Abstract
GATA1 is an essential transcriptional regulator of myeloid hematopoietic differentiation towards red blood cells. During erythroid differentiation, GATA1 forms different complexes with other transcription factors such as LDB1, TAL1, E2A and LMO2 (“the LDB1 complex”) or with FOG1. The functions of GATA1 complexes have been studied extensively in definitive erythroid differentiation; however, the temporal and spatial formation of these complexes during erythroid development is unknown. We applied proximity ligation assay (PLA) to detect, localize and quantify individual interactions during embryonic stem cell differentiation and in mouse fetal liver (FL) tissue. We show that GATA1/LDB1 interactions appear before the proerythroblast stage and increase in a subset of the CD71+/TER119− cells to activate the terminal erythroid differentiation program in 12.5 day FL. Using Ldb1 and Gata1 knockdown FL cells, we studied the functional contribution of the GATA1/LDB1 complex during differentiation. This shows that the active LDB1 complex appears quite late at the proerythroblast stage of differentiation and confirms the power of PLA in studying the dynamic interaction of proteins in cell differentiation at the single cell level. We provide dynamic insight into the temporal and spatial formation of the GATA1 and LDB1 transcription factor complexes during hematopoietic development and differentiation.
Introduction
The first hematopoietic cells appear in yolk sac blood islands on embryonic day 6.5 (E6.5) during mouse development. On E10.5 to E11, definitive hematopoietic stem cells (HSC) appear in the aorta-gonad-mesonephros (AGM) region within the embryo (and the vitelline and umbilical arteries). They migrate to the fetal liver (FL), mature from pre-HSC to HSC, and after moving, reside in the adult bone mar-row.1,2 One of the lineages originating from HSC generates erythroid cells.
GATA1 is one of the essential transcription factors for the erythroid (and megakaryocytic) program. Gata1 knockouts (KO) (Gata1−/−) die between E9.5 to E10 due to a block of differentiation at the proerythroblast stage, leading to the absence of mature red blood cells.3,4 GATA1 can form several complexes to regulate erythroid gene expression.5 Two proteins of particular interest bind directly to GATA1. The first, FOG1 (Friend of GATA1), binds to the N-terminal zinc finger (ZnF) of GATA1 and recruits the chromatin remodeling complex NuRD/MeCP1 and/or the C-terminal binding protein (CTBP) corepressor-containing complex to regulate GATA1 target genes.6 The second is LMO2, which is part of a larger complex7–9 containing the LIM-domain-binding protein 1 (LDB1). LDB1 functions as a scaffold protein to form multiprotein transcription complexes that regulate the differentiation of various cell types. Ldb1 KO (Ldb1−/−) mice die between E9.5 and E10 due to severe defects in a number of developing tissues, including abnormal hematopoietic development.10 This abnormal hematopoiesis is also observed in knockout mouse embryos lacking the LDB1 binding-partners TAL111 or LMO2.12
Despite the knowledge on GATA1 binding partners, it is not known when and where GATA1 complexes form. In order to identify the temporal and spatial appearance of GATA1/FOG1 and GATA1/LDB1 complexes during differentiation, we applied proximity ligation assays (PLA)13 in differentiated mouse embryonic stem (ES) cells and FL cells. We detect the first significant GATA1/LDB1 interaction in CD71+ FL cells. Knockdown (KD) of LDB1 in vitro led to fetal cell death and decreased the CD71+ cell populations, providing functional evidence for its essential role at that stage of erythroid differentiation in normal FL.
Methods
Cell culture and mouse FL collection
Wild-type (WT) and Ldb1−/− mouse ES cells were cultured in DMEM-15% FCS-1% non-essential amino acids-100 units/mL penicillin-100 mg/mL streptomycin-6.3e-4% 2-mercaptoethanol-100 units/mL Esgro. Day 12.5 (D12.5) or D13.5 FL were used for cell sorting, nuclear extraction, or directly embedded in OCT Tissue-Tek (Sakura) for tissue slicing. All animal experiments were performed according to guidelines and protocols that had been approved by an independent committee on the ethical use of experimental animals (DEC).
ES cell differentiation by the hanging drop method
Mouse WT and Ldb1−/− ES cells were differentiated as described.14 On D4, D5 or D9 of ES cell differentiation, embryoid bodies (EB) were collected by flushing with PBS in 50 mL falcon tubes then embedded in the OCT Tissue-Tek.
Flow cytometry analysis and cell sorting
Mouse E12.5 or E13.5 FL cells (infected or not by LDB1 or GATA1 small hairpin RNA [shRNA]) were labeled with CD71-FITC and TER119-PE antibodies and sorted on a FACSAria III (BD Biosciences) into four populations: P1 (CD71−/TER119−), P2 (CD71+/TER119−), P3 (CD71+/TER119+) and P4 (CD71−/TER119+).
Real-time quantitative PCR (RT-qPCR)
Total RNA was isolated from sorted FL cells or trypsin-dissociated EB up to D6 of differentiation with Trizol (Invitrogen). RT-qPCR was performed using SybrGreen (Applied Biosystem) on Bio-Rad CFX96. Rnh1 (ribonuclease inhibitor 1) gene was used as internal control for normalization. Primers are indicated in the Online Supplementary Table S1.
Gene expression profiling by RNA sequencing (RNA-seq)
RNA samples from sorted mouse E12.5 FL cells, P1 to P4, were sequenced and analyzed as described10 using independent biological replicates. Significant (at least ±0.6 log two-fold change and P-value ≤0.05) up- and down-regulated genes were selected. Data are deposited in the Sequence Read Archive (SRA) (Accession Number: SRP158286).
Antibodies
Antibodies are indicated in the Online Supplementary Table S2.
RNA interference
Lentiviral particles for LDB1 were produced as described by Stadhouders R15 using Ldb1 shRNA (shRNA#1: 5′-GGACCAAAGAGATATACCA-3′, shRNA#2: 5′-GACTCTGTGTGATACTA-GA-3′) and Gata1 shRNA (5′-GTTTGGATGCAGCATCTTCTT-3′) with non-targeting shRNA as controls. Lentiviral infected cells were harvested 72 hours after transduction and processed for nuclear extraction.
Protein analysis
Murine erythroleukemia (MEL) cells or EB nuclear extract and immunoprecipitation (IP) were prepared as described16 and size-exclusion chromatography was performed on an AKTA-FPLC apparatus with a Superose-6 10/30 column (Amersham Biosciences). Fractions were precipitated with trichloroacetic acid and analyzed by Western blotting using Odyssey system (LI-COR).
Immunofluorescent staining
MEL or FL cells were stained as described8 and analyzed by confocal microscopy (Leica SP5).
PLA on EB and mouse embryo tissue
10 μm sliced E4, 5, 9 EB or E12.5 mouse fetal tissues were fixed and processed for PLA following the manufacturer’s protocol (Duolink, OLINK) using antibodies indicated in the Online Supplemntary Table S2. PLA signals were visualized by Leica SP5 confocal microscopy and were analyzed using BlobFinder software (Uppsala University, Sweden). Signals contained in or adjacent to nuclei were compared between different groups (n=3). The Kruskal–Wallis test for variance between groups was performed and the Tukey method to counteract multiple comparison errors was applied. Deconvolution of PLA signals and volume analysis was performed using Huygens Suite as published.17
Results
LDB1 complexes start to form at D4 of in vitro ES cell differentiation
We applied PLA18 on sliced in vitro differentiated EB to identify when GATA1 complexes form. This enables low level detection of endogenous protein-protein interaction in situ.
