Abstract
Objective
To explore the value of CT texture analysis (CTTA) for determining coronavirus disease 2019 (COVID-19) severity.
Methods
The clinical and CT data of 81 patients with COVID-19 were retrospectively analyzed. The texture features were extracted using LK2.1. The two-sample t test or Mann–Whitney U test was used to find the significant features. Minimum redundancy and maximum relevance (MRMR) method was performed to find the features with maximum correlation and minimum redundancy. These features were then used to construct a radiomics texture model to discriminate the severe patients using multivariate logistic regression method. Besides, a clinical model was also built. ROC analyses were conducted to evaluate the performance of two models. The correlations of clinical features and textural features were analyzed using the Spearman correlation analysis.
Results
Of the total cases included, 60 were common and 21 were severe. (1) For textural features, 20 radiomics features selected by MRMR showed good performance in discriminating the two groups (AUC > 70%). (2) For clinical features, chi-square tests or Mann–Whitney U tests identified 16 clinical features as significant, and 12 were discriminative (p < 0.05) between two groups analyzed by univariate logistic analysis. Of these, 10 had an AUC > 70%. (3) Prediction models for textural features and clinical features were established, and both showed high predictive accuracy. The AUC values of textural features and clinical features were 0.93 (0.86–1.00) and 0.95 (0.95–0.99), respectively. (4) The Spearman correlation analysis showed that most textural and clinical features had above-moderate correlations with disease severity (> 0.4).
Conclusion
Texture analysis can provide reliable and objective information for differential diagnosis of COVID-19.
Key Points
• CT texture analysis can well differentiate common and severe COVID-19 patients.
• Some textural features showed above-moderate correlations with clinical factors.
• CT texture analysis can provide useful information to judge the severity of COVID-19.
Electronic supplementary material
The online version of this article (10.1007/s00330-020-07012-3) contains supplementary material, which is available to authorized users.
Keywords: Coronavirus, Disease, Tomography, X-rays
Introduction
In December 2019, coronavirus disease 2019 (COVID-19) was discovered in Wuhan, China, and then spread across the country [1–3]. According to Chinese guideline, COVID-19 is clinically divided into four types: mild, common, severe, and critically severe [4]. Compared with healthy individuals, mild COVID-19 shows no obvious CT abnormality, but the other three do. Yang et al [5] reported that the mortality of severe COVID-19 in Wuhan exceeded 60%. Therefore, early and correct evaluation of COVID-19 severity and timely treatment can improve patients’ prognosis.
CT plays an important role in the evaluation of COVID-19. Clinical CT results are made by physicians through analyzing the size, shape, position, and internal density of CT images. However, this analysis fails to capture diagnostic/prognostic information about the lesion. Through, texture analysis, mathematical methods are used to extract meaningful characteristics of an image at different gray levels, so as to quantify the lesion’s heterogeneity [6]. This retrospective study was performed to analyze the correlations among CT texture features, clinical features, and clinical subtypes of COVID-19 and to explore the value of CT texture analysis in determining the severity of COVID-19.
Materials and methods
Patients and ethical approval
Our institutional review board (IRB) waived written informed consent for this retrospective study, which evaluated de-identified data and brought no potential risk to patients. To avert any potential breach of confidentiality, no link between the patients and the researchers was available.
The patients’ data were collected from the First Affiliated Hospital of University of Science and Technology of China and the Affiliated Infectious Disease Hospital. During the period between January 20, 2020 and February 20, 2020, patients were included if they met the following criteria: (1) exhibiting positive results of 2019-nCoV nucleic acids and (2) having undergone chest CT examination during the initial diagnosis (within 3 days after admission). Excluded were those who had no obvious lung CT abnormalities or had pneumonia caused by other common bacterial or viral pathogens. According to the clinical classification criteria, 81 patients were enrolled (60 common cases and 21 severe cases).
Scanning
CT was performed for all patients within 3 days after disease onset, with a 128-slice CT detector (NeuViz 128) without contrast agent. The scanning parameters were as follows: tube voltage, 120 kVp; tube current, 150 mA; rotation time, 0.8 s; slice thickness, 5 mm; slice interval, 5 mm; pitch, 1.2; matrix, 512 × 512; and breath-holding when fully aspirated.
