Abstract
The mitochondrial electron transport chain is a major source of reactive oxygen species (ROS) and is also a target of ROS, with an implied role in the stabilization of hypoxia-inducible factor (HIF) and induction of the AMPK pathway. Here we used varying doses of two agents, Mito-Paraquat and Mito-Metformin, that have been conjugated to cationic triphenylphosphonium (TPP+) moiety to selectively target them to the mitochondrial matrix compartment, thereby resulting in the site-specific generation of ROS within mitochondria. These agents primarily induce superoxide (O2•–) production by acting on complex I. In Raw264.7 macrophages, C2C12 skeletal myocytes, and HCT116 adenocarcinoma cells, we show that mitochondria-targeted oxidants can induce ROS (O2•– and H2O2). In all three cell lines tested, the mitochondria-targeted agents disrupted membrane potential and activated calcineurin and the Cn-dependent retrograde signaling pathway. Hypoxic culture conditions also induced Cn activation and HIF1α activation in a temporally regulated manner, with the former appearing at shorter exposure times. Together, our results indicate that mitochondrial oxidant-induced retrograde signaling is driven by disruption of membrane potential and activation of Ca2+/Cn pathway and is independent of ROS-induced HIF1α or AMPK pathways.
Keywords: Mitochondrial ROS, Calcineurin activation, HIF1α, Retrograde signaling, Hypoxia mediated stress, Macrophage cell line, Mitochondria targeted agents
Highlights
-
•
Mitochondrial ROS (O2.•–, H2O2) induce a stress signaling pathway implicated in many diseases.
-
•
Mitochondrial ROS induce the Ca2+/Cn pathway in addition to HIF1 and AMPK pathways.
-
•
Mitochondria-targeted agents MitoPQ and MitoMet induce the Ca2+/Cn and AMPK pathways.
-
•
Inhibition of complex III by Antimycin A also activates the Ca2+/Cn and AMPK pathways but not HIF1α.
-
•
The three pathways appear to require different thresholds of mitochondrial ROS.
1. Introduction
Mitochondria are the major cellular producers of reactive oxygen species (ROS), generated as byproducts of electron transport-linked ATP synthesis. It is estimated that up to 1–2% of the total oxygen consumed by mitochondria can be converted to superoxide (O2•–) [1]. Altered production of ROS destabilizes mitochondrial redox balance and affects mitochondrial DNA and membrane functions [2]. Mitochondria and mitochondrial ROS are major players in various pathophysiological processes, including neurodegeneration, atherosclerosis, aging, and cancer [3]. Mitochondrial ROS are also involved in reprogramming cells for proliferation [4,5] through altered metabolism and retrograde signaling and are also implicated in other physiologically critical signaling pathways.
As the maintenance of mitochondrial number, morphology, intracellular distribution, and functional integrity is critical, mechanisms are in place to maintain these characteristics in response to changes in developmental, physiological, and environmental conditions [6]. The mitochondrial anterograde response, encompassing changes in nuclear gene expression that affect mitochondrial structure and function, has been known for more than four decades [7]. Mitochondrial retrograde signaling (MtRS) (signaling from the mitochondria-to-the nucleus) was first described in budding yeast in pioneering studies from Ronald Butow's laboratory [8,9]. They showed that mitochondrial DNA (mtDNA) depletion or deletion initiates retrograde (Rtg) signaling, leading to adaptive changes in nuclear gene expression in response to altered mitochondrial function.
Activation of the Rtg pathway appears to be restricted to unicellular eukaryotes. In metazoans, induced nuclear gene expression in response to mitochondrial genetic and metabolic changes have been reported in several mammalian, avian, and nematode cells [[9], [10], [11], [12]]. In support of these observations, multiple mechanisms of MtRS, including the Ca2+/Calcineurin pathway [13], mitochondrial unfolded protein response pathway (mtUPR) [14], mitochondrial ROS-induced AMPK pathway [15], and the hypoxia-induced HIF1α activation pathway have been reported in metazoan cells. In asynchronously proliferating Drosophila eye disk cells, a mutation in the complex I subunit Pdsw gene, or mutation in the cytochrome c oxidase Va (CcOVa) gene, can activate G1-S checkpoint as a downstream effect of increased ROS production and activation of the JNK signaling pathway [16]. It is not clear if these multiple pathways have a common causative factor or they are activated independently to deal with different degrees of mitochondrial dysfunction.
Most recently, mitochondrial defects have also been linked to immune responses [17] and increased life span in mouse models [20]. Apparently, paradoxical roles of mitochondrial ROS in normal physiology or pathology seem to depend on the activation or inhibition of cellular signaling factors. In different cancer cells, the mitochondria-generated superoxide can either promote or inhibit cell proliferation [21]. In addition, mtDNA loss or uncoupling of the electron transport chain (ETC) in mouse myoblast C2C12 cells causes increased tumor invasiveness in xenograft mouse models [22]. With respect to the Ca2+/Cn signaling, our overarching hypothesis is that loss of mitochondrial membrane potential causes increased cytosolic Ca2+, resulting in the activation of Cn [23]. Activation of NF-κB by Cn causes nuclear translocation of the cRel:p50 heterodimer to regulate gene expression in association with C/EBPδ, CREB and NFAT in a nuclear enhanceosome complex [12], thus altering the cellular transcription program.
Several studies have indicated that activation of the HIF1α pathway of mammalian MtRS depends on mitochondrial ROS, as ρ° Hep3B cells lacking mtDNA fail to activate HIF1α under hypoxic growth conditions [24], presumably because complex III of the ETC is a source of ROS production and concurrent stabilization of HIF1α [18,19]. In addition, it has been proposed that respiratory complex III-generated superoxide after conversion to H2O2 by superoxide dismutase (SOD), directly oxidizes the non-heme-bound iron of prolyl hydroxylase (PHD), resulting in PHD inactivation and HIF1α stabilization [24]. Low levels of oxygen in the cytoplasm of highly respiring tissues can also inhibit PHD enzymes, further stabilizing HIF1α protein [25,26]. However, a study by Masson et al. [27] reported that asparaginyl hydroxylase (FIH), an inactivator of HIF, is more sensitive to hydrogen peroxide than is PHD. Thus, the precise mechanism of action of mitochondrial ROS on the HIF pathway may vary depending on the physiological state. An alternate mechanism was proposed on the HIF1α stabilization by increased ROS production at complex III [18,19] of the mitochondrial electron transport chain.
In Caenorhabditis elegans, the mtUPR maintains mitochondrial homeostasis by increasing the protein folding and degradation capacity in response to the accumulation of unfolded and aggregated proteins in mitochondria [14]. Subsequent to its initiation, mtUPR signaling promotes alterations in proteostasis, organelle damage, or induces cellular dysfunction and ultimately promotes nuclear changes that maintain the functional integrity of mitochondria [28,29].
Based on multiple reports, it appears that mitochondrial ROS induced by modulators of mitochondrial function or under hypoxic conditions may induce multiple MtRS pathways, including the HIF pathway [18], the Ca2+/Cn pathway [30] and the AMPK pathway [31]. Here, using hypoxia and mitochondria-targeted agents (MitoPQ and MitoMet) as mitochondrial ROS inducers, we evaluated the time course of Ca2+/Cn activation and HIF1α activation in Raw macrophages, C2C12 myoblasts, and HCT116 colon carcinoma cells. We show that Cn is activated well before the activation of HIF1α and that Cn alone is able to induce MtRS and metabolic changes before the activation of HIF1α or AMPK.
2. Materials and Methods
2.1. Cell culture conditions, treatments and exposure to hypoxia
Murine C2C12 skeletal myoblasts (ATCC CRL1772), Raw264.7 macrophages, human non-small cell lung carcinoma cell H1299, colorectal carcinoma cell line HCT116, p53+/+ and its isogenic p53 deficient HCT116 p53−/− were grown in Dulbecco's modified Eagle's medium (DMEM, Gibco, Life Technologies) as described before [32].
Cells were treated with different mitochondria-targeted agents Mito-Paraquat (MitoPQ) (Fig. S1C) [33], Mito-Metformin (MitoMet) (Fig. S1D) [34] which consist of a decyl triphenylphosphonium (DTPP+) moiety conjugated to paraquat or Metformin and their corresponding controls. MitoPQ control is structurally similar to MitoPQ but does not redox cycle because it has two extra methyl groups on the pyridinium rings that disfavor coplanarity of the rings, which is necessary to stabilize a radical cation produced by one-electron reduction (Fig: S1A). MitoMet control (decyl-TPP; DTPP) (Fig. S1B) has the same aliphatic carbon chain linker without any functional group [35]. Cells were grown under normal oxygen conditions of 150 Torr or 21% O2. Cells grown up to 70–80% confluence under normoxia were latter exposed to hypoxia for 2, 4, 6, 8, and 12 h. Simulation of realistic in vivo hypoxia requires that O2 tension be maintained at less than 8 Torr (1% O2). This hypoxic condition was achieved in a temperature controlled hypoxic chamber by a constant flow of premixed gas that was certified to contain 1.0% of oxygen and 5% CO2 (BOC gases, Murray Hill, NJ) [30].
2.2. Generation of SOD2 expressing stable cell lines
A pCMV6 mammalian expression vector carrying human superoxide dismutase-2 (sod2) cDNA (OriGene Technologies, Inc., CAT#: RC212924) and empty vector (control) were used to generate the overexpressing cell lines. Raw264.7 cells were transfected using FuGENE 6 (Roche Diagnostics, Indianapolis, IN, USA) according to the manufacturer's instructions. After 48 h of transfection, cells were treated with G418 (800 μg/ml) until 14 days, and finally, SOD2 expressing clones were selected from the single colony by immunoblot analysis probed against c-terminally tagged Flag protein with SOD2. From now on, we will call this cell line as R.SOD2 cells.
2.3. SDS-PAGE and western blotting
Whole-cell lysates were prepared by washing the cells with ice-cold PBS followed by lysis with lysis buffer (contain 60 mM Tris-HCl pH 7.5, 150 mM NaCl, 10 mM MgCl2, 2 mM EDTA, 10% glycerol, 1% Triton X-100, 10 mM NaF, 2 mM PMSF, protease inhibitors (1 μg/ml each aprotinin, antipain, pepstatin, leupeptin, 1 mM sodium orthovanadate and 0.1 mM molybdic acid). For immunoblot analysis, proteins were resolved on 7–10% polyacrylamide gels and transferred to the nitrocellulose membrane. The blots were probed with specific antibodies (Supplemental Table S1) and developed with infrared-tagged secondary antibodies and imaged through an Odyssey image scanner and analyzed by Odyssey Infrared Imaging System application software (LiCor Inc., Lincoln, NE).
2.4. Redox blotting of Prx1 and Prx3
The posttranslational redox changes in the key cytosolic and mitochondrial thiol proteins, such as peroxiredoxins (Prx1 and Prx3) were probed by the redox immunoblot as described before [[36], [37], [38]]. Briefly, treated and untreated cells were washed with Hanks’ balanced salt solution and covered with NEM Buffer (0.1 M N-ethylmaleimide, 50 mM NaCl, 40 mM HEPES, pH 7.4, 1 mM phenylmethylsulfonyl fluoride, 1 mM EDTA, 1 mM EGTA, and 10 μg/ml catalase and protease inhibitors) for 2 min. Cells were collected and incubated in NEM buffer for 15 min at room temperature. Cells were pelleted down (5 min, 800 g) and lysed with NEM extract buffer supplemented with CHAPS in a final concentration of 1%. Samples were kept at −80 °C until analysis. NEM reacts with the –SH groups of the reduced forms of PRX to prevent the oxidation during the cell harvesting and lysis. Reduced Prx1 and Prx3 migrate as monomers (22 kDa) in SDS-PAGE, whereas the oxidized proteins migrate as dimers (44 kDa). Lysates were thawed on ice and then centrifuged for 5 min at 8000 g. 20 μg of protein extracts, combined with nonreducing protein loading buffer (62.5 mM Tris–HCl pH 6.8, 10% glycerol, 2% SDS, 0.025% bromophenol blue) were resolved by SDS-PAGE. The blots were probed with anti-Prx1 or anti-Prx3 antibodies (Proteintech Group, Inc., Rosemont, IL), followed by the appropriate IR-conjugated secondary antibody. Images were developed through an Odyssey image scanner and quantified by Odyssey Infrared Imaging System application software (LiCor Inc., Lincoln, NE).
