Abstract
Background & Aims
Loss-of-function variants in nucleotide-binding oligomerization domain-containing protein 2 (NOD2) impair the recognition of the bacterial cell wall component muramyl-dipeptide and are associated with an increased risk for developing Crohn’s disease. Likewise, exposure to antibiotics increases the individual risk for developing inflammatory bowel disease. Here, we studied the long-term impact of NOD2 on the ability of the gut bacterial and fungal microbiota to recover after antibiotic treatment.
Methods
Two cohorts of 20-week-old and 52-week-old wild-type (WT) C57BL/6J and NOD2 knockout (Nod2-KO) mice were treated with broad-spectrum antibiotics and fecal samples were collected to investigate temporal dynamics of the intestinal microbiota (bacteria and fungi) using 16S ribosomal RNA and internal transcribed spacer 1 sequencing. In addition, 2 sets of germ-free WT mice were colonized with either WT or Nod2-KO after antibiotic donor microbiota and the severity of intestinal inflammation was monitored in the colonized mice.
Results
Antibiotic exposure caused long-term shifts in the bacterial and fungal community composition. Genetic ablation of NOD2 was associated with delayed body weight gain after antibiotic treatment and an impaired recovery of the bacterial gut microbiota. Transfer of the postantibiotic fecal microbiota of Nod2-KO mice induced an intestinal inflammatory response in the colons of germ-free recipient mice compared with respective microbiota from WT controls based on histopathology and gene expression analyses.
Conclusions
Our data show that the bacterial sensor NOD2 contributes to intestinal microbial community composition after antibiotic treatment and may add to the explanation of how defects in the NOD2 signaling pathway are involved in the etiology of Crohn’s disease.
Keywords: Antibiotic Treatment, NOD2, IBD, Microbial Resilience
Abbreviations used in this paper: CD, Crohn’s disease; CXCL1, chemokine (C-X-C motif) ligand 1; GEE, generalized estimating equation; GF, germ-free; IBD, inflammatory bowel disease; ITS, internal transcribed spacer; KO, knockout; NOD2, nucleotide-binding oligomerization domain-containing protein 2; OTU, operational taxonomic unit; PCoA, principle coordinate analysis; qPCR, quantitative real-time polymerase chain reaction; rRNA, ribosomal RNA; SPF, specific pathogen free; WT, wild-type
Graphical abstract
Summary.
The nucleotide-binding oligomerization domain-containing protein 2 recognizes bacterial antigens and contributes to intestinal barrier function. We report that bacterial communities of nucleotide-binding oligomerization domain-containing protein 2–deficient mice have an altered recovery from antibiotic perturbation, which elicits mucosal inflammation in recipient germ-free, wild-type mice.
Microbial communities are important for physiological homeostasis of the mammalian gut. Although fluctuations in diet and other environmental factors affect the gut microbiota in diverse ways, an altered composition and function of microbial communities likely contribute to the development and pathogenesis of human inflammatory bowel disease (IBD). Currently, more than 200 distinct genetic risk loci for IBD have been identified through genome-wide association studies.1, 2, 3 Many of the identified genes have been shown to play a role in maintaining intestinal barrier function and potentially are involved in controlling the intestinal microbiota (eg, by affecting levels of antimicrobial peptides, mucus composition, or autophagic clearance of intracellular bacteria).2 Nucleotide-binding oligomerization domain-containing protein 2 (NOD2) was the first identified susceptibility gene for Crohn’s disease (CD).4, 5, 6 As an intracellular recognition protein, NOD2 senses muramyl-dipeptide, which is an essential bacterial cell wall component derived from peptidoglycan. NOD2 is highly expressed by specialized intestinal epithelial cells, in particular Paneth cells, but also by myelomonocytic cells and T cells.7, 8, 9 Remarkably, approximately 20% of all Caucasian CD patients carry a deleterious mutation on at least 1 NOD2 allele.5 In human beings, the 3 main CD risk alleles (rs2066844, rs2066845, and rs2066847) lead to a complete or partial loss of function with impaired recognition of muramyl-dipeptide and diminished activation of the downstream target nuclear factor-κB.10 It is assumed that these genetic variants are contributing to disease manifestation as a consequence of a loss of tolerance toward the gut microbiota. In mice, genetic deletion of NOD2 leads to a broad spectrum of abnormal immune responses and an increased susceptibility to experimental colitis.11, 12, 13 Several studies have reported that NOD2-deficient mice are prone to develop a dysbiotic microbial community with an increase in bacterial taxa related to the Bacteroidetes genus already early in life,14,15 whereas other studies found no genotype-dependent differences in the intestinal microbiome at homeostatic conditions among littermates.16,17 Several reasons may explain the diverging findings, which may include animal housing, diurnal variations, as well as coprophagy and self-grooming among littermates, which could lead to a dominant takeover of the dysbiotic microbiota from mutant mice upon co-housing.
Early life exposure to antibiotics has been identified as an independent risk factor for the development of IBD.18 Numerous studies have shown the disruptive effects of external perturbations such as antibiotics or nutrition shifts can have on the host microbiome.19, 20, 21 However, insights on how host genetic risk factors such as NOD2 deficiency shape the recovery of microbiota in response to a defined perturbation still are missing. We thus performed a short-term antibiotic combination therapy22 in wild-type (WT) and Nod2-knockout (KO) mice to investigate the effect of NOD2 on longitudinal dynamics of the bacterial microbiota after a pulsed perturbation in adult mice. We hypothesized that the bacterial sensor NOD2 may contribute to resilience phenomena of the intestinal microbiome.23,24 In line with this hypothesis, a recent report showed an influence of NOD2 on the outcome of early postnatal antibiotic treatment in mice.17 Because fungi (mycobiome) largely were ignored by previous studies, we also performed longitudinal profiling of fungal communities after antibiotic exposure because it may play an important role for gut community stability.
Here, we show that middle-aged (1-year-old) NOD2-deficient mice suffer from a delayed recovery from antibiotic-induced body weight loss and bacterial dysbiosis. We validate these findings in a second animal facility and age cohort (young adult mice, 20 weeks old). Moreover, germ-free (GF) mice were colonized with the microbiota from Nod2-KO and WT mice at 7 weeks after antibiotic exposure. Colonization with microbiota from Nod2-KO mice resulted in increased colonic inflammation in the recipient WT mice compared with those colonized with the microbiota of WT mice. These results point to an important role of the bacterial sensor NOD2 during perturbation and recovery events of intestinal microbial communities with relevance for intestinal inflammation.
Results
Loss of NOD2 Alters Antibiotic-Induced Body Weight Loss and Recovery From Bacterial Dysbiosis
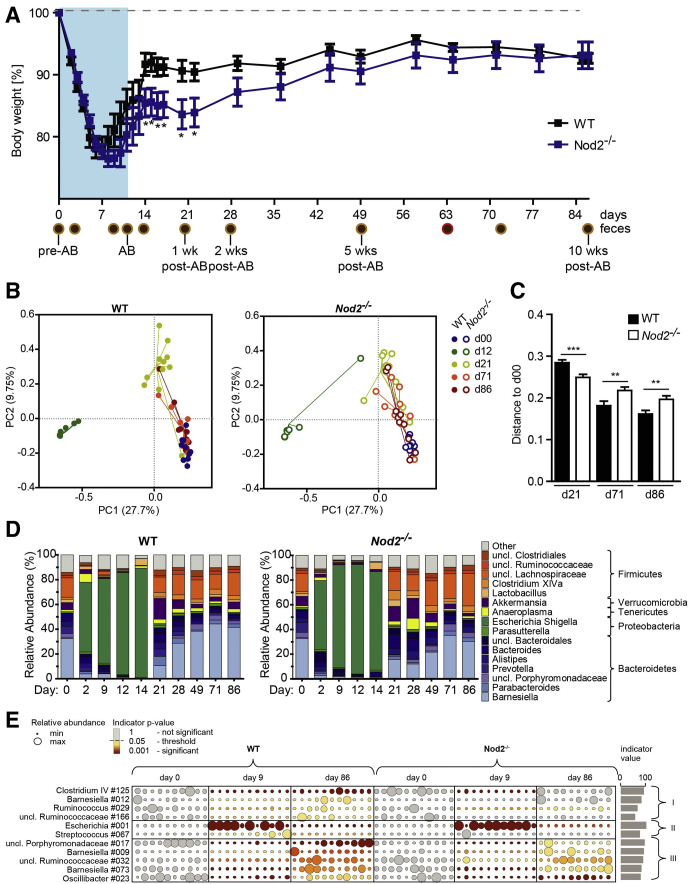
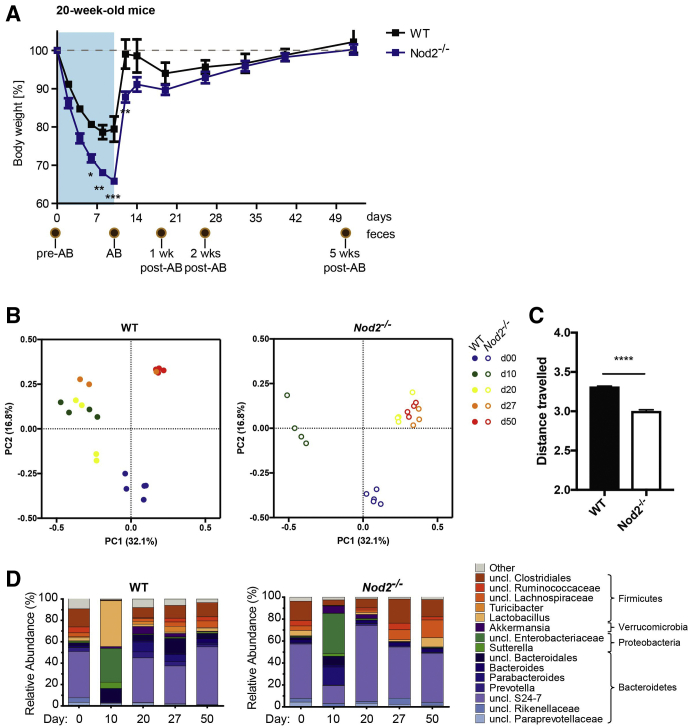
When middle-aged and young WT and Nod2-KO mice (ages, 1 year or 20 weeks, C57BL/6J background) were challenged with a cocktail of broad-spectrum antibiotics (vancomycin, metronidazole, ampicillin, and neomycin22), animals lost substantial weight in both age groups immediately after antibiotic administration (Figures 1A and 2A). However, the kinetics and severity of weight loss differed slightly between age groups. Twenty-week-old animals lost weight more rapidly than 1-year-old mice. Antibiotic treatment had to be stopped on day 10 in 20-week-old-mice for ethical reasons. In contrast, the 1-year-old animals lost weight only until day 6 in the WT and day 8 in the Nod2-KO groups, after which their body weight increased despite continuing antibiotic administration until day 14. Recovery from antibiotic-induced weight loss occurred earlier and faster in WT mice compared with Nod2-KO mice in both age groups (Figures 1A and 2A). All age groups and genotypes showed complete weight recovery on the day they were killed (day 86 for 1-year-old mice and day 50 for 20-week-old mice).
Figure 1.
Loss of NOD2 delayed recovery from antibiotic-induced body weight loss and bacterial dysbiosis in middle aged mice. C57BL/6J (WT) and Nod2-KO mice (age, 52 wk) were administered a combination of broad-spectrum antibiotics for 12 days via drinking water. Mice subsequently were monitored for a total of 86 days to investigate the genotype effect on the gut microbial resilience. Donor microbiota from WT and KO feces collected on day 63 were used to colonize C57BL/6J GF WT mice. (A) Percentage change in body weight of C57BL/6J (WT) and Nod2-KO mice. The percentage of body weight was calculated based on the initial body weight at the time the experiment was started. Data are represented as means ± SEM values (n = 11 WT, n = 10 KO). Brown circles indicate collected fecal pellets and the red circle indicates the feces used for GF colonization. ∗Significant difference (P < .05) between the genotypes evaluated using the Student t test. (B) PCoA using the Bray–Curtis dissimilarity of the fecal bacterial communities across time (days), where principal coordinate (PC)1 explains 27.7% and PC2 explains 9.75% of the data variance. Each dot represents the bacterial composition of an individual fecal sample. Samples are colored by day. Filled circles represent WT and open circles represent Nod2-KO mice. (C) Bray–Curtis distance of samples from d21, d71, and d86 from d0 (baseline). A smaller distance indicates more similar microbial communities. Note the increased distance to d0 in the Nod2-KO compared with the WT, indicating delayed resilience. (D) Relative abundance (%) of 16S-based bacterial community composition across all time points in feces of WT and Nod2-KO mice at the genus level. (E) Bacterial indicator analysis to identify possible key taxa. Bacterial taxa indicating resilience events are depicted in 3 groups: (I) showing a significant difference of indicator values between WT and Nod2-KO (depleted after antibiotic treatment and regained at day 86); (II) comprises 2 indicator taxa that are enriched after antibiotic treatment and again low in abundance at day 86 (E coli/Shigella 001, independent of genotype; Streptococcus 067, significant only in WT); and (III) a group of indicator OTUs being depleted after antibiotic treatment and regained at day 86, which do not formally discriminate between WT and Nod2-KO microbiota. Circle sizes correspond to abundances, each column represents an individual animal, and indicator values shown are obtained from WT animals during the resilience period (days 9–86). Indicator P values are color coded, note that all day 0 abundances are shown as starting points, which are nonsignificant by default. Numbers next to bacterial taxa correspond to the OTU numbers while unclassified taxa are marked by a star. AB, antibiotics; uncl, unclassified.
Figure 2.
Loss of NOD2 alters trajectories of recovery from antibiotic-induced body weight loss and bacterial dysbiosis in 20-week-old mice. The 20-week-old WT and Nod2-KO mice were treated with an antibiotic cocktail for 10 days and were followed up until day 50. (A) Percentage change in body weight of WT and Nod2-KO mice. The percentage body weight was calculated based on initial body weight at the time the experiment started. Data are represented as means ± SEM values (n = 5 WT, n = 5 Nod2-KO). Brown circles indicate collected fecal pellets. ∗Significant difference (P < .05) between the genotypes was evaluated using the Student t test. (B) PCoA using the Bray–Curtis dissimilarity of the fecal bacterial communities across time (days), where PC1 explains 25.0% and PC2 explains 15.6% of the data variance. Each dot represents the bacterial composition of an individual fecal sample. Samples are colored by day. Filled circles represent WT and open circles represent Nod2-KO mice. (C) Cumulative Bray–Curtis distance of samples travelled from d0 to d10, d20, d27 to d50. The distance travelled indicates the degree of change in bacterial composition over time during antibiotic perturbation and recovery. Note the decreased travelled distance in the Nod2-KO compared with the WT. (D) Relative abundance (%) of 16S bacterial community composition across all time points in feces of WT and Nod2-KO mice at genus level. AB, antibiotics; uncl, unclassified.
