Abstract
Ribosomally synthesized and post-translationally modified peptides (RiPPs) are a class of cyclic or linear peptidic natural products with remarkable structural and functional diversity. Recent advances in genomics and synthetic biology, are facilitating us to discover a large number of new ribosomal natural products, including lanthipeptides, lasso peptides, sactipeptides, thiopeptides, microviridins, cyanobactins, linear thiazole/oxazole-containing peptides and so on. In this review, we summarize bioinformatic strategies that have been developed to identify and prioritize biosynthetic gene clusters (BGCs) encoding RiPPs, and the genome mining-guided discovery of novel RiPPs. We also prospectively provide a vision of what genomics-guided discovery of RiPPs may look like in the future, especially the discovery of RiPPs from dominant but uncultivated microbes, which will be promoted by the combinational use of synthetic biology and metagenome mining strategies.
Keywords: Natural products; Ribosomally synthesized and post-translationally modified peptides; RiPPs, Genome mining; Metagenome mining; Synthetic biology
1. Introduction
Microbial secondary metabolites, which have been honed through evolution to provide bacteria with competitive advantages, are a promising source for the discovery of bioactive natural products with medicinal potential. Among them, ribosomally synthesized and post-translationally modified peptides (RiPPs) attract extensive interest from both academic and industrial communities due to their structural variability and functional diversity [1,2]. The manifold chemical space of genetically encoded RiPPs is determined by the encoding nucleotide sequence, which links the diversity of small molecules with the variability of genes. Such a genetically-encoded nature of RiPP enables scientists to readily manipulate the scaffolds of the mature peptides by site-directed mutagenesis and efficiently screen the targets of interests from large libraries even with the volume of 200 million [3].
The minimal components of a RiPP biosynthetic gene cluster (BGC) generally consist of a short precursor peptide, typically includes an N-terminal leader and a C-terminal core peptide, and post-modification (PTM) enzymes. Various PTM enzymes, such as radical SAM enzyme [4], cytochrome P450 [5], Diel-Alderase [6], and ATP-grasp enzyme [7], install distinctive moiety onto the linear precursor peptide to give the mature scaffold, resulting in different classes of RiPPs [8]. Until now, more than 13 representative types of classified and many other unclassified RiPPs have been found in bacteria, fungi, archaea, and plants (Fig. 1) [[8], [9], [10]]. Each class of these RiPPs represents a unique subset of biosynthetic logic, which can be used as a biomarker in targeted genome mining. With the development of new sequencing techniques in the 21st century, genomes or metagenomes have been sequenced on large scales, providing vast opportunities for the genome-mining-based discovery of novel natural products. Various bioinformatics methods such as antiSMASH [11], RiPPMiner [12], PRISM [13], RODEO [14,15] have been developed to predict, deduplicate, and prioritize BGCs, guided by phylogeny closeness, or by chemocentric searches. One classical approach is to use core biosynthetic enzymes as a query to search genomes for homologous molecular machinery. These classical genome mining strategies are usually based on sequence similarity and are robust in finding BGCs with similar biosynthetic routes [16]. However, these phylogeny-based strategies are unable to capture BGCs with convergent enzymes and are easily distracted by divergent homologs of the query.
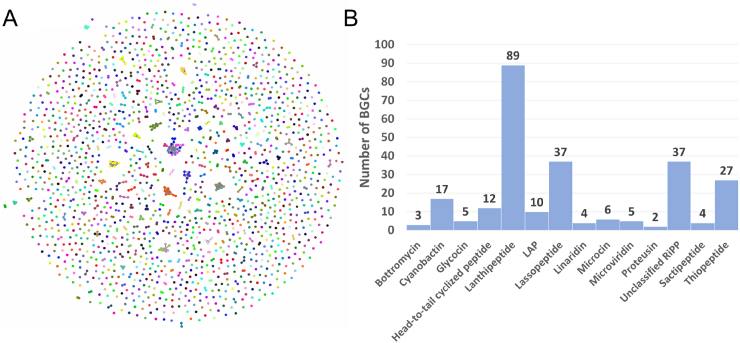
Fig. 1.
Diversity of known RiPP BGCs. A) The network of known RiPPs BGCs from MIBiG, visualizing their diversity. The network is constructive by BiG-SCAPE with default parameters, each node corresponds to one RiPP BGC, similar BGCs are clustered together. B) The numbers of known RiPPs at the family level from MIBiG.
Additionally, the structural and biosynthetic diversity of RiPPs from bacteria make comprehensive mining in large-scale datasets and targeted discovery of these molecules extremely challenging. The era of big data mining is now in full swing, catalyzed by advances in data processing power of bioinformatics algorithms and the development of new artificial intelligence (AI) methods. Various deep learning-based bioinformatics methods such as NeuRiPP [17] and DeepRipps [18] have recently been developed to identify the precursor of RiPPs' BGCs independently of their genetic content. AI strategies that can systematize large volumes of genetic and chemical data and connect genomic information to metabolomic in a high-throughput endeavor are promoting the targeted discovery of RiPPs from large-scale omics datasets. In this review, we recapitulate the developments of genome mining strategies for the discovery of RiPP natural products and introduce representative genomics-guided discovery cases using these mining strategies. Since most of current mining strategies were designed based on bacterial data, herein, we mainly cover genome mining guided discovery of RiPPs from bacteria.
2. Genome mining strategies of RiPPs
In the early date, like other families of natural products, RiPPs were discovered based on bioactivity screening of bacterial secondary metabolites. As sequencing techniques have been rapidly improved, more and more bacterial genome sequences are available and thus provide us an unprecedented chance to study RiPPs at the genetic level. One of the earliest strategies for mining RiPPs is using BLAST [19] to search for certain PTM enzymes from bacterial genomes [20,21]. There were no user-friendly genome mining tools for RiPPs until de Jong et al. developed the first web tool, named BAGEL, to mine for bacteriocin in 2006 [22]. Later on, new BAGEL versions were updated to support mining more RiPPs classes [23,24]. In 2011, Medema et al. developed an integrated genome mining tool antiSMASH that can mine for many different classes of BGCs, including RiPPs, non-ribosomal peptides (NRPS), and polyketides (PKS) [25]. Since then, several genome mining tools have been developed for either comprehensive analysis of BGCs or targeting specific classes of RiPPs following two major strategies (Fig. 2 and Table 1) [11,13,14,23,24,26]. One strategy mainly targeted conserved enzymes of biosynthetic machinery while the other one employed additional algorisms to better identify precursor peptides around PTM enzymes [14,15,23,24] that exclusively represented corresponding RiPPs BGCs. Recently, more sophisticated comparative approaches have been introduced to systematically identify and prioritize BGCs of interest, enabling the targeted discovery of novel RiPPs from the large-scale dataset. In 2017, Mitchell's group adopted a protein-wise comparative tool Enzyme Function Initiative-Enzyme Similarity Tool (EFI-EST) [27], to prioritize novel biosynthetic enzymes [14]. In 2020, Medema's group developed a BGC-wise comparison tool BiG-SCAPE [28], to prioritize novel BGCs at the BGC level. Besides, scientists started to introduce machine learning and deep learning approaches into genome mining of RiPPs by directly targeting precursors [12,17,18], opening a stage of RiPP genome mining powered by artificial intelligence [29]. Over the past two decades, significant advances in bioinformatics have allowed the rapid development of genome mining strategies.
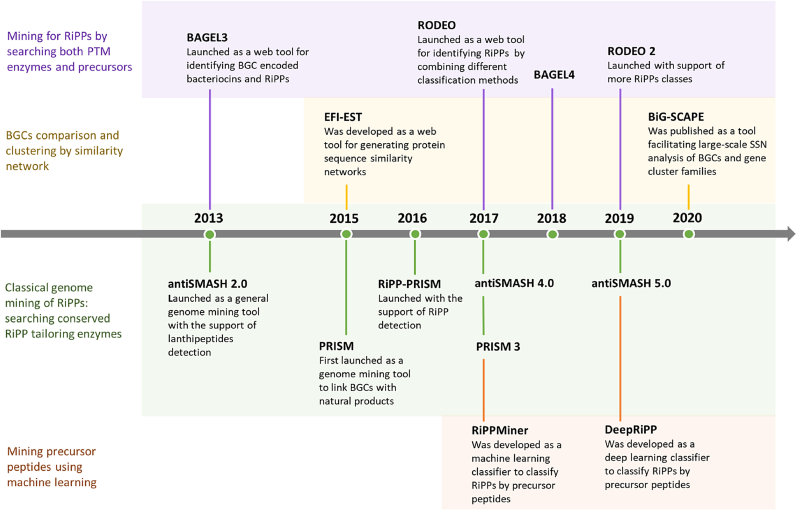
Fig. 2.
The timeline of RiPPs genome mining.
Table 1.
Summary of commonly used and recently developed RiPPs genome mining tools.
| Mining tools | Description | Advantages | Limitations | Methods | Ref |
|---|---|---|---|---|---|
| antiSMASH 5 | Integrated platform for analyzing BGCs and metabolites including RiPPs | Integration of many other bioinformatics tools. Able to analyze and classify more than 50 classes of BGCs. |
Gene context-dependent. |
Rule-based, Hidden Markov model (HMM) |
[11] |
| PRISM 3 | Integrated platform for analyzing BGCs and metabolites including RiPPs | Able to analyze and classify more than 20 classes of BGCs. Able to predict the natural product structures of some types of BGCs. |
Gene context-dependent. | Rule-based, HMM |
[13] |
| BAGEL4 | Combination of direct precursor peptides mining and indirect rule-based BGCs detection of RiPPs. | Able to predict both RiPPs BGCs and their precursor peptides. | Direct mining is searching precursor peptides against Bacteriocins and RiPPs databases, unable to mine for novel RiPPs precursor peptides. | Rule-based, HMM |
[24] |
| RiPPMiner | Machine learning classifier to predict RiPPs structural features based on precursor peptides | Able to classify different classes of RiPPs and predict their cleavage sites by precursor peptides sequences in a gene context-independent manner. | Prediction accuracy is poor for small classes of RiPPs (e.g. Sactipeptides, Linaridins). | Support vector machine (SVM) | [12] |
| RODEO | Mining for RiPPs BGCs and predicting precursor peptides by combination of HMM, heuristic scoring and machine learning. | Accurate prediction of RiPPs precursor peptides and their cleavage sites by context genes. | The current version is limited to some RiPPs classes (e.g. Lanthipeptides, Lasso peptides, etc.). Gene context-dependent. |
Rule-based heuristic scoring, HMM, SVM |
[14] |
| RiPPER | Family-independent identification of RiPPs precursor peptides | Able to identify novel precursor peptides of small RiPP class. | No user friendly webtool is available. Requires prior knowledge of RiPP class to set the parameters. Gene context-dependent. |
Prodigal scoring, HMM |
[38] |
| DeepRiPP | Deep learning-based genome mining | Able to classify RiPPs and predict the cleavage sites by precursor peptides sequences in a gene context-independent manner. | Limited training sets and thus low accuracy for small RiPP class. | Deep neural network | [18] |
2.1. Classical genome mining of RiPPs: searching conserved RiPP tailoring enzymes
Although the structural diversity of each RiPP family is vastly due to their diverse tailoring enzymes, benchmark enzymes of some RiPPs classes such as Ser/Thr dehydratases of lanthipeptide and radical-SAM enzymes of sactipeptides are strikingly conserved [8]. Given these unique features, the most classical and popular mining approach for RiPPs is to focus on finding new members of known classes of RiPPs. Novel chemical structures can be predicted and prioritized by looking for BGCs with shared conserved biosynthetic enzymes but with enough differences in terms of precursor peptide or other tailoring enzymes. For example, to identify BGC of interest from genome data, conserved genes in the RiPPs pathway are used as seed sequences to identify homologs via sequence-based comparison software BLAST. The explosion of genome sequence data in recent years has given rise to many distinguished in silico mining approaches based on conserved RiPPs tailoring enzymes. AntiSMASH [11] and PRISM [13] are two popular integrated platforms of general-propose for BGC prediction and analysis. Powered by rule-based scoring and hidden Markov model (HMM), these two approaches can detect RiPPs classes such as lanthipeptides and lasso peptides, which have distinct and conserved post-modification enzymes. Streptocollin, for instance, is one of the lanthipeptides discovered by traditional genome mining with antiSMASH [16,30]. By searching for lanthipeptides BGCs in Streptomyces collinus Tü 365, authors found a class IV lanthipeptide BGC with high similarity with venezuelin BGC and isolated the product Streptocollin via heterologous expression [30]. However, one major obstacle of classical genome mining is that it can mainly identify RiPPs with PTM logics that are similar to known ones, thus highly limited its ability to reveal BGCs with novel biosynthetic logics [29,31]. In addition, conventional tools such as the early versions of antiSMASH are highly dependent on gene detection algorisms to identify meaningful open reading frames (ORFs) from genome sequences. Thus, this strategy is difficult to find RiPP precursor peptide that is too short in sequence to be identified (after antiSMASH version 4, small ORFs detection logic from RODEO was integrated [32]). Further studies, which take these small ORFs encoding precursors into account, will need to be undertaken.