2.5. Chromatin immunoprecipitation (ChIP) assay
ChIP assays were carried out with cells fixed with 1% formaldehyde using a standard protocol. Immunoprecipitation with anti-HIF1α antibody (Santa Cruz Biotechnology) overnight at 4 °C. For each reaction, 1% of the sample was taken and pre-immune IgG was used as a negative control. Antibody and Chromatin complex were immunoprecipitated by protein A/G agarose beads with salmon sperm DNA (50% Slurry). Eluted DNA-Protein complexes were reversed with 5 M NaCl at 65 °C overnight. Immunoprecipitated DNA was purified and amplified by PCR of HRE binding regions of human or mouse (as described in figures) EPO and VEGF promoters [[39], [40], [41]]. Data were presented as fold enrichment by subtracting the signal with no antibody and expressed as fold increase over the control sample.
2.6. Measurement of ROS production and fluorescent microscopy
ROS generation in whole cells was measured by the Amplex Red hydrogen peroxide/peroxidase assay kit from Thermo Fisher (cat# A22188). In brief, 8.5 × 104 cells were plated per well in the black bottom 96 well plates with phenol-free complete DMEM medium. After different treatments, the media was carefully removed and washed gently with Krebs-Ringer phosphate buffer (KRPG, pH 7.35). KRPG buffer consists of 145 mM NaCl, 5.7 mM sodium phosphate, 4.86 mM KCl, 0.54 mM CaCl2, 1.22 mM MgCl2, 5.5 mM glucose and pH adjusted to 7.35. Then cells were incubated with 50 μl 1 × reaction buffer containing 50 μM Amplex Red, 10 U/ml horseradish peroxidase (HRP) in KRPG buffer for 20 min at room temperature. The method involves horseradish peroxidase (HRP)-catalyzed oxidation of the colorless and nonfluorescent Amplex Red molecule to red-fluorescent resorufin, as described earlier [42]. The fluorescence of resorufin was recorded with an excitation set at 530 nm and emission set at 590 nm in a MicroWin chameleon multilabel plate reader. ROS were also detected using hydroethidine (HE) probe coupled with HPLC-based detection of 2-OH-E+, the superoxide-specific product, and other oxidation products, as described elsewhere [43]. HPLC experiments were performed using an Agilent 1100 system equipped with UV–visible absorption and fluorescence detectors for measuring the reaction products. Cellular oxidants were also measured using MitoSOX Red (Thermo Fisher Scientific, Catalogue #M36008) and following the manufacture's protocol. In brief, treated cells were incubated with 5 μM MitoSOX reagent in Ca/Mg-free HBSS buffer for 10 min in the dark, followed by washing three times with PBS and finally fluorescence was recorded using a microplate reader (Promega Glow Max Discover Plate reader) with excitation/emission maxima of 510/580 nm.
Mitochondrial ROS also detected by using Olympus fluorescent microscope (BX61, Camera Retiga 4000R). Cells were plated and grown on cover slip (5 × 104 cells/well) in 12 well plates and incubated with MitoPQ control and MitoPQ for 16 h at 37 °C. Following incubation, cells were washed with 1 × PBS two times and incubated with 5 μM MitoSOX and 200 nM Mitotracker Green (Thermo Fisher scientific, Catalogue #M7514) in HBSS solution for 10 min in the dark at 37 °C. Next, cells were washed thrice with 1 × PBS and subjected for fluorescent micrograph using TRITC (for MitoSOX) and FITC filter (For Mitotracker Green) using Advanced Metamorph software.
2.7. Aconitase assay
Cell lysates were prepared in 50 mM Tris-Cl (pH 7.4), 1 mM l-cysteine, 0.6 mM MnCl2, 1 mM Na-citrate, and 0.5% Triton X-100 buffer with protease inhibitors (2 mM PMSF, 1 μg/ml each aprotinin, antipain, pepstatin, leupeptin). Aconitase activity was measured with 200 μg of protein in 50 mM Tris-Cl (pH 7.4), 30 mM Na-citrate, 0.6 mM MnCl2, 0.2 mM NADP, 0.2% Triton X-100 with 4 U/ml isocitrate dehydrogenase, by following the formation of NADPH, measured at 340 nm wavelength, for 15–25 min. The time range of linear increase in the absorbance over time was used for the determination of aconitase activity [44,45]. To measure the aconitase activity in hypoxia-treated or following treatment with mitochondrial agents, cells from each treatment group were divided into two portions: One portion was used for immunoblot analysis, and the other was used for colorimetric measurement of aconitase activity.
2.8. Calcineurin assay
Cn activity was measured using the Cn cellular activity assay kit (Enzo Life sciences, Farmingdale, NY, USA) according to the manufacturer's suggested protocol. In brief, the cultured cells were washed and collected by centrifugation in Tris buffer saline (pH 7.4). Cell extracts were prepared by disrupting the cells by lysis buffer (provided) with protease and phosphatase inhibitors (provided). According to the manufacturer's instruction, the phosphopeptide substrate was incubated with cell extract except to phosphate standard curve samples for 10 min at 30 °C in 96 well clear bottom plate. Assays were initiated by adding the calcineurin (40 U/well) or the cell extract (5 μg) and incubated for 30 min at 30 °C. The kit measures free phosphate released from the Cn-specific RII phosphopeptide. Free phosphate released was measured using malachite green at 620 nm. Cn-specific phosphatase activity was calculated by subtracting activity in the presence of EGTA from total phosphatase activity.
2.9. Matrigel invasion assay
Matrigel invasion assays were performed in triplicate for each group using 24 well plate transwell chambers (8 μm pore, Corning, NY). Initially, 100 μl (200 μg/ml) Corning matrigel matrix was used to coat the top part of the membrane. 2.5 × 104 cells/well were seeded for each group with required treatments, as mentioned in Fig. 700 μl DMEM containing 20% FBS was kept in basolateral chambers and incubated for 14–16 h at 37 °C in a 5% CO2 incubator. Cells were then fixed with chilled methanol and incubated for 6 min in hematoxylin solution Gill 3 (Sigma-Aldrich, St. Louis, MO) for cell staining. Non-invading cells were removed from the top of the Matrigel coat layer using a wet cotton swab. Images were taken using an Olympus BX-61 upright microscope (Olympus-Life sciences) with 10 × objective lens. Five random visual fields were selected for counting and represented as the number of invading cells per field.
2.10. Measurement of mitochondrial membrane potential
The mitochondrial membrane potential (ΔΨm) was measured using tetramethylrhodamine ethyl ester (TMRE) (Sigma-Aldrich, St. Louis, MO). In brief, 1 × 106 live cells were washed with PBS at room temperature, re-suspended in 1 ml phenol-free medium without FBS, supplemented with 50 nM TMRE and incubated in the dark for 10 min at 37 °C. As a positive control, 10 μM carbonyl cyanide p-trifluoromethoxyphenylhydrazone (FCCP) was used to disrupt ΔΨm. Fluorescence intensity of TMRE was measured by Cellometer Vision CBA (Nexcelom Biosciences, Lawrence, MA) by setting the filter excitation at 502 nm and emission at 595 nm.
2.11. AMPK assay
The level of activated AMPK in cells was assayed using an ELISA kit (Abcam, ab181422) to measure AMPK-α phosphorylation at Thr172, following the manufacturer's instructions and standards provided in the kit.
2.12. Realtime PCR for mRNA quantification
Total RNA was isolated using TRIzol reagent as per the vendor's protocol (Invitrogen). cDNA was generated from 1 μg of RNA using the cDNA Archives kit from Applied Biosystems (Thermo Fisher Scientific, Waltham, MA), and 25 ng of this cDNA was used as template per reaction. mRNA levels were quantified using SYBR Green (Applied Biosystems) in a Quant Studio 6 Flex real-time PCR system (Applied Biosystems). Data were normalized to β-actin as endogenous control, and relative expression of target genes was expressed using the 2−ΔΔCt method and the used primers listed in supplemental Table S2.
2.13. Cell viability
Raw264.7 cells (8.5 × 104 cells/well) were incubated in 96-well plates. Following different treatments and 16 h of incubation, 20 μL of CellTiter 96® AQueous One Solution Cell Proliferation Assay (MTS; Promega, Madison, WI, USA) reagent was added to each well and then incubated for 2 h at 37 °C. The assay is based on phenazine ethosulfate-mediated reduction of MTS tetrazolium salt to a water-soluble formazan. The absorbance at 490 nm due to the formation of formazan was measured using a microplate reader (Promega Glow Max Discover Plate reader).
2.14. Statistical analysis
Statistical analyses were carried out using Microsoft Excel or GraphPad Prism Software. Statistical significance was determined by an unpaired two-tailed Student's t-test and paired when needed and equality of variance was checked using one-way ANOVA test. All the data for the in vitro experiments have been presented as means ± S.D. of at least three data points from three different experiments. p-values ≤ 0.05 were considered statistically significant and p-values ≤ 0.01 were considered highly significant.
3. Results
3.1. Timeline for induction of nuclear HIF1α and cytoplasmic Cn under hypoxia in Raw264.7 cell line
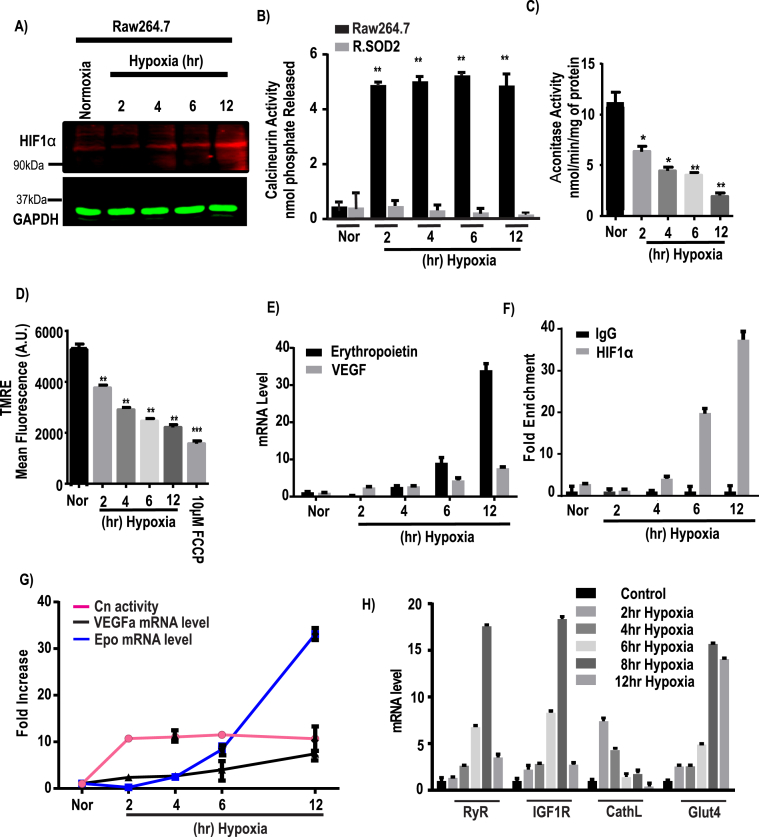
To evaluate the role of mitochondrial ROS in different signaling pathways, we first exposed Raw264.7 cells to hypoxic conditions (1% O2) for different time periods. This treatment resulted in HIF1α protein accumulation starting from about 4 to 6 h and continuing until 12 h of hypoxia (Fig. 1A). In contrast, Cn was activated by over 10-fold as early as 2 h after initiation of hypoxia, and remained at that level for at least 12 h (Fig. 1B). We took a genetic approach to generate stable mitochondrial superoxide dismutase-2 (SOD2) expressing Raw264.7 cell line (as described in Methods, Fig. S2B), which we named R.SOD2 cells. This stable cell line was generated, to see if the scavenging of mitochondrial superoxide can take away the free radical's effect on MtRS. A high increase in Cn activity due to hypoxia was diminished with mitochondrial SOD2 expression (Fig. 1B) proves the involvement of mitochondrial O2•– in MtRS. Mitochondrial aconitase activity is highly sensitive to mitochondrial O2•– and its inactivation has been used as a vital oxidative stress marker [45,46]. Results presented in Fig. 1C show that aconitase activity was inhibited by about 45% after 2 h of hypoxia, and continued to decline steadily until 12 h of hypoxia, suggesting a cumulative effect. The mitochondrial membrane potential, ΔΨm, which is a direct reflection of mitochondrial function, as measured by monitoring the accumulation of TMRE, revealed that ΔΨm was reduced by about 20% by 2 h of hypoxia, and continued to decrease steadily until 12 h of hypoxia. The mitochondrial protonophore FCCP (10 μM), used as a control, caused the maximal disruption of ΔΨm (Fig. 1D). Finally, real-time qPCR data of gene targets of the HIF1α transcription factor shows that erythropoietin and VEGF transcripts are increased gradually up to 6 h of hypoxia and were further pronounced increase at 12 h of hypoxia (Fig. 1E). Moreover, ChIP analysis of the VEGFα promoter demonstrates high levels of HIF1α factor binding by 6 h and continued to increase over the 12 h period of hypoxia (Fig. 1F). Notably, the induction of HIF1α marker genes is markedly slower than the Ca2+/calmodulin dependent activation of Cn (Fig. 1G) and Ca2+/Cn retrograde signaling markers, RyR, IGF1R, and Glut 4 mRNAs were induced by about 2–4 h of hypoxia and reached the maximum level by 6–8 h of hypoxia (Fig. 1H). In contrast, cathepsin L mRNA was induced maximally at 2 h of hypoxia and trended to decline by 6–12 h of hypoxia. Thus, under hypoxic conditions, both pathways are activated, but the kinetics of induction of HIF1α marker genes are markedly slower than the time frame for the initiation of Ca2+/Cn pathway gene expression.
