Abstract
Background
Duchenne Muscular Dystrophy (DMD) is a progressive, fatal neuromuscular disorder caused by mutations in the DMD gene. Emerging antisense oligomer based exon skipping therapy provides hope for the restoration of the reading frame.
Objectives
Population-based DMD mutation database may enable exon skipping to be used for the benefit of patients. Hence, we planned this study to identify DMD gene variants in North Indian DMD cases.
Methods
A total of 100 DMD cases were recruited and Multiplex ligation-dependent probe amplification (MLPA) analysis was performed to obtain the deletion and duplication profile.
Results
Copy number variations (deletion/duplication) were found in 80.85% of unrelated DMD cases. Sixty-eight percent of cases were found to have variations in the distal hotspot region (Exon 45-55) of the DMD gene. Exon 44/45 variations were found to be the most prominent among single exon variations, whereas exon 49/50 was found to be the most frequently mutated locations in single/multiple exon variations. As per Leiden databases, 86.84% cases harboured out-of-frame mutations. Domain wise investigation revealed that 68% of mutations were localized in the region of spectrin repeats. Dp140 isoform was predicted to be absent in 62/76 (81.57%) cases. A total of 45/80 (56.25%) and 23/80 (28.70%) DMD subjects were predicted to be amenable to exon 51 and exon 45 skipping trials, respectively.
Conclusion
A major proportion of DMD subjects (80%) could be diagnosed by the MLPA technique. The data generated from our study may be beneficial for strengthening of mutation database in the North Indian population.
Keywords: Duchenne Muscular Dystrophy (DMD), dystrophin, exon skipping, MLPA, pathogenic variants, neuromuscular disorder
1. Introduction
Duchenne Muscular Dystrophy (DMD) is a rare and incurable disorder caused due to mutations in the DMD gene located on Xp21 loci. DMD gene mutations are the primary cause for pathology and progression of this neuromuscular disorder resulting in the atrophy of muscles [1]. The presence of a single copy of X-chromosome in males increases their susceptibility to the disorder. Most of the mutations found in the DMD gene are deletions/duplications and are non-randomly distributed. Due to deficiency of dystrophin protein, there is a progressive muscular weakness, which causes a reduction in the sarcolemmal elasticity and results in death due to respiratory and cardiac failure, usually in their twenties [2]. Other co-morbidities associated with DMD include scoliosis (curvature of spine), variable degrees of cognitive and neuropsychological alterations [3]. The expression of DMD gene is regulated by a number of internal promoters, which result in different shorter isoforms of dystrophin protein with varying sizes. Clinical heterogeneity in DMD is attributed to differential expression of full length and shorter dystrophin isoforms in various tissues. Full-length dystrophin (Dp427) was reported to be localized in the muscle (Dp427m), cortical and hippocampal neurons (Dp427c) [4] and purkinje isoforms (Dp427p) [5]. The muscle-specific full-length dystrophin protein primarily functions by stabilizing dystrophin-associated protein complex (DAPC). The proximal promoters express Dp260 and Dp116 in the retina and peripheral nerves, respectively [6, 7]. The DMD gene translation through distal promoters results in Dp140, Dp71 and DP40 isoforms, believed to be expressed in the central nervous system (CNS). The regulatory site for Dp140 isoform lies in exon 44/45 and the mutations in this region may result in the loss of Dp140 isoform, reported to be crucial for cognitive function in DMD [8]. Localization of different dystrophin isoforms in crucial body organs suggests its role in the regulation of complex neuromuscular mechanisms. Various domains are implicated in the functional role of dystrophin protein, including actin-binding domain (ABD), central rod like spectrin like repeats (SpR), cystein rich domain (CRD) and C-terminal region (CTR). ABD interacts with actin cytoskeleton, SpRs provides flexibility and stretching to the protein, CRD establishes the interactions with β-dystroglycans; and CTRs interact with other protein components of DAPC [9].
Multiplex polymerase chain reaction (mPCR) techniques developed by Chamberlain [10, 11], Beggs [12] and Kunkel [13] target gene of interest by using traditional PCR-reaction and dominate the genetic testing in many developing countries. However, mPCR was used extensively to screen 18-32 exons of DMD gene [11-13]. Since detection of duplications and other mutations are not possible by the use of mPCR [14-16], multiplex ligation dependent probe amplification (MLPA) has emerged as a useful and robust technique for DMD gene screening [14, 17, 18]. Most of the genetic epidemiology studies of DMD in various regions of Indian sub-continent are carried out by mPCR [19], which limits the detection of most prominent mutations. Globally, majority of the DMD patients were reported to harbor deletions (~68%) followed by small mutations (~20%) and duplications (~ 11%) of one or >1 exons [20]. Asian database shows 72% of mutations as large deletions out of the 1819 subjects submitted to their inventory [21].
With gene therapy trials still awaiting success, steroids are the only means to manage the initial course of the devastating disease. Antisense Oligomer (AOs) based exon-skipping gene therapy is by far the most promising and fast emerging approach to partially restore the reading frame, but highly expensive and unaffordable for the cases with low economic status. The resulting pseudo-expression of functional dystrophin protein through splicing events may render severe symptoms of DMD into a milder phenotype akin to Becker Muscular Dystrophy (BMD). The appropriate strategy for exon skipping requires detailed information about the mutation location for the excision of a minimum number of exons for correcting the reading frame.
By the time exon skipping therapy becomes cost-effective and more technologies emerge with time, there may be an impending requirement to generate a population-specific DMD mutation database. Majority of the DMD mutation data has been reported from the South Indian population [22] which differs from the North Indian population with respect to ethnic background. Moreover, North Indian studies have been dominated by mPCR and thus may under-represent the mutation spectrum of DMD gene. This study is therefore an attempt to not only study the DMD mutations but also to identify the loss of corresponding dystrophin isoforms.
2. Materials and Methods
2.1. Participants
A total of 100 male DMD patients were recruited between 2012-2017 with the help of the Indian Association of Muscular Dystrophy (IAMD) after obtaining informed consent as per Institute Ethics Committee guidelines. No control group was enrolled for genetic investigations. The sample size has been estimated by utilizing significance and power values attributed to the study. The sample size was calculated according to the prevalence of DMD, i.e. 1/3500 males. To achieve the power of 80%, a sample size of ~70 DMD cases was required. Cases with characteristic clinical features of the Duchenne phenotype with early age of onset were included in the study which was followed by genetic confirmation. Cases were also recruited retrospectively with the help of patient support groups. Informed assent and written informed consent were mandatory for inclusion. The cases with BMD or intermediate phenotypes were not considered for inclusion. Moreover, cases with other myopathies were also excluded. The entire study was conducted according to the quality assurance protocols of the Neuroscience Research Lab, acknowledged by the Quality Council of India.
2.2. Isolation of Genomic DNA
Five ml of blood was collected and Peripheral Blood Mononuclear Cells (PBMCs) were isolated through Ficoll density centrifugation. QIAamp DNA Blood Mini kit was used to isolate DNA from the PBMCs or whole blood sample according to the manufacturer’s protocol. DNA was quantified using a UV spectrophotometer (DU730, UV/VIS spectrophotometer, Beckman Coulter). DNA samples with yield ranged from 50-150 ng/μl were used for further analysis. Qualitative analysis of DNA was performed in 0.8% agarose. The DNA samples were then coded and stored at -20ºC.
