Summary
We have developed and integrated several technologies including whole-organ imaging and software development to support an initial precise 3D neuroanatomical mapping and molecular phenotyping of the intracardiac nervous system (ICN). While qualitative and gross anatomical descriptions of the anatomy of the ICN have each been pursued, we here bring forth a comprehensive atlas of the entire rat ICN at single-cell resolution. Our work precisely integrates anatomical and molecular data in the 3D digitally reconstructed whole heart with resolution at the micron scale. We now display the full extent and the position of neuronal clusters on the base and posterior left atrium of the rat heart, and the distribution of molecular phenotypes that are defined along the base-to-apex axis, which had not been previously described. The development of these approaches needed for this work has produced method pipelines that provide the means for mapping other organs.
Subject Areas: Imaging Anatomy, Small Animal Imaging, Rodent Cardiology, Molecular Neuroscience, Cellular Neuroscience, Transcriptomics
Graphical Abstract
Highlights
-
•
Comprehensive single-neuron-scale mapping of the intrinsic cardiac nervous system
-
•
Whole-organ high-throughput imaging and reconstruction at a cellular resolution
-
•
3D anatomical framework for spatially tracked single-neuron molecular phenotypes
-
•
Integrated histology, neuron mapping, and molecular profiles for 3D organ reconstruction
Imaging Anatomy; Small Animal Imaging; Rodent Cardiology; Molecular Neuroscience; Cellular Neuroscience; Transcriptomics
Introduction
In recent years neuroanatomy research in mammalian brain has come to the forefront (e.g., in the NIH Brain Initiative, Blue Brain Project, and Connectome Project), dependent on the development of three-dimensional (3D) digital reference atlases at the cellular scale. This has revealed the complex diversity of the molecular phenotypes of neurons, often showing orderly spatial gradients of neuron types (e.g., work from the David Van Essen laboratory and the Allen Institute). In this report we further develop and extend these approaches to bring them to bear on the rodent intrinsic cardiac nervous system (ICN). Here, we present a comprehensive 3D mapping of rat ICN distribution in the overall histological and ontological context of the heart while demonstrating anatomically specific single-neuron transcriptional gradients. We herein show the development, coordination, and integration of several technologies including whole-organ imaging, software development, precise 3D neuroanatomical mapping, and molecular phenotyping. The development of the approaches needed to acquire these data has produced two method pipelines that can achieve the goals of the National Institutes of Health Common Fund's Stimulating Peripheral Activity to Relieve Conditions (SPARC) program: Comprehensive Functional Mapping of Neuroanatomy of the Heart.
Autonomic control of cardiac function arises from several integrative centers in the central nervous system, but the final level of neural integration controlling cardiac function lies in the ICN. Although the heart is known to possess a significant population of neurons, these have not previously been mapped precisely as to their number or extent/position/distribution while maintaining the histological context of the whole heart. This 3D neuroanatomical information is necessary to understand the connectivity of the neurons of the ICN and to develop their functional circuit organization. Mapping of molecular phenotypes and cell functions must also occur in the anatomical context to ensure a holistic comprehension of ICN.
Prior mapping efforts of ICN in small animals have included: mouse (Li et al., 2010, Li et al., 2014, Rysevaite et al., 2011a), rat (Ai et al., 2007, Cheng et al., 1999, Cheng et al., 2004, Cheng and Powley, 2000), rabbit (Saburkina et al., 2014), guinea pig (Hardwick et al., 2014, Steele et al., 1994), and human (Armour et al., 1997). Some of these prior efforts provided limited mapping. Others represented gross staining of neurons with coarse-grain graphical representations of the areas involved. Although qualitative and gross anatomical descriptions of the ICN have been presented earlier, we present here a comprehensive neurocardiac atlas of the ICN in rat at cellular and molecular levels at a microscopic level.