2.2. Mining BGCs by searching both PTM enzymes and precursors
Precursor peptides are the substrates of the RiPPs biosynthetic machinery, the variation of which could significantly increase the chemical space of RiPPs, even if sharing the same post-modification enzymes. Mining new precursor peptides would be another ideal strategy to discover new RiPPs. However, the small ORFs which encode RiPPs precursors are often neglected by traditional gene annotation algorisms like Prodigal [33] or Glimmer [34] because of their short sequence lengths. Therefore, general-purpose genome mining tools built upon gene annotations such as antiSMASH and PRISM suffer from low accuracy in predicting precursor peptide, especially those with substrate tolerant enzymes that function with multiple precursors [35,36]. BAGEL [24] and RODEO [14] are two genome mining tools designed explicitly for genome mining of RiPPs. BAGEL uses the rule-based strategy to detect the conserved domains of post-modification enzymes, then classified BGCs into different RiPPs classes. It predicts RiPPs precursors by detecting small ORFs nearby core PTM enzymes and blasts them against its core peptide database. In contrast, RODEO adopts a more comprehensive approach that uses HMM and Pfam database to detect RiPPs BGCs, then combines heuristic scoring and machine learning to predict RiPPs precursors. Both tools take the small ORFs that possibly encode RiPPs precursor peptides into account. Thus, they are more reliable for RiPPs mining than canonical genome mining tools. For instance, upon the prediction of precursor peptides by RODEO, Mitchell's group successfully identified a novel lasso peptide citrulassin and a new class of RiPPs, namely ranthipeptides [14,15]. Later on, Walker et al. applied RODEO to more than 100,000 bacterial and archaeal genomes and found nearly 8500 lanthipeptide precursor peptides, including many previously uncharacterized lanthipeptides and potential antibiotics, further demonstrating the power of this method [37]. With the concern that RODEO can only identify RiPPs of well-studied classes, Truman, A.W.'s group developed RiPPER, which was built upon RODEO but with a different logic of finding precursors [38]. Instead of classifying precursors by their sequences, RiPPER finds all small ORFs within ±8 kb distance of the PTM enzyme and scores them by Prodigal, the top 3 scored ORFs are considered as putative precursors. By this method, the authors revealed that the “rare” thioamidated RiPPs were, in fact, largely unexplored and facilitated the discovery of two novel thioamidated RiPPs from Streptomyces varsoviensis. These findings demonstrated the power of the genome mining approach that targets both PTM enzymes and precursors.
In current genome mining approaches, the prediction of RiPPs structure has been automated by the use of various computational tools. However, the identification of RiPP is usually conducted in a case-by-case manner by genetic manipulation or heterologous expression and thus time-consuming. Dorrestein's group developed a computational tool RiPPquest that automated connect natural product genotypes with their corresponding chemotypes from metabolomic data sets [39]. Similar to BAGEL and RODEO, RiPPquest firstly finds the core PTMs of RiPPs and searches for small ORFs around the enzymes. Next, RiPPquest generates an MS/MS peptide database of predicted products and matches with user-provided MS/MS data of microbial extract to identify the candidate RiPPs precursors.
2.3. Mining precursor peptides using machine learning
Classical genome mining via targeting the homology of tailoring enzymes relies heavily on the phylogenetic closeness with the query sequence, thus hinder their ability to target RiPPs of novel family. Nevertheless, using precursor peptide sequences of known RiPPs as queries to mine new RiPPs will result in analogs of known compounds too. Thus, scientists are trying to understand the intrinsic identities of RiPPs precursor peptides and use machine learning to grasp the feature of a small peptide that attributes to the RiPP precursor. RiPPMiner [12] is a machine learning classifier that is capable of differentiating RiPPs precursors from non-precursor small peptides and predicting the cleavage sites of some well-studied RiPPs including lanthipeptides, lasso peptides, and cyanopeptides. Powered by a machine learning method named support vector machine (SVM), RiPPMiner was trained on more than 500 experimentally verified RiPPs for prediction. Authors reported high sensitivity and specificity of its RiPP identification (0.93 and 0.90, respectively) and RiPP classification (0.79 and 0.98, respectively). Nevertheless, limited by the small training dataset and the drawback of the selected machine learning method, the cleavage site prediction was relatively poorly performed, with a precision of only 0.69.
To tackle these problems, researchers have introduced a new artificial intelligence method named deep learning, which is based on artificial neural networks, into RiPPs mining tools. NeuRiPP [17] for instance, combined two famous deep learning models, convolutional neural network (CNN) and long short-term memory (LSTM), to predict the probability of a short peptide as a RiPPs precursor. Another tool, DeepRiPP [18], was recently introduced by Magarvey's group for RiPPs identification and classification. DeepRiPP utilized an advanced transfer-learning deep neural network method named Universal Language Model Fine-tuning (ULMFiT), which was initially developed by the Fastai team in 2018 [40]. The original method has shown significant improvements in solving many problems in the field of natural language processing. Thus, the authors of DeepRiPP tried to adopt this method to predict precursor peptides and identify RiPPs in a gene context-independent manner. Compared to RiPPMiner, DeepRiPP was trained on a much larger dataset containing more than 3000 RiPPs precursors and 3000 non-RiPPs short peptides, making the prediction more reliable. It also took advantage of a previously developed chemical structure prediction algorism--GARLIC [41], to predict RiPPs structures. Combining the metabolomic data from mass spectra, DeepRiPP can automate the process of targeting novel RiPPs from bacteria cultures. Prioritization upon this integrated platform, authors successfully isolated and identified three novel RiPPs from Streptomyces and Flavobacterium [18]. It is envisioned that the integration of artificial intelligent approaches into genome mining will shed light on the vast unknown universe of RiPPs and advance their genomics-guided discovery. However, compared to NRPS and PKS, the number of RiPPs discovered so far is still limited, which hinders the potential of deep learning approaches to find novel RiPPs classes. More studies in both contexts dependent (traditional) and independent (precursor-based) genome mining are still needed to further enhance deep learning methods.
2.4. BGC comparison and clustering by similarity network
Upon sequence similarity-based searching, the targeted identification of RiPPs of interest has become possible by searching for conserved enzymes; however, usually at the cost of finding similar BGCs. To avoid rediscovery of known RiPPs, some research groups have been actively introducing comparative BGC analysis for the discovery of novel BGCs. Comparative BGCs analysis is a post-processing workflow to prioritize BGC of interest. EFI-EST is a recently developed web tool that computes sequence similarity networks (SSNs) of proteins [27]. This tool is able to group similar enzymes by calculating their sequence similarity to distinguish novel RiPPs enzymes from known ones. Applying SSN analysis to precursor peptide identified by RODEO, Mitchell's group significantly expanded the library of putative lasso peptides to >1300 and characterized six new lasso peptides upon prioritization [14]. Aside from comparing a single enzyme or precursor peptide, another approach to assess the novelty of BGCs is to evaluate their phylogenetic distant as well as biosynthetic novelty via correlating multiple genes or entire BGC to the known family. BiG-SCAPE [28] is a recently devised bioinformatic tool that facilitates large-scale SSN analysis of BGCs on a multiple-gene level. This software extends comparative analysis from a single gene to the entire BGC, and it is also capable of analyzing multiple user-submitted BGCs together with known BGCs from the MIBiG database [42]. Furthermore, by taking into account class-specific differences and evolutionary relationships between and within BGCs, the BiG-SCAPE/CORASON platform can easily classify and chart the BGC family from a large-scale dataset. This platform provides researchers an intuitive way of distinguishing novel BGC families from known ones.
3. Genome mining guided discovery of RiPPs
Genome mining is a process of discovering BGCs that conforms to a given biosynthetic logic. Powered by new bioinformatics tools, the recent genome mining strategies allow us to use a supercomputer to act as a high-throughput screening platform to accelerate the discovery of novel natural products from big genomic data. An essential rule in mining any type of RiPP is to find a short precursor peptide (generally less than 150 AA) adjacent to a post-modification enzyme. While for varied RiPPs, the conserved motifs present in the tailoring enzymes or precursors distinguish one from another. Almost all the RiPPs have more than one hallmarks which either present in the BGCs or the adjacent gene context. These biomarkers enabled the successful discovery of RiPPs BGCs of interest using individual or integrated strategies. Below we describe the genomics-guided discovery of novel RiPPs of the different families using genome mining strategies mentioned above.
3.1. Lanthipeptides: a well-established prediction hallmark with high accuracy
Lanthipeptides are a class of lanthionine-containing RiPPs which exhibit a wide range of bioactivities, including antiviral [2,43], antibacterial [44], antifungal [45], and antiallodynic functions [46]. The structurally distinctive feature of lanthipeptides is the thioether crosslink. Such a motif is constructed via the dehydration of serine or threonine followed by conjugate addition with the hydrosulfuric group of cysteine [46] (Fig. 3A and 3B). Based on the mechanism of dehydration and cyclization, lanthipeptides can be divided into four major types. Two proteins individually catalyze the dehydration (LanB) and a Michael-type cyclization (LanC) in class I lanthipeptide biosynthesis. Of note, LanB accomplishes the dehydration by transferring glutamate from glutamyl-tRNAGlu to the β-hydroxyl group of serine or threonine followed by β-elimination [46,47]. Class II features a single protein with both dehydration and cyclization domains (LanM), in which the dehydration is mediated by ATP-dependent phosphorylation of serine or threonine side chain followed by phosphate elimination [36,48]. Class III and IV are featured by their unique phosphorylation-mediated dehydration mechanism, in which donors of the phosphate group are not nucleotide-specific [[49], [50], [51], [52]]. The absence or presence of zinc-binding motif in the cyclization domain further distinguishes class III (without zinc-binding motif) and class IV (containing conserved zinc-binding ligands) lanthipeptides. Since extensive works have been done for discovering novel lanthipeptides, elucidating their biosynthetic gene clusters, and illustrating enzymatic mechanisms of key steps, a variety of unique hallmark genes have been accumulated for PTM enzyme-based genome mining (Fig. 3C).
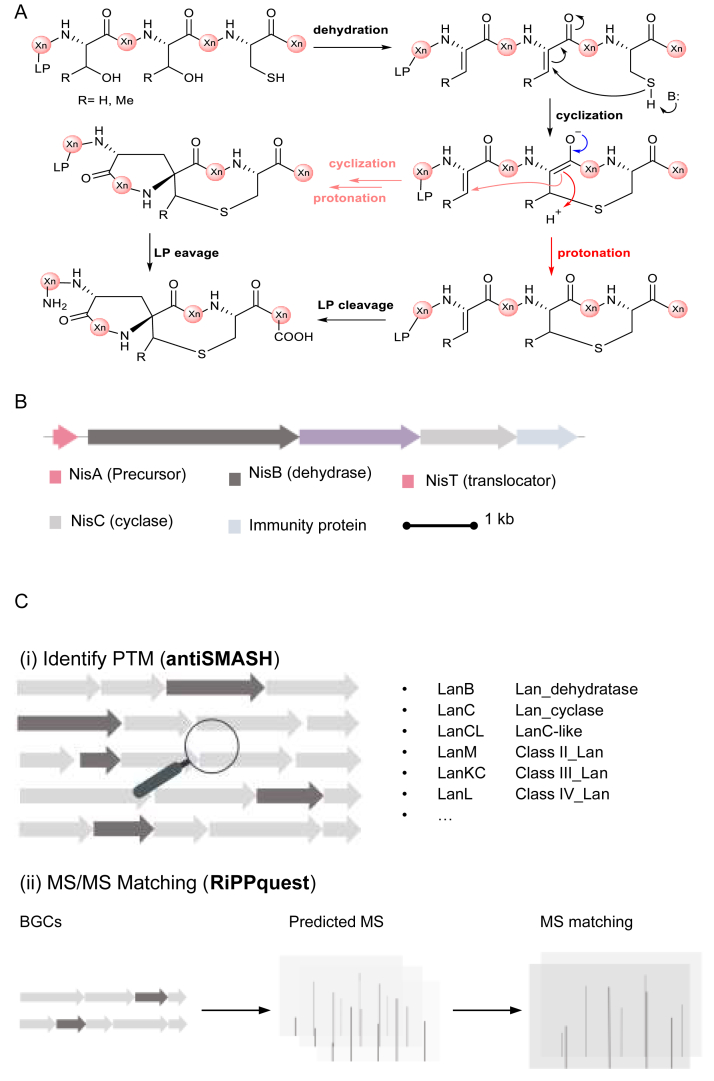
Fig. 3.
Lanthipeptide biosynthetic logic and mining strategy. A) Exemplified enzymatic mechanism of lanthipeptide biosynthesis. LP, leader peptide. Xn, peptide with n residues. B) Nisin biosynthetic gene cluster from Lactococcus lactis. C) Lanthipeptide mining strategy. (i) Tailoring enzyme-based mining mainly focuses on identifying the hallmark of lanthipeptide biosynthesis, such as dehydratase and cyclase. (ii) MS/MS matching connects genotypes and chemotypes. The MS data of predicated mature peptides was aligned with the experimentally obtained MS data to guide target isolation.
