Abstract
Background
The five‐year survival rate of lung adenocarcinoma patients (LUAD) is very low,and the methods of predicting survival are a great obstacle for LUAD therapies. Endothelin receptor type B (EDNRB) gene is associated with tumorigenesis. In this study, we aimed to evaluate the predictive value of EDNRB on LUAD.
Methods
Survival analyses was performed to assess the correlation between EDNRB expression and survival of LUAD patients from the Cancer Genome Atlas (TCGA) and the Chinese Glioma Genome Atlas (CGGA) datasets. Gene set enrichment analysis (GSEA) was conducted to illustrate possible biological functions of EDNRB. Laboratory methods were used to verify the function of EDNRB in LUAD.
Results
The TCGA results showed that a low expression of EDNRB was found in LUAD patients which led to poor outcome and worse survival, compared with the high expression in GSEA results which suggested that expression of EDNRB might be associated with regulation of the ERK pathway. Laboratory results suggested that EDNRB could inhibit the proliferation and migration of LUAD H1299 cells.
Conclusions
EDNRB is a potential prognostic marker for LUAD patients and might exert its functions by regulating the ERK pathway in LUAD.
Keywords: EDNRB, Erk signaling pathway, GSEA, lung adenocarcinoma, TCGA
Introduction
Lung cancer is a malignant tumor of the respiratory system, about 80% of which is non‐small cell lung cancer (NSCLC). 1 With nonspecific early clinical symptoms, the onset of NSCLC is insidious, and most patients are diagnosed at an advanced stage, and the incidence of lung adenocarcinoma (LUAD) accounts for about 30% of NSCLC.2, 3 Relevant studies 4 have pointed out that after treatment, a considerable number of patients with LUAD still have tumor metastasis, and the five‐year survival rate of patients is very low. If the prognosis of patients with LUAD can be evaluated as early as possible, it will be of positive significance for clinical intervention measures to reduce the incidence of end‐point events. However, the sensitivity and accuracy of the methods used to evaluate the prognosis of patients with LUAD are poor.
The endothelin receptor type B (EDNRB) gene is located in q22 of chromosome 13, GenBank ID 1910, with a length of about 24 Kb, including seven exons and six introns. Its promoter contains a CpG island, which is inactivated during hypermethylation. The product of which, EDNRB protein belongs to the G‐protein‐coupled receptor family. It binds to the ligand, endothelin (ET) to transmit extracellular signals. In the process of embryonic development, EDNRB plays an important role in the migration and differentiation of neural crest cells into ganglion cells, which is related to megacolon disease.5, 6 The hypermethylation of EDNRB gene promoter is associated with a variety of tumors, accompanied by the silencing of EDNRB gene expression. EDNRB gene has been found to have low expression in melanoma 7 and breast carcinomas. 8 However, the correlation between EDNRB and LUAD prognosis has not yet been characterized. Therefore, this study sought to determine the role and mechanism of EDNRB in the tumorigenesis and development of LUAD, and provide a theoretical basis for EDNRB as the treatment target of LUAD.
Methods
Cell lines
LUAD H1299 cells were purchased from the ATCC cell bank; RPMI1640 and fetal bovine serum were purchased from GIBCO Company; penicillin/streptomycin, EDNRB carrier, pLEX‐MCS carrier, Cell Counting Kit‐8 (CCK‐8), DMSO reagent and Transwell cells were purchased from Costar company. EDNRB antibody (rabbit antibody, 1:500) and antibody (goat anti‐rabbit IgGHRP and rabbit anti‐human β‐actin IgG, 1:1000) purchased from Santa Cruz Biotechnology.