2.3. Multiplex Ligation Dependent Probe Amplification (MLPA)
2.3.1. Amplification of the Probes
Probe sets P034 and P035 (MRC-Holland, Amsterdam, the Netherlands) were used for detecting mutation in the target region spanning 1-79 exons of DMD gene. To rule out the possibility of disorders with similar phenotypes, the genes namely, SMN1, LMNA, DYSF, MYOT, CAV3, APP and PSEN were also screened (Supplementary Table 1 (660.8KB, pdf) ). MLPA procedure was carried out as per established protocol [14]. Briefly, 50 ng/μl of DNA samples (along with three reference samples) were denatured at 95°C for 1 min, followed by incubation at 60°C for 16-20 h for hybridization reaction. Hybridized probes were ligated by using ligase buffers and DNA ligase enzymes (Fig. 1).
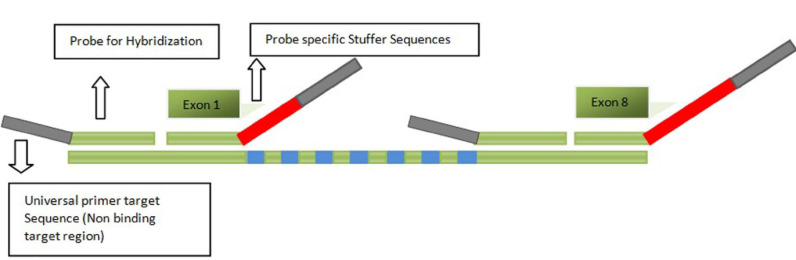
Fig. (1).
Schematic representation of MLPA probe which consists of oligonucleotides for the target region, universal primer binding region and stuffer sequences for unique amplicon length. (A higher resolution / colour version of this figure is available in the electronic copy of the article).
Amplification was performed using FAM modified primer mix and SALSA DNA polymerase. Fragment separation was carried out in the ABI platform through capillary electrophoresis. Coffalyser.NET software (MRC Holland, Amsterdam, the Netherlands) was used to obtain electropherograms. Alternately, GeneMarker software (Softgenetics) was also used to analyze the .fsa data to obtain the ratios. Dosage Quotient (DQ) ratio was generated by comparing the electropherogram of DNA samples of DMD cases with that of reference samples. Affected domains were predicted based on mutation location at the corresponding domain. Dp140 expression was predicted based on mutation upstream and downstream of exon 44, as previously described. The proximal hotspot region of exon 2-20 and distal hotspot region of exon 44-55 were considered. Reading frame concordance was obtained from Leiden databases.
3. Results
3.1. Patient Demography
The demography profile has been provided in Table 1. A total of 100 DMD cases were recruited from various geographical locations of Chandigarh, Delhi, Punjab, Haryana, Himachal Pradesh, Rajasthan and Uttar Pradesh. The majority of them (66%) belonged to the urban habitat. Out of 100 cases, 80% were Hindus and 17% were Sikhs. Six families had two affected children and both the siblings were included in the study. Among these, 3 siblings had deletion, 1 had duplication and 2 could not be detected through MLPA. Hence, there were 94 unrelated DMD cases.
Table 1.
Demographic characteristics of DMD cases (n=100).
| Variables | DMD Group (Cases) |
|---|---|
| Sample Size | 100 |
| Gender | Male (100%) |
| Age | 11.18 ± 3.71 |
| Phenotype | DMD |
| Sibling pairs | 6 Pairs |
| Religion | |
| Hindu | 80 (80%) |
| Sikh | 17 (17%) |
| Others | 3 (3%) |
| Habitat | |
| Rural | 34 (34%) |
| Urban | 66 (66%) |
| Geographical Distribution | |
| Chandigarh | 10 (10%) |
| Punjab | 20 (20%) |
| Delhi | 29 (29%) |
| Himachal Pradesh | 16 (16%) |
| Haryana | 12 (12%) |
| UP | 11 (11%) |
| Rajasthan | 2 (2%) |
3.2. Distribution of Exonic Mutations
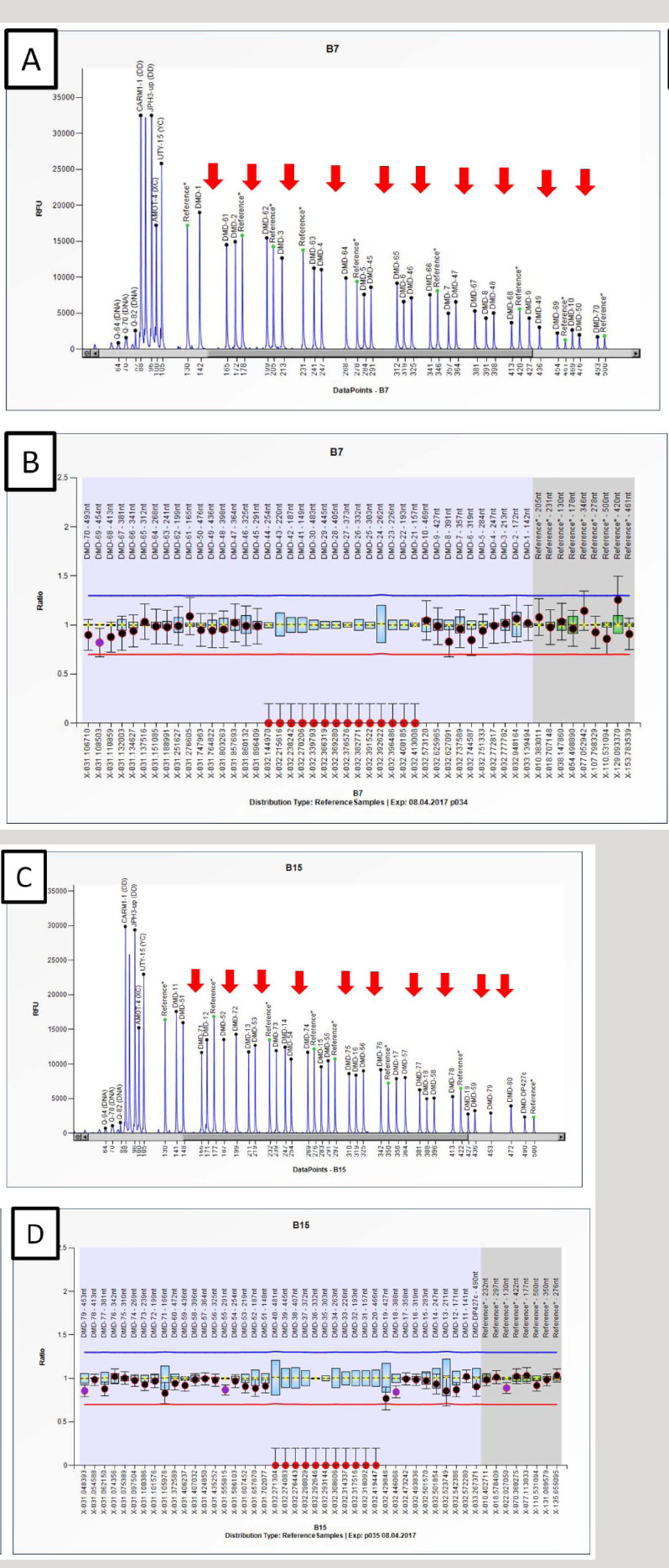
Among the 94 unrelated DMD cases, MLPA detected deletion or duplication in 76/94 (80.85%) cases and no mutation in 18/94 (19.14%) cases. Representative electropherograms and ratio charts have been provided in Fig. (2).
Fig. (2).