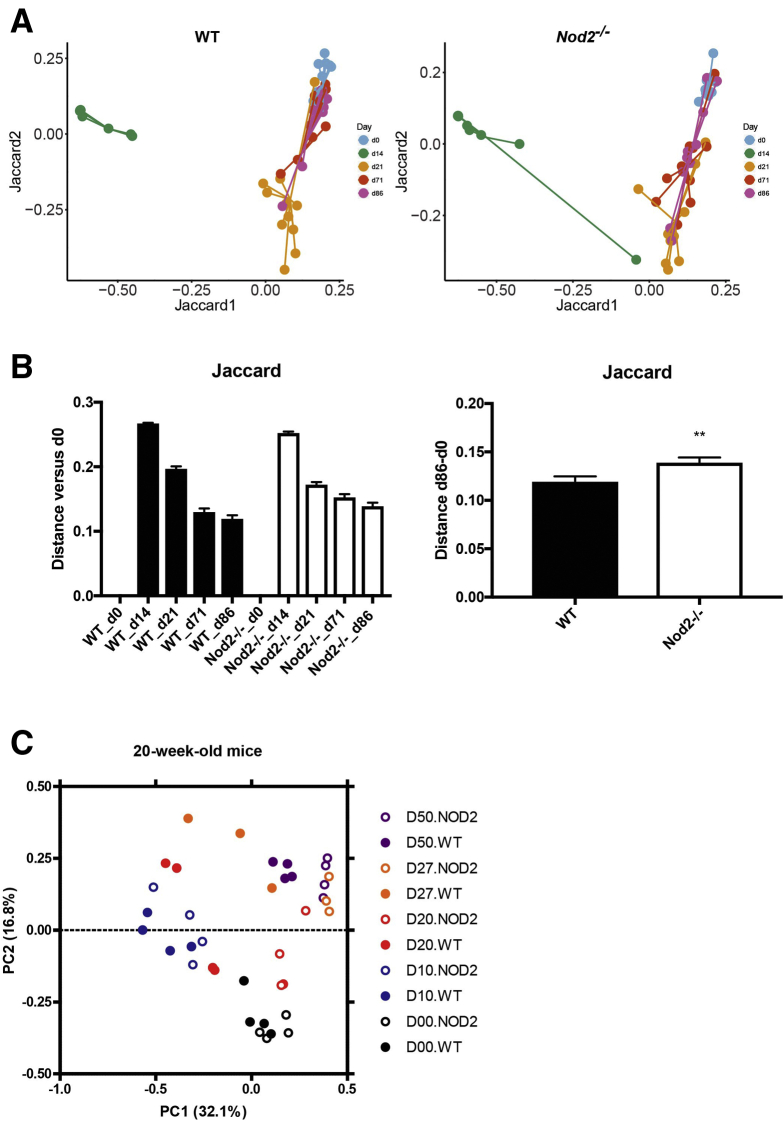
16S ribosomal RNA (rRNA) gene amplicon sequencing was performed on longitudinally sampled fecal material to monitor the temporal development of the gut bacterial composition during and after antibiotic perturbation. First, alterations of β-diversity (ie, bacterial diversity between samples) were studied using principle coordinate analysis (PCoA) based on Bray–Curtis and Jaccard distances of operational taxonomical units (OTUs). A nonparametric analysis of variance (Table 1) comparing both genotypes showed no difference between WT and Nod2-KO mice before antibiotic treatment (day 0 [d0]) in 1-year-old mice (P = .9411) or 20-week-old mice (P = .233). Antibiotic treatment drastically changed the microbiota composition in both genotypes and age groups (Figures 1B and C and 2B and C). After withdrawal of the antibiotics, the individual bacterial communities shifted back toward their initial states in 1-year-old mice, although d86 communities still were significantly different from day 0 (P = .00099). Samples from day 86, however, clustered more closely with day 0 samples in WT animals compared with Nod2-KO mice as measured by Bray–Curtis distance (Figure 1B and C), indicating an impaired recovery in the Nod2-KO animals. This also was confirmed through Jaccard β-diversity distance analyses comparing the distance between day 0 and day 86 for both genotypes, which showed a significantly greater distance among Nod2-KO mice compared with the WT animals (Figure 3, Table 1). In the 20-week-old mice, however, neither the microbiota of the WT nor Nod2-KO returned to its initial state but shifted to a new configuration (Figure 2B, Table 1), suggesting an age- and/or animal house–dependent trajectory of the microbiota during the time course of the experiment. Interestingly in this age group, although already starting from a different composition, the overall changes of the microbiota of Nod2-KO mice were smaller compared with WT animals (Figure 2C), as measured by the total path travelled (sum of the change in distance between time points). The Nod2-KO mice already reached an equilibrium state on day 20, which, however, was significantly different from the respective WT animals (P = .035) (Table 1).
Table 1.
Nonparametric Analysis of Variance on Bray–Curtis Distance in Relation to Bacterial Resilience
| Matrix (comparison) | Psuedo F | P value |
|---|---|---|
| 20-week-old mice | ||
| WTday0–WTday50 | 7.704 | .01000a |
| KOday 0–KOday 50 | 10.607 | .00500a |
| WTday0–KOday0 | 1.273 | .23300 |
| WTday50–KOday50 | 9.156 | .03500 |
| 1-year-old mice | ||
| WTday0–WTday86 | 3.723 | .000999a |
| KOday 0–KOday 86 | 3.608 | .000999a |
| WTday0–KOday0 | 0.606 | .9411 |
| WTday86–KOday86 | 1.955 | .01199a |
Indicates significance.
Figure 3.
Jaccard-based analysis of microbial shifts during and after antibiotic perturbation. (A) PCoA based on the Jaccard metric of days 0, 14, 21, 71, and 86 of 1-year-old WT and Nod2-KO mice. (B) Jaccard distance of samples from d14, d21, d71, and d86 from d0 (baseline). A smaller distance indicates more similar microbial communities. ∗∗P < .005, Mann–Whitney test. (C) PCoA using the Jaccard metric of days 0, 10, 20, and 50 of 20-week-old WT and Nod2-KO mice.
To analyze taxonomic shifts associated with antibiotic treatment and/or with the absence of NOD2, we next performed phylogenetic analyses on the 16S rRNA amplicon data sets. At baseline, the most abundant phyla in both genotypes and age groups were Bacteroidetes and Firmicutes (Figures 1D and 2D). A clear increase in the relative abundance of Proteobacteria was observed during the antibiotic period. Verrucomicrobia showed a greater relative abundance (P values listed in Table 2) early after the antibiotic period in both age groups regardless of genotype (day 20/21), but were found at higher frequency in the 1-year-old animals than in the 20-week-old mice. Recovery phenomena were observed in WT and Nod2-KO mice of both age groups, for example, with a striking contraction of the relative abundance of Proteobacteria after antibiotic treatment. Taken together, these data show a complex influence of NOD2 on microbial recovery processes after antibiotic exposure.
Table 2.
Significance (P value) Determined by GEE Correlation Matrix of Phylum Level Gut Bacteria Comparing Day 0 (Pretreatment) With Time Points Across the Longitudinal Study (Kinetics) or for Genotype (WT vs Nod2-KO)
| Bacteroidetes | Proteobacteria | Verrucomicrobia | Firmicutes | Tenericutes | |
|---|---|---|---|---|---|
| Genotype (WT vs Nod2-KO) | 6.26E-01 | 1.38E-01 | 0.576497 | 8.23E-01 | 5.10E-01 |
| Day 2 vs day 0 | 1.29E-05a | 5.72E-11a | 0.302459 | 2.94E-08a | 3.31E-01 |
| Day 9 vs day 0 | 1.40E-11a | 0.00E+00a | 0.001512a | 1.11E-11a | 8.29E-03a |
| Day 12 vs day 0 | 0.00E+00a | 0.00E+00a | 0.000808a | 3.87E-11a | 9.03E-03a |
| Day 14 vs day 0 | 1.74E-14a | 0.00E+00a | 0.016084a | 2.96E-03a | 4.29E-03a |
| Day 21 vs day 0 | 1.30E-01 | 8.27E-01 | 0.190371 | 3.10E-01 | 3.00E-01 |
| Day 28 vs day 0 | 5.46E-02 | 5.50E-01 | 0.025025a | 8.18E-01 | 2.35E-02a |
| Day 49 vs day 0 | 5.33E-03a | 5.38E-01 | 0.932214 | 2.24E-02a | 1.40E-02a |
| Day 70 vs day 0 | 9.37E-01 | 1.80E-01 | 0.290782 | 5.25E-01 | 7.10E-01 |
| Day 86 vs day 0 | 3.58E-01 | 1.04E-01 | 0.023226a | 1.71E-01 | 2.35E-01 |
Significant at P ≤ .05.
To analyze the difference in recovery processes between NOD2 and WT mice in greater detail (ie, to identify bacterial taxa indicating significant resilience), we next investigated the overall effect size of genotype and antibiotic treatment with regard to bacterial community composition on the phylum and genus levels. Of note, we focused on middle-aged mice from here on because they showed similar microbiota at baseline, and recovery processes were not influenced additionally by age- and/or laboratory-dependent trajectories. We therefore generated a generalized estimating equation (GEE) correlation matrix (Tables 2 and 3). With this method, the overall effect of the genotype (regardless of the time point) or the respective time point (regardless of the genotype) can be shown. At the phylum level, most taxa differed significantly during days 2–14 in comparison with before antibiotic treatment (day 0). However, only Verrucomicrobia remained significantly different on day 86 (Table 3). At the genus level, Barnesiella was significantly discriminating between WT and Nod2-KO microbiota, which resulted from changes of the genus before and early after antibiotics. Prevotella and Clostridium XIVa remained significantly different at all time points compared with day 0, however, comparison between Nod2-KO and WT mice did not reach formal significance in the GEE analysis. To get a more detailed view on the level of OTUs, we next performed an indicator species analysis based on the relative abundances of OTUs in longitudinal comparisons (see the Materials and Methods section for details). The indicator analysis identified 11 bacterial OTUs, which followed differential patterns of resilience over time (Figure 1E, Table 4). These could be categorized further into 3 groups. Group I is characterized by loss of abundance followed by significantly higher recovery in the WT and included 4 OTUs classified as Clostridium IV #125, Barnesiella #2, and Ruminococcaceae #29 and #166. Group II shows 2 OTUs, which were enriched during antibiotic treatment and depleted again upon recovery (Streptococcus #067 is significant only in WT, whereas Escherichia coli/Shigella #001 showed a similar pattern in WT and Nod2-KO). Group III recovered independently of the NOD2 genotype. The results show subtle differences of specific microbial taxa between WT and Nod2-KO animals upon recovery from antibiotic treatment.
Table 3.
Significance (P value) Determined by GEE Correlation Matrix of Genus Level Gut Bacteria Comparing Day 0 (Pretreatment) With Time Points Across the Longitudinal Study (Kinetics) or for Genotype (WT vs Nod2-KO)
| Barnesiella | Parabacteroides | Uncl. porphyromonadaceae | Prevotella | Alistipes | Bacteroides | Uncl. Bacteroidales | Parasutterella | Escherichia Shigella | Anaeroplasma | Akkermansia | Lactobacillus | Clostridium XlVa | Uncl. Lachnospiraceae | Uncl. Ruminococcaceae | Uncl. Clostridiales | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genotype (WT vs Nod2-KO) | 0,045312a | 0,4866 | 2,34E-01 | 2,08E-01 | 2,41E-01 | 2,16E-01 | 0,40942 | 0,69085 | 0,70228 | 0,74512 | 4,95E-01 | 7,01E-01 | 2,27E-01 | 2,23E-01 | 7,72E-01 | 3,62E-01 |
| day 2 vs day 0 | 0a | 0,182 | 1,20E-08a | 8,29E-06a | 2,69E-01 | 1,89E-01 | 0,00702a | 0,55719 | 0a | 0,125309 | 3,87E-02a | 6,14E-01 | 1,83E-05a | 1,94E-13a | 1,95E-08a | 6,03E-01 |
| day 9 vs day 0 | 0a | 0,0396a | 2,22E-16a | 6,63E-10a | 7,44E-15a | 1,67E-06a | 0,21807 | 0,72818 | 0a | 0,000455a | 1,31E-07a | 3,44E-04a | 1,12E-02a | 1,94E-10a | 1,78E-05a | 1,11E-03a |
| day 12 vs day 0 | 0a | 0,1362 | 0,00E+00a | 5,55E-16a | 0,00E+00a | 5,60E-09a | 0,09343 | 0,00278a | 0a | 0,000339a | 1,54E-07a | 8,98E-06a | 6,55E-04a | 1,84E-13a | 8,18E-11a | 2,44E-06a |
| day 14 vs day 0 | 0a | 0,6734 | 0,00E+00a | 0,00E+00a | 0,00E+00a | 1,43E-02a | 0,00563a | 0,46939 | 0a | 0,633845 | 2,43E-05a | 1,22E-01 | 2,26E-06a | 4,35E-14a | 4,24E-12a | 5,31E-07a |
| day 21 vs day 0 | 0,000144a | 0,0241a | 5,32E-07a | 4,81E-02a | 3,98E-01 | 2,56E-01 | 0,13552 | 0,31411 | 0,00301a | 0,142477 | 2,44E-02a | 6,33E-01 | 1,40E-02a | 6,48E-01 | 4,23E-02a | 4,28E-01 |
| day 28 vs day 0 | 0,031842a | 0,0887 | 2,72E-02a | 3,98E-04a | 4,26E-01 | 5,28E-01 | 0,02707a | 0,2149 | 0,05153 | 0,018377a | 1,01E-01 | 7,80E-02 | 9,71E-03a | 9,53E-01 | 3,11E-01 | 6,40E-01 |
| day 49 vs day 0 | 0,535091 | 0,3141 | 5,96E-01 | 3,07E-10a | 9,81E-01 | 1,07E-01 | 0,1357 | 0,86042 | 0,03436a | 0,03386a | 3,19E-01 | 3,92E-02a | 3,62E-03a | 2,77E-01 | 2,70E-01 | 2,02E-02a |
| day 70 vs day 0 | 0,085452 | 0,1169 | 3,41E-01 | 4,62E-07a | 8,92E-04a | 8,38E-03a | 0,09318 | 0,96517 | 0,26435 | 0,765982 | 5,02E-02 | 2,39E-01 | 1,91E-02a | 8,26E-01 | 4,44E-01 | 1,89E-01 |
| day 86 vs day 0 | 0,415301 | 0,1269 | 7,12E-01 | 7,83E-11a | 3,53E-01 | 1,57E-03a | 0,14705 | 0,22099 | 0,68781 | 0,07713 | 1,54E-01 | 1,40E-01 | 2,14E-03a | 1,73E-01 | 1,85E-01 | 8,62E-02 |
uncl, unclassified.
