Abstract
Background:
Diagnosis of Gleason 6 prostate cancer can leave uncertainty about the presence of undetected aggressive disease.
Objective:
To evaluate the utility of a four kallikrein (4K) panel in predicting the presence of high-grade cancer in men on active surveillance.
Design, setting, and participants:
Plasma collected before the first and subsequent surveillance biopsies was assessed for 718 men prospectively enrolled in the multi-institutional Canary PASS trial. Biopsy data were split 2:1 into training and test sets. We developed statistical models that included clinical information and either the 4Kpanel or serum prostate-specific antigen (PSA).
Outcome measurements and statistical analysis:
The endpoint was reclassification to Gleason ≥7. We used receiver operating characteristic (ROC) curve analyses and area under the curve (AUC) to assess discriminatory capacity, and decision curve analysis (DCA) to report clinical net benefit.
Results and limitations:
Significant predictors for reclassification were 4Kpanel (odds ratio [OR] 1.54, 95% confidence interval [CI] 1.31–1.81) or PSA (OR 2.11, 95% CI 1.53–2.91), ≥20% cores positive (OR 2.10, 95% CI 1.33–3.32), two or more prior negative biopsies (OR 0.19, 95% CI 0.04–0.85), prostate volume (OR 0.47, 95% CI 0.31–0.70), and body mass index (OR 1.09, 95% CI 1.04–1.14). ROC curve analysis comparing 4K and base models indicated that the 4Kpanel improved accuracy for predicting reclassification (AUC 0.78 vs 0.74) at the first surveillance biopsy. Both models performed comparably for prediction of reclassification at subsequent biopsies (AUC 0.75 vs 0.76). In DCA, both models showed higher net benefit compared to biopsy-all and biopsy-none strategies. Limitations include the single cohort nature of the study and the small numbers; results should be validated in another cohort before clinical use.
Conclusions:
The 4Kpanel provided incremental value over routine clinical information in predicting high-grade cancer in the first biopsy after diagnosis. The 4Kpanel did not add predictive value to the base model at subsequent surveillance biopsies.
Patient summary:
Active surveillance is a management strategy for many low-grade prostate cancers. Repeat biopsies monitor for previously undetected high-grade cancer. We show that a model with clinical variables, including a panel of four kallikreins, indicates the presence of high-grade cancer before a biopsy is performed.
Keywords: Prostatic neoplasms, Prospective studies, Active surveillance, Biomarker, Kallikrein
1. Introduction
Active surveillance is a management strategy for low-grade, localized prostate cancer that allows men to delay or be spared the potential morbidities of treatment. Cancers that appear to be low-risk at diagnosis are monitored, typically with serial prostate-specific antigen (PSA) measurements, clinical examinations, and repeat prostate biopsies. Intervention is recommended on evidence of a more aggressive tumor, usually based on changes in biopsy characteristics. However, fear of occult high-grade cancer, in part because of the known undersampling of systematic prostate biopsies, has tempered widespread adoption of active surveillance. Even with emerging magnetic resonance imaging (MRI)–based biopsy protocols, there remains uncertainty surrounding the presence of more aggressive disease against a background of apparently low-risk cancer. In addition, the optimal surveillance schedule and triggers for intervention have not been established, resulting in substantial variations in the practice of active surveillance. Prostate biopsy can be painful, anxiety-provoking, expensive, and potentially morbid, so avoiding unnecessary surveillance biopsies is attractive. Methods to reduce the number of biopsies in active surveillance regimens, while maximizing the identification of high-grade cancers that may benefit from treatment, would have substantial clinical utility.
A promising approach to determine active surveillance candidacy and surveillance regimens (eg, more intensive vs less intensive biopsy schedules) involves the addition of biomarker panels to prediction models based on known clinical and demographic variables [1]. Among men suspected of having prostate cancer, a panel of four kallikreins (total PSA [tPSA], free PSA [fPSA], intact PSA [iPSA], and human kallikrein 2 [hK2]) combined with age using a mathematical algorithm improves the prediction of high-grade cancers compared to the PCPT risk calculator or models using tPSA alone [2,3]. Here, we explore the utility of prediction models incorporating the predefined four kallikrein panel algorithm (4Kpanel) to predict the presence of occult high-grade disease in men already diagnosed with Gleason 6 cancer and on active surveillance. We use plasma specimens and data from the prospective, multi-institutional Canary Prostate Active Surveillance Study (PASS).