Electropherogram and ratio chart obtained through coffalyser.NET showing profile of DMD subject with long stretch deletion between Exon 20 to 44. (A-B) Electropherogram and ratio chart showing deletion from Exon 31 to 40 and Exon 20 covered by P035 probemix. (C-D) Electropherogram and ratio chart showing deletion from Exon 41 to 44 and Exon 21 to 30 covered by P034 probemix. Ratio between 0.70-1.30 is considered in the normal range while ratio of 0.00 was considered as deletion (depicted in red dots and arrows). (A higher resolution / colour version of this figure is available in the electronic copy of the article).
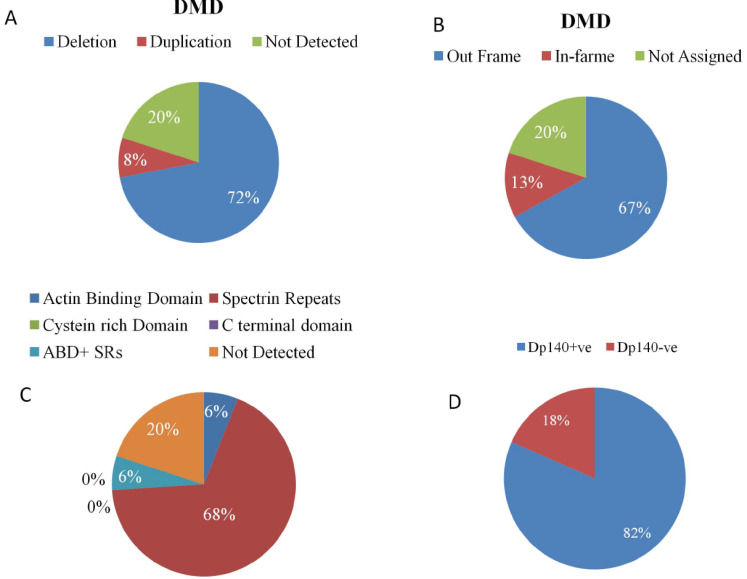
Among these 94 unrelated families, 69/94 (73%) DMD subjects revealed deletions, whereas, in 7/94 (7%), duplications were observed. Among the deletions, single exon deletions (SEDel) were observed only in 16/69 (23.18%) cases. The exon 45 deletion was the most common SEDel; however, the most frequently deleted either as single or multiple exon deletions were exon 49/50. Out-of-frame and in-frame mutations were found in 66/76 (86.84%) and 10/76 (13.15%), respectively. We found exon 45-55 to be the most commonly affected region in 68% of the cases followed by exon 1-20 in 13%. Domain wise analysis revealed that 68% cases harboured mutations in the SpR and 6% in the ABD. In the remaining 6% cases, mutation lesion ranged from the ABD and SpR domains. The spectrum of deletion and duplication profile of DMD gene has been described in Fig. (3) and Table 2.
Fig. (3).
Distribution of mutations in the cases with DMD. Pie diagram showing A) Mutation rate B) Predicted proportion of cases with out-of-frame or in-frame mutations according to Leiden Databases. C) Domain wise distribution of mutations D) Proportion of presence or absence of Dp140 Isoform in DMD cases. (A higher resolution / colour version of this figure is available in the electronic copy of the article).
Table 2.
Distribution and spectrum of DMD gene variations in 94 unrelated DMD cases.
| Variables | N (%) |
|---|---|
| Total DMD cases | 100 |
| MLPA +ve Cases | 80/100 (80%) |
| MLPA -ve Cases | 20/100 (20%) |
| Sibling pairs | 6 |
| Unrelated DMD cases | 94 |
| Copy number variations (CNVs) | 76/94 (80.85%) |
| CNV-ve | 18/94 (19.14%) |
| Proximal Hotspot (exon 2-20) | 11/76 (14.47%) |
| Distal Hotspot (exon 45-55) | 62/76 (81.57%) |
| Dp140+ve | 14/76 (18.42%) |
| Dp140-ve | 62/76 (81.57%) |
| Out of frame | 66/76 (86.84%) |
| In-frame | 10/76 (13.15%) |
| Deletions | 69/94 (73%) |
| Deletions among CNVs | 69/76 (90.78%) |
| Single exon deletion (SEDel) | 16/69 (23.18%) |
| Most common SEDel | Exon 45: 5/69 (7.24) |
| Multiple exon deletion patterns | 53/69 (76.81%) |
| Proximal Deletions | 10/69 (14.49%) |
| Proximal-Distal Deletions | 4/69 (5.79%) |
| Distal Deletions | 57/69 (82.60%) |
| Out-of-frame Deletions | 62/69 (89.85%) |
| In-frame Deletions | 7/69 (10.14%) |
| Most Prominent Exon Deletion | Exon 49/50 |
| Duplications | 7/94 (7%) |
| Duplications among CNVs | 7/76 (9.21%) |
| Single exon Duplications (SEDup) | 1/7 (14.28%) |
| Multiple exon Duplications | 6/7 (86%) |
| Proximal Duplications | 6/7 (86%) |
| Distal Duplications | 1/7 (14.28%) |
| Out-of-frame Duplications | 4/7 (57.14%) |
| In-frame Duplications | 3/7 (4.85%) |
The detailed mutation profile representing mutation type as per Human Genome Variation Society (HGVS) nomenclature and the predicted dystrophin protein expression has been provided in Table 3. DMD cases who did not exhibit any alterations in the copy number status indicated the probability of point mutation in the dystrophin gene. We could not perform further experiments in MLPA negative cases. Furthermore, screening of SMN1, LMNA, DYSF, MYOT, CAV3, APP and PSEN genes did not reveal del/dup in cases with no del/dup in DMD gene (Supplementary Fig. 1 (660.8KB, pdf) ).
Table 3.
Mutation profile of indian duchenne muscular dystrophy cases (n=100).
