Abstract
Mutation is the most powerful driver of change for life on Earth. Pathogenic bacteria utilize mutation as a means to survive strong live-die selective pressures generated by chemical antibiotics. As such, the traditional drug-making pipeline, characterized by significant financial and time investment, is insufficient to keep pace with the rapid evolution of bacterial resistance to structurally fixed and chemically unmalleable antibacterial compounds. In contrast, the genetic diversity and adaptive mutability of the bacteriophage can be leveraged to not only overcome resistance but also used for the development of enhanced traits that increase lytic potential and therapeutic efficacy in relevant host microenvironments. This is the fundamental premise behind Baylor College of Medicine's Tailored Antibacterials and Innovative Laboratories for Phage (Φ) Research (TAILΦR) initiative. In this perspective, we outline the concept, structure, and process behind TAILΦR's attempt to generate a personalized therapeutic phage that addresses the most clinically challenging of bacterial infections.
Keywords: bacteriophage, phage therapy, BCM tailor labs
Introduction
The abundance and diversity of life on Earth is generated by the most powerful driver of evolution—mutation. It is the process from where all extant organisms came. Without mutation, the diversity of plant, fish, mammals, and microbes, or their colors, shapes, adaptations, and chemistries, that collectively constitute our living planet is not possible.
The concept of mutation as a driver of change was elusive. Darwin's theory of natural selection was sufficient to describe the process of change, but he, and his disciples, struggled to explain the mechanism.1 At that time, the concept of a gene was unknown. There was talk of particles but the gardening monk Gregor Mendel, unbeknown to the world, had secretly worked out that something must be inherited in his garden peas; how else can one explain the near-perfect mathematical arrangement of the lengths, colors, and shapes of the peas and stems, those so-called traits, every time there was a parental cross?2 It would take another botanist, Hugo de Vries, to not only “rediscover” in his own work the idea of inheritance but also observe the bizarre appearance of new traits as he bred the evening primrose.3 There must be a “pangene” responsible, and it can change … it can mutate.4 From Drosophila to yeast to mice to genome-wide association studies in humans, the gene and its mutations are now recognized as the fundamental unit of change in all biology.5–7
For millions of years, bacteria have laid siege to Homo sapiens and its hominid predecessors. The very water you drank or food you ate could be your last. Simple wounds became putrid, and infections of the lungs and nasopharynx turned deadly, especially in children. Childbirth brought high rates of infection to both baby and mother. But in the 1900s, three miracles of science pushed bacteria further back than they had ever been pushed.8 Civil engineering and hygiene cleaned our water and food and prevented transmission. Vaccines ameliorated some of the deadliest childhood scourges. Antibiotics, the miracle of miracles, covered the rest. The golden years of antibiotic discovery and synthesis, whereby a hundred different variants built around five or six core structures, fueled an entire industry for a good 50 years.9 Entire companies were built on the success of antibiotics. So confident were we that the age of bacterial dominance over humanity had come to an end that prominent members of the medical community publicly stated so.10
In our temporary triumph over these microscopic creatures, we lost sight of the original and most fundamental of all observations: that life is built on change. The very bacteria we wrote off as slain wield mutability as their greatest asset to undermine our efforts.
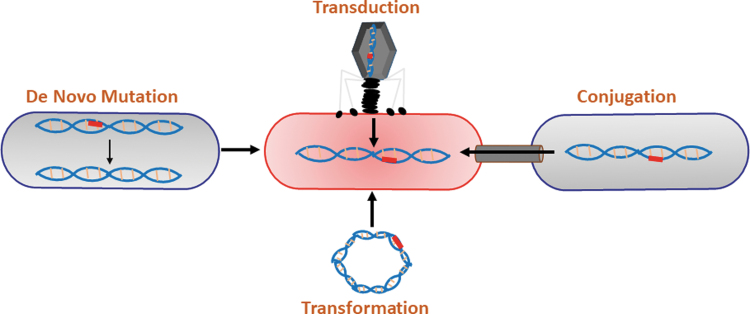
Change they do, and quickly, and by several ways, which is precisely the problem. There are four main mechanisms of mutagenesis in bacteria. The first is de novo mutation, which is the result of an error in the copying of the DNA into the daughter strains (e.g., insertion of an extra base, deletion or loss of a base, and substitution with an incorrect base). The second is transformation, whereby entire pieces of DNA (such as plasmids) are either taken in or ejected out of the cell. The third is conjugation, the bacterial version of sex where a bacterium transfers genetic material to another bacterium through a cell–cell contact. Finally, transduction, the insertion of foreign DNA, usually from viruses or mobile elements, leaves parts of their nucleic acid behind as a kind of primitive parasite. These four intersecting highways, what we can creatively describe as the Mutagenic Tetrasect,8 are constantly paving a new path for bacteria to generate new variants in the face of strong selective pressures (Fig. 1). It is the mutagenic tetrasect that has so deeply threatened the therapeutic impact of antibiotics.
FIG. 1.
The mutagenic tetrasect. The mutagenic tetrasect consists of de novo mutation, transformation, conjugation, and transduction. Each of these means of acquiring new DNA content contributes to the spread, virulence, and resistance to antibiotics of pathogenic bacteria. Reprinted with permission from Maresso, A.W. Bacterial Virulence: A Conceptual Primer, Textbook, 2019.