The present datasets are a revelation of previously unsuspected complexity and diversity of modulators, receptors, and neurotransmitters that is already stimulating new anatomical and functional studies. The present discovery of phenotypical spatial gradients will stimulate connectomic studies to associate these with cardiac targets and functions going forward. Such work will ultimately be important for cardiac electrophysiologists performing ablation procedures. Prior work could not and did not relate cell-scale neuroanatomy and cardiac-scale organ anatomy, which our present results do. There is no prior literature on ICN transcriptomics or localization. This will be invaluable to researchers and clinicians including, but not limited to, autonomic nervous system investigators, vagus nerve cardiac regulation investigators, vagus nerve therapy investigators, cardiologists, and heart anatomists.
Results
Multidisciplinary Approach to Data Acquisition Pipelines
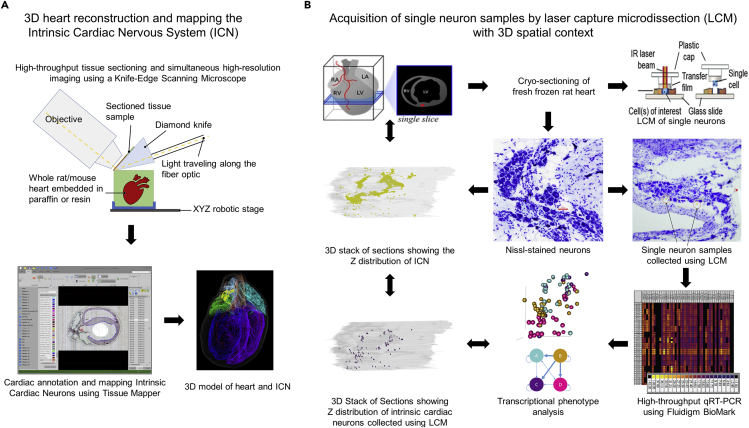
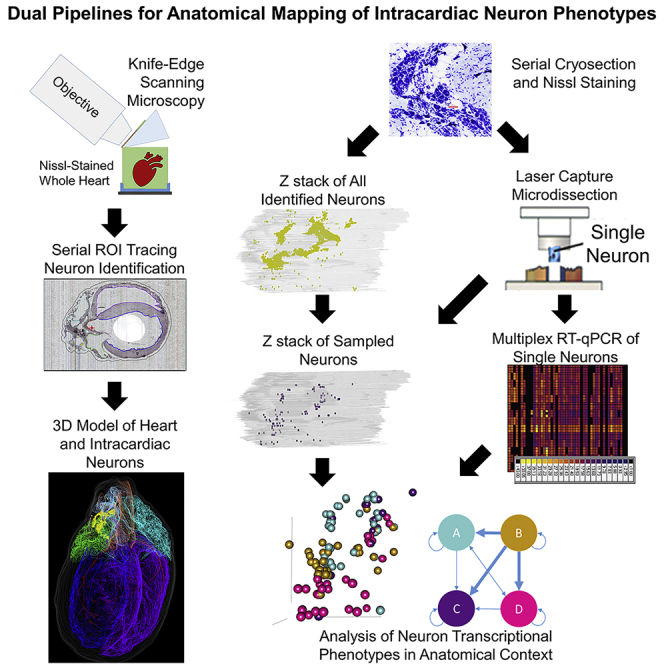
We developed and applied a dual method pipeline to create a comprehensive anatomical map and molecular profile, within the 3D structure, of cardiac neurons in a rat heart. We combined a diverse set of technologies that enabled a geographically distributed network of researchers with distinct skills to coordinate and enable the present results. These data acquisition approaches are graphically represented in the two pipelines in Figure 1. In the pipeline represented in Figure 1A we used knife-edge scanning microscopy (KESM) to image and retain the 3D structure of the heart. In parallel, custom software was used to assemble and compress the resulting ∼750,000 individual images to generate a single image volume. Annotation of the resulting image for mapping the ICN in the histological context of the whole heart was performed with Tissue Mapper, a software created for this purpose.