Region-of-interest segmentation and feature extraction
All images were segmented on the LK2.1 software package (GE Healthcare). First, the images were resampled to voxel size 1 × 1 × 1 mm3, and a Gaussian filter was applied for denoising. Then, the lung was automatically segmented into five lobes, three-dimensional volumes of interest (VOIs) were created for each lobe, and the CT score was calculated [7]. If the automatic segmentation failed to create favorable volumes, an experienced radiologist was assigned to manually delineate the VOIs, and another radiologist to check the segmentation. Any difference in opinions was resolved through negotiation. Pyradiomics was performed to extract features. In total, 1042 features were extracted, including histogram features, gray-level co-occurrence matrix, gray-level size zone matrix (GLSZM), and gray-level run length matrix (GLRLM) features.
Statistical analysis of radiomics and clinical features
All statistical analyses were performed using R (version 3.5.1; www.R-project.org) software. First, for radiomics features and CT score, the independent t test or Wilcoxon test was used. For clinical features described as continuous variables, the Mann–Whitney U test was used for abnormal distributions and t test for normal distributions. For clinical features described as nominal variables, chi-square test or Fisher’s exact test was used. Features with p <0.05 were deemed statistically significant. Second, for radiomics features, univariate logistic analysis was performed to evaluate whether the features were discriminative in two groups (p < 0.05). Then, the minimum redundancy and maximum relevance (MRMR) algorithm was applied to further select the relevant and non-redundant features. For clinical factors, univariate logistic analysis was performed to find the discriminative features (p < 0.05). Third, for both radiomics features and clinical features, backward stepwise multivariate logistic regression selection was performed, and the likelihood ratio test was used to select the subset of the most predictive features and construct the predictive model. Finally, to verify the reliability of texture analysis, 100-fold leave-group-out cross-validation (LGOCV) was performed. Finally, Spearman correlation analyses were performed to evaluate the correlations between the predictive radiomics features and clinical factors.
Results
Eighty-one patients with a mean age of 51.35 ± 14.31 years diagnosed with COVID-19 were included. Of which, 21 (26%) had severe and 60 (74%) had common symptoms.
The CT characteristics included ground-glass opacity (GGO) or GGO combined with fine grid or consolidation. Patients with severe disease exhibited diffuse lesions; in four patients, initial CT images showed involvement in all the segments of both lungs. Most lesions were observed in the peripheral zone of the lung field, particularly the lower lobe. Figures 1 and 2 show representative images of common and severe COVID-19, respectively.
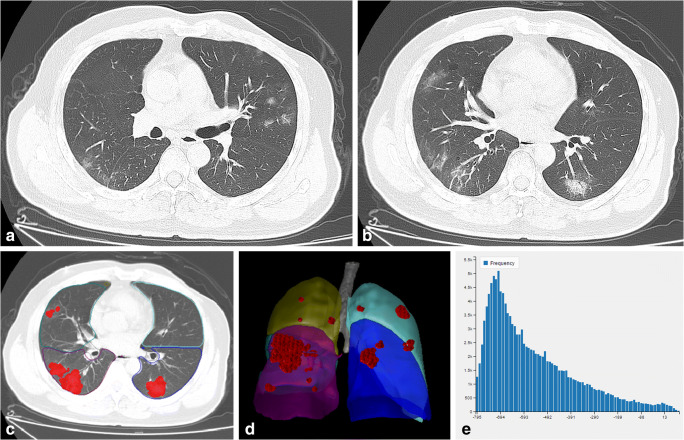
Fig. 1.
A common case of COVID-19. A man aged 53 years presented with a 3-day history of fever, cough, and sputum. a, b On the third day after disease onset, CT imaging revealed pure ground-glass opacity (GGO) and GGO with fine grid in the bilateral lobes. c, d The area of the lesions was delineated on the axial and reconstructed images. e Histogram map of the lesions
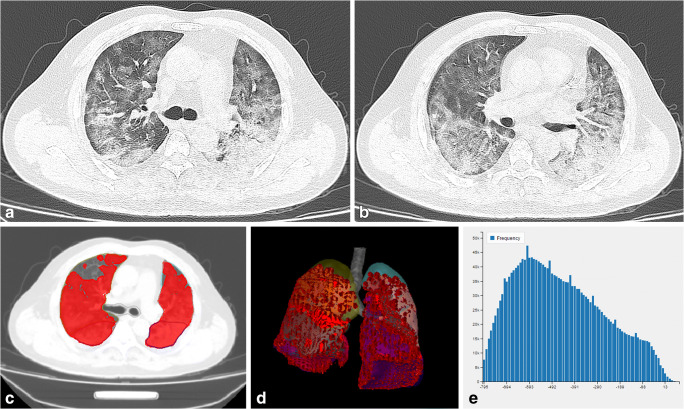
Fig. 2.