Because of mutation, the evolution of bacterial mechanisms that destroy our antibacterial drugs will continue. But there is another confounding, more overlooked, issue also at play; antibiotics do not change. After 10 years of planning, screening, development, medicinal chemistry, testing in animals, testing in humans, approval, large-scale production, and distribution, at a cost of perhaps a billion dollars, that well-designed and hopeful new antibiotic can be introduced into clinical use and a short time later a report may appear that describes a resistant case.11,12 Why? The ability of bacteria to mutate via the tetrasect, combined with their short generation time (in some cases 20 min), means that a rare variant with a slightly better fitness can be selected under life-or-death pressures. Essentially, it is a math problem. Take your microbiome, the microbial ecosystem on and inside you. Every day, it is estimated that you generate 9 million new mutant strains of bacteria in your microbiome,13 yielding potentially 3.5 billion novel mutants per year. There are nearly 8 billion people. In 1 year, that is ∼1018 total mutants, just shy of what astronomers believe are the number of stars in the known universe.14 That calculation does not even mention cows, chickens, pigs, the soil, or all the lakes and oceans on this planet generating bacterial mutants. Of course, most of these mutations are redundant and not all of them are beneficial; but only one is needed. A single change of one letter of the code can produce a novel enzymatic variant that confers an enhanced ability to catalytically destroy an antibiotic.15,16 Inactivation of antibiotics via catalytic mechanisms is just one way that bacteria become resistant. They also can inhibit, block, and pump antibiotics out as well as activate alternative metabolic pathways to circumvent the action of the antibiotic.17
Using their mutagenic talents, bacteria have regained an advantage. The rise of antibiotic resistance over the past 70 years has led to dire predictions for the future, with 10 million deaths by 2050 at a cost of 100 trillion dollars.18 As people live longer and require more and more extreme measures to keep them alive—devices, prosthetics, catheters, implants, and treatments that inherently suppress the immune system (chemotherapy for cancer or immunosuppression for transplants, as examples)—these mutable “opportunists” will continue to infect. The paradigm has changed. What was once a great concern, that we could “catch” something from the outside—a dirty well, a rusty nail, etc.—has now morphed into a concern from “within.” The so-called ESKAPE bacteria (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter spp.) are considered the highest concern for multidrug-resistant bacteria by the U.S. Centers for Disease Control. These ESKAPE pathogens, most of which constitute native microbiome that live on and in us, have become the new plague.19 They use us as carriers, infect our catheters and devices and surgical wounds (many cases being chronic), spring-resistant variants during prolonged antibiotic treatment, and finally lay siege to the body during bacteremia and/or sepsis. They spread in the meantime, lingering in our hospitals, clinics, and nursing homes so that when they infect the next patient, there are limited or no treatment options for the attending clinician. If the drugs we keep throwing at them fall to the power of molecular evolution, what do we do next?
Enter bacteriophage, nature's 2 billion year-old flu for bacteria, a kind of plague for the plague, and its ironic counter-evolutionary masterpiece. The readers of this piece do not need a lengthy description of the biology of phage or their virtues in phage therapy. These excellent reviews cover those topics.20–24 What we will stress here is that phages harbor the largest biological repository of undiscovered antibacterial elements on Earth. Antibiotics are fixed structures. Medicinal chemists can add new substituents to the main frame to produce new compounds, pair antibiotics with adjuvants (e.g., sugars, terminal electron acceptors, beta-lactamase inhibitors), or produce new combinations of existing antibiotics, but the chemical space available to them and the possible combinations of existing drugs and adjuvants are limited. This is not so with nucleic acid, a phage's own secret weapon. The arrangement of all those As, Gs, Cs, and Ts along a linear stretch, combined with an arrangement that produces an even more complex protein with not 4 permutations per slot but 20 (the amino acids), generates possibilities that are so varied that its computation is impractical. The sheer number and mutagenic potential of lytic phage, combined with mutations that can occur in real time and not only be enhanced in the laboratory but selected for, turns the evolutionary table on bacteria. It matters not how the trajectory of history anointed antibiotics over phage in the first pass of this story, but just that we now have the ability to write the next chapter. Critical adherence to new explorations and rigorous science will be the pen.
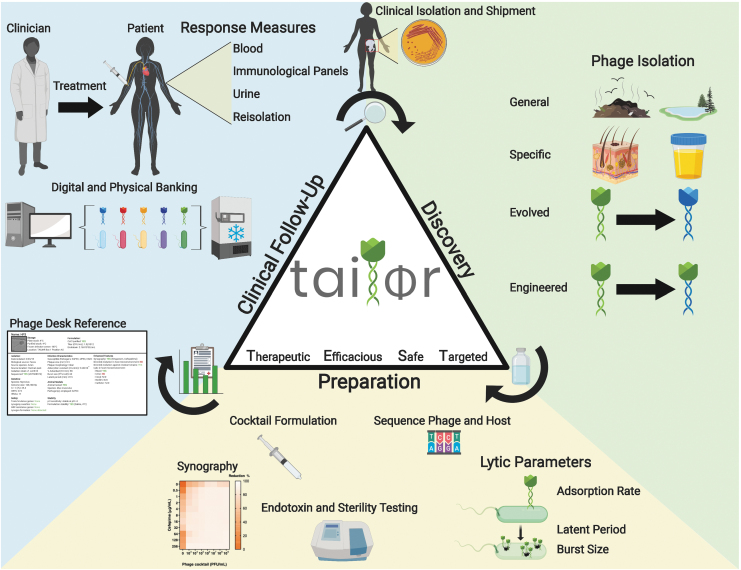
Either we change the way we develop and approve new drugs, or we develop and approve new drugs that change. This is the foundational concept behind Baylor College of Medicine's Tailored Antibacterials and Innovative Laboratories for Phage (Φ) Research (TAILΦR) initiative. TAILΦR is a team of virologists, bacteriologists, and clinicians spanning basic and translational research that support an exploratory scientific program aimed at discovering and preparing the most efficacious therapeutic phages. A basic outline of the program is illustrated in Figure 2, which follows three phases: discovery, preparation, and clinical follow-up.
