Abstract
Background
Circular RNAs (circRNAs) have been demonstrated to act as key regulators in the chemoresistance of human cancers, including breast cancer (BC). Here, we aimed to explore the role of circ‐RNF111 in paclitaxel (PTX) resistance of BC.
Methods
Quantitative real‐time polymerase chain reaction (qRT‐PCR) was employed to determine the expression of circ‐RNF111, microRNA‐140‐5p (miR‐140‐5p) and E2F transcription factor 3 (E2F3) mRNA. The half maximal inhibitory concentration (IC50) of PTX, cell viability, colony formation and cell invasion were assessed by cell counting kit‐8 (CCK‐8) assay, colony formation assay and transwell assay, respectively. Glucose consumption and lactate production were determined by specific kits. A murine xenograft model was established to investigate the role of circ‐RNF111 in PTX resistance of BC in vivo. Dual‐luciferase reporter assay and RNA immunoprecipitation (RIP) assay were performed to verify the relationship between miR‐140‐5p and circ‐RNF111 or E2F3. Western blot assay was conducted to examine the protein level of E2F3.
Results
Circ‐RNF111 was upregulated in PTX‐resistant BC tissues and cells. Circ‐RNF111 knockdown restrained IC50 of PTX, cell viability, colony numbers, cell invasion and glycolysis in PTX‐resistant BC cells in vitro and enhanced PTX sensitivity in vivo. MiR‐140‐5p was a target of circ‐RNF111 and miR‐140‐5p expression was negatively correlated with circ‐RNF111 expression in BC tissues. The effect of circ‐RNF111 knockdown on PTX resistance was rescued by miR‐140‐5p deletion. Additionally, miR‐140‐5p could interact with E2F3 and negatively regulate E2F3 expression. Moreover, miR‐140‐5p suppressed IC50 of PTX, cell viability, colony numbers, cell invasion and glycolysis by targeting E2F3.
Conclusions
Circ‐RNF111 improved PTX resistance of BC by upregulating E2F3 via sponging miR‐140‐5p.
Keywords: Breast cancer, circ‐RNF111, E2F3, miR‐140‐5p, PTX
Introduction
As a common and aggressive cancer, breast cancer (BC) has become the second leading cause of cancer‐related death in females. 1 Although there have been improvements in therapeutic strategies, the prognosis for BC patients is still unsatisfactory. 2 Chemotherapy is an effective treatment method for BC, but the acquisition of drug resistance often leads to treatment failure. 3 , 4 Paclitaxel (PTX) is a frontline chemotherapy drug in the clinic, but becomes less effective over time due to chemoresistance. 5 Hence, development of a novel therapeutic method which can sensitize BC cells to PTX and explore the mechanism of PTX resistance in BC is urgently required.
Circular RNAs (circRNAs) are a series of non‐coding RNAs (ncRNAs) featured by covalent closed loops. 6 Currently, multiple circRNAs have been discovered and identified to participate in the regulation of biological processes. 7 Moreover, emerging evidence has verified that circRNAs are essential mediators in drug resistance of human cancers. For example, circFN1 contributed to cisplatin resistance in gastric cancer (GC) via promoting cell viability and repressing apoptosis. 8 CircEIF6 suppressed tumor growth and cisplatin resistance in thyroid carcinoma. 9 In BC, though several circRNAs were identified to be related to the progression of chemoresistance, such as circ_0025202, 10 circ_0006528 11 and circKDM4C, 12 the studies of circRNAs on drug resistance in BC remain insufficient. Tang et al. declared that circ_0001982 (also termed as circ‐RNF111) functioned as an oncogene in BC. 13 However, the association between circ‐RNF111 and drug resistance has not been addressed.
MicroRNAs (miRNAs) are ncRNAs with ~22 nucleotides which can modulate gene expression through interacting with the 3′ untranslated region (3′UTR) of target mRNA. 14 In BC, it has been documented that diverse miRNAs are associated with chemoresistance in cancers. For example, miR‐200c sensitized BC cells to trastuzumab by binding to ZNF217 and ZEB1. 15 MiR‐487a enhanced the sensitivity of BC cells to mitoxantrone by targeting BCRP/ABCG2. 16 MiR‐140‐5p has been proved to take part in drug resistance process in cancers, such as osteosarcoma, 17 hepatocellular carcinoma 18 and GC. 19 Even so, the function of miR‐140‐5p in PTX resistance in BC remains unknown.
E2F transcription factor 3 (E2F3) locates on chromosome 6p22 and plays vital roles in proliferation and cell cycle process. 20 Recent studies have shown that miRNAs can regulate drug resistance in human cancers by targeting E2F3. For example, miR‐203 improved the sensitivity of glioma cells to temozolomide via interacting with E2F3. 21 MiR‐200b could also target E2F3 to improve the response of human lung adenocarcinoma cells to docetaxel. 22 Nonetheless, whether E2F3 can be target of miR‐140‐5p has not yet been documented.
The aim of our work was to explore the functional roles and potential mechanisms of circ‐RNF111 in PTX resistance in BC, to discover an effective target to counter PTX resistance in BC.
Methods
Tissue collection
A total of 30 PTX‐resistant BC patients and 30 PTX‐sensitive BC patients were enrolled into the study. The patients with worse disease during primary chemotherapy and patients who suffered recurrent disease within six months of completion of primary chemotherapy were called PTX‐resistant patients. The patients without recurrence or with recurrence more than six months after chemotherapy were called PTX‐sensitive patients. Tumor tissues were obtained via surgical resection at China‐Japan Union Hospital of Jilin University and saved at −80°C before use. The research was permitted by the Ethics Committee of China‐Japan Union Hospital of Jilin University and written informed consent were signed by the patients.
Cell culture
Normal breast epithelial cells (MCF‐10A) and BC cells (MCF‐7 and MDA‐MB‐231) were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA). PTX‐resistant BC cells (MCF‐7/PTX and MDA‐MB‐231/PTX) were established by treating MCF‐7 and MDA‐MB‐231 cells with rising concentrations of PTX (Sangon, Shanghai, China) at an initial concentration of 0.5 nM PTX. After the cells had grown steadily at each PTX concentration condition, PTX concentration was increased and the culture continued until the cells had grown steadily in the medium containing 5 nM PTX. The process lasted for three months. All cells were maintained in Roswell Park Memorial Institute 1640 Medium (Thermo Fisher Scientific, Waltham, MA, USA) including 10% fetal bovine serum (FBS; Gibco, Grand Island, NY, USA) and 1% penicillin‐streptomycin (Solarbio; Beijing, China) at a condition of 5% CO2 and 37°C. To maintain the resistance of PTX‐resistant BC cells, the culture medium was added with 5 nM PTX (Sangon) until two weeks before further experiments.
Cell transfection
Small interfering RNA (siRNA) targeting circ‐RNF111 (si‐circ‐RNF111; GTTCCCTCAGGCTTTCCTTAA) and corresponding control (si‐NC; UCACAACCUCCUAGAAAGAGUAGA), the overexpression vector of circ‐RNF111 (circ‐RNF111), the overexpression vector of E2F3 (E2F3) and their control (pcDNA), short hairpin RNA targeting circ‐RNF111 (sh‐circ‐RNF111; GCTGTTCCCTCAGGCTTTCCT) and corresponding control (sh‐NC; UUCAAGAGA), miR‐140‐5p mimics (miR‐140‐5p; 5′‐CCAUAGGGUAAAACCACUGUU‐3′) and its control (miR‐NC; 5′‐UUCUCCGAACGUGUCACGUTT‐3′), miR‐140‐5p inhibitors (anti‐miR‐140‐5p; 5′‐AACCCAUGGAAUUCAGUUCUCA‐3′) and matched control (anti‐miR‐NC; 5′‐CAGUACUUUGUGUAGUACAA‐3′) were synthesized by RIBOBIO (Guangzhou, China). Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) was utilized for cell transfection. Sh‐NC and sh‐circ‐RNF111 were stably transfected into cells, and the other synthetic oligonucleotides or plasmids were transiently transfected into cells.