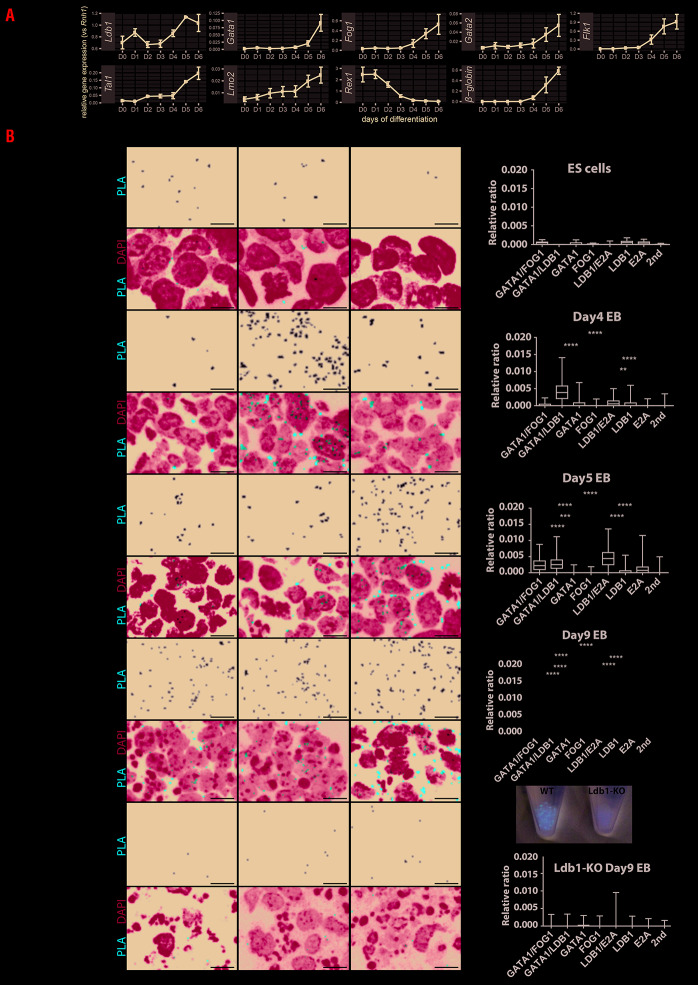
First, we characterized gene expression dynamic for genes of interest during ES cell differentiation (Figure 1A). As expected, the stem cell marker Rex1 is expressed early (day 0 to 2 [D0-D2]) and decreases during differentiation, while β-globin increases at later stages at D5-D6. Thus Ldb1 is expressed both in early and late stages of ES differentiation, and in the erythroid cell lineage at D5-D6. Following differentiation Gata1, Fog1, Gata2, Flk1, Tal1 and Lmo2 expression gradually increases. Of note Gata2 gene induction starts at D4 whereas Gata1 expression is delayed for 24 hours (h) (Kolovos et al. submitted), i.e. the GATA-switch occurs in early embryogenesis.19
Figure 1.
Gene expression and proximity ligation assay on embryonic stem cell differentiation. (AcGene expression during embryonic stem (ES) cell differentiation from day 0 until day 6 (D0-D6) of genes of interest. Standard error of the mean (SEM) from three biological replicates is shown as error bar. (B) Proximity ligation assay (PLA) detection of GATA1/FOG1, GATA1/LDB1 and LDB1/E2A complexes was performed on wild-type ES cell differentiation at D0 (ES cells), D4, D5 and D9, and Ldb1-KO ES cell differentiation at D9. On D9, EB from wild-type (WT) or Ldb1-KO are collected and pictured as shown. PLA signal representing protein-protein interaction was in white on a black background and in Texas-Red in merged pictures together with DAPI in blue. Each red dot represents a fluorescent signal of GATA1 complex formation. The images were analyzed using BlobFinder software38 which quantifies the number of PLA-positive dots in cells. The boxplot (Figure 1B, right panel) shows the density of the dots related to its nuclear area (from three biological replicates). Comparisons of the PLA signals obtained with two primary antibodies versus those obtained in the negative controls were tested in ANOVA and the significance is shown in asterisks. All scale bars represent 10 μm. PLA signals have been quantified on each day and compared with negative controls including GATA1, FOG1, or E2A single-antibody or secondary antibodies alone. Asterisk shows the significant interactions between the two primary antibodies PLA signal and single primary antibody controls. The significance was analyzed with Kruskal-Wallis test as follows, ****: P≤0.0001, ***: P≤0.001, **: P≤0.01, *: P≤0.05. The Tukey method to counteract multiple comparison errors was applied.
PLA representing combinations of transcription factor (TF) interactions (GATA1/LDB1, GATA1/FOG1 and LDB1/E2A) was performed in undifferentiated cells D0, D4, D5 and D9 differentiated WT or Ldb1-KO EB (Figure 1B). Quantification of PLA signals showed that these interactions are absent in ESC, while GATA1/LDB1 and LDB1/E2A interactions already occur at D4 of ES cell differentiation. The GATA1/FOG1 interaction appeared 24 h later at D5. No red blood cells emerged in Ldb1-KO EB at D9 as shown by the lack of red coloration. This correlates with absence of interaction detection by PLA constituting an important control for PLA specificity, in addition to the single probes and secondary antibodies alone used a negative control.
We next characterized the dynamic expression of hematopoietic TF proteins during the time course of ES cell differentiation at D4-5 using Ldb1-KO cells as the control (Online Supplementary Figure S1A). This showed that the different factors already form a complex as determined by immunoprecipitation at D4 and D5 (Online Supplementary Figure S1C-D) using Ldb1-KO cells as the control (Online Supplementary Figure S1E). Online Supplementary Figure S1C shows that LDB1 and FOG1 fail to pull down GATA1 (and vice versa) in D4 differentiated cells, which is likely due to very low amounts of the bridging factor LMO2. Day 5 (Online Supplementary Figure S1D) shows more GATA1, but LMO2 is still undetectable and LDB1 and GATA1 appear to fail pulling down each other. The same is seen for GATA1 and FOG1. Interestingly FOG1 appears to be regulated by (the) LDB1 (complex) as it is present in D5 WT cells, but not in Ldb1-KO cells. This agrees with our observation that LDB1 binds to the Fog1 gene in Flk1 positive cells sorted four days after ES cell differentiation and the reduction of Fog1 expression in Ldb1-KO cells analysed by RNA-Seq.10 The amounts of the proteins involved (directly or indirectly) were too low to allow their detection by immunoprecipitation or mass spectrometry (data not shown), because D4 and D5 differentiated ES cells are a mixture of few hematopoietic cells in the presence of many non-hematopoietic cells. For example, many cells express Oct4 (Online Supplementary Figure S1A) or cardiac genes in cardiac progenitors.20 The complexes are barely detectable by size-exclusion chromatography (Online Supplementary Figure S1B). By contrast, the clear PLA signals (Figure 1B) show the power of PLA to analyze complexes in individual cells. The few PLA signals in undifferentiated ES cells are background since such signals are also detected with single antibody controls.