FIG. 2.
Overview of the discovery, preparation, and clinical follow-up process for TAILΦR. Phase 1 (phage discovery): Phages can be isolated from the environment, targeted samples, and/or selected by their enhanced activities; can be evolved by using a co-culture system to generate phages that are capable of infecting another host strain; and can be engineered genetically to have enhanced features. Phase 2 (phage preparation): To formulate a phage cocktail, phages are characterized based on their lytic parameters (adsorption rate, latent period, and burst size), sequenced, assayed for synergism with antibiotics (synography), and tested for endotoxin levels, sterility, and stability. A report about the qualified phage cocktail is then sent to the attending physician and is used for the analysis and approval by the Food and Drug Administration. If approved, the cocktail can be sent to the physician. Phase 3 (clinical follow-up): TAILΦR assists with examining therapeutic efficacy during and after treatment by determining phage titers from clinical samples, developing new phages if required, and re-testing bacterial isolates for phage sensitivity. Lastly, all discovered phages are catalogued (Phage Desk Reference) and stored for long-term usage. TAILΦR, Tailored Antibacterials and Innovative Laboratories for Phage (Φ) Research.
Phase 1: Phage Discovery
TAILΦR is currently engaged with dozens of clinical cases of complicated bacterial infection. The cases run the gamut of types of infections, including catheter, device, and prosthetic-associated infections, pulmonary infections, intra-abdominal infections, and bacteremia, to name a few. Some common features of typical cases, and U.S. Food and Drug Administration (FDA)-approved compassionate care cases already published25,26 include that the bacterial strains are often resistant to multiple antibiotics (or have developed resistance during treatment),27 the infections are chronic (i.e., the patient has had them for months to years),28 quality of life is substantially diminished (and sometimes, life-threatening),29 there are underlying conditions,30 including immunosuppression,31 and the attending physician has essentially used most options, to no avail, to make the patient infection free.
TAILΦR receives bacterial samples from the clinical microbiology lab that typed the species or strain (usually the most recent isolate), clinical history, and antibiogram data; consults with the physician about the challenges that may be involved with the case; and, if mutually agreed to, moves forward. The expected timeline from phage discovery to application varies depending on whether we have characterized and purified phages, which can be as quickly as 1 month to up to 3 months.
Table 1 shows some of the problematic bacterial pathogens for which TAILΦR has generated phages, excluding multiple pathotypes of Escherichia coli. A description of some of these has been published, but most have not.32 The very first step is screening our library of phages for activity against the patient's strain, starting with plaque assays, followed by efficiency of plating (EOP) and bacterial growth curve analysis.33 At the moment, this process incorporates both manual and automated steps, and it usually takes 2–3 days. If “hits” are observed, the process can move immediately to phase 2. If not, a discovery program is initiated as outlined next. At the moment, TAILΦR'S “hit” rate is about 40%, a number that has steadily increased as the library has grown. The two main advantages of having a hit occur in this step are time and characterization. The time to phage preparation is dramatically shortened, sometimes by weeks or months, if a purified phage is already in the library. This is mainly because the phages in the library are already “vetted” as therapeutic.
Table 1.
Bacterial Pathogens for Which Tailored Antibacterials and Innovative Laboratories for Phage (Φ) Research Have Phages
| Bacterial species targeted by TAILΦR phages |
|---|
| Achromobacter xylosoxidans |
| Enterobacter cloacae |
| Enterococcus faecalis |
| Enterococcus faecium |
| Klebsiella aerogenes |
| Klebsielle pneumoniae |
| Staphylococcus aureus |
| Staphylococcus pseudintermedius |
Note: Escherichia coli includes phages against uropathogenic E. coli, enteroaggregative E. coli, enterohemorrhagic E. coli, and various extraintestinal E. coli of the multidrug-resistant ST131 pandemic clonal group.
TAILΦR, Tailored Antibacterials and Innovative Laboratories for Phage (Φ) Research.