Quantitative real‐time polymerase chain reaction (qRT‐PCR)
Total RNA in tissues and cells was extracted with TRIzol reagent (Invitrogen). Then, 1 μg RNA was reversely transcribed into complementary DNA (cDNA) with M‐MLV Reverse Transcriptase Kit (Promega, Madison, WI, USA) or TaqMan microRNA assay kit (Thermo Fisher Scientific). Next, qRT‐PCR was performed on an ABI 7500 PCR system (Applied Biosystems, Foster City, CA, USA) using the Platinum SYBR Green qPCR SuperMix UDG (Invitrogen). The conditions were: 95°C for 5 minutes; 40 cycles of 95°C for 30 seconds, 60°C for 45 seconds and 72°C for 30 seconds; dissolving curve at 95°C for 90 seconds, 60°C for 180 seconds, and 95°C for 10 seconds. The expression was evaluated using the 2‐ΔΔCt method with normalization to U6 or glyceraldehyde 3‐phosphate dehydrogenase (GAPDH). The primers were: circ‐RNF111: (F: 5′‐TAGCAGTTCCCCAATCCTTG‐3′ and R: 5′‐CACAAATTCCCATCATTCCC‐3′); miR‐140‐5p: (F: 5′‐GAGTGTCAGTGGTTTTACCCT‐3′ and R: 5′‐GCAGGGTCCGAGGTATTC‐3′); E2F3: (F: 5′‐CAGGCTGGTTTCGGAAATGC‐3′ and R: 5′‐TGGACTTCGTAGTGCAGCTC‐3′); GAPDH: (F: 5′‐ATGTTGCAACCGGGAAGGAA‐3′ and R: 5′‐AGGAAAAGCATCACCCGGAG‐3′); U6: (F: 5′‐CTCGCTTCGGCAGCACATATACTA‐3′ and R: 5′‐ACGAATTTGCGTGTCATCCTTGCG‐3′). The experiment was repeated three times.
Cell counting kit‐8 (CCK‐8) assay
PTX resistance in cells was assessed by CCK‐8 assay. First, BC cells or PTX‐resistant BC cells were plated into 96‐well plates (1 × 104 cells/well) and maintained for 24 hours. Indicated synthetic oligonucleotides or vectors were then transfected into the cells. Next, transfected cells were treated with or without 20 mM 2‐deoxy‐glucose (2‐DG; Sangon) for 48 hours. Thereafter, cells were exposed to different doses of PTX (Sangon) and kept for 48 hours. Subsequently, CCK‐8 (5 mg/mL; Solarbio) was added to each well and incubated for another 4 hours. The optical density value at 450 nm was recorded using a microplate reader (Bio‐Rad, Hercules, CA, USA). The relative survival curve was employed to estimate the half maximal inhibitory concentration (IC50). The experiment was repeated three times.
CCK‐8 assay was also performed to detect BC cell viability. In brief, 1 × 104 cells were plated into each well of 96‐well plates and then 10 μL CCK‐8 (Solarbio) was added at indicated time points and kept for an additional 4 hours. The absorbance at 450 nm was measured by a microplate reader (Bio‐Rad). The experiment was repeated three times.
Colony formation assay
PTX‐resistant cells were plated into six‐well plates (500 cells/well). After two weeks, colonies were fixed with 4% paraformaldehyde (Sangon) and stained with 0.5% crystal violet (Solarbio). The stained cells were photographed and counted under a microscope (Olympus, Tokyo, Japan). The experiment was repeated three times.
Transwell assay
A transwell chamber (Corning, Inc., Corning, NY, USA) coated with Matrigel (Solarbio) was employed to test cell invasion. Briefly, transfected PTX‐resistant BC cells (1 × 106 cells) in serum‐free medium were added into the upper chamber and culture medium including 10% FBS (Gibco) was added into the bottom chamber. Then, 24 hours later, invaded cells were fixed with methanol, stained with crystal violet (Solarbio) and observed under an inverted microscope (Olympus). The experiment was repeated three times.
Detection of glucose consumption and lactate production
Glucose assay kit (Sigma‐Aldrich, St. Louis, MO, USA) and lactate assay kit (Sigma‐Aldrich) were utilized to determine the levels of glucose consumption and lactate production in PTX‐resistant BC cells based on the protocols. The experiment was repeated three times.
Murine xenograft model
The following work was approved by the Ethics Committee of Animal Research of China‐Japan Union Hospital of Jilin University. The 6‐week‐old BALB/c nude mice (Shanghai SLAC Laboratory Animals Co., Ltd., Shanghai, China) were divided into four groups (seven mice/group). Sh‐circ‐RNF111 or sh‐NC was stably transfected into MCF‐7/PTX cells. Then, 5 × 105 MCF‐7/PTX cells were injected subcutaneously into the nude mice, and eight days later, the mice were administered with or without 3 mg/kg PTX (Solarbio) every three days. Tumor length and width were monitored every three days and calculated with the equation: (length × width2)/2. After 29 days of injection, the mice were sacrificed and tumors were weighed and saved at −80°C for further qRT‐PCR assay.
Dual‐luciferase reporter assay
The fragments of circ‐RNF111 or 3′UTR of E2F3 including the predicted wild‐type or mutant complementary sequences of miR‐140‐5p were inserted into pmirGLO vector (Promega) to establish luciferase reporter vectors: WT‐circ‐RNF111, MUT‐circ‐RNF111, E2F3 3′UTR‐WT and E2F3 3′UTR‐MUT. Next, the indicated luciferase reporter vector and miR‐140‐5p or miR‐NC were cotransfected into PTX‐resistant BC cells. The luciferase activity was examined by dual‐luciferase reporter assay kit (Promega).
RNA immunoprecipitation (RIP) assay
Magna RIP RNA Binding Protein Immunoprecipitation Kit (Millipore, Bedford, MA, USA) was adopted to confirm the relationship between miR‐140‐5p and circ‐RNF111 or E2F3. Briefly, PTX‐resistant cells were lysed in RIP buffer and incubated with magnetic beads which were conjugated with anti‐Argonaute2 (anti‐Ago2; Abcam, Cambridge, MA, USA) or anti‐immunoglobulin G (anti‐IgG; Abcam). Next, proteinase K (Solarbio) was added to digest the protein. Finally, immunoprecipitated RNA was isolated and the enrichment of circ‐RNF111, miR‐140‐5p and E2F3 was determined by qRT‐PCR.
Western blot assay
Total protein was isolated using RIPA buffer (Beyotime, Shanghai, China) and examined using a BCA protein assay kit (Tiangen, Beijing, China). Equal of proteins were subjected to sodium dodecyl sulfonate‐polyacrylamide gel (SDS‐PAGE; Solarbio) and transferred onto polyvinylidene difluoride membranes (Millipore). Next, the samples were blocked with 5% non‐fat milk for 1 hour at room temperature. Afterward, the samples were incubated with primary antibodies: E2F3 (ab50917; Abcam) or GAPDH (ab181602; Abcam) overnight at 4°C and relevant secondary antibody (ab205719; Abcam) for 2 hours at room temperature. Finally, enhanced chemiluminescence reagent (Vazyme, Nanjing, China) was utilized to visualize the proteins.
Statistical analysis
Data from three independent experiments were displayed as mean ± standard deviation, processed by software GraphPad Prism 7 (GraphPad Inc., La Jolla, CA, USA). Differences were estimated by Student's t‐test or one‐way analysis of variance (ANOVA). The correlation between miR‐140‐5p and circ‐RNF111 or E2F3 was estimated by Spearman's correlation coefficient analysis. P < 0.05 was considered statistically significant.