Prior to the genomic era, identifying BGCs relied heavily on sequence similarity searching, such as BLAST (Fig. 3C). The two-component lantibiotic, haloduracin, was identified from the Bacillus halodurans C-125 by sequence similarity searching of the lantibiotic mersacidin [53] rather than traditional isolation-based identification (Fig. 4). The HalA1 precursor peptide share significant sequence identity with the precursor peptide from the known two-component lantibiotics. However, two LanM genes designated as halM1 and halM2 are the distinctive markers of haloduracin with low similarity with known LanM, suggesting chemical novelty of its encoding products Halα and Halβ (Fig. 4). Both of these two lanthipeptides exhibit bactericidal activity against Lactococcus lactis CNRZ 117 [53]. Similarly, Lichenicidin was discovered by using BLAST searching combined with the initial version of BAGLE [22]. Briefly, 89 LanM homologs were first obtained using BLAST, and then 61 genome sequences were prioritized and further analyzed with BAGLE to identify putative BGCs [20]. Lichenicidin exhibits antimicrobial activity against Listeria monocytogenes, methicillin-resistant Staphylococcus aureus, and vancomycin-resistant Enterococcus strains [20]. In addition to searching homologs of tailoring enzymes, other BGC-associated proteins were also used to mine novel lantibiotic BGCs. For example, the LanT, which is designated as ABC transporter [46] that excretes lanthipeptide after biosynthesis, was used as a marker in identifying putative lanthipeptide BGCs [54]. Another similar case is associated with the use of profile HMM, by which the LanB was examined in the human oral and gut microbiome [55].
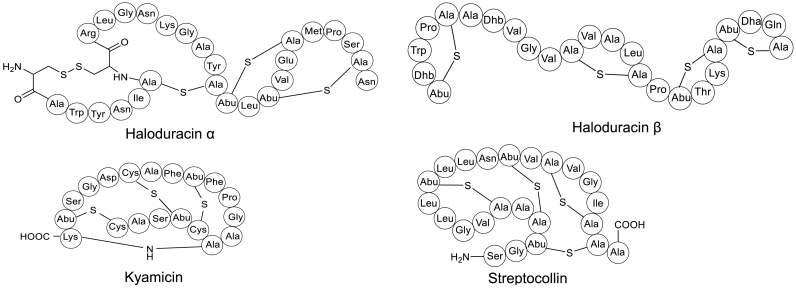
Fig. 4.
Examples of lanthipeptides discovered by genome mining. Haloduracin α and β were isolated from Bacillus halodurans C-125. Kyamicin and streptocollin were discovered from Saccharopolyspora species and Streptomyces collinus Tu 365, respectively. Abu, d-α-aminobutyric acid. Dha, 2,3-didehydroalanine. Dhb, 2,3-didehydrobutyrine.
The increasing number of well-studied biosynthetic logics of lanthipeptides discovery via genome mining, in return, has made a significant contribution to the development of genome mining strategies. For example, the cleavage site motifs of class I to class IV and the tailoring enzymes, including LanB, LanC, LanM, LanKC, and LanL [46], are frequently used as hallmarks in the training prediction model for identifying lanthipeptides or lanthipeptide-like RiPPs, such as antiSMASH75. Upon the prioritization of antiSMASH-based genome mining, streptocollin [16,30] and kyamicin [56] were isolated and structurally identified (Fig. 4). Class IV lanthipeptide Streptocollin was isolated from Streptomyces collinus Tü 365, and its BGC was further confirmed by heterologous expression. Kyamicin, a type B cinnamycin-like lantibiotic antibiotic, was discovered via in situ activation and heterologous expression of a cryptic BGC mined from Saccharopolyspora species.
Informatipeptin was identified as a new class III lanthipeptide from Streptomyces by mass spectrometry-based genome mining using an algorithmic tool named RiPPquest, which was developed by Dorrestein's group [39]. Unlike the traditional genome-guided discovery that requires manual inspection of mass spectrometry data and genetic information, RiPPquest is able to automatically connect natural product genotypes predicted from microbial genome sequences with their corresponding chemotypes from metabolomic data sets (Fig. 3C) [39]. In the trajectory of MS-based genome mining, a set of gene fragments centered at LanC-domain were generated from 16 sequenced Streptomyces strains followed by the prediction of potential precursor smaller than 100 aa [39]. A computed MS/MS spectra dataset of all possible mature peptides were then matched with experimentally generated LC-MS/MS data. Peptide-spectrum matches were scored and molecular network [57] analysis was then used to prioritize compounds of interest.
Traditional natural products discovery methods have been successfully applied to discover many lanthipeptides with varied BGC architectures and tailoring enzymes. The known BGC pool will keep expanding so that prediction confidence in defining an unknown BGC or assigning the function of each ORF in that BGC will increase as well. A straightforward mining tool, like antiSMASH that is supported by RODEO, is becoming more and more reliable in mining lanthipeptides.
3.2. Lasso peptides: from sequence similarity searching to pattern-based matching
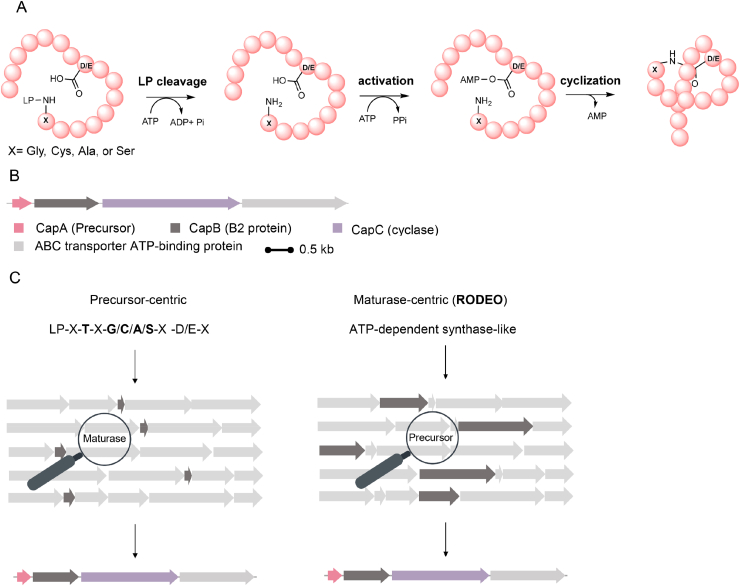
Lasso peptides are a class of cyclotides which features a distinct “threaded lasso” topological folding that distinguishes them from any other RiPPs. Such a structurally complex architecture can be achieved using as few as 20 amino acids: (I) the γ-carboxyl group of glutamate or the β-carboxyl group of aspartate firstly forms an isopeptide bond between the amino group of N-terminal amino acid to give a 7-, 8- or 9- amino acids macrocycle and, (II) the C-terminal of the precursor threads lactam ring to yield a tail and a loop region (Fig. 5A and B) [58]. The number and type of disulfide bond in the mature peptide divide this type of RiPP into four major groups: (i) class I lasso peptide harbors two interlinked disulfide bonds, (ii) class II has no disulfide bonds, (iii) class III contains only one single interlinked disulfide bond and (iv) class IV contains a handcuff disulfide bond [8,14]. The minimal lasso peptide biosynthetic gene cluster always includes a precursor protein (denoted as A protein), a precursor recognition protein with proteolytic activity (B protein), and an ATP-dependent asparagine synthase-like lasso cyclase (C protein) [58]. The conserved patterns present in the precursors and C proteins are the distinctive hallmarks for lasso peptide genome mining.
Fig. 5.
Lasso peptide biosynthetic logic and mining strategy. A) Enzymatic mechanism of lasso peptide biosynthesis. LP, leader peptide. X indicates Gly, Cys, Ala, or Ser residue. B) Capistruin biosynthetic gene cluster from Burkholderia thailandensis E264. C) Representative strategies of lasso peptide mining. The precursor-centric approach is to search putative lasso peptide precursors based on the conserved patterns of known lasso peptides. BGC candidates were identified by searching adjacent tailoring enzymes and ranking the conservativeness based on then conserved motifs, such as Cys-His-Asp catalytic triad for proteases (B protein) and Asp-rich motif for asparagine synthetases (C protein). In contrast, the maturase-centric strategy starts by retrieving tailoring enzymes-containing BGCs followed by searching possible adjacent short peptides, which could be the precursors.
The first genome mining-guided discovery of lasso peptide is the sequence similarity searching based identification of capistruin BGC (Fig. 5B) from Burkholderia thailandensis E264 in 2008 [59], using the most well-studied lasso peptide MccJ25 as a query. BGC of MccJ25 contains a peptidase McjB and a typical C protein McjC that catalyze the maturation of the precursor McjA [60]. By using McjB and McjC as the query sequence [60,61], the authors found corresponding homologs in B. thailandensis and located the putative tailoring enzymes CapB and CapC with overall similarities of 36% and 38%, respectively. Followed by manually analyzing the adjacent genes, the precursor CapA was identified. Heterologous expression of the putative BGC in E. coli resulted in the production of mature lasso peptide Capistruin (Fig. 6A), which showed antimicrobial activity against Burkholderia and Pseudomonas strains [59]. Such a straightforward sequence similarity-based strategy has been effectively applied to identify and prioritize BGCs for the targeted discovery of RiPPs, including lasso peptides.
Fig. 6.
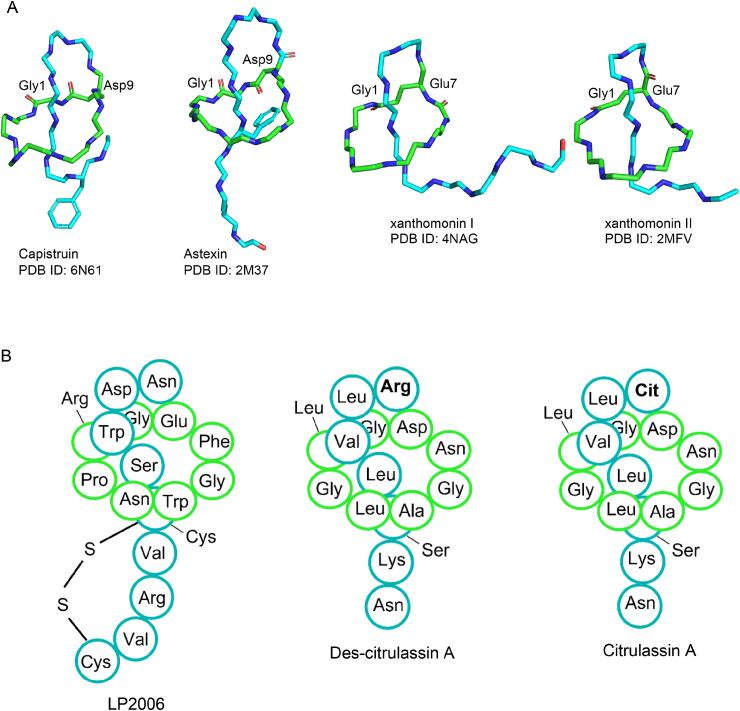
Representative lasso peptides discovered by genome mining. A) Lasso peptides mined by a precursor-centric approach. Astexin (PDB ID 2M37) was isolated from freshwater bacterium Asticcacaulis excentricus. Xanthomonin I and II (PDB ID 4NAG, 2MFV) were derived from Xanthomonas gardneri. The 3D structures of xanthomonin I and II showed here are truncated by four and six residues, respectively. B) LP2006, Des-citrulassin A and Citrulassin A were derived from Nocardiopsis alba NRRL B-24146 and Streptomyces albulus NRRL B-3066, respectively. Arg9 in Des-citrulassin A was modified to citrulline in Citrulassin.
Later in 2012, more comprehensive mining of lasso peptide BGCs was conducted based on conserved motifs of both precursors and maturation enzymes [62]. Based on conservativeness of the penultimate threonine in leader peptides of the known lasso peptides, authors built a preliminary filtering rule for a precursor-centric genome mining, which allows for a more global survey of lasso BGCs. Two known (microcin J25 and capistruin) and nine putative BGCs were used as a MEME (Multiple EM for Motif Elicitation) [63] training set to generate the conserved motifs in McjB/CapB-like and McjC/CapC-like tailoring enzymes. Briefly, four motifs were used as a filter in matching McjB/CapB-related enzymes and three motifs were set as markers for identifying McjC/CapC-like enzyme, including an Asp- and Ser-rich ATP binding pocket [62]. By counting the overall number of conserved motifs present in the predicted precursors and modification enzymes, the potential lasso peptide BGCs were ranked (Fig. 5C). Applying this pattern-based approach, authors conducted a global lasso BGC analysis of 3000 prokaryotic genomes and identified a highly polar lasso peptide astexin-1 with antimicrobial activity against Caulobacter crescentus [62]. A similar pattern-based strategy that searches for the pattern of adenylation domains and tailoring enzymes was also applied to the global genome mining of NRPS from thousands of bacterial genomes, leading to the discovery of novel peptide antibiotics and resistance enzymes [64,65]. Compared to sequence similarity searching, the pattern-based matching is capable of comprehensively analyzing the distribution of a specific type of lasso peptide BGCs among different phyla regardless of their phylogenetic distance. Besides, this strategy can be modified and utilized to mine novel lasso peptides or other types of RiPPs with conserved motifs in precursors or PTM enzymes. For example, a similar approach was used in the genome mining of a unique 7-residues macrolactam ring [66,67]. The McjB protein was used as a query for PSI-BLAST, which resulted in 124 homologs of McjB, and 74 putative precursors were then identified [66]. Of note, 7 out of 74 precursors were found with a Glu residue for potential lasso ring formation at the seventh position, instead of the canonical eighth or ninth position, of the proposed lasso peptide sequence [67]. Heterologous production and crystallization of isolated products revealed Xanthomonins I–III were 7-ring lasso peptides (Fig. 6A).