Figure 1.
Data Acquisition Pipelines
(A) Acquisition of a 3D, accurate organ reference framework using high-resolution collection of histological tissue sections. Once the entire heart is sectioned and imaged, the images are then compiled into an image volume by the TissueMaker software to enable 3D heart reconstruction and ICN mapping with the TissueMapper software.
(B) Acquisition of cresyl violet-stained neuronal samples from fresh heart tissue by cryostat sectioning. Single neurons were identified by position in the ICN and lifted for qPCR or RNA-seq molecular phenotyping.
A second pipeline demonstrates the acquisition of neurons as single-cell-scale samples after cryostat sectioning of the heart to ascertain circuit connectivity and molecular phenotypes using transcriptional profiling (Figure 1B). Then the molecular phenotype(s) of these neurons can be placed in whole heart and ICN anatomical context. Images of these sections, including neuronal positions, are registered and aligned using Tissue Maker to create a whole heart volume with neuronal phenotype data that can be brought into the 3D reference system created by the first approach thus generating single-cell transcriptomics in a robust anatomical context that can incorporate data from multiple subjects for future comparison.
Comprehensive Neuroanatomy of the Rat Heart
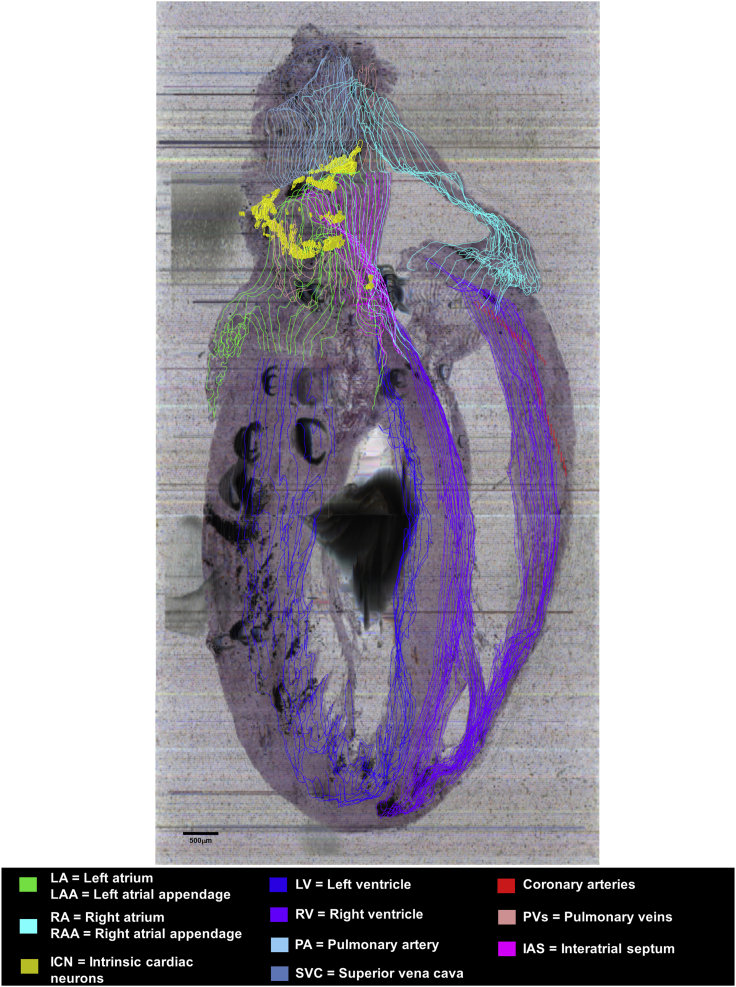
Heart tissue sections were imaged as they were cut using a fourth-generation KESM platform, preserving tissue section morphology and registration to precisely conform to that of the intact, paraffin-embedded organ. The image resolution (0.5 μm per pixel) yields clear visualization of cellular/neuronal-scale histology. The registration and stacking of all the section images is in a format compatible with the purpose-built software, TissueMaker and Tissue Mapper, which supports using the latter to precisely map the position of each neuron and histological features of the heart and blood vessels from each tissue section. We then combine the positions of all neurons and features of cardiac tissues using computer graphics to hold all the data as a precise 3D representation of that specific heart's ICN distribution in the histological context of the heart. The representation is precise and reproducible because both the tissue image acquisition and the mapping of neurons and cardiac features are under sub-micron control. This data acquisition pipeline is presented in Figure 1A. The complete dataset of anatomical mapping is available online via SPARC Data Portal (http://data.sparc.science).