Database analysis
We downloaded the RNA‐seq expression data and clinical information for LUAD from TCGA database (http://cancergenome.nih.gov). Cases with incomplete clinical information were excluded. A total of 996 patients with detailed clinical information were included for subsequent survival analysis. Expression data of EDNRB in different tumors and normal controls were obtained from TCGA and the Genotype‐Tissue Expression (GTEx) databases. Log‐rank test in Kaplan‐Meier analysis was performed to compare the effect of EDNRB expression on prognosis using survival R package. The median expression of EDNRB was used to dichotomize patients into either a high‐EDNRB (top 498 samples) or a low‐EDNRB (last 498 samples) group in survival analyses. P‐values < 0.05 were regarded as the cut‐off for significance.
Gene functional analysis
Gene set enrichment analysis (GSEA) was used to study the possible mechanisms of EDNRB in LUAD patients. All LUAD patients were divided into two groups according to the median expression level of LUAD. The annotated gene sets (c2.cp.kegg.v6.0.symbols.gmt) were chosen as the reference gene sets. |NES| >1, P‐values < 0.05, and FDR < 0.25 were considered as statistically significant.
Cell culture
LUAD H1299 cells were cultured in RPMI‐1640 containing 10% fetal bovine serum. The culture medium contained 1% penicillin/streptomycin. Cells were cultured at 37°C in an incubator with 5% CO2, and subcultured when they had grown to 80% in the petri dish.
Cell transfection
The culture medium was replaced with serum‐free culture medium two hours before transfection. Two 1.5 mL EP tubes were used for transfection. First, 250 μL Opti‐MEM was placed in the EP tubes, and the transfected plasmid DNA and lipofectamine 2000 were in turn added. The tubes were then left to incubate at room temperature for 5 minutes. The two EP tubes were gently mixed together and left to incubate for 20 minutes. The mixture was then added to the petri dish with medium. The medium was replaced after six hours. Then, 24 hours after the transfection, the cells were ready to be used for subsequent experiments. H1299 cells transfected with EDNRB plasmid were the experimental group (H1299/EDNRB), and H1299 cells transfected with pLEX‐MCS plasmid were the control group (H1299/vector). Western blot experiment was used to detect the EDNRB protein in both groups.
Cell proliferation experiment
A CCK‐8 assay was used to study cell proliferation of H1299/EDNRB and H1299/vector cells. The cells were seeded in 96 well plates with 2000 cells per well. The reagent was added on days 0, one, two, three, and four and the optical density (OD) value of the hole detected with a wavelength of 450 nm. The OD value was obtained and statistically analyzed.
Cell scratch test
When the number of H1299/EDNRB and H1299/vector cells had grown to 90%, we used 200 μL pipette tips to mark 300–500 μm of a cellular scratching area. The scratch width was measured and images taken after 24 hours. Before the images were taken, the culture medium was changed into PBS and then returned to the culture medium.
Transwell cell migration experiment
After H1299/EDNRB and H1299/vector cells in petri dishes were digested and centrifuged, the concentration of cells was adjusted to 5 × 108/L. We added 100 mL cell suspension to each chamber. Then, 10% FBS and a series of concentrations of oleic acid to RPMI1640 were added in the lower chambers. After 24 hours, the cells were removed in the upper chamber using a cotton swab, and then fixed with 4% paraformaldehyde for 15 minutes. PBS was then used to rinse and 0.1% crystal violet to stain. Five visual fields were randomly selected under the microscope and photographed.
Results
Expression pattern of EDNRB in tumor patients
The results showed that the expression of EDNRB was downregulated in 17 tumors, such as adenoid cystic carcinoma (ACC), bladder urothelial carcinoma (BLCA), esophageal carcinoma (ESCA), and prostate adenocarcinoma (PRAD). There was a significant difference in the EDNRB expression between the LUAD group and the normal group (P < 0.05) (Fig 1), which indicated that EDNRB might participate in tumorigenesis in various tumors and be a potential prognostic gene marker for LUAD.
Figure 1.