Fig. 1.
Downstream effects of HIF1α and calcineurin A in response to hypoxia. Raw264.7 cells were grown under hypoxia for different time points (0–12h) and each group was divided into two subgroups for immunoblot and biochemical assays. A, a representative immunoblot analysis for HIF1α and GAPDH as loading control, using 40 μg of cell extract in each case. B, calcineurin activity was assayed by using an assay kit as described in the Materials & Methods. C, Aconitase activity assay was performed with 200 μg of total cell lysates as an indicator of oxidative stress and expressed as nmol/min/mg of protein. D, mitochondrial membrane potential ( was measured using TMRE and presented as mean fluorescence intensity (A.U.). E, Hypoxia induced changes in erythropoietin (Epo) and VEGFα mRNA levels. The mRNA levels were monitored by qRT-PCR analysis. Results from 3 independent experiments (n = 3) were normalized to β-actin mRNA levels and expressed as a fold change over the normoxic control. F, Chromatin immunoprecipitation (ChIP) analysis was performed on a HIF1α binding site located on VEGFα promoter at nucleoside −944 to −831 from the transcription start site and enrichment of HIF1α on VEGFα promoter was measured in Raw cells after hypoxic stimulation (n = 3 for all groups). Data represent mean ± S.D. G, the graph represents the fold increase of Cn activity (means ± S.D., n = 3) relative to VEGFα and Epo mRNA levels at each time point of hypoxia after normalization to control. H, mRNA expression profile of different MtRS signaling markers at different time of hypoxia. The mRNA levels were measured by qRT-PCR analysis. Results from 3 independent experiments (n = 3) were normalized to beta actin mRNA levels and expressed as a fold change over the normoxic control. Values showing significant differences from control or individual groups are indicated with an asterisk (*p < 0.05, **p < 0.01).
3.2. Effects of mitochondrially targeted agents on Raw264.7 macrophages
We next explored the impact of mitochondria-targeted agents MitoPQ and MitoMet that were reported previously to induce mitochondrial superoxide generation and affect mitochondrial function in different cells. Similar to other TPP+ conjugated compounds reported in the literature [[33], [34], [35],47], the two mitochondrially targeted agents used in this study accumulate in the mitochondrial matrix to a significantly higher extent than in the cytoplasm and other subcellular compartments [33]. Fig. S1 shows the structures of all four compounds used, i.e., decyl-TPP+ (DTPP+), which is a common control for both mitochondria-targeted PQ and Metformin, MitoPQ-control in which a non-redox cycling bipyridinium moiety attached to DTPP+, MitoPQ, and MitoMet. Previous studies showed that DTPP+ inhibited mitochondrial respiration at high doses [35,48]. We, therefore, used both DTPP+ and MitoPQ-control for the analysis of ROS production and mitochondrial stress signaling as negative controls.
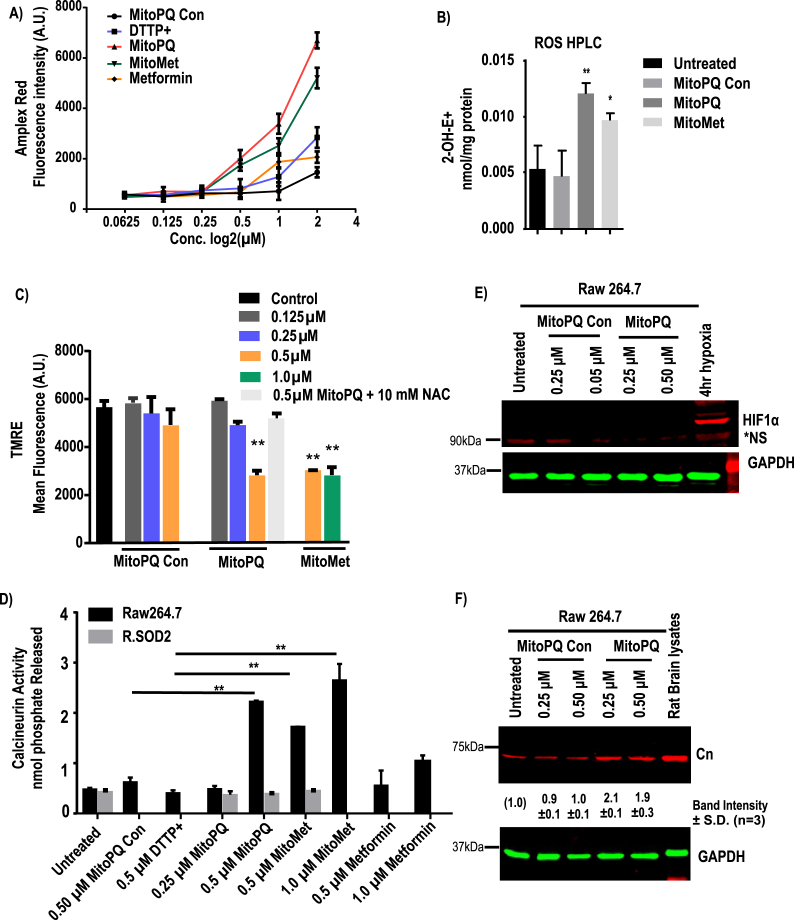
MitoPQ produces mitochondrial ROS by redox recycling at complex I, while MitoMet produces ROS by inhibiting complex I [21,33,49,50]. As shown in Fig. 2A, Raw 264.7 macrophages were treated with different doses of control compounds, and the two test compounds selected based on the dose-dependent inhibition of aconitase (Fig. S2A). Increased production of O2•– leads to higher levels of H2O2 which can be conveniently measured in the extracellular medium using Amplex Red assay (Fig. 2A) by fluorometry. We also used HPLC-based profiling of hydroethidine oxidation products, including the measurement of 2-hydroxyethidium (2-OH-E+), a product of the reaction of hydroethidine with superoxide (Fig. 2B). Additionally, fluorometrically, we measured MitoSOX-derived red fluorescence (Fig. S3A) generated by cellular oxidants in response to treatment. In addition, a fluorescent micrograph of MitoSOX loaded cells with Mitotracker green is presented in Fig. S3B.
Fig. 2.
Effects of mitochondria targeted pro-oxidant MitoPQ on subcellular redox state. ROS generation was assayed in Raw264.7 cells that were either untreated or treated with MitoPQ control, MitoPQ or MitoMet and DTPP + for 16 h at the indicated concentrations. A, ROS generation was measured by Amplex Red assay (n = 3 independent experiments, in duplicate). Fluorescence values were normalized to cell number. B, Effects on superoxide production. Superoxide was measured using hydroethidine, as described in Materials and Methods. Results are mean ± S.D. (n = 3). C, mitochondrial membrane potential was measured using TMRE and represented as mean fluorescent intensity (A.U.) D, calcineurin activity was assayed by calcineurin cellular activity assay kit as nmol of phosphate released. E & F, a representative immunoblot analysis of proteins for HIF1α, calcineurin A and loading control GAPDH following MitoPQ control or MitoPQ treatment (n = 3). Significance was calculated by one-way ANOVA with Tukey's multiple-comparison test, and data are presented as treatment group versus no-treatment control. In all experiments error bars represent standard deviations (*p < 0.05, **p < 0.01). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
We found that both MitoMet and MitoPQ induced higher levels of oxidants when used at 0.5 and 1 μM concentrations than did the two control compounds used (Fig. S3A). While 0.5 μM MitoPQ-control alone produced marginal fluorescence above the untreated control, 0.5 μM MitoMet or MitoPQ produced nearly 3-fold higher fluorescence intensity and 1 μM MitoMet produced an even higher level. In the hydroethidine assay (Fig. 2B), the compounds increased the cellular level of the superoxide-specific 2-OH-E+ product, when compared to treatment with MitoPQ-control.
MitoMet and MitoPQ, when used at the concentration of 1 μM (p ≤ 0.01) and 2 μM (p ≤ 0.01) decreased cell viability to a higher extent (20–30%) than did the two control compounds or metformin alone (Fig. S3C). We also measured mitochondrial membrane potential, ΔΨm, after treatment with the four compounds using the TMRE probe (Fig. 2C). While treatment with MitoPQ control and DTPP+ up to 0.5 μM did not affect ΔΨm, 0.5 μM MitoPQ disrupted the transmembrane potential significantly (Fig. 2C). Additionally, the disruptive effect of MitoPQ was attenuated by the addition of 10 mM NAC (N-acetylcysteine), a known antioxidant. MitoMet at both 0.5 μM and 1 μM levels markedly disrupted ΔΨm (Fig. 2C).
We then investigated the levels of HIF1α by immunoblot analysis, inactivation of cellular aconitase (Fig. S2A), and the Cn assay to assess Ca2+/Cn activation, as described in the Methods section. Cn activity (Fig. 2D) induced by treatment with 0.25 μM and 0.5 μM MitoPQ in a dose-dependent fashion. MitoMet treatments (0.5 μM and 1 μM) also increased the Ca2+/Cn activation in Raw cells. While 0.5 μM metformin by itself did not induce Cn activity, Cn activity marginally induced at 1 μM (Fig. 2D) metformin treatment. Thus, mitochondria-targeted compounds were significantly more effective. We then evaluated the impact of mitochondrial oxidants on the accumulation of HIF1α and Cn proteins following treatment with either MitoPQ-control or MitoPQ (Fig. 2E and F). Immunoblot analysis shows that the HIF1α levels remained nearly undetectable in the total extracts of Raw 264.7 macrophages treated with different concentrations of MitoPQ-control as well as MitoPQ (Fig. 2E). Even 0.5 μM MitoPQ, which induced ROS production, did not show detectable HIF1α induction (Fig. 2E), suggesting that these agents do not stabilize HIF1α. In contrast, and in support of the Cn activity presented in Fig. 2D, the Cn protein level was induced by both 0.25 μM and 0.5 μM MitoPQ (Fig. 2F), but not by MitoPQ-control up to 0.5 μM level tested (Fig. 2F). The Cn activity by mitochondria-targeted agents were reversed to near normal level in SOD2 overexpressing R.SOD2 cells (Fig. 2D).
