Abstract
Background
Long-term antiretroviral therapy (ART) enables people living with HIV (PLW-HIV) to be healthier and live longer; though they remain at greater risk of pneumonia and chronic lung disease than the general population. Lung microbial dysbiosis has been shown to contribute to respiratory disease.
Methods
16S-rRNA gene sequencing on the Miseq-platform and qPCR for typical respiratory pathogens were performed on sputum samples collected from 64 PLW-HIV (median blood CD4 count 676 cells/μL) and 38 HIV-negative participants.
Finding
Richness and α-diversity as well as the relative-abundance (RA) of the major taxa (RA>1%) were similar between both groups. In unweighted-Unifrac ß-diversity, the samples from PLW-HIV showed greater diversity, in contrast to the HIV negative samples which clustered together. Gut bacterial taxa such as Bilophila and members of Enterobacteriaceae as well as pathogenic respiratory taxa (Staphylococcus, Pseudomonas and Klebsiella) were significantly more frequent in PLW-HIV and almost absent in the HIV-negative group. Carriage of these taxa was correlated with the length of time between HIV diagnosis and initiation of ART (Spearman-rho=0·279, p=0·028).
Interpretation
Although the core airway microbiome was indistinguishable between PLW-HIV on effective ART and HIV-negative participants, PLW-HIV's respiratory microbiome was enriched with potential respiratory pathogens and gut bacteria. The observed differences in PLW-HIV may be due to HIV infection altering the local lung microenvironment to be more permissive to harbour pathogenic bacteria that could contribute to respiratory comorbidities. Prompt start of ART for PLW-HIV may reduce this risk.
Keywords: Microbiota, Bacteria/Classification, 16S rRNA sequencing, HIV, Infection, Respiratory
Research in context.
Evidence before this study
Effective antiretroviral therapy (ART) provides HIV control and host immune reconstitution such that people living with HIV (PLW-HIV) on ART are healthier and have a near normal life span. However, PLW-HIV remain at greater risk of pneumonia as well as chronic lung disease including Chronic Obstructive Pulmonary Disease (COPD).
Evidence demonstrates a significant role for the lung microbiome in determining respiratory health, and how microbial dysbiosis contributes to the prognosis of chronic lung disease.
HIV infection impairs pulmonary adaptive and innate immunity which in turn alters the airway microbiome by reducing the richness and alpha diversity of the airway microbial communities, and enriching abundance of some signature bacteria such as Tropheryma whipplei and some taxa such as Prevotella and Veillonella that may contribute to chronic lung inflammation. However, with effective ART these deviations in the microbiome are thought to be alleviated
Added value of this study
Although the core airway microbiome was indistinguishable between PLW-HIV using antiretroviral therapy and a HIV negative comparator population, gut bacteria like Bilophila and members of Enterobacteriaceae and potential respiratory pathogens such as Staphylococcus, Pseudomonas and Klebsiella were more frequently observed in PLW-HIV, while these bacteria were almost absent in the comparator group. Their presence was associated with the amount of time PLW-HIV had spent off ART.
Implications of all the available evidence
The enrichment of the respiratory microbiome with potential respiratory pathogens and gut bacteria may contribute to the greater incidence of pneumonia and chronic lung disease in PLW-HIV despite ART.
Alt-text: Unlabelled box
1. Introduction
Effective antiretroviral therapy (ART) has changed the natural history of HIV infection. People living with HIV (PLW-HIV) and using ART now have near-normal life expectancy [1]. HIV complications have also shifted from acute opportunistic infections and neoplasms to non-AIDS related comorbidities [2]. Despite the reduction in respiratory infections such as Pneumocystis jirovecii pneumonia, bacterial pneumonia and tuberculosis with effective ART [3,4], respiratory illness remains common. PLW-HIV appear at higher risk of developing chronic lung disease such as chronic obstructive pulmonary disease (COPD), pulmonary hypertension and lung cancer [5].
Recent studies have demonstrated that the microbiome has an important role in determining respiratory health and can be a key player in the pathogenesis and prognosis of inflammatory chronic lung disease [6,7]. Through the impact of HIV on innate and adaptive immunity in the lungs, the potential exists to alter the lung microenvironment and affect the homeostasis of the microbial communities within it [8].
Current evidence suggests that, without ART, HIV infection alters the airway microbiome to reduce the richness and alpha diversity (a measurement of the taxa diversity within each sample) [9] of the microbial communities in the airways, and to enrich signature bacteria such as Tropheryma whipplei and some taxa such as Prevotella and Veillonella that may contribute to lung inflammation [10], [11], [12], [13]. Effective ART appears to reverse these changes: data from the Lung HIV Microbiome Project suggests that the lung microbiome in healthy PLW-HIV on ART with preserved blood CD4 counts is similar to uninfected individuals [14]. The importance of the environment on the microbiome (and hence the need for comparator populations) has been shown by Iwai et al. who compared the airway microbiome between two similar cohorts of PLW-HIV admitted to hospital with pneumonia in San Francisco and Uganda. They found that the airway microbiome of each cohort was distinct - which they attributed to local environmental factors [15,16].
In this paper, using 16S rRNA gene sequencing we describe the compositional differences in the airway microbiome of a cohort of adult PLW-HIV with high blood CD4 counts on ART and HIV uninfected participants within the UK. We then add to this data by specifically evaluating the presence of three pathogenic respiratory species with targeted quantitative PCR.
2. Methods
2.1. Study design and population
The present work is part of a prospective observational cohort study conducted in the Royal Free London NHS Foundation Trust comparing the incidence of acute respiratory illness in PLW-HIV and HIV negative individuals. The study was approved by the London Hampstead Research Ethics Committee (14/LO/1409) and registered with the ISRCTN registry (ISRCTN38386321). Participants were recruited between November 2015 and January 2017. PLW-HIV were invited to participate when they attended ambulatory HIV care appointments. The only eligibility criteria were age over 18 years, consent to participate, and absence of symptoms of acute respiratory illness at study entry. HIV negative participants were recruited from Sexual Health clinics and Primary Care, using primary care records to invite potential participants with similar age, gender, and smoking status to the expected characteristics of the HIV positive population sampled. HIV negative participants had their HIV status confirmed by blood test at recruitment. All participants provided written informed consent. Participants were invited to provide a sputum sample for analysis at the baseline visit. Sputum samples that were collected from participants who could expectorate were used in the present analysis. Sputum samples were stored at −80 °C within 3 h.
