Abstract
Despite improvements in prevention and treatment, cervical cancer (CC) still poses a serious threat to women’s health. CHMP4C (chromatin modified protein 4C) is a subunit of the endosomal sorting complex required for transport, which is expressed in both nucleus and cytoplasm. Here, we examined the effect of CHMP4C on the biological behavior of CC cells and the underlying mechanisms. We report that CHMP4C expression is higher in CC tissues, and high CHMP4C expression is associated with lower survival. Up‐regulation of CHMP4C in C‐33A cells accelerates cell proliferation, migration and invasion, whereas down‐regulation of CHMP4C in Ca Ski cells had the opposite effect. Moreover, overexpression of CHMP4C induced activation of the epithelial–mesenchymal transition pathway, whereas depletion of CHMP4C inhibited activation. Our results suggest that CHMP4C contributes to the viability and motility of CC cells by modulating epithelial–mesenchymal transition and may facilitate the identification of novel biomarkers for CC therapy.
Keywords: cervical cancer, chromatin modified protein 4C, EMT, proliferation
Chromatin modified protein 4C is highly expressed in cervical cancer (CC) tissues and cell lines, and contributes to the proliferation, invasion and migration of CC cells by modulating the epithelial–mesenchymal transition. Our findings may facilitate the identification of novel biomarkers for CC therapy.
![]()
Abbreviations
- CC
cervical cancer
- CCK‐8
Cell Counting Kit‐8
- CHMP4C
chromatin modified protein 4C
- EMT
epithelial–mesenchymal transition
- HPV
human papillomavirus
- qRT‐PCR
quantitative real‐time PCR
- SD
standard deviation
Cervical cancer (CC) is one of the most common gynecological malignancies [1, 2]. Every year, more than 260 000 women die of CC, mostly in developing countries [3]. The difference in incidence between countries is mainly due to the different awareness of effective screening or preventive treatment [3]. To date, it has been clarified that long‐term recurrent human papillomavirus (HPV) infection is the maximum risk factor for CC [4, 5]. Although effective screening and prevention methods are available, CC still poses a serious threat to women’s health because of its expensive prices and the backward concepts [6]. Hence it is of great clinical significance to explore the molecular mechanism of CC for improving its early diagnosis and treatment.
Chromatin modified protein 4C (CHMP4C) belongs to the CHMP (chromatin modified protein) family. It is a subunit of the endosomal sorting complex required for transport, which is expressed in both nucleus and cytoplasm [7, 8]. Endosomal sorting complex required for transport is involved in the cytokinesis of daughter cells [9], and its dependent extracellular vesicles play an important role in many processes, including the etiology and progression of cancer [10]. Sadler et al. [11] found that the polymorphism of CHMP4C increased the susceptibility of cancer and may induce tumorigenesis by disrupting the stability of genome. Based on this feature, CHMP4C has been proved to be unbalanced in many cancers, such as human lung cancer [12], ovarian cancer [13], prostate cancer [14], among others. Nevertheless, no studies have investigated whether it contributes to the malignant progression of CC. According to bioinformatics analysis, we found that CHMP4C was also dysregulated in CC, suggesting that regulating the expression of CHMP4C might be a novel strategy to treat CC. Consequently, based on the published literature and the results of bioinformatics analysis, we chose CHMP4C for further research.
This work aimed to verify the relevancy between the expression of CHMP4C and survival rate of patients with CC, and to explore whether the modulation of CHMP4C expression would impact the biological behavior of CC cells, as well as the regulatory pathways, hoping to discover a novel biomarker that can predict the development of CC.
Materials and methods
Data collection
The expression data of CC were downloaded from The Cancer Genome Atlas database (https://www.cancer.gov/about‐nci/organization/ccg/research/structural‐genomics/tcga), including tumor group (n = 306) and normal group (n = 3). These data were used to analyze the differential expression of CHMP4C in CC tissues and the correlation between CHMP4C expression and outcomes of patients with CC.
Cell culture
Human CC cell lines (HeLa, C‐33A and Ca Ski) and normal control cells (Ect1/E6E7) were obtained from the cell bank of the Typical Culture Preservation Committee of the Chinese Academy of Sciences. Cells were routinely cultured in Roswell Park Memorial Institute‐1640 medium with 10% FBS, 100 U·mL−1 penicillin and 0.1 mg·mL−1 streptomycin at 37 °C in 5% CO2.
Cell transfection
siRNA sequences (si‐CHMP4C#1: 5′‐CCAAGAAATCTCAGAAG‐3′; si‐CHMP4C#2: 5′‐GATGGCACACTTTCTAC‐3′ and si‐con: 5′‐CGAACUCACUGGUCUGACC‐3′) were synthesized from Shanghai GenePharma Co., Ltd. (Shanghai, China), which were used for knockdown of CHMP4C. Simultaneously, for overexpression of CHMP4C in CC cell lines, pcDNA3.1‐CHMP4C and corresponding control vector were also purchased from Shanghai GenePharma Co., Ltd. Lipofectamine 2000 Reagent (Invitrogen, Carlsbad, CA, USA) was used for transfection, according to the manufacturer’s protocol.