We use the acronym TEST to describe these phages. The “T” stands for “Therapeutic,” meaning the phages generally meet FDA guidelines for being lytic, purified, and sequenced. Sequencing consists of annotating the genome for genes that may encode bacterial toxins, antibiotic resistance, bacterial virulence factors, or integrases. The “E” stands for “Efficacious.” That is, the phages have potent activity at clearing their host strain in various biomimetic models that we have generated. These biomimetic models act as “medical simulators” in that they allow us to either discover or train phages that work well in these environments. They are designed to simulate the microenvironment of the mammalian host, especially where pathogenic bacteria find a niche and thrive. This includes assessment of phage killing in blood, urine, or a medium designed to mimic the luminal content of the intestine (what we term colonic media), as well as lytic activity in or on cell lines and organotypic cultures, including human intestinal enteroids.34 Other biomimetics include phage efficacy against biofilms formed on plates or catheters and/or drive lines as well as suitable and/or animal models of bacterial infection, including models of gut-derived sepsis, bacteremia, urinary tract infection, and peritonitis. In the latter regard, the most progress has been made with various pathotypes of E. coli.35–37
The “S” stands for “Safe.” In addition to the sequencing data described earlier, the phage preparations are tested in certified onsite laboratories for endotoxin and sterility. When available, some TEST phages have animal safety data. The final “T” stands for “Targeted,” that is, we have a solid idea of the specificity of the phage toward similar and divergent strains of the same species. In some cases, we know the receptor for the phage. This information is useful in the construction of the final cocktail since phages that target different receptors can be paired together. Knowing the receptor can also be useful in directing bacterial evolution for exploitable traits (loss of virulence or restored sensitivity toward antibiotics).38,39 The purpose of generating the TEST criteria is to begin the formal process of classifying the parameters of importance for therapeutic phages. We envision these guidelines will continuously expand as knowledge and experience increases. If we do not have TEST phages for the patient's strain, we initiate the discovery program, which has four components (Fig. 2).
Component 1 aims at isolating novel phages from environmental samples. This is a straightforward approach and consists of normal “phage hunting” from environments where bacteria are found. At the moment, there is a priority for collecting lytic phages against ESKAPE pathogens, preferably those isolated from clinical cases that are multidrug resistant. Environmental samples include the soil from local and state parks and farms, water from puddles, ponds, rivers, the ocean and lakes, and human and animal urine and fecal waste, the latter principally coming from raw sewage.
Component 2 aims at selecting phages from more specific locales, or at selecting phages with enhanced lytic activity in mammalian biomimetic systems that we have generated in the lab. The former consists of searching for phages from animal and human skin, hair or hair follicles, saliva, or mucosal surfaces. The latter consists of applying any samples to systems that mimic human microenvironments, and selecting phages that work specifically and are lytic in those environments. For example, we routinely select (or deselect) phages that demonstrate enhanced activity at human mucosal surfaces such as the gastrointestinal tract or human fluids such as urine or blood or a modified intestinal luminal content (colonic media). Screening also consists of finding phages with optimal biofilm-destroying or -penetrating activity and phages that demonstrate lytic qualities against bacteria on catheters, prosthetics, or drive-line device-related material. Phages that can “edit” certain intestinal pathobionts from a human microflora community are also sought, since they may selectively remove problematic pathobionts (e.g., Escherichia, Klebsiella, Enterobacter, or Staphylococci spp.) to keep the balance of the microbiome intact and to avoid dysbiosis.
Component 3 aims at evolving phages using directed evolution in a co-culture system that TAILΦR scientists have constructed, termed a Tetrastat. Therapeutic phages produced from this process are regarded as “enhanced,” or ePhage. In this regard, three types of ePhage are produced. Type 1 ePhage are those that grow on a propagation host that have been adapted to grow on a closely related, but previously retractable, host strain of the same species. This phage type is useful, because it can take advantage of preexisting and already characterized phages that on first pass do not kill a given target strain of the same species. Type 2 ePhage are those evolved to infect a strain of bacteria that has become resistant to the parental phage. They are useful not only for the reasons stated earlier for type 1 phage but also because they can be used to either retreat an infection that has become resistant to the first treatment or be made into a cocktail with the parental strain that reduces the number of evolutionary routes available to develop resistance. Type 3 ePhage are those evolved to have enhanced infectious properties toward their target bacterial strain under conditions they are expected to perform in, including some of the biomimetics described earlier. This is especially true in environments that lend themselves to chemostat studies such as fluids like blood or urine.
Component 4 aims at using knowledge from components 1–3 described earlier to directly engineer phages to have enhanced features that their parental phages lack. For great reviews on this topic, see Refs.40–42 At the moment, TAILΦR has not generated a phage of this type, but it is one of the several ways that such research may progress as more and more enhanced features are discovered. In this case, it is expected that one or more core phages will serve as the backdrop to add the enhanced component, followed by testing to determine that no other properties of the phages have been altered. There is much precedence in the literature for tail fiber swapping to change specificity.43 One could imagine that this can be extended to other properties deemed useful in therapeutic phages, including reduced interaction with the mammalian immune system (less neutralization or inflammation), increased half-life or locational targeting to tissues or in circulation, decreased dependence on metal concentration, decreased interference with other phages, increased avidity for target or burst sizes, and longer shelf storage times or stability, etc.