A severe case of COVID-19. A man aged 47 years presented with a 7-day history of fever, cough, and sputum. a, b On the third day after disease onset, CT imaging revealed diffuse pure GGO and GGO with fine grid in the bilateral lobes. c, d The area of the lesions was delineated on the axial and reconstructed images. e Histogram map of the lesions
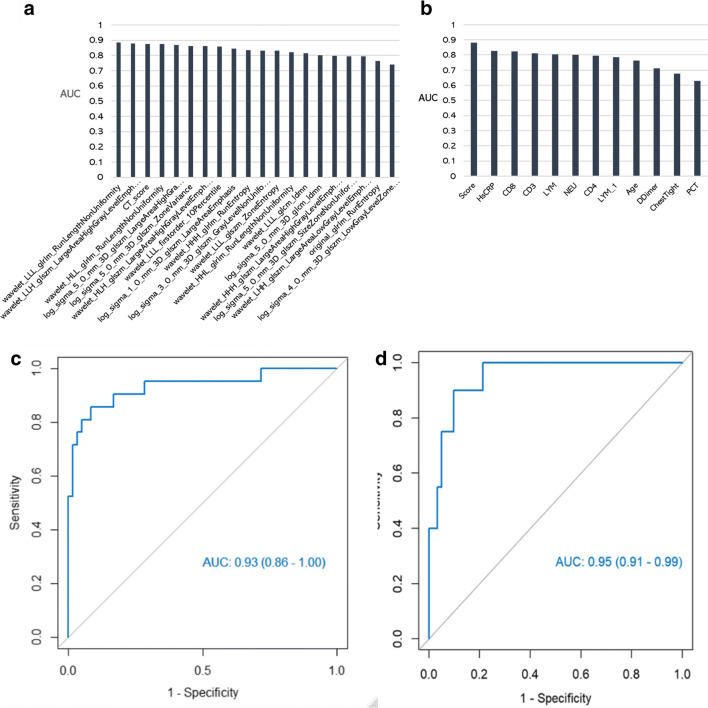
Of the 1042 radiomics features, 511 were statistically significant when assessed by independent t test or Mann–Whitney test (p < 0.05). Of these, 358 were selected by univariate logistic analysis (p < 0.05), then 20 were retained after MRMR analysis. The AUC values of these 20 radiomics features are shown in Fig. 3 a. All of them showed good predictive performance, with AUC values > 70%. Of the clinical factors, 16 features were statistically significant when assessed by the chi-square or Mann–Whitney U test (Table 1), and 12 clinical features were retained after univariate logistic regression (p < 0.05). These 12 features’ AUC values are shown in Fig. 3 b. After backward stepwise multivariate logistic regression and selection with the likelihood ratio test, eight radiomics features (Table 2) and four clinical features (Table 3) were finally retained, and predictive models were constructed. Both the radiomics signature and the clinical model showed favorable predictive accuracy, with AUC values of 0.93 (0.86–1.0) and 0.95 (0.95–0.99), respectively (Fig. 3c, d).
Fig. 3.
a Performance of 20 radiomics features. All showed good performance (AUC > 70%). b AUC values of 12 clinical features. Both the radiomics signature and the clinical model showed favorable predictive accuracy. c The radiomics signature had an AUC value of 0.93 (0.86–1.00). d The corresponding AUC value for the clinical model was 0.95 (0.95–0.99)
Table 1.