RNA extraction and quantitative real‐time PCR
The total RNA of cells was extracted using the TRIzol reagent (Thermo, Waltham, MA, USA) based on the manufacturer’s protocol. After reverse transcription to form cDNA, quantitative real‐time PCR (qRT‐PCR) was carried out to detect the expression of CHMP4C using SYBR Green I (Invitrogen), according to the supplier’s standards. The CHMP4C primers (forward: 5′‐AGACTGAGGAGATGCTGGGCAA‐3′, reverse: 5′‐TAGTGCCTGTAATGCAGCTCGC‐3′) were obtained from GENEWIZ (Suzhou, China). Glyceraldehyde‐3 phosphate dehydrogenase was used as internal reference with sequences (forward: 5′‐TGTGTCCGTCGTGGATCTGA‐3′, reverse: 5′‐CCTGCTTCACCACCTTCTTGA‐3′), and the expression of CHMP4C was calculated with the 2−ΔΔCt method.
Western blotting assay
The radioimmunoprecipitation assay lysate (containing protease inhibitor) was used to crack the cells, and M‐PER Mammalian® Protein Extraction Reagent (Thermo Scientific, Waltham, MA, USA) was used to extract the proteins. The protein concentration was detected using bicinchoninic acid kit. About 20 μg protein was added to each well in the vertical electrophoresis tank, and then electrophoresis was performed with 10% SDS/PAGE. The protein on the gel was transferred to poly(vinylidene fluoride) membrane, followed by blocking with 5% defatted milk powder for 1 h. Then the membrane was incubated overnight at 4 °C with primary antibodies as follows: CHMP4C (1 : 2000; Abcam, Cambridge, UK), E‐cadherin (1 : 500; Abcam, Cambridge, UK), N‐cadherin (1 : 1000; Abcam), Vimentin (1 : 1000; Abcam), Snail (1 : 500; Abcam) and glyceraldehyde‐3 phosphate dehydrogenase (1 : 10 000; Abcam). The next day, the membrane was incubated with horseradish peroxidase–conjugated secondary antibody (Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA) for 1 h at room temperature. After rinsing, electrochemiluminescence developer was added to develop the images. Glyceraldehyde‐3 phosphate dehydrogenase was used as a control to evaluate the relative protein levels.
Cell Counting Kit‐8 and colony formation assay
For Cell Counting Kit‐8 (CCK‐8) assay, cells were seeded into 96‐well plates (1000 cells per well) and cultured with CO2. We assessed the cell viability at 24, 48 and 72 h, following the standard of the CCK‐8 kit (Dojindo Molecular Technologies, Rockville, MD, USA). The absorbance at 450 nm was measured using a microplate reader (Bio‐Rad, Hercules, CA, USA).
The colony formation assay was performed using the methods described in previous studies [15]; 400 cells were seeded into the culture dish and cultured for 1–2 weeks with 5% CO2 at 37 °C. When clones were visible to the naked eye, the cells were fixed with 4% paraformaldehyde and dyed with 0.1% crystal violet dye. Ultimately, numbers of colony were counted.
Transwell assay
Cell invasion was measured by cell‐penetrating matrix gel‐coated membranes. The invasion assay was conducted as described previously [15]. After transfection for 24 h, 1 × 105 cells were added to the upper chamber with serum‐free medium, and 500 mL serum medium used as the chemical attractant was put to the lower chamber. After incubation, the remaining cells in the upper chamber were erased using a cotton swab. The bottom membrane with invaded cells was fixed with 4% paraformaldehyde and dyed with 0.1% crystal violet. Then the randomly selected fields of vision were photographed for counting.
The migration assay was similar to the invasion assay, but it was evaluated by the penetration of cells into the plain membranes.
Statistical analysis
SPSS22.0 statistical analysis software (IBM, Armonk, NY, USA) was used to analyze the experimental data. All the assays were repeated three times, and data were presented as the mean ± standard deviation (SD). The discrepancy between two groups was compared with Student’s t‐test, and one‐way ANOVA and Dunnett posttest were used to compare the multiple groups. The relationship between gene expression and clinical features was assessed using chi‐square test. Kaplan–Meier analysis was used to plot the survival curve, and the difference between groups was measured using log‐rank test. P < 0.05 was considered statistically significant.