As we take such care to represent the exact positions of all the neurons in a precisely preserved heart-organ morphology, we now can interact with the 3D model presenting all the mappings in various formats, scales, or representations to appreciate the neuroanatomy of the ICN, enabling the following analyses:
-
●
As illustrated in Figure 2 the ICN distribution is seen in the context of the whole 3D heart, depicting the unexpectedly extensive distribution of neurons on both posterior atria extending in the superior-inferior dimension from the base of the heart to the coronary sulcus or atrioventricular groove.
-
●
We are also able to view subsets of the ICN in their local heart context by restricting the 3D model using the “partial projection” software tool. For example, Figure 3 is a 3-mm sagittal section representing neurons along the superior-inferior extent of the heart. Figure 4B creates a 3-mm-thick section in the transverse plane that visualizes neurons located on the base/hilum of the heart. In contrast, Figure 5 excludes these neurons showing only the more inferiorly positioned neurons.
-
●
We are able to describe the exact position of ICN neurons in histological sections. Figure 6 consists of single sagittal section views of neurons throughout the superior-inferior extent in a series of sections going from the right to the left side of the heart.
-
●
The digital neuron mapping supports quantitative analyses of neuron distributions and packing density as illustrated in Figure 7.
-
●
These above-mentioned anatomical templates support addition of connectome and molecular identity data. Figure 8 shows the neurons in the digitized high-resolution images of histological sections. Figures 8 and 9 illustrate the strategy used in which we employed the pipeline represented in Figure 1B, to laser capture single neurons for molecular analysis. Figure 10 visualizes the mapped ICN distribution to the distribution of neurons sampled and analyzed for molecular phenotypes. Figures 11, 12, and 13 present analyses of molecular neuronal phenotypes, and demonstrate the gradients and distribution of phenotypes across the extent of the ICN.
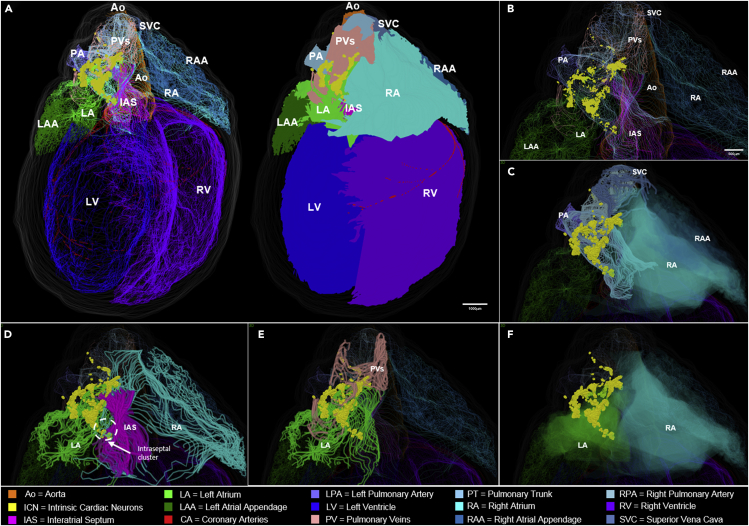
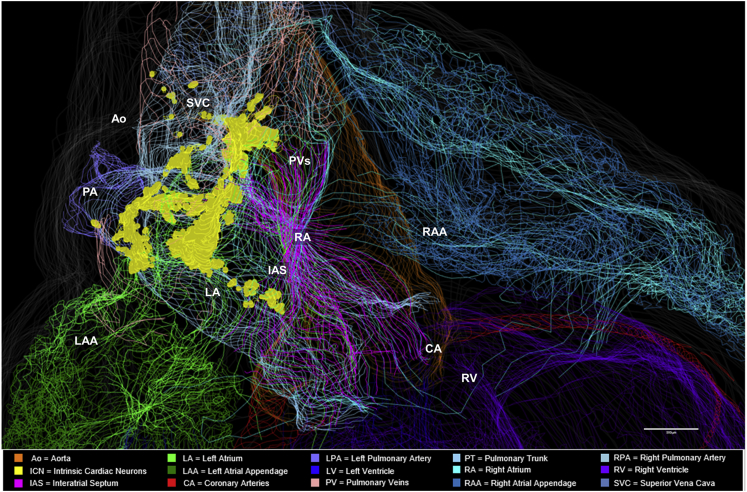
Figure 2.
Posterior View of the 3D Reconstructed Male Rat Heart