Significant at P ≤ .05.
Table 4.
Resilience Indicator OTUs Observed in Response to Genotype
| OTU | WT |
Nod2-/- |
||||
|---|---|---|---|---|---|---|
| Day 0 | Day 9 | Day 86 | Day 0 | Day 9 | Day 86 | |
| Clostridium IV #125 | 3 | 0a | 2a | 2 | 0a | 1 |
| Barnesiella #012 | 7 | 0a | 13a | 0 | 0 | 2 |
| Ruminococcus #029 | 7 | 0a | 4a | 5 | 0 | 1a |
| Unclassified Ruminococcaceae #166 | 1 | 0a | 1a | 1 | 0 | 0 |
| Escherichia #001 | 0 | 1171a | 0a | 0 | 1154a | 1a |
| Streptococcus #067 | 0 | 1a | 0a | 0 | 0 | 0 |
| Unclassified Porphyromonadaceae #017 | 23 | 0a | 13a | 32 | 0a | 20a |
| Barnesiella #009 | 6 | 0a | 73a | 4 | 0a | 24a |
| Unclassified Ruminococcaceae #032 | 6 | 0a | 15a | 5 | 0a | 7a |
| Barnesiella #073 | 4 | 0a | 3a | 3 | 0a | 3a |
| Oscillibacter #023 | 16 | 0a | 7a | 6 | 0a | 10a |
NOTE. Numbers represent relative abundances. Data corresponding to Figure 1E.
Indicative effect.
Emergence of Antibiotic Resistance Occurs Independently of NOD2 Signaling
Exposure to antibiotics may induce resistances.25 To study whether NOD2 plays a role in the emergence of antibiotic resistance, we surveyed the presence and relative abundance of antibiotic resistance genes in the middle-aged cohort by a validated quantitative real-time polymerase chain reaction (qPCR) panel containing 11 genetic elements relevant for the antibiotic cocktail used.26 Antibiotic resistance genes commonly associated to the bacterial phylum Proteobacteria (Type C β-lactamase, β-lactamase, ampicillin resistance) and Firmicutes (vanC2-C3-2 and vanA2 genes, vancomycin resistance) were detected in the fecal microbiota of both WT and Nod2-KO mice (Tables 5 and 6). The number of resistance genes in the fecal microbiota increased significantly during the antibiotic administration and peaked right after (day 14), and on subsequent days (days 21 and 71) (P = .048 vs day 0) (Table 6). However, a difference associated with NOD2 genotype was not observed with the current set of genes.
Table 5.
Assays Used in This Study as Part of the TaqMan-Based Microfluidic Real-Time PCR System to Detect Antibiotic Resistance Genes
| Family/genus | Target | Target gene | Assay ID | Context sequence |
|---|---|---|---|---|
| Enterobacteriaceae Phylum: Proteobacteria |
ESBL | CTX-M group9 | Pa04646127_s1 | GCTTAATCAGCCTGTCGAGATCAAG |
| SHV | Pa04646134_s1 | CGCTTTCCCATGATGAGCACCTTTA | ||
| TEM | Pa04646128_s1 | GCGGCCAACTTACTTCTGACAACGA | ||
| OXA-1 | Pa04646133_s1 | TCATACACCAAAGACGTGGATGCAA | ||
| AmpC resistance | BIL/LAT/CMY | Pa04646135_s1 | TAAGACGTTTAACGGCGTGTTGGGC | |
| ACT/MIR | Pa04646124_s1 | ACCGTTACGCCGCTGATGAAAGCGC | ||
| Carbapenem resistance | VIM | Pa04646155_s1 | GATGGTGATGAGTTGCTTTTGATTG | |
| KPC | Pa04646152_s1 | CCTCGTCGCGGAACCATTCGCTAAA | ||
|
Enterococcus Phylum: Firmicutes |
Vancomycin resistance | vanA2 | Pa04646147_s1 | AGCTACTCCCGCCTTTTGGGTTATT |
| vanB | Pa04646150_s1 | AACTTAACGCTGCGATAGAAGCGGC | ||
| vanC2-C3-2 | Pa04646122_s1 | TTGAGATCGGTTGCGGTATTTTGGG |
AmpC, Type C β-lactamase; ESBL, extended-spectrum β-lactamases.
Table 6.
Summary of Antibiotic Resistance Genes Present in Feces of Individual Mice From Days 0, 2, 14, 21 and 71
| Target gene | Target | Associated phylum | Individual mouse | Day resistance gene was present |
|---|---|---|---|---|
| ACT/MIR | AmpC resistance | Proteobacteria | #46 WT | 14 |
| ACT/MIR | AmpC resistance | Proteobacteria | #55 WT | 71 |
| vanC2-C3-2 | Vancomycin resistance | Firmicutes | #24 WT | 21 and 71 |
| vanA2 | Vancomycin resistance | Firmicutes | #32 KO | 0 |
| vanC2-C3-2 | Vancomycin resistance | Firmicutes | #35 KO | 14 |
| vanC2-C3-2 | Vancomycin resistance | Firmicutes | #11 KO | 21 |
| BIL/LAT/CMY | AmpC resistance | Proteobacteria | #7 KO | 21 |
NOTE. The presence of resistance genes was determined by qPCR assays listed in Table 5.
AmpC, Type C β-lactamase.
The Fungal Gut Communities Change Long Term Irrespective of NOD2 Deficiency
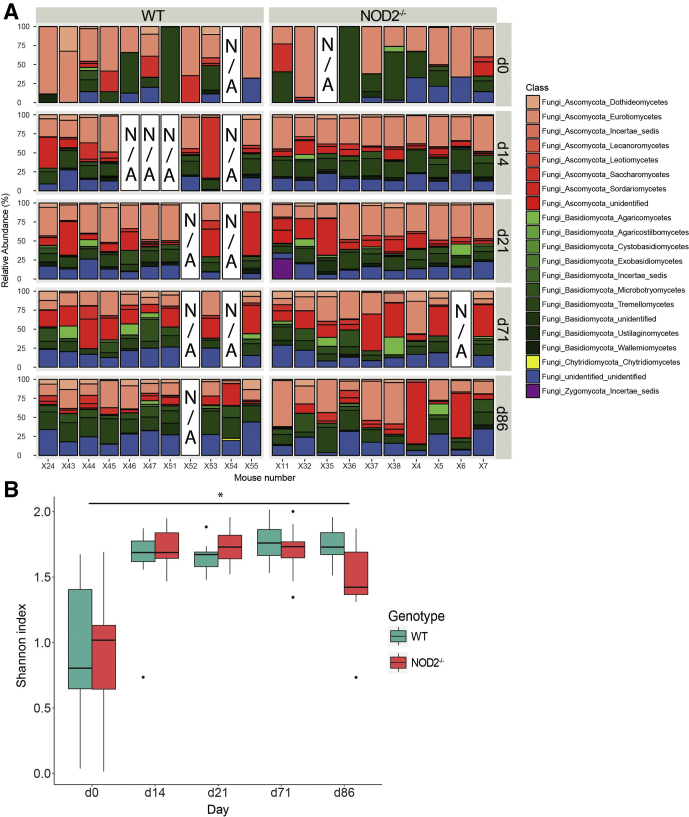
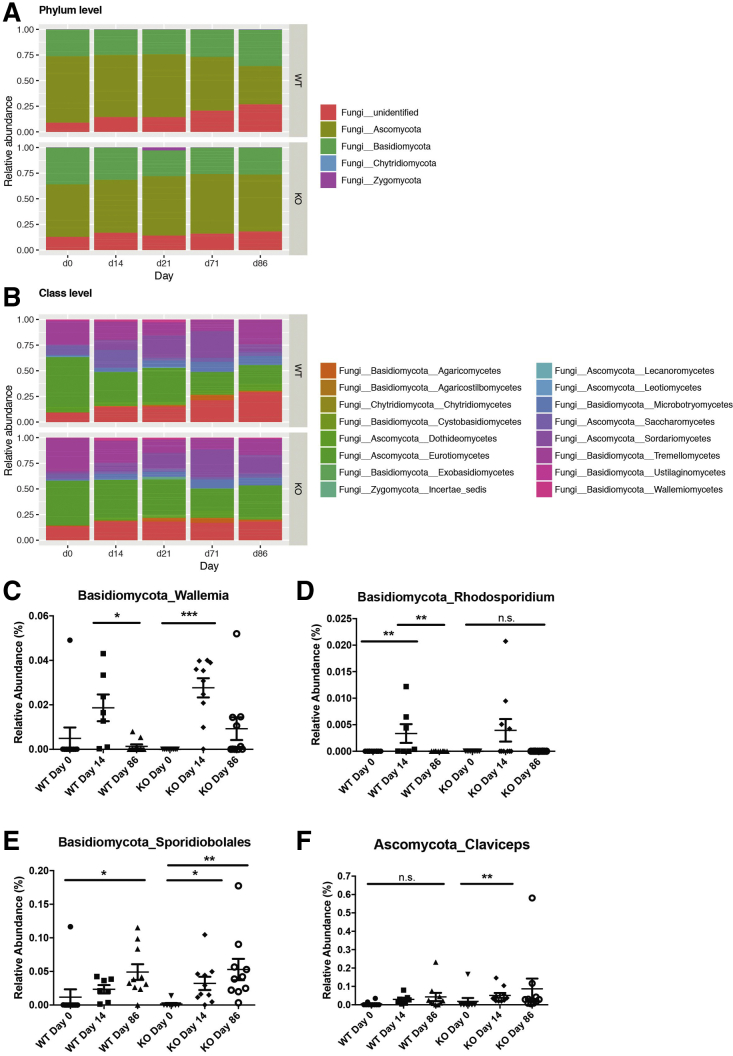
To elucidate the role of NOD2 on additional members of the gut microbiome other than bacteria, we performed amplicon sequencing of the internal transcribed spacer (ITS1) in the middle-aged cohort to assess the composition of the fungal gut communities. PCoA based on the weighted Bray–Curtis distance showed clustering by day rather than by genotype. However, some interindividual heterogeneity was observed with a trend of differing fungal communities at day 0 (Figure 4). Importantly, contrary to the results for bacterial communities, PCoA plots and nonparametric statistics using Bray–Curtis distances showed a near-complete lack of resilience in the fungal communities of both genotypes (Figure 4D). These observations were supported further using unweighted Jaccard-based analyses (Figure 5). The dominant phyla in both genotypes were Ascomycota and Basidiomycota (Figures 6A and 7). To characterize fungal α-diversity across time, Shannon and Chao1 diversity indices were used. Remarkably, during and after antibiotic treatment, α-diversity of the fungal community increased in both genotypes (Figure 6B and 5D). Similar to α-diversity, fungal community shifts continued for the duration of the study with individuals remaining distinct from day 0 (pretreatment) (Figures 6A and 7).
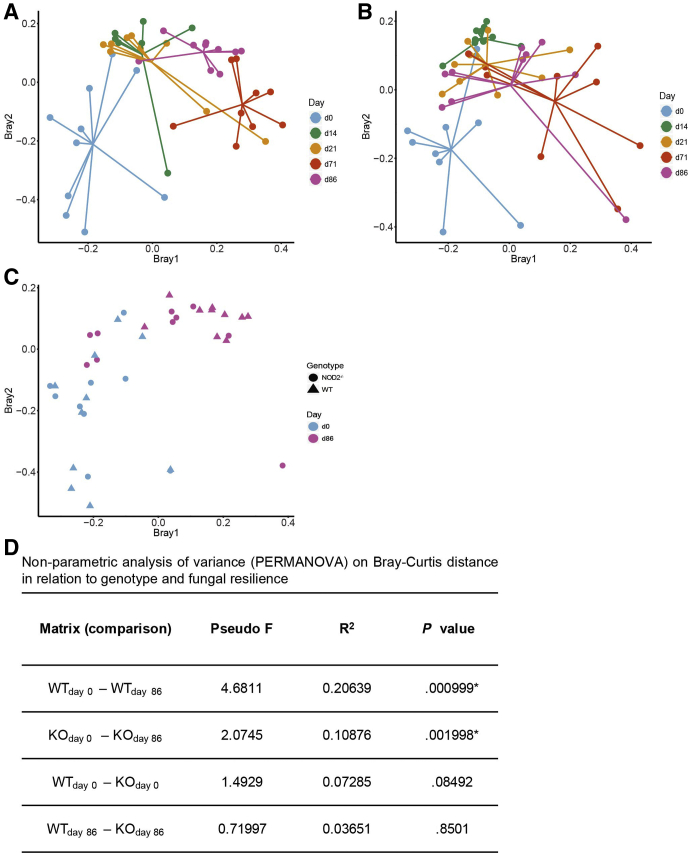
Figure 4.
Weighted β-diversity analysis of fungal communities in 1-year-old mice after antibiotic perturbation. PCoA of fecal fungal composition across time (days 0, 14, 21, 71, 86) in 1-year-old (A) WT and (B) Nod2-KO mice, and (C) combined data for both genotypes only at day 0 (pretreatment) and day 86. (D) Nonparametric analysis of variance (PERMANOVA) on Bray–Curtis distance in relation to genotype and fungal community composition. Pseudo F, Pseudo F ratio as calculated by PERMANOVA. Larger F-ratios indicate more pronounced group separation.