Details on methods are given in the online data supplement. Briefly, sputum samples were treated with Sputasol® (Oxoid, UK) then were heated at 95 °C for 30 min. One millilitre of each sample was centrifuged at 13,000xg for 10 min; the pellets were disrupted with glass beads on Fast-Prep®-24 Instrument for 45 s. Metagenomic DNA was then extracted on the automated DiaSorin® Ixt extraction platform using DiaSorin® Arrow DNA extraction kit.
The bacterial loads of Pseudomonas aeruginosa [17] and Haemophilus influenzae, Moraxella catarrhalis and Streptococcus pneumoniae [18] were quantified using two quantitative-PCRs (qPCR) methods on Qiagen Rotor– gene® 6000 real-time PCR machine (Corbett Research UK, Cambridgeshire, UK). The bacterial load of each tested organism was calculated in colony forming unit (CFU) per mL of sputum and a mean of technical triplicates was taken for each sample. An internal amplification control, SPUD A, was used at final concentration of 0·04 pM to test for PCR inhibition [19].
A sequence library was created by amplification of V3-V4 regions of the bacterial 16S rRNA gene through conventional PCR on the extracted metagenomic DNA using 341 forward primer and 805 reverse primer. Each sample was assigned a unique pair combination of standard Illumina® dual indexed primers (with adaptors attached). The PCR products (577bp amplicons) were cleaned up using Agencourt AMPure XP beads (Beckman Coulter, UK). The samples were pooled in an equimolar ratio at 5 nM into one library which was then checked on bioanalyzer. Sequencing was performed using Illumina MiSeq Platform using costume sequencing primers, MiSeq® Reagent Kit v2 (500 cycles). Additional details on the PCRs and primers sequences are provided in the online data supplement. Sequence data are deposited in the European Nucleotide Archive (ENA), study accession number is PRJEB32399.
An extraction negative control and a no-template PCR control (water) were run alongside each batch of samples throughout the process as negative controls to allow for the evaluation of potential contamination. A laboratory prepared mock community was run as a positive control.
2.2. Bioinformatics and statistical analysis
In bioinformatic analysis we adopted the workflow established by Microbiome helper [20] using QIIME pipeline v1.9.1. [21] where the sequences were clustered based on 97% similarity into Operational Taxonomic Units (OTU) and taxonomic classification was assigned to OTUs using open reference OTU picking against Greengenes database version 13_8. The OTU table was then rarefied per sample to 20,000. Alpha and beta diversity indices were calculated on the rarefied OTU table. The appropriate statistical significance tests were calculated using SPSS v. 23 or QIIME wrapper scripts. STAMP (v2.1.3) [22] was used to visualize the results and explore the OTUs showing significant differences in relative abundance between the two study groups using White's non-parametric t-test [23] and all p-values were corrected using Benjamini–Hochberg False Discovery Rate (FDR) method for multiple comparisons.
2.3. Role of funding source
None of the funders had any role in paper design, data collection, data analysis, interpretation, or writing of the paper.
3. Results
3.1. Clinical and demographic Cohort characteristics
Overall 102 participants out of 217 in the wider cohort provided sputum samples at baseline assessment. There were no significant differences between those who could and couldn't expectorate, with respect to airflow obstruction (p = 0·34), blood CD4 count (p = 0·95), respiratory symptoms (defined by St Georges Respiratory Questionnaire (SGRQ) total score) (p = 0·16), age (p = 0·16), smoking status (p = 0·127) nor HIV status (p = 0·59) as similar proportion of PLW-HIV (45%) and controls (49%) produced sputum. Sputum samples collected from 64 PLW-HIV using ART and 38 HIV negative participants were included (Table 1). All participants were free of symptoms of acute respiratory illness (more details in the online data supplement). The median length of time between HIV diagnosis and starting ART was 1342 days (IQR 3131-448 days).
Table 1.
Demographic and clinical characteristics of participants.
| Characteristics | HIV Positive (n=64) | HIV Negative (n=38) | p-value | |
|---|---|---|---|---|
| Agea(years) | 52 (10·47) | 53 (7·76) | 0·594d | |
| Sexc | Males | 55 (86%) | 29 (76%) | 0·218e |
| Females | 9 (14%) | 9 (24%) | ||
| Sexualityc | Heterosexual | 13 (20%) | 29 (76%) | <0·001e |
| Homosexual | 42 (66%) | 9 (24%) | ||
| Bisexual | 9 (14%) | 0 | ||
| Ethnicityc | Caucasian | 51 (80%) | 36 (95%) | 0·108f |
| Black (African or Caribbean) | 7 (11%) | 0 | ||