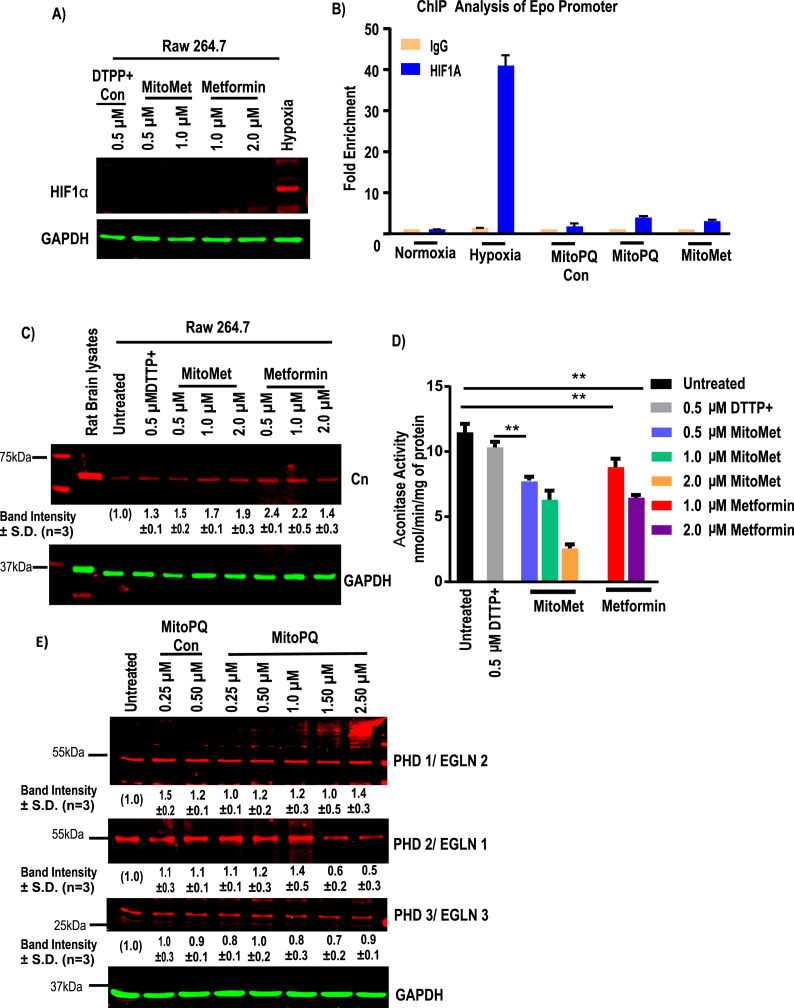
Similar approaches were used to test the effects of MitoMet. Immunoblot analysis shows that neither MitoMet nor metformin induced any detectable HIF1α protein accumulation (Fig. 3A). In support of immunoblot data indicating that HIF1α is not stabilized by MitoPQ, ChIP analysis of Epo promoter DNA revealed that MitoPQ control, MitoMet, as well as MitoPQ, were unable to induce occupancy of HIF1α on the target promoters under in vitro conditions. In contrast, hypoxia-grown cells showed a high level of promoter occupancy by HIF1α (Fig. 3B). Similar to MitoPQ, both MitoMet and unconjugated metformin showed increased Cn protein levels (Fig. 3C). Although not shown, Cn activity was induced under these treatment conditions. In agreement with the Cn induction results (Fig. 3C), the aconitase activity assay shows that MitoMet at 0.5 μM and 1 μM level, as well as unconjugated metformin at 1 μM and 2 μM levels, inhibited aconitase, suggesting induction of oxidative stress (Fig. 3D). In order to validate the generation of mitochondrial oxidant, we used non-reducing SDS-PAGE gel and immunoblotting to check the conversion of reduced peroxiredoxin (22 kDa) to oxidized form (44 kDa). Mitochondrial peroxiredoxin 3 (Prx3) and cytosolic peroxiredoxin 1 (Prx1) are the endogenous indicators of compartmental redox state [51]. Prx3, which scavenges mitochondrial H2O2 [34], changes from reduced monomer to oxidized dimer upon cell treatment with MitoPQ (Fig. S4A) and MitoMet (Fig. S4B) with only moderate effects on the redox state of cytosolic Prx1. More pronounced oxidation effect of Prx3 was observed in MitoPQ treatment when compared with control MitoPQ. 70–80% of Prx3 was oxidized upon the treatment with 0.25 μM and 0.5 μM MitoPQ (Fig. S4C), while about 60% of oxidized Prx3 protein was observed upon treatment with MitoMet (0.5 μM and 1.0 μM), as shown in Fig. S4D. These results indicate that mitochondria are the primary source of ROS upon cell treatment with MitoPQ and MitoMet. However, we did not see any significant changes in the total (oxidized + reduced) amount of Prx1 and Prx3 (Fig. S4 E and F). These results collectively show that both MitoMet and MitoPQ induce oxidant production, inhibit aconitase activity, and induce Cn activity but do not contribute significantly to HIF1α activation.
Fig. 3.
Effects of mitochondria-targeted MitoPQ and MitoMet on subcellular redox state. Raw264.7 cells were treated with either DTPP + or TPP + conjugated metformin, MitoPQ or unconjugated metformin for 16 h at the indicated doses. A, a representative immunoblot analysis of HIF1α and loading control GAPDH (n = 3). B, ChIP analysis was performed for HIF1α binding site on Epo promoter site and enrichment of HIF1α binding was determined (n = 3 for all groups). Data are shown as mean ± S.D. C, a representative immunoblot analysis of Calcineurin A and loading control GAPDH. D Aconitase activity assay was performed with 200 μg of total cell lysates as an indicator of oxidative stress and expressed as nmol/min/mg of protein. (means ± S.D., n = 3). E, a representative immunoblot analysis of cell extracts for PHD1, PHD2, PHD3 and loading control GAPDH following treatment with indicated doses of MitoPQ and MitoPQ control. Significance was calculated by one-way ANOVA with Tukey's multiple-comparison test, and data are presented as treatment group versus no-treatment control. In all experiments error bars represent standard deviations (*p < 0.05, **p < 0.01).
Low O2 levels inhibit the post-translational modification of HIF1α by blocking PHD-mediated ubiquitination and degradation [52]. Different concentrations of MitoPQ treatment in Raw264.7 cells did not change the levels of PHD1 or PHD3 compared to the control untreated cells and cells treated with MitoPQ control (Fig. 3E). On the other hand, PHD2 levels gradually decreased following treatment with relatively high doses (1.5 μM and 2.5 μM) of MitoPQ. These results support our finding that mitochondria-targeted ROS inducers did not stabilize HIF1α for up to 16 h of treatment tested.
3.3. MitoPQ and MitoMet induce calcineurin activity in other cell types as well
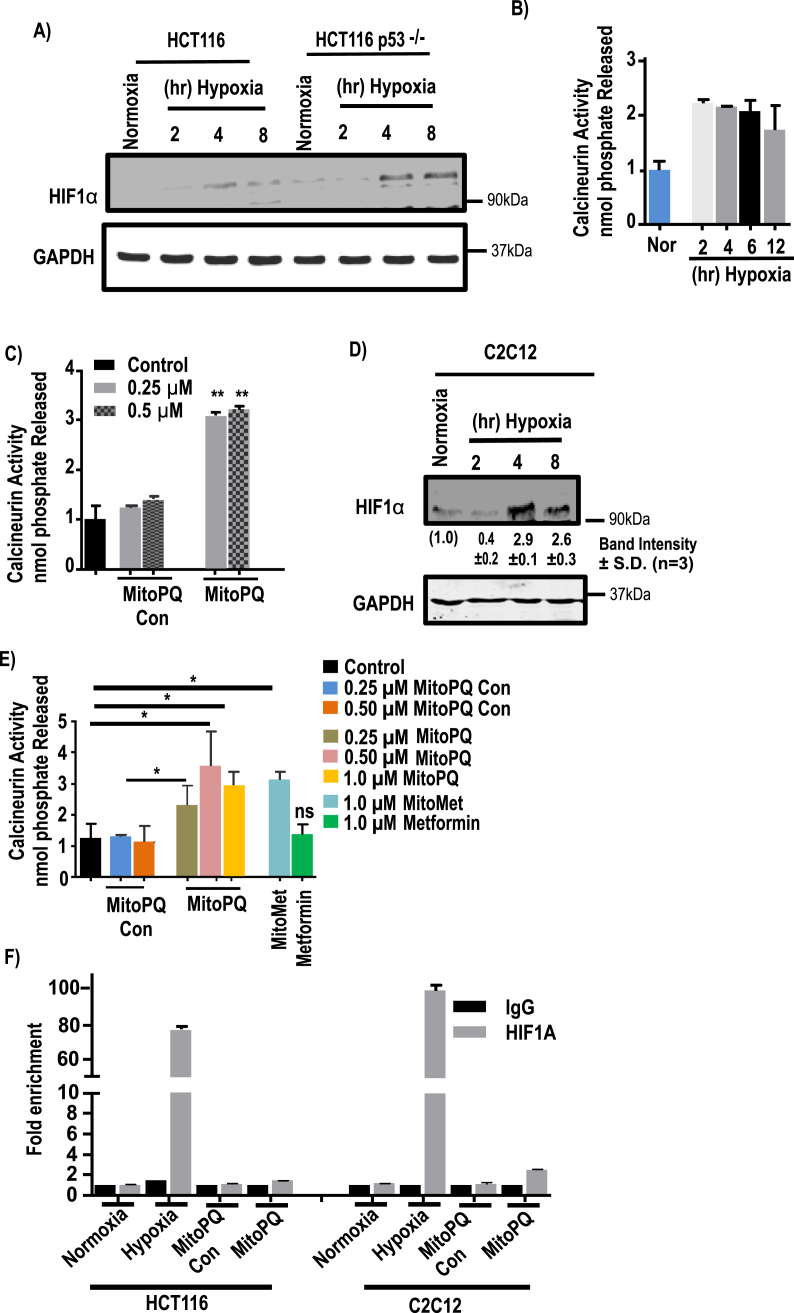
To determine the potential role of the Cn signaling pathway in MtRS, we evaluated the effects of hypoxia (2–8 h) as well as the mitochondria-targeted agents, MitoPQ and MitoMet (16 h) on HCT116 and C2C12 cells. Immunoblot analysis of HCT116 WT and HCT116 p53−/− cells grown under hypoxia for different lengths of time revealed that HIF1α protein starts accumulating at 4 h of hypoxia (Fig. 4A). As previously shown for Raw264.7 macrophages, Cn activity is induced to about 2.2-fold after 2 h of hypoxia and remains at this level until 12 h in HCT116 cells (Fig. 4B). While MitoPQ control at either 0.25 μM or 0.5 μM did not induce Cn activity beyond the untreated control cell level, the same concentrations of MitoPQ induced near 3-fold activity (Fig. 4C). On the other hand, there was no significant HIF1α activation by either MitoPQ control or MitoPQ as shown by the occupancy of Epo promoter DNA sites by HIF1α protein (Fig. 4F, left hand panel). Growth of HCT116 cells under hypoxia for 6 h, however, yields high enrichment of HIF1α by ChIP analysis.
Fig. 4.
