Abstract
The breach of proteostasis, leading to the accumulation of protein aggregates, is a hallmark of ageing and age-associated disorders, up to now well-established in neurodegeneration. Few studies have addressed the issue of dysfunctional cell response to protein deposition also for the cardiovascular system. However, the molecular basis of proteostasis decline in vascular cells, as well as its relation to ageing, are not understood. Recent studies have indicated the associations of Nrf2 transcription factor, the critical modulator of cellular stress-response, with ageing and premature senescence. In this report, we outline the significance of protein aggregation in physiological and premature ageing of murine and human endothelial cells (ECs). Our study shows that aged donor-derived and prematurely senescent Nrf2-deficient primary human ECs, but not those overexpressing dominant-negative Nrf2, exhibit increased accumulation of protein aggregates. Such phenotype is also found in the aortas of aged mice and young Nrf2 tKO mice. Ageing-related loss of proteostasis in ECs depends on Keap1, well-known repressor of Nrf2, recently perceived as a key independent regulator of EC function and protein S-nitrosation (SNO). Deposition of protein aggregates in ECs is associated with impaired autophagy. It can be counteracted by Keap1 depletion, S-nitrosothiol reductant or rapamycin treatment. Our results show that Keap1:Nrf2 protein balance and Keap1-dependent SNO predominate Nrf2 transcriptional activity-driven mechanisms in governing proteostasis in ageing ECs.
Keywords: Ageing, Endothelial cells, Keap1, Nrf2, Protein aggregates, S-nitrosation
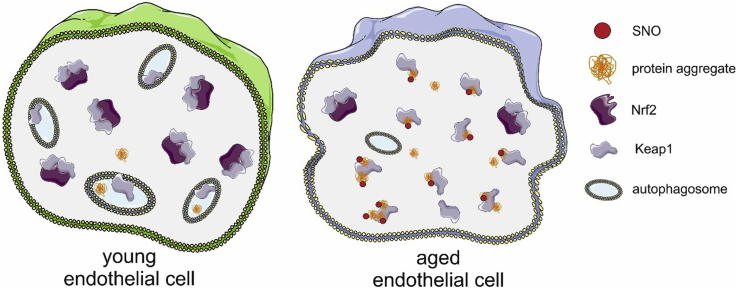
Graphical abstract
Highlights
-
•
Physiological and premature ageing facilitates aggregation of proteins in ECs.
-
•
Loss of proteostasis depends on Keap1-driven S-nitrosation in ageing ECs.
-
•
Keap1:Nrf2 ratio predominates Nrf2 transcriptional activity in proteostasis control.
-
•
Keap1 or SNO depletion, or rapamycin treatment restore proteostasis in ageing ECs.
1. Introduction
Loss of protein homeostasis (proteostasis) is a hallmark of ageing. The gradual accumulation of misfolded and aggregated proteins impairs intracellular processes and cell function, contributing to multiple age-related diseases [[1], [2], [3]]. It also correlates with the functional decline and lifespan reduction in many species, including yeast, nematodes, insects and mammals [4]. Accordingly, the proteomes of long-lived eukaryotic organisms are less prone to aggregate [4,5], and modulation of proteostasis components affects invertebrate and vertebrate health and lifespan [1,6].
The detrimental effect of protein aggregation is best established for neurological disorders [7]. Besides, we start to recognise its role in cardiac disease [8,9]. However, abnormal protein deposition may also cause endothelial cell (EC) dysfunction [10], and the impairment of cellular response to aggregating proteins occurs in atherosclerosis [11] and diabetes [12]. Therefore the cardiovascular system seems to be the second, next to the nervous system, strongly influenced by proteostasis-related pathomechanisms. Still, it has not been elucidated if age favours the formation of protein aggregates in ECs and the molecular mechanisms of proteostasis loss in the vascular system remain uncovered.
Nrf2 (nuclear factor erythroid 2-related factor 2), encoded by the NFE2L2 gene, is a transcription factor controlling homeostatic response to cellular anomalies, including oxidative stress. It has received much attention as one of the significant determinants of ageing [13,14]. Transcriptional modulation of the cellular processes, such as premature senescence, mitochondrial dysfunction, or impaired unfolded protein response, is considered to explain the Nrf2 associations with ageing [15]. Here, we describe another mechanism, which prevails Nrf2 transcriptional activity.
We demonstrate that physiological and premature ageing promotes protein aggregation in murine and human ECs. We show that the accumulation of protein aggregates results from autophagy impairment and is accompanied by a deregulated proteostasis network. Deposition of aggregates relies on protein S-nitrosation driven by Keap1, the thiol-rich redox-sensitive protein, broadly recognised as the repressor of Nrf2 transcriptional activity, recently identified as a critical Nrf2-independent player in EC function [16,17]. Loss of proteostasis in ECs can be prevented either by pharmacological induction of autophagy by rapamycin, or Keap1 depletion or treatment of cells with ascorbate, an S-nitrosothiol reductant.
2. Results
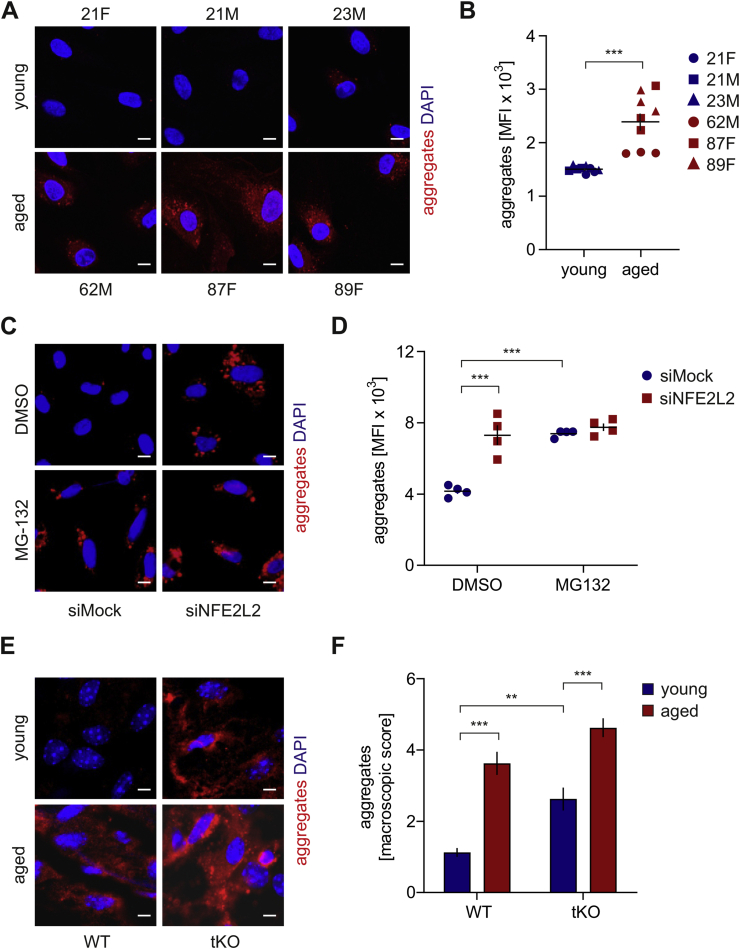
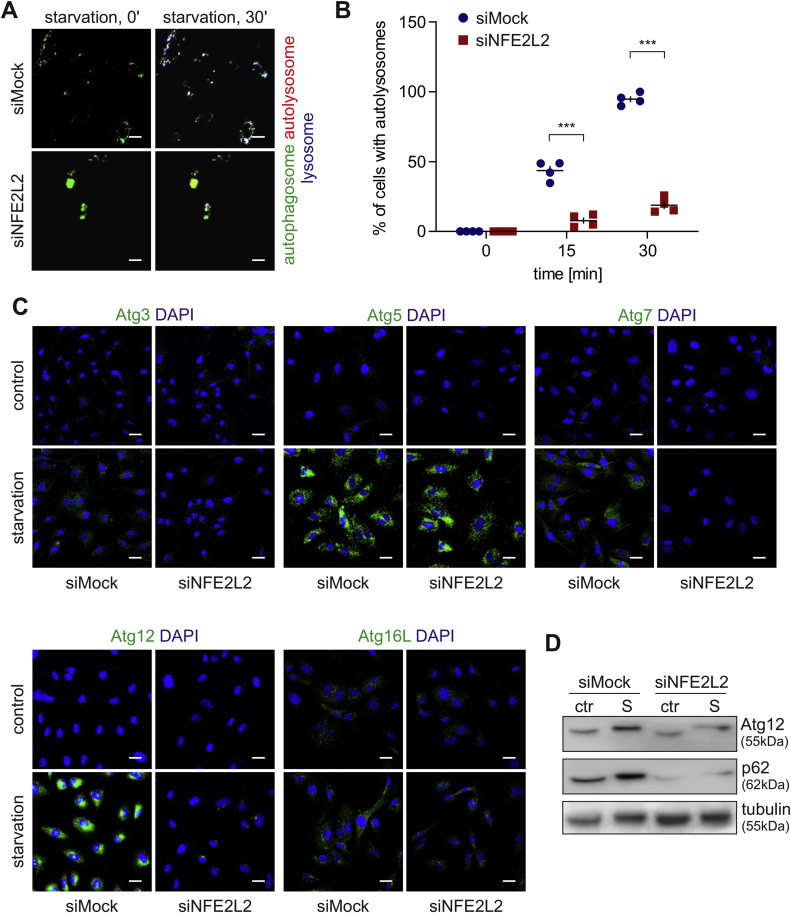
Physiological ageing and premature senescence favour protein aggregation in the human and murine aortic endothelium. To verify if ageing impacts proteostasis in ECs, we inspected the formation of protein aggregates in primary human aortic endothelial cells (HAECs) isolated from donors of different age. A substantial increase in protein aggregation was found in aged HAECs in comparison to young donor-derived cells (Fig. 1A and B). The aggregates were observed in less than 10% and ~95% of young and aged donor-derived cells, respectively. Whereas the accumulation of aggregates was minor in the case of cells isolated from a 62-year-old donor, the octogenarian-derived HAECs displayed a robust incidence of protein aggregation (Fig. 1A and B). We also observed the enhanced formation of aggregates in young donor-derived HAECs devoid of Nrf2 (siNFE2L2) (Fig. 1C, D, S1A), which exhibit premature senescence [16]. The level of protein aggregate deposition was comparable between Nrf2-deficient and proteasome inhibitor MG132-treated HAECs (Fig. 1C and D). Aggregation of proteins was found in the vast majority of siNFE2L2 (90–95%) and very few siMock cells. In contrast, no protein aggregates were observed in young donor-derived HAECs overexpressing dominant-negative Nrf2 (Fig. S1B), which also do not undergo premature senescence [17]. En face staining of aggregates in the aortas of young (8-week old) and aged (12-14-month old) wild-type, and Nrf2 transcriptional knockout (tKO) mice revealed an appreciable increase in their accumulation in aged animals, stronger in Nrf2 tKO mice compared to WT (Fig. 1E, F, S1C). The aggregates were the most abundantly present in the aortic arch and abdominal part of the aorta, but not in the thoracic part. A similar phenotype was also observed in the aortas of young Nrf2 tKO mice (Fig. 1E, F, S1C), which are prematurely senescent [17]. These data demonstrate that both physiological and Nrf2-related premature ageing of human and murine ECs is associated with protein aggregation. Moreover, the phenotype of proteostasis loss is accompanied by the functional angiogenic and proliferative impairment of aged and senescent HAECs (Fig. S1D and E and [16]).
Fig. 1.