Results
High expression of circ‐RNF111 in PTX‐resistant BC tissues and cells
In order to address the function of circ‐RNF111 in PTX resistance of BC, the expression of circ‐RNF111 in PTX‐resistant and PTX‐sensitive BC tissues was first examined using qRT‐PCR assay. As shown in Fig 1a, circ‐RNF111 was highly expressed in PTX‐resistant BC tissues in comparison with PTX‐sensitive BC tissues. Besides, we found that circ‐RNF111 expression was notably increased in MCF‐7 and MDA‐MB‐231 cells when compared to MCF‐10A cells; moreover, circ‐RNF111 was higher expressed in MCF‐7/PTX and MDA‐MB‐231/PTX cells than in MCF‐7 and MDA‐MB‐231 cells (Fig 1b). Collectively, the aberrant expression of circ‐RNF111 might play a role in PTX resistance of BC.
Figure 1.
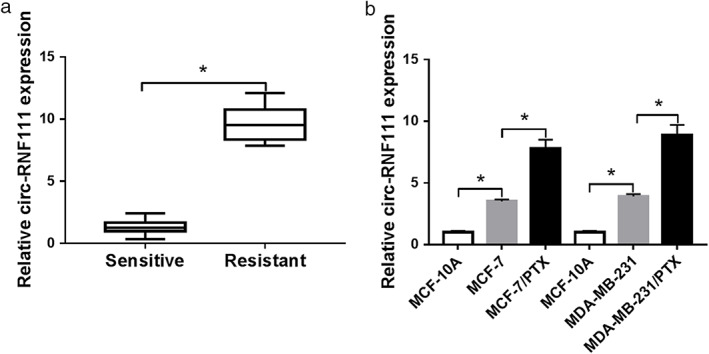
Circ‐RNF111 is upregulated in PTX‐resistant BC tissues and cells. (a) The expression of circ‐RNF111 in PTX‐resistant and PTX‐sensitive BC tissues was measured by qRT‐PCR. (b) The level of circ‐RNF111 in MCF‐10A, MCF‐7, MDA‐MB‐231, MCF‐7/PTX and MDA‐MB‐231/PTX cells was determined through qRT‐PCR assay. *P < 0.05.
Circ‐RNF111 knockdown suppresses PTX resistance, cell viability, colony formation, invasion and glycolysis in PTX‐resistant BC cells
As observed in Fig 2a, IC50 of PTX was increased in MCF‐7/PTX and MDA‐MB‐231/PTX cells compared to MCF‐7 and MDA‐MB‐231 cells, indicating PTX resistance was produced in MCF‐7/PTX and MDA‐MB‐231/PTX cells. Subsequently, to investigate whether circ‐RNF111 was involved in the regulation of PTX resistance in PTX‐resistant BC cells, MCF‐7/PTX and MDA‐MB‐231/PTX cells were transfected with si‐circ‐RNF111 to downregulate the expression of circ‐RNF111. The qRT‐PCR data exhibited that si‐circ‐RNF111 transfection led to a noteworthy reduction in circ‐RNF111 level in MCF‐7/PTX and MDA‐MB‐231/PTX cells compared to corresponding controls (Fig 2b). CCK‐8 assay indicated that the resistance of MCF‐7/PTX and MDA‐MB‐231/PTX cells to PTX was decreased following the knockdown of circ‐RNF111, as demonstrated by decreased IC50 value (Fig 2c). CCK‐8 assay also demonstrated that the viability of MCF‐7/PTX and MDA‐MB‐231/PTX cells was markedly inhibited following the interference of circ‐RNF111 (Fig 2d). Colony formation assay displayed that the colony numbers of MCF‐7/PTX and MDA‐MB‐231/PTX cells were reduced after circ‐RNF111 silencing compared to control groups (Fig 2e). The results of transwell assay showed that cell invasion was conspicuously restrained in MCF‐7/PTX and MDA‐MB‐231/PTX cells transfected with si‐circ‐RNF111 in reference to si‐NC groups (Fig 2f). Subsequently, the effect of circ‐RNF111 on glycolysis was explored by detecting the levels of glucose uptake and lactate production. The data confirmed that circ‐RNF111 silencing caused marked suppression in glucose consumption and lactate production, indicating the glycolysis was repressed by circ‐RNF111 silencing in MCF‐7/PTX and MDA‐MB‐231/PTX cells (Fig 2g,h). To determine the effect of glycolysis on PTX resistance in MCF‐7/PTX and MDA‐MB‐231/PTX cells, cells were exposed to 2‐DG to inhibit glycolysis. As presented in Fig 2i,j, glucose uptake and lactate production were drastically repressed in MCF‐7/PTX and MDA‐MB‐231/PTX cells after 2‐DG treatment compared to control groups. Thereafter, circ‐RNF111 was successfully transfected into MCF‐7/PTX and MDA‐MB‐231/PTX cells to elevate circ‐RNF111 expression (Fig 2k). CCK‐8 assay indicated that IC50 of PTX was increased in MCF‐7/PTX and MDA‐MB‐231/PTX cells after circ‐RNF111 overexpression, while the effect was abrogated by the treatment of 2‐DG, suggesting that the inhibition of glycolysis could restore PTX resistance mediated by circ‐RNF111 in MCF‐7/PTX and MDA‐MB‐231/PTX cells (Fig 2l). In summary, circ‐RNF111 silencing enhanced PTX sensitivity in PTX‐resistant BC cells.
Figure 2.
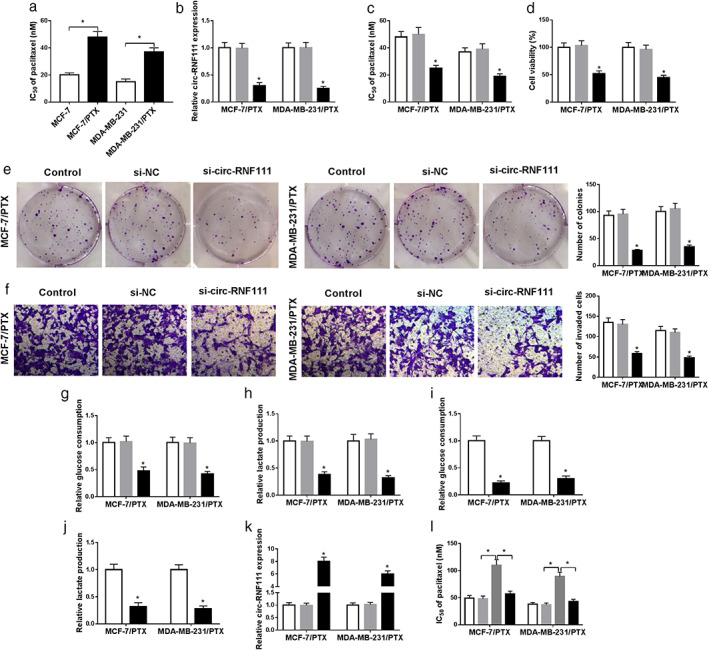
Deficiency of circ‐RNF111 improves PTX sensitivity and suppresses cell viability, colony formation, cell invasion and glycolysis in PTX‐resistant BC cells. (a) IC50 of PTX in MCF‐7/PTX and MDA‐MB‐231/PTX cells was assessed using CCK‐8 assay. (b–g) MCF‐7/PTX and MDA‐MB‐231/PTX cells were treated with or without si‐NC or si‐circ‐RNF111. (b) Circ‐RNF111 level in MCF‐7/PTX and MDA‐MB‐231/PTX cells was detected by qRT‐PCR (c,d) IC50 of PTX and cell viability in MCF‐7/PTX and MDA‐MB‐231/PTX cells were evaluated using CCK‐8 assay. (e) Cell colony formation ability was analyzed by colony formation assay. (f) Cell invasion was tested through transwell assay. (g,h) The levels of glucose consumption and lactate production were determined using relevant kits. (i,j) MCF‐7/PTX and MDA‐MB‐231/PTX cells were treated with or without 2‐DG and then glucose consumption and lactate production were examined with specific kits. (k) MCF‐7/PTX and MDA‐MB‐231/PTX cells were transfected with or without pcDNA or circ‐RNF111 and then circ‐RNF111 level was measured by qRT‐PCR assay. (l) IC50 of PTX in control, pcDNA, circ‐RNF111 and circ‐RNF111 + 2‐DG groups was analyzed by CCK‐8 assay. *P < 0.05. ( ) Control, (
) Control, ( ) si‐NC, (
) si‐NC, ( ) si‐circ‐RNF111; (
) si‐circ‐RNF111; ( ) Control, (
) Control, ( ) 2‐DG; (
) 2‐DG; ( ) Control, (
) Control, ( ) pcDNA, (
) pcDNA, ( ) circ‐RNF111; (
) circ‐RNF111; ( ) Control, (
) Control, ( ) pcDNA, (
) pcDNA, ( ) circ‐RNF111, (
) circ‐RNF111, ( ) si‐circ‐RNF111+2‐DG
) si‐circ‐RNF111+2‐DG
Circ‐RNF111 knockdown suppresses PTX resistance of BC in vivo
To further determine the effect of circ‐RNF111 on PTX resistance of BC in vivo, MCF‐7/PTX cells were stably transfected with sh‐NC or sh‐circ‐RNF111 and the transfection efficiency was determined by qRT‐PCR assay (Fig 3a). Transfected cells were then injected into the mice. The mice were administered with or without 3 mg/kg PTX every three days after eight days of inoculation, and then 29 days later, the mice were sacrificed and tumors were weighed. As shown in Fig 3b,c, tumor volume and weight were distinctly hampered by PTX treatment or circ‐RNF111 knockdown compared to control groups. The data of qRT‐PCR assay implied that the expression of circ‐RNF111 was downregulated in the tumors collected from sh‐circRNF111 and sh‐circ‐RNF111 + PTX groups when compared to control groups (Fig 3c). These results suggested that circ‐RNF111 silencing improved PTX sensitivity of BC in vivo.