Effects of mitochondria-targeted agents on the activation of signaling pathways in HCT116 and C2C12 cells. HCT116, HCT116p53−/− and C2C12 cells were exposed to hypoxia for different time points or treated with different mitochondria-targeted agents for indicated time and concentrations. A, immunoblot analysis of HIF1α and loading control GAPDH for the indicated cell lines. B–C, calcineurin activity was assayed by calcineurin cellular activity assay kit and nmol of phosphate released/mg of protein was plotted (means ± S.D., n = 3). D, immunoblot analysis of C2C12 cells subjected to hypoxia for HIF1α and loading control GAPDH. The blots are representative of three different runs. E, calcineurin activity, with or without treatment with mitochondria-targeted prooxidants in C2C12 cells. Activity was measured by calcineurin activity assay kit and (means ± S.D, n = 3). F, ChIP analysis was performed on a HIF1α binding site of Epo promoter to evaluate promoter occupancy by HIF1α in HCT116 and C2C12 cells after treatment with TPP+, MitoPQ for 16hr or hypoxia exposure for 12 h (n = 3 for all groups). Data represent mean ± S.D. The significance was calculated by one-way ANOVA with Tukey's multiple-comparison test, and data are presented as treatment group versus no-treatment control. In all experiments error bars represent standard deviations (*p < 0.05, **p < 0.01).
Immunoblot analysis also revealed the time dependent accumulation of HIF1α protein in C2C12 myoblasts exposed to hypoxia 4 and 8 h (Fig. 4D). Although not shown, Cn activation in these cells occurs at 2 h of hypoxia. While treatment of C2C12 cells for 16 h with 0.25 μM and 0.5 μM MitoPQ control failed to induce Cn activation above the basal untreated cell level (Fig. 4E), treatment with 0.25 μM and 0.5 μM MitoPQ and 1 μM MitoMet induced Cn activation by 2.5 to 3-fold. The addition of 1 μM unconjugated metformin alone did not induce significant Cn activity. Furthermore, the ChIP analysis of both 0.5 μM MitoPQ-control and MitoPQ failed to show any enrichment of HIF1α binding to the Epo promoter DNA, whereas 4 h hypoxia treated C2C12 cells showed high enrichment of HIF1α binding (Fig. 4F, right hand panel). Together, these results show that while hypoxia induces both Cn and HIF1α activation in time separated fashion, mitochondria-targeted agents failed to show any HIF1α activation under the same experimental conditions that activate Cn.
3.4. Mitochondrial ROS and AMPK activity in Raw264.7 and C2C12 cells
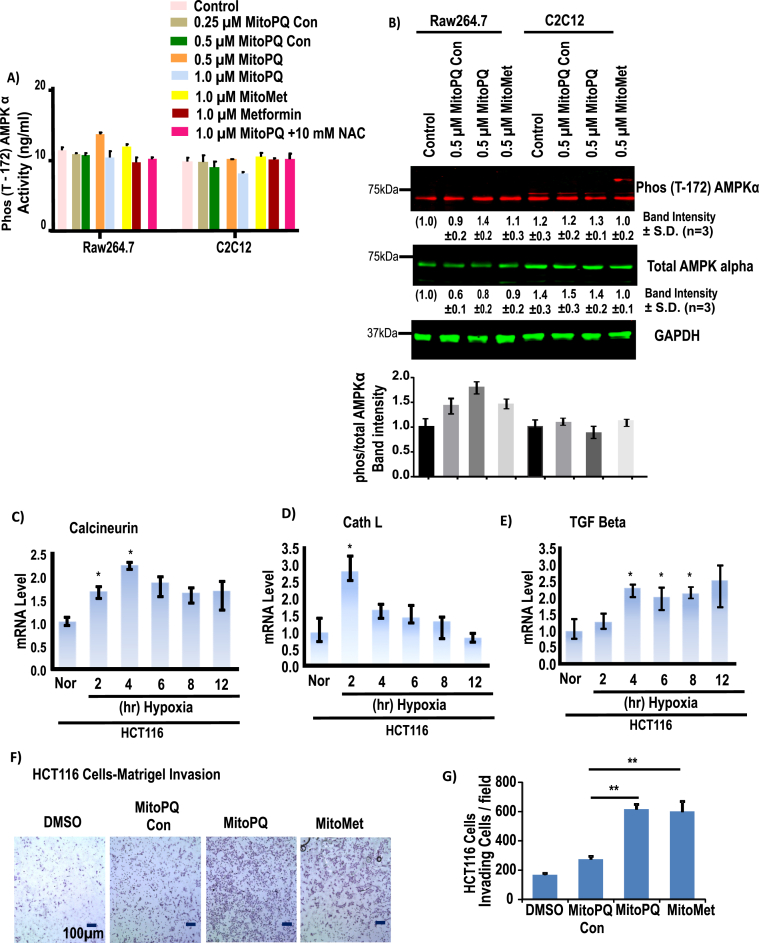
AMPK signaling is an essential pathway for balancing glycolysis and mitochondrial oxidative metabolism and is activated when mitochondrial ATP production goes down [53]. Moreover, in cancer cells under metabolic stress, AMPK can induce ATP generation from non-glucose substrates. H2O2 generation by glucose oxidase promotes AMPK activation. Here, we determined if AMPK activity was induced by mitochondrial oxidants stimulated by MitoPQ, MitoMet and MitoPQ-control. We did not observe any increase in AMPK activity at concentrations of MitoPQ and MitoMet that induced Cn signaling (Fig. 5A). Immunoblot analysis shows no significant increase in AMPKα levels with similar treatment of either C2C12 or Raw264.7 cells (Fig. 5B) and a modest increase in AMPK phosphorylation by drug treatment (Fig. 5B).
Fig. 5.
Effects of pro-oxidants on AMPK activity and Cn pathay. AMPKα (Thr172) activity in Raw264.7 and C2C12 cells were measured following treatment with DTPP + control or TPP + conjugated agents (MitoPQ and MitoMet) for 16 h at the indicated doses. A, Phosphorylated AMPKα level was measured in Raw264.7 cells and C2C12 cells. B, immunoblot analysis of phosphor-AMPKα protein along with GAPDH as loading control. The Bar diagram representing (the band intensity AMPKα Phos/tolal) The blots are representative of three different runs (n = 3). C-E, mRNA expression profile of different MtRS signaling markers at different time of hypoxia in HCT116 cells. The mRNA levels were monitored in qRT-PCR assays. The results from 3 independent experiments and normalized to beta actin mRNA levels and expressed as a fold change over the normoxic control. F, Matrigel invasion of control HCT116 cells treated with or without mito-targeted agents for 16h, before cells were loaded in to the chamber. G, quantitation of invading cells from triplicate experiments as in F. Significance was calculated by one-way ANOVA with Tukey's multiple-comparison test, and data are presented as treatment group versus no-treatment control. In all experiments, error bars represent standard deviations (*p < 0.05, **p < 0.01).
3.4.1. Induction of mitochondrial retrograde signaling markers in response to mitochondria-targeted agents
We next analyzed the marker genes for Cn mediated MtRS, including Cathepsin L and TGF-β mRNAs. Realtime qPCR results show that in HCT116 cells, Cn targets were induced by 2 h of hypoxia and levels were maintained up to 12 h of hypoxia (Fig. 5C). Cathepsin L mRNA was induced by 2 h of hypoxia and came down steadily with longer hours of hypoxia (Fig. 5D). The reason for this decline is unclear. TGF-β mRNA was induced marginally by 2 h of hypoxia but more robustly (2.5 to 3-fold) by 4 h and beyond (Fig. 5E).
It was shown that in C2C12 and other cell types, activation of Cn-mediated MtRS induced invasive behavior, as assessed by invasion through a Matrigel membrane layer [[54], [55], [56]]. When tested with 0.5 μM MitoMet and MitoPQ, HCT116 cells showed high level of invasiveness, while treatment with 0.5 μM MitoPQ-control alone had no significant effect (Fig. 5F and G). These results confirm that mitochondria-targeted oxidants induce MtRS as seen by the induction of biomarkers, including invasive behavior.
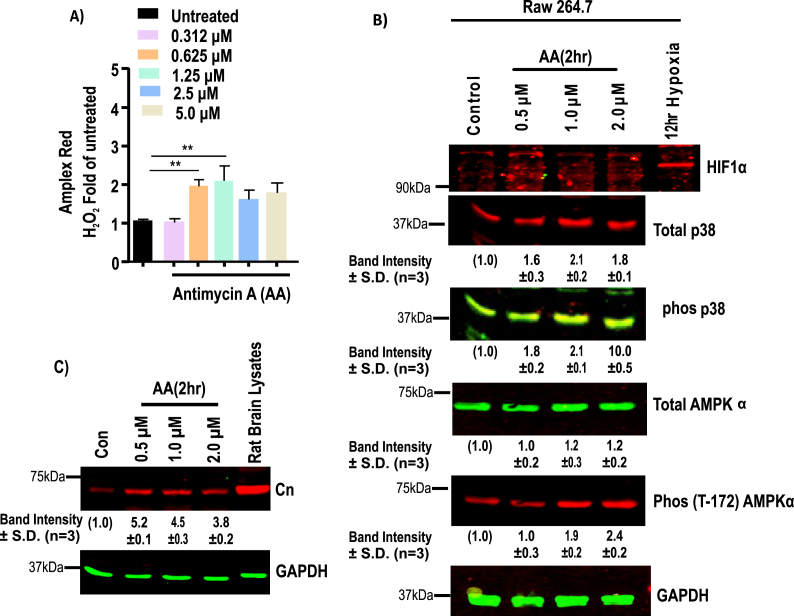
3.5. Complex III mediated ROS and HIF1α accumulations
Antimycin A (AA) is a known inhibitor of cytochrome bc1 complex and promotes ROS production 4 times faster than basal levels in bovine heart mitochondria [57] in vitro. We, therefore, tested the effects of AA on Raw264.7 cells (Fig. 6A). At 0.625 μM and 1.25 μM level, AA significantly induced H2O2 production, as measured by using the Amplex Red assay (Fig. 6A). Treatment with AA caused pronounced oxidation of mitochondrial Prx3 in comparison to cytosolic Prx1, indicating mitochondrial generation of ROS (Fig. S5) in treated Raw264.7 cells. However, AA up to the maximum range of 5.0 μM failed to induce any accumulation of HIF1α protein in Raw264.7 cells (Fig. 6B). As the phosphorylation status of p38 changes in response to ROS generation, we next evaluated the p38 phosphorylation status in AA treated cells. As expected, phosphorylation of p38 gradually increased with increasing concentrations of AA for 2 h (Fig. 6B). At higher concentrations of AA (2.0 μM), phosphorylation of p38 was significantly higher (Fig. 6B). Induction of AMPK phosphorylation was observed at 1.0 μM and 2.0 μM of AA treated cells. In contrast, as shown in Fig. 5B, TPP+-linked mitochondria targeted agents that can initiate ROS production did not have any significant effect on AMPK phosphorylation. In addition, the level of the catalytic subunit of Cn induced by low doses of AA and further decreased with increasing AA doses (Fig. 6C). These results suggest that ROS generated by complex III inhibitors did not stabilize the HIF1α protein under the current experimental conditions though it did activate Ca2+/Cn and AMPK pathways.
Fig. 6.
Effects of Complex III inhibitor on HIF1α. ROS generation in Raw264.7 cells in response to treatment with antimycin A for 2h at the indicated concentrations. A, ROS generation was measured by Amplex Red assay (n = 3 independent experiments, in duplicate). Fluorescent values were normalized to cell number. Significance was calculated by one-way ANOVA with Tukey's multiple-comparison test, and data are presented as treatment group versus no-treatment control. In all experiments error bars represent standard deviations (*p < 0.05, **p < 0.01). B, Representative immunoblots of HIF1α, p38, phosphorylated p38, phosphorylated AMPKα proteins and GAPDH as loading control. C. Immunoblot analysis of Raw 264.7 cell extracts treated with indicated concentrations of AA for 2h. GAPDH was used as a loading control. The blots are representative of three different runs. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Immunoblot in Fig. S6 shows that both MitoPQ and MitoMet did not induce the accumulation of oxidative stress-responsive transcription factor NRF-2 in a significant manner. This is consistent with the results in Figs. S3 and S4.