(A) Whole-heart view showing the context, extent, and distribution of the intrinsic cardiac neurons (ICN), located on superior and posterior surfaces of the atria.
(B–F) A higher-resolution view of the atria and blood vessels that are shown in (B), with various contoured features of heart anatomy that are selectively removed to appreciate the anatomical relationship of the ICN to (C) the pulmonary artery and superior vena cava (SVC), (D) the interatrial septum, (E) the left atrium and pulmonary veins, (F) the right atrium, where clusters #1 and #2 appear to be on the surface of both atria, whereas cluster #3, which is located around the border of the superior vena cava, left atrium, and right atrium, appears on the right atrium. (See also Video S2). Scale bars: panel A, 1000 μm; panels B–F, 500 μm.
Figure 3.
ICN Distribution in a Partial Projection of Sagittal Heart Sections
A partial projection of contours and ICN in a 3-mm-thick sagittal image slab illustrates the distribution of neurons along the superior-inferior extent of the heart. Scale bar: 500 μm.
Figure 4.
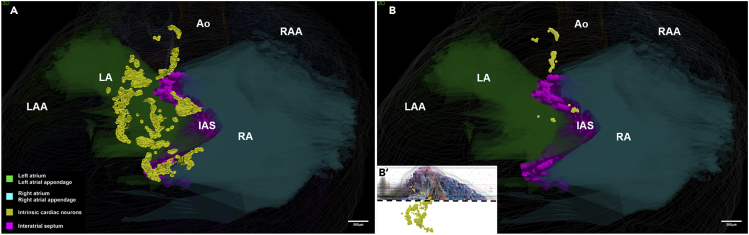
Superior View of the ICN
(A and B′) (A) Viewing the ICN distribution (neurons mapped as yellow dots) looking from the base toward the apex. The left and right distribution of all ICN neurons is discriminated by their relationship to the interatrial septum. Note that most ICN neurons visualized in (A) are not all on the base of the heart but mostly distributed at more inferior-caudal levels of the heart on both atria. (B′) To selectively view those neurons on the base of the heart we took this posterior view of the full ICN and retained only those above the cutoff point indicated by the dotted black line.
(B) Then these neurons are here observed from the superior view of the heart, showing the position of ICN neurons located within the hilum in between the aorta, superior vena cava, and pulmonary artery. (See also Video S3). Scale bars: 500 μm.
Figure 5.
Distribution of More Inferiorly Located ICN
Use of the TissueMapper Partial Projection tool that visualizes the ICN in a 3-mm-thick transverse image slab rotated in a superior view. This illustrates the locations in the transverse plane of section to highlight more inferiorly (caudally) located neurons. Neurons are represented as yellow dots. Scale bar: 500 μm.