Physiological ageing and premature senescence favour protein aggregation in the human and murine aortic endothelium. (A–B) Assessment of the level of protein aggregates in HAECs isolated from young and aged donors. Age and sex of donors are pointed out. Protein aggregates were detected using a fluorescent probe. (A) Representative images of 9 experiments. Scale bar 5 μm. (B) Quantitative analysis by flow cytometry. Student's t-test, n = 9, ***p < 0.001. (C–D) Analysis of protein aggregates in Nrf2-deficient HAECs derived from a young donor. Protein aggregates were detected using a fluorescent probe. The cells were incubated for 24 h with 5 μM MG132, which was used here as a positive control. (C) Representative images of 5 experiments. Scale bar 10 μm. (D) Quantitative analysis of protein aggregates by flow cytometry. Two-way ANOVA + Tukey's post-hoc test, n = 4, ***p < 0.001. (E–F) Assessment of level of protein aggregates in the abdominal aorta of young and aged WT and Nrf2 tKO mice. Protein aggregates were detected using a fluorescent probe. (E) Representative images of 8 animals. Scale bar 10 μm. (F) Scoring of protein aggregation. Two-way ANOVA + Tukey's post-hoc test, n = 8, ***p < 0.001.
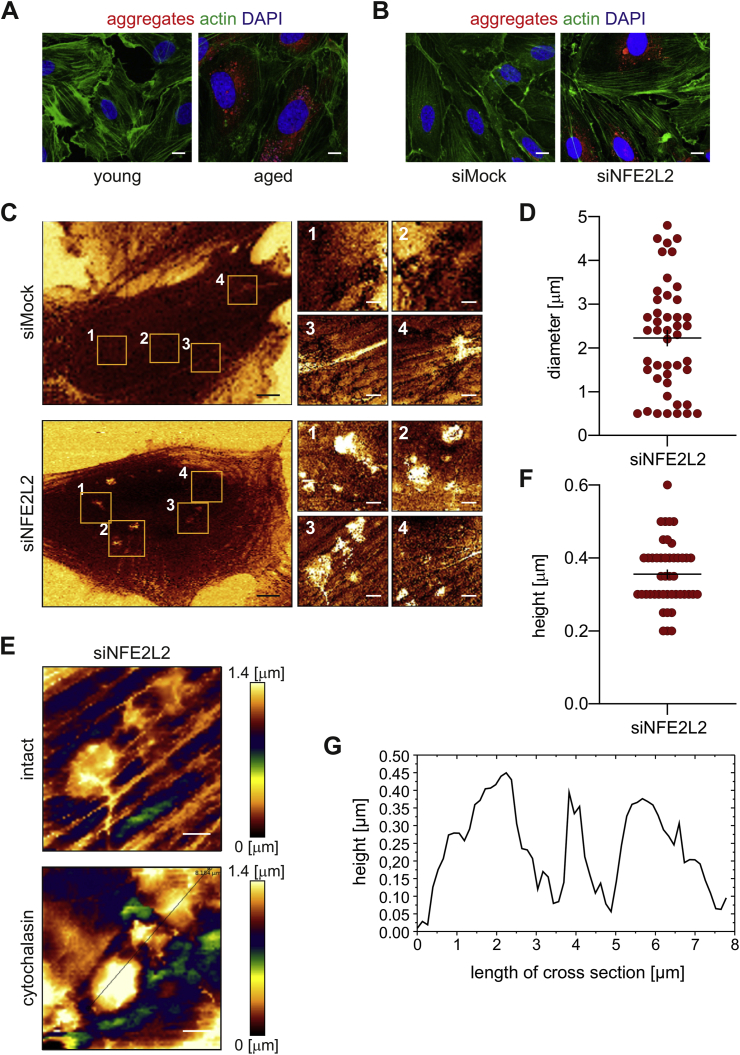
Globular protein aggregates predominate in ECs. The fibrillar form of protein aggregation is the prevailing one in neurodegenerative diseases [18]. In contrast, fluorescent staining of protein aggregates in aged and senescent Nrf2-deficient HAECs showed mainly globular form of aggregates spread throughout the cytoplasm (Fig. 2A and B). Imaging of siMock and siNFE2L2 cells with atomic force microscopy (AFM) confirmed the presence of stiff structures in the cytoplasm of Nrf2-deficient, but not in control cells (Fig. 2C). These structures appeared as globules of 2.23 ± 0.19 μm in diameter (Fig. 2D), localised beneath the actin fibres (Fig. 2E), which may reflect larger protein aggregates or clusters of protein aggregates visualised by the fluorescent probe (Fig. 1, Fig. 2B). The AFM profiling of the globule surface (Fig. 2F and G) indicated that their height is 0.36 ± 0.01 μm.
Fig. 2.
Globular protein aggregates predominate in ECs. (A–B) High-resolution images of protein aggregates in (A) HAECs isolated from young and aged donors, (B) Nrf2-deficient young donor-derived HAECs. Representative images of 3 experiments. Green - actin, red - aggregates, blue - nuclei. Scale bar 2 μm. (C–G) Analysis of cellular topography and stiffness of siMock and siNFE2L2 young donor-derived HAECs using atomic force microscopy. (C) Representative images of cellular topography. n = 3, black scale bar 5 μm, white scale bar 1 μm. (D) Measurement of the diameter of globule cytoplasmatic structures visualised in siNFE2L2 young donor-derived HAECs. n = 45. (E) Stiffness map of the globular objects visualised in siNFE2L2 young donor-derived HAECs, obtained before and after the treatment with cytochalasin. Representative images. n = 3, scale bar 1 μm. (F–G) The AFM profiling of the globule surface. (F) Assessment of the height of the globule structures visualised in siNFE2L2 young donor-derived HAECs. n = 45. (G) Exemplary profile of the globule structures. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
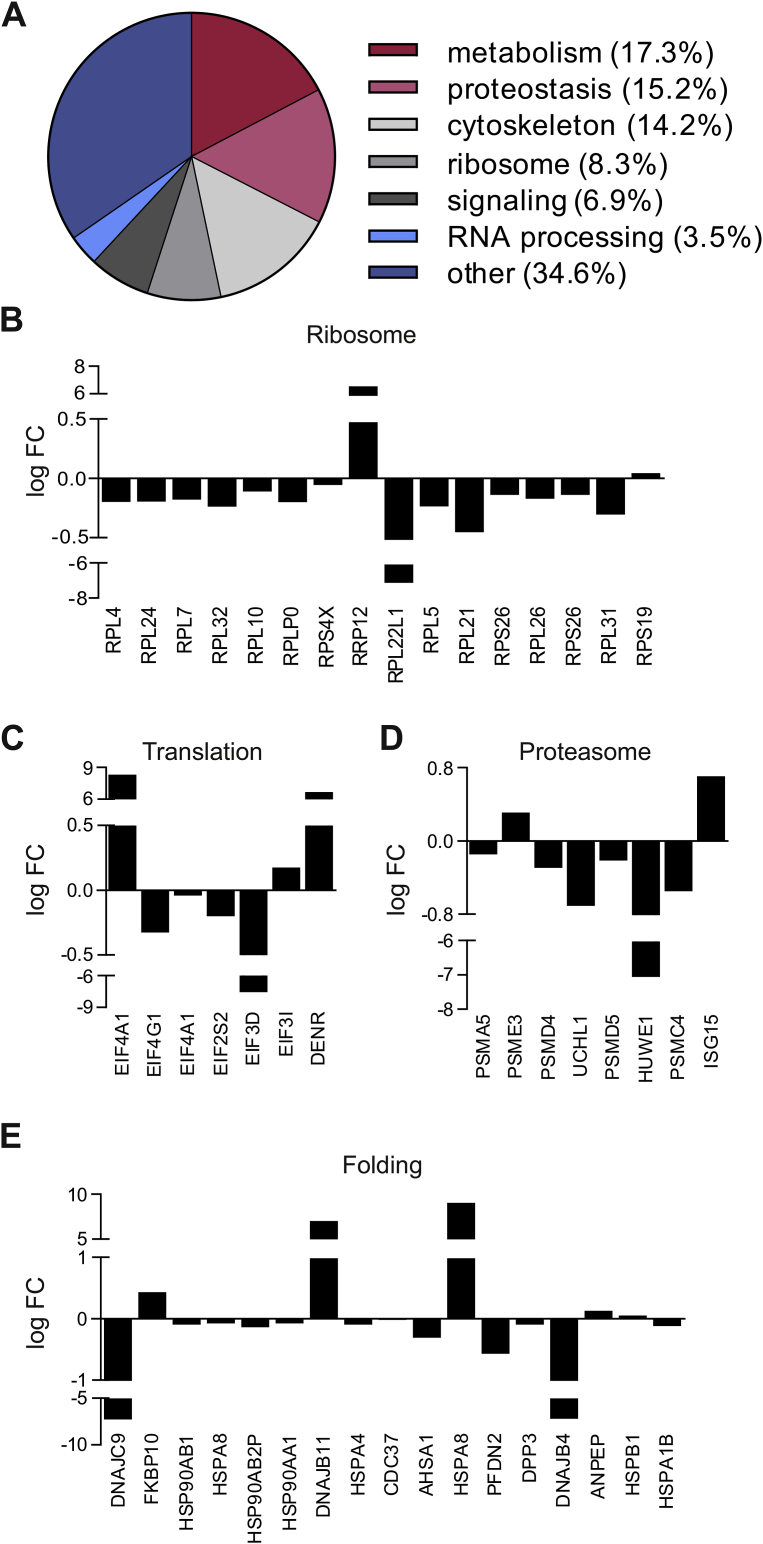
Nrf2 deficiency impairs basal and starvation-induced autophagy in ECs. Mass spectrometry proteome analysis of the soluble fraction of proteins from Mock and NFE2L2 RNAi-transfected young donor-derived HAECs [16] revealed substantial deregulation among the mediators of protein homeostasis (Fig. 3A). The differences between siMock and siNFE2L2 proteome profiles were observed in case of proteins building up ribosomes, involved in translation, protein folding and degradation via proteasome (Fig. 3B–E), which could account for the increased number of aggregates seen in Nrf2-deficient cells. Since the clearance of protein aggregates, irrespective of their origin, occurs mainly through autophagy [19,20], we hypothesised that enhanced aggregation of proteins in Nrf2-deficient cells results from the autophagy impairment. Real-time cell imaging revealed that the depletion of Nrf2 hampers the starvation-induced formation of autolysosomes. Moreover, Nrf2-deficient HAECs displayed fewer but bigger autophagosomes (LC3-GFP) at the basal state, which might be caused by their clustering (Fig. 4A and B). The level of crucial autophagy regulators, Atg3, Atg7, Atg12 and p62 (Fig. 4C, D, S2A, B), but not Atg5 and Atg16L (Fig. 4C, S2A), were significantly diminished in basal conditions and/or upon 3 h starvation in siNFE2L2 HAECs in comparison to Mock-transfected cells. These results show that the proteostasis network and the process of starvation-induced autophagy are impaired in HAECs devoid of Nrf2. Moreover, the deposition of protein aggregates in siNFE2L2 cells triggers no autophagy flux.
Fig. 3.
Mass spectrometry profiling of control and Nrf2-deficient ECs. (A) Functional classification of proteins with a significantly (p < 0.05) altered level in siNFE2L2 young donor-derived HAECs in comparison to Mock-transfected cells. A soluble-fraction of proteins was analysed by mass spectrometry. FC – fold change. n = 5, Mann-Whitney U test. (B-E) Proteostasis-related proteins with a significantly different level in siNFE2L2 young donor-derived HAECs in comparison to Mock-transfected cells. Whole-cell lysates were analysed by mass spectrometry. FC – fold change. n = 5, Mann-Whitney U test.
Fig. 4.