| Asian | 2 (3%) | 1 (3%) | ||
| Others | 4 (6%) | 1 (3%) | ||
| Educationc | University Degree | 28 (46%) | 20 (54%) | 0·912f |
| Secondary Education or equivalent qualifications | 21(34%) | 10(28%) | ||
| No Qualification | 4 (7%) | 2 (5%) | ||
| Others | 8 (13%) | 5 (14%) | ||
| BMIa | 26·18 (4·93) | 25·04 (2·91) | 0·789d | |
| Self-reported Comorbiditiesc | Asthma | 8 (21%) | 5 (36%) | 0·292f |
| COPD | 2 (5%) | 1 (7%) | 1·000f | |
| Cancer | 7 (18%) | 1 (7%) | 0·665f | |
| Heart Disease | 5 (13%) | 0 (0%) | 0·309f | |
| Diabetes | 4 (10%) | 0 (0%) | 0·563f | |
| Smokingc | Current Smoker | 25 (39%) | 4 (11%) | 0·009e |
| Ex-smoker | 21 (33%) | 16 (43%) | ||
| Never smoked | 18 (28%) | 17 (46%) | ||
| Recreational drug use (ever)c | 48 (75%) | 20 (57%) | 0·067e | |
| Recreational drug use (last 3 months)c | 25 (39%) | 5 (14%) | 0·007e | |
| CD4 count (cells/µL)b | 676 (880-467) | |||
| Nadir CD4 (cells/µL)b | 225 (371-107) | |||
| CD8 count (cells/µL)b | 1030 (1377-723) | |||
| CD4/CD8 Ratiob | 0·67 (0.92-0·43) | |||
| Subjects with Blood HIV Viral load < 40 copies/mLc | 54 (84%) | |||
| Blood HIV Viral load >40 copies/mLc Viral load (copies/mL) b | 10 (16%) 122 (418-71) | |||
| Prescribed Antiretroviral therapy c | 63 (100%) | |||
| Prescribed Antiretroviral therapy (years)b | 10·5 (15-5) | |||
| Prescribed Antiretroviral therapy over 10 yearsc | 32 (53%) | |||
| FEV1 (L)b | 3·28 (3·79-2·77) | 3·62 (4·09-2·84) | 0·132d | |
| FEV1 z-scoreb | -0·53 (-1·18-0·07) | -0·35 (-0·85-0·32) | 0·115d | |
| FVC (L)b | 4·30 (4·74-3·37) | 4·61 (5·27-3·78) | 0·061d | |
| FVC % predictedb | 93% (99·8-81·2) | 98% (110-98) | 0·008d | |
| Airflow Obstructiong,c | 10 (16%) | 2 (5%) | 0·202f | |
| Restrictive Airflowh,c | 14 (22%) | 3 (8%) | 0·062e | |
| Inhaled respiratory medicationc | 10 (16%) | 4 (11%) | 0·500e | |
| ß2 Agonistc | 5 (8%) | 2 (6%) | 1·00f | |
| ICSc | 4 (7%) | 0 (0%) | 0·293f | |
Mean (SD)
Median (IQR)
n (%)
p-value by Mann-Whitney Test
p-value by Chi square
p-value by Fisher Exact Test
FEV1/FVC <0·7
FEV1/FVC ≥0·7 and FVC< 80% predicted
BMI: Body Mass Index
FEV1: Forced Expiratory volume in 1 sec
FVC: Forced Vital Capacity
ICS: Inhaled Corticosteroids
Fifty-three percent of PLW-HIV had been taking ART for over 10 years and 84% had undetectable HIV plasma load (VL) (<40 copies/mL). Ten participants (16%) had detectable VL (median 122 copies/mL, IQR 418-71). The median blood CD4 count was 676 cells/ µL (IQR 880-467 cells/µL); whilst the nadir CD4 was 225 cells/µL (IQR 371-107 cells/µL) of which 44% had nadir CD4 < 200 cells/µL.
No significant differences were found between the two groups regarding age which ranged between 30 and 74 years old, sex, ethnicity, educational level, body mass index (BMI), and self-reported co-morbidities; e.g. asthma COPD, heart disease, cancer and diabetes. However, current tobacco smoking and recent use of recreational drugs were significantly higher in PLW-HIV where the prevalence was 3·5 and 3 times that of controls respectively.
The spirometry results were normal for most participants; though, the percentage of FVC predicted was significantly lower in the HIV positive group (Median 93% vs 98% in the comparator group, p<0·01). Although no statistically significant differences were detected in terms of obstructive airflow which we defined as FEV1/FVC <0·7, or restrictive airflow which we defined as FEV1/FVC >0·7 and FVC< 80% predicted, airflow obstruction was three times higher among PLW-HIV.
3.2. Bacterial communities composition
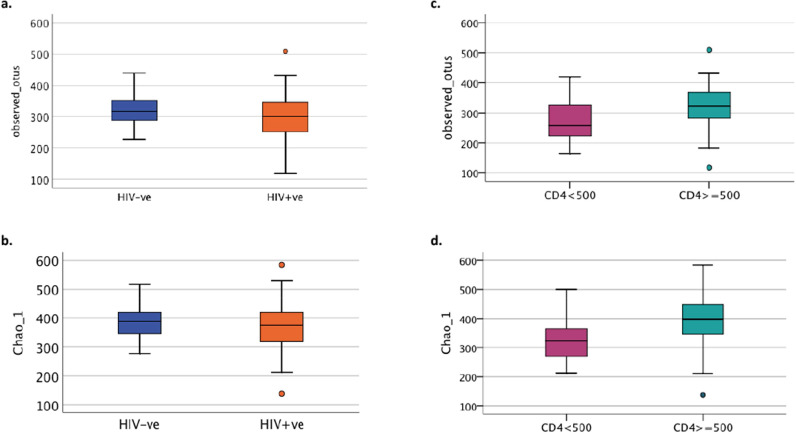
All richness and alpha-diversity indices such as number of observed OTUs or Chao1 (Fig. 1a and 1b respectively) as well as PD whole tree, Fisher alpha, Shannon and Simpson diversity indices (data not shown) were not significantly different between PLW-HIV and HIV uninfected participants. However, within the HIV positive group the number of observed OTUs, Chao1 (Fig. 1c and 1d respectively), PD whole tree and Fisher alpha (data not shown) were significantly lower in participants whose blood CD4 count was <500 cells/µL.
Fig. 1.
Comparison of Richness and alpha diversity of microbial communities in sputum with respect to HIV status and CD4 blood count (a) Richness measured by number of observed OTU (p=0·247, t-test) and (b) α-diversity by Chao1 (p=0·216, t-test) in HIV negative and HIV positive groups, Within the HIV positive group (c) the richness by number of observed OTU (p=0·026, t-test) and (d) α-diversity by Chao1 (p=0·013, t-test) were significantly lower in participants with CD4 count less than 500 cells/µL. Group size: 38 HIV-ve group and 64 HIV +ve group; 22 CD4<500 and 42 CD4≥ 500.