In summary, PLA monitors the dynamic changes of different protein complexes even of low amounts present in a subset of cells. The PLA signals in D5 EB appear to distinguish a subpopulation of cells, suggesting that some specification is already in progress towards hematopoietic cells in the mixed three-dimensional cell aggregates.
The GATA1 complexes (GATA1/LDB1 and GATA1/FOG1) are observed in mouse E12.5 FL cells
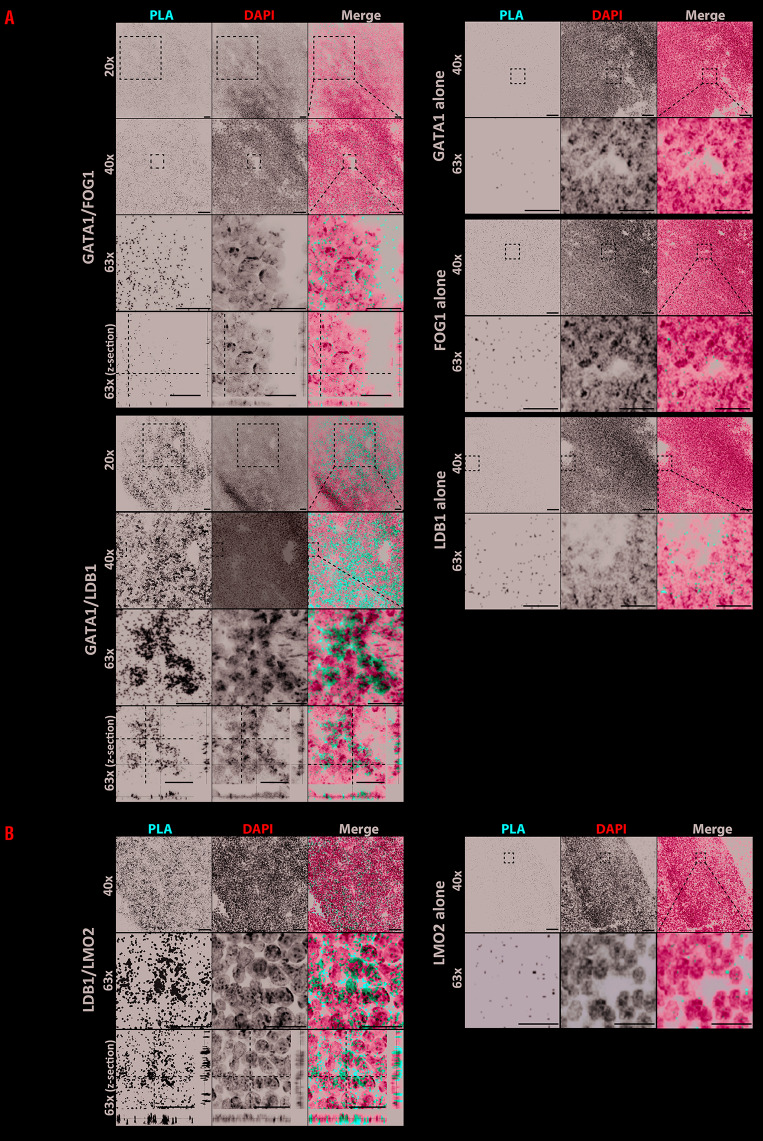
(Pre-)HSC move to the FL at embryonic day E10.5 to 11.5. We applied PLA on FL sections at E12.5 to under-stand the temporal appearance of the two GATA1 complexes in definitive blood cells. Figure 2 shows the comparison of the GATA1/LDB1 and GATA1/FOG1 complexes, together with negative controls of GATA1, FOG1 or LDB1 single-primary antibody. Although FL tissue is com-pact and single cells can be difficult to distinguish, clearly some cells contained very dense GATA1/LDB1 PLA signals when compared to surrounding cells (Figure 2A). Close up images show that the signal co-localizes with the DAPI staining (of note, those cells have little cytoplasm relative to the size of the nucleus) and that cells with no signal (next to strong positive ones) include FL endothelial cells which are expected to be negative for GATA1. A similar result was found in fetal aorta (not shown). Specific PLA signals were also detected for LDB1/LMO2, which is part of the same GATA1/LDB1 complex (Figure 2B) suggesting GATA1/LDB1 and LDB1/LMO2 and by inference the GATA1/LDB1/LMO2 complex are present at a high level in a subpopulation of cells. This is in accordance with our previous co-immunoprecipitation and ChIP-sequencing co-localisation data of these factors in MEL and in Flk1+ cells sorted four days after ES cell differentiation.5,8,10 In contrast, GATA1/FOG1 signals appear in similar numbers of cells but are less abundant/weaker and more evenly distributed (Figure 2, upper panel A). In agreement with the PLA results, immunofluorescent staining for individual LDB1, GATA1 and FOG1 proteins in FL sections showed higher co-expression of GATA1 and LDB1 in a subpopulation of cells than GATA1 and FOG1 (Online Supplementary Figure S2), although it should be noted these are signals from different antibodies (see Methods).
Figure 2.
Proximity ligation assay of GATA1 complexes in E12.5 fetal liver tissue. (A) Proximity ligation assay (PLA) for GATA1/FOG1 and GATA1/LDB1 interactions was performed on sections of mouse fetal liver (FL) tissue at embryonic day 12.5 (E12.5), together with GATA1, FOG1 or LDB1 single-primary antibody negative controls. PLA protein interactions were visualized in red, and DAPI staining in blue was used to visualize the nucleus (scale bars, 50 μm in 20x and 40x). For each inter-action zoom-in pictures corresponding to the white square area are also shown (scale bars, 20 μm in 63x). Z-stack images of each protein combination and fluorescent channels were projected by Maxi-Projection algorithm. (B) PLA for GATA1/LMO2 was performed as in Panel A using LMO2 antibodies replacing FOG1 antibodies. Scale bars represent 50 μm apart fromscale bars in zoom in picures bars which represent 10 μm.
FL contains erythroid cells at different stages of differentiation and the PLA results suggest that the LDB1 complex, is more important at particular stages of erythroid cells in agreement with data showing that GATA1 increases before the end stage of erythroid differentiation8 primarily in the LDB1 complex.