Nrf2 deficiency impairs starvation-induced autophagy in ECs. (A–B) Monitoring of formation of autolysosomes in siMock and siNFE2L2 young donor-derived HAECs. Cells were transduced with Tandem GFP-RFP LC3B sensor. (A) Representative images of 4 experiments. Scale bar 10 μm. (B) Quantitative analysis. Three-way ANOVA + Tukey's post-hoc test, n = 4, ***p < 0.001. (C) Immunofluorescent staining of Atg3, Atg5, Atg7, Atg12 and Atg16L in starved siMock and siNFE2L2 young donor-derived HAECs. Representative images of 3–5 experiments. Scale bar 20 μm. (D) Assessment of Atg12 and p62 level in starved siMock and siNFE2L2 young donor-derived HAECs. Whole-cell lysates were analysed by immunoblotting. α-tubulin served as a loading control. A representative example of 3 experiments. S – starvation.
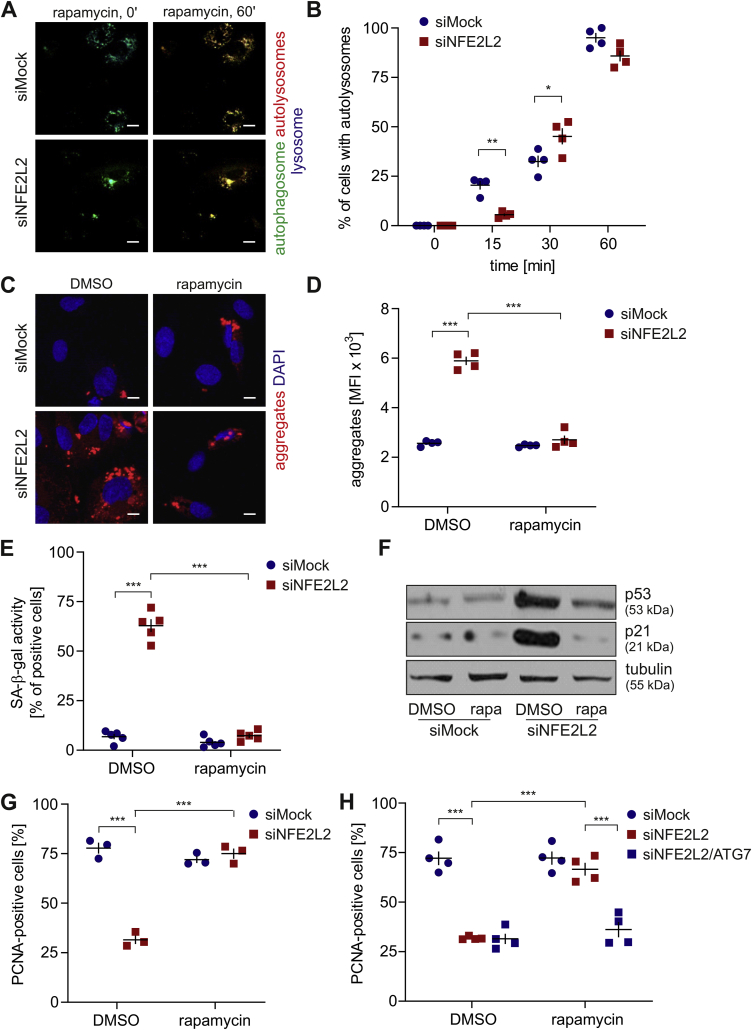
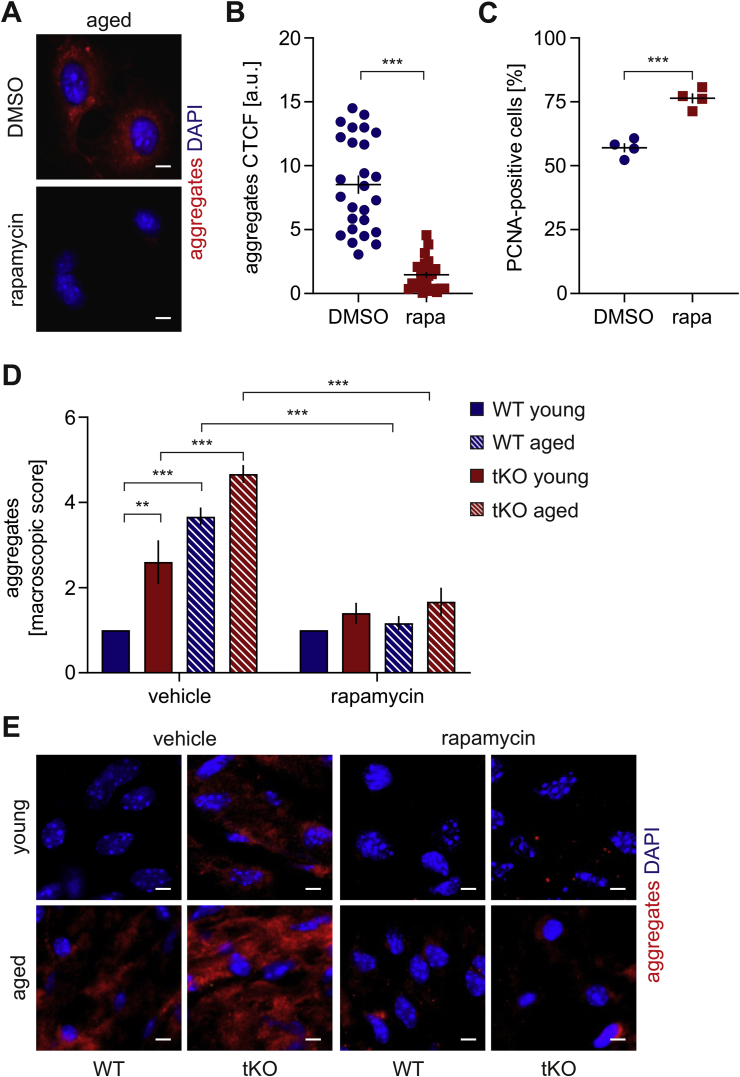
Rapamycin reverses the senescent phenotype of Nrf2-deficient ECs through the induction of autophagy and protein aggregate clearance. To further elucidate the autophagy status in Nrf2-deficient cells, we incubated them with rapamycin, the mTOR inhibitor and autophagy inducer [21]. Stimulation of cells with a non-toxic concentration of rapamycin induced autolysosome formation regardless of the Nrf2 level within 1 h (Fig. 5A and B). Rapamycin diminished the level of protein aggregates in HAECs lacking Nrf2 to control degree (Fig. 5C and D). Moreover, it reversed the senescent phenotype of siNFE2L2 cells assessed by the measurement of senescence-associated β-galactosidase (SA-β-gal) activity (Fig. 5E), further confirmed by decreased p21 and p53 protein levels (Fig. 5F, S3A) and restored the cell proliferation (Fig. 5G). To verify if rapamycin exerted its positive effect through autophagy induction, ATG7, a crucial regulator of autophagy [22], was knocked down in siNFE2L2 HAECs (Fig. S1A). Atg7 depletion abolished the positive effects of rapamycin, as verified by the percentage of PCNA-positive cells (Fig. 5H). Rapamycin also had a positive impact on proteostasis in aged-donor derived HAECs decreasing the number of aggregates (Fig. 6A and B) and enhancing their proliferation potential (Fig. 6C). Similar results were obtained in the in vivo study, where treatment of animals with rapamycin induced aggregate clearance in the aorta of Nrf2 tKO and ageing animals (Fig. 6D and E). These data show the interdependence of impaired autophagy, protein aggregation and ageing or senescent phenotype of ECs.
Fig. 5.
Rapamycin reverses the senescent phenotype of Nrf2-deficient Ecs through the induction of autophagy and protein aggregate clearance. (A–B) Monitoring of formation of autolysosomes in siMock and siNFE2L2 young donor-derived HAECs treated with 10 nM rapamycin for 24 h. DMSO was used as solvent control. Cells were transduced with Tandem GFP-RFP LC3B sensor. (A) Representative images of 4 experiments. Scale bar 10 μm. (B) Quantitative analysis. Three-way ANOVA + Tukey's post-hoc test, n = 4, *p < 0.05, **p < 0.01. (C–D) Analysis of the level of protein aggregates in siMock and siNFE2L2 young donor-derived HAECs treated with 10 nM rapamycin for 24 h. DMSO was used as solvent control. Protein aggregates were detected using a fluorescent probe. (C) Representative images of 4 experiments. Scale bar 10 μm. (D) Quantitative analysis of protein aggregates by flow cytometry. Two-way ANOVA + Tukey's post-hoc test, n = 4, ***p < 0.001. (E) Assessment of SA-β-gal activity in siMock and siNFE2L2 young donor-derived HAECs treated with 10 nM rapamycin for 24 h. DMSO was used as solvent control. Two-way ANOVA + Tukey's post-hoc test, n = 5, ***p < 0.001. (F) Comparison of p53 and p21 level in siMock and siNFE2L2 young donor-derived HAECs treated with 10 nM rapamycin for 24 h. DMSO was used as solvent control. Whole-cell lysates were analysed by immunoblotting. α-tubulin served as a loading control. A representative example of 3 independent experiments. (G) Assessment of PCNA, proliferating cell nuclear antigen, level in siMock and siNFE2L2 young donor-derived HAECs treated with 10 nM rapamycin for 24 h. DMSO was used as solvent control. Quantitative data. Two-way ANOVA + Tukey's post-hoc test, n = 3, ***p < 0.001. (H) Assessment of PCNA, proliferating cell nuclear antigen, level in siMock, siNFE2L2 and siATG7-transfected young donor-derived HAECs treated with 10 nM rapamycin for 24 h. DMSO was used as solvent control. Quantitative data. Three-way ANOVA + Tukey's post-hoc test, n = 4, ***p < 0.001.
Fig. 6.
Effect of rapamycin on protein aggregation and proliferation capacity in aged cells. (A–B) Assessment of level of protein aggregates in HAECs isolated from the aged donor and treated with 10 nM rapamycin for 24 h. Protein aggregates were detected using a fluorescent probe. (A) Representative images of 4 experiments. Scale bar 10 μm. (B) Quantitative analysis. Corrected total cell fluorescence (CTCF). Student's t-test, ***p < 0.001. (C) Assessment of PCNA, proliferating cell nuclear antigen, level in HAECs isolated from the aged donor and treated with 10 nM rapamycin for 24 h. DMSO was used as solvent control. Student's t-test, n = 4, ***p < 0.001. (D–E) En face assessment of the level of protein aggregates in the abdominal aorta of young and aged WT and Nrf2 tKO mice. Protein aggregates were detected using a fluorescent probe. (D) Scoring of protein aggregation. Two-way ANOVA + Tukey's post-hoc test, n = 6, ***p < 0.001. (E) Representative images of 6 animals. Scale bar 10 μm.