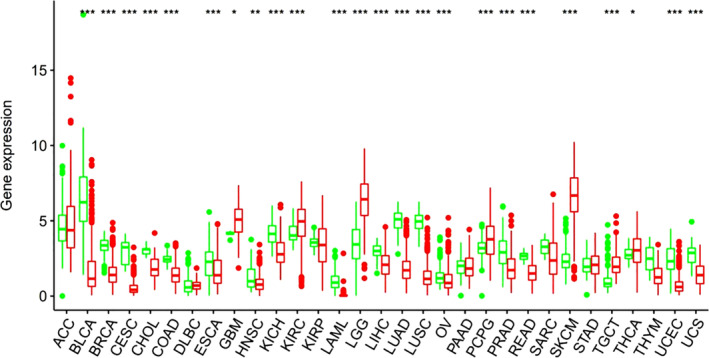
Profile of endothelin receptor type B (EDNRB) gene expression and Kaplan‐Meier plots in TCGA RNA‐seq dataset ( ) Normal, (
) Normal, ( ) Tumor.
) Tumor.
Relationship between expression of EDNRB and overall survival of patients with LUAD
We then inspected the correlation between EDNRB expression and survival in LUAD patients. Kaplan‐Meier plots demonstrated EDNRB expression was significantly associated with survival (P = 0.034) (Fig 2). This result indicated that LUAD patients with a high EDNRB expression might have better survival and prognosis.
Figure 2.
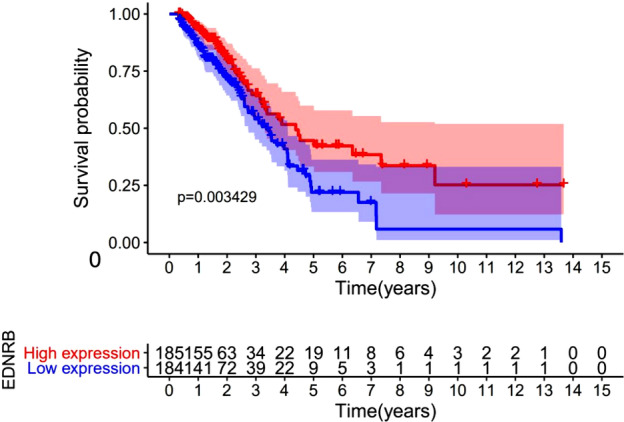
The survival analyses of patients with different expression level of endothelin receptor type B (EDNRB) ( ) high expression, (
) high expression, ( ) low expression.
) low expression.
EDNRB might be associated with ERK pathway
To illustrate the possible biological functions and pathways of EDNRB in LUAD, we performed GSEA using TCGA datasets. The regulation of ERK and PI3K‐Akt pathway were top two enriched in TCGA datasets (Table 1, Fig 3). These results suggested that the expression of EDNRB might be strongly associated with regulation of the ERK pathway.
Table 1.
Correlation between endothelin receptor type B (EDNRB) and signal pathway of lung adenocarcinoma (LUAD)
| Signal pathway | Enrichment | P‐value |
|---|---|---|
| Pathways in cancer | −0.454 272 | 0.001003 |
| ERK signaling pathway | −0.472 453 | 0.001025 |
| PI3K‐Akt signaling pathway | −0.508 594 | 0.001015 |
| RAS signaling pathway | −0.606 667 | 0.001056 |
| Regulation of actin cytoskeleton | −0.472 949 | 0.001056 |
| Proteoglycans in cancer | −0.481 285 | 0.001056 |
| Focal adhesion | −0.580 193 | 0.001057 |
| Rap1 signaling pathway | −0.571 431 | 0.001059 |
| Cytokine‐cytokine receptor interaction | −0.514 928 | 0.001066 |
| Chemokine signaling pathway | −0.545 630 | 0.001066 |
Figure 3.