2. Patients and methods
2.1. Study cohort
This study included men from Canary PASS, a multicenter, prospective study enrolling men on active surveillance [4]. Participants in PASS consented to specimen collection as part of the PASS protocol (clinicaltrials.gov NCT00756665), which was approved by institutional review boards at participating sites. The PASS protocol includes monitoring at clinic visits every 6 mo, with the first ≥10-core prostate needle biopsy at 6–12 mo, the second at 24 mo after cancer diagnosis, and subsequent biopsies every 2 yr. Specimens, including EDTA plasma, were collected at study entry and every 6-mo clinic visit, and were stored at −70°C until use. In February 2015, 1170 participants were enrolled in PASS at nine sites throughout North America. Of these, 956 participants had an on-study biopsy, of whom 877 had Gleason 3 + 3 disease at study entry, 771 had not used 5α-reductase inhibitors, and EDTA plasma collected before biopsy was available for 753 men. Participants with missing prostate volume or ratio of positive to total biopsy cores were excluded from the modeling (n = 35); the remaining 718 men, who had 1111 biopsies, were included in this study.
2.2. Laboratory methods
Blood was collected in K2EDTA vacutainers, inverted, centrifuged at 1600 × g, and frozen at −70°C within 4 h of collection. Frozen plasma was stored until shipment on dry ice to OPKO Labs (Nashville, TN, USA) for analysis. The analysis laboratory was blinded to all specimen and clinical information. Specimens were thawed immediately before analysis. tPSA, fPSA, iPSA, and hK2 were measured [2].
2.3. Study design and analyses
The objective of the analyses was to determine whether a model using clinical predictors and kallikrein data collected after diagnosis of Gleason 6 cancer, but before surveillance biopsy, can predict high-grade cancer in the surveillance biopsy. Sequential surveillance biopsies were considered as two groups: (1) the initial biopsy after cancer diagnosis (sometimes called confirmatory biopsy) and (2) all subsequent surveillance biopsies. Biopsy data were split 2:1 into training and test sets matched by outcome.
The primary outcome was reclassification from Gleason score 6 to Gleason score ≥7. A value for the 4Kpanel was calculated with tPSA, fPSA, iPSA, hk2, and age using locked down coefficients developed before the study was conducted [3]. This combination of the four kallikreins is the same as in the commercial 4Kscore. However, the commercial 4Kscore is a model containing the 4Kpanel and clinical data available before cancer diagnosis, and is calibrated for a patient before diagnosis. Because we evaluated the kallikreins in a cohort already diagnosed with cancer, we developed a new model that included the 4Kpanel and clinical information available after a diagnosis of cancer, and calibrated to an active surveillance population. Additional clinical predictors considered in modeling included age, body mass index (BMI), race (African American or other), digital rectal examination (DRE) results, number of previous biopsies after diagnosis, number of negative biopsies after diagnosis, core ratio (ratio of biopsy cores containing cancer to total cores) from previous biopsy, maximum core ratio among all previous biopsies, months since diagnosis, and prostate volume (prostate size measured closest to the time of sampling and imputed within 2 yr). Either the 4Kpanel (logit scale) or clinical serum PSA (log-transformed) was used in models. Prediction models were built using data in the training set, and then clinical performance was assessed using the testing set. We followed the principles set forth by the US Food and Drug Administration critical path initiative, using an established biomarker with analytic validity for the intent of clinical validation in the intended use population [7]. Furthermore, we followed reporting recommendations for tumor marker prognostic studies (REMARK) [8] and the Tumor Marker Utility Grading System [9] in reporting the clinical utility of the biomarker panel.
2.3.1. Model building
Data from initial and subsequent biopsy groups were combined for model development. Interaction terms between biopsy group (initial vs subsequent surveillance biopsy) and other variables were evaluated to investigate whether effects may differ for an initial biopsy and a subsequent biopsy. Logistic regression was used to fit the models, with robust variance to account for the correlation among multiple biopsies on the same patient. Forward stepwise model selection procedures were implemented. Variable selection criteria included p < 0.15, area under the receiver operating characteristic(ROC) curve(AUC) ≥0.005, or quasi-likelihood under the independence model criterion (QIC) with threshold of zero [5]. Final models were compared to identify variables that were robust to selection procedures. We first identified a full model including clinical predictors and 4Kpanel, and then a base model with serum PSA substituted for the 4Kpanel. In some clinics, prostate volume may not be reliably available, so models without prostate volume were fitted sequentially.