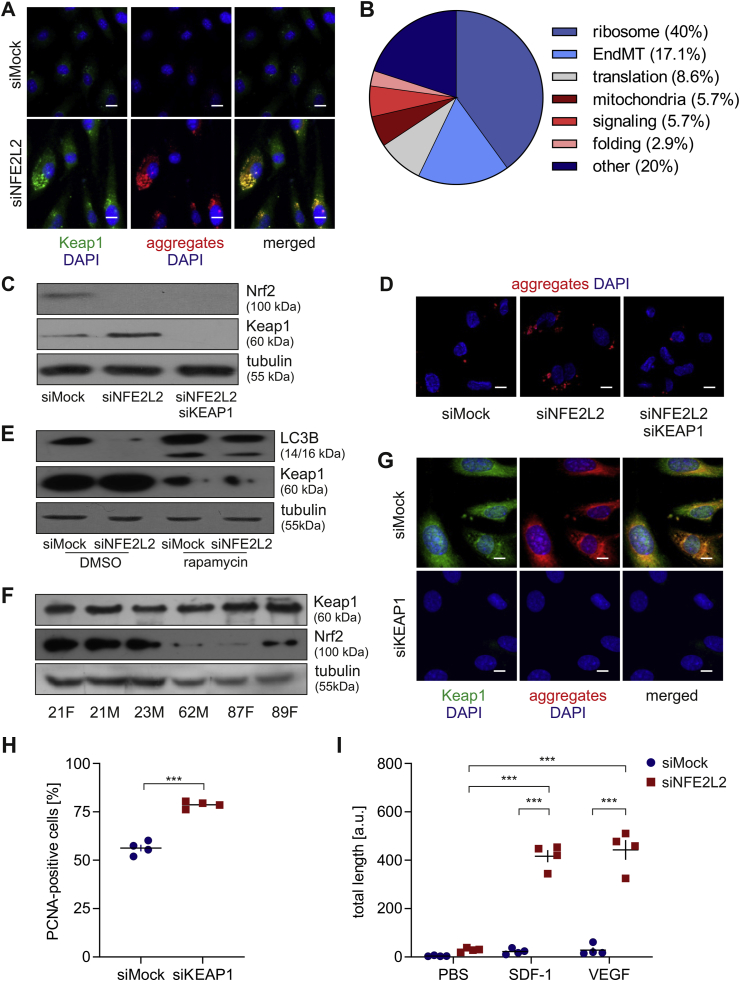
Age- and senescence-related protein aggregation and autophagy impairment depend on Keap1. Recently we have found that dysfunction of ECs devoid of Nrf2 results from the overabundance of Keap1, and is independent of Nrf2 transcriptional activity. Keap1, unrestrained by Nrf2, induces massive protein S-nitrosation (SNO), prevents podosome assembly and angiogenesis, and triggers premature senescence in ECs [16,17]. In the present study, Keap1 level was much higher in Nrf2-deficient HAECs comparing to control cells, which corroborates our previous data [16,17]. We noticed that it was distributed partly in foci and partly evenly in the cytoplasm (Fig. 7A). The globular protein aggregates were observed to colocalise with Keap1 in siNFE2L2 cells (Pearson's r = 0.874) (Fig. 7A), which may indicate the crucial role of Keap1 in the formation or maintenance of those structures. Mass spectrometry analysis of an aggregating fraction of proteins in siNFE2L2 young donor-derived HAECs compared to siMock control revealed that more than 60% of them are involved in translation and protein folding and also build up ribosomes (Fig. 7B, Table 1). Interestingly, all of them either contain the ETGE-like motif and/or increased content of amino acids promoting intrinsic disorder propensity [23] (Table 1), which may facilitate their binding to Keap1 [[24], [25], [26]]. Of note, simultaneous silencing of KEAP1 in siNFE2L2 HAECs (Fig. 7C, S4A) abrogated protein aggregation (Fig. 7D, S4B), restored p62 level (Figs. S5A and B), starvation-induced Atg12 and Atg7 levels (Figs. S5C and D), and functional autophagy (Figs. S5E and F). Moreover, protein aggregation was increased upon adenoviral-mediated overexpression of Keap1 in Nrf2-deficient ECs. Such effect, though to the lower extent, was also visible in Keap1 overexpressing Mock-transfected cells (Fig. S6A). In accordance, the level of Keap1 was significantly reduced in rapamycin-treated cells, which showed the increased conversion of LC3B (Fig. 7E, S6B) and aggregate clearance (5C, D). Analysis of Keap1 and Nrf2 level in young and aged-donor derived HAECs revealed that Keap1/Nrf2 ratio robustly elevates with age (Fig. 7F, S7A). It may suggest the insufficient saturation of Keap1 by Nrf2 in aged ECs. Depletion of Keap1 reverted protein aggregation, increased proliferation potential and the angiogenic response of aged-donor derived HAECs (Fig. 7G–I, S7B). These data show that the loss of proteostasis in physiologically and prematurely ageing ECs depends on Keap1.
Fig. 7.
Age- and senescence-related protein aggregation depends on Keap1. (A) Colocalisation of Keap1 and protein aggregates in siMock and siNFE2L2 young donor-derived HAECs. Immunofluorescent staining. Green – Keap1, red – protein aggregates, blue – nuclei. Representative images of 5 experiments. Scale bar 10 μm. (B) Functional classification of proteins aggregating differentially in siNFE2L2 comparing to siMock young donor-derived HAECs. An aggregating fraction of proteins was analysed by mass spectrometry. (C) Comparison of Nrf2 and Keap1 protein level in siMock, siNFE2L2 and siNFE2L2/KEAP1 young donor-derived HAECs. Whole-cell lysates were analysed by immunoblotting. α-tubulin served as a loading control. A representative example of 4 experiments. (D) Analysis of the level of protein aggregates in siMock, siNFE2L2 and siNFE2L2+siKEAP1 young donor-derived HAECs. Protein aggregates were detected using a fluorescent probe. Representative images of 5 experiments. Scale bar 5 μm. (E) Comparison of Keap1, p62 and LC3B level in siMock and siNFE2L2 young donor-derived HAECs treated with 10 nM rapamycin for 24 h. DMSO was used as solvent control. Whole-cell lysates were analysed by immunoblotting. α-tubulin served as a loading control. A representative example of 4 experiments. (F) Comparison of Keap1 and Nrf2 level in HAECs isolated from young and aged donor. Age and sex of donors are pointed out. Whole-cell lysates were analysed by immunoblotting. α-tubulin served as a loading control. A representative example of 3 experiments. (G) Assessment of Keap1 and protein aggregate level in aged donor-derived HAECs transfected with siMock or siKEAP1. Immunofluorescent staining. Green – Keap1, red – protein aggregates, blue – nuclei. Representative images of 4 experiments. Scale bar 10 μm. (H) Assessment of PCNA, proliferating cell nuclear antigen, level in siMock and siKEAP1 aged donor-derived HAECs. Student's t-test, n = 4, ***p < 0.001. (I) Effect of KEAP1 knockdown on SDF-1- and VEGF-induced angiogenic response in aged donor-derived HAECs – 3D fibrin bead assay. Cells were seeded on beads, immersed in fibrin gel and treated with PBS, SDF-1 or VEGF for 3 days. Two-way ANOVA + Tukey's post-hoc test, n = 4, ***p < 0.001. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Table 1.
Proteins aggregating in siNFE2L2 young donor-derived HAECs. Whole-cell lysates were analysed by mass spectrometry. For each amino acid. the percentage higher or equal to that seen in IDPs [23] has been bolded.
| Annotation | Protein | FC siNFE2L2/siMock | ETGE-like DLG | % P | % E | % S | % Q | % K |
|---|---|---|---|---|---|---|---|---|
| KRT2 | Keratin, type II cytoskeletal 2 epidermal | 13,87438 | – | 1.3 | 6.3 | 13.1 | 4.9 | 5.5 |
| KRT1 | Keratin, type II cytoskeletal 1 | 9,102523 | – | 1.1 | 5.7 | 14.0 | 5.6 | 4.3 |
| IKBIP | Inhibitor of nuclear factor kappa-B kinase-interacting protein | 7,788155 | ETNE, EKVE | 2.0 | 7.7 | 10.0 | 5.1 | 11.1 |
| KRT10 | Keratin, type I cytoskeletal 10 | 6,788999 | EITE | 0.5 | 7.5 | 15.1 | 4.5 | 3.9 |
| RPL8 | 60S ribosomal protein L8 | 5,896294 | – | 5.4 | 3.9 | 3.1 | 1.9 | 10.1 |
| RPL13A | 60S ribosomal protein L13a | 5,525886 | – | 4.4 | 4.4 | 1.5 | 3.0 | 13.8 |
| COX5A | Cytochrome c oxidase subunit 5A, mitochondrial | 3,777531 | ETDE | 6.7 | 7.3 | 4.0 | 2.0 | 4.7 |
| DDX5 | Probable ATP-dependent RNA helicase DDX5 | 3,344207 | EKDE | 5.0 | 5.7 | 5.4 | 4.9 | 5.5 |
| KRT9 | Keratin, type I cytoskeletal 9 | 3,225101 | EDFE | 0.5 | 6.7 | 14.0 | 5.3 | 4.2 |
| EIF2S1 | Eukaryotic translation initiation factor 2 subunit 1 | 3,143379 | EQLE, ESME, EKIE | 3.2 | 12.1 | 5.4 | 2.2 | 6.7 |
| RPS3A | 40S ribosomal protein S3a | 3,080348 | – | 2.7 | 5.7 | 4.2 | 4.5 | 14.4 |
| RPS4X | 40S ribosomal protein S4, X isoform | 2,934382 | – | 5.3 | 3.4 | 3.0 | 1.1 | 10.3 |
| RPL7 | 60S ribosomal protein L7 | 2,869711 | EGVE | 3.6 | 6.9 | 2.4 | 2.0 | 13.7 |
| RPL26 | 60S ribosomal protein L26 | 2,52743 | ETIE | 3.4 | 6.9 | 6.2 | 5.5 | 16.6 |
| GLIPR2 | Golgi-associated plant pathogenesis-related protein 1 | 2,509817 | – | 4.5 | 7.1 | 10.4 | 5.2 | 9.7 |
| RPS20 | 40S ribosomal protein S20 | 2,230851 | – | 5.9 | 13.4 | 5.9 | 1.7 | 10.9 |
| RPS9 | 40S ribosomal protein S9 | 2,17632 | – | 3.6 | 6.7 | 3.6 | 3.6 | 9.8 |
| FARSA | Phenylalanine--tRNA ligase alpha subunit | 2,158905 | EGEE, ERSE | 5.5 | 8.7 | 7.3 | 4.7 | 5.5 |
| RPS27A | Ubiquitin-40S ribosomal protein S27a | 2,114257 | – | 3.8 | 6.4 | 4.5 | 3.8 | 16.0 |
| NT5E | 5-nucleotidase | 2,05089 | EFDE | 4.7 | 5.1 | 6.6 | 3.0 | 6.1 |
| FN1 | Fibronectin; Anastellin; Ugl-Y1; Ugl-Y2; Ugl-Y3 | 1,849641 | EDGE, EGDE, EAEE | 7.8 | 5.9 | 8.1 | 5.3 | 3.1 |
| RPL18 | 60S ribosomal protein L18 | 1,807441 | – | 7.4 | 2.7 | 4.3 | 2.7 | 13.8 |
| EEF1G | Elongation factor 1-gamma | 1,774063 | EMDE, EKRE | 5.9 | 9.2 | 5.7 | 5.0 | 7.3 |
| RPL31 | 60S ribosomal protein L31 | 1.764461 | – | 4.8 | 6.4 | 2.4 | 0.8 | 12.8 |
| PTRF | Polymerase I and transcript release factor | 1.734553 | EITE. EAVE. EVNE. EAPE. EGEE. EVEE | 3.8 | 14.4 | 7.4 | 3.3 | 10.0 |
| PPIB | Peptidyl-prolyl cis-trans isomerase B | 1.693058 | EGME | 3.7 | 5.6 | 4.2 | 0.9 | 12.0 |
| RPS14 | 40S ribosomal protein S14 | 1.666266 | – | 4.6 | 6.0 | 6.0 | 3.3 | 7.9 |
| LMNA | Prelamin-A/C; Lamin-A/C | 1.609112 | EDDE. EGEE | 2.3 | 10.7 | 10.4 | 6.0 | 6.0 |
| H3F3 | Histone H3 | 1.578381 | EASE | 4.4 | 5.1 | 4.4 | 5.9 | 9.6 |
| RPL6 | 60S ribosomal protein L6 | 1.572808 | EITE | 6.3 | 5.2 | 4.9 | 3.1 | 18.1 |