4. Discussion
MtRS, also called mitochondrial stress signaling, plays a role in many pathophysiological processes including cancer [58], aging, neurodegeneration [12], and even in innate immunity [59]. More recently, mitochondrial stress has also been implicated in mitohormesis, a process of preconditioning cells or tissues against oxidative stress-induced injury or mitochondrial mutations [47,60]. There are several effector pathways elicited by MtRS, including the AMPK pathway [61] that can promote insulin resistance and reflects the metabolic state of cells [62], the HIF1α pathway, which is implicated in cancer initiation and progression and also in other diseases, mitochondrial UPR signaling that has been implicated in resistance to pathogenic infection [2], and the Ca2+/Cn signaling pathway which is involved in tumor development and cardiac dysfunction. This study provides a comparative view of three different signaling pathways initiated by mitochondrial dysfunction.
Recently, mitochondrial ROS have received special attention as a causative factor in many pathological processes through activation of MtRS [13,63]. Some studies suggest that ROS-induced signaling invariably involves both HIF1α and AMPK activation [19,31]. However, in our laboratory, we have shown that the disrupted ΔΨm can also induce the Ca2+/Cn signaling pathway [13] in multiple modes of mitochondrial dysfunction, including reduced mtDNA content [23], cytochrome c oxidase disruption [54], disruption of IKBβ, which supports Cn-mediated NFκβ activation [64], and hypoxia [30]. This pathway activates multiple transcription factors and causes a change in nuclear gene expression program [9]. As it currently remains unclear how or if the HIF1α, AMPK, and Ca2+/Cn activation pathways are related, we undertook a comparative analysis of the three MtRS pathways under hypoxic conditions and following stimulation of mitochondrial oxidants. We did not evaluate the mtUPR pathway since the threshold of oxidative stress needed for mtUPR activation and the transcription factors involved are quite different from the Ca2+/Cn [9], HIF1α [19] and AMPK pathways [31]. Nor did we evaluate pathways leading to mitohormesis, which can be induced by very low levels (<0.1 μM) of MitoPQ to provide protection against ischemia-induced myocardial dysfunction [47].
Using hypoxia and mitochondria-targeted agents (MitoPQ and MitoMet) as inducers of mitochondrial ROS [33,34], we have carefully evaluated the dose and time-dependent activation of the three pathways, i.e., Ca2+/Cn, HIF1α and AMPK and their target gene expression to understand the time-frame of their activation and the relationship, if any, between these pathways. Several tangible mechanisms have been proposed for the role of mitochondrial ROS in HIF1α activation and AMPK activation [19,65] though there exist differences of opinions on each of the pathways. In the case of ROS mediated Ca2+/Cn activation, which also can get activated by proline dependent ROS [66], we believe that the direct attack of ROS on mitochondrial inner membrane components leads to disruption of ΔΨm and an associated increase in cytoplasmic Ca2+. In support of this possibility, we have shown that myocardial ischemia/reperfusion and/or hypoxia induce selective degradation of CcO subunits I, IVi1 and Vb, causing disruption of ΔΨm [61]. Similarly, Lesnefski's group has shown that hypoxia and mitochondrial ROS induce peroxidation of cardiolipin that is important for CcO function [65]. Other mitochondrial reactive species such as NO and CO can also affect CcO function by direct binding to the heme components of CcO [67], suggesting the involvement of other species in disturbing membrane integrity and induction of stress signaling [68]. Our results with hypoxia (Fig. 1, Fig. 4) clearly show that activation of Cn occurs by 2–4 h of hypoxia, while the activation of HIF1α, as tested by the most sensitive ChIP analysis, is a significantly delayed process. The same is true with AMPK level (Fig. 5A). Thus, we believe that the Ca2+/Cn pathway, with its distinct molecular targets, is the most sensitive to oxidant-mediated upregulation. Furthermore, some studies suggest that calcium/calmodulin dependent protein kinase II activity is also induced under cellular ROS environment [69,70]. Finally, it is clear from our current and published data [32] that Ca2+/Cn pathway functions independently of the other two pathways and, in most studies involving mitochondrial ROS, one is expected to see the compounding effects on all three pathways.
The use of mitochondria-targeted agents MitoMet and MitoPQ [33,34] in three different cell lines clearly supports the above conclusion. First, both agents at sub-toxic levels induce Ca2+/Cn but failed to induce detectable HIF1α or AMPK activation (Fig. 2, Fig. 3, Fig. 5A). As shown in Fig. 1, Fig. 5, these agents also induce the activation of Ca2+/Cn-dependent stress target genes (RyR, Glut4, IGF1R, TGFβ, etc.) and in vitro Matrigel invasion by C2C12 cells, which is a gold standard biomarker of retrograde signaling through Ca2+/Cn activation. These results suggest that the HIF1α activation and AMPK activation require a higher threshold of mitochondrial ROS production than that is achieved by these mitochondria-targeted agents. The results with these drugs are of particular significance because of their ability to act more selectively on mitochondrial targets. In addition, since they form a steep gradient with the highest concentration in the mitochondrial matrix compartment, they cause significantly less damage in other subcellular compartments.
ETC complex III is another critical site for ROS production [65]. We tested the effects of complex III inhibitor AA on HIF1α activation and AMPK activation. Complex III inhibition by AA led to AMPK activation, as seen by the increase in phosphorylated AMPKα and its dependent p38 (Fig. 6). However, AA at high levels (up to 5 μM) did not induce HIF1α protein accumulation or inhibit PDH level (Fig. 6) under the present experimental conditions.
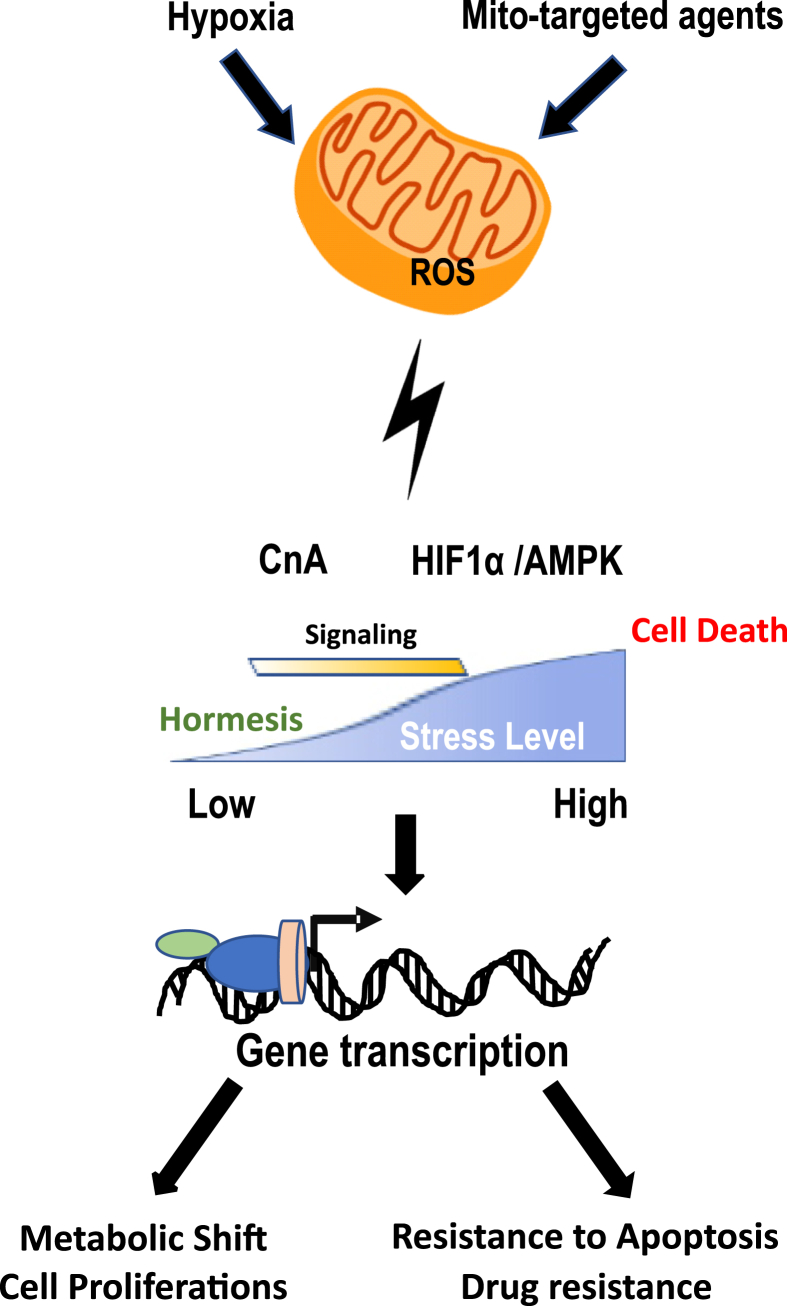
In summary, this study, compares the effects of different modes and doses of mitochondrial ROS on the three mitochondrial stress induced signaling pathways. Based on our results, we conclude that Ca2+/Cn pathway is the most sensitive, as it is activated even at low thresholds of mitochondrial ROS (see Fig. 7). In that regard, the Ca2+/Cn pathway is likely to be important in communicating the dynamic aspects of mitochondrial metabolic and energy status with the nuclear compartment. However, under higher oxidative stress conditions such as hypoxia, all three stress signaling pathways may be activated and function concordantly to influence the cellular response to stress. The ability of very low levels (0.1 μM) MitoPQ-induced ROS to provide cardioprotection by inducing mitohormesis [47], while high levels (1–2 μM) of MitoMet inhibiting cell proliferation [34], supports the dose-dependent nature of these varied MtRS signaling cascades.
Fig. 7.
Schematic diagram of retrograde signaling in response to Mitochondrial ROS. A schematic presentation of mitochondrial ROS induced retrograde signaling which affects multiple cellular process. The relative levels of mitochondrial ROS needed for the activation of each pathway is only relative and arbitrary units.