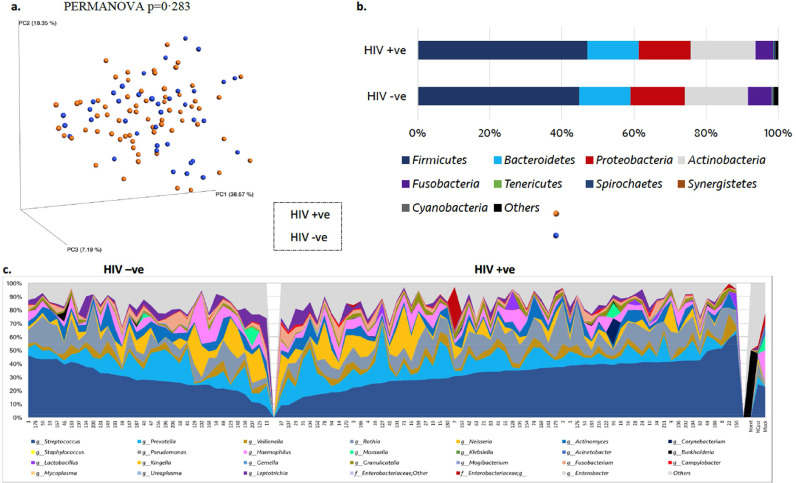
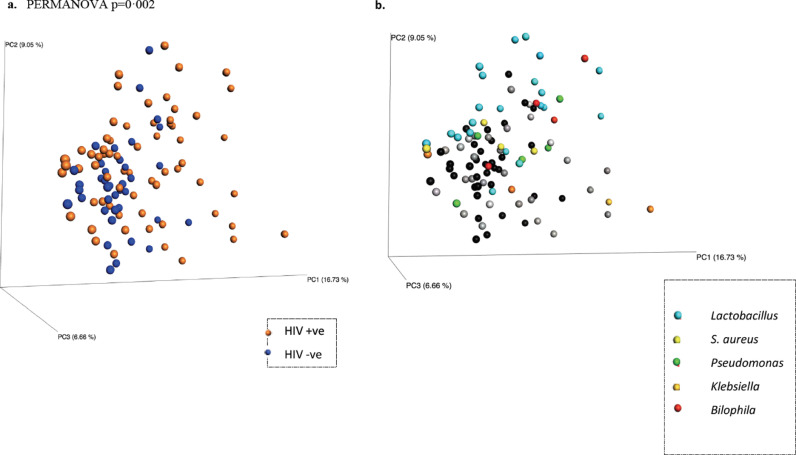
In principal co-ordinate analysis (PCoA) of weighted Unifrac ß-diversity index (Fig. 2a), the samples did not cluster by HIV status (PERMANOVA p = 0·283). In addition, the core taxa constituting the microbiome profiles in sputum at both the phylum and genus level (Fig. 2b and 2c respectively) were very similar between the HIV positive and negative groups, and no significant differences in the relative abundances (RA) of the highly abundant taxa whose RA>1% were detected. Out of 782 OTUs that were classified into 252 taxa, only 20 were present at RA >1%, and they were present in all samples. Nevertheless, the samples were distinguished based on the HIV infection status in unweighted Unifrac ß-diversity index (PERMANOVA p = 0·002), which unlike the weighted Unifrac is more sensitive to the presence or absence of taxa regardless of the weight of their relative abundances. In PCoA of unweighted Unifrac, the controls comparator group formed a cluster while PLW-HIV the HIV positive group were more dispersed, depending on the presence of some minor taxa (Fig. 3).
Fig. 2.
Microbiome Profile of HIV Negative and Positive groups (a) PCoA of weighted Unifrac ß-diversity index showing no separation with respect to the HIV status (p=0·283, PERMANOVA) (b) microbiome profile at the phylum level (p>0·05, White non-parametric t-test) (c) microbiome profile at the genus level. NCext: extraction negative control, NCpcr: no-template PCR negative control, Mock: laboratory prepared mock community. Group size: 38 HIV-ve group and 64 HIV +ve group.
Fig. 3.
PCoA plot of unweighted Unifrac ß-diversity (p=0.002, PERMANOVA). a) With respect to HIV status, Blue: HIV –ve group (n=38), orange: HIV positive (n=64). b) With respect to the presence of pathogenic taxa: S. aureus (yellow), Pseudomonas (green), Klebsiella (orange), Lactobacillus (light blue), Bilophila: (red), otherwise the free HIV-ve samples are black and the free HIV positive samples are grey
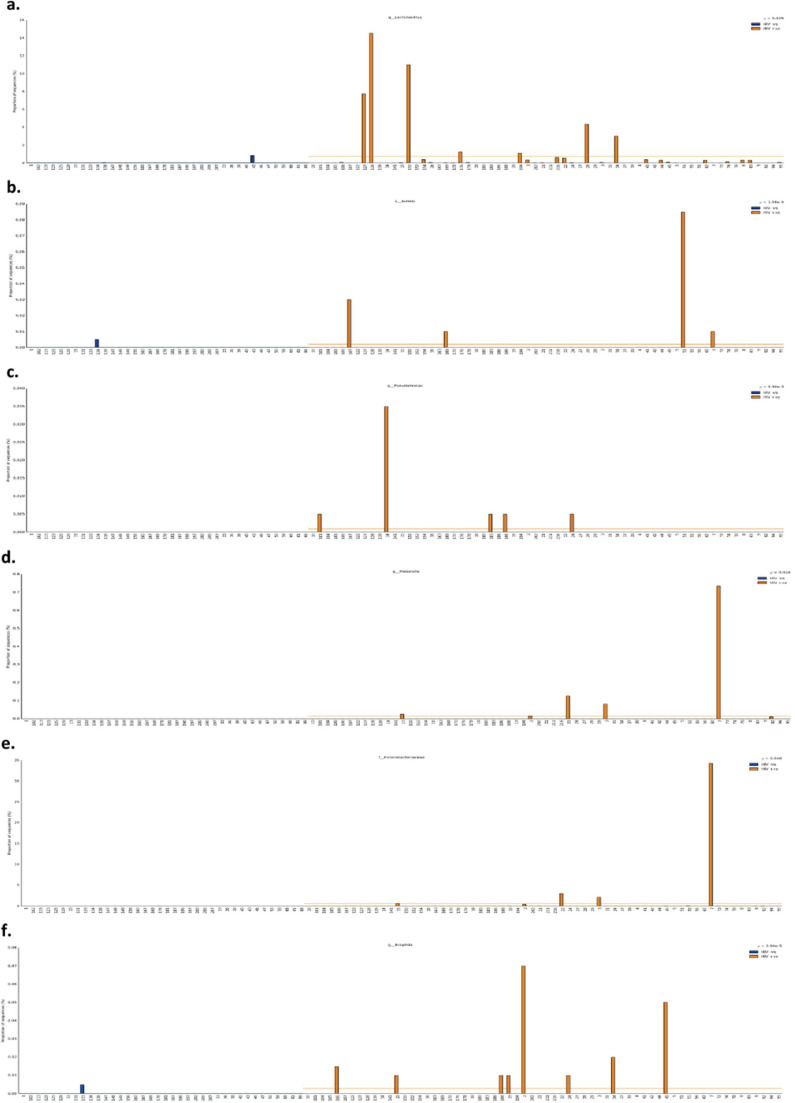
Looking more closely at the microbiome profile, some low abundant taxa such as Staphylococcus aureus (p = 0·0016), Pseudomonas (p = 0·0093), genus Lactobacillus (p = 0·029), Klebsiella (p = 0·024), Bilophila (p < 0·0001), and family Enterobacteriaceae (p = 0·04) were significantly more prevalent and abundant in the PLW-HIV compared to the HIV negative comparator group (Fig. 4). Staphylococcus aureus (OTU 1040220) and Pseudomonas (OTU 144452), were present in 8% (n = 5) of PLW-HIV, and Klebsiella (OTU 144814) in 9% (n = 6). In contrast, these OTUs were completely absent in controls. Bilophila (OTU 359872) was detected in 13% (n = 8) of HIV positive and 3% (n = 1) of HIV negative group. The relative abundance of both genus Lactobacillus and family Enterobacteriaceae taxa were significantly higher in PLW-HIV – being present in 66% (n = 42) and 30% (n = 19) respectively of PLW-HIV samples, 45% (n = 17) and 18% (n = 7) in the HIV negative group. Lactococcus (OTU 557570) was present in 5% (n = 3) and 8% (n = 3) of the PLW-HIV and HIV negative comparators respectively, though was significantly more abundant in the former (p = 0·0068) (Fig. S1 online data supplement).