Figure 3.
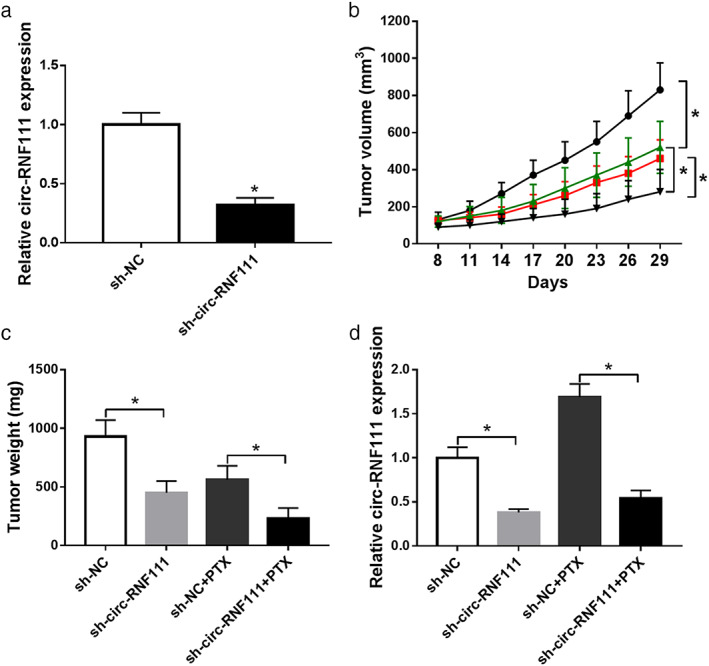
Downregulation of circ‐RNF111 inhibits PTX resistance in vivo. (a) Sh‐RNF111 or sh‐NC was transfected into MCF‐7/PTX cells and then the level of circRNF111 was measured by qRT‐PCR assay. (b–d) The mice were divided into four groups: sh‐NC, sh‐circRNF111, sh‐NC + PTX and sh‐circ‐RNF111 + PTX. (b) Tumor volume was examined every 3 days after 8 days of inoculation ( ) sh‐NC, (
) sh‐NC, ( ) sh‐circRNF111, (
) sh‐circRNF111, ( ) sh‐NC + PTX, (
) sh‐NC + PTX, ( ) sh‐circ‐RNF111 + PTX. (c) Tumor weight was measured after 29 days of inoculation. (d) Circ‐RNF111 level in the tumor samples collected from the mice was detected by qRT‐PCR assay. *P < 0.05.
) sh‐circ‐RNF111 + PTX. (c) Tumor weight was measured after 29 days of inoculation. (d) Circ‐RNF111 level in the tumor samples collected from the mice was detected by qRT‐PCR assay. *P < 0.05.
Circ‐RNF111 negatively modulates miR‐140‐5p expression by direct interaction in PTX‐resistant BC cells
In order to explore the potential mechanism of circ‐RNF111 in regulating PTX resistance of BC, starBase v2.0 software was employed to query the potential target of circ‐RNF111. The data showed that miR‐140‐5p contained the potential complementary sequences of circ‐RNF111 (Fig 4a). To verify this prediction, dual‐luciferase reporter assay and RIP assay were conducted. The results of dual‐luciferase reporter assay indicated that miR‐140‐5p transfection effectively suppressed the luciferase activity of WT‐circ‐RNF111 reporter vector in both MCF‐7/PTX and MDA‐MB‐231/PTX cells compared to that in miR‐NC transfected cells, but no obvious change was observed in MUT‐circ‐RNF111 groups (Fig 4b,c). RIP assay showed that miR‐140‐5p and circ‐RNF111 were all prominently elevated in Ago2 immunoprecipitates in MCF‐7/PTX and MDA‐MB‐231/PTX cells relative to IgG control groups (Fig 4d,e). As we expected, miR‐140‐5p was weakly expressed in PTX‐resistant BC tissues in reference to PTX‐sensitive BC tissues (Fig 4f). The level of miR‐140‐5p in MCF‐7 and MDA‐MB‐231 cells was lower than in MCF‐10A cells; moreover, miR‐140‐5p expression in MCF‐7/PTX and MDA‐MB‐231/PTX cells was lower than in MCF‐7 and MDA‐MB‐231 cells (Fig 4g). As analyzed by Spearman's correlation coefficient analysis, miR‐140‐5p expression was inversely correlated with circ‐RNF111 expression in BC tissues (Fig 4h). Subsequently, to further determine the regulatory relationship between circ‐RNF111 and miR‐140‐5p, MCF‐7/PTX and MDA‐MB‐231/PTX cells were transfected with si‐circ‐RNF111, circ‐RNF111 or corresponding controls. The data of qRT‐PCR assay exhibited that si‐circ‐RNF111 transfection markedly decreased the level of circ‐RNF111 and increased the level of miR‐140‐5p in both MCF‐7/PTX and MDA‐MB‐231/PTX cells, while circ‐RNF111 transfection showed the opposite results (Fig 4i,j). All these results illustrated that circ‐RNF111 directly targeted miR‐140‐5p and negatively regulated miR‐140‐5p expression in PTX‐resistant BC cells.
Figure 4.