In contrast to the precursor-centric matching [62], Mitchell's group developed a tailoring-enzyme-based tracing by using RODEO (rapid ORF description and evaluation online), which also mines genes adjacent for the prediction of precursor peptide [14]. In this combinational strategy, the authors first manually curated amino acid sequences of 28 known lasso cyclases, followed by BLAST-P against the NCBI-nr database. When the top 1000 hits were obtained, RODEO was then used to search the gene context for the precursor prediction. The lasso peptide BGC was defined by the presence of genes encoding proteins that match the Pfam HMMs for lasso cyclase (C protein), leader peptidase (B protein), a RiPP recognition element (RRE), and a precursor peptide [14]. A total number of 1419 potential lasso peptide BGCs were identified based on the initial 28 known BGCs, which significantly expand the chemical diversity of lasso peptides. Upon precursors annotation and ranking using a combination of heuristic scoring, motif analysis, and machine learning, a list of high-scoring 1315 precursors were subjected to SSN analysis [27] to visualize their diversity, distribution, and discovery status for BGC prioritization. By examining RODEO predicted BGCs in the public database USDA-ARS (http://nrrl.ncaur.usda.gov/), six BGCs were prioritized and led to the discovery of 5 new lasso peptides, citrulassin A, lagmysin, LP2006, anantin B1 and moomysin [14] (Fig. 6B).
3.3. ω-ester bond containing peptides: a phylogenetic tree-guided profiling approach
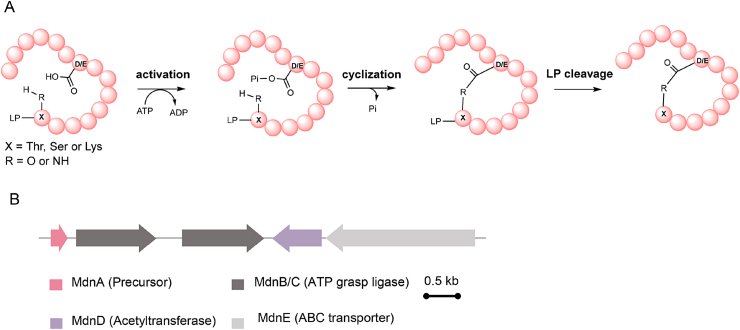
ω-ester bond containing peptides (OEPs) are a class of RiPPs that contain an intramolecular ω-amide or ω-ester bond [[68], [69], [70], [71]]. The formation of ω-amide or ω-ester bond adopts general acid-base chemistry and is similar to that in lasso peptide biosynthesis, both of which are ATP-dependent condensation. The significant differences here are that (i) unlike the formation of AMP-precursor intermediate in lasso peptide maturation, the precursor peptides in OEPs are activated by phosphorylation of the carboxyl side chain of glutamate or aspartate and (ii) condensation exhibits a side-to-side manner in OEP rather than a side-to-end pattern in lasso peptides (Fig. 7A) [58,72]. Side-to-side connection in OEPs is capable of creating topologically complex multi-cyclotides. The first OEP, microviridin, was discovered and isolated from Cyanobacterium Microcystis viridis in 1990 [73]. Until 2008, the biosynthetic pathway of microviridin was identified via analyzing the producer strains M. aeruginosa NIES298 and Microcystis UOWOCCMRC genomic DNA and screening fosmid libraries (Fig. 8) [69,74]. Heterologous production in E. coli proofed that such a tricyclic depsipeptide is ribosomally synthesized and modified by a stand-alone ATP-grasp-type ligase (Fig. 7B) [75].
Fig. 7.
OEPs biosynthetic logic. A) Mechanism of ATP-grasp enzyme-catalyzed ω-ester amide bond formation. X indicates Thr, Ser, or Lys residue. B) Microviridin biosynthetic gene cluster from Microcystis aeruginosa NIES-298.
Fig. 8.
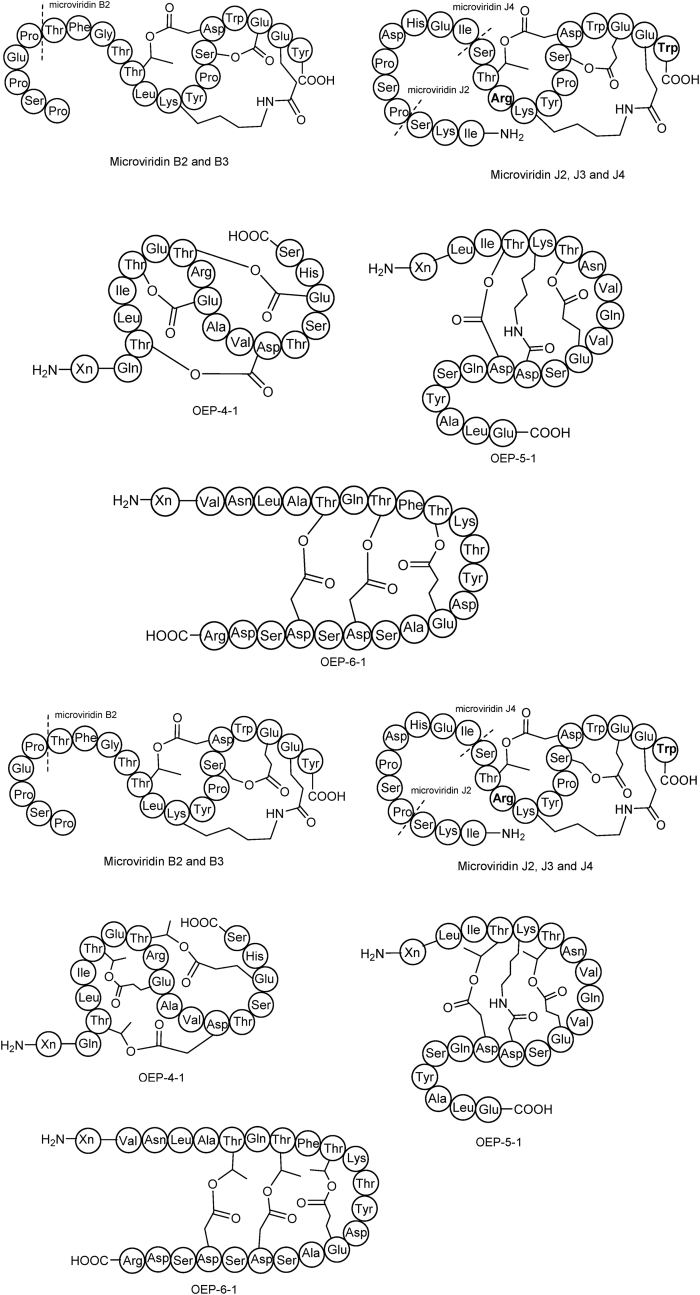
Microviridins and other OEPs discovered by genome mining. Microviridins were isolated via heterologous expression of mdnABCDE gene cluster in E. coli. Arg in C-terminal Trp was highlighted in bold, indicating the differences between microviridin B and microviridin J in their cyclic region. OEP-4-1, OEP-5-1 and OEP-6-1 BGCs were derived from Sphingobacteriales bacterium 44–61, Vibrio sp. JCM 18905 and Chryseobacterium greenlandense UMB34, respectively. The mature peptides were obtained via heterologous expression of the corresponding gene clusters in E. coli.
Based on the deciphered OEPs BGCs, more and more OEPs with diverse topology have been discovered. Seokhee Kim's group disclosed more than 1500 OEPs BGCs and comprehensively analyzed their distributions by SSN- and phylogenetic tree-guided approach [7]. Their initial workflow is similar to that in RODEO, i.e., they manually collected four known OEP BGCs [[70], [71], [72],74] and compiled a list of ATP-grasp enzymes as PSI-BLAST (position-specific iterative basic local alignment) input which returned 5276 unique homology enzymes under the thresh hold of 10∧-35 [7]. An evolutional tree was then constructed and revealed a significant clue for their following mining, i.e., the topologically diverse four know OEPs were catalyzed by four phylogenetically divergent ATP-grasp enzymes [7]. This finding further supported that novel OEPs could be unveiled by retrieving the precursors that adjacent to ATP-grasp proteins and grouping them into different clades in the phylogenetic tree. Phylogenetic tree-guided profiling enabled the identification of 12 groups of OEPs (Fig. 8), including an unprecedented BGC in which a single ATP-grasp enzyme is capable of catalyzing the formation of ω-amide and ω-ester bonds simultaneously [7]. Mining novel enzymes from a phylogenetic perspective is relatively straightforward and powerful, because functionally divergent enzymes among a superfamily are always distributed in different clades. For many other kinds of natural products, such as terpenes, this strategy can also work well in mining novel terpene synthases [76,77]. However, phylogenetic tree-guided approaches may not be suitable for mining RiPPs of novel family. Because strategies based on phylogeny will inevitably generate homologous results, which restrict their capability to find novel RiPPs classes.
3.4. Sactipeptides and ranthipeptides: a radical SAM-centric approach
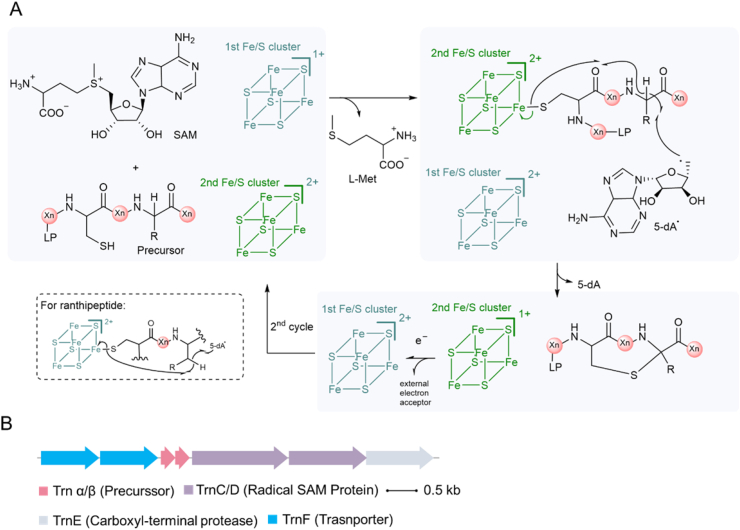
Sactipeptides refer to sulf-linked-to-alpha-carbon peptides. Although both lanthipeptides and sactipeptides contain intramolecular thioether bond, the enzymatic mechanisms involved in the C–S bond formation are different. Lanthipeptidic thioether is formed by a Michael-type addition between a nucleophilic cysteine and an electrophilic β-carbon of the dehydrated residue [46]. Whereas Cα-S bond in sactipeptides is formed via a radical approach-catalyzed by the radical S-adenosylmethionine (SAM) enzymes (RaS) [4,78]. In many other cases, RaS may install a Cβ-S or a Cγ-S thioether linkage [15,79]. In order to structurally distinguish these radical non-α thioether peptides from sactipeptides, they are named as ranthipeptides [15]. The mechanism of C–S bond formation is postulated to be a radical-mediated manner, i.e., (i) 5′-deoxyadenosyl (5-dA) radical is firstly generated through the reductive cleavage of SAM by [4-Fe-4S] cluster of RaS enzyme, (ii) 5′-dA radial is then capable of abstracting a hydrogen atom from α-, β- or γ-carbon, (iii) thioether bridge is formed via intramolecular attack (Fig. 9A) [[80], [81], [82]]. Currently, seven sactipeptides with confirmed BGCs were isolated: subtilosin A [81], thurincin H [75,83], thuricin C/D [84], thuricin Z [85], ruminococcin C [86,87] and sporulation killing factor (Fig. 9B) [80]. The typical components of sactipeptides or ranthipeptides BGC consist of a precursor peptide, a radical SAM protein, a peptidase, and (or) a transporter protein. The varied function of RaS bestows the diverse thioether linkage pattern in mature peptides. Thus, RaS is the most frequently used hallmark in genome mining for sactipeptides and ranthipeptides.
Fig. 9.
Proposed radical-based mechanism of carbon-sulfur bond formation in the biosynthesis of sactipeptides and ranthipeptides. A. In the initial step, 5′-deoxyadenosyl (5-dA) radical is generated through the reductive cleavage of SAM by the first [4Fe-4S] cluster, highlighted as dark cyan. Then, a hydrogen atom was abstracted by 5-dA radical from the precursor peptide, which was coordinated by the second [4Fe-4S] cluster, as highlighted by dark green. Meanwhile, one electron was transferred onto the second cluster via intramolecular attack, leading to the C-S bond formation. The second [4Fe-4S] cluster then transfers one electron to the first [4Fe-4S] cluster to regenerate. For ranthipeptides, side chain hydrogen atom is abstracted, as showed in the dashed box. Two [4Fe-4S] clusters are involved in the overall transformation. Xn, peptide with n residues. LP, leader peptide. B. Thurincin CD biosynthetic gene cluster from Bacillus thuringiensis strain DPC6431.