GATA1/LDB1 complex is highly localized in early erythroid differentiating cells in sorted FL
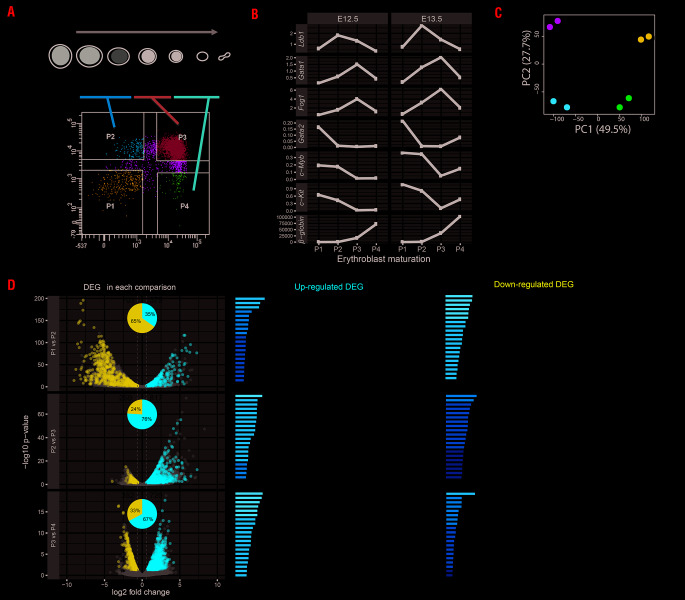
In order to identify the cells containing high PLA signals, E12.5 or E13.5 FL cells were sorted using glycophorin (TER119) and transferrin receptor (CD71) antibodies into four populations: P1 (CD71−/TER119−), P2 (CD71+/TER119−), P3 (CD71+/TER119+) and P4 (CD71−/TER119+) in order to separate different stages from proerythroblasts to orthochromatic erythroblasts21,22 (Figure 3A).
Figure 3.
Fetal liver cell sorting and RNA-sequencing data analysis. (A) Schematic description of erythroblast development. Fetal liver (FL) cells are sorted into four populations based on membrane markers TER119 and CD71. Different stages of erythroblasts are indicated into P2 to P4 populations. The C-KIT positive cell population (P1, CD71−/TER119−) represents precursors or other lineage cell types present before the initiation of erythroblast maturation. Proerythroblasts/colony forming uniterythroid [CFU-E] cells) express high level of CD71 and low level of TER119 (P2, CD71+/TER119−). Following erythroblast maturation, TER119 expression increases (P3, CD71+/TER119+). When the differentiation reaches the orthochromatic stage, they loose CD71 expression (P4, CD71−/TER119+). Pro: immature proerythroblast; Baso: basophilic erythroblast; Poly: polychromatic erythroblast; Orth: orthochromatic erythroblast; Ret: reticulocytes; Ery: erythrocyte. Gray bars represent the changes of c-Kit, Cd71 and Ter119 gene expression. The darker color of the bar represents higher expression for the indicated gene. (B) Quantitative PCR on indicated genes in E12.5 or E13.5 sorted FL cells (E12.5 and E13.5 were used as duplicates) as described in Panel A. Relative expression values are calculated by comparing to control gene Rnh1. (C) Principle component analysis of RNA-sequencing data from two replicates of each P1 to P4 sorted FL cells as described in (A) (D) The total number of down-regulated or up-regulated differentially expressed genes (DEG) are shown for each comparison in a Volcanoplot, in which x-axis represents log two-fold change and y-axis represents log 10 adjusted P-value. Significant DEG are shown in blue for down-regulated and red for up-regulated. Gray dots represent the non-significant genes. The ratio of down- or up-regulated DEG in each comparison is shown in pie-chart inserted in the Volcanoplot. Top 20 gene ontology terms for down- or up-regulated DEG in each comparison are shown.
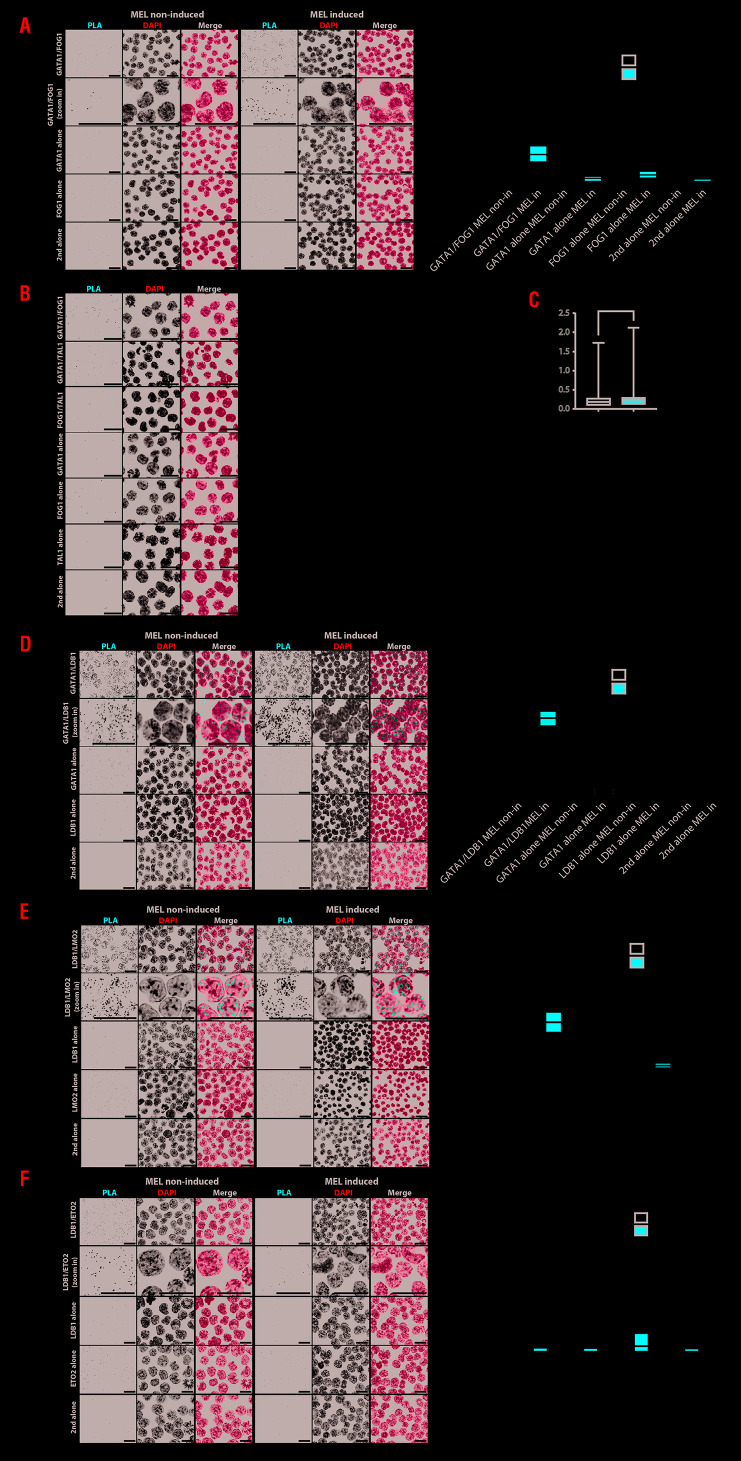
First, gene expression was measured in the four populations (Figure 3B). At early stages (P1 and P2), expression of Ldb1, Gata1 and Fog1 starts increasing followed by a decrease at P3 for Ldb1 and at P4 for Gata1 and Fog1, which follow each other. Gata2, c-Kit and c-Myb genes are expressed highly in P1 which contains precursor cells (and other cell types), their expression decreases during differentiation, whereas the β-globin gene increased dramatically from P3 to P4. The CD71/TER119 sorted cells were used for RNA-seq analysis in two independent biological replicates for each P1 to P4 population of E12.5 FL cells. Principal component analysis (Figure 3C) shows that biological replicates of each population cluster and can be separated from each other. As expected, the RNA-seq result in those populations (Online Supplementary Figure S3) is very similar to the genes analysed by qPCR (Figure 3B and Online Supplementary Figure S3). There is an increase of expression of the transcription factors Ldb1, Gata1 and Fog1 from P1 to P2 (together with E2A) and continuing to P3 for Gata1 and Fog1 (together with Klf1). Gata2, c-myb and c-kit expression is inversely correlated and follows the same trend as Eto2 and Irf2bp2, with a decrease peaking at P3 followed by erythroid specific markers such as β-globin, Alas2 and Gypa and the transcription factor Lmo2. The result shows that the sorting method clearly separates the different stages of the erythroid cells. The significantly differentially expressed genes (±0.6 log log two-fold change and P-value ≤0.05) between the populations is shown in the Online Supplementary Table S3. Of note Ldb1 expression presents a two-fold increase between P1 and P2, but is not included in the Online Supplementary Table S3 due to the threshold parameters used in our study to detect the strongest differentially expressed genes. In P1 and P2, 65% of the downregulated genes are enriched for e.g. neutrophil degranulation, cytokine production and hemostasis, i.e. genes crucial for other cell types (Figure 3D). The up-regulated genes (35%) represent essential functions for erythroid genes, e.g. erythrocyte homeostasis, porphyrinsynthesis and cell cycle genes as cells at this stage are still replicating. During mid- (P2 to P3) or late (P3 to P4) differentiation, the majority (76% and 67%) of upregulated genes have erythroid differentiation bio-functions. Down-regulated genes, (24% and 33%) show functions like lymphocyte differentiation, B-cell differentiation and lymphocyte migration. At