Epidemiological history and clinical data of 81confirmed patients
| Variable | Sample | class 1 | class 2 | Statistics | p value |
| Gender | |||||
| male | 47 | 32 (52.46%) | 15 (75.00%) | 3.142 | 0.076 |
| female | 34 | 29 (47.54%) | 5 (25.00%) | ||
| Epidemiology | |||||
| Epidemiology1 | 24 | 20 (32.79%) | 4 (20.00%) | – | 0.037 |
| Epidemiology2 | 17 | 8 (13.11%) | 9 (45.00%) | ||
| Epidemiology3 | 16 | 14 (22.95%) | 2 (10.00%) | ||
| Epidemiology4 | 24 | 19 (31.15%) | 5 (25.00%) | ||
| Cough | |||||
| NO | 18 | 13 (21.31%) | 5 (25.00%) | 0.001 | 0.973 |
| yes | 63 | 48 (78.69%) | 15 (75.00%) | ||
| expectoration | |||||
| NO | 58 | 40 (65.57%) | 18 (90.00%) | 4.42 | 0.036 |
| yes | 23 | 21 (34.43%) | 2 (10.00%) | ||
| dyspnea | |||||
| no | 80 | 61 (100.00%) | 19 (95.00%) | – | 0.247 |
| yes | 1 | 0 (0.00%) | 1 (5.00%) | ||
| Fatigue | |||||
| no | 60 | 45 (73.77%) | 15 (75.00%) | 0.012 | 0.913 |
| yes | 21 | 16 (26.23%) | 5 (25.00%) | ||
| nausea and vomiting | |||||
| no | 80 | 61 (100.00%) | 19 (95.00%) | – | 0.247 |
| Yes | 1 | 0 (0.00%) | 1 (5.00%) | ||
| diarrhea | |||||
| no | 73 | 58 (95.08%) | 15 (75.00%) | 4.754 | 0.029 |
| yes | 8 | 3 (4.92%) | 5 (25.00%) | ||
| Tight chest | |||||
| no | 62 | 52 (85.25%) | 10 (50.00%) | 8.551 | 0.003 |
| yes | 19 | 9 (14.75%) | 10 (50.00%) | ||
| thoracodynia | |||||
| no | 78 | 58 (95.08%) | 20 (100.00%) | 0.0 | 0.571 |
| yes | 3 | 3 (4.92%) | 0 (0.00%) | ||
| shake | |||||
| no | 74 | 57 (93.44%) | 17 (85.00%) | 0.501 | 0.479 |
| yes | 7 | 4 (6.56%) | 3 (15.00%) | ||
| PCT | |||||
| no | 25 | 23 (37.70%) | 2 (10.00%) | 5.984 | 0.05 |
| yes | 56 | 38 (62.3%) | 18 (90.00%) | ||
| Age | 81 | 43.00 (29.00, 51.00) | 56.50 (44.45, 68.00) | −3.499 | < 0.001 |
| temperature | 81 | 36.90 (36.50, 37.30) | 36.70 (36.34, 37.39) | 0.964 | 0.335 |
| heartrate | 81 | 85.00 (78.00, 90.60) | 85.00 (82.00, 95.10) | −1.227 | 0.22 |
| WBC | 81 | 5.21 ± 1.81 | 5.50 ± 1.47 | −0.651 | 0.517 |
| NEU% | 81 | 64.54 ± 13.70 | 79.52 ± 10.94 | −4.442 | < 0.001 |
| LYM% | 81 | 26.80 (17.72, 35.19) | 12.45 (8.23, 18.90) | 4.085 | < 0.001 |
| Mon% | 81 | 7.50 (5.64, 8.36) | 5.60 (4.59, 7.61) | 1.922 | 0.055 |
| NEU | 81 | 3.15 (2.12, 4.20) | 4.61 (3.42, 5.31) | −2.125 | 0.034 |
| LYM | 81 | 1.29 (0.87, 1.69) | 0.74 (0.40, 1.07) | 3.806 | < 0.001 |
| Mon | 81 | 0.36 (0.27, 0.46) | 0.33 (0.24, 0.51) | 0.274 | 0.784 |
| ESR | 81 | 30.10 (25.85, 49.60) | 30.10 (23.30, 63.21) | −0.027 | 0.978 |
| MYT | 81 | 30.00 (28.00, 34.90) | 35.50 (29.45, 73.00) | −2.256 | 0.024 |
| hsCRP | 81 | 8.50 (1.56, 22.40) | 55.35 (16.59, 98.27) | −4.37 | < 0.001 |
| ALT/AST | 81 | 0.78 (0.63, 1.12) | 0.95 (0.67, 1.34) | −1.035 | 0.301 |
| CK | 81 | 86.40 (54.41, 144.64) | 106.90 (73.39, 192.00) | −1.04 | 0.298 |
| DDimer | 81 | 0.20 (0.09, 0.29) | 0.28 (0.22, 0.57) | −2.826 | 0.005 |
| CD4 | 81 | 513.00 (376.00, 828.20) | 271.50 (123.70, 338.95) | 3.926 | < 0.001 |
| CD8 | 81 | 350.00 (215.20, 638.48) | 152.12 (103.07, 213.90) | 4.315 | < 0.001 |
| CD3 | 81 | 927.00 (632.50,1593.40) | 449.00 (247.75, 594.10) | 4.14 | < 0.001 |
| CD4_CD8 | 81 | 1.41 (1.22, 1.79) | 1.51 (0.88, 2.06) | 0.0 | 1.0 |
| score | 81 | 8.00 (3.00, 12.00) | 16.00 (14.45, 18.55) | −5.076 | < 0.001 |
Table 2.