| Patient | Age | HGVS Nomenclature | Genetic Mutation | Dystrophin Protein and Isoform |
ORF
Prediction |
Domain Dp140-ve | CNS Dp140 Isoform | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| P-1 | 9 | c.6291-?_6438+?del | Del Exon 44 | Dp427(M/L/C/P), Dp260, Dp140 | OF | SPR | Dp140-ve | ||||
| P-2 | 9 | c.7661-?_8027+?del | Del Exon 53-54 | Dp427(M/L/C/P), Dp260, Dp140 | OF | SPR | Dp140-ve | ||||
| P-3 | 8 | Not Assigned | No Del/Dup | Not Assigned | NA | NA | NA | ||||
| P-4 | 4 | c.94-?_3603+?del | Del Exon 03-26 | Dp427(M/L/C/P) | IF | ABD, SPR | Dp140+ve | ||||
| P-5 | 7 | c.6615-?_7309+?del | Del Exon 46-50 | Dp427(M/L/C/P), Dp260, Dp140 | OF | SPR | Dp140-ve | ||||
| P-6 | 9 | Not Assigned | No Del/Dup | Not Assigned | NA | NA | NA | ||||
| P-7 | 8 | c.6439-?_6614+?del | Del Exon 45 | Dp427(M/L/C/P), Dp260, Dp140 | OF | SPR | Dp140-ve | ||||
| P-8 | 13 | c.265-?_649+?del | Del Exon 05-07 | Dp427(M/L/C/P) | OF | ABD | Dp140+ve | ||||
| P-9 | 12 | Not Assigned | No Del/Dup | Not Assigned | NA | NA | NA | ||||
| P-10 | 9 | c.6439-?_6912+?del | Del Exon 45-47 | Dp427(M/L/C/P), Dp260, Dp140 | IF | SPR | Dp140-ve | ||||
| P-11@ | 13 | c.94-?_264+?dup | Dup 3-4 | Dp427(M/L/C/P) | IF | ABD | Dp140+ve | ||||
| P-12@ | 12 | c.94-?_264+?dup | Dup 3-4 | Dp427(M/L/C/P) | IF | ABD | Dp140+ve | ||||
| P-13 | 11 | c.6291-?_6438+?dup | Dup 44 | Dp427(M/L/C/P), Dp260, Dp140 | OF | SPR | Dp140-ve | ||||
| P-14 | 10 | c.6913-?_7660+?del | Del Exon 48-52 | Dp427(M/L/C/P), Dp260, Dp140 | OF | SPR | Dp140-ve | ||||
| P-15 | 6 | c.7099-?_7660+?del | Del Exon 49-52 | Dp427(M/L/C/P), Dp260, Dp140 | OF | SPR | Dp140-ve | ||||
| P-16 | 11 | c.650-?_960+?del | Del Exon 08-09 | Dp427(M/L/C/P) | OF | ABD, SPR | Dp140+ve | ||||
| P-17 | 12 | c.6615-?_6912+?del | Del Exon 46-47 | Dp427(M/L/C/P), Dp260, Dp140 | OF | SPR | Dp140-ve | ||||
| P-18 | 10 | c.6913-?_7660+?del | Del Exon 48-52 | Dp427(M/L/C/P), Dp260, Dp140 | OF | SPR | Dp140-ve | ||||
| P-19 | 10 | Not Assigned | No Del/Dup | Not Assigned | NA | NA | NA | ||||
| P-20 | 15 | c.6615-?_7200+?del | Del Exon 46-49 | Dp427(M/L/C/P), Dp260, Dp140 | OF | SPR | Dp140-ve | ||||
| P-21 | 9 | c.6615-?_7200+?del | Del Exon 46-49 | Dp427(M/L/C/P), Dp260, Dp140 | OF | SPR | Dp140-ve | ||||
| P-22 | 8 | c.2804-?_3603+?dup | Dup Exon 22-26 | Dp427(M/L/C/P) | OF | SPR | Dp140+ve | ||||
| P-23 | 8 | Not Assigned | No Del/Dup | Not Assigned | NA | NA | NA | ||||
| P-24 | 14 | c.6439-?_7200+?del | Del Exon 45-49 | Dp427(M/L/C/P), Dp260, Dp140 | IF | SPR | Dp140-ve | ||||
| P-25 | 8 | c.7310-?_7542+?del | Del Exon 51 | Dp427(M/L/C/P), Dp260, Dp140 | OF | SPR | Dp140-ve | ||||
| P-26 | 8 | c.2169-?_5922+?dup | Dup Exon 18-41 | Dp427(M/L/C/P), Dp260 | OF | SPR | Dp140+ve | ||||
| P-27 | 8 | Not Assigned | No Del/Dup | Not Assigned | NA | NA | NA | ||||
| P-28 | 11 | c.7201-?_7309+?del | Del Exon 50 | Dp427(M/L/C/P), Dp260, Dp140 | OF | SPR | Dp140-ve | ||||
| P-29 | 6 | c.6439-?_7660+?del | Del Exon 45-52 | Dp427(M/L/C/P), Dp260, Dp140 | OF | SPR | Dp140-ve | ||||
| P-30 | 10 | c.6291-?_6438+?del | Del EXON 44 | Dp427(M/L/C/P), Dp260, Dp140 | OF | SPR | Dp140-ve | ||||
| P-31# | 8 | c.7099-?_7660+?del | Del Exon 49-52 | Dp427(M/L/C/P), Dp260, Dp140 | OF | SPR | Dp140-ve | ||||
| P-32# | 13 | c.7099-?_7660+?del | Del Exon 49-52 | Dp427(M/L/C/P), Dp260, Dp140 | OF | SPR | Dp140-ve | ||||
| P-33 | 16 | c.6913-?_7309+?del | Del Exon 48-50 | Dp427(M/L/C/P), Dp260, Dp140 | OF | SPR | Dp140-ve | ||||
| P-34 | 14 | c.1150-?_7200+?del | Del Exon 11-49 | Dp427(M/L/C/P), Dp260, Dp140 | IF | SPR | Dp140-ve | ||||
| Patient | Age | HGVS Nomenclature | Genetic Mutation | Dystrophin Protein and Isoform |
ORF Prediction |
Domain Dp140-ve | CNS Dp140 Isoform | ||||
| P-35 | 11 | c.6439-?_6614+?del | Del Exon 45 | Dp427(M/L/C/P), Dp260, Dp140 | OF | SPR | Dp140-ve | ||||
| P-36 | 9 | c.6118-?_6290+?del | Del Exon 43 | Dp427(M/L/C/P), Dp260 | OF | SPR | Dp140+ve | ||||
| P-37 | 14 | c.6439-?_7660+?del | Del Exon 45-52 | Dp427(M/L/C/P), Dp260, Dp140 | OF | SPR | Dp140-ve | ||||
| P-38$ | 9 | Not Assigned | No Del/Dup | Not Assigned | NA | NA | NA | ||||
| P-39 | 11 | c.1150-?_7200+?del | Del Exon 11-49 | Dp427(M/L/C/P), Dp260, Dp140 | IF | SPR | Dp140-ve | ||||
| P-40 | 9 | c.6439-?_7660+?del | Del Exon 45-52 | Dp427(M/L/C/P), Dp260, Dp140 | OF | SPR | Dp140-ve | ||||
| P-41 | 10 | c.7201-?_7309+?del | Del Exon 50 | Dp427(M/L/C/P), Dp260, Dp140 | OF | SPR | Dp140-ve | ||||
| P-42 | 8 | Not Assigned | No Del/Dup | Not Assigned | NA | NA | NA | ||||
| P-43 | 7 | c.6615-?_7542+?del | Del Exon 46-51 | Dp427(M/L/C/P), Dp260, Dp140 | OF | SPR | Dp140-ve | ||||
| P-44 | 13 | c.6439-?_7660+?del | Del Exon 45-52 | Dp427(M/L/C/P), Dp260, Dp140 | OF | SPR | Dp140-ve | ||||
| P-45 | 10 | c.6439-?_7660+?del | Del Exon 45-52 | Dp427(M/L/C/P), Dp260, Dp140 | OF | SPR | Dp140-ve | ||||
| P-46 | 5 | c.7099-?_7660+?del | Del Exon 49-52 | Dp427(M/L/C/P), Dp260, Dp140 | OF | SPR | Dp140-ve | ||||
| P-47 | 9 | c.6291-?_6438+?del | Del Exon 44 | Dp427(M/L/C/P), Dp260, Dp140 | OF | SPR | Dp140-ve | ||||
| P-48 | 12 | c.6439-?_6614+?del | Del Exon 45 | Dp427(M/L/C/P), Dp260, Dp140 | OF | SPR | Dp140-ve | ||||
| P-49$ | 14 | Not Assigned | No Del/Dup | Not Assigned | NA | NA | NA | ||||
| P-50 | 13 | Not Assigned | No Del/Dup | Not Assigned | NA | NA | NA | ||||
| P-51 | 15 | c.6615-?_8217+?del | Del Exon 46-55 | Dp427(M/L/C/P), Dp260, Dp140, Dp116 | OF | SPR | Dp140-ve | ||||