Results
CHMP4C was highly expressed in CC tissues and cell lines
CHMP4C has been reported to be imbalanced in many cancers, but whether CHMP4C expression is associated with CC is still unknown. First, the data of control tissues (n = 3) and CC tissues (n = 306) were downloaded from The Cancer Genome Atlas database to evaluate the differential expression of CHMP4C. It can be seen from Fig. 1A that CHMP4C was highly expressed in CC tissues compared with the control tissues (P < 0.01). Subsequently, qRT‐PCR assay was performed to detect the expression of CHMP4C in CC cell lines. The data indicated that the expression of CHMP4C was up‐regulated in different degrees in the CC cell lines (HeLa, C‐33A and Ca Ski) compared with normal Ect1/E6E7 cells (Fig. 1B, P < 0.01). Furthermore, C‐33A cells with the highest relative expression were selected for knockdown experiments, whereas Ca Ski cells were chosen for overexpression experiments, which showed relatively low expression.
Fig. 1.
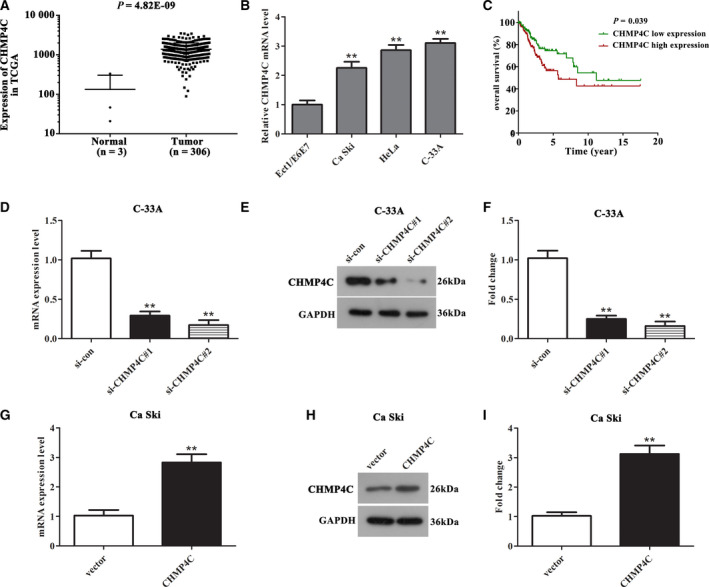
CHMP4C was highly expressed in CC tissues and cell lines, which caused poor outcomes. (A) The expression of CHMP4C in CC tissues was significantly higher than that in healthy control tissues (P < 0.001). (B) The mRNA expression level of CHMP4C was up‐regulated in CC cell lines (HeLa, C‐33A and Ca Ski) compared with that in normal control cells Ect1/E6E7. (C) The higher the expression of CHMP4C, the lower the survival rate of patients with CC (P < 0.001). (D–F) The CHMP4C expression was obviously declined in C‐33A cells after transfection with si‐CHMP4C#1 and si‐CHMP4C#2 compared with that in the si‐con group. (G–I) The expression of CHMP4C was markedly elevated in Ca Ski cells after transfection with pcDNA3.1‐CHMP4C compared with that in the vector group. All experiments were repeated three times, and the error bars represent SD. **P < 0.01.
High CHMP4C expression caused poor outcomes and was associated with the age of patients with CC
To further analyze whether high CHMP4C expression leads to unoptimistic prognosis, we divided the patients into high‐CHMP4C‐expression and low‐CHMP4C‐expression groups based on the median level, and analyzed the survival rate of the high‐ and low‐expression groups with Kaplan–Meier survival analysis. The results indicated that the higher the CHMP4C expression, the shorter the survival time (Fig. 1C, P = 0.039).
Then the relationship between the expression of CHMP4C gene and clinical features was evaluated using chi‐square test. The statistical results in Table 1 demonstrated that the expression of CHMP4C was related to the age of CC patients (P = 0.036), and we can clearly observe that patients with age older than 60 years have higher ratio of overexpressed CHMP4C.
Table 1.
The correlation between CHMP4C expression and clinical features of patients with CC. M, metastasis; N, regional lymph node; T, tumor.
| Characteristics | Expression of CHMP4C | P‐value | |
|---|---|---|---|
| Low | High | ||
| Age, years | 0.036* | ||
| <60 | 127 | 112 | |
| ≥60 | 25 | 40 | |
| Grade | 0.795 | ||
| G1 + G2 | 76 | 77 | |
| G3 | 61 | 58 | |
| Clinical stage | 0.515 | ||
| I + II | 119 | 112 | |
| III + IV | 31 | 35 | |
| Pathologic T | 0.150 | ||
| T1 + T2 | 107 | 104 | |
| T3 + T4 | 11 | 19 | |
| Pathologic N | 0.633 | ||
| N0 | 66 | 67 | |
| N1 | 32 | 28 | |
| Pathologic: M | 0.615 | ||
| M0 | 60 | 56 | |
| M1 | 6 | 4 | |
P < 0.05.
Moreover, we conducted a Cox regression analysis to investigate whether CHMP4C can be used as an independent predictor for CC prognosis (Table 2). Univariate analysis showed that CHMP4C expression, clinical stage, Pathologic T, Pathologic M and Pathologic N can be used as prognostic factors for CC. Furthermore, multivariate analysis showed that only Pathologic T can be used as an independent prognostic factor for CC.