Multivariate logistic of the predictive radiomics features
| VarName | OR(95%CI) | p value | AUC |
| (Intercept) | 0.14(0.04–0.37) | 0.00 | |
| log_sigma_1_0_mm_3D_glszm_LargeAreaEmphasis | 0.03(0.00–0.42) | 0.03 | 0.86 |
| log_sigma_5_0_mm_3D_glszm_SizeZoneNonUniformity | 0.23(0.03–1.34) | 0.12 | 0.80 |
| CT_score | 3.20(0.73–20.92) | 0.17 | 0.87 |
| wavelet_HHL_glrlm_RunLengthNonUniformity | 129.08(2.19–3637.51) | 0.04 | 0.82 |
| wavelet_HLL_glrlm_RunLengthNonUniformity | 0.00(0.00–0.40) | 0.05 | 0.87 |
| wavelet_HLH_glszm_LargeAreaHighGrayLevelEmphasis | 41.03(2.95–1498.64) | 0.03 | 0.86 |
| log_sigma_5_0_mm_3D_glszm_LargeAreaHighGrayLevelEmphasis | 14.23(0.70–809.09) | 0.13 | 0.87 |
| log_sigma_3_0_mm_3D_glszm_GrayLevelNonUniformity | 60.43(1.82–7167.65) | 0.04 | 0.83 |
Table 3.
Multivariate logistic of the predictive clinical features
| VarName | OR(95%CI) | p value | AUC |
| (Intercept) | 0.05(0.001–0.20 | 0.00 | |
| score | 2.45(0.99–7.59) | 0.08 | 0.88 |
| CD8 | 0.06(0.001–0.51 | 0.03 | 0.82 |
| hsCRP | 3.77(1.48–12.73) | 0.01 | 0.83 |
| Tight chest | 2.52(1.15–6.19) | 0.03 | 0.68 |
The sensitivity, specificity, and accuracy were calculated based on the Youden index (Table 4). Both the clinical model and radiomics signature showed good performance in discriminating patients with common and severe COVID-19. In the clinical model, the accuracy, sensitivity, and specificity values were 0.91, 0.81, and 0.95, respectively; in the radiomics signature, these values were 0.90, 0.90, and 0.90, respectively.
Table 4.
The sensitivity, specificity, accuracy of clinical model and radiomcs signature
| threshold | Accuracy | Sensitivity | Specificity | Pos Pred Value | Neg Pred Value | |
| Clinical model | −0.77 | 0.90 | 0.9 | 0.90 | 0.75 | 0.96 |
| Radiomics signature | 0.04 | 0.91 | 0.81 | 0.95 | 0.85 | 0.93 |
The mean accuracy, sensitivity, and specificity values of the 100-fold LGOCV are shown in (Table 5). Both the clinical and radiomics models showed good stability, indicating that the texture analysis was valuable for discriminating common and severe COVID-19 patients, and that the results were not due to overfitting.
Table 5.