these stages (P3 to P4), cell cycle functions are suppressed as erythroid cells enter the terminal differentiation and proceed to enucleation. Next GATA1/LDB1 PLA was applied on the four sorted populations (Figure 4A), showing high signals in P2 and P3. Quantification confirmed that P2 had the highest density of GATA1/LDB1 interaction signals per nuclear area (Figure 4B). Interaction between the LDB1 and GATA1 proteins appears already shortly before the proerythroblast stage. It is most abundant in the P2 population (most likely colony forming unit-erythroid [CFU-E] cells) and basophilic erythroblasts but decreases during the final stages of differentiation in vivo. We applied PLA to detect the interaction between GATA1/FOG1, GATA1/LDB1, GATA1/TAL1, LDB1/LMO2 and LDB1/ETO2 in MEL cells, which mimic FL cells. Uninduced MEL cells represent proerythroblasts, i.e. part of the P1 and P2 population, while induced MEL cells represent P3 and further differentiated populations.9, 28,29 Figure 5A shows the detection of the GATA1/FOG1 interaction by PLA using single-primary antibody and secondary antibody alone as controls. Quantification of the PLA signals in nuclei show a significant increase of GATA1/FOG1 interaction after MEL cell differentiation. An additional negative control experiment to further demonstrate the specificity of the PLA assay was a TAL1/FOG1 interaction which is known not to be formed.5 Quantification of the different PLA signals confirmed the absence of TAL1/FOG1 interaction detection in MEL cells and is comparable to the one of the single probe GATA1 only control, thereby supporting the specificity of positive PLA signals (Figure 5B). This control on non-inter-acting highly expressed proteins (TAL1 and FOG1 in MEL cells)5 also ruled out a potential threshold effect of PLA i.e. where ligations may be more likely when the relevant TF partners are expressed highly. Similar quantification of PLA signals was performed in the cytoplasm (Figure 5B). It was much lower than that observed in the nucleus and does not increase upon differentiation as seen in the nuclei. Of note the increase of signal was not due to an increase of signal volume between the differentiated and undifferentiated states (Figure 5C). In MEL cells, the LDB1 complex binds its target genes during erythroid differentiation,23 e.g. the α- and β-globin locus bind the LDB1 complex in differentiated cells resulting in upregulation,24–27 due to the loss of the repressor ETO2 (encoding by Cbfa2t3) from the complex.9 We quantified the PLA signals for GATA1/LDB1, LDB1/LMO2 and LDB1/ETO2 interactions (Figure 5D-F) and confirmed not only an increase of both GATA1/LDB1, and LDB1/LMO2 interactions upon differentiation but further confirmed that the LDB1/ETO2 interaction is lost during differentiation in MEL9 (Figure 5F), supporting its role as a negative regulator during erythroid differentiation where ETO2 and IRF2BP2 with the NCOR1/SMRT co-repressor complex suppress the expression of typical erythroid genes such as Klf1 which is needed to express β-globin and Gypa genes.8,26
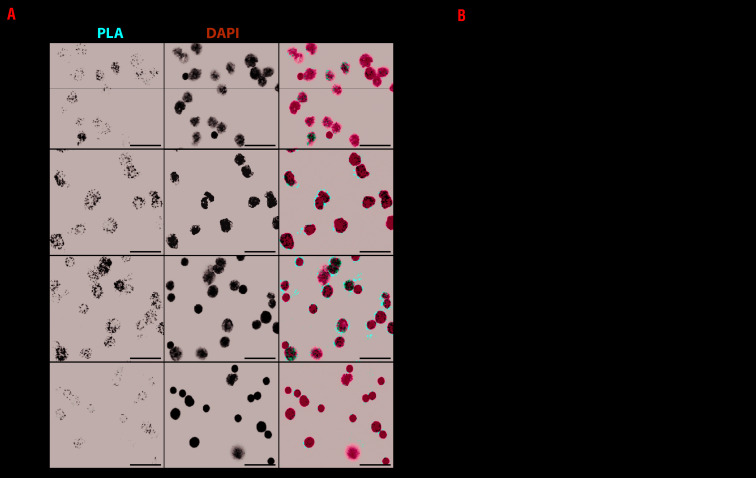
Figure 4.
GATA1/LDB1 proximity ligation assay on sorted fetal liver cells. (A) GATA1/LDB1 proximity ligation assay (PLA) on the four sorted cell populations as described in Figure 3A. PLA signal is in red and nucleus in blue. Scale bar represents 20 μm. (B) Quantification of GATA1/LDB1 PLA signals from three biological replicates (total number of dots in one cell vs. nucleus area) and boxplot comparison among the sorted four cell populations. ****Indicates the significance (P≤0.0001) between any of two cell populations. The statistical significance is determined by Kruskal-Wallis test (Tukey method).
Figure 5.
Proximity ligation assay on GATA1 complexes in MEL cells. (A) GATA1/FOG1 proximity ligation assay (PLA) on non-induced and induced MEL cells. PLA signal representing protein-protein interaction in white in a black background and in red in merged pictures together with DAPI in blue. GATA1 or LDB1 antibody alone was also performed as negative controls. All scale bars represent 20 μm. The significance from three biological replicates was analyzed with Kruskal-Wallis test as ****: P≤ 0.0001 and *: P≤0.05. The statistical significance is determined by Kruskal-Wallis test (Tukey method). (B) PLA detection of GATA1/FOG1 and GATA1/TAL1 interaction compared to non-existing interaction FOG1/TAL1 in MEL non-induced cells. The PLA signal is separated into relative ratio in nucleus or cytoplasm. ****Indicates the significance (P≤0.0001) between indicated interactions and is determined by Kruskal-Wallis test (Tukey method). (C) The volume of individual GATA1/LDB1 PLA signals from 53 MEL non-induced and 56 induced cells is calculated following deconvolution and shown in boxplot. P-values of both statistical analysis of parametric (t-test) and non-parametric (Mann-Whitney) are 0.827 and 0.674, respectively. (D) GATA1/LDB1 PLA on MEL cells as in (A) (E) LDB1/LMO2 PLA on MEL cells as in Panel A. Panel F: LDB1/ETO2 PLA on MEL cells as in Panel A.