2.3.2. Model validation
Calibration plots were used to gauge the goodness of fit of each model. We used ROC analyses and AUC to assess the discriminatory capacity of a model for separating patients with and without reclassification. Decision curve analysis (DCA) was used to report the clinical net benefit of each model compared to biopsy-all and biopsy-none strategies [6]. The potential clinical impact was illustrated by plotting the number of cancers missed versus the number of biopsies avoided per 1000 individuals. To illustrate the clinical consequence of each model, we report the number of biopsies that could be avoided and the number of Gleason ≥7 cancers that might be missed if a risk-based threshold is applied as a criterion for biopsy. All evaluations were conducted on the initial biopsy and subsequent biopsy groups separately and combined. Confidence intervals (CIs) and significance tests were calculated using the bootstrap resampling procedure to account for within-subject correlations. All analyses were conducted using R version 3.1.1 (www.r-project.org).
3. Results
Of the 718 men in this study, there were 478 participants in the initial biopsy group for whom kallikreins were assayed: 319 in the training set (60 [18.8%] with Gleason ≥7) and 159 in the test set (34 [21.4%] with Gleason ≥7; Table 1). In bivariate analyses, prostate volume, ratio of positive to total cores, and the 4Kpanel were significantly associated with grade reclassification. There were 444 participants (of whom 204 were also in the initial biopsy group) with 633 subsequent surveillance biopsies, 422 in the training set (70 [17%] with Gleason ≥7; Table 2) and 211 in the test set (31 [15%] with Gleason ≥7; Supplementary Table 1). Biopsies in this group ranged from the second to eighth after diagnosis, and most patients had Gleason score 6 or no cancer at their surveillance biopsies, varying slightly across biopsy number.
Table 1 –
Characteristics for 478 participants with kallikreins assayed before the initial surveillance biopsy after diagnosis for combined Gleason score <7 versus ≥7 for the training and test cohorts
| Characteristics | Training set | Test set | ||||
|---|---|---|---|---|---|---|
| Gleason <7 | Gleason ≥7 | p value | Gleason <7 | Gleason ≥7 | p value | |
| Sample size (n) | 259 | 60 | 125 | 34 | ||
| Age at diagnosis (yr) | 63 (58–67) | 64 (60–68) | 0.109 | 64 (58–68) | 64 (57–67) | 0.876 |
| Body mass index (kg/m2) | 27 (25–30) | 28 (25–33) | 0.116 | 27 (25–29) | 28 (26–31) | 0.305 |
| Race | ||||||
| Non-African American | 248 (96) | 56 (93) | 121 (97) | 29 (85) | ||
| African American | 11 (4) | 4 (7) | 0.646 | 4 (3) | 5 (15) | 0.522 |
| Time from diagnosis (mo) | 12.0 (8.4–14.1) | 12.7 (8.6–14.8) | 0.237 | 12.2 (8.8–14.0) | 12.6 (10.3–17.6) | 0.189 |
| Digital rectal examination | ||||||
| Normal | 238 (92) | 55 (92) | 118 (94) | 30 (88) | ||
| Abnormal | 21 (8) | 5 (8) | 0.971 | 7 (6) | 4 (12) | 0.031 |
| Prostate volume (cm3) | 41.0 (30.0–56.5) | 35.5 (25.0–50.0) | 0.041 | 40.0 (30.0–51.0) | 30.0 (24.0–42.8) | 0.006 |
| Positive:total core ratio | 0.08 (0.08–0.17) | 0.17 (0.08–0.20) | <0.001 | 0.08 (0.08–0.17) | 0.17 (0.17–0.25) | <0.001 |
| Clinical serum PSA (ng/ml) | 4.60 (2.91–6.40) | 4.81 (4.35–6.42) | 0.108 | 4.56 (3.11–6.24) | 5.65 (4.58–7.88) | 0.024 |
| 4Kpanel (logit) | 0.21 (0.08–0.29) | 0.32 (0.16–0.44) | <0.001 | 0.20 (0.07–0.28) | 0.36 (0.18–0.53) | <0.001 |
PSA = prostate-specific antigen.