| P-52 | 13 | Not Assigned | No Del/Dup | Not Assigned | NA | NA | NA | ||||
| P-53 | 16 | c.1150-?_7098+?del | Del Exon 11-48 | Dp427(M/L/C/P), Dp260, Dp140 | IF | SPR | Dp140-ve | ||||
| P-54 | 19 | c.6291-?_8217+?dup | Dup 44-55 | Dp427(M/L/C/P), Dp260, Dp140, Dp116 | OF | SPR | Dp140-ve | ||||
| P-55 | 17 | c.6439-?_7660+?del | Del Exon 45-52 | Dp427(M/L/C/P), Dp260, Dp140 | OF | SPR | Dp140-ve | ||||
| P-56 | 18 | c.6439-?_6614+?del | Del Exon 45 | Dp427(M/L/C/P), Dp260, Dp140 | OF | SPR | Dp140-ve | ||||
| P-57 | 16 | c.94-?_1482+?dup | Dup Exon 3-12 | Dp427(M/L/C/P) | IF | ABD,SPR | Dp140+ve | ||||
| P-58 | 19 | Not Assigned | No Del/Dup | Not Assigned | NA | NA | NA | ||||
| P-59 | 10 | c.1332-?_2380+?del | Del Exon 12- 19 | Dp427(M/L/C/P) | OF | SPR | Dp140+ve | ||||
| P-60 | 10 | c.7099-?_7309+?del | Del Exon 49-50 | Dp427(M/L/C/P), Dp260, Dp140 | OF | SPR | Dp140-ve | ||||
| P-61 | 11 | c.6913-?_7660+?del | Del Exon 48-52 | Dp427(M/L/C/P), Dp260, Dp140 | OF | SPR | Dp140-ve | ||||
| P-62 | 12 | c.265-?_649+?dup | Dup Exon 5-7 | Dp427(M/L/C/P) | OF | ABD | Dp140+ve | ||||
| P-63 | 14 | c.6439-?_6614+?del | Del Exon 45 | Dp427(M/L/C/P), Dp260, Dp140 | OF | SPR | Dp140-ve | ||||
| P-64 | 8 | c.6913-?_7660+?del | Del Exon 48-52 | Dp427(M/L/C/P), Dp260, Dp140 | OF | SPR | Dp140-ve | ||||
| P-65 | 15 | c.7099-?_7660+?del | Del Exon 49-52 | Dp427(M/L/C/P), Dp260, Dp140 | OF | SPR | Dp140-ve | ||||
| P-66 | 17 | c.6615-?_7309+?del | Del Exon 46-50 | Dp427(M/L/C/P), Dp260, Dp140 | OF | SPR | Dp140-ve | ||||
| P-67 | 19 | Not Assigned | No Del/Dup | Not Assigned | NA | NA | NA | ||||
| P-68 | 3 | c.7310-?_7542+?del | Del of Exon 51 | Dp427(M/L/C/P), Dp260, Dp140 | OF | SPR | Dp140-ve | ||||
| P-69 | 19 | c.6913-?_7309+?del | Del Exon 48-50 | Dp427(M/L/C/P), Dp260, Dp140 | OF | SPR | Dp140-ve | ||||
| Patient | Age | HGVS Nomenclature | Genetic Mutation | Dystrophin Protein and Isoform |
ORF Prediction |
Domain Dp140-ve | CNS Dp140 Isoform | ||||
| P-70 | 9 | c.6615-?_7310+?del | Del Exon 46-51 | Dp427(M/L/C/P), Dp260, Dp140 | OF | SPR | Dp140-ve | ||||
| P-71 | 15 | c.94-?_2949+?del | Del Exon 03-22 | Dp427(M/L/C/P) | IF | ABD,SPR | Dp140+ve | ||||
| P-72 | 14 | c.6439-?_7660+?del | Del Exon 45-52 | Dp427(M/L/C/P), Dp260, Dp140 | OF | SPR | Dp140-ve | ||||
| P-73 | 15 | Not Assigned | No Del/Dup | Not Assigned | NA | NA | NA | ||||
| P-74 | 11 | c.2381-?_7660+?del | Del Exon 49-52 | Dp427(M/L/C/P), Dp260, Dp140 | OF | SPR | Dp140-ve | ||||
| P-75& | 13 | c.7099-?_6438+?del | Del Exon 20-44 | Dp427(M/L/C/P), Dp260, Dp140 | OF | SPR | Dp140-ve | ||||
| P-76& | 9 | c.7099-?_6438+?del | Del Exon 20-44 | Dp427(M/L/C/P), Dp260, Dp140 | OF | SPR | Dp140-ve | ||||
| P-77 | 7 | c.6615-?_7310+?del | Del Exon 46-51 | Dp427(M/L/C/P), Dp260, Dp140 | OF | SPR | Dp140-ve | ||||
| P-78 | 7 | c.7310-?_8217+?del | Del Exon 51-55 | Dp427(M/L/C/P), Dp260, Dp140, Dp116 | OF | SPR | Dp140-ve | ||||
| P-79 | 12 | c.6615-?_8027+?del | Del Exon 46-54 | Dp427(M/L/C/P), Dp260, Dp140 | IF | SPR | Dp140-ve | ||||
| P-80 | 6 | c.7099-?_7660+?del | Del Exon 49-52 | Dp427(M/L/C/P), Dp260, Dp140 | OF | SPR | Dp140-ve | ||||
| P-81 | 9 | Not Assigned | No Del/Dup | Not Assigned | NA | NA | NA | ||||
| P-82 | 12 | c.650-?_3603+?del | Del Exon 8-26 | Dp427(M/L/C/P) | OF | ABD, SPR | Dp140+ve | ||||
| P-83 | 11 | c.650-?_1812+?del | Del Exon 08-15 | Dp427(M/L/C/P) | OF | ABD, SPR | Dp140+ve | ||||
| P-84 | 4 | c.7201-?_7660+?del | Del Exon 50-52 | Dp427(M/L/C/P), Dp260, Dp140 | OF | SPR | Dp140-ve | ||||
| P-85 | 11 | c.7543-?_8027+?del | Del Exon 52-54 | Dp427(M/L/C/P), Dp260, Dp140 | OF | SPR | Dp140-ve | ||||
| P-86 | 6 | Not Assigned | No Del/Dup | Not Assigned | NA | NA | NA | ||||
| P-87 | 17 | c.6291-?_6438+?del | Del Exon 44 | Dp427(M/L/C/P), Dp260, Dp140 | OF | SPR | Dp140-ve | ||||
| P-88 | 7 | c.7543-?_7660+?del | Del Exon 52 | Dp427(M/L/C/P), Dp260, Dp140 | OF | SPR | Dp140-ve | ||||
| P-89^ | 14 | Not Assigned | No Del/Dup | Not Assigned | NA | NA | NA | ||||
| P-90^ | 10 | Not Assigned | No Del/Dup | Not Assigned | NA | NA | NA | ||||
| P-91 | 10 | c.7201-?_8027+?del | Del Exon 50-54 | Dp427(M/L/C/P), Dp260, Dp140 | OF | SPR | NA | ||||
| P-92* | 14 | c.94-?_264+?del | Del Exon 3-4 | Dp427(M/L/C/P) | IF | ABD | Dp140+ve | ||||
| P-93 | 8 | c.6439-?_8027+?del | Del Exon 45-54 | Dp427(M/L/C/P), Dp260, Dp140 | OF | SPR | Dp140-ve | ||||
| P-94 | 6 | c.7099-?_7660+?del | Del Exon 49-52 | Dp427(M/L/C/P), Dp260, Dp140 | OF | SPR | Dp140-ve | ||||
| P-95* | 19 | c.94-?_264+?del | Del Exon 3-4 | Dp427(M/L/C/P) | IF | ABD | Dp140+ve | ||||
| P-96 | 19 | c.6615-?_6912+?del | Del Exon 46-47 | Dp427(M/L/C/P), Dp260, Dp140 | OF | SPR | Dp140-ve | ||||
| P-97 | 9 | c.7201-?_7309+?del | Del Exon 50 | Dp427(M/L/C/P), Dp260, Dp140 | OF | SPR | Dp140-ve | ||||
| P-98 | 11 | Not Assigned | No Del/Dup | Not Assigned | NA | NA | NA | ||||
| P-99 | 10 | Not Assigned | No Del/Dup | Not Assigned | NA | NA | NA | ||||
| P-100 | 11 | c.7310-?_7872+?del | Del Exon 51-53 | Dp427(M/L/C/P), Dp260, Dp140 | OF | SPR | Dp140-ve | ||||
Abbreviations: Dp: Dystrophin Protein, Dp260, Dp140, Dp116, Dp71: Short Dystrophin isoforms (260, 140, 116 and 71 in kiloDaltons), M/L/C/P: Muscle/ lymphoblastoid cells/Cortex/Purkinje Cells, Del: Deletion, Dup: Duplication, ORF: Open reading frame, OF: Out of frame, IF: In-frame, ABD: Actin binding domain, SPR: Spectrin Repeats. @,#,$,&,^, *: Sibling groups.