Acknowledgements
We thank the Avadhani lab members for useful suggestions and discussions and Dr. Leslie King for critically reading and editing the paper. This work was in part supported by NIH RO1 Grants GM-34883 and AA022986 and an endowment from the Harriet Ellison Woodward trust to NGA and grants from the Medical Research Council UK (MC_U105663142) and by a Wellcome Trust Investigator award (110159/Z/15/Z) to MPM.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2020.101606.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Cadenas E., Davies K.J. Mitochondrial free radical generation, oxidative stress, and aging. Free Radic. Biol. Med. 2000;29(3-4):222–230. doi: 10.1016/s0891-5849(00)00317-8. PubMed PMID: 11035250. [DOI] [PubMed] [Google Scholar]
- 2.Droge W. Free radicals in the physiological control of cell function. Physiol. Rev. 2002;82(1):47–95. doi: 10.1152/physrev.00018.2001. PubMed PMID: 11773609. [DOI] [PubMed] [Google Scholar]
- 3.Harman D. Aging: a theory based on free radical and radiation chemistry. J. Gerontol. 1956;11(3):298–300. doi: 10.1093/geronj/11.3.298. PubMed PMID: 13332224. [DOI] [PubMed] [Google Scholar]
- 4.Sies H. Hydrogen peroxide as a central redox signaling molecule in physiological oxidative stress: oxidative eustress. Redox Biol. 2017;11:613–619. doi: 10.1016/j.redox.2016.12.035. PubMed PMID: 28110218; PubMed Central PMCID: PMCPMC5256672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Galluzzi L., Vitale I., Abrams J.M., Alnemri E.S., Baehrecke E.H., Blagosklonny M.V. Molecular definitions of cell death subroutines: recommendations of the Nomenclature Committee on Cell Death 2012. Cell Death Differ. 2012;19(1):107–120. doi: 10.1038/cdd.2011.96. PubMed PMID: 21760595; PubMed Central PMCID: PMCPMC3252826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Battersby B.J., Richter U. Why translation counts for mitochondria - retrograde signalling links mitochondrial protein synthesis to mitochondrial biogenesis and cell proliferation. J. Cell Sci. 2013;126(Pt 19):4331–4338. doi: 10.1242/jcs.131888. PubMed PMID: 24013545. [DOI] [PubMed] [Google Scholar]
- 7.Attardi G., Schatz G. Biogenesis of mitochondria. Annu. Rev. Cell Biol. 1988;4:289–333. doi: 10.1146/annurev.cb.04.110188.001445. PubMed PMID: 2461720. [DOI] [PubMed] [Google Scholar]
- 8.Parikh V.S., Morgan M.M., Scott R., Clements L.S., Butow R.A. The mitochondrial genotype can influence nuclear gene-expression in yeast. Science. 1987;235(4788):576–580. doi: 10.1126/science.3027892. PubMed PMID: WOS:A1987F783400022. [DOI] [PubMed] [Google Scholar]
- 9.Butow R.A., Avadhani N.G. Mitochondrial signaling: the retrograde response. Mol. Cell. 2004;14(1):1–15. doi: 10.1016/s1097-2765(04)00179-0. PubMed PMID: 15068799. [DOI] [PubMed] [Google Scholar]
- 10.da Cunha F.M., Torelli N.Q., Kowaltowski A.J. Mitochondrial retrograde signaling: triggers, pathways, and outcomes. Oxid. Med. Cell Longev. 2015;2015:482582. doi: 10.1155/2015/482582. PubMed PMID: 26583058; PubMed Central PMCID: PMCPMC4637108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang H., Morais R. Up-regulation of nuclear genes in response to inhibition of mitochondrial DNA expression in chicken cells. Biochim. Biophys. Acta. 1997;1352(3):325–334. doi: 10.1016/s0167-4781(97)00035-3. PubMed PMID: 9224956. [DOI] [PubMed] [Google Scholar]
- 12.Guha M., Avadhani N.G. Mitochondrial retrograde signaling at the crossroads of tumor bioenergetics, genetics and epigenetics. Mitochondrion. 2013;13(6):577–591. doi: 10.1016/j.mito.2013.08.007. PubMed PMID: 24004957; PubMed Central PMCID: PMCPMC3832239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Biswas G., Anandatheerthavarada H.K., Zaidi M., Avadhani N.G. Mitochondria to nucleus stress signaling: a distinctive mechanism of NFkappaB/Rel activation through calcineurin-mediated inactivation of IkappaBbeta. J. Cell Biol. 2003;161(3):507–519. doi: 10.1083/jcb.200211104. PubMed PMID: 12732617; PubMed Central PMCID: PMCPMC2172940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haynes C.M., Yang Y., Blais S.P., Neubert T.A., Ron D. The matrix peptide exporter HAF-1 signals a mitochondrial UPR by activating the transcription factor ZC376.7 in C. elegans. Mol. Cell. 2010;37(4):529–540. doi: 10.1016/j.molcel.2010.01.015. PubMed PMID: 20188671; PubMed Central PMCID: PMCPMC2846537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harman D. The biologic clock: the mitochondria? J. Am. Geriatr. Soc. 1972;20(4):145–147. doi: 10.1111/j.1532-5415.1972.tb00787.x. PubMed PMID: 5016631. [DOI] [PubMed] [Google Scholar]
- 16.Owusu-Ansah E., Yavari A., Mandal S., Banerjee U. Distinct mitochondrial retrograde signals control the G1-S cell cycle checkpoint. Nat. Genet. 2008;40(3):356–361. doi: 10.1038/ng.2007.50. PubMed PMID: 18246068. [DOI] [PubMed] [Google Scholar]
- 17.Wang D., Wang Y., Argyriou C., Carriere A., Malo D., Hekimi S. An enhanced immune response of Mclk1(+)/(-) mutant mice is associated with partial protection from fibrosis, cancer and the development of biomarkers of aging. PloS One. 2012;7(11) doi: 10.1371/journal.pone.0049606. PubMed PMID: 23166727; PubMed Central PMCID: PMCPMC3498213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chandel N.S., Maltepe E., Goldwasser E., Mathieu C.E., Simon M.C., Schumacker P.T. Mitochondrial reactive oxygen species trigger hypoxia-induced transcription. Proc. Natl. Acad. Sci. U. S. A. 1998;95(20):11715–11720. doi: 10.1073/pnas.95.20.11715. PubMed PMID: 9751731; PubMed Central PMCID: PMCPMC21706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chandel N.S., McClintock D.S., Feliciano C.E., Wood T.M., Melendez J.A., Rodriguez A.M. Reactive oxygen species generated at mitochondrial complex III stabilize hypoxia-inducible factor-1alpha during hypoxia: a mechanism of O2 sensing. J. Biol. Chem. 2000;275(33):25130–25138. doi: 10.1074/jbc.M001914200. PubMed PMID: 10833514. [DOI] [PubMed] [Google Scholar]
- 20.Lapointe J., Hekimi S. Early mitochondrial dysfunction in long-lived Mclk1+/- mice. J. Biol. Chem. 2008;283(38):26217–26227. doi: 10.1074/jbc.M803287200. PubMed PMID: 18635541; PubMed Central PMCID: PMCPMC3258865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kalyanaraman B., Cheng G., Hardy M., Ouari O., Bennett B., Zielonka J. Teaching the basics of reactive oxygen species and their relevance to cancer biology: mitochondrial reactive oxygen species detection, redox signaling, and targeted therapies. Redox Biol. 2018;15:347–362. doi: 10.1016/j.redox.2017.12.012. PubMed PMID: 29306792; PubMed Central PMCID: PMCPMC5756055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tang W., Chowdhury A.R., Guha M., Huang L., Van Winkle T., Rustgi A.K. Silencing of IkBbeta mRNA causes disruption of mitochondrial retrograde signaling and suppression of tumor growth in vivo. Carcinogenesis. 2012;33(9):1762–1768. doi: 10.1093/carcin/bgs190. PubMed PMID: 22637744; PubMed Central PMCID: PMCPMC3514893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Biswas G., Adebanjo O.A., Freedman B.D., Anandatheerthavarada H.K., Vijayasarathy C., Zaidi M. Retrograde Ca2+ signaling in C2C12 skeletal myocytes in response to mitochondrial genetic and metabolic stress: a novel mode of inter-organelle crosstalk. EMBO J. 1999;18(3):522–533. doi: 10.1093/emboj/18.3.522. PubMed PMID: 9927412; PubMed Central PMCID: PMCPMC1171145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bell E.L., Klimova T.A., Eisenbart J., Moraes C.T., Murphy M.P., Budinger G.R. The Qo site of the mitochondrial complex III is required for the transduction of hypoxic signaling via reactive oxygen species production. J. Cell Biol. 2007;177(6):1029–1036. doi: 10.1083/jcb.200609074. PubMed PMID: 17562787; PubMed Central PMCID: PMCPMC2064363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jaakkola P., Mole D.R., Tian Y.M., Wilson M.I., Gielbert J., Gaskell S.J. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science. 2001;292(5516):468–472. doi: 10.1126/science.1059796. PubMed PMID: 11292861. [DOI] [PubMed] [Google Scholar]
- 26.Ivan M., Kondo K., Yang H., Kim W., Valiando J., Ohh M. HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science. 2001;292(5516):464–468. doi: 10.1126/science.1059817. PubMed PMID: 11292862. [DOI] [PubMed] [Google Scholar]
- 27.Masson N., Singleton R.S., Sekirnik R., Trudgian D.C., Ambrose L.J., Miranda M.X. The FIH hydroxylase is a cellular peroxide sensor that modulates HIF transcriptional activity. EMBO Rep. 2012;13(3):251–257. doi: 10.1038/embor.2012.9. PubMed PMID: 22310300; PubMed Central PMCID: PMCPMC3323130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haynes C.M., Fiorese C.J., Lin Y.F. Evaluating and responding to mitochondrial dysfunction: the mitochondrial unfolded-protein response and beyond. Trends Cell Biol. 2013;23(7):311–318. doi: 10.1016/j.tcb.2013.02.002. PubMed PMID: 23489877; PubMed Central PMCID: PMCPMC3700555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rugarli E.I., Langer T. Mitochondrial quality control: a matter of life and death for neurons. EMBO J. 2012;31(6):1336–1349. doi: 10.1038/emboj.2012.38. PubMed PMID: 22354038; PubMed Central PMCID: PMCPMC3321185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Srinivasan S., Avadhani N.G. Hypoxia-mediated mitochondrial stress in RAW264.7 cells induces osteoclast-like TRAP-positive cells. Ann. N. Y. Acad. Sci. 2007;1117:51–61. doi: 10.1196/annals.1402.067. PubMed PMID: 18056037; PubMed Central PMCID: PMCPMC4446731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chae S., Ahn B.Y., Byun K., Cho Y.M., Yu M.H., Lee B. A systems approach for decoding mitochondrial retrograde signaling pathways. Sci. Signal. 2013;6(264):rs4. doi: 10.1126/scisignal.2003266. PubMed PMID: 23443683. [DOI] [PubMed] [Google Scholar]
- 32.Chowdhury A.R., Long A., Fuchs S.Y., Rustgi A., Avadhani N.G. Mitochondrial stress-induced p53 attenuates HIF-1alpha activity by physical association and enhanced ubiquitination. Oncogene. 2017;36(3):397–409. doi: 10.1038/onc.2016.211. PubMed PMID: 27345397; PubMed Central PMCID: PMCPMC5192009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Robb E.L., Gawel J.M., Aksentijevic D., Cocheme H.M., Stewart T.S., Shchepinova M.M. Selective superoxide generation within mitochondria by the targeted redox cycler MitoParaquat. Free Radic. Biol. Med. 2015;89:883–894. doi: 10.1016/j.freeradbiomed.2015.08.021. PubMed PMID: 26454075. [DOI] [PubMed] [Google Scholar]