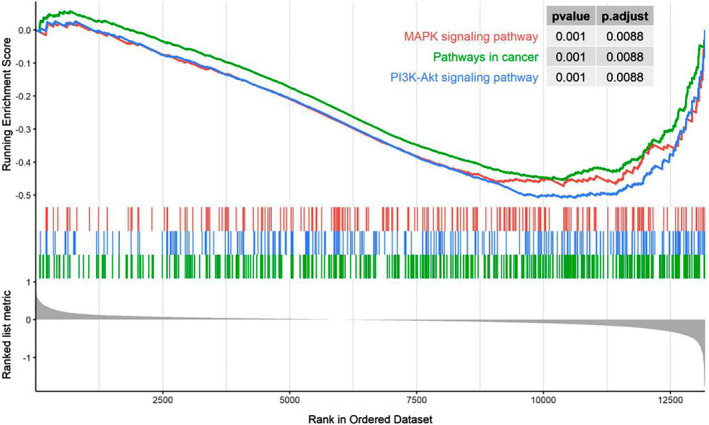
Functional investigation of endothelin receptor type B (EDNRB) in lung adenocarcinoma (LUAD) ( ) MAPK signaling pathway, (
) MAPK signaling pathway, ( ) pathways in cancer, (
) pathways in cancer, ( ) PI3K‐Akt signaling pathway.
) PI3K‐Akt signaling pathway.
EDNRB could inhibit proliferation of H1299 cells
In order to verify the role of EDNRB in the carcinogenesis and development of LUAD, CCK‐8 method was used to detect the proliferation of H1299/EDNRB and H1299/vector. The results showed that the trend appeared from 48 hours, and the difference was the most significant at 96 hours (P < 0.05). The proliferation of H1299/EDNRB cells was weaker than that of H1299/vector cells, indicating that EDNRB could inhibit the proliferation of LUAD (Fig 4a).
Figure 4.
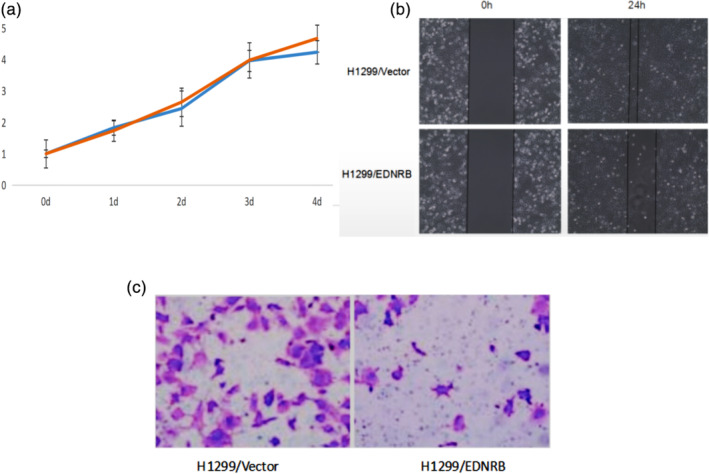
Laboratory methods were used to verify the function of endothelin receptor type B (EDNRB) to lung adenocarcinoma (LUAD). (a) The CCK‐8 method was used to detect the proliferation of H1299/EDNRB and H1299/vector cells ( ) H1299/EDNRB, (
) H1299/EDNRB, ( ) H1299/vector. (b) The effect of EDNRB overexpression on H1299 cell migration (cell scratch test). (c) The effect of EDNRB overexpression on H1299 cell migration (transwell experiment).
) H1299/vector. (b) The effect of EDNRB overexpression on H1299 cell migration (cell scratch test). (c) The effect of EDNRB overexpression on H1299 cell migration (transwell experiment).
EDNRB could inhibit migration of H1299 cells
In order to determine the effect of EDNRB on the migration of H1299 cells, a cell scratch test and transwell cell migration experiment were used to detect the migration of H1299/EDNRB and H1299/vector. The cell scratch test results showed that the migration speed of H1299/EDNRB cells was significantly slower than that of H1299/vector cells (0.36 ± 0.02, 0.77 ± 0.04, P < 0.05) (Fig 4b). The results of the transwell cell migration experiment showed that the migration ability of H1299/EDNRB was significantly weaker than that of H1299/vector cells (21.11 ± 3.85, 54.49 ± 9.42, P < 0.05) (Fig 4c). These results showed that EDNRB inhibited the migration of H1299 cells.