Data are presented as median (interquartile range) for continuous variables and as n (%) for categorical variables.
Table 2 –
Biopsy characteristics at each sequential surveillance biopsy after diagnosis for 558 participants in the training set
| Parameter | Initial biopsy First | Subsequent surveillance biopsies | ||||||
|---|---|---|---|---|---|---|---|---|
| Second | Third | Fourth | Fifth | Sixth | Seventh | Eighth | ||
| Biopsies (n) | 319 | 246 | 108 | 34 | 20 | 10 | 3 | 1 |
| CR for previous biopsya | ||||||||
| Median (IQR) | 0.08 (0.08) | 0.07 (0.17) | 0.08 (0.17) | 0.06 (0.12) | 0.06 (0.12) | 0 (0.07) | 0.11 (0.06) | 0 (0) |
| Missing, n (%) | 0 | 5 (2) | 5 (5) | 0 | 0 | 0 | 0 | 0 |
| Median MCRb (IQR) | 0.08 (0.08) | 0.11 (0.08) | 0.13 (0.15) | 0.17 (0.13) | 0.10 (0.17) | 0.14 (0.15) | 0.17 (0.08) | 0.17 (0.00) |
| Negative biopsiesc, n (%) | ||||||||
| 0 | 319 (100) | 145 (59) | 44 (41) | 10 (29) | 4 (20) | 1 (10) | 1 (33) | 0 |
| 1 | 0 | 101 (41) | 38 (35) | 13 (38) | 6 (30) | 3 (30) | 2 (67) | 0 |
| 2 | 0 | 0 | 26 (24) | 6 (18) | 3 (15) | 1 (10) | 0 | 1 (100) |
| 3 | 0 | 0 | 0 | 5 (15) | 2 (10) | 3 (30) | 0 | 0 |
| 4 | 0 | 0 | 0 | 0 | 5 (25) | 2 (20) | 0 | 0 |
| Median PV, cm3 (IQR) | 41.0 (26.5) | 38.0 (27.0) | 41.0 (27.0) | 48.5 (25.0) | 59.5 (36.5) | 43.5 (27.8) | 41.0 (19.5) | 97.0 (0.0) |
| Biopsy GS, n (%) | ||||||||
| Negative | 107 (34) | 95 (39) | 38 (35) | 11 (32) | 8 (40) | 6 (60) | 2 (67) | 0 |
| 6 | 152 (48) | 108 (44) | 48 (45) | 21 (62) | 10 (50) | 3 (30) | 1 (33) | 1 (100) |
| 7 | 58 (18) | 42 (17) | 21 (19) | 2 (6) | 2 (10) | 1 (10) | 0 | 0 |
| 8 | 1 (0) | 1 (0) | 1 (1) | 0 | 0 | 0 | 0 | 0 |
| 9 | 1 (0) | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
CR = core ratio; IQR = interquartile range; MCR = maximum CR; PV = prostate volume; GS = Gleason score.
CR is defined as the number of biopsy cores containing cancer divided by the total number of biopsy cores in the previous biopsy.
MCR among all previous biopsies.
Number of surveillance biopsies in which no cancer was found.
In the full clinical model (Table 3) including the 4Kpanel, significant predictors for reclassification were BMI (odds ratio [OR] 1.09, 95% CI 1.04–1.14], >20% of cores positive in the prior biopsy (OR 2.10, 95% CI 1.33–3.32), a history of two or more biopsies negative for cancer (OR 0.19, 95% CI 0.04–0.85), prostate volume (per fold increase, OR 0.47, 95% CI 0.31–0.70), and 4Kpanel (OR 1.5, 95% CI 1.31–1.81). In the clinical model with serum PSA replacing the 4Kpanel, PSA was significantly associated with reclassification (per fold increase, OR 2.11, 95% CI 1.53–2.91) and age was not. In models that did not include prostate volume, the effects were similar for covariates left in the model (Supplementary Table 2). Model calibration in the test set showed predicted probabilities of reclassification closely matching the empirical rates (Supplementary Fig. 1).