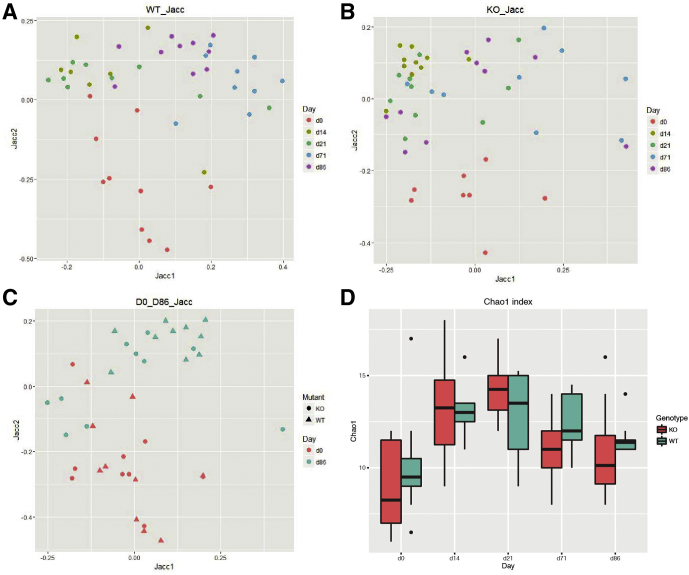
Figure 5.
Unweighted β-diversity analysis of fungal communities in 1-year-old mice after antibiotic perturbation. PCoA of fecal fungi composition in 1-year-old (A) WT across time, (B) Nod2-KO across time, and (C) both genotypes at day 0 (pretreatment) and day 86 based on Jaccard (Jacc) distances. (D) Chao1 index showing fungal richness in WT and KO across time. Time points (days 14, 21, and 71) in both genotypes differ significantly from day 0. P < .05; Student t test.
Figure 6.
The fungal gut communities change long-term irrespective of NOD2 deficiency in 1-year-old mice after antibiotic perturbation. (A) ITS1 fungal relative abundance (%) of class level community composition in all middle-aged Nod2-KO and WT individuals across time (days). N/A indicates the sample was unavailable. (B) Shannon index showing fungal diversity at class level in WT and Nod2-KO across time (days). P < .05; Student t test.
Figure 7.
Abundance of selected fungal taxa relative abundance (%) of fungal taxa in 1-year-old WT and Nod2-KO mice at (A) phylum and (B) class levels. (C–F) Relative abundance (%) of the genera (C) Wallemia, (D) Rhodosporidium, (E) Sporidiobolales, and (F) Claviceps in 1-year-old feces of WT and Nod2-KO across time (days 0, 14, 86). ∗P < .05, ∗∗P < .005, and ∗∗∗P < .0005.
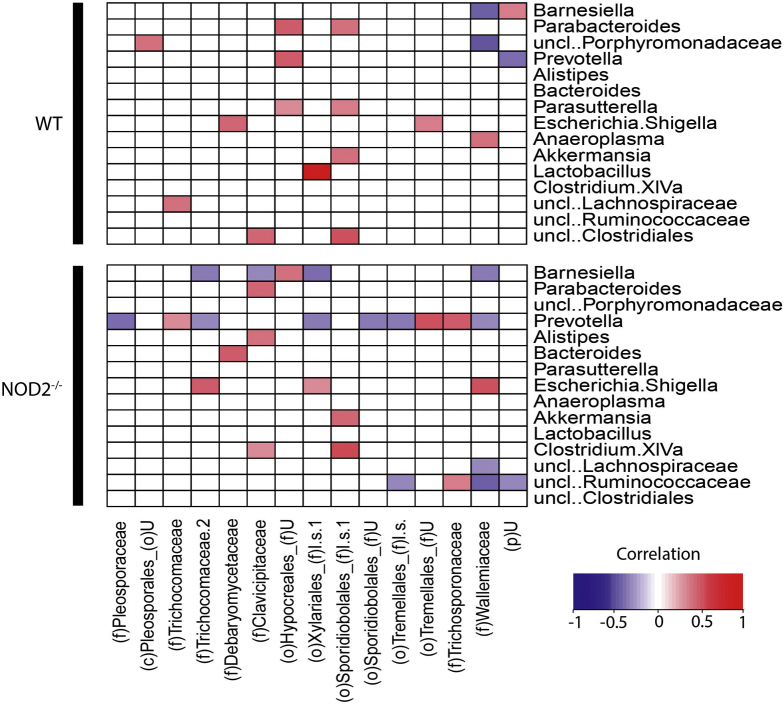
We then investigated whether specific bacterial taxa were associated to members of the mycobiome across all time points in relation to NOD2 genotype. Notably, a higher number of negative bacteria–fungi correlations were observed in Nod2-KO compared with WT mice, including the bacterial genera Barnesiella, Prevotella, and Ruminococcaceae (Figure 8). In particular, Prevotella had negative correlations to various fungal taxa, which prompted us also to investigate levels of Prevotella and fungal abundance in the colonization experiment in GF mice.
Figure 8.
Fungal bacteria correlation after antibiotic perturbation. Correlation analysis between fungi and bacteria in feces of 1-year-old WT and Nod2-KO mice. The blue-to-red gradient indicates negative and positive correlations, respectively. White spaces indicate no correlations were observed. P < .05 was considered as the significance threshold as determined by the Student t test. uncl, unclassified; p, phylum; c, class; o, order; f, family; uncl, unclassified.
The Postantibiotic Nod2-KO Microbiota Elicits a Proinflammatory Response Upon Transfer to GF WT Mice
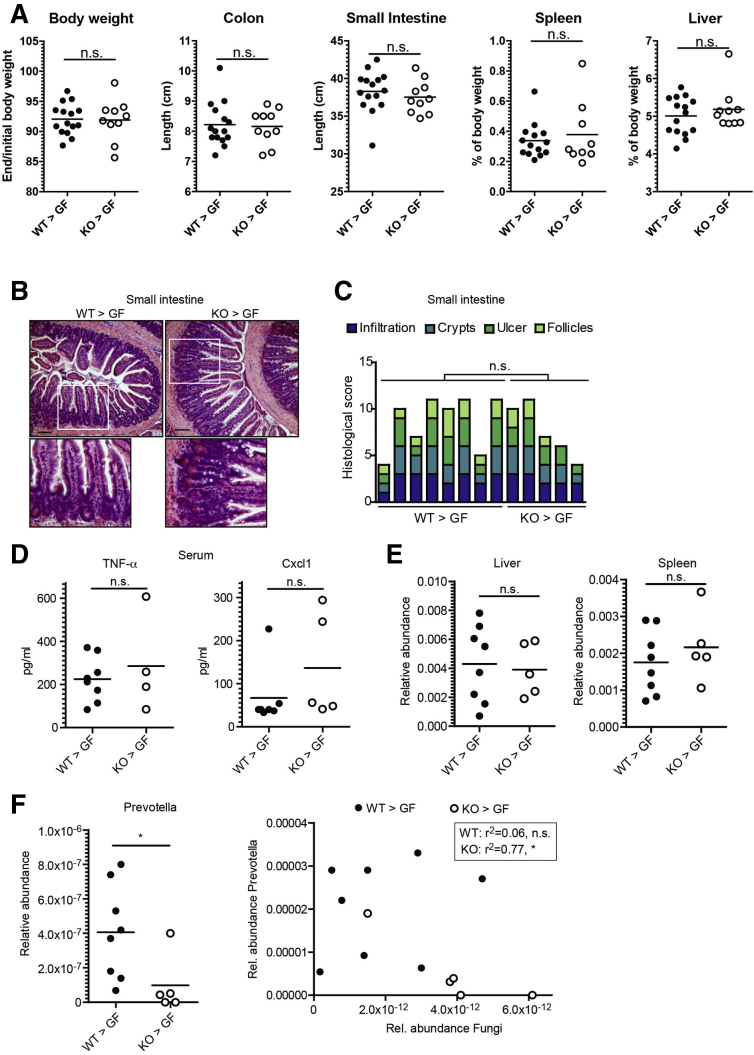
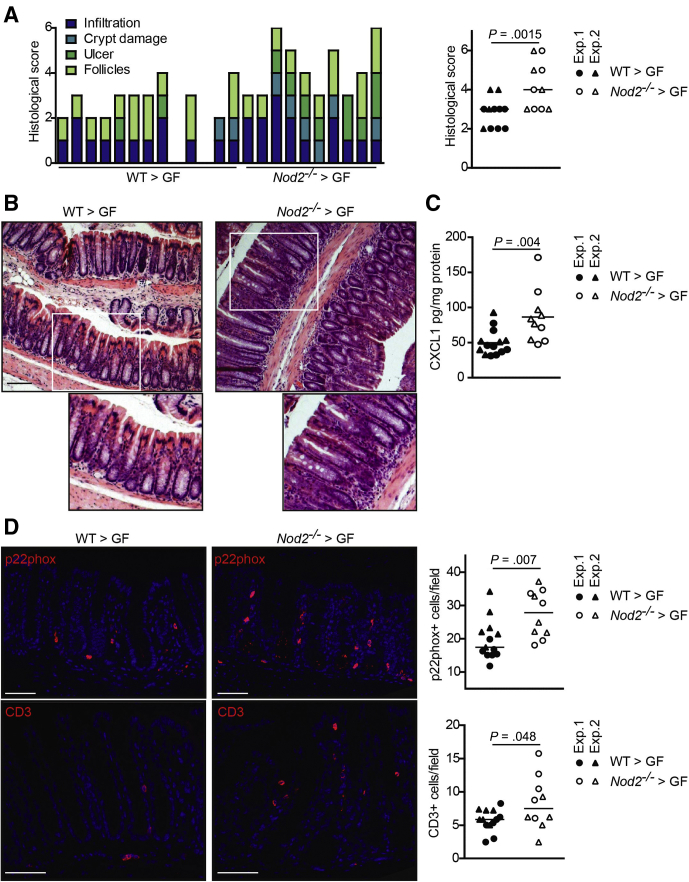
To test whether the NOD2-dependent differences in microbial resilience lead to functional physiological consequences, in particular altered intestinal inflammation, we transferred the fecal microbial communities of donor WT and Nod2-KO mice into adult 15-week-old GF WT recipients (WT>GF and NOD2>GF, respectively) in 2 independent transfer experiments. Mice were killed 17 days after initial colonization. In both sets of mice, there were no significant differences between WT>GF and NOD2>GF mice in body weight or organ characteristics (ie, colon length, small intestinal length, spleen, or liver weight), which are known to potentially be affected by intestinal inflammatory responses (Figure 9A). Histologic analysis of the colon showed higher levels of mucosal inflammation in NOD2>GF compared with WT>GF mice as indicated by a significantly increased histologic score (Figure 10A and B) (P = .01). Moreover, colonic levels of the proinflammatory chemokine chemokine (C-X-C motif) ligand 1 (CXCL1) were significantly higher in NOD2>GF compared with WT>GF mice (Figure 10C) (P = .004). In line with this observation, the numbers of p22 phagocyte b-cytochrome-positive phagocytes were higher in NOD2>GF compared with WT>GF mice (Figure 10D, upper panel) (P = .007). P22 phagocyte b-cytochrome is a component of the reduced nicotinamide adenine dinucleotide phosphate oxidase complex involved in the antimicrobial oxidative burst of neutrophils and macrophages. Levels of CD3–T-cell coreceptor–positive cells also were increased significantly in NOD2>GF mice (Figure 10D, lower panel) (P = .048). Taken together, these data indicate that transfer of the dysbiotic postantibiotic NOD2 microbiota elicited an increased proinflammatory response in the colon of GF recipient mice in comparison with the postantibiotic WT microbiota. Inflammation also was detectable in the small intestine, however, this was independent of the NOD2 genotype (Figure 9B–D). To determine whether the observed significant negative correlations between Prevotella and fungal taxa in the postantibiotic microbiota also could be transferred to the colonized recipient mice, we measured the abundance of Prevotella and fungi by reverse-transcription qPCR. Prevotella were more abundant in the WT>GF than in NOD2>GF mice (Figure 9G). Moreover, the abundance of Prevotella and fungi were correlated inversely in the NOD2>GF mice (P = .04), but not in the WT>GF recipients. These data therefore indicate a functional transfer of the postantibiotic, Nod2-KO–dependent microbiota into the recipient GF mice.
Figure 9.
Absence of small intestinal and systemic signs of inflammation after transfer of dysbiotic postantibiotic microbiota of Nod2-KO mice. Fecal transfer of postantibiotic microbiota from WT and Nod2-KO donor mice into GF WT recipients. GF WT mice were colonized for 17 days with feces collected from day 63 from WT and Nod2-KO mice (49 days after antibiotic treatment). (A) Change in body weight expressed as the ratio of the end/start of the experiment (ie, 17 days after and before colonization), and organ measures (length of colon and small intestine, weights of spleen and liver) at the end of the experiment. (B) Representative H&E staining of paraffin-embedded small intestinal sections. Scale bar: 100 μm. (C) Histologic scoring of the small intestine. Each bar represents an individual mouse. (D) Levels of tumor necrosis factor (TNF)-α and Cxcl1 encoding chemokine (C-X-C motif) ligand 1 in serum of the colonized mice as measured by enzyme-linked immunosorbent assay. (E) Quantification of bacterial load in the liver and spleen of the colonized mice measured by reverse-transcription qPCR. (F) Abundance of Prevotella (left panel) and correlation of abundances between fungi and Prevotella (right panel) in stool samples of the colonized mice as by reverse-transcription qPCR. Rel., relative.
Figure 10.
The dysbiotic postantibiotic microbiota of Nod2-KO mice elicits an intestinal inflammatory response in the colon of recipient WT mice. GF WT mice were colonized with fecal material collected on day 63 from middle-aged WT and Nod2-KO mice and then were killed 17 days after colonization. (A) Histologic scoring of colon sections. Each bar represents an individual mouse (P = .0015). (B) Representative pictures of H&E-stained colon sections. Scale bar: 100 μm. (C) Levels of the proinflammatory cytokine CXCL1 in colon lysates as measured by enzyme-linked immunosorbent assay (P = .004). (D) Representative immunofluorescence pictures and quantification of p22 phagocyte b-cytochrome (p22phox) (marker for neutrophils and activated macrophages; upper panel; P = .007) and CD3 (marker for mature T lymphocytes; lower panel; P = .048) of colonic sections from the colonized mice. Scale bar: 100 μm. Every dot in the dot plots represents an individual recipient mouse. Horizontal lines indicate the group mean. Data are combined from 2 independent colonization experiments as indicated by circles and triangles. Exp., experiment.