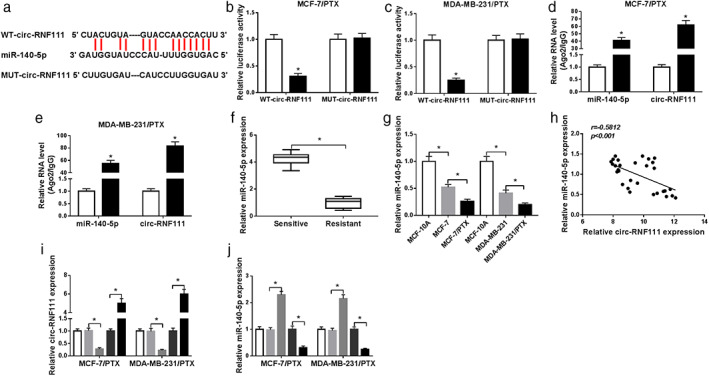
Circ‐RNF111 negatively regulates miR‐140‐5p expression by directly targeting. (a) The potential binding sites between circ‐RNF111 and miR‐140‐5p were predicted by starBase v2.0. (b,c) The luciferase activity in MCF‐7/PTX and MDA‐MB‐231/PTX cells co‐transfected with WT‐circ‐RNF111/MUT‐circ‐RNF111 and miR‐140‐5p/miR‐NC was examined by dual‐luciferase reporter assay. (d,e) The levels of miR‐140‐5p and circ‐RNF111 in IgG or Ago2 immunoprecipitates were measured by qRT‐PCR. (f) MiR‐140‐5p expression in PTX‐resistant BC tissues and PTX‐sensitive BC tissues was determined by qRT‐PCR. (g) MiR‐140‐5p expression in MCF‐10A, MCF‐7, MDA‐MB‐231, MCF‐7/PTX and MDA‐MB‐231/PTX cells was determined by qRT‐PCR. (h) The correlation between circ‐RNF111 and miR‐140‐5p in BC tissues was analyzed by Spearman's correlation coefficient analysis. (i,j) The levels of circ‐RNF111 and miR‐140‐5p in MCF‐7/PTX and MDA‐MB‐231/PTX cells transfected with si‐NC, si‐circ‐RNF111, pcDNA or circ‐RNF111 were measured by qRT‐PCR assay. *P < 0.05. ( ) miR‐NC, (
) miR‐NC, ( ) miR‐140‐5p; (
) miR‐140‐5p; ( ) IgG, (
) IgG, ( ) Ago2; (
) Ago2; ( ) Control, (
) Control, ( ) si‐NC, (
) si‐NC, ( ) si‐circ‐RNF111, (
) si‐circ‐RNF111, ( ) pcDNA, (
) pcDNA, ( ) circ‐RNF111
) circ‐RNF111
Inhibitory effects of circ‐RNF111 knockdown on PTX resistance, cell viability, colony formation, invasion and glycolysis ameliorated by miR‐140‐5p inhibition in PTX‐resistant BC cells
Based on the above experimental results, we speculated if circ‐RNF111 could regulate PTX resistance of BC by targeting miR‐140‐5p. Thus, we assigned MCF‐7/PTX and MDA‐MB‐231/PTX cells into five groups: control, si‐NC, si‐circ‐RNF111, si‐circ‐RNF111 + anti‐miR‐NC and si‐circ‐RNF111 + anti‐miR‐140‐5p. Transfection efficiency was tested by qRT‐PCR and the data showed that the upregulation of miR‐140‐5p caused by circ‐RNF111 knockdown was reversed by anti‐miR‐140‐5p transfection in both MCF‐7/PTX and MDA‐MB‐231/PTX cells (Fig 5a). As illustrated by CCK‐8 assay, colony formation assay and transwell assay, the suppressive roles of circ‐RNF111 silencing in PTX resistance, cell viability, colony formation ability and cell invasion were effectively weakened following the deletion of miR‐140‐5p in both MCF‐7/PTX and MDA‐MB‐231/PTX cells (Fig 5b–e). In addition, the inhibitory impacts on glucose consumption and lactate production mediated by circ‐RNF111 knockdown were partly overturned by miR‐140‐5p inhibition in MCF‐7/PTX and MDA‐MB‐231/PTX cells (Fig 5f,g). Furthermore, anti‐miR‐140‐5p was transfected into MCF‐7/PTX and MDA‐MB‐231/PTX cells to downregulate miR‐140‐5p expression (Fig 5h). CCK‐8 assay displayed that miR‐140‐5p downregulation apparently elevated IC50 value of PTX, while the administration of 2‐DG reversed the impact (Fig 5i). Taken together, circ‐RNF111 knockdown repressed PTX resistance via interacting with miR‐140‐5p in PTX‐resistant BC cells.
Figure 5.
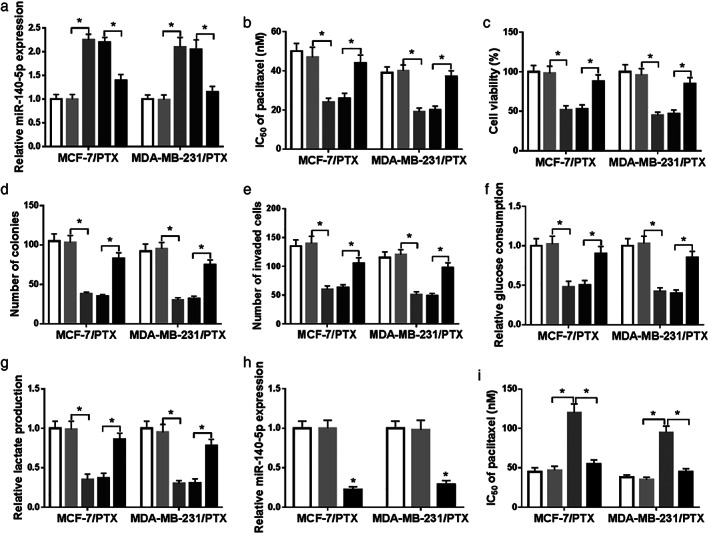
Circ‐RNF111 contributes to PTX resistance by targeting miR‐140‐5p. (a–f) MCF‐7/PTX and MDA‐MB‐231/PTX cells were divided into five groups: control, si‐NC, si‐circ‐RNF111, si‐circ‐RNF111 + anti‐miR‐NC and si‐circ‐RNF111 + anti‐miR‐140‐5p. (a) The expression of miR‐140‐5p in MCF‐7/PTX and MDA‐MB‐231/PTX cells was determined by qRT‐PCR. (b–e) IC50 of PTX, cell viability, colony numbers and cell invasion in MCF‐7/PTX and MDA‐MB‐231/PTX cells were evaluated by CCK‐8, colony formation and transwell assays, respectively. (f,g) Glucose uptake and lactate production in MCF‐7/PTX and MDA‐MB‐231/PTX cells were analyzed by specific kits. (h) The expression of miR‐140‐5p in anti‐miR‐NC or anti‐miR‐140‐5p transfected MCF‐7/PTX and MDA‐MB‐231/PTX cells was measured by qRT‐PCR. (i) MCF‐7/PTX and MDA‐MB‐231/PTX cells were untreated or treated with anti‐miR‐NC, anti‐miR‐140‐5p or anti‐miR‐140‐5p + 2‐DG and then IC50 of PTX was estimated by CCK‐8 assay. *P < 0.05. ( ) control, (
) control, ( ) si‐NC, (
) si‐NC, ( ) si‐circ‐RNF111, (
) si‐circ‐RNF111, ( ) si‐circ‐RNF111 + anti‐miR‐NC, (
) si‐circ‐RNF111 + anti‐miR‐NC, ( ) si‐circ‐RNF111 + anti‐miR‐140‐5p; (
) si‐circ‐RNF111 + anti‐miR‐140‐5p; ( ) Control, (
) Control, ( ) anti‐miR‐NC, (
) anti‐miR‐NC, ( ) anti‐miR‐140‐5p; (
) anti‐miR‐140‐5p; ( ) Control, (
) Control, ( ) anti‐miR‐NC, (
) anti‐miR‐NC, ( ) anti‐miR‐140‐5p, (
) anti‐miR‐140‐5p, ( ) anti‐miR‐140‐5p + 2‐DG
) anti‐miR‐140‐5p + 2‐DG
E2F3 is a direct target gene of miR‐140‐5p
Through further searching online in starBase v2.0, we discovered that E2F3 might be a target gene of miR‐140‐5p (Fig 6a). Dual‐luciferase reporter assay indicated that cotransfection of miR‐140‐5p and E2F3 3′UTR‐WT led to a distinct reduction in the luciferase activity in MCF‐7/PTX and MDA‐MB‐231/PTX cells compared to miR‐NC and E2F3 3′UTR‐WT cotransfected groups, whereas the luciferase activity was not changed in E2F3 3′UTR‐MUT groups (Fig 6b,c). RIP assay suggested that miR‐140‐5p and E2F3 were all enriched in Ago2‐containing beads in MCF‐7/PTX and MDA‐MB‐231/PTX cells compared to IgG control groups, further confirming the interaction between miR‐140‐5p and E2F3 (Fig 6d,e). We also determined the mRNA and protein levels of E2F3 in PTX‐resistant and PTX‐sensitive BC tissues. As we observed in Fig 6f,g, E2F3 mRNA and protein levels were raised in PTX‐resistant BC tissues relative to PTX‐sensitive BC tissues. Moreover, we found that E2F3 protein level in MCF‐7 and MDA‐MB‐231 cells was higher than in MCF‐10A cells and lower than in MCF‐7/PTX and MDA‐MB‐231/PTX cells (Fig 6h). Spearman's correlation coefficient analysis indicated that there was an inverse correlation between miR‐140‐5p and E2F3 mRNA in BC tissues (Fig 6i). These findings demonstrated that E2F3 was a target of miR‐140‐5p.