Table 2.
Cox regression analysis of CHMP4C as a predictor of prognosis of CC. CI, confidence interval; HR, hazard ratio; M, metastasis; N, regional lymph node; T, tumor.
| Variables | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|
| P‐value | HR | 95% CI | P‐value | HR | 95% CI | |
| CHMP4C expression(low/high) | 0.041* | 1.638 | 1.021–2.628 | 0.260 | 1.800 | 0.647–5.013 |
| Clinical stage(I + II/III + IV) | 0.001* | 2.286 | 1.397–3.743 | 0.215 | 0.332 | 0.058–1.895 |
| Pathologic T (T1 + T2/T3 + T4) | 0.000* | 3.613 | 1.907–6.846 | 0.015* | 6.442 | 1.434–28.930 |
| Pathologic M (M0/M1) | 0.020* | 3.671 | 1.229–10.962 | 0.989 | 0.000 | 0.000 |
| Pathologic N (N0/N1 + N2 + N3) | 0.003* | 2.807 | 1.408–5.593 | 0.77 | 2.545 | 0.904–7.168 |
| Age (<60/≥60 years) | 0.054 | 1.641 | 0.991–2.715 | |||
| Grade (G1 + G2/G3 + G4) | 0.681 | 0.896 | 0.530–1.513 | |||
P < 0.05.
Inhibition of CHMP4C in C‐33A cells and overexpression of CHMP4C in Ca Ski cells
To achieve knockdown and overexpression of CHMP4C, we transfected si‐ CHMP4C#1 and si‐CHMP4C#2 into C‐33A cells, respectively, and transfected pcDNA3.1‐CHMP4C into Ca Ski cells. After 24 h, the mRNA and protein levels of CHMP4C in the transfected cells were detected using qRT‐PCR and western blot assay. It can be seen from Fig. 1D–F that transfection with si‐CHMP4C#1 or si‐CHMP4C#2 can obviously reduce the mRNA and protein levels of CHMP4C in C‐33A cells, and si‐CHMP4C#2 showed a higher knockdown efficiency (P < 0.01). Meanwhile, as presented in Fig. 1G–I, the CHMP4C expression in Ca Ski cells was markedly increased after transfection with pcDNA3.1‐CHMP4C (P < 0.01).
Knockdown of CHMP4C can inhibit malignant biological behavior of C‐33A cells, whereas overexpression of CHMP4C showed an opposite effect in Ca Ski cells
Next, we conducted functional experiments to measure whether CHMP4C contributes to the malignant biological behavior of CC cells. From the CCK‐8 assay, we can see that down‐regulation of CHMP4C can decrease the viability of C‐33A cells compared with control (Fig. 2A, P < 0.01), whereas the viability of Ca Ski cells transfected with pcDNA3.1‐CHMP4C was markedly enhanced compared with that transfected with vector (Fig. 2B, P < 0.01). The similar results can be seen from the colony formation assay. The colony number was obviously decreased in the si‐CHMP4C group compared with that in the si‐con group (Fig. 2C,D, P < 0.01), whereas up‐regulation of CHMP4C in Ca Ski cells can promote the capability to form colonies (Fig. 2E,F, P < 0.01).
Fig. 2.
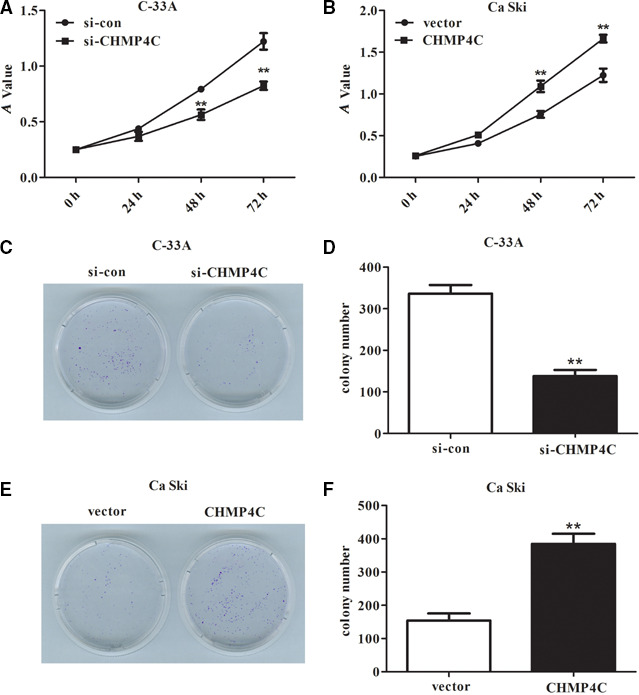
Knockdown of CHMP4C in C‐33A cells inhibited proliferation, and up‐regulation of CHMP4C in Ca Ski cells accelerated proliferation. (A) CCK‐8 assay revealed that deletion of CHMP4C can decrease the cell viability in C‐33A cells. (B) The cell viability was markedly enhanced in Ca Ski cells after overexpression of CHMP4C. (C, D) Colony formation assay revealed that down‐regulation of CHMP4C can reduce the ability of cells to form colonies. (E, F) Up‐regulation of CHMP4C can increase significantly the number of colonies formed compared with that in the vector group. All experiments were repeated three times, and the error bars represent SD. **P < 0.01. A, absorbance.