Phase 2: Preparation
Once phages are discovered, their basic infection characteristics are determined, including their EOP, adsorption rate, and burst size. The phages and their host strains are sent for sequencing. Phage lysates are sometimes suitable for this, but often purified phages are needed. Purified phages are prepared by scaling up lysis in several liters of batch culture, precipitating with polyethylene glycol and sodium chloride (NaCl), concentrating in a cesium chloride (CsCl) gradient, and dialyzing out the CsCl in a modified Tris Buffer.44 Genomic DNA derived from these purified preparations yield great coverage depth (500 × or greater), which allows for insights beyond the phage's genomic composition, including the presence or absence of any host-derived prophages that may have co-purified with the phage.45
It is preferable to use the patient's infecting strain as the propagating and purification host. This ensures that the resulting phages that are derived are given the best opportunity to succeed. In some cases, it may be advantageous to use related host bacterial strains that are known to be free of genes that encode virulence factors or bacterial toxins, that is, more defined and thoroughly vetted host strains. It must be determined though that the resulting phages are as active on the patient's strain as the propagating, safe surrogate. The use of defined bacterial strains ensures that these toxins or virulence factors do not make their way into the final purified preparations.
Initially, new phages are selected for further characterization based on their plaque traits, with a focus on those that form large, clear plaques with “halos” indicative of strongly lytic attributes.33 In general, purified phages are included in the cocktail (usually three or four phages) if (1) EOP ≥0.1 (when compared with the strain on which the phage was originally isolated) for efficient scale-up during purification32,46; (2) sequence analysis determines they are devoid of undesirable elements such as antibiotic resistance and toxin genes, integrases, and attachment sites, and NOT identical to any other phage in the preparation; (3) they do NOT interfere with other phages in the preparation as determined by colony-forming units from bacterial killing assays in culture; and (4) the titers are high (≥1010 plaque-forming unit/mL). The objective is to provide enough phages to overcome both dilution and clearance by the patient's circulatory and immune systems, respectively, and reach the infection site(s) with virulent titers (multiplicity of infections capable of killing the bacteria). We typically aim at providing 109 PFU per dose for intravenous (IV) administration. Phage dosing efficacy cannot be applied broadly, as the pharmacokinetics of each phage and cocktail will vary between patients, administration routes, and infection type.47 However, 109 PFU per IV dose does appear to be efficacious in reported clinical cases,48 and is an accepted dose for compassionate use (personal experience with investigational new drug [IND] applications).
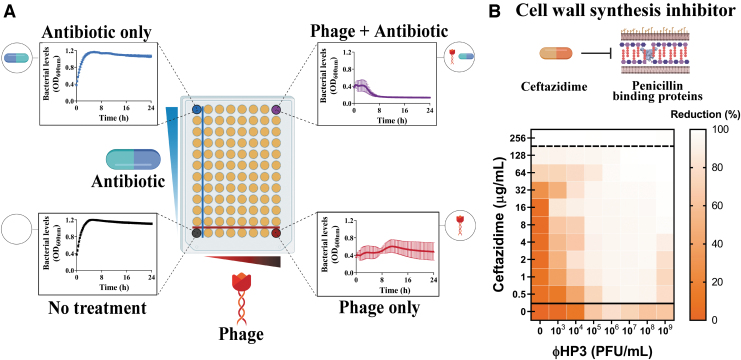
The individual phages or a cocktail are then examined for synergy, additivity, or antagonism when combined with antibiotics to which the bacterial strain may be sensitive to, or that the attending physician determines is the best option given a clinical assessment of the situation. At TAILΦR, we have developed a high-throughput optically based assay that queries two orders of antibiotic concentration that blanket the minimum inhibitory concentration against seven orders of phage titer, in triplicate, in one system. We term the process “Synography,” and the resulting data, represented as an easy-to-read heat map, a “synogram” (Fig. 3). Multiple antibiotics, and from different classes, can be assessed in this manner. Synography can also be performed in environments where the combinatorial treatment is expected to be efficacious, including blood and urine. Not only do these studies allow for the best pairing of antibiotics and phages to be determined, but they also provide guidance for the dose of each and the likelihood that it will work in the host microenvironment.
FIG. 3.
Phage-antibiotic combinatorial testing via synography. (A) Diagram of the plate layout showing on the Y-plane increasing concentrations of antibiotics and on the X-plane increasing concentrations of phage. (B) A typical synogram here showing Escherichia coli phage (ΦHP3) and ceftazidime. Killing activity is represented as a heat map with the greatest percent reduction of bacterial density of viability represented by lighter regions and the least percent reduction in bacteria represented by increasing color (in this case, orange). In synography, the heat-map is indicative of optimal concentrations of phages and antibiotics that are most effective. Such data can guide clinicians in making scientifically driven and informed decisions about the optimal phages and antibiotics to use during treatment.
Multiple phages are then made into cocktails based on the criteria mentioned, a standard practice for phage therapy. This approach is believed to be one way to avoid resistance and increase efficacy. The cocktail undergoes four main tests. The first is another determination of titers to demonstrate virulence against the patient's bacterial strain. The second is an Limulus Amebocyte Lysate chromogenic assay to determine endotoxin concentration. The U.S. FDA sets the acceptable level of endotoxin units (EUs) for IV administration at 5 EU/(kg·h). The third test is United States Pharmacopeia 71 (USP71) sterility cultures. The latter two determinations are performed by accredited and certified laboratories at either Baylor College of Medicine or affiliates. Finally, the titers are monitored under desired formulated conditions (e.g., diluted in saline and stored at 4°C in syringes or other delivery device) to determine preparation stability for as long as necessary, usually throughout the course of treatment. If the cocktail is sterile, contains little to no endotoxin, and titers remain stable under desired conditions, a formalized report is prepared and sent to the attending physician. This report is used in the IND or emergency IND application that the attending physician sends to the FDA for analysis and approval. The FDA weighs in on the data and case and may require further information or tests, or may approve the application. If approved, the cocktail is either formulated at TAILΦR into doses to be administrated by the physician or sent to the physician's compounding pharmacy for formulation.