Figure 6.
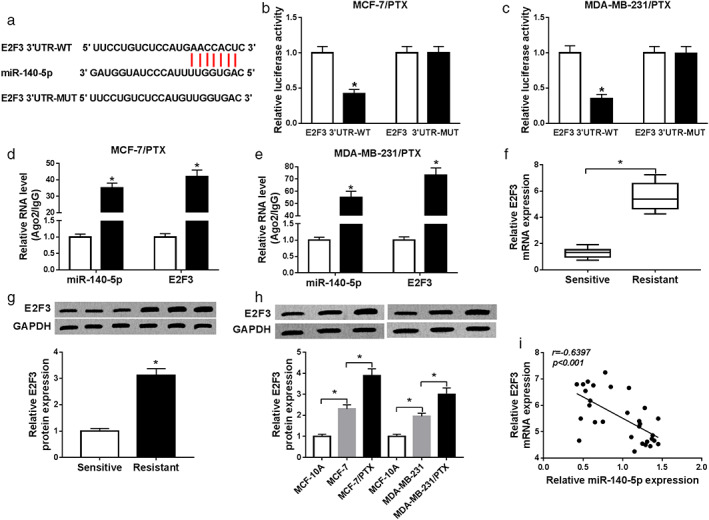
MiR‐140‐5p directly interacts with E2F3. (a) The predicted binding sites between miR‐140‐5p and E2F3 were presented. (b,c) The interaction between miR‐140‐5p and E2F3 was verified by dual‐luciferase reporter assay. (d,e) The enrichment of miR‐140‐5p and E2F3 in the IgG or Ago2 immunoprecipitates was determined by qRT‐PCR after RIP assay. (f,g) The mRNA and protein levels of E2F3 in PTX‐resistant and PTX‐sensitive BC tissues were examined by qRT‐PCR assay and western blot assay, respectively. (h) The protein level of E2F3 in MCF‐10A, MCF‐7, MDA‐MB‐231, MCF‐7/PTX and MDA‐MB‐231/PTX cells was measured by western blot assay. (i) The correlation between miR‐140‐5p and E2F3 mRNA in BC tissues was analyzed by Spearman's correlation coefficient analysis. *P < 0.05. ( ) miR‐NC, (
) miR‐NC, ( ) miR‐140‐5p; (
) miR‐140‐5p; ( ) IgG, (
) IgG, ( ) Ago2
) Ago2
MiR‐140‐5p overexpression restrains PTX resistance, cell viability, colony formation, cell invasion and glycolysis in PTX‐resistant BC cells by binding to E2F3
Subsequently, we transfected miR‐NC, miR‐140‐5p, miR‐140‐5p + pcDNA or miR‐140‐5p + E2F3 into MCF‐7/PTX and MDA‐MB‐231/PTX cells to investigate the association between miR‐140‐5p and E2F3 in regulating PTX resistance. As shown in Fig 7a,b, miR‐140‐5p transfection evidently decreased the protein level of E2F3 in MCF‐7/PTX and MDA‐MB‐231/PTX cells, while the influence was ameliorated by E2F3 transfection. CCK‐8 assay, colony formation and transwell assay indicated that PTX resistance, cell viability, colony numbers and cell invasion were all impeded by miR‐140‐5p overexpression in MCF‐7/PTX and MDA‐MB‐231/PTX cells, while the elevation of E2F3 partly overturned the impacts (Fig 7c–f). Moreover, miR‐140‐5p overexpression apparently reduced the levels of glucose uptake and lactate production in MCF‐7/PTX and MDA‐MB‐231/PTX cells, while E2F3 upregulation abolished the effects (Fig 7g,h). Additionally, the effect of glycolysis on E2F3‐induced PTX resistance in MCF‐7/PTX and MDA‐MB‐231/PTX cells was determined. The data showed that 2‐DG treatment rescued the promotional role of E2F3 overexpression in PTX resistance (Fig 7i). All these data implicated that E2F3 overexpression reversed the inhibitory effect of miR‐140‐5p on PTX resistance in PTX‐resistant BC cells.
Figure 7.
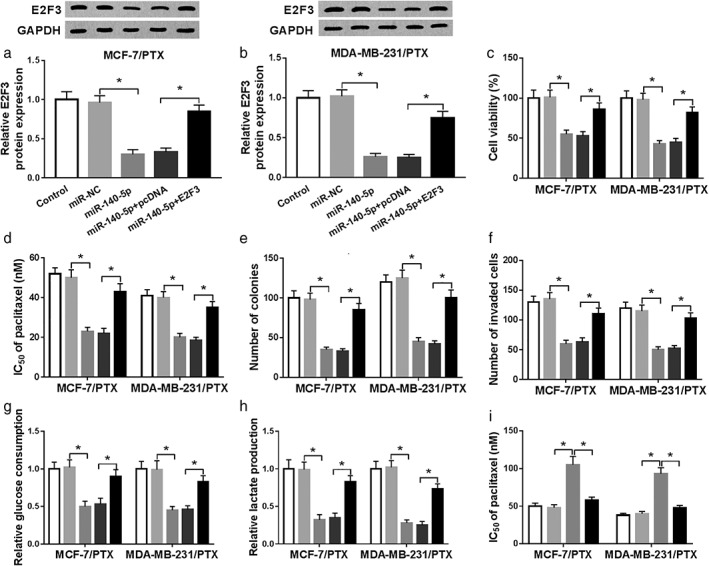
MiR‐140‐5p enhances PTX sensitivity via interacting with E2F3. (a–g) MCF‐7/PTX and MDA‐MB‐231/PTX cells were divided into five groups: control, miR‐NC, miR‐140‐5p, miR‐140‐5p + pcDNA and miR‐140‐5p + E2F3. (a,b) The protein level of E2F3 in MCF‐7/PTX and MDA‐MB‐231/PTX cells was examined by western blot assay. (c–f) PTX resistance, cell viability, colony numbers and cell invasion in MCF‐7/PTX and MDA‐MB‐231/PTX cells were assessed by CCK‐8, colony formation and transwell assays, respectively. (g,h) The levels of glucose consumption and lactate production were examined by relevant kits. (i) MCF‐7/PTX and MDA‐MB‐231/PTX cells were untreated or treated with pcDNA, E2F3 or E2F3 + 2‐DG and then IC50 of PTX was evaluated by CCK‐8 assay. *P < 0.05. ( ) control, (
) control, ( ) miR‐NC, (
) miR‐NC, ( ) miR‐140‐5p, (
) miR‐140‐5p, ( ) miR‐140‐5p + pcDNA, (
) miR‐140‐5p + pcDNA, ( ) miR‐140‐5p + E2F3; (
) miR‐140‐5p + E2F3; ( ) Control, (
) Control, ( ) pcDNA, (
) pcDNA, ( ) E2F3, (
) E2F3, ( ) E2F3 + 2‐DG
) E2F3 + 2‐DG
Circ‐RNF111 silencing downregulates E2F3 expression by sponging miR‐140‐5p
To further investigate the regulatory relationship among circ‐RNF111, miR‐140‐5p and E2F3, MCF‐7/PTX and MDA‐MB‐231/PTX were transfected with si‐NC, si‐circ‐RNF111, si‐circ‐RNF111 + anti‐miR‐NC or si‐circ‐RNF111 + anti‐miR‐140‐5p. The analysis of E2F3 protein level showed that circ‐RNF111 knockdown caused a distinct reduction in E2F3 protein level in MCF‐7/PTX and MDA‐MB‐231/PTX cells, while miR‐140‐5p inhibition restored the reduction (Fig 8a,b). These data demonstrated that circ‐RNF111 promoted E2F3 expression through targeting miR‐140‐5p in PTX‐resistant BC cells.