Cross validation of clinical model and radiomcs signature
| Group | Accuracy | Sensitivity | Specificity | |
| Clinical model | Training | 0.97 | 0.98 | 0.97 |
| Test | 0.90 | 0.93 | 0.81 | |
| Radiomics signature | Training | 0.95 | 0.96 | 0.91 |
| test | 0.89 | 0.92 | 0.80 |
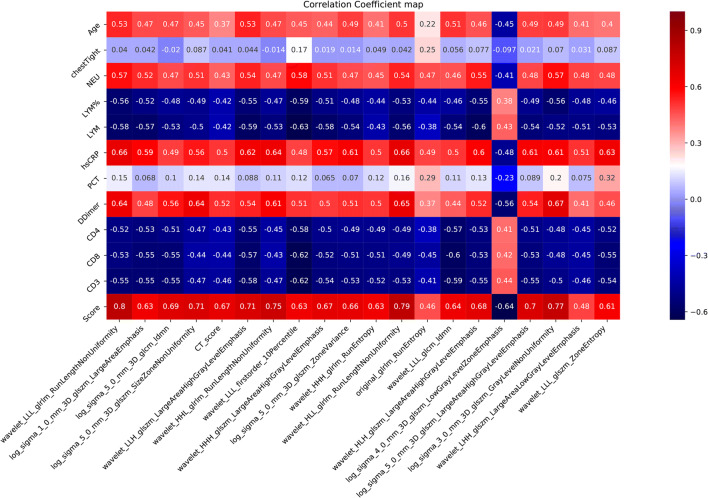
The correlations between the predictive radiomics and clinical features are shown in Fig. 4. The association was defined as strong (r = 0.7–1), moderate (r = 0.4–0.69), or low (r = 0.1–0.39). The prediction score showed strong correlations with wavelet_LLL_glrlm_RunLengthNonUniformity (r = 0.80, p < 0.0001), log_sigma_5_0_mm_3D_glszm_SizeZoneNonUniformity (r = 0.71, p < 0.0001), wavelet_HHL_glrlm_RunLengthNonUniformity (r = 0.75, p < 0.0001), and wavelet_HLL_glrlm_RunLengthNonUniformity (r = 0.79, p < 0.001). Most other clinical features and radiomics features showed moderate correlations (Fig. 4).
Fig. 4.
Correlations between the predictive radiomics and clinical features. There is a strong association between the correlation scores and radiomics features
Discussion
Chest CT can be used to make rapid diagnosis and assess the severity of COVID-19 patients [8]. On CT images, COVID-19 may be manifested with GGO, GGO combined with grid, and/or consolidation, all changing rapidly [9]. We can judge the severity of the disease by the lesion range on the CT image, the results of which may be subjective and inaccurate.
In CT texture analysis, image features of different gray levels can be filtered out. An image’s pixel and spatial parameters are used to quantitatively extract pathophysiological features of the lesions that cannot be recognized by naked eyes and to reveal the internal heterogeneity between tissues [10–12]. Texture analysis is objective in that it evaluates lesion with gray level on the image [13].
COVID-19 is an acute and highly infectious disease [12]. Early prediction and timely treatment can improve patients’ prognosis. The clinical stages of COVID-19 have been rarely classified by texture analysis based on CT images. In this study, a radiomics texture model and a clinical model were established for 81 patients to early predict the clinical stages of COVID-19, and a correlational study was conducted to reveal the associations between CT and clinical variables in common and severe patients. Both radiomics and clinical models showed favorable predictive accuracy, with an AUC of 0.93 (0.86–1.0) and 0.95 (0.95–0.99), respectively. The prediction model based on CT text features achieved a sensitivity of 0.80 and a specificity of 0.95, which means that CT texture analysis can early accurately assess disease severity.
It has been reported that coronavirus acts mainly on lymphocytes, especially T lymphocytes, as SARS-CoV does. Therefore, a decrease in lymphocytes can be used as a reference index for clinical diagnosis of new coronavirus infection [14–17]. CD3 represents the total count of T lymphocytes, and CD4 or CD8 represents that of their subsets. Among the clinical features measured in this research, the percentage and absolute count of lymphocytes and the counts of CD3, CD4, and CD8 in the severe group decreased significantly and showed high diagnostic value to differentiate patients of different COVID-19 severity levels (all AUC > 70%). These results suggest that the lymphocyte count in the severe group was much lower than that in the common group. Feng et al [18] used a semi-quantitative scoring system to quantitatively estimate the pulmonary involvement of lung lesions. The inflammation score was correlated with the severity. Our results showed that the inflammation score was also effective in the differential diagnosis between the two groups (AUC = 88%).