3.3. Predicted Loss of Short Dystrophin Isoforms and Target Range for Exon Skipping
Mutation profile analysis indicated a loss of short dystrophin isoforms in DMD. Out of 76 cases, 14% cases were predicted to have affected full-length dystrophin protein. Retina specific Dp260 isoform was predicted to be affected in 66% cases. CNS specific Dp140 isoform was predicted to be absent in 64% DMD cases. Moreover, in 3% cases, Dp116 was also affected. Mutation data revealed that exon 51 skipping would be effective in 56.25% DMD cases. However, based on the frequency of exonic mutations 28.75% DMD cases could be amenable to Exon 45 skipping (Table 4).
Table 4.
Amenability of exon skipping in Indian DMD subjects.
| Amenability to Exon Skipping | DMD Cases | Dystrophin Isoform Restored | Target |
|---|---|---|---|
| Exon 51 | 45/80 | Dp160, Dp71, 40 | Muscle and CNS |
| Exon 45 | 23/80 | Dp71, 40 | Muscle and CNS |
| Exon 43 | 14/80 | Dp140, 71, 40 | Muscle, CNS, Kidney |
| Exon 28 | 14/80 | Dp260, Dp140, 71, 40 | Muscle and CNS, Kidney and retina |
4. Discussion
The analysis of mutation spectrum has revealed mutations in 80.85% DMD cases with 73% deletions and 7% duplications in unrelated families. Previous Indian studies have reported 75% [16] and 86.6% [22] mutation detection rate by MLPA. Spectrin repeat domain was found to be affected in the majority of cases. Based on the mutation location, Dp140 was found to be prominently affected in our study group, which may explain cognitive and behavioral abnormalities in DMD. Similarly, alterations in other dystrophin isoforms including Dp116 and Dp260 are crucial for understanding the disease spectrum because the absence of Dp260 isoform in the retina is believed to affect rod pathway signaling in electroretinogram studies [23]. Analysis of other genes involved in myopathies and cognitive impairment revealed the monogenic effect of DMD gene mutation in the development of Duchenne phenotype in our cohort.
In our study, 81.57% DMD subjects revealed mutations in the 45-55 hotspot regions (Table 2); however, exon 2-20 mutations were detected only in 14.47% DMD cases. Various studies have reported exon 2-20 and 45-55 regions to be the hotspot regions for DMD gene mutation falling under the rod domain (SpR) while exon 56-79 mutations are rare [24-26]. The mutation frequencies and hotspot regions in our study cohort were found to be similar to the global spectrum of dystrophin gene mutations; however, we did not examine the junctional breakpoints. Breakpoints need to be confirmed by sanger/next-generation sequencing approaches for confirming the eligibility of DMD cases. Our study suggests the hotspot region of Exon 45-55 and Exon 2-20 as a prominent target site for multi-exon targeting of Phosphorodiamidate morpholino oligomers (PMOs).
With overall 23.18% cases with single exon deletions, our study reports exon 44/45 to be the equally prominent single-exon copy number rearrangements. This location was reported to be varied in Asian populations. In this context, Exon 50 has been reported to be the most prominent SEDel
found in Singapore, exon 49/50 in Japanese and exon 51 in Vietnamese populations [27], which pertains to Dp140 isoform. In a study conducted in 112 DMD cases of the South Indian region, deletions were detected in 73% of the cases. SEDels were detected in 20.4% (23.18% in our study) and in contrast to our study, the most common SEDel was exon 50 with 38.5% cases (exon 44/45 in our study) [28]. Though the most prominently mutated single exons differed in both studies, yet both locations are pertinent to Dp140 isoform. SEdels need to be confirmed by alternate methods like PCR and sequencing. However, we could not validate the SEdel/dup by sequencing which may have revealed potential polymorphism or point mutation in the probe binding site. In a study conducted in the East Indian region, although the deletion rate of 65.7% was reported, deletion in the distal hot spot exonic region was noted in 82.61% of cases and that in the proximal hotspot, region was 10.87% [29]. In a South Indian MLPA based study, Vengalil et al. reported deletions and duplications in 91% and 9% cases, respectively from a cohort of 279 DMD subjects [30] and reported exon 50 deletion to be the most common mutation.
Most prominently mutated exons in our study, i.e. exon 44/45, are also considered to have a breakpoint in the dystrophin gene [31, 32]. It is important to discuss the evolutionary significance of this exonic location. An admixture of dystrophin exon 44 regions between the Neanderthal genome and expanding Homo sapiens nearly between 80 and 50 thousand years ago, was reported [33]. In addition, the prevalence of DMD in the African black population (1/250,000) being less than the UK (1/40,000) suggests the role of admixture in the instability of the loci [34].
Bhattacharya et al. reported ABD and CRD as a hub of mutational events [9]; however, our cohort mainly represented SpR domains. In view of frequently encountered mutation of exon 49 and 50, Exon 51 skipping approach might benefit a large proportion [45/80 (56.25%)] of our study group. Before exon-skipping therapeutics took shape, Wilton in 1999 showed using mdx mouse model, that AONs can remove the mutation in the exon resulting in increased dystrophin production by its repetitive administration [35].
Antisense oligonucleaotide (AO) based exon skipping therapies are the most promising approach to treat DMD. PMOs are important AOs with the ability to skip multiple exons. Eteplirsen (exondys51) became the first FDA approved exon skipping therapy for DMD in the United States [36]. PMO based Exondys 51 or Eteplirsen is the first FDA approved therapy applicable for 13% of DMD based on a <1% increase in pseudo-expression of dystrophin. Despite a limited improvement in dystrophin expression, multi-exon skipping potentially increases the amenability to 80-90% of DMDs [37]. For exon-skipping therapeutics to be successfully applied, it is important to populate the genetic database with information about dystrophin isoforms and define its distribution pattern corresponding to various organ systems. German human genome database, which shows 2982 mutations occurring in DMD gene [38], is an example of such databases. Similarly, in France, more than 13,500 registrations were made from 31 different countries, by generating a mutation database in 2015.
Conclusion
This study updates the existing DMD gene mutation spectrum in the Indian population. Besides strengthening the mutation databases, amenability to exon skipping trials and impaired cognitive functions associated with Dp140 isoform could be predicted through the type and location of mutation. Since, early institution of treatment benefits in terms of enhanced life expectancy and reduced morbidities, a newborn screening program for DMD will be of paramount importance in countries like India with a high prevalence of the disorder. Genetic counseling, prenatal and carrier screening are crucial for the prevention and management of DMD. However, awareness in the medical fraternity and general population; and empowering patient support groups may be beneficial to reduce the disease burden.