Figure 8.
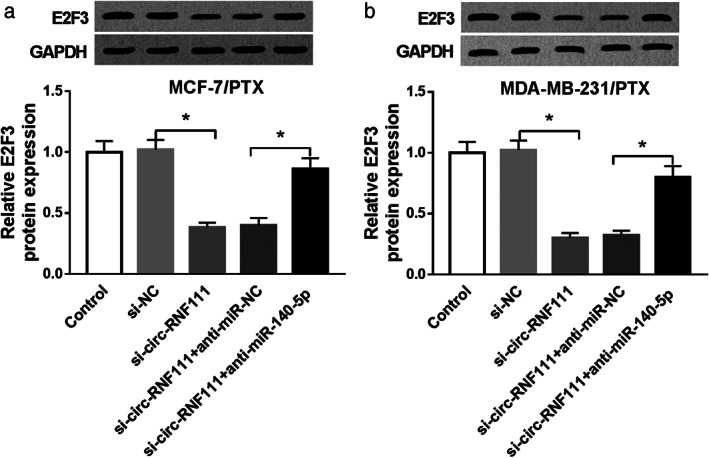
Circ‐RNF111 knockdown suppresses E2F3 expression through miR‐140‐5p. (a,b) MCF‐7/PTX and MDA‐MB‐231/PTX were assigned to control, si‐NC, si‐circ‐RNF111, si‐circ‐RNF111 + anti‐miR‐NC and si‐circ‐RNF111 + anti‐miR‐140‐5p groups, and then the protein level of E2F3 was measured by western blot assay. *P < 0.05.
Discussion
Mounting evidence has demonstrated that chemoresistance is produced in BC cells and chemoresistance is a major obstacle for BC treatment. 23 CircRNAs have been demonstrated to be involved in chemoresistance in cancers; however, their effects on drug resistance in BC are poorly studied. Herein, we observed that circ‐RNF111 was abnormally increased in PTX‐resistant BC tissues and cells. Through function and mechanism analysis, circ‐RNF111 was found to facilitate the resistance of PTX‐resistant BC cells to PTX via regulating miR‐140‐5p/E2F3 axis.
Some circRNAs has been proven to be dysregulated and take part in regulating drug resistance in BC. For instance, Sang et al. disclosed that circ_0025202 was weakly expressed in BC, and repressed cell growth, colony formation and motility and facilitated apoptosis and tamoxifen sensitivity via targeting miR‐182‐5p. 10 Gao et al. implicated that circ_0006528 level was raised and contributed to adriamycin resistance by sponging miR‐7‐5p in adriamycin‐resistant BC cells. 11 Liang et al. declared that circKDM4C was reduced in BC and hampered BC growth, metastasis and doxorubicin resistance through binding to miR‐548p. 12 These findings illustrate that circRNAs plays dual roles in tumor growth and chemoresistance in BC. Herein, we explored the effect of circ‐RNF111 on the resistance of BC to PTX. Our data indicated that circ‐RNF111 was notably increased in PTX‐resistant BC tissues and cells. Functionally, circ‐RNF111 deficiency sensitized PTX‐resistant BC cells to PTX and impeded cell viability, colony formation and invasion. To meet the energy and biomolecules needed for tumor cell growth, tumor cells tend to display deranged metabolism. 24 Accumulating evidence has addressed that increased glycolysis contributed to drug resistance in BC. 25 , 26 Therefore, we explored whether circ‐RNF111 altered the glycolysis process. We found that circ‐RNF111 knockdown resulted in a marked reduction in glucose consumption and lactate production in PTX‐resistant BC cells, indicating glycolysis was repressed. Furthermore, 2‐DG treatment hampered glycolysis and thereby restored circ‐RNF111‐induced PTX resistance in PTX‐resistant BC cells. In addition, circ‐RNF111 silencing blocked tumor growth and improved PTX sensitivity in BC in vivo. All these results demonstrated that circ‐RNF111 promoted the resistance of BC to PTX.
CircRNAs can serve as miRNA sponges to alter their functions. 27 In this work, circRNF111 was verified as a sponge of miR‐140‐5p and negatively modulated miR‐140‐5p expression. Yu et al. reported that miR‐140‐5p participated in regulating cell growth and 5‐Fluorouracil resistance in GC by recognizing NDRG3. 19 Lu et al. demonstrated that miR‐140‐5p was reduced and its elevation restrained autography and drug resistance in melphalan‐resistant multiple myeloma cells. 28 Meng et al. suggested that miR‐140‐5p bound to HMGN5 to inhibit drug resistance of osteosarcoma. 17 Moreover, miR‐140‐5p was low expressed and functioned as a suppressor of tumor progression and glycolysis in BC. 29 , 30 Herein, low expression of miR‐140‐5p in PTX‐resistant BC was observed. The elevation of miR‐140‐5p enhanced PTX sensitivity and suppressed cell viability, colony formation, cell invasion and glycolysis in PTX‐resistant BC cells. The deletion of miR‐140‐5p partly overturned the impact of circ‐RNF111 deficiency on PTX resistance in PTX‐resistant BC cells, indicating that circ‐RNF111 could regulate PTX resistance by targeting miR‐140‐5p.
A growing body of evidence has revealed that E2F3 is involved in drug resistance of cancers via being targeted by some miRNAs, such as miR‐210‐3p, 31 miR‐433 32 and miR‐203. 21 In the research, we found that E2F3 was a target of miR‐140‐5p and was elevated in PTX‐resistant BC. E2F3 overexpression restored the inhibitory impact of miR‐140‐5p on PTX resistance, cell viability, colony formation, cell invasion and glycolysis in PTX‐resistant BC cells.
In summary, our study unraveled that high expression of circ‐RNF111 contributed to PTX resistance, cell growth, invasion and glycolysis in PTX‐resistant BC cells. Moreover, a novel regulatory network circ‐RNF111/miR‐140‐5p/E2F3 axis was discovered, which might provide a promising therapeutic strategy for PTX resistance in BC patients.
Disclosure
The authors declare that they have no financial conflicts of interest.
Acknowledgment
None.
