed]
- 177.Protein-protein interaction prediction via Collective Matrix Factorization Qian Xu, Evan Wei Xiang, Qiang Yang 2010 IEEE International Conference on Bioinformatics and Biomedicine (BIBM) (2010-12) https://doi.org/csnv5m DOI: 10.1109/bibm.2010.5706537
- 178.A network embedding model for pathogenic genes prediction by multi-path random walking on heterogeneous network Bo Xu, Yu Liu, Shuo Yu, Lei Wang, Jie Dong, Hongfei Lin, Zhihao Yang, Jian Wang, Feng Xia BMC Medical Genomics (2019-12) https://doi.org/ggmrpq DOI: 10.1186/s12920-019-0627-z · PMID: 31865919 · PMCID: PMC6927107 [DOI] [PMC free article] [PubMed]
- 179.Predicting gene-disease associations from the heterogeneous network using graph embedding Xiaochan Wang, Yuchong Gong, Jing Yi, Wen Zhang 2019 IEEE International Conference on Bioinformatics and Biomedicine (BIBM) (2019-11) https://doi.org/ggmrpj DOI: 10.1109/bibm47256.2019.8983134
- 180.Network embedding-based representation learning for single cell RNA-seq data Xiangyu Li, Weizheng Chen, Yang Chen, Xuegong Zhang, Jin Gu, Michael Q. Zhang Nucleic Acids Research (2017-08-28) https://doi.org/ggmrpg DOI: 10.1093/nar/gkx750 · PMID: 28977434 · PMCID: PMC5737094 [DOI] [PMC free article] [PubMed]
- 181.Neuro-symbolic representation learning on biological knowledge graphs Mona Alshahrani, Mohammad Asif Khan, Omar Maddouri, Akira R Kinjo, Núria Queralt-Rosinach, Robert Hoehndorf Bioinformatics (2017-04-25) https://doi.org/gbv6vm DOI: 10.1093/bioinformatics/btx275 · PMID: 28449114 · PMCID: PMC5860058 [DOI] [PMC free article] [PubMed]
- 182.Deep Learning the Protein Function in Protein Interaction Networks Kire Trivodaliev, Martin Josifoski, Slobodan Kalajdziski Communications in Computer and Information Science (2018) https://doi.org/ggmrpd DOI: 10.1007/978-3-030-00825-3_16
- 183.Detection of protein complexes from multiple protein interaction networks using graph embedding Xiaoxia Liu, Zhihao Yang, Shengtian Sang, Hongfei Lin, Jian Wang, Bo Xu Artificial Intelligence in Medicine (2019-05) https://doi.org/ggmrpf DOI: 10.1016/j.artmed.2019.04.001 · PMID: 31164203 [DOI] [PubMed]
- 184.Large-scale structural and textual similarity-based mining of knowledge graph to predict drug–drug interactions Ibrahim Abdelaziz, Achille Fokoue, Oktie Hassanzadeh, Ping Zhang, Mohammad Sadoghi Journal of Web Semantics (2017-05) https://doi.org/gcrwk3 DOI: 10.1016/j.websem.2017.06.002.