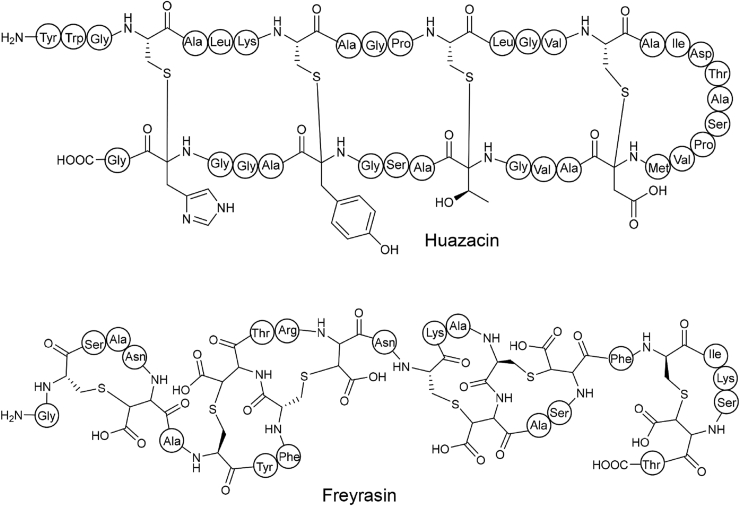
Applying a similar RODEO-based strategy used in lasso peptide discovery, Mitchell's group performed comprehensive mining of RaS enzymes, identified hundreds of canonical Cα-S bond sactipeptides BGCs and several BGC families of non-α thioether RiPPs [15]. The overall mining was radical SAM-centric, i.e., a list of 4600 sactipeptide-related RaS proteins was first retrieved from InterPro 72.0 database [88] using four experimentally confirmed and two putative sactipeptide-related RaS sequences as PSI-BLAST input. The cognate precursors were identified by RODEO 2.0 and subsequent SSN analysis of precursor revealed four popular but uncharacterized sactipeptide groups. Among them, Huazacin was identified from B. thuringiensis (Fig. 10) [15], which was also identified but named as Thuricin Z by Zhang's group at the same time [85]. Notably, the SSN analysis related the RaS of Huazacin BGC to QhpD, a previously characterized quinohemoprotein amine dehydrogenase but not a RiPP maturase. QhpD is known to catalyze several post-translational modifications, including the Cβ- and Cγ-S linkage between Cys-Asp and Cys-Glu, respectively [89,90]. Thus, the unexpected relatedness between RaS and QhpD indicated a new RaS family that may catalyze non-α thioether bond formation [15]. Furthermore, the identification of freyrasin via heterologous expression supported the Cβ-S cross-linkage in freyrasin (Fig. 10) [15]. The genome mining guided discovery of Cβ-S linkage ranthipeptides may provide an important indication that protein or peptide-related PTM enzymes, just like QhpD, could be used as indicators in genome mining of novel RiPPs.
Fig. 10.
Representative sacti- and ranthipeptides discovered by genome mining. Huazacin was isolated from Bacillus thuringiensis serovar huazhongensis. Freyrasin belongs to ranthipeptide which contains six Cβ-S bonds formed between Cys and Asp residues.
3.5. Thioamidated RiPPs and thiopeptide
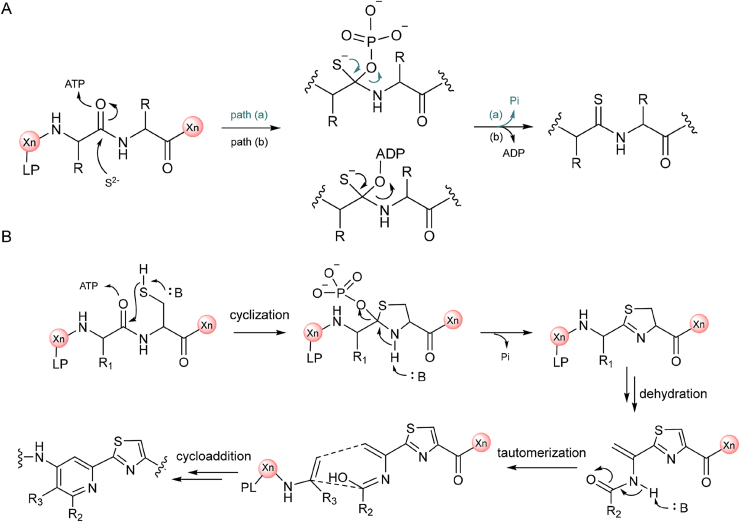
Thioamidated peptides are exceptionally rare in nature. Currently, only a few thioamidation processes have been disclosed, including the biosynthesis of RiPPs thiopeptin [91,92] and thioviridamides [93,94], non-ribosomal peptides costhioamide [95] and post-translational modification of methyl-coenzyme M reductase [96]. In vitro reconstitution of methyl-coenzyme M reductase thioamidation demonstrated that sulfur atom was introduced by ATP-dependent YcaO enzyme (Fig. 11A) [97] and TfuA-like partner protein, which was proposed to serve as precursor peptide recognition element (RRE) in the maturation process [98]. YcaO enzyme and TfuA-like RRE are thus frequently used as hallmarks for genome mining of thioamidated RiPPs, as exemplified by the discovery of Thiostreptamide S4 and Thiovarsolins A-D described below.
Fig. 11.
Proposed enzymatic mechanisms of YcaO-catalyzed thioamide bond and thiazole ring formation in the post modification of RiPPs. A) ATP-dependent YcaO-catalyzed thioamidated RiPPs biosynthesis. B) Mechanisms of thiazole ring and pyridine formation. In the cyclization step, Cys residue can be replaced by Ser or Thr to give other azoline motifs.
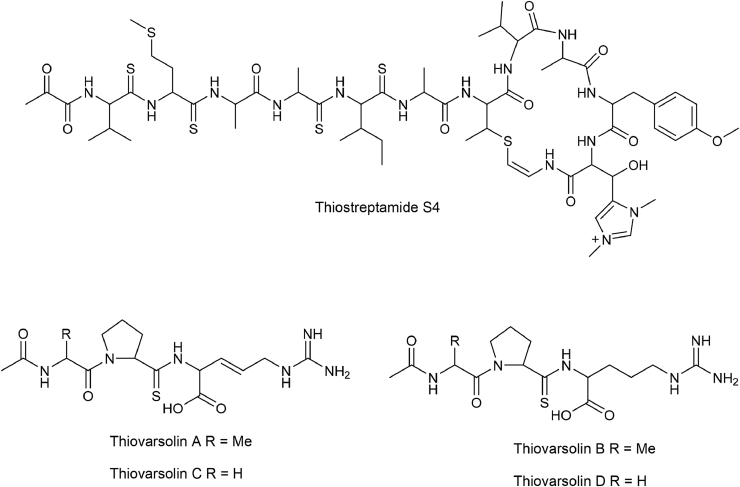
Thioviridamide, which features five contiguous thioamide bonds, was first isolated from Streptomyces olivoviridis culture broth in the course of screening for antitumor agents [93]. Cloning and heterologous expression of the thioviridamide BGC confirmed its ribosomally-synthetic pathway [99]. Thiostreptamide S4 is a thioviridamide-like molecule that was identified by BLAST using the YcaO domain of thioviridamide BGC (Fig. 12) [94]. Thiostreptamide S4 contains four thioamide groups, which are subtly different from thioviridamide. Bioactivity assay revealed that Thiostreptamide S4 exhibited potent antiproliferative activity on tumor cell lines [94]. Besides using YcaO for genome mining, a RiPPER-based comprehensive analysis of TfuA-like protein against available genome data revealed that thioamidated RiPPs were widely spread in actinobacteria. Among those putative candidates, Thiovarsolin BGC was cloned from Streptomyces varsoviensis and heterologously expressed in Streptomyces. coelicolor M1146, which resulted in the identification of a series of linear thioamidated RiPPs Thiovarsolin A-D [38].(Fig. 12).
Fig. 12.
Representative thioamidated RiPPs discovered by genome mining. Thiostreptamide S4 was isolated from Streptomyces olivoviridis NA005001. Thiovarsolin was identified by heterologous expression of Thiovarsolin BGC derived from Streptomyces varsoviensis.
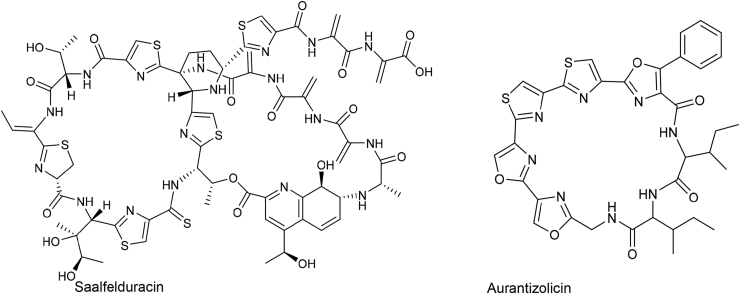
Thiopeptides are a class of azoline ring-rich macrocycles. The biosynthetic machinery is highly conserved among all reported thiopeptides: (i) an Ocin/ThiF-dependent YcaO cyclodehydrates Cys and Ser/Thr to form azoline moiety [97], (ii) a LanB-like dehydratase then dehydrates remained Ser or Thr to yield dehydroalanine and, (iii) an intermolecular [4 + 2] cycloaddition enzyme catalyzes the formation of six-membered pyridine (Fig. 11B) [6,97]. The [4 + 2] Diels−Alderase is the distinctive feature of thiopeptide so that it can be used as a hallmark in genome mining. By targeting the Diels−Alderase via RODEO, Mitchell's group identified several novel thiopeptides and expanded the members of this family [100]. One of the representatives is saalfelduracin (Fig. 13), which is a hybrid of three classes of RiPPs: linear azole-containing peptides, lanthipeptides, and thioamide-containing RiPPs. Aurantizolicin was identified by a classical integrated approach RiPP-PRISM [26]. In addition to the thiazole ring, aurantizolicin contains additional multiple oxazole rings that distinguish it from other thiopeptides (Fig. 13). Other members of aurantizolicin-like RiPPs were classified into YM-216391 [101,102] family.
Fig. 13.
Representative thiopeptides discovered by genome mining. Saalfelduracin was produced by strain Amycolatopsis saalfeldensis NRRL B-24474. Aurantizolicin was isolated from the fermentation broth of Streptomyces aurantiacus JA 4570.
3.6. Other RiPPs: a case-specific biomarker in targeted mining
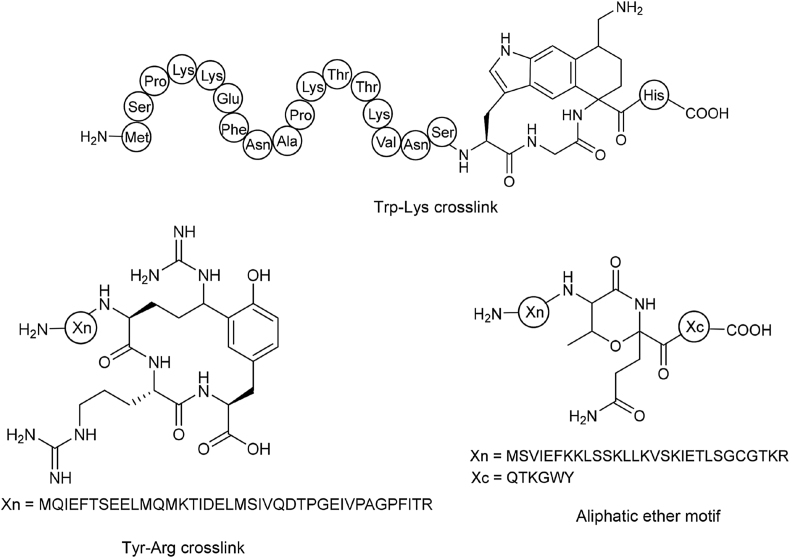
For biosynthesis of well-known lasso peptide, OEPs, lanthipeptides, and sactipeptides, there are either unequivocal conserved residues in precursor peptides or conserved domains in PTM enzymes or both. For some new RiPPs family with little known cases, however, no hallmark gene is available for genome mining. For instance, C–C crosslink [[103], [104], [105]] and aliphatic ether-containing RiPPs [106]. Unlike those well-studied RiPPs families, the enzymes responsible for the C–C or aliphatic ether bond installation of RiPPs are rare and yet to know. Radical SAM enzyme PqqE is capable of catalyzing the C–C crosslink between γ-C of glutamate and tyrosine of pyrroloquinoline quinone cofactor [107] and is the first reported radical SAM enzyme that can modify a linear peptide via C–C cross-coupling. The discovery of Pep1357C [108], renamed as streptide [104] later, is a representative example of the C–C crosslinked RiPPs. Streptide features a lysine-to-tryptophan crosslink and was initially isolated from Streptococcus thermophilus, a gram-positive non-pathogenic strain. In the disclosed streptide biosynthetic gene cluster, a quorum sensing (QS) operon was found in the upstream of precursor peptide [104,108]. Quorum sensing, or cell-cell communication, is the regulation of gene expression in response to fluctuations in cell density [109]. Inactivation of QS operon abolished the Pep1357C biosynthesis in Streptococcus thermophilus [108]. The Cooccurrence of QS operon and Pep1357C-type BGC revealed an inherent and evolutionarily important role of Pep1357C peptide. Inspired by the correlation of QS system and streptide BGC, Seyedsayamdost's group explored 2875 streptococcal genomes and identified 667 putative RiPPs BGCs via pattern-based mining, which contains both SAM and conserved QS locus [105]. 592 streptide-like BGCs were then verified manually and found to be mainly distributed in 16 different groups. Among these16 different groups, a single radical SAM enzyme WgkB was proved to regio- and stereospecifically catalyze two C–C bonds formation between four inactivated positions, resulting in a unique tetrahydro [5,6]benzindole moiety (Fig. 14) [105]. The detailed mechanism regarding how one single-electron oxidant (the 5 dA·) accomplishes a four-electron involving cyclization remains unclear. Another radical SAM enzyme RrrB was reported to cyclize the Cδ of the arginine side chain and ortho-position of tyrosine (Fig. 14) [103]. In addition to C–C crosslink formation, QS system-guided mining also led the discovery of aliphatic ether bond-containing RiPPs, which is formed between the threonine side chain and α-carbon of glutamine (Fig. 14) [106]. This is a rare post-modification which has not been found in other types of RiPPs families. The question regarding why the QS system is adjacent to these RiPPs BGCs and what the biological role of these structurally novel RiPPs awaits further exploration. Nevertheless, the adjacent QS system is a promising specific biomarker in targeted mining RiPPs.