Transwell assay was performed to detect the influence of knockdown or overexpression of CHMP4C on cell migration and invasion. It can be seen from Fig. 3A,B that C‐33A cells transfected with si‐CHMP4C resulted in a marked reduction on the number of invaded and migrated cells compared with the si‐con group (P < 0.01), whereas overexpression of CHMP4C in Ca Ski cells showed opposite results (Fig. 3C,D, P < 0.01). Taken together, these results illustrated that CHMP4C functioned as an oncogene in the malignant biological behavior of CC cells, thus possibly being involved in the development of CC tumors.
Fig. 3.
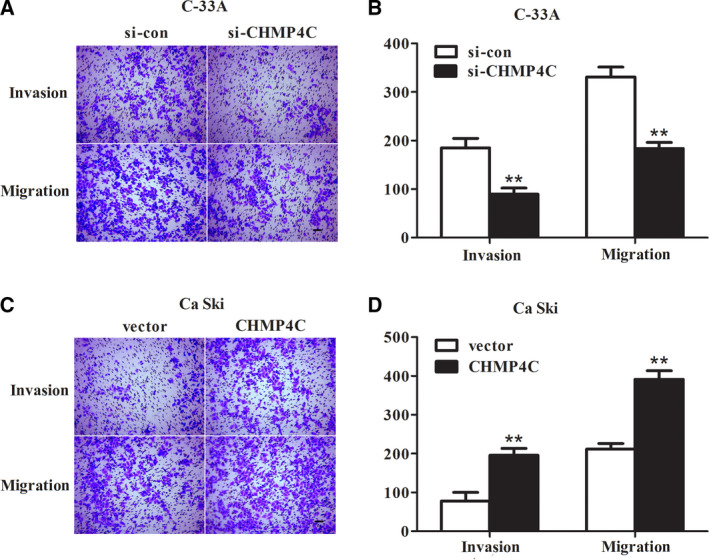
Deletion of CHMP4C suppressed invasion and migration of C‐33A cells, and overexpression of CHMP4C promoted invasion and migration of Ca Ski cells. (A, B) Through the Transwell assay, we found that knockdown of CHMP4C showed a significant inhibition on invasion and migration in C‐33A cells. (C, D) The number of invading and migrating cells was significantly increased after Ca Ski cells transfected with pcDNA3.1‐CHMP4C compared with that in the vector group. Scale bars: 200 μm (A, C). All experiments were repeated three times, and the error bars represent SD. **P < 0.01.
Epithelial–mesenchymal transition pathway mediates the regulation of CHMP4C on the malignant biological behavior of CC cells
Epithelial–mesenchymal transition (EMT) is a key event that promotes the migration and invasion of resting tumor cells [16]. Thus, we performed western blotting assay to detect the expression of EMT‐related proteins in C‐33A and Ca Ski cells, to assess whether the EMT pathway mediates the regulation of CHMP4C on CC cells malignant phenotype. The results in Fig. 4A,B indicated that knockdown of CHMP4C can increase the protein expression of E‐cadherin, whereas the expressions of N‐cadherin, Vimentin and Snail decline (P < 0.01). Meanwhile, overexpression of CHMP4C in Ca Ski cells presented the opposite outcomes (Fig. 4C,D, P < 0.01). Because the absence of E‐cadherin is considered to be a marker of EMT, these results illustrated that inhibition of CHMP4C can inhibit the EMT of CC cells. These data prompted a possibility that CHMP4C may contribute to the regulation of CC malignant progression through regulating the EMT pathway.
Fig. 4.