Phase 3: Clinical Follow-Up
The final phase consists of banking any new phages or strains that resulted from the process, cataloging all information, and assisting with an examination of therapeutic efficacy during and after treatment. The latter can consist of determining phage titers in clinical samples such as serum or urine, determining the host response to phage treatment, re-testing new bacterial isolates for phage sensitivity as treatment progresses, and developing new phages if resistance emerges.
Phage libraries and banking
All phages discovered by the TAILΦR team are catalogued and stored. Cataloguing consists of a sort of “Phage Desk Reference,” analogous to a Physician's Desk reference that lists the properties of medical compounds. Basic infection parameters, genome size and annotation, date and place of location, and so on are assembled in a searchable system (Fig. 2). In our hands, plate lysates in phage buffer (Tris Buffer with 100 mM NaCl, 10 mM MgCl2, pH 8.0) are suitable for short-term storage up to a year at 4°C. Phages may be propagated for an extended period by creating new plate lysates from these stocks. Long-term storage for years at −80°C is typically achieved by either adding 25% glycerol to the plate lysates or flash-freezing infected bacterial hosts in 15% glycerol.49 In any case, periodic monitoring of all stocks informs the selection of a suitable storage method for each phage. TAILΦR not only builds libraries against each species (and various strains) of ESKAPE bacteria but also generates “institution-focused” libraries that target strains circulating in clinics or hospitals. These libraries are the first choice of screening for strains that emerge from patients at those institutions.
This leaves us with the question, what is left to accomplish? Much more would be the answer. TAILΦR is only the beginning of the process and what we learn here will help shape the future of these personalized endeavors, much of which we hope is adopted by institutions around the world. Some of the drawbacks to these personalized approaches that we have learned at the time of this writing include the lack of standardization of each hospital's compounding pharmacy, the length of time it takes to find phages against more uncommon bacterial pathogens, and the length of time it takes to prove the preparation is sterile.
Regarding the first point, there is a requirement for common language and procedure for making the final phage preparations (compounding) that has universal agreement and applicability. Some compounding entities have more leeway as to what they accept, and others are very regimented. To some degree there may be concern about liability at this step; however, standardization and universal agreement in procedure and the use of common sense and accepted practices will go a long way to alleviating fears.
Regarding the second point (phages against uncommon pathogens), it is often observed that some phages can be readily found in one species but another species yields not many phages at all. It will be important to develop procedures that are enriching for these more difficult phages to find. A good example is phages against S. aureus, a common nosocomial pathogen with multidrug resistance. Raw sewage, for example, rarely yields lytic phages but sampling of human or canine hair (in our hands) has been quite productive. This makes sense given that S. aureus is a resident of hair follicles. Thus, standardization of such practices will also be required, but the old adage “that one finds phage where bacteria are” seems to be a guiding principle. Each phage requires a unique search.
Finally, some regulatory agencies require that the final preparation be proven to be sterile, which is reasonable. However, the method often accepted (that of USP71 testing) requires >2 weeks of no growth to be considered a “pass.” This length of time can be risky for the therapy, especially when the infection may be worsening. Time is often of the essence and if acceptable procedures can be developed that prove the preparation is sterile without having to wait more than a few days, this would be ideal. It is also reasonable to expect that if a central stock solution of phages is proven to be sterile, then aliquoting that stock into useable doses under good manufacturing practices will not require re-testing of sterility. This step saves both time and money. These are just a few of the challenges that TAILΦR will address in the next year.
A challenging, but courageous, future
All things change, including medical therapies and delivery of medical care. The increased precision and personalization permitted by appreciation of the human genome, the microbiome, and cancer biology emphatically demonstrate that the silver-bullet approach to medicine is outdated. Each human being is vastly more different from another than previously believed, and this variety is reflected in the spectrum of human diseases. The clones of cancer cells that emerge as resistant to the chemotherapy, driven by the unique mutations they acquire, will need a unique medicine to combat them. Mutation teaches us that personalized medicine is more than a fancy term, it is the inevitable future. Adaptable T cell therapies and mutation-correcting clustered regularly interspaced short palindromic repeats elements are recent examples of this concept in action. It seems justified, then, that one of the more mutable and adaptable entities on the planet, that of bacteria, will require medicines that too can adapt. With respiratory viruses such as severe acute respiratory syndrome coronavirus 2 and drug-resistant bacteria using these tactics against us, it seems that infectious disease medicine should be the front line for the testing of the personalized medicine approach. These tailored approaches—which are streamlined, controlled, and most importantly, adaptable—may offer physicians and scientists the best strategy to stymie the mutagenic tetrasect. As phage biologists often say, “there's a phage for that.”