Fig. 4.
The Relative Abundance of (a) genus Lactobacillus (p=0•029) (b) S. aureus (p=0•0016) (c) genus Pseudomonas (p=0•0093) (d) genus Klebsiella (p=0•024) (e) Family Enterobacteriaceae (p=0•04) (f) genus Bilophila (p<0•0001) in the individual sputum samples. Group size: 38 HIV-ve group (blue) and 64 HIV +ve group (orange), p-values by White non-parametric t-test, corrected by Benjamini-Hochberg FDR method.
Using the Nucleotide BLAST analysis tool against the NCBI 16S ribosomal RNA database [24], the top hits with the maximum scores identified the following OTUs: 1040220, 144452, and 144814 as 99% identical to S. aureus, Ps. aeruginosa, and Klebsiella pneumoniae strains respectively. None of these OTUs or taxa were detected in the negative controls.
The presence of one or more of the pathogenic bacterial taxa; S. aureus, Pseudomonas, Klebsiella and Bilophila and some OTUs belonging to the Enterobacteriaceae family was associated with longer time between HIV diagnosis and commencement of ART.
(Spearman-rho=0·279, p = 0·028). (Fig. S9 online data supplement). There was no significant association found between presence of these taxa and the value of the nadir CD4 count.
Smoking (adonis p = 0·001) and recreational drug use (adonis p = 0·001) were significant covariates in principle component analysis of both weighted and unweighted Unifrac ß-diversity index (details in the online data supplement Figs. S1–S3). But none of them were explanatory covariates for the above reported taxa apart from Lactobacillus (Figs. S5 and S6 online data supplement).
3.3. Load and prevalence of airway bacteria using q-PCR
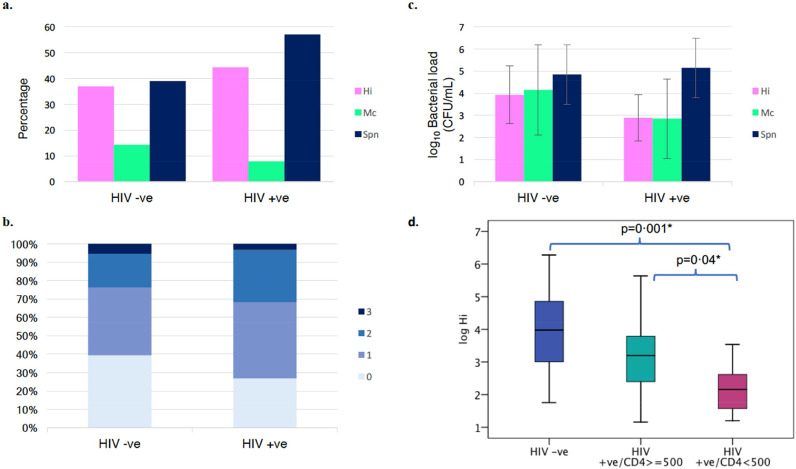
The prevalence of the three bacteria: S. pneumoniae (Spn), H. influenzae (Hi) and M. catarrhalis (Mc) were similar within the two study groups (Fig. 5a). In PLW-HIV S. pneumoniae and H. influenzae were detected in 57% and 44% respectively with both organisms being identified together in 25%). This compares to the HIV negative comparator group, where Spn was present in 39% and Hi in 37%, with both organisms present together in 16%. M. catarrhalis was the least prevalent in both groups (detected in 8% of PLW-HIV and 14% in HIV negatives). The co-existence of the three respiratory pathogens was similar in both groups in less than 5% (Fig. 5b).
Fig. 5.
Prevalence and loads of pathogenic airway bacteria; H. influenzae (Hi), M. catarrhalis (Mc), S. pneumoniae (Spn). (a) Prevalence of the three airway bacteria within the two study groups (p=0·488, Chi squared test) (b) Co-existence of the three bacteria in the sputum samples in each group (p=0·469, Fisher Exact test) (c) Mean bacterial load of each of the three bacteria as determined by the multiplex q-PCR (p=0·867, MANOVA). (d) Comparison of H. influenzae loads between HIV positive and negative groups (p=0·008, t-test) and within the HIV positive group between participants with CD4 count ≥500 cells/µL and whose CD4 count <500 cells/µL (p=0·035, t-test) (p=0·005 by ANOVA) *LSD post hoc test. Bacterial Loads are presented in CFU/ mL of original sputum sample for each bacterium, means were calculated for each study group. Samples which gave negative results for a given bacteria were excluded from the analysis. Error bars show ±1 SD; Group size: 38 HIV-ve group and 64 HIV +ve group.