Acknowledgements
We acknowledge Dr. Mitali Mukerjee and their team at the Institute of Genomics and Integrated Biology for providing resources and assistance. We thank Ms. Sanjana Goyal, President, Indian Association of Muscular Dystrophy for providing patient resources. We thank Ms. Rajdeep Kaur, Ph.D. Scholar, PGIMER, for her help in revising the manuscript.
SUPPLEMENTARY MATERIAL
Supplementary material is available on the publisher’s website along with the published article.
Ethics Approval and Consent to Participate
The study was approved by the Institutional Ethics Committee of PGIMER, Chandigarh, India vide approval Int/IEC/ 2015/732.
Human and Animal Rights
No animals were used for the study that are the basis of this research. All experimental protocols on patients were followed according to the guidelines of Institutional Ethics Committee of Postgraduate Institute of Medical Education and Research, Chandigarh, India.
Consent for Publication
Written informed consent was obtained before the recruitment of study subjects.
Availability of Data and Materials
The data that support the findings of this study are available from the corresponding author, [A.A.], upon reasonable request.
Funding
Funding support was provided by the Department of Atomic Energy, Mumbai, Government of India [Sanction No: 37(1)/14/53/2014-BRNS]. Fellowship support was provided by the Indian Council of Medical Research (ICMR).
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
Authors’ Contributions
AA: Conceptualization, management of the study, editing & Final approval the manuscript, RT: Co-conceptualization under supervision, Data Acquisition and analysis as PhD student, Experiments, writing the manuscript: SK: Experiment Assistance PK: Assistance in Data Acquisition MM: Co-investigator in grant application FM: Experiments related to Capillary sequencing: AD: Inter-laboratory validation of the data.
References
- 1.Hoffman E.P., Brown R.H., Jr, Kunkel L.M. Dystrophin: the protein product of the Duchenne muscular dystrophy locus. Cell. 1987;51(6):919–928. doi: 10.1016/0092-8674(87)90579-4. [DOI] [PubMed] [Google Scholar]
- 2.Petrof B.J., Shrager J.B., Stedman H.H., Kelly A.M., Sweeney H.L. Dystrophin protects the sarcolemma from stresses developed during muscle contraction. Proc. Natl. Acad. Sci. USA. 1993;90(8):3710–3714. doi: 10.1073/pnas.90.8.3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mercuri E., Muntoni F. Muscular dystrophies. Lancet. 2013;381(9869):845–860. doi: 10.1016/S0140-6736(12)61897-2. [DOI] [PubMed] [Google Scholar]
- 4.Nudel U., Zuk D., Einat P., Zeelon E., Levy Z., Neuman S., Yaffe D. Duchenne muscular dystrophy gene product is not identical in muscle and brain. Nature. 1989;337(6202):76–78. doi: 10.1038/337076a0. [DOI] [PubMed] [Google Scholar]
- 5.Holder E., Maeda M., Bies R.D. Expression and regulation of the dystrophin Purkinje promoter in human skeletal muscle, heart, and brain. Hum. Genet. 1996;97(2):232–239. doi: 10.1007/BF02265272. [DOI] [PubMed] [Google Scholar]
- 6.D’Souza V.N., Nguyen T.M., Morris G.E., Karges W., Pillers D.A., Ray P.N. A novel dystrophin isoform is required for normal retinal electrophysiology. Hum. Mol. Genet. 1995;4(5):837–842. doi: 10.1093/hmg/4.5.837. [DOI] [PubMed] [Google Scholar]
- 7.Byers T.J., Lidov H.G., Kunkel L.M. An alternative dystrophin transcript specific to peripheral nerve. Nat. Genet. 1993;4(1):77–81. doi: 10.1038/ng0593-77. [DOI] [PubMed] [Google Scholar]
- 8.Bardoni A., Felisari G., Sironi M., Comi G., Lai M., Robotti M., Bresolin N. Loss of Dp140 regulatory sequences is associated with cognitive impairment in dystrophinopathies. Neuromuscul. Disord. 2000;10(3):194–199. doi: 10.1016/S0960-8966(99)00108-X. [DOI] [PubMed] [Google Scholar]
- 9.Simanti B.A.D., Angshuman B. Domain wise distribution of mutations in dystrophin protein and duchenne muscular dystrophy. Gene Technol. 2015;4(3):128. [Google Scholar]
- 10.Chamberlain J.S., Gibbs R.A., Ranier J.E., Nguyen P.N., Caskey C.T. Deletion screening of the Duchenne muscular dystrophy locus via multiplex DNA amplification. Nucleic Acids Res. 1988;16(23):11141–11156. doi: 10.1093/nar/16.23.11141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chamberlain J. S. 1990.
- 12.Beggs A.H., Koenig M., Boyce F.M., Kunkel L.M. Detection of 98% of DMD/BMD gene deletions by polymerase chain reaction. Hum. Genet. 1990;86(1):45–48. doi: 10.1007/BF00205170. [DOI] [PubMed] [Google Scholar]
- 13.Kunkel L., Snyder J., Beggs A., Boyce F., Feener C. Searching for dystrophin gene deletions in patients with atypical presentations. Etiology of human disease at the DNA level. New York: Raven; 1991. pp. 51–60. [Google Scholar]
- 14.Schouten J.P., McElgunn C.J., Waaijer R., Zwijnenburg D., Diepvens F., Pals G. Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res. 2002;30(12):e57. doi: 10.1093/nar/gnf056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vorstman J.A., Jalali G.R., Rappaport E.F., Hacker A.M., Scott C., Emanuel B.S. MLPA: a rapid, reliable, and sensitive method for detection and analysis of abnormalities of 22q. Hum. Mutat. 2006;27(8):814–821. doi: 10.1002/humu.20330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murugan S., Chandramohan A., Lakshmi B.R. Use of multiplex ligation-dependent probe amplification (MLPA) for Duchenne muscular dystrophy (DMD) gene mutation analysis. Indian J. Med. Res. 2010;132:303–311. [PubMed] [Google Scholar]
- 17.Lalic T., Vossen R.H., Coffa J., Schouten J.P., Guc-Scekic M., Radivojevic D., Djurisic M., Breuning M.H., White S.J., den Dunnen J.T. Deletion and duplication screening in the DMD gene using MLPA. Eur. J. Hum. Genet. 2005;13(11):1231–1234. doi: 10.1038/sj.ejhg.5201465. [DOI] [PubMed] [Google Scholar]
- 18.Dastur R.S., Kachwala M.Y., Khadilkar S.V., Hegde M.R., Gaitonde P.S. Identification of deletions and duplications in the Duchenne muscular dystrophy gene and female carrier status in western India using combined methods of multiplex polymerase chain reaction and multiplex ligation-dependent probe amplification. Neurol. India. 2011;59(6):803–809. doi: 10.4103/0028-3886.91355. [DOI] [PubMed] [Google Scholar]
- 19.Nalini A., Polavarapu K., Preethish-Kumar V. Muscular dystrophies: An Indian scenario. Neurol. India. 2017;65(5):969–970. doi: 10.4103/neuroindia.NI_733_17. [DOI] [PubMed] [Google Scholar]