- 185.Matrix Factorization-Based Prediction of Novel Drug Indications by Integrating Genomic Space Wen Dai, Xi Liu, Yibo Gao, Lin Chen, Jianglong Song, Di Chen, Kuo Gao, Yongshi Jiang, Yiping Yang, Jianxin Chen, Peng Lu Computational and Mathematical Methods in Medicine (2015) https://doi.org/gb58g8 DOI: 10.1155/2015/275045 · PMID: 26078775 · PMCID: PMC4452507 [DOI] [PMC free article] [PubMed]
- 186.Abstract eLife Sciences Publications, Ltd https://doi.org/gf4fdb DOI: 10.7554/elife.26726.001
- 187.Drug-Target Interaction Prediction with Graph Regularized Matrix Factorization Ali Ezzat, Peilin Zhao, Min Wu, Xiao-Li Li, Chee-Keong Kwoh IEEE/ACM Transactions on Computational Biology and Bioinformatics (2017-05-01) https://doi.org/ggmrrp DOI: 10.1109/tcbb.2016.2530062 · PMID: 26890921 [DOI] [PubMed]
- 188.Predicting Drug-Target Interaction Using Deep Matrix Factorization Hafez Eslami Manoochehri, Mehrdad Nourani 2018 IEEE Biomedical Circuits and Systems Conference (BioCAS) (2018-10) https://doi.org/ggmrrn DOI: 10.1109/biocas.2018.8584817
- 189.Network-based prediction of drug–target interactions using an arbitrary-order proximity embedded deep forest Xiangxiang Zeng, Siyi Zhu, Yuan Hou, Pengyue Zhang, Lang Li, Jing Li, L Frank Huang, Stephen J Lewis, Ruth Nussinov, Feixiong Cheng Bioinformatics (2020-01-23) https://doi.org/ggmrrk DOI: 10.1093/bioinformatics/btaa010 · PMID: 31971579 [DOI] [PMC free article] [PubMed]
- 190.DrPOCS: Drug Repositioning Based on Projection Onto Convex Sets Yin-Ying Wang, Chunfeng Cui, Liqun Qi, Hong Yan, Xing-Ming Zhao IEEE/ACM Transactions on Computational Biology and Bioinformatics (2019-01-01) https://doi.org/ggmrrq DOI: 10.1109/tcbb.2018.2830384 · PMID: 29993698 [DOI] [PubMed]
- 191.Neighborhood Regularized Logistic Matrix Factorization for Drug-Target Interaction Prediction Yong Liu, Min Wu, Chunyan Miao, Peilin Zhao, Xiao-Li Li PLOS Computational Biology (2016-02-12) https://doi.org/ggmrrw DOI: 10.1371/journal.pcbi.1004760 · PMID: 26872142 · PMCID: PMC4752318. [DOI] [PMC free article] [PubMed]
- 192.Predicting drug-target interactions by dual-network integrated logistic matrix factorization Ming Hao, Stephen H. Bryant, Yanli Wang Scientific Reports (2017-01-12) https://doi.org/ggmrrj DOI: 10.1038/srep40376 · PMID: 28079135 · PMCID: PMC5227688 [DOI] [PMC free article] [PubMed]
- 193.Drug–Disease Association and Drug-Repositioning Predictions in Complex Diseases Using Causal Inference–Probabilistic Matrix Factorization Jihong Yang, Zheng Li, Xiaohui Fan, Yiyu Cheng Journal of Chemical Information and Modeling (2014-08-22) https://doi.org/f6hpb4 DOI: 10.1021/ci500340n · PMID: 25116798 [DOI] [PubMed]
- 194.Predicting drug-disease associations by using similarity constrained matrix factorization Wen Zhang, Xiang Yue, Weiran Lin, Wenjian Wu, Ruoqi Liu, Feng Huang, Feng Liu BMC Bioinformatics (2018-06-19) https://doi.org/ggmrrt DOI: 10.1186/s12859-018-2220-4 · PMID: 29914348 · PMCID: PMC6006580 [DOI] [PMC free article] [PubMed]
- 195.Deep mining heterogeneous networks of biomedical linked data to predict novel drug–target associations Nansu Zong, Hyeoneui Kim, Victoria Ngo, Olivier Harismendy Bioinformatics (2017-04-18) https://doi.org/gbqjgx DOI: 10.1093/bioinformatics/btx160 · PMID: 28430977 · PMCID: PMC5860112 [DOI] [PMC free article] [PubMed]
- 196.Scalable and Accurate Drug–target Prediction Based on Heterogeneous Bio-linked Network Mining Nansu Zong, Rachael Sze Nga Wong, Victoria Ngo, Yue Yu, Ning Li Cold Spring Harbor Laboratory (2019-02-03) https://doi.org/ggmrrm DOI: 10.1101/539643
- 197.Drug Similarity Integration Through Attentive Multi-view Graph Auto-Encoders Tengfei Ma, Cao Xiao, Jiayu Zhou, Fei Wang arXiv (2018-04-28) https://arxiv.org/abs/1804.10850v1