Table 3 –
Summary of fitted models including clinical variables + serum PSA or 4Kpanel in the training set
| Variable | PSA + full clinical model | 4K + full clinical model | ||
|---|---|---|---|---|
| OR (95% CI) | p value | OR (95% CI) | p value | |
| Age | 1.03 (1.00–1.06) | 0.068 | ||
| Body mass index | 1.11 (1.06–1.16) | <0.001 | 1.09 (1.04–1.14) | <0.001 |
| Positive ore ratio >0.2 | 2.19 (1.39–3.44) | 0.001 | 2.10 (1.33–3.32) | 0.001 |
| Negative biopsies >2 | 0.19 (0.04–0.80) | 0.023 | 0.19 (0.04–0.85) | 0.029 |
| Log(prostate volume) | 0.31 (0.20–0.48) | <0.001 | 0.47 (0.31–0.70) | <0.001 |
| Log(PSA) | 2.11 (1.53–2.91) | <0.001 | ||
| 4Kpanel | 1.54 (1.31–1.81) | <0.001 | ||
PSA = prostate-specific antigen; OR = odds ratio; CI = confidence interval.
ROC curve analysis (Table 4, Supplementary Fig. 2) comparing the full model with the 4Kpanel and the full clinical model with serum PSA indicated that the 4Kpanel significantly improved the accuracy for predicting reclassification (AUC 0.78 vs 0.74) in the initial surveillance biopsy, with a significant incremental value in AUC of 0.04 (95% CI 0.003–0.09). In a model without prostate volume, the incremental value in AUC was 0.07 (95% CI 0.02–0.11). The 4Kpanel did not improve prediction of reclassification in subsequent biopsies relative to PSA (AUC 0.75 vs 0.76).
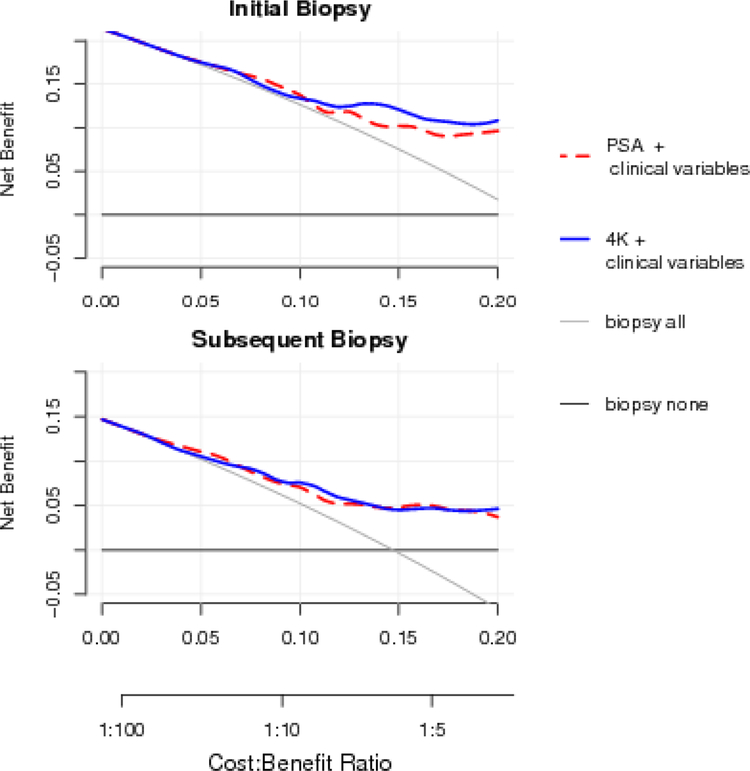
Similar findings were observed in DCA. Compared to a clinical model with serum PSA, the model with 4Kpanel showed a higher net benefit for the initial surveillance biopsy, but there was no benefit for subsequent biopsies. All models showed substantial gain in net benefit compared with the biopsy-all and biopsy-none strategies across a range of plausible cost and benefit ratios (Fig. 1 and Supplementary Fig. 3).
The clinical consequences, or the number of biopsies and the number of high-grade cancers that could be avoided or delayed per 1000 patients, were illustrated based on prediction models with the 4Kpanel or PSA (Table 5). For example, using a model with the 4Kpanel and a clinical rule of only performing an initial surveillance biopsy in patients whose risk of high-grade cancer exceeded 10%, 252 biopsies would be avoided, 19 of which would contain high-grade cancer as defined by any pattern 4 disease, and zero biopsies with primary Gleason 4. Comparing the two models at the same numbers of biopsies avoided (Supplementary Fig. 4) shows that the 4K model appears to miss fewer higher-grade cancers while avoiding the same number of initial biopsies.