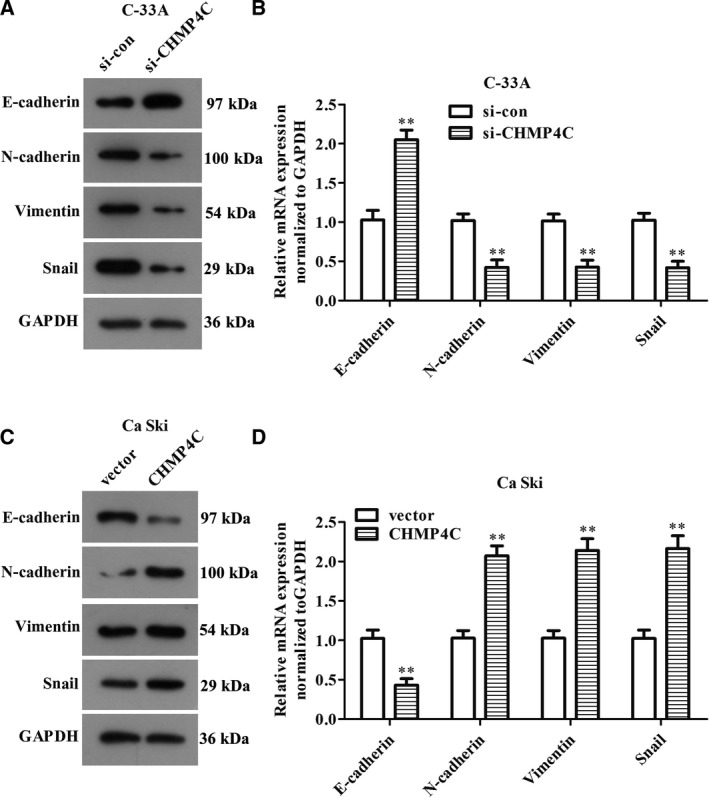
CHMP4C mediates the occurrence of EMT in CC cells. (A, B) Down‐regulation of CHMP4C suppresses the EMT pathway in C‐33A cells. (C, D) Up‐regulation of CHMP4C promotes the EMT pathway in Ca Ski cells. All experiments were repeated three times, and the error bars represent SD. **P < 0.01.
Discussion
The occurrence and development of CC is a complex process of multifactor regulation [17]. Currently, the incidence of CC shows a trend of youth [18]. In spite of some progress being made in the potential mechanism of the occurrence and malignant progression of CC, the prognosis of current treatment is still poor [19], so it is an urgent problem to find an effective therapeutic target. In this study, we first found that the expression of CHMP4C was elevated in both CC tissues and cell lines. High CHMP4C expression leads to poor outcomes and is associated with the age of patients with CC. Subsequently, functional experiment indicated that CHMP4C promotes proliferation, migration and invasion of CC cells, in part by activating EMT.
The function of CHMP4C in cell dynamics and endocytogenesis has been widely reported [20, 21, 22]. Based on the role of CHMP4C in a variety of cell functions, research has begun to focus on whether it contributes to the progression of cancer. It has been found that CHMP4C has abnormal expression in cancer. Nikolova et al. [23] found for the first time that CHMP4C was highly expressed in ovarian cancer tissues, and its high expression may be related to the poor outcomes of ovarian cancer. It is worth noting that CHMP4C has been identified as a new susceptibility gene for ovarian cancer [24]. In addition, Li et al. [12] demonstrated that CHMP4C can promote cell survival and enhance radiation resistance of tumor cells in human non‐small cell lung cancer cells, which functioned as a carcinogenic gene. Most importantly, it has been found that HPV E6/E7 plays an important role in HPV‐induced carcinogenesis [5]. Inhibition of HPV E6/E7 expression results in a significant increase in exosomes, whereas CHMP4C stimulation can promote exosome production and induce its apoptosis [25]. Because HPV infection is a risk factor for CC, we speculated that CHMP4C may also be involved in the pathogenesis and progression of CC. Our data showed that CHMP4C was highly expressed in CC and was associated with poor prognosis, which can be used as a prognostic factor for CC. Further functional experiments indicated that CHMP4C accelerated the malignant biological behavior of CC cells, and it functioned as an oncogene in CC.
EMT is considered to be the key factor to accelerate tumor invasion and migration [16], which can be regulated by many factors, such as miRNA, mRNA, long noncoding RNA [26], etc. Activation of the EMT pathway in tumor cells can promote the development of almost all malignant tumor‐related features [27]. Deletion of E‐cadherin and overexpression of vimentin are hallmarks of EMT activation [28], and miRNA can function in the regulation of CC by regulating E‐cadherin [29]. Some studies have shown that the loss of epithelial features (E‐cadherin, etc.) and the acquisition of mesenchymal properties (N‐cadherin, vimentin, snail, etc.) can improve the invasion of tumors [30]. Notably, high expression of snail is closely related to the deterioration of human cancer [31]. Extensive studies have confirmed that EMT is involved in the malignant progression of many cancers, such as colon cancer [32], ovarian serous cancer [33], breast cancer [34], among others. So far, studies have also reported the association between the progression of CC and EMT. EMT accelerates the invasion and metastasis of primary CC, and plays an important role in the lymph node metastasis of CC [35]. Our findings revealed that up‐regulation of CHMP4C in CC cells can activate the EMT, and thus promote the invasion and migration of CC cells. Knockdown of CHMP4C presented the opposite outcomes. Combined with the data in this article, we concluded that overexpression of CHMP4C induces the occurrence of EMT in CC cells, thereby accelerating cell invasion and migration.
In summary, our study found for the first time that CHMP4C was highly expressed in CC, and its high expression caused poor outcomes. Furthermore, CHMP4C may promote cell proliferation, invasion and migration by activating the EMT pathway. These findings prompted a novel strategy for CC therapy; however, the potential application in clinical treatment still needs further exploration.