| ANPEP | Aminopeptidase N | 1.529331 | EFEGE. EPTE. ENKE. ELVE. ECEE | 4.8 | 6.3 | 8.1 | 4.1 | 4.7 |
| RPS13 | 40S ribosomal protein S13 | 1.483448 | – | 5.3 | 3.3 | 7.3 | 2.6 | 11.9 |
| DDOST | Dolichyl-diphosphooligosaccharide--protein glycosyltransferase 48 kDa subunit | 1.435433 | DLG. EFDE | 5.3 | 4.6 | 9.4 | 3.1 | 4.6 |
| DAD1 | Dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit DAD1 | 1.416131 | – | 3.5 | 2.7 | 10.6 | 4.4 | 1.8 |
| EHD1 | EH domain-containing protein 1 | 1.390275 | EAEE. EFSE | 6.0 | 7.5 | 4.9 | 3.7 | 8.2 |
| SLC25A3 | Phosphate carrier protein. mitochondrial | 1.33242 | EMPE | 6.6 | 4.7 | 6.6 | 3.3 | 6.9 |
| HSPA6 | Heat shock 70 kDa protein 6 | 1.328892 | EEVE. EEYE. DLG | 3.9 | 8.1 | 5.3 | 4.7 | 6.1 |
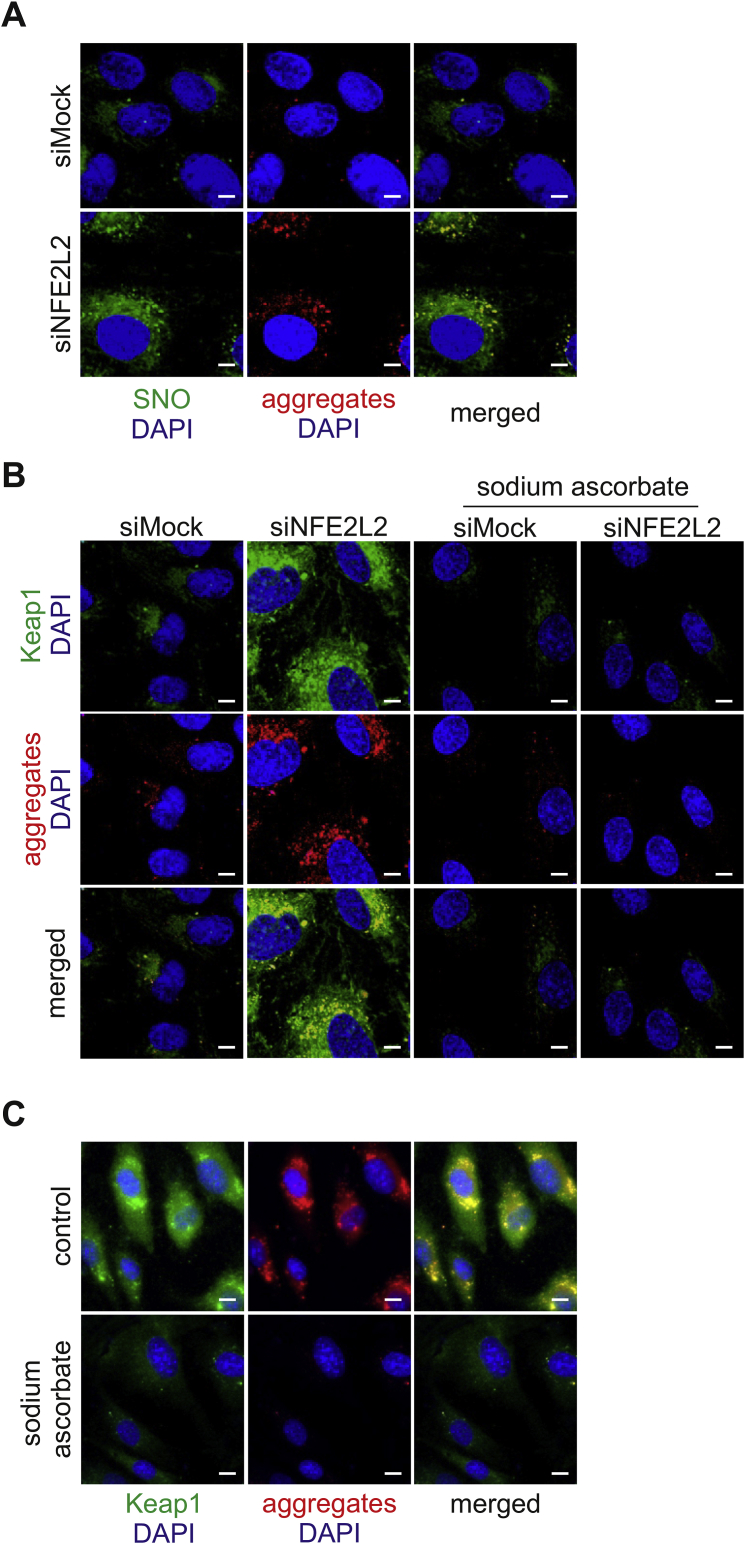
Protein aggregation is dependent on Keap1-driven S-nitrosation. Recently we found that Keap1 controls SNO and together with NO synthase and GAPDH forms a mammalian enzymatic complex necessary for this modification [17]. Moreover, SNO is known to interfere with the proteostasis network in cells, inhibit autophagy and promote protein aggregation [27,28]. Interestingly, in Nrf2-deficient ECs, we observed that globular protein aggregates colocalise with S-nitrosated proteins (Pearson's r = 0.821) (Fig. 8A). Treatment of siNFE2L2 cells with sodium ascorbate (SA, 1 mM, 24 h), the S-nitrosothiol reductant [29], eliminated protein aggregates. Moreover, it reduced the level of Keap1 in Nrf2-deficient ECs (Fig. 8B, S8A). It suggests that protein aggregation may depend on Keap1-related protein S-nitrosation in siNFE2L2 ECs. To further corroborate those findings in physiological ageing, we incubated old-donor derived HAECs with SA. In accordance with premature senescence model, we observed a decrease in protein aggregates and Keap1 level in response to SA treatment (Fig. 8C, S8B). Overall, these results imply that the mechanism of protein aggregation in Nrf2-deficient and physiologically ageing cells might be related to Keap1-dependent S-nitrosation. Both, aggregation of SNO-modified proteins as well as regulation of proteostatic machinery by SNO can be considered as the potential molecular basis of Keap1-dependent protein aggregation.
Fig. 8.
Protein aggregation is dependent on Keap1-driven S-nitrosation. (A) Colocalisation of protein aggregates and S-nitrosated proteins in siMock and siNFE2L2 young donor-derived HAECs. Immunofluorescent staining. Green – S-nitrosated proteins, red – protein aggregates, blue – nuclei. Representative images of 3 experiments. Scale bar 5 μm. (B) Colocalisation of Keap1 and protein aggregates in siMock and siNFE2L2 young donor-derived HAECs incubated with 1 mM sodium ascorbate (SA) for 24 h to reduce S-nitrosated cysteines. Immunofluorescent staining. Green – Keap1, red – protein aggregates, blue – nuclei. Representative images of 3 experiments. Scale bar 5 μm. (C) Colocalisation of Keap1 and protein aggregates in aged donor-derived HAECs incubated with 1 mM sodium ascorbate (SA) for 24 h to reduce S-nitrosated cysteines. Immunofluorescent staining. Green – Keap1, red – protein aggregates, blue – nuclei. Representative images of 4 experiments. Scale bar 10 μm. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3. Discussion
Loss of proteostasis is one of the characteristics of ageing cells. The decline in chaperone availability and activity, and dysfunction of proteolytic systems, leading together to the accumulation of protein aggregates, are the primary cellular mechanisms determining the proteostatic imbalance in ageing [1,30]. Such relation is well-established mostly in age-related neurological disorders [31]. Apart from neurons, age-related deposition of protein inclusions was also shown in the heart [9] and muscle fibres [32]. It remains elusive, however, how ageing affects proteostasis in the vascular system, particularly in ECs, and how it influences the function of blood vessels. It has been found that the abnormal response to unfolded proteins may have an impact on the pathogenesis of atherosclerosis [11] and diabetes [[33], [34], [35]], both directly related to endothelial dysfunction. Taking into account that the cardiovascular system is dramatically affected by ageing, potential dysfunction of proteostatic mechanisms in aged vascular cells is likely.
The striking correlation between Nrf2 and lifespan observed in mammals, flies and nematodes [14] gives a great incentive to elucidate its molecular basis. The substantial decrease in transactivation of Nrf2 target genes is pointed out as the leading cause of ageing and impairment of cellular function [36]. The role of Nrf2 in the context of proteostasis is mostly attributed to transcriptional regulation of genes belonging to the ubiquitin-proteasome system and autophagy [15,[37], [38], [39], [40]]. Moreover, modulation of unfolded protein response by Nrf2 through the effect on reactive oxygen species has also been proposed [15]. Our findings indicate that both physiological and premature Nrf2-related ageing favours the accumulation of protein aggregates in ECs and highlight the primary role of Keap1 in this process. Remarkably, this role is not related to the Keap1-dependent regulation of Nrf2 activity.
We observed that Keap1 colocalises with globular protein aggregates in aged and Nrf2-deficient young donor-derived HAECs. Notably, Keap1 was shown to colocalise with ubiquitin-positive aggregates in HEK293 cells [41], p62-positive inclusions in autophagy-defective hepatocytes [42], and with p62-positive LC3-negative aggregates in muscle biopsies from human autophagic vacuolar myopathy (AVM) [43]. The functional meaning of Keap1/aggregate colocalisation might be, however, bimodal. In basal conditions in cells with the normal level of Nrf2, Keap1 assists in ubiquitin aggregate clearance through autophagy, where it interacts with p62 and LC3. Thus, in this case, the depletion of Keap1 leads to autophagy impairment and increased deposition of protein aggregates [41]. In the setting of autophagy disorder, like in AVM induced by hydroxychloroquine or colchicine (autophagy inhibiting drugs used for rheumatologic disorders and gout), Keap1 is sequestered by p62-positive protein aggregates, which triggers Nrf2 release and its hyperactivation [43]. A similar mechanism was found in the autophagy-defective livers [42]. In the case of aged ECs and young donor-derived Nrf2-deficient ECs, which both are characterised by a substantial deposition of protein aggregates, the ratio of cytoplasmic Keap1 and Nrf2 is heavily imbalanced too. Therefore we postulate that in ageing cells and cells with defective autophagy, Keap1 is not saturated by Nrf2, does not undergo degradation and can serve as a cytoplasmic protein hub, sequestering proteins and inducing their accumulation. As our data show, this process can be counteracted by either Keap1 depletion or autophagy induction by rapamycin. Under this view, in the cataractous lens, which is an age-related disorder caused by excessive protein aggregation [44], Keap1 level increases [45].
Keap1 is a protein possessing a broad interactome [46]. Its binding partners contain sequences resembling the ETGE motif, less often DLG motif, which are responsible for their direct interaction with Keap1 [25,46]. Moreover, the Kelch domain of Keap1 preferentially binds intrinsically disordered proteins (IDPs) [25]. Therefore, we suppose that the process of protein sequestration by free Keap1 in the cytoplasm might be related to its high affinity to IDPs [25,26]. Of note, almost all proteins identified in aggregates in Nrf2-deficient cells contain an increased number of intrinsic disorder-promoting amino acid residues [23]. Many of them possess motifs resembling ETGE. Although the conserved segment for interactions with the Kelch domain of Keap1 is GE, usually EXGE, and it is valid for majority of verified Keap1 partners [47], the direct interactions of Keap1 dependent on ELKE and ENKE motifs were shown too [48]. Binding of IDPs may be the mechanism of protein sequestration by Keap1 in Nrf2-deficient ECs and possibly further aggregation.