Table 5 –
Clinical consequences showing the number of biopsies that could be avoided for initial surveillance biopsy or subsequent surveillance biopsy
| HGC probability | Biopsies | High-grade cancers | Primary Gleason 4 cancers | |||
|---|---|---|---|---|---|---|
| Performed | Avoided | Found | Missed | Found | Missed | |
| Initial surveillance biopsy | ||||||
| Biopsy all | 1000 | 0 | 214 | 0 | 44 | 0 |
| Initial biopsy: risk by clinical variables + PSA | ||||||
| >5% | 943 (896–970) | 57 (30–104) | 214 (157–284) | 0 (0–24) | 44 (21–88) | 0 (0–24) |
| >10% | 761 (689–821) | 239 (179–311) | 201 (146–270) | 13 (3–45) | 44 (21–88) | 0 (0–24) |
| >15% | 509 (432–586) | 491 (414–568) | 164 (114–229) | 50 (26–96) | 38 (17–80) | 6 (1–35) |
| Initial biopsy: risk by clinical variables + 4K | ||||||
| >5% | 956 (912–979) | 44 (21–88) | 214 (157–284) | 0 (0–24) | 44 (21–88) | 0 (0–24) |
| >10% | 748 (676–809) | 252 (191–324) | 195 (141–263) | 19 (6–54) | 44 (21–88) | 0 (0–24) |
| >15% | 522 (445–598) | 478 (402–555) | 182 (130–250) | 31 (14–71) | 44 (21–88) | 0 (0–24) |
| Subsequent surveillance biopsies | ||||||
| Biopsy all | 1000 | 0 | 147 | 0 | 47 | 0 |
| Risk by clinical variables + PSA | ||||||
| >5% | 844 (789–886) | 156 (114–211) | 147 (105–201) | 0 (0–18) | 47 (26–85) | 0 (0–18) |
| >10% | 692 (627–750) | 308 (250–373) | 133 (93–185) | 14 (5–41) | 43 (23–79) | 5 (1–26) |
| >15% | 445 (380–513) | 555 (487–620) | 109 (74–158) | 38 (19–73) | 43 (23–79) | 5 (1–26) |
| Risk by clinical variables + 4K | ||||||
| >5% | 848 (794–890) | 152 (110–206) | 142 (101–196) | 5 (1–26) | 47 (26–85) | 0 (0–18) |
| >10% | 654 (588–715) | 346 (285–412) | 133 (93–185) | 14 (5–41) | 47 (26–85) | 0 (0–18) |
| >15% | 408 (344–475) | 592 (525–656) | 100 (66–147) | 47 (26–85) | 38 (19–73) | 9 (3–34) |
HGC = high-grade cancer.
Results are presented as the number (95% confidence interval) per 1000 men.
4. Discussion
In this study using a prospectively enrolled multi-institutional cohort of men on active surveillance, we show that addition of a panel of four kallikrein markers to a model that includes clinical information can significantly improve prediction of the outcome in the first surveillance biopsy. Both models performed comparably for prediction of reclassification in subsequent biopsies. Importantly, in DCA both models showed a higher net benefit compared to biopsy-all and biopsy-none strategies. Lastly, we showed that the 4Kpanel added to currently available clinical metrics and how the results impact clinical management.
There is a growing body of evidence that true Gleason 6 prostate cancer is indolent and will not cause harm if left untreated [10–12]. This knowledge is balanced by the known undersampling in prostate needle biopsies, and while some have advocated that select Gleason 3 + 4 cancers may undergo surveillance, level 1 clinical trial data and treatment guidelines generally recommend treatment of higher-grade cancers, including Gleason 3 + 4 disease [13,14]. Our efforts focus on developing tools for use after diagnosis of Gleason 6 prostate cancer to provide a higher degree of certainty that no occult high-grade cancer was missed at diagnosis. More accurate tools would not only support the practice of active surveillance but could also promote less intensive monitoring regimens.