In conclusion, the GATA1/LDB1 complex starts to be formed just before the proerythroblast stage and activates erythroid specific genes of erythroid differentiation in vivo, when it looses ETO2.
LDB1 KD results in loss of the erythroid cell population
We examined the importance of the GATA1/LDB1 complex in fetal erythropoiesis at E12.5 by three independent (partial) KD rather than a (lethal) KO using anti-LDB1 shRNA (shLDB1_1 and shLDB1_2) and an anti-GATA1 shRNA (shGATA1). Treatment with an empty vector pLL3.7 shRNA or scrambled shRNA (Scr) was used as the controls. The level of LDB1 or GATA1 protein relative to valosin containing protein (VCP) decreased in the FL cells from D1 to D3 in the KD (Online Supplementary Figure S4). On D3, the cells were sorted by fluorescence-activated cell sorting (FACS) using TER119 and CD71 expression and the relative ratio normalized versus pLL3.7 control (ctrl), in each population is shown in the Online Supplementary Figure S4B. LDB1 protein decreased by ~60% at D3 (54.3%, for shRNA#1 and 64.3% for shRNA#2 compared to ctrl) which resulted in a ±50% decrease in the P2, P3 and P4 population of cells (see above). In three additional LDB1 KD, a pLL3.7 plasmid containing EGFP was used to determine the RNA levels in GFP+ cells versus the total cells (“All cells”). As expected, the “All cells” result shows a similar ratio in each population as was observed in Figure 3A. The GFP+ cells representing the really infected cells, showed a strong KD effect in P2 to P3 compared to pLL3.7 control, when the level of LDB1 appears highest (Figure 3A). The GATA1 KD showed a decrease of GATA1 protein of ~43.5% at D3 (relative to Scr ctrl) resulting in a ±40% decrease in the P3 population while majority of cells are P1. This result is consistent with the phenotype observed in inducible Gata1 KO mice showing an over-representation of cells in P1 and an under-representation of cells in P3 and P4).28 The difference of the KD effect in LDB1 and GATA1 therefore correlates with their expression in those populations (Figure 3B). It is impossible to conclude whether the decrease in the LDB1 complex has an effect before the proerythroblast stage, because the number of progenitors committed to the erythroid lineage in the P1 is not known, but the 40-50% decrease in P2 to P4 suggests that there is a decrease in the number of proerythroblasts that is propagated to the later cell compartments. Of note, the cells (P2 to P4) were strongly affected by the KD of LDB1 or GATA1 and were dying.
Discussion
Our study shows a modulation of the levels of the protein complexes from the early stages of erythroid differentiation of the systems that we used (erythroid differentiation from embryonic stem cells, MEL cells, primary erythroid cell populations from mouse FL and FL tissue sections). These diverse systems represent different stages and therefore heterogeneous systems of hematopoietic development and differentiation. They show that the same complex is formed at these quite different stages, although these experiments do not directly show that the formation of the complex is essential at all these stages and one system might not directly extrapolate to the other. However previous data using KO or KD experiments of individual components of the complex lead to defective hematopoiesis/erythropoiesis.9,10,29 We therefore conclude that GATA1/LDB1 complex formation is essential in these diverse systems and provide novel insight in GATA1 complexes. The result describes the sequential emergence of GATA1/LDB1, GATA1/FOG1 and LDB1/E2A complexes in early stage ES cell differentiation, suggesting dynamic changes of the complexes and their function taking place during the blast colony-forming cell (BL-CFC) stage similar to the hemangioblast stage in vivo. GATA1 as part of the LDB1 complex is absent in undifferentiated ES cells and only appears after a few days of differentiation, while GATA2 is expressed and is part of the LDB1 complex in the very early stages of differentiation where it activates GATA119 in a feed forward type system (Kolovos et al., in revision), which is in agreement with the data on the function of GATA1 and GATA2 in mice or differentiating ES cells.4,30
GATA2 is present during early erythropoiesis and binds to the Gata1 gene to activate its expression.19 Gata1 expression in turn represses Gata2 expression via the FOG1/MeCP1 complex, while activating its own expression. This "GATA switch" represents a forward drive towards late stage erythroid differentiation through changes in gene expression.19,31 GATA2 regulates important proliferation genes of stem or progenitor cells whereas GATA1 also regulates the final erythroid fate through the expression of erythroid specific genes. A similar process operates early during embryonic development. GATA2 is present before GATA1 during ES cells differentiation to the hematopoietic lineage. The GATA2/LDB1 complex is present in very low concentration binding a small set of hematopoietic specific transcription factors in a co-factor dependent manner (PK, CA-S and FG, manuscript in revision). Unfortunately, the low level of GATA2 and poor quality of its antibodies prevent a PLA signal although the GATA switch must start early as we identified the GATA1/LDB1 and LDB1/E2A complexes already in D4 EB. PLA does not show which cell population contains the GATA1/LDB1 and LDB1/E2A complexes in D4 EB, but it is known that LDB1 is expressed in BL-CFC.10 The cells expressing GATA1 and LDB1 in D4 EB are differentiating hematopoietic cells. We know from ChIP-seq data that the two factors already form a complex at this stage targeting a core set of genes enriched for categories related to differentiation/quantity of blood/hematopoietic progenitor cells (Kolovos et al., manuscript in preparation). Moreover, we described in Mylona et al. 201310 that blast colony-forming cells deficient for Ldb1 (i.e. Flk1+ cells sorted four days after ES cell differentiation) are unable to differentiate to the hematopoietic lineages with a severe reduction in the number of blast colony forming cells and their failure to give rise to blast colonies.