Intriguingly, the globular aggregates, which colocalise with Keap1 in Nrf2-deficient ECs, seem to consist of SNO-modified proteins. A decrease in protein aggregates in response to SA treatment in siNFE2L2 and aged-donor derived cells suggests that ageing-related protein aggregation may depend on SNO, and possibly it represents a mechanism on how Keap1 triggers protein aggregation. Since the incubation of cells with SA was done 48 h after siRNA transfection, we may conclude that SA induced the clearance of protein aggregates in siNFE2L2 cells. Ascorbate was suggested to induce autophagy in cancer cells [49,50] by the mechanism which was not recognised. In view of our results, we may propose that Keap1-dependent S-nitrosation blocks the process of autophagy, thus facilitating protein aggregation. Depletion of S-nitrosation by SA or Keap1 knockdown rescue the ability of cells to induce autophagy-dependent clearance of protein aggregates.
The critical question raised by this study is whether the predominant mechanism of ageing-associated impairment of autophagy-dependent on Keap1 is related to its repressive influence on Nrf2 (and thus Nrf2 transcriptional activity) or other, Nrf2-independent, Keap1 functions. The studies have convincingly shown that p62, ubiquitin and Trim16, modulating autophagy, are regulated by Nrf2 on the transcriptional level [39,40]. Though, we believe that this mechanism is essential in oxidative stress-induced autophagy. Based on the results presented in our manuscript, we postulate that in prematurely senescent and physiologically ageing endothelial cells the shortage of Nrf2 protein resulting in Keap1 overabundance and increased protein S-nitrosation, rather than decreased Nrf2 transcriptional activity, is responsible for the presence of protein aggregates and inhibition of basal state autophagy. This conclusion is further supported by the lack of protein aggregates in DN Nrf2 overexpressing HAECs and Keap1-depleted siNFE2L2 cells. Silencing of Keap1 in Nrf2-deficient cells restored the level of p62, Atg12 and Atg7. It also rescued functional autophagy. Also, we observed that rapamycin induced autophagy and aggregate clearance, despite lack of Nrf2 or its low level in siNFE2L2 young or aged donor-derived cells, respectively.
In this study, we give new insights into the regulation of Nrf2/Keap1 axis in ageing, emphasising the significance of the maintenance of Nrf2:Keap1 balance. We show that Keap1, unrestrained by Nrf2 in the cytoplasm, is liable for protein aggregation in physiologically and prematurely ageing ECs. These results have further strengthened the conviction that Nrf2 serves not only as a transcription factor but also plays a fundamental role as a repressor of Keap1, as we have outlined in our recent studies [16,17].
4. Materials and methods
Animals. Nrf2-transcriptional knockout (tKO) C57BL/6 mice, initially developed by Prof. Masayuki Yamamoto as described previously [51], were kindly provided by Prof. Antonio Cuadrado (Universidad Autonoma de Madrid) together with WT mice. The mice were crossed back to C57BL/6J strain every 10 generations in our animal facility. 8-week old (referred to as young) or 12-14-month-old (aged) male and female animals were used in the study. Rapamycin was administered at dose 4 mg/kg i.p. for 14 days, every day; vehicle-treated mice received 5% Tween-80, 5% Kolliphor. All experimental procedures were approved by the Second Local Ethics Committee for Animal Experiments in Krakow (No. 133/2018) and followed the guidelines from Directive 2010/63/EU of the European Parliament on the protection of animals used for scientific purposes.
Cell culture. In the experiments, we used a model of human aortic endothelial cells (HAECs), which were obtained from donors of different age and sex. The cell lines were provided by PromoCell and Gibco. The cells were grown in Endothelial Basal Medium (EBM-2) (Lonza) supplemented with EGM-2MV SingleQuot Kit Supplements & Growth Factors (Lonza) and 10% foetal bovine serum (FBS) (EURx). Cells were cultured at 37 °C in a humidified incubator in 5% CO2 atmosphere. Upon reaching ~80–90% confluence, the cells were passaged using 0.25% trypsin-0.05% EDTA (1x) phenol red solution (Gibco). The cells used in all experiments were between passages 5 and 8. To exclude the impact of cell culture on cellular phenotype, when assessing the impact of donor age, the cells were not kept in culture for more than 2 passages. Senescence of cells was routinely checked by SA-β-gal staining.
Transfection with siRNA. 24 h before transfection, 1.5x104 cells per well were seeded onto 24-well plate. Transfections of HAECs were performed using 50 nM siRNA targeted against human NFE2L2 (Life Technologies, s9493), KEAP1 (Life Technologies, s18983), ATG7 (Life Technologies, s20652) or scrambled siRNA (Life Technologies, 4390846) using Lipofectamine™ 2000 Transfection Reagent (Life Technologies) in Opti-MEM I Reduced Serum medium (Life Technologies). The further analysis and treatment of cells were performed at 48 h after transfection.
Transduction of HAECs with adenoviral vectors. The transductions with adenoviral vectors AdNFE2L2DN or AdGFP were performed at a multiplicity of infection 50. The transductions with adenoviral vectors AdKEAP1 or AdGFP were performed at a multiplicity of infection 10 at 24 h after transfection with Mock or NFE2L2-siRNA. The details on DN Nrf2 are described in our recent paper [17]. After 24 h of incubation, medium with vectors was removed and fresh EGM-2MV 10% FBS complete medium was added for the next 24 h.
Quantitative Real-Time PCR. Total RNA was isolated using RNeasy Mini Kit (Qiagen), according to the manufacturer's description. The RNA was eluted with 30 μL of nuclease-free water. Reverse transcription reaction was performed using High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific, USA) according to the vendor's protocol. Quantitative RT-PCR was carried out in a StepOnePlus real-time PCR system (Applied Biosystems) in a mixture containing SYBR Green PCR Master Mix (Sigma), specific primers and 30 ng of cDNA in a total volume of 15 μL. Primers were designed using the NCBI primer design tool “Primer-BLAST” or purchased from Sigma. The EEF2 housekeeping gene was used as a reference. Quantitative analysis of gene expression was carried out using the ΔΔCt protocol. Primer sequences: EEF2 F: TGAGCACACTGGCATAGAGGC, R: GACATCACCAAGGGTGTGCAG; NFE2L2 F: TTGAGCAAGTTTGGGAGGAGCTA, R: GGAGAGGATGCTGCTGAAGG; ATG7 F: TGTGCCTCACCAGGTTCTTG, R: CTGCTCATAAGGGCAGGGAG.
Western blotting. Total protein was isolated using RIPA buffer (50 mM Tris-HCl pH 8.0, 150 mM NaCl, 1% NP-40, 0.5% Na-deoxycholate, 0.1% SDS) with protease inhibitors (Roche). Western blotting was performed according to standard procedures. Protein concentration was determined by BCA method (bicinchoninic acid method). 30 μg of protein was denatured, and SDS-PAGE was performed, followed by the wet electrotransfer. The membranes were blocked in 5% non-fat milk for 1 h and incubated overnight with primary antibodies at 4 °C. Then they were washed with TBS containing 0.1% Tween and incubated for 1 h with HRP-linked secondary antibodies (Cell Signalling Technology). The visualisation was performed using Millipore Western Substrate using X-ray films. Membranes were probed with antibodies against Keap1 (Clone 144, Millipore), Nrf2 (H-300, Santa Cruz Biotechnology), p21 (Cell Signaling Technology), p53 (Cell Signaling Technology), LC3B (Cell Signaling Technology), p62 (Cell Signaling Technology), Atg12 (Cell Signaling Technology) and a-tubulin (Sigma).
Immunofluorescent stainings. For immunofluorescent stainings, cells were seeded on coverslips. After treatment, they were fixed with 80% methanol for 10 min in -20 °C. Then, the slides were washed with PBS 3 times and the cells were blocked with 3% BSA at room temperature. The cells were overnight incubated with primary antibodies recognising Atg3, Atg5, Atg7, Atg12, Atg16L (Cell Signaling Technology) or Keap1 (Clone 144, Millipore). The next day, slides were washed, incubated with Alexa Fluor-conjugated secondary antibody (Life Technologies), counterstained with DAPI (Sigma Aldrich) and mounted in Dako Mounting Medium (Dako). High-resolution images were taken using a meta laser scanning confocal microscope (LSM-880; Carl Zeiss).
Assessment of cell proliferation. Cells were washed with PBS, fixed with 4% paraformaldehyde for 8 min, incubated in 99% EtOH for 2 min, washed with PBS, and permeabilised in 0.2% Triton X-100 in PBS for 20 min. After blocking in 10% goat serum in PBS for 1 h, cells were incubated with anti-PCNA antibody (Dako; 1:200) diluted in 1% goat serum at 4 °C overnight. Next, the secondary antibody conjugated with Alexa Fluor 568 (1:500) was added for 2 h and cells were incubated with Hoechst 33342 for 5 min to stain the nuclei. High-resolution images were taken using a meta laser scanning confocal microscope (LSM-880; Carl Zeiss).
Detection of SA-β-gal activity. Cells were fixed for 3 min with 4% formaldehyde, washed twice with PBS and incubated overnight with staining solution (5 mM potassium ferricyanide, 5 mM potassium ferrocyanide, 150 mM NaCl, 2 mM MgCl2, 1 mg/mL X-gal in citrate buffer pH 6). Next day, cells were washed with PBS and stained with Hoechst 33342 to detect nuclei. Images were taken in the brightfield and fluorescence using a fluorescent microscope (Nikon).
Detection of protein aggregates. Protein aggregates were detected with the use of fluorescent dye Proteostat™ (Enzo). For immunofluorescent stainings, the cells were seeded on coverslips. After treatment, they were fixed with 80% methanol for 10 min in -20 °C. Then, the slides were washed with PBS 3 times and the cells were blocked with 3% BSA at room temperature. The cells were stained with the dye (dilution 1:2000) overnight at 4 °C and counterstained with DAPI (Sigma Aldrich) and phalloidin (Cytoskeleton) to visualise nucleus and actin, respectively. The next day, slides were washed and mounted in Dako Mounting Medium (Dako). High-resolution images were taken using a meta laser scanning confocal microscope (LSM-880; Carl Zeiss).
For quantitative analysis with flow cytometry, the cells were detached with trypsin, washed with PBS, permeabilised with ice-cold methanol for 10 min and stained for 30 min with the Proteostat™ Dye. Before collection on the flow cytometer, the cells were washed with PBS. The cells were collected in PE-Texas Red channel using BD Fortessa flow cytometer.
En face staining of protein aggregates. For en face immunostaining, the mice were perfused with 1% PFA. The whole aorta was removed, gently cleaned of perivascular fat and fixed for 15 min with 4% paraformaldehyde (PFA). Next, aortas were incubated for 3 h in blocking permeabilising buffer (0.3% Triton X-100, 5% goat serum, 1% BSA) and then incubated overnight with the Proteostat™ Dye (Enzo). After that, the aortas were washed, and the nuclei were counterstained with DAPI (Sigma-Aldrich). After washing, the aortas were opened, intima facing up and mounted with Dako Fluorescence Mounting Medium (Dako). High-resolution images were taken using a meta laser scanning confocal microscope (LSM-880; Carl Zeiss). The assessment of protein aggregation was performed via macroscopic scoring: 1 – no aggregates; 2 – aggregates visible in up to 20% of the cellular surface (mostly around ER), 3 – aggregates up to 40%; 4 – up to 60%; 5 – up to 90%.
Life cell imaging. At 24 h after transfection, cells were transduced with Premo™ Autophagy Tandem Sensor RFP-GFP-LC3B according to manufacturer's recommendations. The cells were analysed at 48 h after transduction under a meta laser scanning confocal microscope (LSM-880; Carl Zeiss).