Discussion
EDNRB gene encodes a G‐protein‐coupled receptor‐mediated endothelin, inducing development and transformation of the neural crest cell‐specific lineage. Recently, evidence has shown that there is reduced EDNRB expression in cancer cells when compared to normal cells. Silencing of EDNRB expression has also been shown in nasopharyngeal carcinoma, prostate cancer, melanoma and esophageal carcinoma.9, 10, 11, 12 A study by Chen et al. from Taiwan reported that EDNRB was downregulated in lung cancer patients. 13 In this study, abnormal low expression of EDNRB gene was detected in 26 of the 79 lung cancer patients and concluded that the EDNRB gene may have the characteristics of a tumor suppressor gene in lung cancer. The study illustrated the relationship between EDNRB and lung cancer, but its biological role and possible mechanism were not focused upon.
In our study, we confirmed EDNRB as a suppressor gene for LUAD and observed the role of EDNRB in LUAD. The survival analyses of patients with different expression levels of EDNRB based on TCGA LUAD data also indicated that EDNRB was a potential prognostic marker for LUAD. The survival of high EDNRB expression cases was significantly better than that of low EDNRB cases. This result indicated that EDNRB might be a predictor for LUAD patients and play a key role during prognosis of tumor patients. In other reports, the results were similar with our findings. For example, Tao et al. 14 reported that low expression of EDNRB was associated with the proliferation of gastric cancer cells. One study suggested that low expression of EDNRB was related to a poor prognosis in breast cancer. 15 Therefore, we carried out an in‐depth study on the EDNRB gene in LUAD. After construction of EDNRB overexpression H1299 cell line, we carried out experiments related to cell proliferation and migration. The experimental results showed that EDNRB inhibited the proliferation and migration of H1299 cells. Combined, these results suggest that EDNRB might be a necessary suppressor gene and could inhibit the proliferation and migration of LUAD.
Another important finding in our study was that EDNRB was closely related to the ERK pathway. Based on GSEA using TCGA datasets, regulation of the ERK pathway was the most significantly enriched in TCGA datasets. These results suggested that the expression of EDNRB might be strongly associated with regulation of the ERK pathway. The ERK signaling pathway is a classic mitogen‐activated protein kinase (MAPK) signaling pathway, which can be activated by RAS, PKC, etc and participates in cell metastasis and invasion. The activation of ERK can promote the migration and invasion of cancer cells. 16 It has also been reported that ERK activation can promote the migration and invasion of lung cancer. 17 EDNRB may participate in the process of ERK on LUAD, which provides a possible theoretical basis for EDNRB to regulate the invasion and migration of LUAD.
In conclusion, EDNRB is a potential prognostic marker for LUAD patients and might exert its functions by regulating the ERK pathway in LUAD. Further investigations should focus on specific clinical events, biological behaviors and potential mechanism of EDNRB in the regulation of LUAD, which may allow a better understanding of LUAD pathogenesis and provide a theoretical basis for EDNRB.
Disclosure
The authors have no conflicts of interest to declare.
Acknowledgments
This work was supported by the Key Medical Projects of Jiangsu Province (ZDXKB2016020), the Southeast University Projects (2018yy‐jccx003) and Jiangsu Social Development project (BE2018711).