The mean bacterial load of H. influenzae was lower in PLW-HIV compared to controls; the difference between the means of the two groups was −1·04 log10 (p = 0·008). Within the HIV positive group, the mean bacterial load of H. influenzae in participants with blood CD4 count <500 cells/µL was significantly lower by 0·95 log10 than those with a CD4 count ≥500 cells/µL (p = 0·035) (Fig. 5d). The mean bacterial load of M. catarrhalis was slightly but not significantly lower by 1·3 log10 in the HIV positive group compared to controls. S. pneumoniae was the most populous in both groups with a mean bacterial load (SD) of 5 (1·3) log10 CFU/mL which was similar within the two groups (Fig. 5c).
The presence of P. aeruginosa was confirmed using qPCR method. P. aeruginosa was detected in 8.7% of HIV positive group and was completely absent in the HIV negative comparator group; however, the detected loads were too low, less than the method's sensitivity (limit of quantification (LOQ) 500 CFU/mL) in most cases (data not shown).
4. Discussion
The primary aim of this study compared the airway microbiome of a UK cohort of PLW-HIV using effective ART with good blood CD4 counts, compared to an HIV negative comparator group. Our results support the findings of similar US cohort studies,[14,25] in that the core microbiome in the two populations was very similar in terms of the relative abundance of the major taxa, and the richness and α-diversity of the sputum microbial communities. However, in principal coordinate analysis (PCoA) of unweighted Unifrac ß-diversity index, the samples from PLW-HIV were more dispersed, whereas those from the HIV negative comparator group were more similar and clustered together.
We find, for the first time, potentially important differences in the prevalence of some minor taxa (relative abundance <1%) that may have a role in the pathogenesis of lung disease such as Staphylococcus, Pseudomonas and Klebsiella being detected significantly more frequently in a subset of PLW-HIV (around 8·5%), yet were almost absent in controls.
Although the 16S rRNA sequencing suggest that these pathogenic OTUs were minor taxa, the result of the mock community, run alongside the samples as a positive control for sequencing (Fig. S7 online data supplement), indicated that there might be some negative bias against the amplification of 16S rRNA gene, especially from Klebsiella and Pseudomonas. In line with our results, these pathogenic taxa are not normally detected in the healthy airway microbiome and are more associated with inflammatory chronic lung disease and acute pneumonic infections.[6,7,26]
Lactobacillus and Lactococcus were significantly more abundant in PLW-HIV - a similar observation to that reported by Twigg et al and Yang et al.[11,27,28] Other taxa which were enriched in the sputum of some PLW-HIV are regarded as gut bacteria. These include Bilophila and members of family Enterobacteriaceae. Similarly in the lung HIV Microbiome Project Lozupone et al. reported the enrichment of bronchoalveolar lavage from PLW-HIV with Tropheryma whipplei which is a gut bacterium and the etiological agent of Whipple's disease and whose relative abundance was reduced following effective ART.[13] A finding also noted by Twigg et al. [11]. The enrichment of the lung microbiome with gut bacteria may be explained by the known breakdown of the gut mucosal barrier in HIV infection leading to increased translocation of gut microbes and microbial products into the systemic circulation through which they can eventually lodge and find a favorable ecological niche in the lungs of PLW-HIV [10,11,29].
The enrichment of the respiratory microbial communities with gut and/or potential pathogenic respiratory bacteria was in subsets of PLW-HIV, and it may also explain the dispersal of samples from PLW-HIV in the PCoA of unweighted Unifrac ß-diversity (Fig. 3). We also found a positive correlation between the presence of these taxa in the sputum and the length of time between HIV diagnosis and starting ART. However, this observation needs to be confirmed by a suitably- powered future study.
Previous studies have associated respiratory microbiome enrichment with Prevotella and Veillonella and the enhanced alveolar inflammation that may contribute to chronic lung pathology in PLW-HIV [7,27,30]. In contrast, numerous studies have linked these two genera with lung health where they were promptly replaced by members of Proteobacteria or Firmicutes in chronic lung disease [7,31,32]. In our cohort, the relative-abundances of these two genera were not significantly different in the sputum samples of both HIV positive and negative groups. In a Canadian cohort, these two genera were less abundant in bronchoscopic specimens from PLW-HIV compared to HIV negative controls [33]. Therefore, the role of Prevotella and Veillonella species in respiratory microbiome generally, and in PLW-HIV specifically, remain controversial. More work is needed to understand the interactions and dynamic changes of these two genera with the immune tone and ART in HIV infected population [34].
Our primary analysis was to compare the airway microbiome between PLW-HIV and HIV uninfected subjects, however, microbiome data are complex and multidimensional. To address this many parameters and indices were applied to summarise and compare the data. The alpha and beta diversity indices were more dispersed within PLW-HIV group compared to the HIV negative comparator group. Variation in the peripheral blood CD4 count may account for the observed dispersion in PLW-HIV. To test this the HIV positive group was stratified with respect to the peripheral CD4 count. CD4 was found to be a significant covariate in unweighted UniFrac ß-diversity (p = 0·01) within this group. Furthermore, α-diversity was significantly lower in PLW-HIV with lower CD4 count. A similar observation was reported by Twigg et al. [27].
Recent epidemiological data showed that although the incidence of pneumonia has been reduced in the ART era, PLW-HIV are at greater risk of bacterial pneumonia - a major cause of morbidity and mortality in this population [35]. S. pneumoniae followed by S. aureus and H. influenzae are the most frequently identified etiologic agents of pneumonia in PLW-HIV.[26] It was, therefore, important to specifically detect the prevalence and load of these typical respiratory bacterial pathogens in our cohort using a specific qPCR method as the resolution of 16S rRNA sequencing is often restricted to the genus level for most taxa and sputum contains high proportions of Streptococcus viridans and commensal Haemophilus. Quantitative PCR results demonstrated that S. pneumoniae followed by H. influenzae were the most prevalent in the PLW-HIV. This is consistent with our previous findings using the same method in a similar cohort [36]. There were no significant differences in the loads and prevalence of M. catarrhalis and S. pneumoniae in both HIV positive and negative groups; however, S. pneumoniae was slightly more prevalent in the HIV positive group though this study lacked the power to prove a significant difference.