Conflict of interest
The authors declare no conflict of interest.
Author contributions
S‐LL, MW and QL performed the experiments. S‐LL and MW analyzed the data. S‐LL and Q‐QC wrote the manuscript. S‐LL, Q‐QC and QL reviewed and edited the manuscript. All authors read and approved the final manuscript.
References
- 1. Torre LA, Islami F, Siegel RL, Ward EM and Jemal A (2017) Global cancer in women: burden and trends. Cancer Epidemiol Biomarkers Prev 26, 444–457. [DOI] [PubMed] [Google Scholar]
- 2. Shafabakhsh R, Reiter RJ, Mirzaei H, Teymoordash SN and Asemi Z (2019) Melatonin: a new inhibitor agent for cervical cancer treatment. J Cell Physiol 234, 21670–21682. [DOI] [PubMed] [Google Scholar]
- 3. Tsu V and Jeronimo J (2016) Saving the World's women from cervical cancer. N Engl J Med 374, 2509–2511. [DOI] [PubMed] [Google Scholar]
- 4. Bosch FX, Lorincz A, Munoz N, Meijer CJ and Shah KV (2002) The causal relation between human papillomavirus and cervical cancer. J Clin Pathol 55, 244–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sadri Nahand J, Moghoofei M, Salmaninejad A, Bahmanpour Z, Karimzadeh M, Nasiri M, Mirzaei HR, Pourhanifeh MH, Bokharaei‐Salim F, Mirzaei H and et al (2020) Pathogenic role of exosomes and microRNAs in HPV‐mediated inflammation and cervical cancer: a review. Int J Cancer 146, 305–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mailhot Vega RB, Balogun OD, Ishaq OF, Bray F, Ginsburg O and Formenti SC (2019) Estimating child mortality associated with maternal mortality from breast and cervical cancer. Cancer 125, 109–117. [DOI] [PubMed] [Google Scholar]
- 7. Petsalaki E, Dandoulaki M and Zachos G (2018) The ESCRT protein Chmp4c regulates mitotic spindle checkpoint signaling. J Cell Biol 217, 861–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Petsalaki E and Zachos G (2018) CHMP4C: A novel regulator of the mitotic spindle checkpoint. Mol Cell Oncol 5, e1445944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lata S, Schoehn G, Solomons J, Pires R, Gottlinger HG and Weissenhorn W (2009) Structure and function of ESCRT‐III. Biochem Soc Trans 37 (Pt 1), 156–160. [DOI] [PubMed] [Google Scholar]
- 10. Juan T and Furthauer M (2018) Biogenesis and function of ESCRT‐dependent extracellular vesicles. Semin Cell Dev Biol 74, 66–77. [DOI] [PubMed] [Google Scholar]
- 11. Sadler JBA, Wenzel DM, Williams LK, Guindo‐Martinez M, Alam SL, Mercader JM, Torrents D, Ullman KS, Sundquist WI and Martin‐Serrano J(2018) A cancer‐associated polymorphism in ESCRT‐III disrupts the abscission checkpoint and promotes genome instability. Proc Natl Acad Sci U S A 115, E8900–e8908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li K, Liu J, Tian M, Gao G, Qi X, Pan Y, Ruan J and Liu C and Su X(2015) CHMP4C disruption sensitizes the human lung cancer cells to irradiation. Int J Mol Sci 17, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gusev A, Lawrenson K, Lin X, Lyra PC Jr, Kar S, Vavra KC, Segato F, Fonseca MAS, Lee JM, Pejovic T et al (2019) A transcriptome‐wide association study of high‐grade serous epithelial ovarian cancer identifies new susceptibility genes and splice variants. Nat Genet 51, 815–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fujita K, Kume H, Matsuzaki K, Kawashima A, Ujike T, Nagahara A, Uemura M, Miyagawa Y, Tomonaga T and Nonomura N (2017) Proteomic analysis of urinary extracellular vesicles from high Gleason score prostate cancer. Sci Rep 7, 42961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yang PP, Yu XH and Zhou J (2020) Tryptophanyl‐tRNA synthetase (WARS) expression in uveal melanoma – possible contributor during uveal melanoma progression. Biosci Biotechnol Biochem 84, 471–480. [DOI] [PubMed] [Google Scholar]
- 16. Yeung KT and Yang J (2017) Epithelial‐mesenchymal transition in tumor metastasis. Mol Oncol 11, 28–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mi Y, Wang L, Zong L, Pei M, Lu Q and Huang P (2014) Genetic variants in microRNA target sites of 37 selected cancer‐related genes and the risk of cervical cancer. PLoS One 9, e86061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet‐Tieulent J and Jemal A (2015) Global cancer statistics, 2012. CA Cancer J Clin 65, 87–108. [DOI] [PubMed] [Google Scholar]