Fig. 14.
Other Radical SAM enzyme-catalyzed RiPPs identified by a quorum sensing-based approach. Streptide-like Trp-Lys and Tyr-Arg crosslink containing RiPPs were respectively found in Streptococcus ferus DSM 20646 and Streptococcus. suis LSS38. The aliphatic ether motif-containing RiPP was identified from Streptococcus suis.
3.7. Emerging universal hallmark: RiPP recognition element
RiPP recognition element (RRE) is regarded as a peptide-binding domain within the RiPP biosynthetic enzymes. Although the specific function and presence of RRE are not well studied, more and more pieces of evidence have supported its prevalence among RiPPs or RiPPs-related biosynthesis. One of the most convincing cases is the PQQ biosynthesis, in which RRE is a stand-alone open reading frame encoded by gene pqqD [110]. A similar architecture is also found in lasso peptides [111] and streptide-like RiPP [105]. Whereas, for other cases, including sactipeptides, RRE may fuse with post-modification enzymes either in the N-terminal or in the C terminal [4]. A comprehensive analysis of RRE by using profile hidden Markov models revealed that RRE is present in more than 50% prokaryotic RiPPs [112,113], including class I lanthipeptides, radical SAM-catalyzed sactipeptides and thiopeptides. RREs are structurally similar but highly divergent in primary sequences. Overall, currently reported RRE domains adopt a similar folding manner: three to four consecutive α-helices followed by three to four consecutive β-sheet, or reverse. Although conserved residues cannot be found among these RRE using routine sequence alignment, such as BLAST, their similarities in 3D structure may be a hidden hallmark that can be developed for RiPPs genome mining. For instance, Medema, van Wezel, and Mitchell's groups recently developed a tool named RRE-finder to discover RREs in protein sequences for the finding of RiPP BGCs [114], which uses pHMMs and secondary structure predictors such as HHpred [112] to address on the 3D structure similarity.
4. Conclusions and perspectives
With the development of new sequencing techniques, more and more bacterial genomes and metagenomes have been sequenced, providing an excellent chance to harness their genetic potential for natural product discovery. In the meantime, the demand for more advanced biosynthetic analysis promotes a large variety of bioinformatics tools for genome mining. Genome mining strategies are now commonplace in natural product discovery. Traditional genome mining of RiPPs based on the distinct PTM enzymes has facilitated the targeted discovery of many RiPPs from different classes over the decades. Tools such as BAGEL and RODEO that were explicitly designed for RiPPs genome mining provided more detailed information and showed more confidence in finding precursor peptides than general genome mining tools. However, these traditional methods rely on the sequence similarity of well-studied proteins of cultivated microbes and thus suffer from the bias issue when applying to metagenome data. Additionally, they are conducted in a gene context-dependent manner and limit their utilization to well-assembled genome data only. The limitations of these genome mining approaches hinder the mining of RiPPs BGCs from the complex microbiome due to the amount, assembling quality, and complexity of metagenomic data.
Nowadays, the increase of DNA sequences of bacterial genomes, especially, metagenomes is staggering. However, mining metagenomic data for natural product discovery is particularly challenging. Most of the current RiPPs genome mining tools described above are not fully-applicable for metagenomic data. Although sophisticated techniques have been developed to assemble the short DNA sequences in the metagenome, fragmentation often occurs from variable coverage of shotgun sequencing. The fragmentation impedes genome mining tools to find the post-modification enzymes or precursors in RiPPs BGCs, or even hinder BGC detection itself. The recent success of deep learning approaches in image recognition and natural language processing has inspired researchers in the fields of genome mining. Deep learning-based genome mining can grasp the hidden essence of RiPPs precursors in a gene context-independent manner. Therefore, deep learning-based strategies require merely the sequences of RiPPs precursors, which are more obtainable from poorly assembled genomes (e.g., metagenome). In a trade-off, however, their prediction accuracies are lower than traditional methods with predefined rules. Currently, deep learning in RiPPs genome mining is still nascent and suffered from bias caused by small data size. As more sophisticated models are invented and more disclosed RiPPs biosynthetic gene clusters are available for training the prediction models, the prediction confidence of one specific type of RiPP would be significantly improved. Owing to data size requirements by deep learning models, the insufficiency in the number of rare RiPPs families will gradually become another bottleneck of applying deep learning in genome mining of novel BGCs. Therefore, researchers in chemistry and biology are expected to collaborate more with those in bioinformatics and computer science. Retaining the virtuous circle of inventing new methods and applying them to discover more RiPPs is envisioned to be the best way to develop new genome mining approaches.
As conventional fermentation-based discovery from cultivated microbe is dwindling, the exploration of the untapped microbiome has risen as a major focal point for new drug discovery. Although so far, no sophisticated metagenome mining tool has been developed specifically for RiPPs, scientists are trying to adapt existing alternating methods to enable metagenome mining for RiPPs from the unexplored microbiome. Recent advances in microbial genomics, metabolomics, and synthetic biology are facilitating us to explore the large microbial world present that is not yet cultured, which represents an unprecedented opportunity for natural product discovery. RiPPs have attracted intense interest owing to their structural and functional diversity and the predictability of the biosynthetic logic of the genetically encoded assembly lines that produce them. The small size of RiPP BGCs (5–15 kb) makes their assembly or synthesis more practical and economical. Their immense genetic and biochemical diversity is only beginning to be appreciated and is becoming a rich source of novel chemical entities for the discovery of more potent drugs. Full leverage the genetic potential of RiPPs for drug discovery will require new efficient discovery approaches and productive interplay between analytical chemistry, computational biology, and synthetic biology. In the future, the integrative application of advanced interdisciplinary technologies, including metabolomics, genomics, AI, and synthetic biology, will promote interdisciplinary collaborations for RiPPs-based drug discovery (Fig. 15). For example, we can envision that the combinational use of deep learning and synthetic biology tools in the discovery stage not only tremendously increases the chance of identification of new antibiotic leads but also unlocks unknown chemical language encoded within the microbiome in shaping the ecosystem or human health. Although bias in the training set is inevitable for current AI-based mining strategies, their capability of identifying RiPPs precursors from metagenome data is promising. Metagenome mining, coupled with cutting-edge synthetic biology strategies, can harness the chemical potential of the untapped microbiome, which offers a new venue for the unexplored RiPPs library with medicinal potential.
Fig. 15.
The flowchart of future RiPPs (meta)genome mining approach.
Acknowledgment
Z. Zhong, B. He, and Y-X Li acknowledge a partial financial support from the National Key R & D Program of China (2018YFA0903200) and Southern Marine Science and Engineering Guangdong Laboratory (Guangzhou) (SMSEGL20SC01). J. Li acknowledges a partial financial support by a National Institutes of Health (NIH) grant P20GM103641 and a National Science Foundation EPSCoR Program OIA-1655740.
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
References
- 1.Imai Y., Meyer K.J., Iinishi A., Favre-Godal Q., Green R. A new antibiotic selectively kills Gram-negative pathogens. Nature. 2019;576:459–464. doi: 10.1038/s41586-019-1791-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang X., Lennard K.R., He C., Walker M.C., Ball A.T. A lanthipeptide library used to identify a protein-protein interaction inhibitor. Nat Chem Biol. 2018;14:375–380. doi: 10.1038/s41589-018-0008-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Delivoria D.C., Chia S., Habchi J., Perni M., Matis I. Bacterial production and direct functional screening of expanded molecular libraries for discovering inhibitors of protein aggregation. Sci Adv. 2019;5 doi: 10.1126/sciadv.aax5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mahanta N., Hudson G.A., Mitchell D.A. Radical S-adenosylmethionine enzymes involved in RiPP biosynthesis. Biochemistry. 2017;vol. 56:5229–5244. doi: 10.1021/acs.biochem.7b00771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reisberg S.H., Gao Y., Walker A.S., Helfrich E.J.N., Clardy J. Total synthesis reveals atypical atropisomerism in a small-molecule natural product. tryptorubin A Science. 2020;367:458–463. doi: 10.1126/science.aay9981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bogart J.W., Bowers A.A. Thiopeptide pyridine synthase TbtD catalyzes an intermolecular formal aza-diels-alder reaction. J Am Chem Soc. 2019;141:1842–1846. doi: 10.1021/jacs.8b11852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee H., Choi M., Park J.U., Roh H., Kim S. Genome mining reveals high topological diversity of omega-ester-containing peptides and divergent evolution of ATP-grasp macrocyclases. J Am Chem Soc. 2020;142:3013–3023. doi: 10.1021/jacs.9b12076. [DOI] [PubMed] [Google Scholar]
- 8.Arnison P.G., Bibb M.J., Bierbaum G., Bowers A.A., Bugni T.S. Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat Prod Rep. 2013;30:108–160. doi: 10.1039/c2np20085f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ongpipattanakul C., Nair S.K. Biosynthetic proteases that catalyze the macrocyclization of ribosomally synthesized linear peptides. Biochemistry. 2018;vol. 57:3201–3209. doi: 10.1021/acs.biochem.8b00114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Veer S.J., Kan M.W., Craik Cyclotides D.J. From structure to function. Chem Rev. 2019;119:12375–12421. doi: 10.1021/acs.chemrev.9b00402. [DOI] [PubMed] [Google Scholar]
- 11.Blin K., Shaw S., Steinke K., Villebro R., Ziemert N. antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019;47:W81–W87. doi: 10.1093/nar/gkz310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Agrawal P., Khater S., Gupta M., Sain N., Mohanty RiPPMiner D. A bioinformatics resource for deciphering chemical structures of RiPPs based on prediction of cleavage and cross-links. Nucleic Acids Res. 2017;45:W80–W88. doi: 10.1093/nar/gkx408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Skinnider M.A., Merwin N.J., Johnston C.W., Magarvey N.A. PRISM 3: expanded prediction of natural product chemical structures from microbial genomes. Nucleic Acids Res. 2017;45:W49–W54. doi: 10.1093/nar/gkx320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tietz J.I., Schwalen C.J., Patel P.S., Maxson T., Blair P.M. A new genome-mining tool redefines the lasso peptide biosynthetic landscape. Nat Chem Biol. 2017;13:470–478. doi: 10.1038/nchembio.2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hudson G.A., Burkhart B.J., DiCaprio A.J., Schwalen C.J., Kille B. Bioinformatic mapping of radical S-Adenosylmethionine-Dependent ribosomally synthesized and post-translationally modified peptides identifies new calpha, cbeta, and cgamma-linked thioether-containing peptides. J Am Chem Soc. 2019;141:8228–8238. doi: 10.1021/jacs.9b01519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ziemert N., Alanjary M., Weber T. The evolution of genome mining in microbes - a review. Nat Prod Rep. 2016;33:988–1005. doi: 10.1039/c6np00025h. [DOI] [PubMed] [Google Scholar]