References
- 1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019; 69 (1): 7–34. [DOI] [PubMed] [Google Scholar]
- 2. Sutter SA, Slinker A, Balumuka DD, Mitchell KB. Surgical management of breast cancer in Africa: A continent‐wide review of intervention practices, barriers to care, and adjuvant therapy. J Glob Oncol 2017; 3 (2): 162–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Caetano‐Pinto P, Jansen J, Assaraf YG, Masereeuw R. The importance of breast cancer resistance protein to the kidneys excretory function and chemotherapeutic resistance. Drug Resist Updat 2017; 30: 15–27. [DOI] [PubMed] [Google Scholar]
- 4. Broxterman HJ, Gotink KJ, Verheul HM. Understanding the causes of multidrug resistance in cancer: A comparison of doxorubicin and sunitinib. Drug Resist Updat 2009; 12 (4–5): 114–26. [DOI] [PubMed] [Google Scholar]
- 5. Akiyama M, Sowa Y, Taniguchi T e a. Three combined treatments, a novel HDAC inhibitor OBP‐801/YM753, 5‐fluorouracil, and paclitaxel, induce G(2) phase arrest through the p38 pathway in human ovarian cancer cells. Oncol Res 2017; 25 (8): 1245–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hentze MW, Preiss T. Circular RNAs: splicing's enigma variations. EMBO J 2013; 32 (7): 923–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhong Y, Du Y, Yang X et al Circular RNAs function as ceRNAs to regulate and control human cancer progression. Mol Cancer 2018; 17 (1): 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Huang XX, Zhang Q, Hu H e a. A novel circular RNA circFN1 enhances cisplatin resistance in gastric cancer via sponging miR‐182‐5p. J Cell Biochem 2020. 10.1002/jcb.29641. [DOI] [PubMed] [Google Scholar]
- 9. Liu F, Zhang J, Qin L e a. Circular RNA EIF6 (Hsa_circ_0060060) sponges miR‐144‐3p to promote the cisplatin‐resistance of human thyroid carcinoma cells by autophagy regulation. Aging 2018; 10 (12): 3806–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sang Y, Chen B, Song X e a. circRNA_0025202 regulates tamoxifen sensitivity and tumor progression via regulating the miR‐182‐5p/FOXO3a axis in breast cancer. Mol Ther 2019; 27 (9): 1638–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gao D, Zhang X, Liu B e a. Screening circular RNA related to chemotherapeutic resistance in breast cancer. Epigenomics 2017; 9 (9): 1175–88. [DOI] [PubMed] [Google Scholar]
- 12. Liang Y, Song X, Li Y e a. circKDM4C suppresses tumor progression and attenuates doxorubicin resistance by regulating miR‐548p/PBLD axis in breast cancer. Oncogene 2019; 38 (42): 6850–66. [DOI] [PubMed] [Google Scholar]
- 13. Tang YY, Zhao P, Zou TN e a. Circular RNA hsa_circ_0001982 promotes breast cancer cell carcinogenesis through decreasing miR‐143. DNA Cell Biol 2017; 36 (11): 901–8. [DOI] [PubMed] [Google Scholar]
- 14. He L, Hannon GJ. MicroRNAs: Small RNAs with a big role in gene regulation. Nat Rev Genet 2004; 5 (7): 522–31. [DOI] [PubMed] [Google Scholar]
- 15. Bai WD, Ye XM, Zhang MY e a. MiR‐200c suppresses TGF‐beta signaling and counteracts trastuzumab resistance and metastasis by targeting ZNF217 and ZEB1 in breast cancer. Int J Cancer 2014; 135 (6): 1356–68. [DOI] [PubMed] [Google Scholar]
- 16. Ma MT, He M, Wang Y e a. MiR‐487a resensitizes mitoxantrone (MX)‐resistant breast cancer cells (MCF‐7/MX) to MX by targeting breast cancer resistance protein (BCRP/ABCG2). Cancer Lett 2013; 339 (1): 107–15. [DOI] [PubMed] [Google Scholar]
- 17. Meng Y, Gao R, Ma J e a. MicroRNA‐140‐5p regulates osteosarcoma chemoresistance by targeting HMGN5 and autophagy. Sci Rep 2017; 7 (1): 416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ye J, Zhang R, Du X et al Long noncoding RNA SNHG16 induces sorafenib resistance in hepatocellular carcinoma cells through sponging miR‐140‐5p. Onco Targets Ther 2019; 12: 415–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yu J, Shen J, Qiao X e a. SNHG20/miR‐140‐5p/NDRG3 axis contributes to 5‐fluorouracil resistance in gastric cancer. Oncol Lett 2019; 18 (2): 1337–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. van den Heuvel S, Dyson NJ. Conserved functions of the pRB and E2F families. Nat Rev Mol Cell Biol 2008; 9 (9): 713–24. [DOI] [PubMed] [Google Scholar]
- 21. Tang G, Wu J, Xiao G et al MiR‐203 sensitizes glioma cells to temozolomide and inhibits glioma cell invasion by targeting E2F3. Mol Med Rep 2015; 11 (4): 2838–44. [DOI] [PubMed] [Google Scholar]
- 22. Feng B, Wang R, Song HZ, Chen LB. MicroRNA‐200b reverses chemoresistance of docetaxel‐resistant human lung adenocarcinoma cells by targeting E2F3. Cancer 2012; 118 (13): 3365–76. [DOI] [PubMed] [Google Scholar]
- 23. Cherdyntseva NV, Litviakov NV, Denisov EV, Gervas PA, Cherdyntsev ES. Circulating tumor cells in breast cancer: Functional heterogeneity, pathogenetic and clinical aspects. Exp Oncol 2017; 39 (1): 2–11. [PubMed] [Google Scholar]
- 24. DeBerardinis RJ, Lum JJ, Hatzivassiliou G et al The biology of cancer: Metabolic reprogramming fuels cell growth and proliferation. Cell Metab 2008; 7 (1): 11–20. [DOI] [PubMed] [Google Scholar]
- 25. La Ferla M, Lessi F, Aretini P et al ANKRD44 gene silencing: A putative role in Trastuzumab resistance in Her2‐like breast cancer. Front Oncol 2019; 9: 547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhao Y, Liu H, Liu Z e a. Overcoming trastuzumab resistance in breast cancer by targeting dysregulated glucose metabolism. Cancer Res 2011; 71 (13): 4585–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kulcheski FR, Christoff AP, Margis R. Circular RNAs are miRNA sponges and can be used as a new class of biomarker. J Biotechnol 2016; 238: 42–51. [DOI] [PubMed] [Google Scholar]
- 28. Lu D, Yang C, Zhang Z, Cong Y, Xiao M. Knockdown of Linc00515 inhibits multiple myeloma autophagy and chemoresistance by upregulating miR‐140‐5p and downregulating ATG14. Cell Physiol Biochem 2018; 48 (6): 2517–27. [DOI] [PubMed] [Google Scholar]
- 29. He Y, Deng F, Zhao S e a. Analysis of miRNA‐mRNA network reveals miR‐140‐5p as a suppressor of breast cancer glycolysis via targeting GLUT1. Epigenomics 2019; 11 (9): 1021–36. [DOI] [PubMed] [Google Scholar]
- 30. Lu Y, Qin T, Li J e a. MicroRNA‐140‐5p inhibits invasion and angiogenesis through targeting VEGF‐A in breast cancer. Cancer Gene Ther 2017; 24 (9): 386–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jin Y, Wei J, Xu S, Guan F, Yin L, Zhu H. miR2103p regulates cell growth and affects cisplatin sensitivity in human ovarian cancer cells via targeting E2F3. Mol Med Rep 2019; 19 (6): 4946–54. [DOI] [PubMed] [Google Scholar]
- 32. Weiner‐Gorzel K, Dempsey E, Milewska M e a. Overexpression of the microRNA miR‐433 promotes resistance to paclitaxel through the induction of cellular senescence in ovarian cancer cells. Cancer Med 2015; 4 (5): 745–58. [DOI] [PMC free article] [PubMed] [Google Scholar]