Discussion
The present study investigated the influence of NOD2 on resilience phenomena of the gut microbiota after broad-spectrum antibiotic perturbation in mice. Supporting previous literature using a number of vertebrate hosts including human beings,27, 28, 29 we show that a single antibiotic treatment severely disrupted the intestinal bacterial and fungal communities, and that after antibiotic exposure a recovery of microbial communities could be observed. Among middle-aged WT mice, the microbial equilibrium clearly shifted back to its initial state. In younger WT animals from another animal facility, the microbiota showed a temporal trajectory to a different equilibrium, which could either reflect ongoing maturation processes or differences of housing conditions. Interindividual variation within each experiment clearly point to stochastic events, which act on restoration of community composition after such catastrophic events. The absence of NOD2 delayed recovery of the bacterial communities in the middle-aged cohort and interfered with microbiota trajectories in the second experiment, indicating that NOD2 has an influence on recovery processes independently of housing environment. Transfer of the postantibiotic microbiota of middle-aged Nod2-KO and WT mice into GF WT recipient mice showed an increased proinflammatory potential of the transferred Nod2-KO microbiota in 2 independent colonization experiments. The delayed recovery in Nod2-KO mice therefore may have opened up a window of opportunity for pathobionts.
Numerous studies have reported an increased abundance of Proteobacteria and Enterobacteriaceae in IBD patients.30, 31, 32 In our study, a relative abundance of Proteobacteria, namely E coli/Shigella, increased significantly during antibiotic treatment. This effect is from the vancomycin component of the antibiotic cocktail, which has been shown previously to promote an expansion of Proteobacteria33,34 including Enterobacteriaceae and E coli.33 Vancomycin targets gram-positive bacteria and therefore may have created a niche availability to members of the gram-negative phylum Proteobacteria (eg, by disruption of mechanisms of bacterial cross-feeding), which in turn may have led to an increase in microbiota-liberated sugars, promoting the growth of opportunistic pathogens.33,35,36 It is important to note that the observed enrichment of Enterobacteriaceae is an important indicator of dysbiosis during antibiotic treatment but was not unique to NOD2 deficiency. This antibiotic-induced change in function of entire communities may be harmful for the health of the host, for example, by colonization of known opportunistic pathogens24 or bloom of otherwise unrecognized bacterial taxa, which may lead to monodominance community states in the gut.37 Similar to our study, the postantibiotic Proteobacteria-dominated state was present in both NOD2-proficient and NOD2-deficient animals and readily reversible after cessation of antibiotics in both groups, we have no evidence for succession events, in which transient Proteobacteria might be followed by other dominant taxa. It will be interesting to follow-up such events on a strain level, in which ancestral strains of similar taxa might be replaced by other strains with different evolved functions.38,39
Contrary to several previous studies by us and others in mice and human beings, we did not observe an unequivocal increase in Bacteroidetes at the phylum level in the Nod2-KO compared with WT before antibiotic treatment in the 2 animal houses, and the microbiota of Nod2-KO and WT animals did not differ at the β-diversity level at the beginning of the experiment.14,15,40, 41, 42, 43, 44, 45 These data, however, are in line with other previous studies reporting no NOD2 genotype-dependent differences in the intestinal microbiome under homeostatic conditions.16,17 Several factors may explain the observations. First, 3 of the published studies, including our own, showed the strongest differences between Nod2-KO and WT bacterial communities in ileal content or in mucosal tissue samples, whereas the fecal microbiota were analyzed here. Second, all experiments presented here were performed in different animal houses (because facilities were moved both in Kiel and Lille) compared with our previous studies, which is why we cannot fully exclude potential influences of even subtle changes in environmental factors such as standards of hygiene, food, and bedding. Despite the lack of NOD2-dependent microbiota differences before the antibiotic treatment, both cohorts of Nod2-KO animals showed an influence of the KO on the postantibiotic trajectories of their bacterial communities. This phenotype is supported by a recent study showing an impaired microbial resilience and an establishment of genotype-dependent microbiota differences after early postnatal antibiotic treatment.17 We can only speculate about potential mechanisms of how NOD2 deficiency translates into altered recovery of bacterial communities because our experimental set-up aimed at longitudinal follow-up evaluation and did not include early time points for analysis of the host side (eg, differential activation of innate immune pathways). From other murine studies, however, we know that a number of mechanisms relevant to host–bacteria interaction in the gut of NOD2-deficient animals are disturbed, including decreased expression of antimicrobial peptides and proteins,11,46,47 as well as altered cytokine/chemokine levels,12,42 or autophagy/endoplasmic reticulum stress signals.48, 49, 50 Two recent studies identified further interactions between the microbiota and NOD2 that modulate immune responses and susceptibility to intestinal inflammation. Kim et al51 showed that adjuvant activity of cholera toxin during vaccination critically depends on recognition of symbiotic bacteria through NOD2 and subsequent production of interleukin 1β. Caruso et al52 described that the functional absence of the 2 CD risk genes NOD2 and phagocyte reduced nicotinamide adenine dinucleotide phosphate oxidase lead to increased susceptibility to the gram-negative bacterium Mucispirillium schaedleri owing to impairment of neutrophil recruitment and bacterial clearance in a murine model. More detailed studies investigating the impact of such alterations on microbial community dynamics thus are warranted to understand the exact impairment of signals or selective principles, which are critically influenced by NOD2 function.
In addition to selection mechanisms of the host, other nonbacterial members of the intestinal microbiota could play a role in shaping the bacterial communities. Because overgrowth of fungi in the gut is promoted through the use of antibiotics53 and gut inflammation also can promote proliferation of fungi,54 we hypothesized that the observed changes between NOD2 genotypes could be associated with different shifts in the mycobiome. We observed in the middle-aged cohort that fungal community diversity significantly increased in both genotypes after antibiotic treatment. Interestingly, the enhanced diversity was sustained during the course of the experiment (ie, despite the recovery of bacterial microbiota), pointing to potential long-term effects of single-antibiotic courses. Although NOD2 did not appear to affect the mycobiome composition in our experimental setting, it still could be possible that the inflammatory response to the increased diversity of fungal taxa could be altered in the NOD2-deficient situation. Because loss of NOD2 has been shown to lead to a faulty recognition of fungi,55 this mechanism could contribute to the observed weight-loss phenotype and delayed bacterial resilience. Notably, we detected negative correlations of various fungal taxa to the bacterial genus Prevotella, which generally is associated with a healthy intestinal function.15,56 Our findings are supported by a study investigating the role of diet and microbial interactions on gut fungi that also identified numerous correlations between Prevotella and fungal taxa as possible means of influencing host health.57
To formally test whether the alterations of the postantibiotic microbiota, which were licensed by the absence of a functional NOD2 signaling pathway, were able to induce an inflammatory phenotype, we colonized WT GF recipients with the fecal microbiota of Nod2-KO and WT donor mice and monitored histologic changes, infiltration of immune cells, and the induction of CXCL1/KC (keratinocyte chemoattractant) as a mucosal inflammation marker.58,59 Indeed, in 2 independent experiments, transfer of the postantibiotic microbiota from middle-aged Nod2-KO donors was able to induce an increased inflammatory response in comparison with the respective WT donors. The results are in line with other studies, which showed transmission of susceptibility to intestinal inflammation42 or metabolic syndrome60 by fecal transfer from dextran sodium sulfate or high-fat challenged Nod2-KO mice to WT recipients, respectively. A recent study by Goethel et al17 described a similar phenomenon in Nod2-KO mice in which antibiotic treatment of NOD2-deficient animals early in life led to a higher susceptibility to a second inflammatory hit, in this case dextran sodium sulfate colitis, later in life. Mechanistically, the observed increase of CXCL1 may result from a breach of the intestinal barrier, which is reflected by increased histologic subscores for epithelial damage and ulcers. In turn, CXCL1 is a potent chemoattractant,61 which may explain the observed increase of p22 phagocyte b-cytochrome-positive cells in the lamina propria.
In summary, resilience of bacterial communities might be a critical and under-recognized factor to host health,21,62 and our results emphasize the need for more longitudinal studies to identify consequences of these dynamic shifts and the influences of genotypic variation. We want to point out that our results are derived solely from mice, which may not fully reflect the human being situation. However, we clearly show that microbial resilience to antibiotic treatments can have considerable individual variation and that the genotype of the respective host contributes to the microbial response to antibiotics. Consequently, microbial perturbations and current treatments need further evaluation in human individuals at genetic risk with standardized procedures and sampling times. Antibiotics often are used in the management of IBD patients because of several different reasons (eg, infections). Our findings of the NOD2-dependent proinflammatory potential of a postantibiotic microbiota therefore may add another layer to the complex disease etiology. Genotype-adjusted supplementation with lost bacteria after antibiotic therapy thus may represent a potential intervention strategy.
Materials and Methods
Specific Pathogen–Free Mice
All animal experiments were approved by the local animal safety review board of the federal ministry of Schleswig Holstein and conducted according to national and international laws and policies (V 312-72241.121-33 [95-8/11] and V242-62324/2016 [97-8/16]). Specific pathogen–free (SPF) animals were housed in mouse facilities at the Christian Albrechts University of Kiel (Germany) and the Institute Pasteur de Lille (France) (accreditation C59-350009). A single NOD2-deficient male mouse was crossed to a C57BL/6J female to obtain heterozygous offspring. Heterozygous mice (F1) then were used to generate WT and Nod2-KO breeder pairs (F2). Male offspring of the next 2 generations then were used for the experiment and maintained in single cages under SPF conditions. At the onset of the study (Figures 1A and 2A, day 0), mice were approximately 1 year (middle-aged) or 20 weeks (young adult) old. Fecal pellets were collected immediately from mice throughout the experiment and stored at -80°C. Mice were monitored and weighed regularly, and killed at the conclusion of the study, at which point organ and tissue samples were collected and stored at -80°C.
Antibiotic Treatment
SPF C57BL/6J WT and Nod2-KO mice were treated with a cocktail of broad-spectrum antibiotics composed of ampicillin (1 g/L), vancomycin (500 mg/L), neomycin (1 g/L), and metronidazole (1 g/L) (Sigma Aldrich)22,63 for 12 days (1-year-old mice) or 10 days (20-week-old mice). Antibiotics were freshly prepared and administered ad libitum via drinking water in light protected bottles.
Colonization of GF Mice
GF animals were housed in mouse facilities at the Max-Planck-Institute for Evolutionary Biology in Plön, Germany. Fifteen-week-old female C57BL/6J mice were maintained under GF conditions as described previously.64,65 Briefly, mice were kept under a 12-hour light cycle and fed an autoclaved chow diet ad libitum (V1124-927; ssniff, Soest, Germany). Colonization of GF mice by fecal microbiota transfer was performed as described previously.66 Briefly, 1 frozen fecal pellet (∼50 mg) of SPF Nod2-KO or C57BL/6J WT control mice from day 63 was resuspended in 1.2 mL sterile phosphate-buffered saline and GF mice were gavaged orally with 200 μL of freshly prepared suspension. Two to 3 recipient GF mice were colonized per fecal sample of each 3 Nod2-KO and WT control mice, and co-housed in individually ventilated cages (Green Line; Tecniplast [Hohenpeiβenberg, Germany]) for 17 days without opening the cages. In total, 35 GF mice were colonized in 2 independent experiments (n = 15 in experiment 1, n = 20 in experiment 2). Donors were selected based on cages (each mouse was from a different cage) and PCoA clustering (to avoid taking individuals that were more distant than the approximate average to reflect the overall variation in the groups). On day 17 after colonization, mice were killed by cervical dislocation. Blood was collected from the vena porta and tissue biopsy specimens were collected, snap frozen, and stored at -80°C.
16S Amplicon Sequencing
Bacterial profiling using 16S amplicon sequencing was performed as described previously.67 Briefly, DNA from fecal pellets was isolated using the PowerSoil DNA Isolation Kit (Qiagen, Hilden, Germany) according to the manufacturer’s directions. Individual amplicons were tagged with specific multiplex identifier barcodes and pooled for library construction before sequencing. The 16S rRNA gene variable region V3–V4 was amplified and sequenced on an Illumina (San Diego, CA) MiSeq with 2 × 300 base pairs. Sequencing reads initially were processed for quality control and downstream analysis using Mothur68 and MacQIIME v1.9.1.69 Paired-end sequences were assembled to V3–V4 contigs. Only sequences with less than 450 base pairs, a homopolymer count of 6 and zero ambiguities, with a mean quality score ≥25 were considered for further alignment with the SILVA reference database. Initially processed sequences with the SILVA reference database that did not align to the target region of the 16S rRNA gene were discarded from analysis. Chimeric sequences were detected using the UCHIME algorithms and were removed from the analysis. Reads matching to eukaryotic, chloroplast, or mitochondrial sequences were excluded. All samples were rarified to 1190 reads per sample to eliminate sampling bias and uneven depth of coverage. β-diversity was assessed by Bray–Curtis and Jaccard metrics,68 and statistical significance was studied with Adonis/nonparametric analysis of variance using Vegan package in R (v 3.2.5). Distances of all samples were measured in comparison with day 0 to assess the amount of change from baseline. A smaller distance value indicated greater similarity. A paired Wilcoxon test was performed for time wise analysis and a nonparametric Mann–Whitney test was performed for genotype comparison using GraphPad 7. To determine the proportional abundance of microbiota at the phylum and genus level, we used a GEE model.70 To identify bacterial taxa indicating significant resilience, taxa were selected by their ability to show resilience (indicator species for day 0 vs day 9 and day 9 vs day 86, while not being an indicator for day 0 vs day 86). Indicator species are defined as an OTU with an indicator value within the 95th percentile (of all OTUs indicator values), and an indicator P value of ≤ .05. After applying these criteria, 11 resulting bacterial indicator taxa were grouped into 3 categories. Data for days 2, 12, 14, 21, 28, 49, and 71 were omitted for visualization purposes.