- 20.Aartsma-Rus A., Ginjaar I.B., Bushby K. The importance of genetic diagnosis for Duchenne muscular dystrophy. J. Med. Genet. 2016;53(3):145–151. doi: 10.1136/jmedgenet-2015-103387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bladen C.L., Salgado D., Monges S., Foncuberta M.E., Kekou K., Kosma K., Dawkins H., Lamont L., Roy A.J., Chamova T., Guergueltcheva V., Chan S., Korngut L., Campbell C., Dai Y., Wang J., Barišić N., Brabec P., Lahdetie J., Walter M.C., Schreiber-Katz O., Karcagi V., Garami M., Viswanathan V., Bayat F., Buccella F., Kimura E., Koeks Z., van den Bergen J.C., Rodrigues M., Roxburgh R., Lusakowska A., Kostera-Pruszczyk A., Zimowski J., Santos R., Neagu E., Artemieva S., Rasic V.M., Vojinovic D., Posada M., Bloetzer C., Jeannet P.Y., Joncourt F., Díaz-Manera J., Gallardo E., Karaduman A.A., Topaloğlu H., El Sherif R., Stringer A., Shatillo A.V., Martin A.S., Peay H.L., Bellgard M.I., Kirschner J., Flanigan K.M., Straub V., Bushby K., Verschuuren J., Aartsma-Rus A., Béroud C., Lochmüller H. The TREAT-NMD DMD Global Database: analysis of more than 7,000 Duchenne muscular dystrophy mutations. Hum. Mutat. 2015;36(4):395–402. doi: 10.1002/humu.22758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Polavarapu K., Preethish-Kumar V., Sekar D., Vengalil S., Nashi S., Mahajan N.P., Thomas P.T., Sadasivan A., Warrier M., Gupta A., Arunachal G., Debnath M., Keerthipriya M.S., Pradeep-Chandra-Reddy C., Puttegowda A., John A.P., Tavvala A., Gunasekaran S., Sathyaprabha T.N., Chandra S.R., Kramer B., Delhaas T., Nalini A. Mutation pattern in 606 Duchenne muscular dystrophy children with a comparison between familial and non-familial forms: a study in an Indian large single-center cohort. J. Neurol. 2019;266(9):2177–2185. doi: 10.1007/s00415-019-09380-3. [DOI] [PubMed] [Google Scholar]
- 23.Ricotti V., Jägle H., Theodorou M., Moore A.T., Muntoni F., Thompson D.A. Ocular and neurodevelopmental features of Duchenne muscular dystrophy: a signature of dystrophin function in the central nervous system. Eur. J. Hum. Genet. 2016;24(4):562–568. doi: 10.1038/ejhg.2015.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koenig M., Hoffman E.P., Bertelson C.J., Monaco A.P., Feener C., Kunkel L.M. Complete cloning of the Duchenne muscular dystrophy (DMD) cDNA and preliminary genomic organization of the DMD gene in normal and affected individuals. Cell. 1987;50(3):509–517. doi: 10.1016/0092-8674(87)90504-6. [DOI] [PubMed] [Google Scholar]
- 25.Den Dunnen J.T., Grootscholten P.M., Bakker E., Blonden L.A., Ginjaar H.B., Wapenaar M.C., van Paassen H.M., van Broeckhoven C., Pearson P.L., van Ommen G.J. Topography of the Duchenne muscular dystrophy (DMD) gene: FIGE and cDNA analysis of 194 cases reveals 115 deletions and 13 duplications. Am. J. Hum. Genet. 1989;45(6):835–847. [PMC free article] [PubMed] [Google Scholar]
- 26.Wang X., Wang Z., Yan M., Huang S., Chen T-J., Zhong N. Similarity of DMD gene deletion and duplication in the Chinese patients compared to global populations. Behav. Brain Funct. 2008;4(1):20. doi: 10.1186/1744-9081-4-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lai P-S. 2002. [Google Scholar]
- 28.Swaminathan B., Shubha G.N., Shubha D., Murthy A.R., Kiran Kumar H.B., Shylashree S., Gayathri N., Jamuna R., Jain S., Purushottam M., Nalini A. Duchenne muscular dystrophy: a clinical, histopathological and genetic study at a neurology tertiary care center in Southern India. Neurol. India. 2009;57(6):734–738. doi: 10.4103/0028-3886.59468. [DOI] [PubMed] [Google Scholar]
- 29.Basak J., Dasgupta U.B., Mukherjee S.C., Das S.K., Senapati A.K., Banerjee T.K. Deletional mutations of dystrophin gene and carrier detection in eastern India. Indian J. Pediatr. 2009;76(10):1007–1012. doi: 10.1007/s12098-009-0214-y. [DOI] [PubMed] [Google Scholar]
- 30.Vengalil S., Preethish-Kumar V., Polavarapu K., Mahadevappa M., Sekar D., Purushottam M., Thomas P.T., Nashi S., Nalini A. Duchenne muscular dystrophy and becker muscular dystrophy confirmed by multiplex ligation-dependent probe amplification: genotype-phenotype correlation in a large cohort. J. Clin. Neurol. 2017;13(1):91–97. doi: 10.3988/jcn.2017.13.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prior T.W., Bridgeman S.J. Experience and strategy for the molecular testing of Duchenne muscular dystrophy. J. Mol. Diagn. 2005;7(3):317–326. doi: 10.1016/S1525-1578(10)60560-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee B.L., Nam S.H., Lee J.H., Ki C.S., Lee M., Lee J. Genetic analysis of dystrophin gene for affected male and female carriers with Duchenne/Becker muscular dystrophy in Korea. J. Korean Med. Sci. 2012;27(3):274–280. doi: 10.3346/jkms.2012.27.3.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yotova V., Lefebvre J.F., Moreau C., Gbeha E., Hovhannesyan K., Bourgeois S., Bédarida S., Azevedo L., Amorim A., Sarkisian T., Avogbe P.H., Chabi N., Dicko M.H. Kou’ Santa Amouzou, E.S.; Sanni, A.; Roberts-Thomson, J.; Boettcher, B.; Scott, R.J.; Labuda, D. An X-linked haplotype of Neandertal origin is present among all non-African populations. Mol. Biol. Evol. 2011;28(7):1957–1962. doi: 10.1093/molbev/msr024. [DOI] [PubMed] [Google Scholar]
- 34.Ballo R., Viljoen D., Beighton P. Duchenne and Becker muscular dystrophy prevalence in South Africa and molecular findings in 128 persons affected. S. Afr. Med. J. 1994;84(8 Pt 1):494–497. [PubMed] [Google Scholar]
- 35.Mendell J.R., Sahenk Z., Rodino-Klapac L.R. Clinical trials of exon skipping in Duchenne muscular dystrophy. Expert Opin. Orphan Drugs. 2017;5(9):683–690. doi: 10.1080/21678707.2017.1366310. [DOI] [Google Scholar]
- 36.Aartsma-Rus A., Krieg A.M. FDA approves eteplirsen for duchenne muscular dystrophy: the next chapter in the eteplirsen saga. Nucleic Acid Ther. 2017;27(1):1–3. doi: 10.1089/nat.2016.0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Echigoya Y., Yokota T. Skipping multiple exons of dystrophin transcripts using cocktail antisense oligonucleotides. Nucleic Acid Ther. 2014;24(1):57–68. doi: 10.1089/nat.2013.0451. [DOI] [PubMed] [Google Scholar]
- 38.Zhu Y., Gan J., Luo J., Zheng X., Wei S., Hu D. Splicing mutation of a gene within the Duchenne muscular dystrophy family. Genetics Mol Res: GMR. 2016;15(2) doi: 10.4238/gmr.15028258. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material is available on the publisher’s website along with the published article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author, [A.A.], upon reasonable request.