A panel of four kallikreins, when combined in a mathematical algorithm, improves the prediction of newly diagnosed high-grade (Gleason ≥7) cancer [3]. This panel of markers also improved long-term prediction of metastatic disease among men with PSA ≥2 in a Swedish cohort [15]. In this study, we asked whether the same panel of markers [3] improved the prediction of high-grade disease in surveillance biopsies of men already diagnosed with Gleason 6 cancer. We found that when the kallikreins were assessed before the initial surveillance biopsy (sometimes called the confirmatory biopsy), the 4Kpanel provided incremental benefit for prediction of high-grade cancer (Gleason ≥7) over the clinical factors that are available at diagnosis. Specifically, depending on the choice from the various cutpoints that are based on the risk of high-grade disease, a substantial number of biopsies could be avoided while minimizing the number of missed high-grade cancers, few of which had primary pattern 4. The 4Kpanel was not of value over PSA for the prediction of reclassification in subsequent biopsies after the first surveillance biopsy. We found that the impact of other biopsy information, primarily volume of core involvement in previous biopsies and the number of previous negative biopsies, carries such a statistical weight in modeling that the impact of the 4Kpanel is minimized. For example, if a patient had low-volume disease at the initial surveillance biopsy or had subsequent negative biopsies after the initial diagnosis, then these factors were highly protective against biopsy reclassification at subsequent biopsy. It should be noted that our analysis of these subsequent biopsies used the 4Kpanel from the plasma sample that was closest to the subsequent biopsy, not necessarily the plasma sample from study entry, which could be months or years earlier than the subsequent biopsy.
We included serum PSA and prostate volume separately in our models instead of calculating PSA density, as we find a better model fit when the variables enter the model independently. While transurethral ultrasound prostate volume measurements may suffer from imprecision [16], statistical models that included prostate volume appeared to provide slightly improved predictive performance (AUC for all groups 0.77 with volume vs 0.75 without volume). Furthermore, prostate volume is a strong predictor of finding higher-grade cancers, with larger prostates being protective, as previously reported [17].
This study has limitations that merit mention. First, the model was developed and tested in the same cohort and with relatively limited numbers that resulted in wide confidence intervals and minor differences between the training and test sets. The results should clearly be validated in other cohort before clinical application. However, we expect that our results will be similar to those found in a community setting, as PASS is a multicenter center study that represents a broad spectrum of men utilizing active surveillance. Similarly, as PASS is primarily a Caucasian cohort, the findings of this study may not be generalizable to African American patients. Another limitation is that the serum PSA measurements used were obtained as part of standard clinical care, and the local site assays may differ from the one used with the 4Kpanel. Thus, the comparative modeling using PSA versus 4Kpanel may have slightly different tPSA values, with caution suggested for comparisons between the models. Lastly, as the use of imaging such as multiparametric MRI (mpMRI) is increasing, we do not have MRI data for most of our participants and recognize the potential value of future studies incorporating results from mpMRI and biomarkers in active surveillance.
5. Conclusions
The 4Kpanel was significantly associated with reclassification at the first surveillance biopsy, providing incremental value over routine clinical information, and the 4K model performed significantly better than the base model in this group. The 4Kpanel did not add predictive value to a PSA clinical model for biopsy decision-making for men at subsequent surveillance biopsies. This work aims to provide clinical validation of a biomarker that will help determine those men who have or will develop aggressive prostate cancer, allowing for the accurate determination of those men who may avoid or delay the burden of immediate treatment safely, while concurrently identifying men who may optimally benefit from early treatment.
Supplementary Material
Fig. 1 –
Decision curve analysis for full models with serum Prostate-specific antigen (PSA) or with the 4Kpanel. Strategies for biopsying all men (biopsy all) or no men (biopsy none) are also shown. The line with the highest net benefit at any particular threshold probability for biopsy (x-axis) will yield the best clinical results.
Table 4 –
Results of final regression models for reclassification
| Base model | Area under the curve (95% confidence interval) | ||
|---|---|---|---|
| 4K + clinical model | PSA + clinical model | Difference | |
| Full clinical model | |||
| Initial biopsy | 0.783 (0.691–0.871) | 0.740 (0.652–0.828) | 0.043 (0.003–0.086) |
| Subsequent biopsy | 0.754 (0.657–0.838) | 0.755 (0.653–0.841) | −0.001 (−0.037–0.041) |
| Clinical model without prostate volume | |||
| Initial biopsy | 0.748 (0.654–0.840) | 0.678 (0.579–0.774) | 0.069 (0.016–0.114) |
| Subsequent biopsy | 0.738 (0.633–0.825) | 0.718 (0.611–0.810) | 0.02 (−0.023–0.07) |
PSA = prostate-specific antigen.