These cells appear between ES cell differentiation D3.75 to D4.25 expressing vascular endothelial growth factor receptor-2 (VEGFR2, i.e. FLK1). The absence of LDB1 results in less BL-CFC and failure to generate hematopoietic and endothelial lineages.10 Of its targets Gata2, Scl/Tal1, Runx1 and Gif1b are down-regulated showing that the complex is essential for activation of early embryonic hematopoiesis in agreement with our observation here that the GATA1/LDB1 interaction already takes place at this early stage. It has been shown that a KO of Gata1 in ES cells did not affect the formation of clonogenic progenitors in chimeric in vitro differentiation, and Gata1 KO colonies contained phenotypically normal macrophages, neutrophils and megakaryocytes,3,4 suggesting that GATA1 is not yet essential at this early stage. Importantly GATA2 is essential for the generation of FLK1+ BL-CFC during in vitro ES cell differentiation32 and Gata2−/− embryos die at E11.5 with severe anemia.30
LDB1 IP experiments in undifferentiated ES cells show that a GATA1/LDB1 complex is absent, but appears at low levels in nuclear extracts (NE) from D4 and D5 ES cell EB (Online Supplementary Figure S1C-D). The expression of FOG1, LDB1 and GATA1 increases from D4 to D5, but is still very low and the binding partner LDB1 could not detected by IP in a GATA1 pulldown (Online Supplementary Figure S1). The E2A protein was detected in the LDB1 IP only at D5, but again the band is very weak. We also applied size-exclusion chromatography to distinguish the different GATA1 complexes. The expression of LMO2 protein increases at D4 (Online Supplementary Figure S1B), most of it is in the fractions of 37-39, but some is present in fractions 22-24 indicating that some complex has formed. In MEL cells, i.e. a much later stage of development and differentiation, most LMO2 is in the fractions overlapping with LDB1 (Online Supplementary Figure S1B), showing they are in the same complex consistent with previous results.8 Although the expression of GATA1 and partners is low at D4-5 ES cell differentiation, we could detect the formation of the different complexes by PLA due to its ability to detect 102-fold or 106-fold lower protein concentrations than ELISA or WB, respectively.33
GATA1/FOG1 regulates genes that are less important for erythroid specific functions in the undifferentiated stage, e.g. pro-erythroblasts. We originally postulated a switch from GATA1/FOG1, repressing alternative lineage genes to GATA1/GFI1B repressing proliferation related genes during differentiation.5 In MEL non-induced cells, GFI1B and LDB1 can also be detected in an ETO2 pull-down, indicating that LDB1 complexes containing ETO2 and/or GFI1B proteins suppress archetypical erythroid genes primed for the onset of terminal erythroid differentiation.26 Indeed, RNA-seq data shows that Cbfa2t3 (encoding ETO2) and Irf2bp2 expression decreased from P1 to P2 suggesting these genes mainly function in P1 to P2, similar to the stage of non-induced MEL cells. Meanwhile, Klf1 increased its expression and reached the peak at P3, together with the increase of typical erythroid specific genes (Online Supplementary Figure S3). This result indicates that clear erythroid differentiation starts at P2. Our PLA result on sorted FL cells show that the GATA1/LDB1 interaction peaks in CD71+/TER119− cells at a relatively early stage of erythropoietic differentiation, like undifferentiated MEL cells, when many erythroid genes are still suppressed, until ETO2 disappears from the complex turning on typical erythroid genes. Other LDB1 complex regulated genes such as c-myb remain suppressed, because they no longer bind the (activating) LDB1 complex through an as yet unresolved mechanism (Giraud et al., unpublished data).
We show that PLA can detect endogenous level of GATA1/LDB1 and GATA1/FOG1 interactions in FL tissues and we located the GATA1/LDB1 interaction in a specific cell type in situ. Whether it corresponds to E7.5 yolk-sac derived primitive cells, E8.5/E9 yolk sac and placenta derived intermediate erythroid-myeloid,34,35 or HSC derived definitive progenitors is still an open question.
From PLA images a number of transcription factor complexes seem to be located outside the nuclei. The studied factors are certainly formed in the cytoplasm and travel to the nuclei; whether a fraction of interacting factors remains in the cytoplasm is a possibility. The extensive set of complementary experiments that would aim at studying the existence of interactions in the cytoplasm, such as cellular compartment fractionation followed by IP would be similarly questionable, as (like for PLA) it would be difficult to exclude the possibility of the nuclear extract fractions leaking out in the cytoplasmic fractions. We performed an additional control by quantifying the PLA signals in the cytoplasmic area of MEL cells for the positive GATA1/FOG1 and the non-existing TAL1/FOG1 interactions and associated negative controls (single probes and secondary antibodies only), and observed that non-existing TAL1/FOG1 interaction detection presents a lower level to the existing GATA1/FOG1 and GATA1/TAL1 in the cytoplasm (Figure 5B). These differences suggest the existence of a certain level of transcription factor interaction in the cytoplasmic fraction and the amount of signal in TAL1/FOG1 sets the level of background of the PLA technique. It should also be noted that the level of PLA signals from different interactions cannot be compared directly, since PLA is dependent on antibody quality. Therefore, like other immuno-based technic PLA present a certain level of background, easily quantifiable by using single probe, secondary antibodies only, protein mutant and/or non-existing interaction detection. PLA can detect an interaction up to a 40 nm distance36 and enables super high resolution immunofluorescence microscopy, which can distinguish molecules at a similar distance37. Although PLA cannot detect real-time protein-protein interaction, quantification of PLA signals of different GATA1 complexes in sequential stages of ES cell differentiation improves our understanding on the temporal changes of these complexes.
This study therefore reveals that PLA is a powerful tool to examine dynamic protein/protein interactions and their dynamics in differentiating erythroid cells and demonstrates that it provides an excellent alternative for cells in which the abundance of proteins is too low to perform standard co-IP experiments. Our study revealed that PLA can be used to detect very low amount of essential GATA1 complexes emerging at early time point of ES cell differentiation and later on FL tissue, the site of definitive erythropoiesis. In addition we show that the increase followed by a decrease of expression of GATA1 and LDB1 affects a number of genes differentially, for example the expression of the c-myb gene which is regulated by the LDB1 complex15 decreases during differentiation, while the expression of the β-globin genes, which are also dependent on the LDB1 complex increases. Further studies will be needed to understand how these differences are regulated.