Limitations of our work include the potential risk of contamination of the sputum with microbes from the upper respiratory tract and oral cavity. Quality assessment was not performed on sputum samples to evaluate pharyngeal contamination. Nevertheless, our main finding relates to the detection of some pathogenic and gut bacterial taxa, that are not normally oral taxa, in samples only from PLW-HIV. Also, samples from other body sites were not available; therefore, we could not compare the microbial signature between upper and lower respiratory compartments and the gut. We relied on the participant's ability to expectorate sputum; and this is likely to be simpler for people who smoke or have underlying lung disease – raising the possibility of selection bias when comparing our study population to the wider HIV community. Current tobacco smoking and recreational drug use were significantly more frequently identified in our HIV positive group which are also common in other HIV cohort studies. Both factors were found to be significant covariates in ß-diversity indices. Confounder analysis is a challenge in microbiome studies [37,38]. Since these two variables are inherent in HIV population, adjustment by propensity score analysis matching [39] in a relatively small sample size would result in a diminished HIV negative comparator group. Therefore, the confounding effect of smoking and recreational drug use could not be completely excluded in our data. Nevertheless, the taxa contributing to the observed significant differences in our data based on the smoking status and recreational drug use, were all oral taxa; and apart from Lactobacillus which was also differentially abundant between HIV positive and negative groups (Figure S5 online data supplement), none was related to our main finding where potential pathogenic taxa (S. aureus, Pseudomonas, Klebsiella) and gut taxa (Enterobacteriaceae and Bilophila) were enriched in terms of prevalence and/or abundance in PLW-HIV in comparison to the HIV negative comparator group. Applying regression analysis would be invalid in this case as the former two covariates and HIV status are strongly correlated. Further studies on a larger sample size would allow adjustment for the potential confounders.
In conclusion, the core microbiome in the airways of people living with HIV who are on effective ART and clinically well managed, is indistinguishable from the general population in terms of the major bacterial taxa and alpha diversity. However, HIV infection may alter the local lung microenvironment to be more permissive to harbor potential respiratory pathogenic bacteria such Staphylococcus, Pseudomonas and Klebsiella, enabling gut bacteria including Bilophila and members of family Enterobacteriaceae to occupy ecological niches in the lower respiratory tracts of PLW-HIV. The carriage of these pathogenic bacteria may contribute to respiratory complications, such as bacterial pneumonia and inflammatory chronic lung disease. The mechanism underlying the perturbation of the respiratory microbiome is unclear, though is likely to relate to the interaction between the host, HIV and the environment. Our findings emphasis the importance of protecting the lung long-term in PLW-HIV through prompt initiation of ART.
Authors’ contributions
SR and JB processed the samples and did the laboratory work, SR performed the bioinformatics and data analyses, SR, TMcH, MJ, JH, ML and DS interpreted the data, JB & EP collected the samples and clinical data, ML is the principal investigator. SR wrote the first draft of the manuscript with contributions from all other authors. All authors read and approved the final version of the manuscript.
Declaration of Competing Interest
There were no conflicts of interests.
Acknowledgments
Acknowledgments
The first author would like to thank the Missions sector in the Egyptian Ministry of Higher Education and the British Council Egypt for funding her PhD as part of the Newton-Mosharafa scheme.
Funding Statement
This study was supported by grants from the National Institute for Health Research (DRF-2015-08-210) and British HIV Association. SR's PhD studies were funded by Newton-Mosharafa Egypt-UK Scheme.
Footnotes
Funding: National Institute for Health Research and British HIV Association.
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.eclinm.2020.100413.
Appendix. Supplementary materials
References
- 1.van Sighem AI, Gras LA, Reiss P, Brinkman K, de Wolf F. Life expectancy of recently diagnosed asymptomatic HIV-infected patients approaches that of uninfected individuals. AIDS. 2010;24:1527–1535. doi: 10.1097/QAD.0b013e32833a3946. [DOI] [PubMed] [Google Scholar]
- 2.George MP, Singh V, Gladwin MT. Noninfectious and nonneoplastic conditions associated with human immunodeficiency virus infection. Semin Respir Crit Care Med. 2016;37:289–302. doi: 10.1055/s-0036-1572560. [DOI] [PubMed] [Google Scholar]
- 3.Gupta RK, Rice B, Brown AE. Does antiretroviral therapy reduce HIV-associated tuberculosis incidence to background rates? A national observational cohort study from England, Wales, and Northern Ireland. Lancet HIV. 2015;2:e243–e251. doi: 10.1016/S2352-3018(15)00063-6. [DOI] [PubMed] [Google Scholar]
- 4.Sogaard OS, Lohse N, Gerstoft J. Hospitalization for pneumonia among individuals with and without HIV infection, 1995–2007: a Danish population-based, nationwide cohort study. Clin Infect Dis. 2008;47:1345–1353. doi: 10.1086/592692. [DOI] [PubMed] [Google Scholar]
- 5.Grubb JR, Moorman AC, Baker RK, Masur H. The changing spectrum of pulmonary disease in patients with HIV infection on antiretroviral therapy. ADIS. 2006;20(8):1095–1107. doi: 10.1097/01.aids.0000226949.64600.f9. 12. [DOI] [PubMed] [Google Scholar]
- 6.O'Dwyer DN, Dickson RP, Moore BB. The lung microbiome, immunity, and the pathogenesis of chronic lung disease. J Immunol. 2016;196:4839–4847. doi: 10.4049/jimmunol.1600279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dickson RP, Erb-Downward JR, Martinez FJ, Huffnagle GB. The microbiome and the respiratory tract. Annu Rev Physiol. 2016;78:481–504. doi: 10.1146/annurev-physiol-021115-105238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shenoy MK, Lynch SV. Role of the lung microbiome in HIV pathogenesis. Curr Opin HIV AIDS. 2018;13:45–52. doi: 10.1097/COH.0000000000000427. [DOI] [PubMed] [Google Scholar]
- 9.Gotelli NJ, Chao A. Measuring and estimating species richness, species diversity, and biotic similarity from sampling data. In: Levin SA, editor. Encyclopedia of biodiversity. second ed. Elsevier Academic Press; Waltham, MA: 2013. pp. 195–211. editor. [Google Scholar]