Confidence intervals were calculated with bootstrap accounting for correlations among individuals.
Funding/Support and role of the sponsor:
This work was supported by the Department of Defense (PC130355), the Canary Foundation, and the Institute for Prostate Cancer Research. The sponsors played a role in the design and conduct of the study, and collection, management, and analysis of the data.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Financial disclosures: Daniel W. Lin certifies that all conflicts of interest, including specific financial interests and relationships and affiliations relevant to the subject matter or materials discussed in the manuscript (eg, employment/affiliation, grants or funding, consultancies, honoraria, stock ownership or options, expert testimony, royalties, or patents filed, received, or pending), are the following: Yan Dong is an employee of OPKO. The remaining authors have nothing to disclose.
References
- 1.Ankerst DP, Xia J, Thompson IM Jr, et al. Precision medicine in active surveillance for prostate cancer: development of the Canary-Early Detection Research Network active surveillance biopsy risk calculator. Eur Urol 2015;68:1083–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parekh DJ, Punnen S, Sjoberg DD, et al. A multi-institutional prospective trial in the USA confirms that the 4Kscore accurately identifies men with high-grade prostate cancer. Eur Urol 2015;68:464–70. [DOI] [PubMed] [Google Scholar]
- 3.Bryant RJ, Sjoberg DD, Vickers AJ, et al. Predicting high-grade cancer at ten-core prostate biopsy using four kallikrein markers measured in blood in the ProtecT study. J Natl Cancer Inst. 2015;107(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Newcomb LF, Thompson IM Jr, Boyer HD, et al. Outcomes of active surveillance for clinically localized prostate cancer in the prospective, multi-institutional Canary PASS cohort. J Urol 2016;195:313–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pan W Akaike’s information criterion in generalized estimating equations. Biometrics 2001;57:120–5. [DOI] [PubMed] [Google Scholar]
- 6.Vickers AJ, Elkin EB. Decision curve analysis: a novel method for evaluating prediction models. Med Decis Making 2006;26:565–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Food US and Administration Drug. Innovation or stagnation: challenge and opportunity on the critical path to new medical products. Washington, DC: US Department of Health and Human Services; 2004. [Google Scholar]
- 8.McShane LM, Altman DG, Sauerbrei W, et al. Reporting recommendations for tumor marker prognostic studies (REMARK). J Natl Cancer Inst 2005;97:1180–4. [DOI] [PubMed] [Google Scholar]
- 9.Hayes DF, Bast RC, Desch CE, et al. Tumor marker utility grading system: a framework to evaluate clinical utility of tumor markers. J Natl Cancer Inst 1996;88:1456–66. [DOI] [PubMed] [Google Scholar]
- 10.Ross HM, Kryvenko ON, Cowan JE, Simko JP, Wheeler TM, Epstein JI. Do adenocarcinomas of the prostate with Gleason score (GS) ≤6 have the potential to metastasize to lymph nodes? Am J Surg Pathol 2012;36:1346–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eggener SE, Badani K, Barocas DA, et al. Gleason 6 prostate cancer: translating biology into population health. J Urol 2015;194:626–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carter HB, Partin AW, Walsh PC, et al. Gleason score 6 adenocarcinoma: should it be labeled as cancer? J Clin Oncol 2012;30:4294–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilt TJ, Brawer MK, Jones KM, et al. Radical prostatectomy versus observation for localized prostate cancer. N Engl J Med 2012;367:203–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mohler JL, Kantoff PW, Armstrong AJ, et al. Prostate cancer, version 1.2014. J Natl Compr Canc Netw 2013;11:1471–9. [DOI] [PubMed] [Google Scholar]
- 15.Stattin P, Vickers AJ, Sjoberg DD, et al. Improving the specificity of screening for lethal prostate cancer using prostate-specific antigen and a panel of kallikrein markers: a nested case-control study. Eur Urol 2015;68:207–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rodriguez E Jr, Skarecky D, Narula N, Ahlering TE. Prostate volume estimation using the ellipsoid formula consistently underestimates actual gland size. J Urol 2008;179:501–3. [DOI] [PubMed] [Google Scholar]
- 17.Roobol MJ, van Vugt HA, Loeb S, et al. Prediction of prostate cancer risk: the role of prostate volume and digital rectal examination in the ERSPC risk calculators. Eur Urol 2012;61:577–83. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.