Authors' Contributions
The concept of this article derives from a project designed by A.L.T., C.G.L., S.I.G., J.R.C., K.C.S., and A.W.M. The article was drafted by A.W.M. The article was shaped by A.L.T. and C.G.L. The article was extensively reviewed by A.L.T., C.G.L., H.S.H., E.R.H., B.W.T., R.F.R., and A.W.M. All coauthors have reviewed and approved the article before submission.
Author Disclosure Statement
No competing financial interests exist.
Funding Information
This work is supported in part by grant from US Veterans Affairs (VA I01-RX002595, VA CIN 13-413), the Roderick D. MacDonald Research Fund at Baylor St. Luke's Medical Center, the Mike Hogg Foundation, and Seed Funds from Baylor College of Medicine Seed Funds.
References
- 1. Darwin C. On the origin of species by means of natural selection, or the preservation of favoured races in the struggle for life. London: John Murray; 1859;502. [PMC free article] [PubMed] [Google Scholar]
- 2. Abbott S, Fairbanks DJ. Experiments on plant hybrids by Gregor Mendel. Genetics. 2016;204:407–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. de Vries H, Farmer JB, Darbishire AD. The Mutation Theory; Experiments and Observations on the Origin of Species in the Vegetable Kingdom Chicago, IL: Open Court Publishing Company. 1910. https://www.biodiversitylibrary.org/item/16402 (last accessed March20, 2020)
- 4. Gager CS, de Vries H. Intracellular Pangenesis United States: Open Court Publishing Company;2014;270. ISBN: 9780875482095. Last accessed June11, 2020
- 5. Hales KG, Korey CA, Larracuente AM, et al. . Genetics on the fly: A primer on the drosophila model system. Genetics. 2015;201:815–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wheeler DA, Wang L. From human genome to cancer genome: The first decade. Genome Res. 2013;23:1054–1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhu YO, Siegal ML, Hall DW, et al. . Precise estimates of mutation rate and spectrum in yeast. Proc Natl Acad Sci U S A. 2014;111(22):E2310-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Maresso AW. The mutagenic tetrasect. In: Maresso AW; ed. Bacterial Virulence: A Conceptual Primer. Cham: Springer International Publishing, 2019; pp. 59–71 [Google Scholar]
- 9. Maresso AW. Antibiotics …and their destruction. In: Bacterial Virulence: A Conceptual Primer. Maresso Anthony W. (eds), Cham: Springer International Publishing, 2019, pp. 195–212 [Google Scholar]
- 10. Pier GB. On the greatly exaggerated reports of the death of infectious diseases. Clin Infect Dis. 2008;47:1113–1114 [DOI] [PubMed] [Google Scholar]
- 11. Zaman S Bin, Hussain MA, Nye R, et al. . A review on antibiotic resistance: Alarm bells are ringing. Cureus. 2017;9:e1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kolář M, Urbánek K, Látal T. Antibiotic selective pressure and development of bacterial resistance. Int J Antimicrob Agents. 2001;17:357–363 [DOI] [PubMed] [Google Scholar]
- 13. Zhao S, Lieberman TD, Poyet M, et al. . Adaptive evolution within gut microbiomes of healthy people. Cell Host Microbe. 2019;25:656..e8–667.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cole GHA. Wandering stars: About planets and exo-planets: An introductory notebook. Singapore: World Scientific Publishing Company;2006:504. ISBN: 9781783260683
- 15. Palzkill T. Structural and mechanistic basis for extended-spectrum drug-resistance mutations in altering the specificity of TEM, CTX-M, and KPC β-lactamases. Front Mol Biosci. 2018;5:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Patel MP, Fryszczyn BG, Palzkill T. Characterization of the global stabilizing substitution A77V and its role in the evolution of CTX-M β-Lactamases. Antimicrob Agents Chemother. 2015;59:6741–6748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Munita JM, Arias CA. Mechanisms of antibiotic resistance. Microbiol Spectr. 2016;4:1–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Santos MA de O, Vianna MF, Nishino LK, et al. Tackling drug-resistant infections globally: Final report and recommendations. 2016. https://amr-review.org/sites/default/files/160525_Final paper_with cover.pdf Last accessed March20, 2020
- 19. Centers for Disease Control and Prevention. Antibiotic Resistance Threats in the United States, 2019. Atlanta, GA: CDC, 2019. www.cdc.gov/DrugResistance/Biggest-Threats.html Last accessed March20, 2020
- 20. Gordillo Altamirano FL, Barr JJ. Phage therapy in the postantibiotic era. Clin Microbiol Rev. 2019;32:1–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kortright KE, Chan BK, Koff JL, et al. . Phage therapy: A renewed approach to combat antibiotic-resistant bacteria. Cell Host and Microbe. 2019;25(2):219–232 [DOI] [PubMed] [Google Scholar]
- 22. Salmond GPC, Fineran PC. A century of the phage: Past, present and future. Nat Rev Microbiol. 2015;13(12):777–786 [DOI] [PubMed] [Google Scholar]
- 23. Ofir G, Sorek R. Contemporary phage biology: From classic models to new insights. Cell. 2018;172:1260–1270 [DOI] [PubMed] [Google Scholar]