- 10.Lawani MB, Morris A. The respiratory microbiome of HIV-infected individuals. Expert Rev Anti Infect Ther. 2016;14:719–729. doi: 10.1080/14787210.2016.1206469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Twigg HL, 3rd, Weinstock GM, Knox KS. Lung microbiome in human immunodeficiency virus infection. Transl Res. 2017;179:97–107. doi: 10.1016/j.trsl.2016.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Presti RM, Flores SC, Palmer BE. Mechanisms underlying HIV-associated noninfectious lung disease. Chest. 2017;152:1053–1060. doi: 10.1016/j.chest.2017.04.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lozupone C, Cota-Gomez A, Palmer BE. Widespread colonization of the lung by Tropheryma whipplei in HIV infection. Am J Respir Crit Care Med. 2013;187:1110–1117. doi: 10.1164/rccm.201211-2145OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beck JM, Schloss PD, Venkataraman A. Multicenter comparison of lung and oral microbiomes of HIV-infected and HIV-uninfected individuals. Am J Respir Crit Care Med. 2015;192:1335–1344. doi: 10.1164/rccm.201501-0128OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iwai S, Fei M, Huang D. Oral and airway microbiota in HIV-infected pneumonia patients. J Clin Microbiol. 2012;50:2995–3002. doi: 10.1128/JCM.00278-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iwai S, Huang D, Fong S. The lung microbiome of Ugandan HIV-infected pneumonia patients is compositionally and functionally distinct from that of San Franciscan patients. PLoS One. 2014;9 doi: 10.1371/journal.pone.0095726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stressmann FA, Rogers GB, Marsh P. Does bacterial density in cystic fibrosis sputum increase prior to pulmonary exacerbation. J Cyst Fibros. 2011;10:357–365. doi: 10.1016/j.jcf.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 18.Garcha DS, Thurston SJ, Patel AR. Changes in prevalence and load of airway bacteria using quantitative PCR in stable and exacerbated COPD. Thorax. 2012;67:1075–1080. doi: 10.1136/thoraxjnl-2012-201924. [DOI] [PubMed] [Google Scholar]
- 19.Nolan T, Hands RE, Ogunkolade W, Bustin SA. SPUD: a quantitative PCR assay for the detection of inhibitors in nucleic acid preparations. Anal Biochem. 2006;351:308–310. doi: 10.1016/j.ab.2006.01.051. [DOI] [PubMed] [Google Scholar]
- 20.Comeau AM, Douglas GM, Langille MG. Microbiome helper: a custom and streamlined workflow for microbiome research. mSystems. 2017;2 doi: 10.1128/mSystems.00127-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Caporaso JG, Kuczynski J, Stombaugh J. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parks DH, Tyson GW, Hugenholtz P, Beiko RG. STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics. 2014;30:3123–3124. doi: 10.1093/bioinformatics/btu494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.White JR, Nagarajan N, Pop M. Statistical methods for detecting differentially abundant features in clinical metagenomic samples. PLoS Comput Biol. 2009;5:10. doi: 10.1371/journal.pcbi.1000352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.National Center for Biotechnology Information (NCBI) [database on the Internet] National Center for Biotechnology Information; 1988. National library of medicine (US)https://www.ncbi.nlm.nih.gov/ [accessed 16 Feb 2018]. Available from: [Google Scholar]
- 25.Cribbs SK, Uppal K, Li S. Correlation of the lung microbiota with metabolic profiles in bronchoalveolar lavage fluid in HIV infection. Microbiome. 2016;4:3. doi: 10.1186/s40168-016-0147-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gordin FM, Roediger MP, Girard PM. Pneumonia in HIV-infected persons: increased risk with cigarette smoking and treatment interruption. Am J Respir Crit Care Med. 2008;178:630–636. doi: 10.1164/rccm.200804-617OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Twigg HL, 3rd, Knox KS, Zhou J. Effect of advanced HIV infection on the respiratory microbiome. Am J Respir Crit Care Med. 2016;194:226–235. doi: 10.1164/rccm.201509-1875OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang L, Dunlap DG, Qin S. Alterations in oral microbiota in HIV are related to decreased pulmonary function. Am J Respir Crit Care Med. 2019;4 doi: 10.1164/rccm.201905-1016OC. 201905-1016OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brenchley JM, Price DA, Schacker TW. Microbial translocation is a cause of systemic immune activation in chronic HIV infection. Nat Med. 2006;12:1365–1371. doi: 10.1038/nm1511. [DOI] [PubMed] [Google Scholar]
- 30.Segal LN, Alekseyenko AV, Clemente JC. Enrichment of lung microbiome with supraglottic taxa is associated with increased pulmonary inflammation. Microbiome. 2013;1:2049–2618. doi: 10.1186/2049-2618-1-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rofael SAD, McHugh TD, Troughton R. Airway microbiome in adult survivors of extremely preterm birth (The EPICure Study) Eur Respir J. 2018;21:01225–02018. doi: 10.1183/13993003.01225-2018. [DOI] [PubMed] [Google Scholar]
- 32.Hilty M, Burke C, Pedro H. Disordered microbial communities in asthmatic airways. Plos One. 2010;5:e8578. doi: 10.1371/journal.pone.0008578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu S, Tsai A, Sze MA. Decreased microbiome diversity in the HIV small airway epithelium. Respir Res. 2018;19:140. doi: 10.1186/s12931-018-0835-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Segal LN, Dickson RP. The lung microbiome in HIV. getting to the HAART of the host-microbe interface. Am J Respir Crit Care Med. 2016;194:136–137. doi: 10.1164/rccm.201602-0280ED. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sogaard OS, Lohse N, Gerstoft J. Mortality after hospitalization for pneumonia among individuals with HIV, 1995-2008: a Danish cohort study. PLoS One. 2009;4:e7022. doi: 10.1371/journal.pone.0007022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nimmo C, Capocci S, Honeyborne I. Airway bacteria and respiratory symptoms are common in ambulatory HIV-positive UK adults. Eur Respir J. 2015;46:1208–1211. doi: 10.1183/13993003.00361-2015. [DOI] [PubMed] [Google Scholar]
- 37.Allaband C, McDonald D, Vazquez-Baeza Y. Microbiome 101: studying, analyzing, and interpreting gut microbiome data for clinicians. Clin Gastroenterol Hepatol. 2019;17:218–230. doi: 10.1016/j.cgh.2018.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim D, Hofstaedter CE, Zhao C. Optimizing methods and dodging pitfalls in microbiome research. Microbiome. 2017;5:52. doi: 10.1186/s40168-017-0267-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Littnerova S, Jarkovsky J, Parenica J, Pavlik T, Spinar J, Dusek L. Why to use propensity score in observational studies? Case study based on data from the Czech clinical database AHEAD 2006–09. Cor et Vasa. 2013;55:e383–ee90. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.