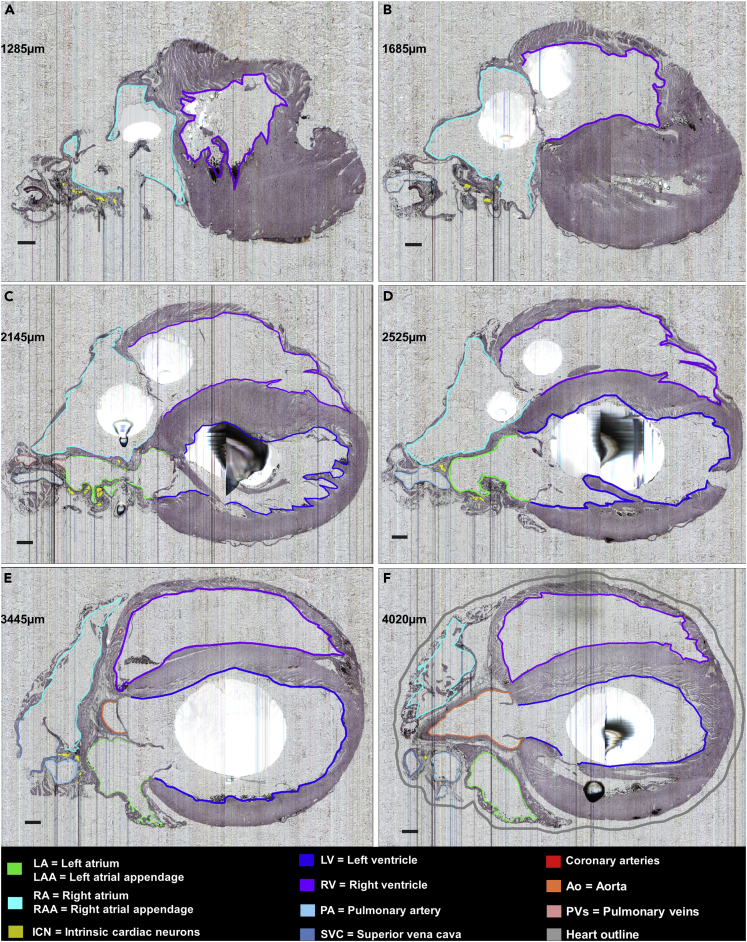
Figure 6.
ICN Distribution at Six Different Semi-sagittal Levels of the Heart
(A–F) Histological sampling of the sagittal sections extending from the right to the left side of the heart at the levels indicated in microns in each panel. The neurons are mapped with yellow dots. The contours help to contextualize the distribution of ICN relative to other features of the heart. Sections are 5 μm, images are at 0.5 μm x-y resolution. The black blobs are artifacts of uneven paraffin embedding. Scale bar: 500 μm.
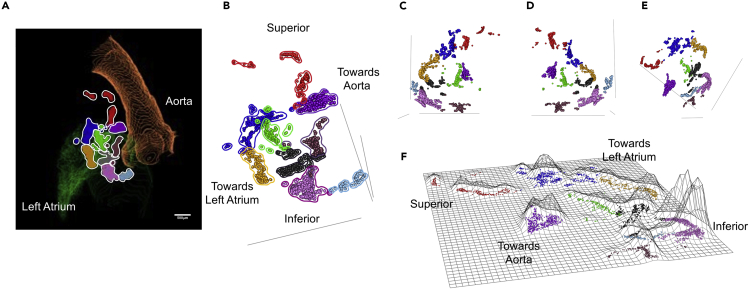
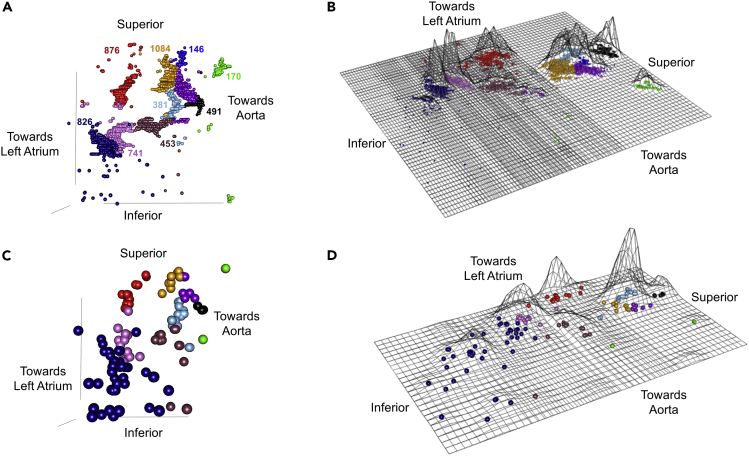
Figure 7.