Fungal ITS1 Amplicon Sequencing
Fungal profiling was performed using ITS1 (located between 18S and 5.8S rRNA genes) amplicon sequencing in a 2-step PCR approach on fecal DNA. The initial PCR was performed using the primers ITS1-F(F) (5’-CTTGGTCATTTAGAGGAAGTAA-3’) and ITS2(R) (5’-GCTGCGTTCTTCATCGATGC-3’)71,72 to amplify the fungal ITS1 region. The 25-μL PCR mixture contained 12.5 μL Master Mix (Phusion HotStart Flex 2× Master Mix; New England Biolabs, Ipswich, MA), 1 μL of each primer (1.25 μmol/L), 7.75 μL PCR clean water, 0.75 μL dimethyl sulfoxide, and 2 μL DNA template. Reactions were held at 94°C for 2 minutes, with amplification proceeding for 40 cycles, with 1 cycle consisting of 94°C for 15 seconds, 55°C for 30 seconds, and 72°C for 45 seconds; a final extension was performed for 10 minutes at 72°C. In a second PCR linker sequence, a sample specific index and an Illumina adapter were attached. The 25-μL PCR mixture contained 12.5 μL Master Mix (Phusion HotStart, Flex 2× Master Mix; New England Biolabs), 0.5 μL of each primer (2.25 μmol/L), 8.75 μL PCR clean water, 0.75 μL dimethyl sulfoxide, and 2 μL of first-round product. Reactions were held at 94°C for 2 minutes, with amplification proceeding for 20 cycles, with 1 cycle consisting of 94°C for 15 seconds, 55°C for 30 seconds, and 72°C for 45 seconds; a final extension was performed for 10 minutes at 72°C. Correct amplification was monitored by agarose gel electrophoresis. Samples were pooled and paired-end sequenced (2 × 300 base pairs) on an Illumina MiSeq. Sequencing resulted in 7,990,874 pairs of reads. The PIPITS pipeline was used for fungal community analysis.73 Read-pairs were joined with PEAR.74 Assembled reads then were quality-filtered with FASTQ_QUALITY_FILTER (FASTX-Toolkit) (Hannon, available from: http://hannonlab.cshl.edu). Files then were converted into FASTA format with FASTQ_TO_FASTA (FASTX-Toolkit). The highly variable ITS1 subregion was extracted from sequences using the software ITSx (version 1.0.11).75 Of the 7,849,824 input sequences, 7,334,632 contained an ITS1 region. ITS1 sequences were clustered into OTUs with VSEARCH (Rognes, available from: https://github.com/torognes/vsearch) at 97% sequence similarity including chimera detection and removal by UCHIME. A total of 7,244,071 reads passing quality control then were used to assign taxonomic classifications with the RDP Classifier against the UNITE fungal ITS reference data set76 and to generate an OTU abundance table with a confidence threshold of 97% sequence identity. Three samples of WT at day 14 were discarded because of poor quality and a low number of reads. α-Diversity estimation for diversity and richness-based methods Shannon and Chao1 index was performed using in-house scripts in R (v 3.2.5). Correlations were calculated between fungi and bacteria at the genus level using the Pearson correlation with a cut-off mean >0.5% relative abundance. The correlation differences in genotype over time then were compared for each combination from fungi and bacteria (P < .05, t test), of which 46 were found to be significant. A heatmap visualizing the correlations was generated by R (package pheatmap).
Detection of Antibiotic Resistance Genes Using Real-Time qPCR
Antibiotic resistance genes present in the bacterial community of SPF mice were assessed by microfluidic real-time PCR. Eleven antibiotic resistance predesigned TaqMan real-time PCR gene expression assays (Thermo Fisher Scientific Waltham, MA) were selected to target genes conferring vancomycin resistance in Enterococcus, and Type C β-lactamase, extended-spectrum β-lactamases, and carbapenem resistance in Enterobacteriaceae (Table 5). A total of 84 fecal DNA samples from WT and Nod2-KO mice from days 0, 14, 21, and 71 were assayed (10 KO and 11 WT per time point). PCRs were performed in a 10-μL volume with 4.5 μL of TaqMan Fast Universal PCR Master Mix (Applied Biosystems), 0.5 μL TaqMan assay, 1 μL fecal DNA template, and 4 μL PCR water. Assays were run on a ViiA 7 real-time PCR system (Applied Biosystems, Waltham, MA) using the following cycling conditions: 95°C for 20 seconds, followed by 45 cycles of 95°C for 1 second, and 60°C for 20 seconds. All assays were performed in duplicate with a positive control of bacterial DNA and a negative control of water. Real-time PCR data were analyzed with ViiA 7 RUO software, using the ΔΔCT-method as previously published.77 A Fisher exact test was performed to determine nonrandom significant associations between the genotypes, days, and antibiotics.
Histopathology
After cervical dislocation, the entire small intestine and colon were excised, flushed with ice-cold PBS, cut longitudinally into 2 equal parts, and rolled up from the distal to the proximal end. The liver also was excised. Tissues were fixed in 10% formalin (Th Geyer, Renningen, Germany) overnight at 4°C. Paraffin sections were cut and stained with H&E stain. The small intestine and colon were scored as a combined score of inflammatory cell infiltration and tissue damage, as described previously by 2 independent blinded observers.78 Slides were visualized by an AxioImager Z1 microscope (Zeiss), and pictures were captured by a digital camera system (Axiocam HrC/HrM; Zeiss, Jena, Germany) using the semi-automated image analysis software (Axio Visision version 08/2013).
Enzyme-Linked Immunosorbent Assay
A segment of the colon and liver was weighed and diluted in a ratio of 20 μL RIPA buffer (including proteases) per milligram of tissue weight. The tissue then was lysed in a tissue lyser (Qiagen, Hilden, Germany) for 1 minute at 25 Hz, followed by centrifugation at 16,000 × g for 15 minutes at 4°C. The amount of protein was measured by bicinchonic acid. Serum, colon, and liver lysates were assayed for cytokines using commercially available kits for chemokine (C-X-C motif) ligand 1 and tumor necrosis factor-α (RnD, Minneapolis, MN) according to the manufacturer’s protocols. Cytokine levels of colon and liver were normalized to protein content.
Real-Time PCR Quantification of the Microbiota
DNA from feces, liver, and spleen was extracted as described. Real-time PCRs were performed on a 7900HT Fast Real-Time PCR system (Applied Biosystems) in duplicates using 16S rRNA gene-specific primers targeting all Eubacteria or Prevotella and fungal-specific primers for relative quantification (Table 7). Reactions consisted of 0.5 μL TaqMan gene expression assay, 4.5 μL TaqMan master mix (Thermo Fisher Scientific), 2.5 μL RNase free water, and 2.5 μL of the DNA sample (1 ng/μL), for a total of 10 μL in each well. The amplification program consisted of the following: (1) pre-incubation at 50°C for 2 minutes, and 95°C for 10 minutes; (2) 45 cycles of denaturation at 95°C for 15 seconds and annealing at the appropriate temperature for 1 minute; and (3) 1 cycle at 95°C for 15 seconds, 53°C for 15 seconds, and 95°C for 15 seconds. For spleen and liver, bacterial load was normalized to expression levels of β-actin (assay ID: Hs99999903_m1) using the standard-curve method. Statistical analysis was performed using GraphPad Prism (version 7; GraphPad Software, San Diego, CA), and statistical significance was evaluated by the Mann–Whitney U test. P values less than .05 were considered statistically significant. Measurements were performed using at least 5 biological replicates.
Table 7.
16S rRNA Gene-Specific Primers for Relative Quantification of Bacteria and Fungi
| Primer | Sequence 5’ to 3’ | Probe 5’ to 3’ | Reference |
|---|---|---|---|
| All bacteria, Bac338_F | ACTCCTACGGGAGGCAG | TGCCAGCAGCCGCGGTAATAC | PMID: 9687486 |
| All bacteria, Bac805_R | GACTACCAGGGTATCTAATCC | ||
| Bacteroidetes, Bac 32F | AACGCTAGCTACAGGCTTAACA | CAATATTCCTCACTGCTGCCTCCCGTA | PMID: 11010920 |
| Bacteroidetes, BactR | ACGCTACTTGGCTGGTTCA | ||
| Firmicutes, 8F | AGAGTTTGATCCTGGCTCAG | CTGATGGAGCAACGCCGCGT | PMID: 10461381 |
| Firmicutes, 534R | ATTACCGCGGCTGCTGG | PMID: 9687486 | |
| Prevotalla, F_Bacter 11 | CCTWCGATGGATAGGGGTT | AAGGTCCCCCACATTG | PMID: 19302550 |
| Prevotalla, R_Bacter 08 | CACGCTACTTGGCTGGTTCAG | ||
| FungiQuant-F | GGRAAACTCACCAGGTCCAG | TGGTGCATGGCCGTT | PMID: 23136846 |
| FungiQuant-R | GSWCTATCCCCAKCACGA |
Availability of Data and Materials
All relevant data are included within the manuscript files. Raw sequencing data included in this study were uploaded to European Bioinformatics Institute (EBI)'s European Nucleotide Archive (ENA) under accession number PRJEB21817 (http://www.ebi.ac.uk/ena/data/view/PRJEB21817).
Statistical Analysis
Biostatistical analyses were performed using GraphPad Prism (version 7) software (GraphPad, Inc, La Jolla, CA), Mothur, MacQIIME v1.9.1, or R (v 3.2.5). Specific comparisons and analyses are described in the individual Materials and Method sections. Differences between the groups were considered significant at P < .05 and the data are presented as means ± SEM. All authors had access to the study data and reviewed and approved the final manuscript.
Acknowledgments
The authors thank Christian Kautz for providing the assays for identifying antibiotic-resistance genes. The authors also thank Karina Greve, Dorina Oelsner, Melanie Vollstedt, Maren Reffelmann, Melanie Nebendahl, Sabine Kock, Stefanie Baumgarten, Tatjana Schmidtke, and Jan Schubert for excellent technical assistance.
Footnotes
Author contributions Jacqueline Moltzau Anderson, Simone Lipinski, Felix Sommer, and Philip Rosenstiel designed the research; Simone Lipinski, Olivier Boulard, and Maren Falk-Paulsen managed the specific pathogen free mouse breeding, monitored the mice, and collected fecal pellets; Sven Künzel and John F. Baines managed the germ-free mouse breeding and provided and maintained the germ-free mouse facility; Simone Lipinski and Felix Sommer performed the germ-free mouse experiment and collected samples; Jacqueline Moltzau Anderson, Simone Lipinski, Maren Falk-Paulsen, Stephanie T. Stengel, and Ateequr Rehman conducted the wet laboratory experiments; Jacqueline Moltzau Anderson, Simone Lipinski, Olivier Boulard, Ateequr Rehman, Maren Falk-Paulsen, Stephanie T. Stengel, Konrad Aden, Felix Sommer, Mathias Chamaillard, and Philip Rosenstiel analyzed and interpreted the data; Jacqueline Moltzau Anderson, Wei-Hung Pan, Ateequr Rehman, Felix Sommer, and Richa Bharti performed the bioinformatics analyses; Jacqueline Moltzau Anderson, Simone Lipinski, Wei-Hung Pan, Felix Sommer, and Robert Häsler prepared the figures; Mathias Chamaillard and Philip Rosenstiel obtained funding; and Jacqueline Moltzau Anderson, Simone Lipinski, Felix Sommer, Mathias Chamaillard, and Philip Rosenstiel co-wrote the manuscript.
Conflicts of interest The authors disclose no conflicts.
Funding This work was supported by the International Max Planck Research Schools of the Max-Planck-Institute for Evolutionary Biology and Christian-Albrechts-University Kiel, the Origin and Function of MetaorganismsCollaborative Research Centre 1182 (C2), the Precision Medicine in Chronic InflammationExcellence Cluster 2167, the Inflammation at InterfacesExcellence Cluster 306 of the Deutsche Forschungsgemeinschaft and the Schleswig-Holstein Excellence Chair Program (P.R. and J.F.B.), and by Institut national de la santé et de la recherche médicale (M.C.) and by the Fondation pour la Recherche Médicale grant (grant number DEQ20130326475 to M.C.).