Atomic force microscopy. Measurements were conducted using the Nanowizard 3 AFM instrument (JPK Instruments). Round glass coverslips with cells were placed in a liquid cell (BioCell, JPK Instruments) and analysed at 37 °C. For live-cell measurements, Hank's balanced Salt Solution (Gibco) was used. For time‐dependent AFM measurements, a group of two to five cells was selected and investigated for 1–3 h. Each experiment was repeated at least three times, and a single representative experimental data set was presented. AFM data were performed using in force versus distance (FD)–based imaging mode (QI; JPK Instruments) allowing for high-resolution imaging of living cells in a time-dependent manner. The methodology enabling tracking the dynamic changes in cellular structures was described in detail before [52,53]. Briefly, single FD-curve is performed in every pixel point of the image and then translated into images of topography and stiffness. Topographical images were translated from the selected trigger force. A linear fit to the selected part of the FD curve (usually corresponding to 100 nm of maximum indentation) was used to calculate stiffness [52]. Experiments were performed using commercially available cantilevers of a radius of 25 nm on cantilevers with a spring constant of 0.1 N/m (SCM-PIC-V2; Bruker). The loading force used varied from 1 to 3.5 nN and was adjusted to obtain a clear contrast of cytoskeletal filaments. The captured images of topography and stiffness were analysed using JPK Data Processing Software. Raw AFM images were subject to minimum processing (plane fitting, contrast corrections) by employing the JPK Processing Software.
Mass spectrometry analysis. Proteome profiling of soluble fraction of siMock and siNFE2L2 cells was described in our previous paper [16]. For the mass spectrometry analysis of protein aggregates, protein was collected using 50 mM Tris (pH 8.0), 0.5% Triton X-100, 150 mM NaCl, 1 mM EDTA, 10% glycerol. After 15 min, the samples were centrifuged at 14 000g for 30 min in 4 °C. The supernatant was considered a soluble fraction, and the remaining pellet was dissolved in 1% SDS buffer and further centrifuged. After this step, the supernatant was regarded as consisting of proteins form an aggregated fraction. The proteins were precipitated with chloroform/methanol. Precipitated proteins after sonication in BioRuptor (30/30 s, setting: high) were processed as described previously [16].
Detection of S-nitrosation by biotin switch assay. The procedure was performed as described in our previous paper. For immunofluorescence detection of S-nitrosation proteins, streptavidin conjugated to Alexa Fluor 488 (Life Technologies) was used. The protein aggregates were detected using Proteostat ™ Dye (Enzo). High-resolution images were taken using a meta laser scanning confocal microscope (LSM-880; Carl Zeiss).
Treatment of cells. HAECs were incubated for 24 h with 10 nM rapamycin (Sigma) or 1 mM sodium ascorbate (SA) (Sigma) in EGM-2MV medium containing 10% FBS. To induce starvation, the cells were incubated with Earl's Balanced Salt Solution (EBSS) for 3 h.
Statistical analysis. All experiments were performed in duplicates and were repeated at least three times. Data are presented as mean ± SEM. Statistical assessment was done with t-test or Mann-Whitney U test for two-group comparisons or by analysis of variance (ANOVA), followed by a Tukey posthoc test for multiple comparisons. Mann-Whitney test was used for the analysis of mass spectrometry data. Differences were accepted as statistically significant for p < 0.05.
Declaration of competing interest
The authors declare no conflict of interest.
Acknowledgements
This work was supported by the National Science Centre grant SONATA BIS No. 2016/22/E/NZ3/00405 (AGP) and the project for PhD students and Young Scientists FBBB K/DSC/005440 (AK). DK acknowledges financial support from the National Science Centre Poland under the ETIUDA doctoral scholarship based on the decision number DEC-2019/32/T/NZ3/00326. We thank Marek Szymonski from the Institute of Physics, Jagiellonian University for providing the AFM system and Pawel Hermanowicz from Malopolska Center of Biotechnology for his assistance with confocal microscopy imaging. We acknowledge the technical help of Adrian Podkowa from the Faculty of Biochemistry, Biophysics and Biotechnology of the Jagiellonian University. Faculty of Biochemistry, Biophysics and Biotechnology and Malopolska Centre of Biotechnology of the Jagiellonian University and Centre the Molecular Biophysics of CNRS in Orleans, France are supported by the International Associated Laboratory (LIA) grant from CNRS and Jagiellonian University. The equipment used was sponsored in part by the Centre for Preclinical Research and Technology (CePT), a project co-sponsored by European Regional Development Fund and Innovative Economy, The National Cohesion Strategy of Poland. The graphical abstract was created using images from Servier Medical Art (http://smart.servier.com).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2020.101572.
Abbreviations
- AFM
atomic force microscopy
- Atg
autophagy-related protein
- ECs
endothelial cells
- HAEC
human aortic endothelial cells
- IDP
intrinsically disordered protein
- Keap1
Kelch-like ECH-associated protein 1
- mTOR
mammalian target of rapamycin
- Nrf2
nuclear factor erythroid 2-related factor 2
- PBS
phosphate-buffered saline
- PCNA
proliferating cell nuclear antigen
- PFA
paraformaldehyde
- SA
sodium ascorbate
- SA-β-gal
senescence-associated β galactosidase
- SNO
S-nitrosation
- tKO
transcriptional knockout
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Fig. S1. (A) Expression of NFE2L2 and ATG7 genes in young donor-derived HAECs transfected with siRNA. EEF2 gene served as a reference gene. One-way ANOVA + Tukey's post-hoc test, n = 4, **p < 0.01, ***p < 0.001. (B) Analysis of the level of protein aggregates in AdGFP and AdNFE2L2DN young donor-derived HAECs. Protein aggregates were detected using a fluorescent probe. Representative images of 3 experiments. Scale bar 10 μm. (C) Negative control for the assessment of protein aggregates in en face projection of murine aorta. Scale bar 10 μM. (D) Analysis of SDF-1- and VEGF-induced angiogenic response in young and aged donor-derived HAECs – 3D fibrin bead assay. Cells were seeded on beads, immersed in fibrin gel and treated with PBS, SDF-1 or VEGF for 3 days. Two-way ANOVA + Tukey's post-hoc test, n = 12, ***p < 0.001. (E) Assessment of PCNA, proliferating cell nuclear antigen, level in aged and young donor-derived HAECs. Age and sex of donors are pointed out. Student's t-test, n = 9, ***p < 0.001.
Fig. S2. (A) Quantitative analysis of immunofluorescent stainings. Fig. 4C. Corrected total cell fluorescence (CTCF), n = 180, two-way ANOVA + Tukey's post-hoc test, ***p < 0.001. (B) Densitometric analysis of western blotting. Fig. 4D n = 3, two-way ANOVA + Tukey's post-hoc test, ***p < 0.001.
Fig. S3. Densitometric analysis of western blotting. Fig. 5F n = 3, two-way ANOVA + Tukey's post-hoc test, ***p < 0.001.
Fig. S4. (A) Densitometric analysis of western blotting. Fig. 7C n = 4, one-way ANOVA + Tukey's post-hoc test, *p < 0.05, **p < 0.01. (B) Quantitative analysis of immunofluorescent stainings. Fig. 7D. Corrected total cell fluorescence (CTCF), n = 26, one-way ANOVA + Tukey's post-hoc test, ***p < 0.001.
Fig. S5. (A-B) Comparison of p62 protein level in siMock, siNFE2L2 and siNFE2L2/KEAP1 young donor-derived HAECs. Whole-cell lysates were analysed by immunoblotting. α-tubulin served as a loading control. (A) A representative example and (B) quantitative analysis of 4 experiments. One-way ANOVA + Tukey's post-hoc test, ***p < 0.001. (C–D) Immunofluorescent staining of Atg12 and Atg7 in starved siMock, siNFE2L2 and siNFE2L2/siKEAP1 young donor-derived HAECs. Representative images of 3 experiments. Scale bar 10 μm. (E–F) Monitoring of formation of autolysosomes in siMock, siNFE2L2 and siNFE2L2/siKEAP1 young donor-derived HAECs. Cells were transduced with Tandem GFP-RFP LC3B sensor. (E) Representative images of 3 experiments. Scale bar 10 μm. (F) Quantitative analysis. Three-way ANOVA + Tukey's post-hoc test, n = 3, ***p < 0.001.
Fig. S6. (A) Analysis of the level of protein aggregates in siMock and siNFE2L2 young donor-derived HAECs, in which Keap1 was overexpressed using adenoviral vectors. Protein aggregates were detected using a fluorescent probe. (A) Representative images of 3 experiments and quantitative analysis. Scale bar 5 μm. Corrected total cell fluorescence (CTCF). n = 53–57, two-way ANOVA + Tukey's post-hoc test, ***p < 0.001. (B) Densitometric analysis of western blotting. Fig. 7E. Two-way ANOVA + Tukey's post-hoc test, n = 4, ***p < 0.001.
Fig. S7. (A) Densitometric analysis of western blotting. Fig. 7F, n = 9, Student's t-test, ***p < 0.001. (B) Quantitative analysis of immunofluorescent staining. Fig. 7G. Corrected total cell fluorescence (CTCF). Student's t-test, n = 59–67, ***p < 0.001.
Fig. S8. (A) Quantitative analysis of immunofluorescent staining. Fig. 8B. Corrected total cell fluorescence (CTCF). Two-way ANOVA + Tukey's post-hoc test, n = 80–85, ***p < 0.001. (B) Quantitative analysis of immunofluorescent staining. Fig. 8C. Corrected total cell fluorescence (CTCF). Two-way ANOVA + Tukey's post-hoc, n = 75–84, ***p < 0.001.