- 198.Modeling polypharmacy side effects with graph convolutional networks Marinka Zitnik, Monica Agrawal, Jure Leskovec Bioinformatics (2018-06-27) https://doi.org/gfgn55 DOI: 10.1093/bioinformatics/bty294 · PMID: 29949996 · PMCID: PMC6022705 [DOI] [PMC free article] [PubMed]
- 199.DrugBank: a knowledgebase for drugs, drug actions and drug targets David S. Wishart, Craig Knox, An Chi Guo, Dean Cheng, Savita Shrivastava, Dan Tzur, Bijaya Gautam, Murtaza Hassanali Nucleic Acids Research (2007-11-29) https://doi.org/d3qqpj DOI: 10.1093/nar/gkm958 · PMID: 18048412 · PMCID: PMC2238889 [DOI] [PMC free article] [PubMed]
- 200.The human disease network K.-I. Goh, M. E. Cusick, D. Valle, B. Childs, M. Vidal, A.-L. Barabasi Proceedings of the National Academy of Sciences (2007-05-14) https://doi.org/bt6qvc DOI: 10.1073/pnas.0701361104 · PMID: 17502601 · PMCID: PMC1885563 [DOI] [PMC free article] [PubMed]
- 201.Evaluation of knowledge graph embedding approaches for drug-drug interaction prediction in realistic settings Remzi Celebi, Huseyin Uyar, Erkan Yasar, Ozgur Gumus, Oguz Dikenelli, Michel Dumontier BMC Bioinformatics (2019-12) https://doi.org/ggmrrv DOI: 10.1186/s12859-019-3284-5 · PMID: 31852427 · PMCID: PMC6921491 [DOI] [PMC free article] [PubMed]
- 202.Drug-Drug Interaction Prediction Based on Knowledge Graph Embeddings and Convolutional-LSTM Network Md. Rezaul Karim, Michael Cochez, Joao Bosco Jares, Mamtaz Uddin, Oya Beyan, Stefan Decker Proceedings of the 10th ACM International Conference on Bioinformatics, Computational Biology and Health Informatics - BCB ’19 (2019) https://doi.org/ggmrrs DOI: 10.1145/3307339.3342161
- 203.Mining Electronic Health Records using Linked Data. David J Odgers, Michel Dumontier AMIA Joint Summits on Translational Science proceedings. AMIA Joint Summits on Translational Science (2015-03-23) https://www.ncbi.nlm.nih.gov/pubmed/26306276 PMID: 26306276 · PMCID: PMC4525267 [PMC free article] [PubMed]
- 204.Applying linked data principles to represent patient’s electronic health records at Mayo clinic Jyotishman Pathak, Richard C. Kiefer, Christopher G. Chute Proceedings of the 2nd ACM SIGHIT symposium on International health informatics - IHI ’12 (2012) https://doi.org/fzm2p7 DOI: 10.1145/2110363.2110415
- 205.PDD Graph: Bridging Electronic Medical Records and Biomedical Knowledge Graphs via Entity Linking Meng Wang, Jiaheng Zhang, Jun Liu, Wei Hu, Sen Wang, Xue Li, Wenqiang Liu arXiv (2017-07-17) https://arxiv.org/abs/1707.05340v2
- 206.Diagnosis Code Assignment Using Sparsity-Based Disease Correlation Embedding Sen Wang, Xiaojun Chang, Xue Li, Guodong Long, Lina Yao, Quan Z. Sheng IEEE Transactions on Knowledge and Data Engineering (2016-12-01) https://doi.org/f9cgtv DOI: 10.1109/tkde.2016.2605687
- 207.EMR-based medical knowledge representation and inference via Markov random fields and distributed representation learning Chao Zhao, Jingchi Jiang, Yi Guan arXiv (2017-09-20) https://arxiv.org/abs/1709.06908v1 [DOI] [PubMed]
- 208.Attention Is All You Need Ashish Vaswani, Noam Shazeer, Niki Parmar, Jakob Uszkoreit, Llion Jones, Aidan N. Gomez, Lukasz Kaiser, Illia Polosukhin arXiv (2017-06-12) https://arxiv.org/abs/1706.03762v5
- 209.Neural Machine Translation by Jointly Learning to Align and Translate Dzmitry Bahdanau, Kyunghyun Cho, Yoshua Bengio arXiv (2014-09-01) https://arxiv.org/abs/1409.0473v7
- 210.Learning the Graphical Structure of Electronic Health Records with Graph Convolutional Transformer Edward Choi, Zhen Xu, Yujia Li, Michael W. Dusenberry, Gerardo Flores, Yuan Xue, Andrew M. Dai arXiv (2019-06-11) https://arxiv.org/abs/1906.04716v3
- 211.The probability of edge existence due to node degree: a baseline for network-based predictions Michael Zietz, Daniel S. Himmelstein, Kyle Kloster, Christopher Williams, Michael W. Nagle, Blair D. Sullivan, Casey S. Greene anubot (2020-03-05) https://greenelab.github.io/xswap-manuscript/ [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.