References
- 1.Ellinghaus D., Jostins L., Spain S.L., Cortes A., Bethune J., Han B., Park Y.R., Raychaudhuri S., Pouget J.G., Hubenthal M., Folseraas T., Wang Y., Esko T., Metspalu A., Westra H.J., Franke L., Pers T.H., Weersma R.K., Collij V., D'Amato M., Halfvarson J., Jensen A.B., Lieb W., Degenhardt F., Forstner A.J., Hofmann A., Schreiber S., Mrowietz U., Juran B.D., Lazaridis K.N., Brunak S., Dale A.M., Trembath R.C., Weidinger S., Weichenthal M., Ellinghaus E., Elder J.T., Barker J.N., Andreassen O.A., McGovern D.P., Karlsen T.H., Barrett J.C., Parkes M., Brown M.A., Franke A. Analysis of five chronic inflammatory diseases identifies 27 new associations and highlights disease-specific patterns at shared loci. Nat Genet. 2016;48:510–518. doi: 10.1038/ng.3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chu H., Khosravi A., Kusumawardhani I.P., Kwon A.H., Vasconcelos A.C., Cunha L.D., Mayer A.E., Shen Y., Wu W.L., Kambal A., Targan S.R., Xavier R.J., Ernst P.B., Green D.R., McGovern D.P., Virgin H.W., Mazmanian S.K. Gene-microbiota interactions contribute to the pathogenesis of inflammatory bowel disease. Science. 2016;352:1116–1120. doi: 10.1126/science.aad9948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jostins L., Ripke S., Weersma R.K., Duerr R.H., McGovern D.P., Hui K.Y., Lee J.C., Schumm L.P., Sharma Y., Anderson C.A., Essers J., Mitrovic M., Ning K., Cleynen I., Theatre E., Spain S.L., Raychaudhuri S., Goyette P., Wei Z., Abraham C., Achkar J.P., Ahmad T., Amininejad L., Ananthakrishnan A.N., Andersen V., Andrews J.M., Baidoo L., Balschun T., Bampton P.A., Bitton A., Boucher G., Brand S., Buning C., Cohain A., Cichon S., D'Amato M., De Jong D., Devaney K.L., Dubinsky M., Edwards C., Ellinghaus D., Ferguson L.R., Franchimont D., Fransen K., Gearry R., Georges M., Gieger C., Glas J., Haritunians T., Hart A., Hawkey C., Hedl M., Hu X., Karlsen T.H., Kupcinskas L., Kugathasan S., Latiano A., Laukens D., Lawrance I.C., Lees C.W., Louis E., Mahy G., Mansfield J., Morgan A.R., Mowat C., Newman W., Palmieri O., Ponsioen C.Y., Potocnik U., Prescott N.J., Regueiro M., Rotter J.I., Russell R.K., Sanderson J.D., Sans M., Satsangi J., Schreiber S., Simms L.A., Sventoraityte J., Targan S.R., Taylor K.D., Tremelling M., Verspaget H.W., De Vos M., Wijmenga C., Wilson D.C., Winkelmann J., Xavier R.J., Zeissig S., Zhang B., Zhang C.K., Zhao H., International I.B.D.G.C., Silverberg M.S., Annese V., Hakonarson H., Brant S.R., Radford-Smith G., Mathew C.G., Rioux J.D., Schadt E.E., Daly M.J., Franke A., Parkes M., Vermeire S., Barrett J.C., Cho J.H. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hugot J.P., Chamaillard M., Zouali H., Lesage S., Cezard J.P., Belaiche J., Almer S., Tysk C., O'Morain C.A., Gassull M., Binder V., Finkel Y., Cortot A., Modigliani R., Laurent-Puig P., Gower-Rousseau C., Macry J., Colombel J.F., Sahbatou M., Thomas G. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature. 2001;411:599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- 5.Ogura Y., Bonen D.K., Inohara N., Nicolae D.L., Chen F.F., Ramos R., Britton H., Moran T., Karaliuskas R., Duerr R.H., Achkar J.P., Brant S.R., Bayless T.M., Kirschner B.S., Hanauer S.B., Nunez G., Cho J.H. A frameshift mutation in NOD2 associated with susceptibility to Crohn's disease. Nature. 2001;411:603–606. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- 6.Hampe J., Cuthbert A., Croucher P.J., Mirza M.M., Mascheretti S., Fisher S., Frenzel H., King K., Hasselmeyer A., MacPherson A.J., Bridger S., van Deventer S., Forbes A., Nikolaus S., Lennard-Jones J.E., Foelsch U.R., Krawczak M., Lewis C., Schreiber S., Mathew C.G. Association between insertion mutation in NOD2 gene and Crohn's disease in German and British populations. Lancet. 2001;357:1925–1928. doi: 10.1016/S0140-6736(00)05063-7. [DOI] [PubMed] [Google Scholar]
- 7.Rosenstiel P., Fantini M., Brautigam K., Kuhbacher T., Waetzig G.H., Seegert D., Schreiber S. TNF-alpha and IFN-gamma regulate the expression of the NOD2 (CARD15) gene in human intestinal epithelial cells. Gastroenterology. 2003;124:1001–1009. doi: 10.1053/gast.2003.50157. [DOI] [PubMed] [Google Scholar]
- 8.Lala S., Ogura Y., Osborne C., Hor S.Y., Bromfield A., Davies S., Ogunbiyi O., Nunez G., Keshav S. Crohn's disease and the NOD2 gene: a role for Paneth cells. Gastroenterology. 2003;125:47–57. doi: 10.1016/s0016-5085(03)00661-9. [DOI] [PubMed] [Google Scholar]
- 9.Inohara N., Ogura Y., Fontalba A., Gutierrez O., Pons F., Crespo J., Fukase K., Inamura S., Kusumoto S., Hashimoto M., Foster S.J., Moran A.P., Fernandez-Luna J.L., Nunez G. Host recognition of bacterial muramyl dipeptide mediated through NOD2. Implications for Crohn's disease. J Biol Chem. 2003;278:5509–5512. doi: 10.1074/jbc.C200673200. [DOI] [PubMed] [Google Scholar]
- 10.Nakagome S., Mano S., Kozlowski L., Bujnicki J.M., Shibata H., Fukumaki Y., Kidd J.R., Kidd K.K., Kawamura S., Oota H. Crohn's disease risk alleles on the NOD2 locus have been maintained by natural selection on standing variation. Mol Biol Evol. 2012;29:1569–1585. doi: 10.1093/molbev/mss006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kobayashi K.S., Chamaillard M., Ogura Y., Henegariu O., Inohara N., Nunez G., Flavell R.A. Nod2-dependent regulation of innate and adaptive immunity in the intestinal tract. Science. 2005;307:731–734. doi: 10.1126/science.1104911. [DOI] [PubMed] [Google Scholar]
- 12.Watanabe T., Kitani A., Murray P.J., Strober W. NOD2 is a negative regulator of Toll-like receptor 2-mediated T helper type 1 responses. Nat Immunol. 2004;5:800–808. doi: 10.1038/ni1092. [DOI] [PubMed] [Google Scholar]
- 13.Philpott D.J., Sorbara M.T., Robertson S.J., Croitoru K., Girardin S.E. NOD proteins: regulators of inflammation in health and disease. Nat Rev Immunol. 2014;14:9–23. doi: 10.1038/nri3565. [DOI] [PubMed] [Google Scholar]
- 14.Rehman A., Sina C., Gavrilova O., Häsler R., Ott S., Baines J., Schreiber S., Rosenstiel P. Nod2 is essential for temporal development of intestinal microbial communities. Gut. 2011;60:1354–1362. doi: 10.1136/gut.2010.216259. [DOI] [PubMed] [Google Scholar]
- 15.Mondot S., Barreau F., Al Nabhani Z., Dussaillant M., Le Roux K., Dore J., Leclerc M., Hugot J.P., Lepage P. Altered gut microbiota composition in immune-impaired Nod2(-/-) mice. Gut. 2012;61:634–635. doi: 10.1136/gutjnl-2011-300478. [DOI] [PubMed] [Google Scholar]
- 16.Robertson S.J., Zhou J.Y., Geddes K., Rubino S.J., Cho J.H., Girardin S.E., Philpott D.J. Nod1 and Nod2 signaling does not alter the composition of intestinal bacterial communities at homeostasis. Gut Microbes. 2013;4:222–231. doi: 10.4161/gmic.24373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goethel A., Turpin W., Rouquier S., Zanello G., Robertson S.J., Streutker C.J., Philpott D.J., Croitoru K. Nod2 influences microbial resilience and susceptibility to colitis following antibiotic exposure. Mucosal Immunol. 2019;12:720–732. doi: 10.1038/s41385-018-0128-y. [DOI] [PubMed] [Google Scholar]
- 18.Ananthakrishnan A.N. Epidemiology and risk factors for IBD. Nat Rev Gastroenterol Hepatol. 2015;12:205–217. doi: 10.1038/nrgastro.2015.34. [DOI] [PubMed] [Google Scholar]
- 19.Jakobsson H.E., Jernberg C., Andersson A.F., Sjolund-Karlsson M., Jansson J.K., Engstrand L. Short-term antibiotic treatment has differing long-term impacts on the human throat and gut microbiome. PLoS One. 2010;5 doi: 10.1371/journal.pone.0009836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jernberg C., Lofmark S., Edlund C., Jansson J.K. Long-term ecological impacts of antibiotic administration on the human intestinal microbiota. ISME J. 2007;1:56–66. doi: 10.1038/ismej.2007.3. [DOI] [PubMed] [Google Scholar]
- 21.Ng K.M., Aranda-Diaz A., Tropini C., Frankel M.R., Van Treuren W., O'Laughlin C.T., Merrill B.D., Yu F.B., Pruss K.M., Oliveira R.A., Higginbottom S.K., Neff N.F., Fischbach M.A., Xavier K.B., Sonnenburg J.L., Huang K.C. Recovery of the gut microbiota after antibiotics depends on host diet, community context, and environmental reservoirs. Cell Host Microbe. 2019;26:650–665 e4. doi: 10.1016/j.chom.2019.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rakoff-Nahoum S., Paglino J., Eslami-Varzaneh F., Edberg S., Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–241. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 23.Botton S., van Heusden M., Parsons J.R., Smidt H., van Straalen N. Resilience of microbial systems towards disturbances. Crit Rev Microbiol. 2006;32:101–112. doi: 10.1080/10408410600709933. [DOI] [PubMed] [Google Scholar]
- 24.Shade A., Peter H., Allison S.D., Baho D.L., Berga M., Burgmann H., Huber D.H., Langenheder S., Lennon J.T., Martiny J.B., Matulich K.L., Schmidt T.M., Handelsman J. Fundamentals of microbial community resistance and resilience. Front Microbiol. 2012;3:417. doi: 10.3389/fmicb.2012.00417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.MacLean R.C., San Millan A. The evolution of antibiotic resistance. Science. 2019;365:1082–1083. doi: 10.1126/science.aax3879. [DOI] [PubMed] [Google Scholar]
- 26.Hasler R., Kautz C., Rehman A., Podschun R., Gassling V., Brzoska P., Sherlock J., Grasner J.T., Hoppenstedt G., Schubert S., Ferlinz A., Lieb W., Laudes M., Heinsen F.A., Scholz J., Harmsen D., Franke A., Eisend S., Kunze T., Fickenscher H., Ott S., Rosenstiel P., Schreiber S. The antibiotic resistome and microbiota landscape of refugees from Syria, Iraq and Afghanistan in Germany. Microbiome. 2018;6:37. doi: 10.1186/s40168-018-0414-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ott S.J., Musfeldt M., Wenderoth D.F., Hampe J., Brant O., Folsch U.R., Timmis K.N., Schreiber S. Reduction in diversity of the colonic mucosa associated bacterial microflora in patients with active inflammatory bowel disease. Gut. 2004;53:685–693. doi: 10.1136/gut.2003.025403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hildebrand F., Nguyen T.L., Brinkman B., Yunta R.G., Cauwe B., Vandenabeele P., Liston A., Raes J. Inflammation-associated enterotypes, host genotype, cage and inter-individual effects drive gut microbiota variation in common laboratory mice. Genome Biol. 2013;14:R4. doi: 10.1186/gb-2013-14-1-r4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blaser M.J. Antibiotic use and its consequences for the normal microbiome. Science. 2016;352:544–545. doi: 10.1126/science.aad9358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Knights D., Silverberg M.S., Weersma R.K., Gevers D., Dijkstra G., Huang H., Tyler A.D., van Sommeren S., Imhann F., Stempak J.M., Vangay P., Al-Ghalith G.A., Russell C., Sauk J., Knight J., Daly M.J., Huttenhower C., Xavier R.J. Complex host genetics influence the microbiome in inflammatory bowel disease. Genome Med. 2014;6:107. doi: 10.1186/s13073-014-0107-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gevers D., Kugathasan S., Denson L.A., Vazquez-Baeza Y., Van Treuren W., Ren B., Schwager E., Knights D., Song S.J., Yassour M., Morgan X.C., Kostic A.D., Luo C., Gonzalez A., McDonald D., Haberman Y., Walters T., Baker S., Rosh J., Stephens M., Heyman M., Markowitz J., Baldassano R., Griffiths A., Sylvester F., Mack D., Kim S., Crandall W., Hyams J., Huttenhower C., Knight R., Xavier R.J. The treatment-naive microbiome in new-onset Crohn's disease. Cell Host Microbe. 2014;15:382–392. doi: 10.1016/j.chom.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morgan X.C., Tickle T.L., Sokol H., Gevers D., Devaney K.L., Ward D.V., Reyes J.A., Shah S.A., LeLeiko N., Snapper S.B., Bousvaros A., Korzenik J., Sands B.E., Xavier R.J., Huttenhower C. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012;13:R79. doi: 10.1186/gb-2012-13-9-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rooks M.G., Veiga P., Wardwell-Scott L.H., Tickle T., Segata N., Michaud M., Gallini C.A., Beal C., van Hylckama-Vlieg J.E., Ballal S.A., Morgan X.C., Glickman J.N., Gevers D., Huttenhower C., Garrett W.S. Gut microbiome composition and function in experimental colitis during active disease and treatment-induced remission. ISME J. 2014;8:1403–1417. doi: 10.1038/ismej.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carvalho F.A., Koren O., Goodrich J.K., Johansson M.E., Nalbantoglu I., Aitken J.D., Su Y., Chassaing B., Walters W.A., Gonzalez A., Clemente J.C., Cullender T.C., Barnich N., Darfeuille-Michaud A., Vijay-Kumar M., Knight R., Ley R.E., Gewirtz A.T. Transient inability to manage proteobacteria promotes chronic gut inflammation in TLR5-deficient mice. Cell Host Microbe. 2012;12:139–152. doi: 10.1016/j.chom.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ubeda C., Pamer E.G. Antibiotics, microbiota, and immune defense. Trends Immunol. 2012;33:459–466. doi: 10.1016/j.it.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Faber F., Tran L., Byndloss M.X., Lopez C.A., Velazquez E.M., Kerrinnes T., Nuccio S.P., Wangdi T., Fiehn O., Tsolis R.M., Baumler A.J. Host-mediated sugar oxidation promotes post-antibiotic pathogen expansion. Nature. 2016;534:697–699. doi: 10.1038/nature18597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hildebrand F., Moitinho-Silva L., Blasche S., Jahn M.T., Gossmann T.I., Huerta-Cepas J., Hercog R., Luetge M., Bahram M., Pryszlak A., Alves R.J., Waszak S.M., Zhu A., Ye L., Costea P.I., Aalvink S., Belzer C., Forslund S.K., Sunagawa S., Hentschel U., Merten C., Patil K.R., Benes V., Bork P. Antibiotics-induced monodominance of a novel gut bacterial order. Gut. 2019;68:1781–1790. doi: 10.1136/gutjnl-2018-317715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Elhenawy W., Tsai C.N., Coombes B.K. Host-specific adaptive diversification of Crohn's disease-associated adherent-invasive Escherichia coli. Cell Host Microbe. 2019;25:301–312 e5. doi: 10.1016/j.chom.2018.12.010. [DOI] [PubMed] [Google Scholar]