- 34.Cheng G., Zielonka J., Ouari O., Lopez M., McAllister D., Boyle K. Mitochondria-targeted analogues of metformin exhibit enhanced antiproliferative and radiosensitizing effects in pancreatic cancer cells. Canc. Res. 2016;76(13):3904–3915. doi: 10.1158/0008-5472.CAN-15-2534. PubMed PMID: 27216187; PubMed Central PMCID: PMCPMC4930686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reily C., Mitchell T., Chacko B.K., Benavides G., Murphy M.P., Darley-Usmar V. Mitochondrially targeted compounds and their impact on cellular bioenergetics. Redox Biol. 2013;1(1):86–93. doi: 10.1016/j.redox.2012.11.009. PubMed PMID: 23667828; PubMed Central PMCID: PMCPMC3647698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cheng G., Zielonka M., Dranka B., Kumar S.N., Myers C.R., Bennett B. Detection of mitochondria-generated reactive oxygen species in cells using multiple probes and methods: potentials, pitfalls, and the future. J. Biol. Chem. 2018;293(26):10363–10380. doi: 10.1074/jbc.RA118.003044. PubMed PMID: 29739855; PubMed Central PMCID: PMCPMC6028982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Myers C.R. Enhanced targeting of mitochondrial peroxide defense by the combined use of thiosemicarbazones and inhibitors of thioredoxin reductase. Free Radic. Biol. Med. 2016;91:81–92. doi: 10.1016/j.freeradbiomed.2015.12.008. PubMed PMID: 26686468. [DOI] [PubMed] [Google Scholar]
- 38.Cox A.G., Pullar J.M., Hughes G., Ledgerwood E.C., Hampton M.B. Oxidation of mitochondrial peroxiredoxin 3 during the initiation of receptor-mediated apoptosis. Free Radic. Biol. Med. 2008;44(6):1001–1009. doi: 10.1016/j.freeradbiomed.2007.11.017. PubMed PMID: 18164270. [DOI] [PubMed] [Google Scholar]
- 39.Wang F., Zhang R., Beischlag T.V., Muchardt C., Yaniv M., Hankinson O. Roles of Brahma and Brahma/SWI2-related gene 1 in hypoxic induction of the erythropoietin gene. J. Biol. Chem. 2004;279(45):46733–46741. doi: 10.1074/jbc.M409002200. PubMed PMID: 15347669. [DOI] [PubMed] [Google Scholar]
- 40.Yeo E.J., Cho Y.S., Kim M.S., Park J.W. Contribution of HIF-1alpha or HIF-2alpha to erythropoietin expression: in vivo evidence based on chromatin immunoprecipitation. Ann. Hematol. 2008;87(1):11–17. doi: 10.1007/s00277-007-0359-6. PubMed PMID: 17712557. [DOI] [PubMed] [Google Scholar]
- 41.Liang Y., Li X.Y., Rebar E.J., Li P., Zhou Y., Chen B. Activation of vascular endothelial growth factor A transcription in tumorigenic glioblastoma cell lines by an enhancer with cell type-specific DNase I accessibility. J. Biol. Chem. 2002;277(22):20087–20094. doi: 10.1074/jbc.M201766200. PubMed PMID: 11912213. [DOI] [PubMed] [Google Scholar]
- 42.Bansal S., Srinivasan S., Anandasadagopan S., Chowdhury A.R., Selvaraj V., Kalyanaraman B. Additive effects of mitochondrion-targeted cytochrome CYP2E1 and alcohol toxicity on cytochrome c oxidase function and stability of respirosome complexes. J. Biol. Chem. 2012;287(19):15284–15297. doi: 10.1074/jbc.M111.314062. PubMed PMID: 22396533; PubMed Central PMCID: PMCPMC3346148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zielonka J., Zielonka M., Sikora A., Adamus J., Joseph J., Hardy M. Global profiling of reactive oxygen and nitrogen species in biological systems: high-throughput real-time analyses. J. Biol. Chem. 2012;287(5):2984–2995. doi: 10.1074/jbc.M111.309062. PubMed PMID: 22139901; PubMed Central PMCID: PMCPMC3270955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gardner P.R., Nguyen D.D., White C.W. Aconitase is a sensitive and critical target of oxygen poisoning in cultured mammalian cells and in rat lungs. Proc. Natl. Acad. Sci. U. S. A. 1994;91(25):12248–12252. doi: 10.1073/pnas.91.25.12248. PubMed PMID: 7991614; PubMed Central PMCID: PMCPMC45414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gardner P.R. Aconitase: sensitive target and measure of superoxide. Methods Enzymol. 2002;349:9–23. doi: 10.1016/s0076-6879(02)49317-2. PubMed PMID: 11912933. [DOI] [PubMed] [Google Scholar]
- 46.Velsor L.W., Kariya C., Kachadourian R., Day B.J. Mitochondrial oxidative stress in the lungs of cystic fibrosis transmembrane conductance regulator protein mutant mice. Am. J. Respir. Cell Mol. Biol. 2006;35(5):579–586. doi: 10.1165/rcmb.2005-0473OC. PubMed PMID: 16763223; PubMed Central PMCID: PMCPMC2643276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Antonucci S., Mulvey J.F., Burger N., Di Sante M., Hall A.R., Hinchy E.C. Selective mitochondrial superoxide generation in vivo is cardioprotective through hormesis. Free Radic. Biol. Med. 2019;134:678–687. doi: 10.1016/j.freeradbiomed.2019.01.034. PubMed PMID: 30731114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schibler J., Tomanek-Chalkley A.M., Reedy J.L., Zhan F., Spitz D.R., Schultz M.K. Mitochondrial-targeted decyl-triphenylphosphonium enhances 2-deoxy-D-glucose mediated oxidative stress and clonogenic killing of multiple myeloma cells. PloS One. 2016;11(11) doi: 10.1371/journal.pone.0167323. PubMed PMID: 27902770; PubMed Central PMCID: PMCPMC5130279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Owen M.R., Doran E., Halestrap A.P. Evidence that metformin exerts its anti-diabetic effects through inhibition of complex 1 of the mitochondrial respiratory chain. Biochem. J. 2000;348 Pt 3:607–614. PubMed PMID: 10839993; PubMed Central PMCID: PMCPMC1221104. [PMC free article] [PubMed] [Google Scholar]
- 50.Ota S., Horigome K., Ishii T., Nakai M., Hayashi K., Kawamura T. Metformin suppresses glucose-6-phosphatase expression by a complex I inhibition and AMPK activation-independent mechanism. Biochem. Biophys. Res. Commun. 2009;388(2):311–316. doi: 10.1016/j.bbrc.2009.07.164. PubMed PMID: 19664596. [DOI] [PubMed] [Google Scholar]
- 51.Winterbourn C.C. Biological production, detection, and fate of hydrogen peroxide. Antioxidants Redox Signal. 2018;29(6):541–551. doi: 10.1089/ars.2017.7425. PubMed PMID: 29113458. [DOI] [PubMed] [Google Scholar]
- 52.Viscomi C., Bottani E., Civiletto G., Cerutti R., Moggio M., Fagiolari G. In vivo correction of COX deficiency by activation of the AMPK/PGC-1alpha axis. Cell Metabol. 2011;14(1):80–90. doi: 10.1016/j.cmet.2011.04.011. PubMed PMID: 21723506; PubMed Central PMCID: PMCPMC3130927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Srinivasan S., Guha M., Dong D.W., Whelan K.A., Ruthel G., Uchikado Y. Disruption of cytochrome c oxidase function induces the Warburg effect and metabolic reprogramming. Oncogene. 2016;35(12):1585–1595. doi: 10.1038/onc.2015.227. PubMed PMID: 26148236; PubMed Central PMCID: PMCPMC4703574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wallace D.C. Mitochondria and cancer. Nat. Rev. Canc. 2012;12(10):685–698. doi: 10.1038/nrc3365. PubMed PMID: 23001348; PubMed Central PMCID: PMCPMC4371788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Amuthan G., Biswas G., Zhang S.Y., Klein-Szanto A., Vijayasarathy C., Avadhani N.G. Mitochondria-to-nucleus stress signaling induces phenotypic changes, tumor progression and cell invasion. EMBO J. 2001;20(8):1910–1920. doi: 10.1093/emboj/20.8.1910. PubMed PMID: 11296224; PubMed Central PMCID: PMCPMC125420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Guha M., Srinivasan S., Sheehan M.M., Kijima T., Ruthel G., Whelan K. Esophageal 3D organoids of MPV17(-/-) mouse model of mitochondrial DNA depletion show epithelial cell plasticity and telomere attrition. Oncotarget. 2019;10(58):6245–6259. doi: 10.18632/oncotarget.27264. PubMed PMID: 31692873; PubMed Central PMCID: PMCPMC6817447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Drose S., Brandt U. The mechanism of mitochondrial superoxide production by the cytochrome bc1 complex. J. Biol. Chem. 2008;283(31):21649–21654. doi: 10.1074/jbc.M803236200. PubMed PMID: 18522938. [DOI] [PubMed] [Google Scholar]
- 58.West A.P., Shadel G.S., Ghosh S. Mitochondria in innate immune responses. Nat. Rev. Immunol. 2011;11(6):389–402. doi: 10.1038/nri2975. PubMed PMID: 21597473; PubMed Central PMCID: PMCPMC4281487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yun J., Finkel T. Mitohormesis. Cell Metabol. 2014;19(5):757–766. doi: 10.1016/j.cmet.2014.01.011. PubMed PMID: 24561260; PubMed Central PMCID: PMCPMC4016106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hinchy E.C., Gruszczyk A.V., Willows R., Navaratnam N., Hall A.R., Bates G. Mitochondria-derived ROS activate AMP-activated protein kinase (AMPK) indirectly. J. Biol. Chem. 2018;293(44):17208–17217. doi: 10.1074/jbc.RA118.002579. PubMed PMID: 30232152; PubMed Central PMCID: PMCPMC6222118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fazakerley D.J., Minard A.Y., Krycer J.R., Thomas K.C., Stockli J., Harney D.J. Mitochondrial oxidative stress causes insulin resistance without disrupting oxidative phosphorylation. J. Biol. Chem. 2018;293(19):7315–7328. doi: 10.1074/jbc.RA117.001254. PubMed PMID: 29599292; PubMed Central PMCID: PMCPMC5950018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lin Y.F., Haynes C.M. Metabolism and the UPR(mt) Mol. Cell. 2016;61(5):677–682. doi: 10.1016/j.molcel.2016.02.004. PubMed PMID: 26942672; PubMed Central PMCID: PMCPMC4779188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen Y., Azad M.B., Gibson S.B. Superoxide is the major reactive oxygen species regulating autophagy. Cell Death Differ. 2009;16(7):1040–1052. doi: 10.1038/cdd.2009.49. PubMed PMID: 19407826. [DOI] [PubMed] [Google Scholar]
- 64.Waypa G.B., Marks J.D., Guzy R., Mungai P.T., Schriewer J., Dokic D. Hypoxia triggers subcellular compartmental redox signaling in vascular smooth muscle cells. Circ. Res. 2010;106(3):526–535. doi: 10.1161/CIRCRESAHA.109.206334. PubMed PMID: 20019331; PubMed Central PMCID: PMCPMC2856085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lesnefsky E.J., Tandler B., Ye J., Slabe T.J., Turkaly J., Hoppel C.L. Myocardial ischemia decreases oxidative phosphorylation through cytochrome oxidase in subsarcolemmal mitochondria. Am. J. Physiol. 1997;273(3 Pt 2):H1544–H1554. doi: 10.1152/ajpheart.1997.273.3.H1544. PubMed PMID: 9321848. [DOI] [PubMed] [Google Scholar]
- 66.Rivera A., Maxwell S.A. The p53-induced gene-6 (proline oxidase) mediates apoptosis through a calcineurin-dependent pathway. J. Biol. Chem. 2005;280(32):29346–29354. doi: 10.1074/jbc.M504852200. PubMed PMID: 15914462. [DOI] [PubMed] [Google Scholar]
- 67.Cleeter M.W., Cooper J.M., Darley-Usmar V.M., Moncada S., Schapira A.H. Reversible inhibition of cytochrome c oxidase, the terminal enzyme of the mitochondrial respiratory chain, by nitric oxide. Implications for neurodegenerative diseases. FEBS Lett. 1994;345(1):50–54. doi: 10.1016/0014-5793(94)00424-2. PubMed PMID: 8194600. [DOI] [PubMed] [Google Scholar]
- 68.Brookes P.S., Levonen A.L., Shiva S., Sarti P., Darley-Usmar V.M. Mitochondria: regulators of signal transduction by reactive oxygen and nitrogen species. Free Radic. Biol. Med. 2002;33(6):755–764. doi: 10.1016/s0891-5849(02)00901-2. PubMed PMID: 12208364. [DOI] [PubMed] [Google Scholar]
- 69.Howe C.J., Lahair M.M., McCubrey J.A., Franklin R.A. Redox regulation of the calcium/calmodulin-dependent protein kinases. J. Biol. Chem. 2004;279(43):44573–44581. doi: 10.1074/jbc.M404175200. PubMed PMID: 15294913. [DOI] [PubMed] [Google Scholar]
- 70.Erickson J.R., Joiner M.L., Guan X., Kutschke W., Yang J., Oddis C.V. A dynamic pathway for calcium-independent activation of CaMKII by methionine oxidation. Cell. 2008;133(3):462–474. doi: 10.1016/j.cell.2008.02.048. PubMed PMID: 18455987; PubMed Central PMCID: PMCPMC2435269. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.