We used LGOCV to verify the reliability of the multivariate logistic regression model, and the results showed that both the imaging and clinical models had good stability. In the training and validation sets, the average accuracy, sensitivity, and specificity values resulting from 100-fold cross-validation were above 0.8, indicating that the results of the model were not caused by overfitting. The results of 100-fold cross-validation in the imaging model showed that the logarithmic transformation characteristics of log_sigma_3_0_mm_3D_glszm_GrayLevelNonUniformity and log_sigma_1_0_mm_3D_glszm_LargeAreaEmphasis appeared more than 50 times during the 100 times cross-validations (Supplementary Materials). The glszm_GrayLevelNonUniformity variable was used to measure the variability of gray-level intensity values in the image, with lower values indicating higher homogeneity of intensity values. Further, glszm_LargeAreaEmphasis was used to measure the distribution of large-sized area zones, with greater values indicating more large-sized area zones and more coarse textures. And the wavelet characteristics include wavelet_HHL_glrlm_RunLengthNonUniformity and wavelet_HLH_glszm_LargeAreaHighGrayLevelEmphasis, of which wavelet_glrlm_RunLengthNonUniformity measures the similarity of run lengths throughout the image, with a lower value indicating more homogeneity among run lengths in the image, and wavelet_HLH_glszm_LargeAreaHighGrayLevelEmphasis measures the proportion in the image of the joint distribution of larger-sized zones with higher gray level. Besides, the CT score also showed more than 50 times, which means these features had high stability and diagnostic values of the severity of the patients. In the clinical model, inflammation score, hsCRP, CD3, PCT, and other factors showed more than 50 times during the 100 times cross-validation (Supplementary Materials), indicating that these parameters were highly correlated with the severity of COVID-19. In addition, the Spearman correlation analysis was performed to evaluate the correlation between significant imaging and clinical features. The results showed that most of the clinical features (e.g., lymphocyte ratio; absolute value; CD3, CD4, and CD8 counts; CRP; and D dimer) had moderate correlations with the imaging features (> 0.4). The strong correlation was found between inflammatory score and partial wavelet transform features and region size matrix GLSZM features (> 0.7), indicating that these image features are closely related to disease severity and can be used for clinical type classification of the COVID-19 patients.
This study has some limitations. Firstly, this study is retrospective, and the sample size is small, especially in the severe group, with only 21 cases. Secondly, some cases have small ground-glass lesions that may be missed when the ROI is automatically delineated. In future studies, it is necessary to increase the number and type of samples. Thirdly, in clinical practice, COVID-19 should be differentiated from other pulmonary diseases or other pneumonias. But, we did not apply CTTA to differentially diagnose COVID-19 in this study. Finally, our results may have had some bias. In this study, we did not consider the effect of patient’s underlying disease, such as chronic respiratory disease (emphysema or interstitial pneumonia, etc.), on the severity judgment. We will establish a multicenter study to explore the value of CT texture analysis in the differential diagnosis of COVID-19.
In summary, texture analysis can provide reliable and objective information for differential diagnosis of COVID-19.
Electronic Supplementary Material
(DOCX 57.9 kb)
Abbreviations
- COVID-19
Coronavirus disease 2019
- CTTA
CT texture analysis
- GGO
Ground-glass opacity
- GLRLM
Gray-level run length matrix
- GLSZM
Gray-level size zone matrix
- LGOCV
Leave-group-out cross-validation
- MRMR
Minimum redundancy and maximum relevance
- VOI
Volume of interest
Funding information
This study has received funding by the Fundamental Research Funds for the Central Universities (WK9110000002).
Compliance with ethical standards
Guarantor
The scientific guarantor of this publication is Wei Wei, MD.
Conflict of interest
One of the authors of this manuscript (Ya-qiong Ge) is an employee of GE Healthcare. The remaining authors declare no relationships with any companies whose products or services may be related to the subject matter of the article.
Statistics and biometry
Yaqiong Ge kindly provided statistical advice for this manuscript, and no complex statistical methods were necessary for this paper.
Informed consent
Written informed consent was waived by the institutional review board.