Identification of Neuronal Clusters in the Rat heart
Nine clusters of neurons were identified using the partitioning around medoids (PAM) algorithm, where identified clusters are shown in different colors.
(A) Visualization of mapped neurons in their 3D orientation in TissueMapper; contours show the aorta (orange) as well as the left atrium (green).
(B) Visualization of mapped neurons in their 3D orientation.
(C–E) Visualization of mapped neurons in their 3D orientation rotated to show different points of view.
(F) Flat-mount projection of mapped neurons where the height of the contours are proportional to the density of neurons. The orientation in (E) matches the flat-mount projection in (F). (See also Video S5). Scale bar: 500 μm.
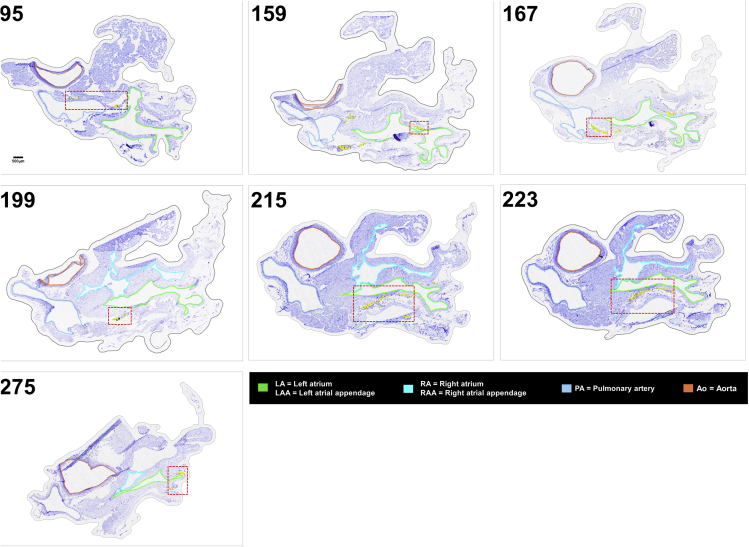
Figure 8.
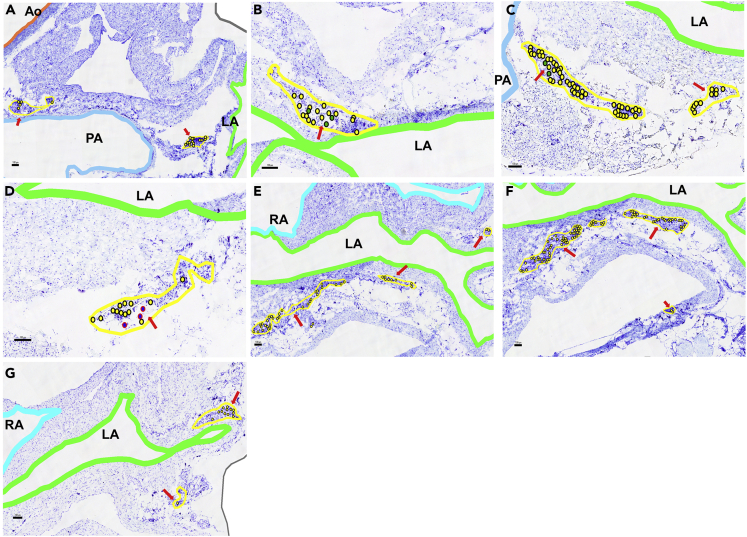
Candidate Sections of Female Rat Heart B to Contextualize the Laser Capture Microdissected Intrinsic Cardiac Neurons
Each of the candidate sections provide additional context for the seven levels of laser capture microdissected neurons. The red boxed areas are the regions of interest that are zoomed in the following figure. Scale bar: 500 μm.