- 17.de Los Santos NeuRiPP E.L.C. Neural network identification of RiPP precursor peptides. Sci Rep. 2019;9:13406. doi: 10.1038/s41598-019-49764-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Merwin N.J., Mousa W.K., Dejong C.A., Skinnider M.A., Cannon M.J. DeepRiPP integrates multiomics data to automate discovery of novel ribosomally synthesized natural products. Proc Natl Acad Sci U S A. 2020;117:371–380. doi: 10.1073/pnas.1901493116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Begley M., Cotter P.D., Hill C., Ross R.P. Identification of a novel two-peptide lantibiotic, lichenicidin, following rational genome mining for LanM proteins. Appl Environ Microbiol. 2009;75:5451–5460. doi: 10.1128/AEM.00730-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sudek S., Haygood M.G., Youssef D.T., Schmidt E.W. Structure of trichamide, a cyclic peptide from the bloom-forming cyanobacterium Trichodesmium erythraeum, predicted from the genome sequence. Appl Environ Microbiol. 2006;72:4382–4387. doi: 10.1128/AEM.00380-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Jong A., van Hijum S.A., Bijlsma J.J., Kok J., Kuipers Bagel O.P. A web-based bacteriocin genome mining tool. Nucleic Acids Res. 2006;34:W273–W279. doi: 10.1093/nar/gkl237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Heel A.J., de Jong A., Montalban-Lopez M., Kok J., Kuipers Bagel3 O.P. Automated identification of genes encoding bacteriocins and (non-)bactericidal posttranslationally modified peptides. Nucleic Acids Res. 2013;41:W448–W453. doi: 10.1093/nar/gkt391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Heel A.J., de Jong A., Song C., Viel J.H., Kok J. BAGEL4: a user-friendly web server to thoroughly mine RiPPs and bacteriocins. Nucleic Acids Res. 2018;46:W278–W281. doi: 10.1093/nar/gky383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Medema M.H., Blin K., Cimermancic P., de Jager V., Zakrzewski P. antiSMASH: rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res. 2011;39:W339–W346. doi: 10.1093/nar/gkr466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Skinnider M.A., Johnston C.W., Edgar R.E., Dejong C.A., Merwin N.J. Genomic charting of ribosomally synthesized natural product chemical space facilitates targeted mining. Proc Natl Acad Sci U S A. 2016;113:E6343–E6351. doi: 10.1073/pnas.1609014113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gerlt J.A., Bouvier J.T., Davidson D.B., Imker H.J., Sadkhin B. Enzyme Function Initiative-Enzyme Similarity Tool (EFI-EST): a web tool for generating protein sequence similarity networks. Biochim Biophys Acta. 2015;1854:1019–1037. doi: 10.1016/j.bbapap.2015.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Navarro-Munoz J.C., Selem-Mojica N., Mullowney M.W., Kautsar S.A., Tryon J.H. A computational framework to explore large-scale biosynthetic diversity. Nat Chem Biol. 2020;16:60–68. doi: 10.1038/s41589-019-0400-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weber T., Kim H.U. The secondary metabolite bioinformatics portal: computational tools to facilitate synthetic biology of secondary metabolite production. Synth Syst Biotechnol. 2016;1:69–79. doi: 10.1016/j.synbio.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iftime D., Jasyk M., Kulik A., Imhoff J.F., Stegmann E. Streptocollin, a type IV lanthipeptide produced by Streptomyces collinus Tu 365. Chembiochem. 2015;16:2615–2623. doi: 10.1002/cbic.201500377. [DOI] [PubMed] [Google Scholar]
- 31.Weber T. In silico tools for the analysis of antibiotic biosynthetic pathways. Int J Med Microbiol. 2014;304:230–235. doi: 10.1016/j.ijmm.2014.02.001. [DOI] [PubMed] [Google Scholar]
- 32.Blin K., Wolf T., Chevrette M.G., Lu X., Schwalen C.J. antiSMASH 4.0-improvements in chemistry prediction and gene cluster boundary identification. Nucleic Acids Res. 2017;45:W36–W41. doi: 10.1093/nar/gkx319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hyatt D., Chen G.L., Locascio P.F., Land M.L., Larimer F.W. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinf. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Delcher A.L., Bratke K.A., Powers E.C., Salzberg S.L. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics. 2007;23:673–679. doi: 10.1093/bioinformatics/btm009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mukherjee S., van der Donk W.A. Mechanistic studies on the substrate-tolerant lanthipeptide synthetase. ProcM J Am Chem Soc. 2014;136:10450–10459. doi: 10.1021/ja504692v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thibodeaux C.J., Ha T., van der Donk W.A. A price to pay for relaxed substrate specificity: a comparative kinetic analysis of the class II lanthipeptide synthetases ProcM and HalM2. J Am Chem Soc. 2014;136:17513–17529. doi: 10.1021/ja5089452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Walker M.C., Mitchell D.A., van der Donk W.A. 2020. Precursor peptide-targeted mining of more than one hundred thousand genomes expands the lanthipeptide natural product family bioRxiv. 2020.2003.2013.990614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Santos-Aberturas J., Chandra G., Frattaruolo L., Lacret R., Pham T.H. Uncovering the unexplored diversity of thioamidated ribosomal peptides in Actinobacteria using the RiPPER genome mining tool. Nucleic Acids Res. 2019;47:4624–4637. doi: 10.1093/nar/gkz192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mohimani H., Kersten R.D., Liu W.T., Wang M., Purvine S.O. Automated genome mining of ribosomal peptide natural products. ACS Chem Biol. 2014;9:1545–1551. doi: 10.1021/cb500199h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Howard J., Ruder S. arXiv e-prints; 2018. Universal Language model fine-tuning for text classification. arXiv:1801. [Google Scholar]
- 41.Dejong C.A., Chen G.M., Li H., Johnston C.W., Edwards M.R. Polyketide and non-ribosomal peptide retro-biosynthesis and global gene cluster matching. Nat Chem Biol. 2016;12:1007–1014. doi: 10.1038/nchembio.2188. [DOI] [PubMed] [Google Scholar]
- 42.Kautsar S.A., Blin K., Shaw S., Navarro-Munoz J.C., Terlouw B.R. MIBiG 2.0: a repository for biosynthetic gene clusters of known function. Nucleic Acids Res. 2020;48:D454–D458. doi: 10.1093/nar/gkz882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ferir G., Petrova M.I., Andrei G., Huskens D., Hoorelbeke B. The lantibiotic peptide labyrinthopeptin A1 demonstrates broad anti-HIV and anti-HSV activity with potential for microbicidal applications. PloS One. 2013;8 doi: 10.1371/journal.pone.0064010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chatterjee C., Paul M., Xie L., van der Donk W.A. Biosynthesis and mode of action of lantibiotics. Chem Rev. 2005;105:633–684. doi: 10.1021/cr030105v. [DOI] [PubMed] [Google Scholar]
- 45.Mohr K.I., Volz C., Jansen R., Wray V., Hoffmann J. Pinensins: the first antifungal lantibiotics. Angew Chem Int Ed Engl. 2015;54:11254–11258. doi: 10.1002/anie.201500927. [DOI] [PubMed] [Google Scholar]
- 46.Repka L.M., Chekan J.R., Nair S.K., van der Donk W.A. Mechanistic understanding of lanthipeptide biosynthetic enzymes. Chem Rev. 2017;117:5457–5520. doi: 10.1021/acs.chemrev.6b00591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Garg N., Salazar-Ocampo L.M., van der Donk W.A. In vitro activity of the nisin dehydratase NisB. Proc Natl Acad Sci U S A. 2013;110:7258–7263. doi: 10.1073/pnas.1222488110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chatterjee C., Miller L.M., Leung Y.L., Xie L., Yi M. Lacticin 481 synthetase phosphorylates its substrate during lantibiotic production. J Am Chem Soc. 2005;127:15332–15333. doi: 10.1021/ja0543043. [DOI] [PubMed] [Google Scholar]
- 49.Wang H., van der Donk W.A. Biosynthesis of the class III lantipeptide catenulipeptin. ACS Chem Biol. 2012;7:1529–1535. doi: 10.1021/cb3002446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jungmann N.A., van Herwerden E.F., Hugelland M., Sussmuth R.D. The supersized class III lanthipeptide stackepeptin displays motif multiplication in the core peptide. ACS Chem Biol. 2016;11:69–76. doi: 10.1021/acschembio.5b00651. [DOI] [PubMed] [Google Scholar]
- 51.Krawczyk B., Voller G.H., Voller J., Ensle P., Sussmuth Curvopeptin R.D. A new lanthionine-containing class III lantibiotic and its co-substrate promiscuous synthetase. Chembiochem. 2012;13:2065–2071. doi: 10.1002/cbic.201200417. [DOI] [PubMed] [Google Scholar]
- 52.Voller G.H., Krawczyk J.M., Pesic A., Krawczyk B., Nachtigall J. Characterization of new class III lantibiotics--erythreapeptin, avermipeptin and griseopeptin from Saccharopolyspora erythraea. Streptomyces avermitilis and Streptomyces griseus demonstrates stepwise N-terminal leader processing Chembiochem. 2012;13:1174–1183. doi: 10.1002/cbic.201200118. [DOI] [PubMed] [Google Scholar]
- 53.McClerren A.L., Cooper L.E., Quan C., Thomas P.M., Kelleher N.L. Discovery and in vitro biosynthesis of haloduracin, a two-component lantibiotic. Proc Natl Acad Sci U S A. 2006;103:17243–17248. doi: 10.1073/pnas.0606088103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Singh M., Sareen D. Novel LanT associated lantibiotic clusters identified by genome database mining. PloS One. 2014;9 doi: 10.1371/journal.pone.0091352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Walsh C.J., Guinane C.M., Pw O.T., Cotter P.D. A Profile Hidden Markov Model to investigate the distribution and frequency of LanB-encoding lantibiotic modification genes in the human oral and gut microbiome. PeerJ. 2017;5:e3254. doi: 10.7717/peerj.3254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vikeli E., Widdick D.A., Batey S.F.D., Heine D., Holmes N.A. In situ activation and heterologous production of a cryptic lantibiotic from an african plant ant-derived Saccharopolyspora species. Appl Environ Microbiol. 2020;vol. 86 doi: 10.1128/AEM.01876-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nguyen D.D., Wu C.H., Moree W.J., Lamsa A., Medema M.H. MS/MS networking guided analysis of molecule and gene cluster families. Proc Natl Acad Sci U S A. 2013;110:E2611–E2620. doi: 10.1073/pnas.1303471110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hegemann J.D., Zimmermann M., Xie X., Marahiel M.A. Lasso peptides: an intriguing class of bacterial natural products. Acc Chem Res. 2015;48:1909–1919. doi: 10.1021/acs.accounts.5b00156. [DOI] [PubMed] [Google Scholar]
- 59.Knappe T.A., Linne U., Zirah S., Rebuffat S., Xie X. Isolation and structural characterization of capistruin, a lasso peptide predicted from the genome sequence of Burkholderia thailandensis E264. J Am Chem Soc. 2008;130:11446–11454. doi: 10.1021/ja802966g. [DOI] [PubMed] [Google Scholar]
- 60.Duquesne S., Destoumieux-Garzon D., Zirah S., Goulard C., Peduzzi J. Two enzymes catalyze the maturation of a lasso peptide in Escherichia coli. Chem Biol. 2007;14:793–803. doi: 10.1016/j.chembiol.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 61.Yan K.P., Li Y., Zirah S., Goulard C., Knappe T.A. Dissecting the maturation steps of the lasso peptide microcin J25 in vitro Chembiochem. 2012;13:1046–1052. doi: 10.1002/cbic.201200016. [DOI] [PubMed] [Google Scholar]
- 62.Maksimov M.O., Pelczer I., Link A.J. Precursor-centric genome-mining approach for lasso peptide discovery. Proc Natl Acad Sci U S A. 2012;109:15223–15228. doi: 10.1073/pnas.1208978109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bailey T.L., Williams N., Misleh C., Li Meme W.W. Discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006;34:W369–W373. doi: 10.1093/nar/gkl198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li Y.X., Zhong Z., Hou P., Zhang W.P., Qian P.Y. Resistance to nonribosomal peptide antibiotics mediated by D-stereospecific peptidases. Nat Chem Biol. 2018;14:381–387. doi: 10.1038/s41589-018-0009-4. [DOI] [PubMed] [Google Scholar]
- 65.Li Y.X., Zhong Z., Zhang W.P., Qian P.Y. Discovery of cationic nonribosomal peptides as Gram-negative antibiotics through global genome mining. Nat Commun. 2018;9:3273. doi: 10.1038/s41467-018-05781-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hegemann J.D., Zimmermann M., Zhu S., Klug D., Marahiel M.A. Lasso peptides from proteobacteria. Genome mining employing heterologous expression and mass spectrometry Biopolymers. 2013;100:527–542. doi: 10.1002/bip.22326. [DOI] [PubMed] [Google Scholar]