References
- 1.Kaushik S., Cuervo A.M. Proteostasis and aging. Nat. Med. 2015;21:1406–1415. doi: 10.1038/nm.4001. [DOI] [PubMed] [Google Scholar]
- 2.Korovila I., Hugo M., Castro J.P., Weber D., Höhn A., Grune T., Jung T. Proteostasis, oxidative stress and aging. Redox Biol. 2017;13:550–567. doi: 10.1016/j.redox.2017.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hipp M.S., Kasturi P., Hartl F.U. The proteostasis network and its decline in ageing. Nat. Rev. Mol. Cell Biol. 2019;20:421–435. doi: 10.1038/s41580-019-0101-y. [DOI] [PubMed] [Google Scholar]
- 4.Vilchez D., Saez I., Dillin A. The role of protein clearance mechanisms in organismal ageing and age-related diseases. Nat. Commun. 2014;5:5659. doi: 10.1038/ncomms6659. [DOI] [PubMed] [Google Scholar]
- 5.Tartaglia G.G., Pellarin R., Cavalli A., Caflisch A. Organism complexity anti-correlates with proteomic β-aggregation propensity. Protein Sci. Publ. Protein Soc. 2005;14:2735–2740. doi: 10.1110/ps.051473805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sands W.A., Page M.M., Selman C. Proteostasis and ageing: insights from long‐lived mutant mice. J. Physiol. 2017;595:6383–6390. doi: 10.1113/JP274334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Agbas A. Trends of protein aggregation in neurodegenerative diseases. Neurochem. Basis Brain Funct. Dysfunct. 2018 doi: 10.5772/intechopen.81224. [DOI] [Google Scholar]
- 8.McLendon P.M., Robbins J. Proteotoxicity and cardiac dysfunction. Circ. Res. 2015;116:1863–1882. doi: 10.1161/CIRCRESAHA.116.305372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ayyadevara S., Mercanti F., Wang X., Mackintosh S.G., Tackett A.J., Prayaga S.V.S., Romeo F., Shmookler Reis R.J., Mehta J.L. Age- and hypertension-associated protein aggregates in mouse heart have similar proteomic profiles. Hypertension. 2016;67:1006–1013. doi: 10.1161/HYPERTENSIONAHA.115.06849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pluquet O., Pourtier A., Abbadie C. The unfolded protein response and cellular senescence. A review in the theme: cellular mechanisms of endoplasmic reticulum stress signaling in health and disease. Am. J. Physiol. Cell Physiol. 2015;308:C415–C425. doi: 10.1152/ajpcell.00334.2014. [DOI] [PubMed] [Google Scholar]
- 11.Amodio G., Moltedo O., Faraonio R., Remondelli P. Targeting the endoplasmic reticulum unfolded protein response to counteract the oxidative stress-induced endothelial dysfunction. Oxid. Med. Cell. Longev. 2018:4946289. doi: 10.1155/2018/4946289. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mukherjee A., Morales-Scheihing D., Butler P.C., Soto C. Type 2 diabetes as a protein misfolding disease. Trends Mol. Med. 2015;21:439–449. doi: 10.1016/j.molmed.2015.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sykiotis G.P., Bohmann D. Stress-activated cap'n’collar transcription factors in aging and human disease. Sci. Signal. 2010;3 doi: 10.1126/scisignal.3112re3. re3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kloska D., Kopacz A., Piechota-Polanczyk A., Nowak W.N., Dulak J., Jozkowicz A., Grochot-Przeczek A. Nrf2 in aging - focus on the cardiovascular system. Vasc. Pharmacol. 2019;112:42–53. doi: 10.1016/j.vph.2018.08.009. [DOI] [PubMed] [Google Scholar]
- 15.Schmidlin C.J., Dodson M.B., Madhavan L., Zhang D.D. Redox regulation by NRF2 in aging and disease. Free Radic. Biol. Med. 2019;134:702–707. doi: 10.1016/j.freeradbiomed.2019.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kloska D., Kopacz A., Cysewski D., Aepfelbacher M., Dulak J., Jozkowicz A., Grochot-Przeczek A. Nrf2 sequesters Keap1 preventing podosome disassembly: a quintessential duet moonlights in endothelium. Antioxid. Redox Signal. 2019;30:1709–1730. doi: 10.1089/ars.2018.7505. [DOI] [PubMed] [Google Scholar]
- 17.Kopacz A., Klóska D., Proniewski B., Cysewski D., Personnic N., Piechota-Polańczyk A., Kaczara P., Zakrzewska A., Forman H.J., Dulak J., Józkowicz A., Grochot-Przęczek A. Keap1 controls protein S-nitrosation and apoptosis-senescence switch in endothelial cells. Redox Biol. 2020;28:101304. doi: 10.1016/j.redox.2019.101304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Soto C., Pritzkow S. Protein misfolding, aggregation, and conformational strains in neurodegenerative diseases. Nat. Neurosci. 2018;21:1332–1340. doi: 10.1038/s41593-018-0235-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lamark T., Johansen T. Aggrephagy: selective disposal of protein aggregates by macroautophagy. Int. J. Cell Biol. 2012 doi: 10.1155/2012/736905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu K., den Brave F., Jentsch S. Pathway choice between proteasomal and autophagic degradation. Autophagy. 2017;13:1799–1800. doi: 10.1080/15548627.2017.1358851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weichhart T. mTOR as regulator of lifespan, aging, and cellular senescence: a mini-review. Gerontology. 2018;64:127–134. doi: 10.1159/000484629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xiong J. Atg7 in development and disease: panacea or Pandora's Box? Protein Cell. 2015;6:722–734. doi: 10.1007/s13238-015-0195-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Uversky V.N. The alphabet of intrinsic disorder. Intrinsically Disord. Proteins. 2013;1 doi: 10.4161/idp.24684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hast B.E., Goldfarb D., Mulvaney K.M., Hast M.A., Siesser P.F., Yan F., Hayes D.N., Major M.B. Proteomic analysis of ubiquitin ligase KEAP1 reveals associated proteins that inhibit NRF2 ubiquitination. Canc. Res. 2013;73:2199–2210. doi: 10.1158/0008-5472.CAN-12-4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cino E.A., Killoran R.C., Karttunen M., Choy W.-Y. Binding of disordered proteins to a protein hub. Sci. Rep. 2013;3:2305. doi: 10.1038/srep02305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Do T.N., Choy W.-Y., Karttunen M. Binding of disordered peptides to Kelch: insights from enhanced sampling simulations. J. Chem. Theor. Comput. 2016;12:395–404. doi: 10.1021/acs.jctc.5b00868. [DOI] [PubMed] [Google Scholar]
- 27.Tegeder I. Nitric oxide mediated redox regulation of protein homeostasis. Cell. Signal. 2019;53:348–356. doi: 10.1016/j.cellsig.2018.10.019. [DOI] [PubMed] [Google Scholar]
- 28.Haldar S.M., Stamler J.S. S-Nitrosylation at the interface of autophagy and disease. Mol. Cell. 2011;43:1–3. doi: 10.1016/j.molcel.2011.06.014. [DOI] [PubMed] [Google Scholar]
- 29.Wolhuter K., Whitwell H.J., Switzer C.H., Burgoyne J.R., Timms J.F., Eaton P. Evidence against stable protein S-nitrosylation as a widespread mechanism of post-translational regulation. Mol. Cell. 2018;69:438–450. doi: 10.1016/j.molcel.2017.12.019. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.López-Otín C., Blasco M.A., Partridge L., Serrano M., Kroemer G. The hallmarks of aging. Cell. 2013;153:1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Knowles T.P.J., Vendruscolo M., Dobson C.M. The amyloid state and its association with protein misfolding diseases. Nat. Rev. Mol. Cell Biol. 2014;15:384–396. doi: 10.1038/nrm3810. [DOI] [PubMed] [Google Scholar]
- 32.Askanas V., Engel W.K., Nogalska A. Inclusion body myositis: a degenerative muscle disease associated with intra-muscle fiber multi-protein aggregates, proteasome inhibition, endoplasmic reticulum stress and decreased lysosomal degradation. Brain Pathol. Zurich Switz. 2009;19:493–506. doi: 10.1111/j.1750-3639.2009.00290.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deng J., Lu P.D., Zhang Y., Scheuner D., Kaufman R.J., Sonenberg N., Harding H.P., Ron D. Translational repression mediates activation of nuclear factor kappa B by phosphorylated translation initiation factor 2. Mol. Cell Biol. 2004;24:10161–10168. doi: 10.1128/MCB.24.23.10161-10168.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hotamisligil G.S. Inflammation and metabolic disorders. Nature. 2006;444:860–867. doi: 10.1038/nature05485. [DOI] [PubMed] [Google Scholar]
- 35.Lee J., Ozcan U. Unfolded protein response signaling and metabolic diseases. J. Biol. Chem. 2013 doi: 10.1074/jbc.R113.534743. jbc.R113.534743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gao Y., Yan Y., Huang T. Human age-related cataracts: epigenetic suppression of the nuclear factor erythroid 2-related factor 2-mediated antioxidant system. Mol. Med. Rep. 2015;11:1442–1447. doi: 10.3892/mmr.2014.2849. [DOI] [PubMed] [Google Scholar]
- 37.Cui T., Lai Y., Janicki J.S., Wang X. Nuclear factor erythroid-2 related factor 2 (Nrf2)-mediated protein quality control in cardiomyocytes. Front. Biosci. Landmark Ed. 2016;21:192–202. doi: 10.2741/4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pajares M., Cuadrado A., Rojo A.I. Modulation of proteostasis by transcription factor NRF2 and impact in neurodegenerative diseases. Redox Biol. 2017;11:543–553. doi: 10.1016/j.redox.2017.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jena K.K., Kolapalli S.P., Mehto S., Nath P., Das B., Sahoo P.K., Ahad A., Syed G.H., Raghav S.K., Senapati S., Chauhan S., Chauhan S. TRIM16 controls assembly and degradation of protein aggregates by modulating the p62-NRF2 axis and autophagy. EMBO J. 2018;37 doi: 10.15252/embj.201798358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tang Z., Hu B., Zang F., Wang J., Zhang X., Chen H. Nrf2 drives oxidative stress-induced autophagy in nucleus pulposus cells via a Keap1/Nrf2/p62 feedback loop to protect intervertebral disc from degeneration. Cell Death Dis. 2019;10:510. doi: 10.1038/s41419-019-1701-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fan W., Tang Z., Chen D., Moughon D., Ding X., Chen S., Zhu M., Zhong Q. Keap1 facilitates p62-mediated ubiquitin aggregate clearance via autophagy. Autophagy. 2010;6:614–621. doi: 10.4161/auto.6.5.12189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Komatsu M., Kurokawa H., Waguri S., Taguchi K., Kobayashi A., Ichimura Y., Sou Y.-S., Ueno I., Sakamoto A., Tong K.I., Kim M., Nishito Y., Iemura S., Natsume T., Ueno T., Kominami E., Motohashi H., Tanaka K., Yamamoto M. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat. Cell Biol. 2010;12:213–223. doi: 10.1038/ncb2021. [DOI] [PubMed] [Google Scholar]
- 43.Duleh S., Wang X., Komirenko A., Margeta M. Activation of the Keap1/Nrf2 stress response pathway in autophagic vacuolar myopathies. Acta Neuropathol. Commun. 2016;4 doi: 10.1186/s40478-016-0384-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moreau K.L., King J.A. Protein misfolding and aggregation in cataract disease and prospects for prevention. Trends Mol. Med. 2012;18:273–282. doi: 10.1016/j.molmed.2012.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Palsamy P., Ayaki M., Elanchezhian R., Shinohara T. Promoter demethylation of Keap1 gene in human diabetic cataractous lenses. Biochem. Biophys. Res. Commun. 2012;423:542–548. doi: 10.1016/j.bbrc.2012.05.164. [DOI] [PubMed] [Google Scholar]
- 46.Kopacz A., Kloska D., Forman H.J., Jozkowicz A., Grochot-Przeczek A. Beyond repression of Nrf2: an update on Keap1. Free Radic. Biol. Med. 2020 doi: 10.1016/j.freeradbiomed.2020.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Karttunen M., Choy W.-Y., Cino E.A. Prediction of binding energy of Keap1 interaction motifs in the Nrf2 antioxidant pathway and design of potential high-affinity peptides. J. Phys. Chem. B. 2018;122:5851–5859. doi: 10.1021/acs.jpcb.8b03295. [DOI] [PubMed] [Google Scholar]
- 48.Yang C., Tan Y.-X., Yang G.-Z., Zhang J., Pan Y.-F., Liu C., Fu J., Chen Y., Ding Z.-W., Dong L.-W., Wang H.-Y. Gankyrin has an antioxidative role through the feedback regulation of Nrf2 in hepatocellular carcinoma. J. Exp. Med. 2016;213:859–875. doi: 10.1084/jem.20151208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Du J., Martin S.M., Levine M., Wagner B.A., Buettner G.R., Wang S., Taghiyev A.F., Du C., Knudson C.M., Cullen J.J. Mechanisms of ascorbate-induced cytotoxicity in pancreatic cancer. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2010;16:509–520. doi: 10.1158/1078-0432.CCR-09-1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen P., Yu J., Chalmers B., Drisko J., Yang J., Li B., Chen Q. Pharmacological ascorbate induces cytotoxicity in prostate cancer cells through ATP depletion and induction of autophagy. Anti Canc. Drugs. 2012;23:437–444. doi: 10.1097/CAD.0b013e32834fd01f. [DOI] [PubMed] [Google Scholar]
- 51.Itoh K., Chiba T., Takahashi S., Ishii T., Igarashi K., Katoh Y., Oyake T., Hayashi N., Satoh K., Hatayama I., Yamamoto M., Nabeshima Y. An nrf2/small maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem. Biophys. Res. Commun. 1997;236:313–322. doi: 10.1006/bbrc.1997.6943. [DOI] [PubMed] [Google Scholar]
- 52.Zapotoczny B., Szafranska K., Owczarczyk K., Kus E., Chlopicki S., Szymonski M. Atomic force microscopy reveals the dynamic morphology of fenestrations in live liver sinusoidal endothelial cells. Sci. Rep. 2017;7:7994. doi: 10.1038/s41598-017-08555-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zapotoczny B., Szafranska K., Kus E., Braet F., Wisse E., Chlopicki S., Szymonski M. Tracking fenestrae dynamics in live murine liver sinusoidal endothelial cells. Hepatol. Baltim. Md. 2019;69:876–888. doi: 10.1002/hep.30232. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.