Ethical approval
Institutional review board approval was obtained.
Methodology
• retrospective
• diagnostic or prognostic study
• performed at one institution
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.WHO, Novel coronavirus – China, Jan 12 (2020) (Accessed Jan 19, 2020), http://www.who.int/csr/don/12-january-2020-novel-coronavirus-china/en/
- 2.Chen NS, Zhou M, Dong X, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.World Health Organization. WHO Director-Generals remarks at the media briefing on 2019-nCoV on 11 February 2020 [EB/OL]. https://www.who.int/dg/speeches/detail/who-director-general-s-remarks-at-the-media-briefing-on-2019-ncov-on-11-february-2020
- 4.General Office of National Health Committee. Office of State Administration of Traditional Chinese Medicine. Notice on the issuance of a programme for the diagnosis and treatment of novel coronavirus (2019-nCoV) infected pneumonia (trial sixth edition) (2020-02-18) [EB/OL]. http://yzs.satcm.gov.cn/zhengcewenjian/2020-02-19/13221.html. (In Chinese)
- 5.Yang X, Yu Y, Xu J et al (2020) Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir Med. 10.1016/S2213-2600(20)30079-5 [DOI] [PMC free article] [PubMed]
- 6.Castellano G, Bonilha L, Li LM, Cendes F. Texture analysis of medical images. Clin Radiol. 2004;59:1061–1069. doi: 10.1016/j.crad.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 7.Chang YC, Yu CJ, Chang SC, et al. Pulmonary sequelae in convalescent patients after severe acute respiratory syndrome: evaluation with thin-section CT. Radiology. 2005;236:1067–1075. doi: 10.1148/radiol.2363040958. [DOI] [PubMed] [Google Scholar]
- 8.Wang YH, Dong CJ, Hu Y et al (2020) Temporal changes of CT findings in 90 patients with COVID-19 pneumonia: a longitudinal study. Radiology. 10.1148/radiol.2020200843 [DOI] [PMC free article] [PubMed]
- 9.Pan Y, Guan H, Zhou S et al (2020) Initial CT findings and temporal changes in patients with the novel coronavirus pneumonia (2019-nCoV): a study of 63 patients in Wuhan. Eur Radiol. 10.1007/s00330-020-06731-x [DOI] [PMC free article] [PubMed]
- 10.Lubner MG, Smith AD, Sandrasegaran K, Sahani DV, Pickhardt PJ. CT texture analysis: definitions, applications, biologic correlates, and challenges. Radiographics. 2017;37:1483–1503. doi: 10.1148/rg.2017170056. [DOI] [PubMed] [Google Scholar]
- 11.Muller H, Michoux N, Bandon D, Geissbuhler A. A review of content-based image retrieval systems in medical applications-clinical benefits and future directions. Int Med Inform. 2004;73:1–23. doi: 10.1016/j.ijmedinf.2003.11.024. [DOI] [PubMed] [Google Scholar]
- 12.Ondimu SN, Murase H. Effect of probability-distance based Markovian texture extraction on discrimination in biological imaging. Comput Electron Agric. 2008;63:2–12. doi: 10.1016/j.compag.2008.01.007. [DOI] [Google Scholar]
- 13.Gillies RJ, Kinahan PE, Hricak H. Radiomics: images are more than pictures, they are data. Radiology. 2016;278:563–577. doi: 10.1148/radiol.2015151169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu X, Chen P, Wang J, et al. Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Sci China Life Sci. 2020;673:457–460. doi: 10.1007/s11427-020-1637-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koo HJ, Lim S, Choe J, Choi SH, Sung H, Do KH. Radiographic and CT features of viral pneumonia. Radiographics. 2018;38:719–739. doi: 10.1148/rg.2018170048. [DOI] [PubMed] [Google Scholar]
- 16.Huang CL, Wang YM, Li XW, Ren LL, Zhao JP, Hu Y. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu Z, Shi L, Wang YJ et al (2020) Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir Med. 10.1016/s2213-2600(20)30076-x [DOI] [PMC free article] [PubMed]
- 18.Feng P, Ye TH, Peng S et al (2020) Time course of lung changes on chest CT during recovery from 2019 novel coronavirus (COVID-19) pneumonia. Radiology. 10.1148/radiol.2020200370
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 57.9 kb)