- 39.Zhao S., Lieberman T.D., Poyet M., Kauffman K.M., Gibbons S.M., Groussin M., Xavier R.J., Alm E.J. Adaptive evolution within gut microbiomes of healthy people. Cell Host Microbe. 2019;25:656–667 e8. doi: 10.1016/j.chom.2019.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Frank D.N., St Amand A.L., Feldman R.A., Boedeker E.C., Harpaz N., Pace N.R. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Manichanh C., Rigottier-Gois L., Bonnaud E., Gloux K., Pelletier E., Frangeul L., Nalin R., Jarrin C., Chardon P., Marteau P., Roca J., Dore J. Reduced diversity of faecal microbiota in Crohn's disease revealed by a metagenomic approach. Gut. 2006;55:205–211. doi: 10.1136/gut.2005.073817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Couturier-Maillard A., Secher T., Rehman A., Normand S., De Arcangelis A., Haesler R., Huot L., Grandjean T., Bressenot A., Delanoye-Crespin A., Gaillot O., Schreiber S., Lemoine Y., Ryffel B., Hot D., Nunez G., Chen G., Rosenstiel P., Chamaillard M. NOD2-mediated dysbiosis predisposes mice to transmissible colitis and colorectal cancer. J Clin Invest. 2013;123:700–711. doi: 10.1172/JCI62236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ubeda C., Bucci V., Caballero S., Djukovic A., Toussaint N.C., Equinda M., Lipuma L., Ling L., Gobourne A., No D., Taur Y., Jenq R.R., van den Brink M.R., Xavier J.B., Pamer E.G. Intestinal microbiota containing Barnesiella species cures vancomycin-resistant Enterococcus faecium colonization. Infect Immun. 2013;81:965–973. doi: 10.1128/IAI.01197-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scanlan P.D., Shanahan F., O'Mahony C., Marchesi J.R. Culture-independent analyses of temporal variation of the dominant fecal microbiota and targeted bacterial subgroups in Crohn's disease. J Clin Microbiol. 2006;44:3980–3988. doi: 10.1128/JCM.00312-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ramanan D., Tang M.S., Bowcutt R., Loke P., Cadwell K. Bacterial sensor Nod2 prevents inflammation of the small intestine by restricting the expansion of the commensal Bacteroides vulgatus. Immunity. 2014;41:311–324. doi: 10.1016/j.immuni.2014.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wehkamp J., Harder J., Weichenthal M., Schwab M., Schaffeler E., Schlee M., Herrlinger K.R., Stallmach A., Noack F., Fritz P., Schroder J.M., Bevins C.L., Fellermann K., Stange E.F. NOD2 (CARD15) mutations in Crohn's disease are associated with diminished mucosal alpha-defensin expression. Gut. 2004;53:1658–1664. doi: 10.1136/gut.2003.032805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rosenstiel P., Till A., Schreiber S. NOD-like receptors and human diseases. Microbes Infect. 2007;9:648–657. doi: 10.1016/j.micinf.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 48.Cooney R., Baker J., Brain O., Danis B., Pichulik T., Allan P., Ferguson D.J., Campbell B.J., Jewell D., Simmons A. NOD2 stimulation induces autophagy in dendritic cells influencing bacterial handling and antigen presentation. Nat Med. 2010;16:90–97. doi: 10.1038/nm.2069. [DOI] [PubMed] [Google Scholar]
- 49.Travassos L.H., Carneiro L.A., Ramjeet M., Hussey S., Kim Y.G., Magalhaes J.G., Yuan L., Soares F., Chea E., Le Bourhis L., Boneca I.G., Allaoui A., Jones N.L., Nunez G., Girardin S.E., Philpott D.J. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat Immunol. 2010;11:55–62. doi: 10.1038/ni.1823. [DOI] [PubMed] [Google Scholar]
- 50.Keestra-Gounder A.M., Byndloss M.X., Seyffert N., Young B.M., Chavez-Arroyo A., Tsai A.Y., Cevallos S.A., Winter M.G., Pham O.H., Tiffany C.R., de Jong M.F., Kerrinnes T., Ravindran R., Luciw P.A., McSorley S.J., Baumler A.J., Tsolis R.M. NOD1 and NOD2 signalling links ER stress with inflammation. Nature. 2016;532:394–397. doi: 10.1038/nature17631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim D., Kim Y.M., Kim W.U., Park J.H., Nunez G., Seo S.U. Recognition of the microbiota by Nod2 contributes to the oral adjuvant activity of cholera toxin through the induction of interleukin-1beta. Immunology. 2019;158:219–229. doi: 10.1111/imm.13105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Caruso R., Mathes T., Martens E.C., Kamada N., Nusrat A., Inohara N., Nunez G. A specific gene-microbe interaction drives the development of Crohn's disease-like colitis in mice. Sci Immunol. 2019;4 doi: 10.1126/sciimmunol.aaw4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dollive S., Chen Y.Y., Grunberg S., Bittinger K., Hoffmann C., Vandivier L., Cuff C., Lewis J.D., Wu G.D., Bushman F.D. Fungi of the murine gut: episodic variation and proliferation during antibiotic treatment. PLoS One. 2013;8 doi: 10.1371/journal.pone.0071806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jawhara S., Thuru X., Standaert-Vitse A., Jouault T., Mordon S., Sendid B., Desreumaux P., Poulain D. Colonization of mice by Candida albicans is promoted by chemically induced colitis and augments inflammatory responses through galectin-3. J Infect Dis. 2008;197:972–980. doi: 10.1086/528990. [DOI] [PubMed] [Google Scholar]
- 55.Wagener J., Malireddi R.K., Lenardon M.D., Koberle M., Vautier S., MacCallum D.M., Biedermann T., Schaller M., Netea M.G., Kanneganti T.D., Brown G.D., Brown A.J., Gow N.A. Fungal chitin dampens inflammation through IL-10 induction mediated by NOD2 and TLR9 activation. PLoS Pathog. 2014;10 doi: 10.1371/journal.ppat.1004050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Juste C., Kreil D.P., Beauvallet C., Guillot A., Vaca S., Carapito C., Mondot S., Sykacek P., Sokol H., Blon F., Lepercq P., Levenez F., Valot B., Carre W., Loux V., Pons N., David O., Schaeffer B., Lepage P., Martin P., Monnet V., Seksik P., Beaugerie L., Ehrlich S.D., Gibrat J.F., Van Dorsselaer A., Dore J. Bacterial protein signals are associated with Crohn's disease. Gut. 2014;63:1566–1577. doi: 10.1136/gutjnl-2012-303786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hoffmann C., Dollive S., Grunberg S., Chen J., Li H., Wu G.D., Lewis J.D., Bushman F.D. Archaea and fungi of the human gut microbiome: correlations with diet and bacterial residents. PLoS One. 2013;8 doi: 10.1371/journal.pone.0066019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kolachala V.L., Vijay-Kumar M., Dalmasso G., Yang D., Linden J., Wang L., Gewirtz A., Ravid K., Merlin D., Sitaraman S.V. A2B adenosine receptor gene deletion attenuates murine colitis. Gastroenterology. 2008;135:861–870. doi: 10.1053/j.gastro.2008.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Aden K., Tran F., Ito G., Sheibani-Tezerji R., Lipinski S., Kuiper J.W., Tschurtschenthaler M., Saveljeva S., Bhattacharyya J., Hasler R., Bartsch K., Luzius A., Jentzsch M., Falk-Paulsen M., Stengel S.T., Welz L., Schwarzer R., Rabe B., Barchet W., Krautwald S., Hartmann G., Pasparakis M., Blumberg R.S., Schreiber S., Kaser A., Rosenstiel P. ATG16L1 orchestrates interleukin-22 signaling in the intestinal epithelium via cGAS-STING. J Exp Med. 2018;215:2868–2886. doi: 10.1084/jem.20171029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rodriguez-Nunez I., Caluag T., Kirby K., Rudick C.N., Dziarski R., Gupta D. Nod2 and Nod2-regulated microbiota protect BALB/c mice from diet-induced obesity and metabolic dysfunction. Sci Rep. 2017;7:548. doi: 10.1038/s41598-017-00484-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shea-Donohue T., Thomas K., Cody M.J., Aiping Z., Detolla L.J., Kopydlowski K.M., Fukata M., Lira S.A., Vogel S.N. Mice deficient in the CXCR2 ligand, CXCL1 (KC/GRO-alpha), exhibit increased susceptibility to dextran sodium sulfate (DSS)-induced colitis. Innate Immun. 2008;14:117–124. doi: 10.1177/1753425908088724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sommer F., Anderson J.M., Bharti R., Raes J., Rosenstiel P. The resilience of the intestinal microbiota influences health and disease. Nat Rev Microbiol. 2017;15:630–638. doi: 10.1038/nrmicro.2017.58. [DOI] [PubMed] [Google Scholar]
- 63.Vijay-Kumar M., Aitken J.D., Carvalho F.A., Cullender T.C., Mwangi S., Srinivasan S., Sitaraman S.V., Knight R., Ley R.E., Gewirtz A.T. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science. 2010;328:228–231. doi: 10.1126/science.1179721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sommer F., Adam N., Johansson M.E., Xia L., Hansson G.C., Backhed F. Altered mucus glycosylation in core 1 O-glycan-deficient mice affects microbiota composition and intestinal architecture. PLoS One. 2014;9 doi: 10.1371/journal.pone.0085254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pan W.H., Sommer F., Falk-Paulsen M., Ulas T., Best P., Fazio A., Kachroo P., Luzius A., Jentzsch M., Rehman A., Muller F., Lengauer T., Walter J., Kunzel S., Baines J.F., Schreiber S., Franke A., Schultze J.L., Backhed F., Rosenstiel P. Exposure to the gut microbiota drives distinct methylome and transcriptome changes in intestinal epithelial cells during postnatal development. Genome Med. 2018;10:27. doi: 10.1186/s13073-018-0534-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sommer F., Stahlman M., Ilkayeva O., Arnemo J.M., Kindberg J., Josefsson J., Newgard C.B., Frobert O., Backhed F. The gut microbiota modulates energy metabolism in the hibernating brown bear Ursus arctos. Cell Rep. 2016;14:1655–1661. doi: 10.1016/j.celrep.2016.01.026. [DOI] [PubMed] [Google Scholar]
- 67.Kiouptsi K., Jackel S., Pontarollo G., Grill A., Kuijpers M.J.E., Wilms E., Weber C., Sommer F., Nagy M., Neideck C., Jansen Y., Ascher S., Formes H., Karwot C., Bayer F., Kollar B., Subramaniam S., Molitor M., Wenzel P., Rosenstiel P., Todorov H., Gerber S., Walter U., Jurk K., Heemskerk J.W.M., van der Vorst E.P.C., Doring Y., Reinhardt C. The microbiota promotes arterial thrombosis in low-density lipoprotein receptor-deficient mice. MBio. 2019;10 doi: 10.1128/mBio.02298-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schloss P.D., Westcott S.L., Ryabin T., Hall J.R., Hartmann M., Hollister E.B., Lesniewski R.A., Oakley B.B., Parks D.H., Robinson C.J., Sahl J.W., Stres B., Thallinger G.G., Van Horn D.J., Weber C.F. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Werner JJ. http://www.wernerlab.org/software/macqiime.
- 70.Hanley J.A., Negassa A., Edwardes M.D., Forrester J.E. Statistical analysis of correlated data using generalized estimating equations: an orientation. Am J Epidemiol. 2003;157:364–375. doi: 10.1093/aje/kwf215. [DOI] [PubMed] [Google Scholar]
- 71.Gardes M., Bruns T.D. ITS primers with enhanced specificity for basidiomycetes--application to the identification of mycorrhizae and rusts. Mol Ecol. 1993;2:113–118. doi: 10.1111/j.1365-294x.1993.tb00005.x. [DOI] [PubMed] [Google Scholar]
- 72.White T, Bruns T, Lee S, Taylor J, Innis M, Gelfand D, Sninsky J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. Academic Press, Cambridge, MA 1990:315–322.
- 73.Gweon H.S., Oliver A., Taylor J., Booth T., Gibbs M., Read D.S., Griffiths R.I., Schonrogge K. PIPITS: an automated pipeline for analyses of fungal internal transcribed spacer sequences from the Illumina sequencing platform. Methods Ecol Evol. 2015;6:973–980. doi: 10.1111/2041-210X.12399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang J., Kobert K., Flouri T., Stamatakis A. PEAR: a fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics. 2014;30:614–620. doi: 10.1093/bioinformatics/btt593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bengtsson-Palme J., Ryberg M., Hartmann M., Branco S., Wang Z., Godhe A., De Wit P., Sánchez-García M., Ebersberger I., de Sousa F., Amend A., Jumpponen A., Unterseher M., Kristiansson E., Abarenkov K., Bertrand Y.J.K., Sanli K., Eriksson K.M., Vik U., Veldre V., Nilsson R.H. Improved software detection and extraction of ITS1 and ITS2 from ribosomal ITS sequences of fungi and other eukaryotes for analysis of environmental sequencing data. Methods Ecol Evol. 2013;4:914–919. [Google Scholar]
- 76.Nilsson R.H., Tedersoo L., Ryberg M., Kristiansson E., Hartmann M., Unterseher M., Porter T.M., Bengtsson-Palme J., Walker D.M., de Sousa F., Gamper H.A., Larsson E., Larsson K.H., Koljalg U., Edgar R.C., Abarenkov K. A comprehensive, automatically updated fungal ITS sequence dataset for reference-based chimera control in environmental sequencing efforts. Microbes Environ. 2015;30:145–150. doi: 10.1264/jsme2.ME14121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 78.Adolph T.E., Tomczak M.F., Niederreiter L., Ko H.J., Bock J., Martinez-Naves E., Glickman J.N., Tschurtschenthaler M., Hartwig J., Hosomi S., Flak M.B., Cusick J.L., Kohno K., Iwawaki T., Billmann-Born S., Raine T., Bharti R., Lucius R., Kweon M.N., Marciniak S.J., Choi A., Hagen S.J., Schreiber S., Rosenstiel P., Kaser A., Blumberg R.S. Paneth cells as a site of origin for intestinal inflammation. Nature. 2013;503:272–276. doi: 10.1038/nature12599. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are included within the manuscript files. Raw sequencing data included in this study were uploaded to European Bioinformatics Institute (EBI)'s European Nucleotide Archive (ENA) under accession number PRJEB21817 (http://www.ebi.ac.uk/ena/data/view/PRJEB21817).