- 19. Ghasemi F, Shafiee M, Banikazemi Z, Pourhanifeh MH, Khanbabaei H, Shamshirian A, Amiri Moghadam S, ArefNezhad R, Sahebkar A, Avan A et al (2019) Curcumin inhibits NF‐kB and Wnt/β‐catenin pathways in cervical cancer cells. Pathol Res Pract 215, 152556. [DOI] [PubMed] [Google Scholar]
- 20. Petsalaki E, Dandoulaki M and Zachos G (2018) Chmp4c is required for stable kinetochore‐microtubule attachments. Chromosoma 127, 461–473. [DOI] [PubMed] [Google Scholar]
- 21. Capalbo L, Mela I, Abad MA, Jeyaprakash AA, Edwardson JM and D'Avino PP (2016) Coordinated regulation of the ESCRT‐III component CHMP4C by the chromosomal passenger complex and centralspindlin during cytokinesis. Open Biol 6, 160248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Carlton JG, Caballe A, Agromayor M and Kloc M and Martin‐Serrano J(2012) ESCRT‐III governs the Aurora B‐mediated abscission checkpoint through CHMP4C. Science 336, 220–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nikolova DN, Doganov N, Dimitrov R, Angelov K, Low SK, Dimova I, Toncheva D, Nakamura Y and Zembutsu H (2009) Genome‐wide gene expression profiles of ovarian carcinoma: Identification of molecular targets for the treatment of ovarian carcinoma. Mol Med Rep. 2, 365–384. [DOI] [PubMed] [Google Scholar]
- 24. Pharoah PD, Tsai YY, Ramus SJ, Phelan CM, Goode EL, Lawrenson K, Buckley M, Fridley BL, Tyrer JP, Shen H et al (2013) GWAS meta‐analysis and replication identifies three new susceptibility loci for ovarian cancer. Nat Genet. 45, 362–370, 370e1‐2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Honegger A, Leitz J, Bulkescher J, Hoppe‐Seyler K and Hoppe‐Seyler F(2013) Silencing of human papillomavirus (HPV) E6/E7 oncogene expression affects both the contents and the amounts of extracellular microvesicles released from HPV‐positive cancer cells. Int J Cancer 133, 1631–1642. [DOI] [PubMed] [Google Scholar]
- 26. Xu Q, Deng F, Qin Y, Zhao Z, Wu Z, Xing Z, Ji A and Wang QJ (2016) Long non‐coding RNA regulation of epithelial‐mesenchymal transition in cancer metastasis. Cell Death Dis. 7 (6), e2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ye X and Weinberg RA (2015) Epithelial‐mesenchymal plasticity: a central regulator of cancer progression. Trends Cell Biol 25, 675–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Oft M, Peli J, Rudaz C, Schwarz H, Beug H and Reichmann E (1996) TGF‐beta1 and Ha‐Ras collaborate in modulating the phenotypic plasticity and invasiveness of epithelial tumor cells. Genes Dev 10, 2462–2477. [DOI] [PubMed] [Google Scholar]
- 29. Nahand JS, Taghizadeh‐Boroujeni S, Karimzadeh M, Borran S, Pourhanifeh MH, Moghoofei M, Bokharaei‐Salim F, Karampoor S, Jafari A, Asemi Z et al (2019) microRNAs: New prognostic, diagnostic, and therapeutic biomarkers in cervical cancer. J Cell Physiol 234, 17064–17099. [DOI] [PubMed] [Google Scholar]
- 30. Polyak K and Weinberg RA (2009) Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer 9, 265–273. [DOI] [PubMed] [Google Scholar]
- 31. Cho ES, Kang HE, Kim NH and Yook JI (2019) Therapeutic implications of cancer epithelial‐mesenchymal transition (EMT). Arch Pharm Res 42, 14–24. [DOI] [PubMed] [Google Scholar]
- 32. Nfonsam VN, Nfonsam LE, Chen D, Omesiete PN, Cruz A, Runyan RB and Jandova J (2019) COMP gene coexpresses with EMT genes and is associated with poor survival in colon cancer patients. J Surg Res 233, 297–303. [DOI] [PubMed] [Google Scholar]
- 33. Yan H, Li H, Silva MA, Guan Y, Yang L, Zhu L, Zhang Z, Li G and Ren C (2019) LncRNA FLVCR1‐AS1 mediates miR‐513/YAP1 signaling to promote cell progression, migration, invasion and EMT process in ovarian cancer. J Exp Clin Cancer Res 38, 356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gao X, Liu X, Lu Y, Wang Y, Cao W, Liu X, Hu H and Wang H (2019) PIM1 is responsible for IL‐6‐induced breast cancer cell EMT and stemness via c‐myc activation. Breast Cancer. 26, 663–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Qureshi R, Arora H and Rizvi MA (2015) EMT in cervical cancer: its role in tumour progression and response to therapy. Cancer Lett 356(2 Pt B), 321–331. [DOI] [PubMed] [Google Scholar]