- 67.Hegemann J.D., Zimmermann M., Zhu S., Steuber H., Harms K., Xanthomonins I.-, III A new class of lasso peptides with a seven-residue macrolactam ring. Angew Chem Int Ed Engl. 2014;53:2230–2234. doi: 10.1002/anie.201309267. [DOI] [PubMed] [Google Scholar]
- 68.Rohrlack T., Christoffersen K., Hansen P.E., Zhang W., Czarnecki O. Isolation, characterization, and quantitative analysis of Microviridin J, a new Microcystis metabolite toxic to Daphnia. J Chem Ecol. 2003;29:1757–1770. doi: 10.1023/a:1024889925732. [DOI] [PubMed] [Google Scholar]
- 69.Ziemert N., Ishida K., Liaimer A., Hertweck C., Dittmann E. Ribosomal synthesis of tricyclic depsipeptides in bloom-forming cyanobacteria. Angew Chem Int Ed Engl. 2008;47:7756–7759. doi: 10.1002/anie.200802730. [DOI] [PubMed] [Google Scholar]
- 70.Lee H., Park Y., Kim S. Enzymatic cross-linking of side chains generates a modified peptide with four hairpin-like bicyclic repeats. Biochemistry. 2017;56:4927–4930. doi: 10.1021/acs.biochem.7b00808. [DOI] [PubMed] [Google Scholar]
- 71.Lee C., Lee H., Park J.-U., Kim S. Introduction of bifunctionality into the multidomain architecture of the ω-ester-containing peptide plesiocin. Biochemistry. 2020;59:285–289. doi: 10.1021/acs.biochem.9b00803. [DOI] [PubMed] [Google Scholar]
- 72.Roh H., Han Y., Lee H., Kim S. A topologically distinct modified peptide with multiple bicyclic core motifs expands the diversity of microviridin-like peptides. Chembiochem. 2019;20:1051–1059. doi: 10.1002/cbic.201800678. [DOI] [PubMed] [Google Scholar]
- 73.Ishitsuka M.O., Kusumi T., Kakisawa H., Kaya K., Watanabe Microviridin M.M. A novel tricyclic depsipeptide from the toxic cyanobacterium Microcystis viridis. J Am Chem Soc. 1990;112:8180–8182. doi: 10.1021/ja00178a060. [DOI] [Google Scholar]
- 74.Philmus B., Christiansen G., Yoshida W.Y., Hemscheidt T.K. Post-translational modification in microviridin biosynthesis. Chembiochem. 2008;9:3066–3073. doi: 10.1002/cbic.200800560. [DOI] [PubMed] [Google Scholar]
- 75.Sit C.S., van Belkum M.J., McKay R.T., Worobo R.W., Vederas J.C. The 3D solution structure of thurincin H, a bacteriocin with four sulfur to alpha-carbon crosslinks. Angew Chem Int Ed Engl. 2011;50:8718–8721. doi: 10.1002/anie.201102527. [DOI] [PubMed] [Google Scholar]
- 76.Ye Y., Minami A., Mandi A., Liu C., Taniguchi T. Genome mining for sesterterpenes using bifunctional terpene synthases reveals a unified intermediate of di/sesterterpenes. J Am Chem Soc. 2015;137:11846–11853. doi: 10.1021/jacs.5b08319. [DOI] [PubMed] [Google Scholar]
- 77.Tang M.-C., Lin H.-C., Li D., Zou Y., Li J. Discovery of unclustered fungal indole diterpene biosynthetic pathways through combinatorial pathway reassembly in engineered yeast. J Am Chem Soc. 2015;137:13724–13727. doi: 10.1021/jacs.5b06108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fluhe L., Marahiel M.A. Radical S-adenosylmethionine enzyme catalyzed thioether bond formation in sactipeptide biosynthesis. Curr Opin Chem Biol. 2013;17:605–612. doi: 10.1016/j.cbpa.2013.06.031. [DOI] [PubMed] [Google Scholar]
- 79.Caruso A., Bushin L.B., Clark K.A., Martinie R.J., Seyedsayamdost M.R. Radical approach to enzymatic beta-thioether bond formation. J Am Chem Soc. 2019;141:990–997. doi: 10.1021/jacs.8b11060. [DOI] [PubMed] [Google Scholar]
- 80.Fluhe L., Burghaus O., Wieckowski B.M., Giessen T.W., Linne U. Two [4Fe-4S] clusters containing radical SAM enzyme SkfB catalyze thioether bond formation during the maturation of the sporulation killing factor. J Am Chem Soc. 2013;135:959–962. doi: 10.1021/ja310542g. [DOI] [PubMed] [Google Scholar]
- 81.Fluhe L., Knappe T.A., Gattner M.J., Schafer A., Burghaus O. The radical SAM enzyme AlbA catalyzes thioether bond formation in subtilosin A. Nat Chem Biol. 2012;8:350–357. doi: 10.1038/nchembio.798. [DOI] [PubMed] [Google Scholar]
- 82.Lanz N.D., Booker S.J. Identification and function of auxiliary iron-sulfur clusters in radical SAM enzymes. Biochim Biophys Acta. 2012;1824:1196–1212. doi: 10.1016/j.bbapap.2012.07.009. [DOI] [PubMed] [Google Scholar]
- 83.Mozolewska M.A., Sieradzan A.K., Niadzvedstki A., Czaplewski C., Liwo A. Role of the sulfur to alpha-carbon thioether bridges in thurincin H. J Biomol Struct Dyn. 2017;35:2868–2879. doi: 10.1080/07391102.2016.1234414. [DOI] [PubMed] [Google Scholar]
- 84.Rea M.C., Sit C.S., Clayton E., O'Connor P.M., Whittal R.M. Thuricin CD, a posttranslationally modified bacteriocin with a narrow spectrum of activity against Clostridium difficile. Proc Natl Acad Sci U S A. 2010;107:9352–9357. doi: 10.1073/pnas.0913554107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mo T.L., Ji X.J., Yuan W., Mandalapu D., Wang F.T. Angewandte Chemie-International Edition; 2019. Thuricin Z: a narrow-spectrum sactibiotic that targets the cell membrane. [DOI] [PubMed] [Google Scholar]
- 86.Chiumento S., Roblin C., Kieffer-Jaquinod S., Tachon S., Lepretre C. Ruminococcin C, a promising antibiotic produced by a human gut symbiont. Sci Adv. 2019;5 doi: 10.1126/sciadv.aaw9969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Balty C., Guillot A., Fradale L., Brewee C., Boulay M. Ruminococcin C, an anti-clostridial sactipeptide produced by a prominent member of the human microbiota Ruminococcus gnavus. J Biol Chem. 2019;294:14512–14525. doi: 10.1074/jbc.RA119.009416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Finn R.D., Attwood T.K., Babbitt P.C., Bateman A., Bork P. InterPro in 2017-beyond protein family and domain annotations. Nucleic Acids Res. 2017;45:D190–D199. doi: 10.1093/nar/gkw1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Satoh A., Kim J.K., Miyahara I., Devreese B., Vandenberghe I. Crystal structure of quinohemoprotein amine dehydrogenase from Pseudomonas putida. Identification of a novel quinone cofactor encaged by multiple thioether cross-bridges. J Biol Chem. 2002;277:2830–2834. doi: 10.1074/jbc.M109090200. [DOI] [PubMed] [Google Scholar]
- 90.Datta S., Mori Y., Takagi K., Kawaguchi K., Chen Z.W. Structure of a quinohemoprotein amine dehydrogenase with an uncommon redox cofactor and highly unusual crosslinking. Proc Natl Acad Sci U S A. 2001;98:14268–14273. doi: 10.1073/pnas.241429098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hensens O.D., Albers-Schonberg G. Total structure of the highly modified peptide antibiotic components of thiopeptin. J Antibiot (Tokyo) 1983;36:814–831. doi: 10.7164/antibiotics.36.814. [DOI] [PubMed] [Google Scholar]
- 92.Puar M.S., Ganguly A.K., Afonso A., Brambilla R., Mangiaracina P. Sch 18640. A new thiostrepton-type antibiotic. J Am Chem Soc. 1981;103:5231–5233. doi: 10.1021/ja00407a047. [DOI] [Google Scholar]
- 93.Hayakawa Y., Sasaki K., Adachi H., Furihata K., Nagai K. Thioviridamide, a novel apoptosis inducer in transformed cells from Streptomyces olivoviridis. J Antibiot (Tokyo) 2006;59:1–5. doi: 10.1038/ja.2006.1. [DOI] [PubMed] [Google Scholar]
- 94.Frattaruolo L., Lacret R., Cappello A.R., Truman A.W. A genomics-based approach identifies a thioviridamide-like compound with selective anticancer activity. ACS Chem Biol. 2017;12:2815–2822. doi: 10.1021/acschembio.7b00677. [DOI] [PubMed] [Google Scholar]
- 95.Dunbar K.L., Dell M., Molloy E.M., Kloss F., Hertweck C. Reconstitution of iterative thioamidation in closthioamide biosynthesis reveals tailoring strategy for nonribosomal peptide backbones. Angew Chem Int Ed Engl. 2019;58:13014–13018. doi: 10.1002/anie.201905992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Nayak D.D., Mahanta N., Mitchell D.A., Metcalf W.W. Post-translational thioamidation of methyl-coenzyme M reductase, a key enzyme in methanogenic and methanotrophic Archaea. Elife. 2017;6 doi: 10.7554/eLife.29218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Burkhart B.J., Schwalen C.J., Mann G., Naismith J.H., Mitchell D.A. YcaO-dependent posttranslational amide activation: biosynthesis, structure, and function. Chem Rev. 2017;117:5389–5456. doi: 10.1021/acs.chemrev.6b00623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mahanta N., Liu A., Dong S., Nair S.K., Mitchell D.A. Enzymatic reconstitution of ribosomal peptide backbone thioamidation. Proc Natl Acad Sci U S A. 2018;115:3030–3035. doi: 10.1073/pnas.1722324115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Izawa M., Kawasaki T., Hayakawa Y. Cloning and heterologous expression of the thioviridamide biosynthesis gene cluster from Streptomyces olivoviridis. Appl Environ Microbiol. 2013;79:7110–7113. doi: 10.1128/AEM.01978-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Schwalen C.J., Hudson G.A., Kille B., Mitchell D.A. Bioinformatic expansion and discovery of thiopeptide antibiotics. J Am Chem Soc. 2018;140:9494–9501. doi: 10.1021/jacs.8b03896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Pei Z.F., Yang M.J., Li L., Jian X.H., Yin Y. Directed production of aurantizolicin and new members based on a YM-216391 biosynthetic system Org. Biomol Chem. 2018;16:9373–9376. doi: 10.1039/c8ob02665c. [DOI] [PubMed] [Google Scholar]
- 102.Jian X.H., Pan H.X., Ning T.T., Shi Y.Y., Chen Y.S. Analysis of YM-216391 biosynthetic gene cluster and improvement of the cyclopeptide production in a heterologous host. ACS Chem Biol. 2012;7:646–651. doi: 10.1021/cb200479f. [DOI] [PubMed] [Google Scholar]
- 103.Caruso A., Martinie R.J., Bushin L.B., Seyedsayamdost M.R. Macrocyclization via an arginine-tyrosine crosslink broadens the reaction scope of radical S-adenosylmethionine enzymes. J Am Chem Soc. 2019;141:16610–16614. doi: 10.1021/jacs.9b09210. [DOI] [PubMed] [Google Scholar]
- 104.Schramma K.R., Bushin L.B., Seyedsayamdost M.R. Structure and biosynthesis of a macrocyclic peptide containing an unprecedented lysine-to-tryptophan crosslink. Nat Chem. 2015;7:431–437. doi: 10.1038/nchem.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bushin L.B., Clark K.A., Pelczer I., Seyedsayamdost M.R. Charting an unexplored streptococcal biosynthetic landscape reveals a unique peptide cyclization motif. J Am Chem Soc. 2018;140:17674–17684. doi: 10.1021/jacs.8b10266. [DOI] [PubMed] [Google Scholar]
- 106.Clark K.A., Bushin L.B., Seyedsayamdost M.R. Aliphatic ether bond formation expands the scope of radical SAM enzymes in natural product biosynthesis. J Am Chem Soc. 2019;141:10610–10615. doi: 10.1021/jacs.9b05151. [DOI] [PubMed] [Google Scholar]
- 107.Barr I., Latham J.A., Iavarone A.T., Chantarojsiri T., Hwang J.D. Demonstration that the radical S-adenosylmethionine (SAM) enzyme PqqE catalyzes de Novo carbon-carbon cross-linking within a peptide substrate PqqA in the presence of the peptide chaperone PqqD. J Biol Chem. 2016;291:8877–8884. doi: 10.1074/jbc.C115.699918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ibrahim M., Guillot A., Wessner F., Algaron F., Besset C. Control of the transcription of a short gene encoding a cyclic peptide in Streptococcus thermophilus: a new quorum-sensing system? J Bacteriol. 2007;189:8844–8854. doi: 10.1128/JB.01057-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bassler B.L. Small talk. Cell-to-cell communication in bacteria Cell. 2002;109:421–424. doi: 10.1016/s0092-8674(02)00749-3. [DOI] [PubMed] [Google Scholar]
- 110.Latham J.A., Iavarone A.T., Barr I., Juthani P.V., Klinman J.P. PqqD is a novel peptide chaperone that forms a ternary complex with the radical S-adenosylmethionine protein PqqE in the pyrroloquinoline quinone biosynthetic pathway. J Biol Chem. 2015;290:12908–12918. doi: 10.1074/jbc.M115.646521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Chekan J.R., Ongpipattanakul C., Nair S.K. Steric complementarity directs sequence promiscuous leader binding in RiPP biosynthesis. Proc Natl Acad Sci U S A. 2019;116:24049–24055. doi: 10.1073/pnas.1908364116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Soding J., Biegert A., Lupas A.N. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244–W248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Burkhart B.J., Hudson G.A., Dunbar K.L., Mitchell D.A. A prevalent peptide-binding domain guides ribosomal natural product biosynthesis. Nat Chem Biol. 2015;11:564–570. doi: 10.1038/nchembio.1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kloosterman A.M., Shelton K.E., van Wezel G.P., Medema M.H., Mitchell Rre-Finder D.A. 2020. A genome-mining tool for class-independent RiPP discovery bioRxiv. 2020.2003.2014.992123. [DOI] [PMC free article] [PubMed] [Google Scholar]