Footnotes
Check the online version for the most updated information on this article, online supplements, and information on authorship & disclosures: www.haematologica.org/content/105/7/1802
References
- 1.Dzierzak E, Speck NA. Of lineage and legacy: the development of mammalian hematopoietic stem cells. Nat Immunol. 2008;9(2):129-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Medvinsky A, Rybtsov S, Taoudi S. Embryonic origin of the adult hematopoietic system: advances and questions. Development. 2011;138(6):1017-1031. [DOI] [PubMed] [Google Scholar]
- 3.Takahashi S, Onodera K, Motohashi H, et al. Arrest in primitive erythroid cell development caused by promoter-specific disruption of the GATA-1 gene. J Biol Chem. 1997;272(19):12611-12615. [DOI] [PubMed] [Google Scholar]
- 4.Pevny L, Lin CS, D'Agati V, Simon MC, Orkin SH, Costantini F. Development of hematopoietic cells lacking transcription factor GATA-1. Development. 1995; 121(1):163-172. [DOI] [PubMed] [Google Scholar]
- 5.Rodriguez P, Bonte E, Krijgsveld J, et al. GATA-1 forms distinct activating and repressive complexes in erythroid cells. EMBO J. 2005;24(13):2354-2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Snow JW, Orkin SH. Translational isoforms of FOG1 regulate GATA1-interacting complexes. J Biol Chem. 2009;284(43):29310-29319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wadman I, Li J, Bash RO, et al. Specific in vivo association between the bHLH and LIM proteins implicated in human T cell leukemia. EMBO J. 1994;13(20):4831-4839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meier N, Krpic S, Rodriguez P, et al. Novel binding partners of Ldb1 are required for haematopoietic development. Development. 2006;133(24):4913-4923. [DOI] [PubMed] [Google Scholar]
- 9.Soler E, Andrieu-Soler C, de Boer E, et al. The genome-wide dynamics of the binding of Ldb1 complexes during erythroid differentiation. Genes Dev. 2010;24(3):277-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mylona A, Andrieu-Soler C, Thongjuea S, et al. Genome-wide analysis shows that Ldb1 controls essential hematopoietic genes/pathways in mouse early development and reveals novel players in hematopoiesis. Blood. 2013;121(15):2902-2913. [DOI] [PubMed] [Google Scholar]
- 11.Shivdasani RA, Mayer EL, Orkin SH. Absence of blood formation in mice lacking the T-cell leukaemia oncoprotein tal-1/SCL. Nature. 1995;373(6513):432-434. [DOI] [PubMed] [Google Scholar]
- 12.Warren AJ, Colledge WH, Carlton MB, Evans MJ, Smith AJ, Rabbitts TH. The oncogenic cysteine-rich LIM domain protein rbtn2 is essential for erythroid development. Cell. 1994;78(1):45-57. [DOI] [PubMed] [Google Scholar]
- 13.Fredriksson S, Gullberg M, Jarvius J, et al. Protein detection using proximity-dependent DNA ligation assays. Nat Biotechnol. 2002;20(5):473-477. [DOI] [PubMed] [Google Scholar]
- 14.Gutierrez L, Lindeboom F, Ferreira R, et al. A hanging drop culture method to study terminal erythroid differentiation. Exp Hematol. 2005;33(10):1083-1091. [DOI] [PubMed] [Google Scholar]
- 15.Stadhouders R, Thongjuea S, Andrieu-Soler C, et al. Dynamic long-range chromatin interactions control Myb proto-oncogene transcription during erythroid development. EMBO J. 2012;31(4):986-999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Boer E, Rodriguez P, Bonte E, et al. Efficient biotinylation and single-step purification of tagged transcription factors in mammalian cells and transgenic mice. Proc Natl Acad Sci U S A. 2003; 100(13):7480-7485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van de Corput MP, de Boer E, Knoch TA, et al. Super-resolution imaging reveals three-dimensional folding dynamics of the beta-globin locus upon gene activation. J Cell Sci. 2012;125(Pt 19):4630-4639. [DOI] [PubMed] [Google Scholar]
- 18.Soderberg O, Gullberg M, Jarvius M, et al. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat Methods. 2006;3(12):995-1000. [DOI] [PubMed] [Google Scholar]
- 19.Grass JA, Boyer ME, Pal S, Wu J, Weiss MJ, Bresnick EH. GATA-1-dependent transcriptional repression of GATA-2 via disruption of positive autoregulation and domain-wide chromatin remodeling. Proc Natl Acad Sci U S A. 2003;100(15):8811-8816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Caputo L, Witzel HR, Kolovos P, et al. The Isl1/Ldb1 complex orchestrates genome-wide chromatin organization to instruct differentiation of multipotent cardiac progenitors. Cell Stem Cell. 2015;17(3):287-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Socolovsky M, Nam H, Fleming MD, Haase VH, Brugnara C, Lodish HF. Ineffective erythropoiesis in Stat5a(-/-)5b(-/-) mice due to decreased survival of early erythroblasts. Blood. 2001;98(12):3261-3273. [DOI] [PubMed] [Google Scholar]
- 22.Liu Y, Pop R, Sadegh C, Brugnara C, Haase VH, Socolovsky M. Suppression of Fas-FasL coexpression by erythropoietin mediates erythroblast expansion during the erythropoietic stress response in vivo. Blood. 2006;108(1):123-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brand M, Ranish JA, Kummer NT, et al. Dynamic changes in transcription factor complexes during erythroid differentiation revealed by quantitative proteomics. Nat Struct Mol Biol. 2004;11(1):73-80. [DOI] [PubMed] [Google Scholar]
- 24.Song SH, Hou C, Dean A. A positive role for NLI/Ldb1 in long-range beta-globin locus control region function. Mol Cell. 2007;28(5):810-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Anguita E, Hughes J, Heyworth C, Blobel GA, Wood WG, Higgs DR. Globin gene activation during haemopoiesis is driven by protein complexes nucleated by GATA-1 and GATA-2. EMBO J. 2004;23(14):2841-2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stadhouders R, Cico A, Stephen T, et al. Control of developmentally primed erythroid genes by combinatorial co-repressor actions. Nat Commun. 2015;6:8893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goardon N, Lambert JA, Rodriguez P, et al. ETO2 coordinates cellular proliferation and differentiation during erythropoiesis. EMBO J. 2006;25(2):357-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gutierrez L, Tsukamoto S, Suzuki M, et al. Ablation of Gata1 in adult mice results in aplastic crisis, revealing its essential role in steady-state and stress erythropoiesis. Blood. 2008;111(8):4375-4385. [DOI] [PubMed] [Google Scholar]
- 29.Li L, Jothi R, Cui K, et al. Nuclear adaptor Ldb1 regulates a transcriptional program essential for the maintenance of hematopoietic stem cells. Nat Immunol. 2011;12(2):129-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsai FY, Keller G, Kuo FC, et al. An early haematopoietic defect in mice lacking the transcription factor GATA-2. Nature. 1994; 371(6494):221-226. [DOI] [PubMed] [Google Scholar]
- 31.Kaneko H, Shimizu R, Yamamoto M. GATA factor switching during erythroid differentiation. Curr Opin Hematol. 2010; 17(3):163-168. [DOI] [PubMed] [Google Scholar]
- 32.Lugus JJ, Chung YS, Mills JC, et al. GATA2 functions at multiple steps in hemangioblast development and differentiation. Development. 2007;134(2):393-405. [DOI] [PubMed] [Google Scholar]
- 33.Gustafsdottir SM, Schallmeiner E, Fredriksson S, et al. Proximity ligation assays for sensitive and specific protein analyses. Anal Biochem. 2005;345(1):2-9. [DOI] [PubMed] [Google Scholar]
- 34.Alvarez-Silva M, Belo-Diabangouaya P, Salaun J, Dieterlen-Lievre F. Mouse placenta is a major hematopoietic organ. Development. 2003;130(22):5437-5444. [DOI] [PubMed] [Google Scholar]
- 35.McGrath KE, Palis J. Hematopoiesis in the yolk sac: more than meets the eye. Exp Hematol. 2005;33(9):1021-1028. [DOI] [PubMed] [Google Scholar]
- 36.Zatloukal B, Kufferath I, Thueringer A, Landegren U, Zatloukal K, Haybaeck J. Sensitivity and specificity of in situ proximity ligation for protein interaction analysis in a model of steatohepatitis with Mallory-Denk bodies. PLoS One. 2014;9(5):e96690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zeng Z, Ma J, Xi P, Xu C. Joint tagging assisted fluctuation nanoscopy enables fast high-density super-resolution imaging. J Biophotonics. 2018;11(9):e201800020. [DOI] [PubMed] [Google Scholar]
- 38.Allalou A, Wahlby C. BlobFinder, a tool for fluorescence microscopy image cytometry. Comput Methods Programs Biomed. 2009; 94(1):58-65. [DOI] [PubMed] [Google Scholar]