- 24. Abedon ST, Kuhl SJ, Blasdel BG, et al. . Phage treatment of human infections. Bacteriophage. 2011;1:66–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hatfull GF, Dedrick RM, Guerrero-Bustamante CA, et al. . Engineered bacteriophages for treatment of a patient with a disseminated drug-resistant Mycobacterium abscessus. Nat Med. 2019;25(5):730–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nir-Paz R, Gelman D, Khouri A, et al. . Successful treatment of antibiotic-resistant, poly-microbial bone infection with bacteriophages and antibiotics combination. Clin Infect Dis. 2019;69(11):2015–2018 [DOI] [PubMed] [Google Scholar]
- 27. Schooley RT, Biswas B, Gill JJ, et al. . Development and use of personalized bacteriophage-based therapeutic cocktails to treat a patient with a disseminated resistant Acinetobacter baumannii infection. Antimicrob Agents Chemother. 2017;61:e00954-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Duplessis C, Biswas B, Hanisch B, et al. . Refractory pseudomonas bacteremia in a 2-year-old sterilized by bacteriophage therapy. J Pediatric Infect Dis Soc. 2018;7:253–256 [DOI] [PubMed] [Google Scholar]
- 29. Aslam S, Pretorius V, Lehman SM, et al. . Novel bacteriophage therapy for treatment of left ventricular assist device infection. J Hear Lung Transplant. 2019;38:475–476 [DOI] [PubMed] [Google Scholar]
- 30. Lavergne S, Hamilton T, Biswas B, et al. . Phage therapy for a multidrug-resistant Acinetobacter baumannii craniectomy site infection. Open Forum Infect Dis. 2018;5(4):ofy064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Aslam S, Courtwright AM, Koval C, et al. . Early clinical experience of bacteriophage therapy in 3 lung transplant recipients. Am J Transplant. 2019;19:2631–2639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gibson SB, Green SI, Liu CG, et al. . Constructing and characterizing bacteriophage libraries for phage therapy of human infections. Front Microbiol. 2019;10:1–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Clokie MRJ, Kropinski AM. Bacteriophages: Methods and protocols. Isolation, Characterization, and Interactions. Volume 1. United Kingdom: Humana Press;2009:308 Last accessed June11, 2020 [Google Scholar]
- 34. Rajan A, Vela L, Zeng XL, et al. . Novel segment- and host-specific patterns of enteroaggregative Escherichia coli adherence to human intestinal enteroids. MBio. 2018;9:1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Green SI, Kaelber JT, Ma L, et al. . Bacteriophages from ExPEC reservoirs kill pandemic multidrug-resistant strains of clonal group ST131 in animal models of bacteremia. Sci Rep. 2017;7:1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ma L, Green SI, Trautner BW, et al. . Metals enhance the killing of bacteria by bacteriophage in human blood. Sci Rep. 2018;8:1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Green SI, Ajami NJ, Ma L, et al. . Murine model of chemotherapy-induced extraintestinal pathogenic Escherichia coli translocation. Infect Immun. 2015;83:3243–3256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chatterjee A, Johnson CN, Luong P, et al. . Bacteriophage resistance alters antibiotic-mediated intestinal expansion of enterococci. Infect Immun. 2019;87(6):e00085-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chan BK, Sistrom M, Wertz JE, et al. . Phage selection restores antibiotic sensitivity in MDR Pseudomonas aeruginosa. Sci Rep. 2016;6:1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chen Y, Batra H, Dong J, et al. . Genetic engineering of bacteriophages against infectious diseases. Front Microbiol. 2019;10:1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Brown R, Lengeling A, Wang B. Phage engineering: How advances in molecular biology and synthetic biology are being utilized to enhance the therapeutic potential of bacteriophages. Quant Biol. 2017;5:42–54 [Google Scholar]
- 42. Pires DP, Cleto S, Sillankorva S, et al. . Genetically engineered phages: A review of advances over the last. Microbiol Mol Biol Rev. 2016;80:523–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yehl K, Yang AC, Torres MDT, et al. . Engineering phage host-range and suppressing bacterial resistance through phage tail fiber article engineering phage host-range and suppressing bacterial resistance through phage tail fiber mutagenesis. Cell. 2019;179:459–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Azeredo J, Sillankorva S. Bacteriophage Therapy—From Lab to Clinical Practice. New York: Huaman Press, 2018 [Google Scholar]
- 45. Philipson CW, Voegtly LJ, Lueder MR, et al. . Characterizing phage genomes for therapeutic applications. Viruses. 2018;10:188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mirzaei MK, Nilsson AS. Isolation of phages for phage therapy: A comparison of spot tests and efficiency of plating analyses for determination of host range and efficacy. PLoS One. 2015;10(3):e0118557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Van Belleghem JD, Dąbrowska K, Vaneechoutte M, et al. . Interactions between bacteriophage, bacteria, and the mammalian immune system. Viruses. 2019;11:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. El Haddad L, Harb CP, Gebara MA, et al. . A systematic and critical review of bacteriophage therapy against multidrug-resistant ESKAPE organisms in humans. Clin Infect Dis. 2019;69:167–178 [DOI] [PubMed] [Google Scholar]
- 49. Golec P, Dabrowski Kamil K, Hejnowicz MS, et al. . A reliable method for storage of tailed phages. J Microbiol Methods. 2011;84:486–489 [DOI] [PubMed] [Google Scholar]