References
- 1. Liu H, Wang M, Hu K et al Research Progress of the resistance mechanism of non‐small cell lung cancer to EGFR‐TKIs. Chin J Lung Cancer 2013; 16 (10): 535–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kono M, Allen PK, Lin SH et al Incidence of second malignancy after successful treatment of limited‐stage small‐cell lung cancer and its effects on survival. J Thorac Oncol 2017; 12 (11): 1696–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Huang CY, Chen BH, Chou WC, Yang CT, Chang JW. Factors associated with the prognosis and long‐term survival of patients with metastatic lung adenocarcinoma:A retrospective analysis. J Thorac Dis 2018; 10 (4): 2070–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Masashi I, Makoto S, Naoto I et al Expression of the GLI family genes is associated with tumor progression in advanced lung adenocarcinoma. World J Surg Oncol 2014; 12 (1): 253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Feinberg AP, Ohlsson R, Henikoff S et al The epigenetic progenitor origin of human cancer. Nat Rev Genet 2006; 7 (1): 21–33. [DOI] [PubMed] [Google Scholar]
- 6. Ludvíková M, Pesta M, Holubec L et al New aspects of tumor pathobiology. Cesk Patol 2009; 45 (4): 94–9. [PubMed] [Google Scholar]
- 7. Spica T, Fargnoli M, Hetet G et al EDNRB gene variants and melanoma risk in two southern European populations. Clin Exp Dermatol 2011; 36 (7): 782–7. [DOI] [PubMed] [Google Scholar]
- 8. Burstein M, Tsimelzon A, Hilsenbeck S et al Abstract P4‐06‐01: Expression and DNA copy number profiling suggest novel therapeutic approaches for triple negative breast cancer subtypes. Cancer Res 2013; 73 (Suppl. 24): P4‐06‐01. [Google Scholar]
- 9. Cheng W, Ngan R, Kwong D et al Abstract 2298: Methylated gene markers discovered in nasopharyngeal carcinoma by differential methylation hybridization (DMH) in high density CpG Island DNA chips. Cancer Res 2014; 74 (Suppl. 19): 2298–8. [Google Scholar]
- 10. Bastian P, Palapattu G, Rogers C et al CpG island hypermethylation profile in the serum of men with clinically localized and hormone refractory metastatic prostate cancer. J Urol 2008; 179 (2): 529–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Asundi J, Lacap J, Clark S et al MAPK pathway inhibition enhances the efficacy of an anti‐endothelin B receptor drug conjugate by inducing target expression in melanoma. Mol Cancer Ther 2014; 13 (6): 1599–610. [DOI] [PubMed] [Google Scholar]
- 12. Nones K, Waddell N, Wayte N et al Genomic catastrophes frequently arise in esophageal adenocarcinoma and drive tumorigenesis. Nat Commun 2014; 5 (5): 5224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen SC, Lin CY, Chen YH et al Aberrant promoter methylation of EDNRB in lung cancer in Taiwan. Oncol Rep 2006; 15 (1): 167–72. [PubMed] [Google Scholar]
- 14. Tao K, Wu C, Wu K et al Quantitative analysis of promoter methylation of the EDNRB gene in gastric cancer. Med Oncol 2012; 29 (1): 107–12. [DOI] [PubMed] [Google Scholar]
- 15. Benatar T, Amemiya Y, Yang W et al Abstract P5‐05‐01: IGFBP‐7 reduces growth of xenografted breast tumors in mice and inhibits breast cancer cell proliferation and migration through the MEK‐ERK pathway. Cancer Res 2010; 70 (Suppl. 24): P5‐05‐01. [Google Scholar]
- 16. Miglietta A, Bozzo F, Bocca C et al Conjugated linoleic acid induces apoptosis in MDA‐MB‐231 breast cancer cells through ERK/MAPK signalling and mitochondrial pathway. Cancer Lett 2006; 234 (2): 149–57. [DOI] [PubMed] [Google Scholar]
- 17. Wang J, Huang S. Fisetin inhibits the growth and migration in the A549 human lung cancer cell line via the ERK1/2 pathway. Exp Therap Med 2018; 15 (3): 2667–73. [DOI] [PMC free article] [PubMed] [Google Scholar]