Figure 9.
Enlarged Sections of LCM Lifted Intrinsic Cardiac Neurons from Six Different Levels in the Female Rat Heart B
(A) Section 95: Small ganglia located between the aorta, pulmonary artery, and left atrium; all neurons marked yellow.
(B) Section 159: neurons are lifted from a medium ganglia near the left atrium and are grouped in the Z1 molecular cluster; cells marked green.
(C) Section 167: Z1 neurons that are located around the left atrium and pulmonary artery; cells marked green.
(D) Section 199: LCM-sampled neurons that were characterized as the Z2 group were part of a small ganglia; cells marked blue.
(E) Section 215: Neurons were sampled from ganglia near the left atrium and right atrium.
(F) Section 223: LCM-sampled neurons from a large cluster near the left atrium.
(G) Section 275: LCM-sampled neurons around the left atrium. Scale bars: 100 μm. Red arrows mark the locations of relevant neurons.
Figure 10.
Identification of Neuronal Clusters in the Rat Heart B
(A–D) Visualization of mapped neurons (A) and sampled neurons (C) in their 3D orientation. Flat-mount projection of mapped neurons (B) and sampled neurons (D) where the heights of the contours are proportional to the density of neurons. Ten clusters of neurons were identified using the partitioning around medoids (PAM) algorithm, where identified clusters are shown in different colors. (See also Video S6).
Figure 11.
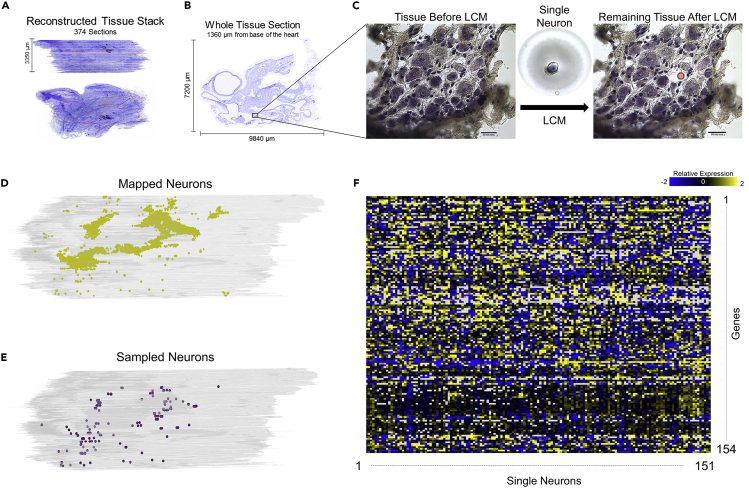
Process of Collecting and Mapping Neurons from the Rat Heart
(A and B) (A) A rat heart was sectioned at 20 μm, imaged, and put into a 3D stack in TissueMaker. The red outline shows a specific section shown in (B).
(C–E) (C) The selected region from (B) is shown in a magnified view before and after laser capture microdissection, where the single neuron that has been collected can be seen on the LCM cap (middle). Scale bar: 50 μm. Distribution of mapped neurons (D) and sampled neurons (E) in the context of their 3D location as seen in TissueMapper.
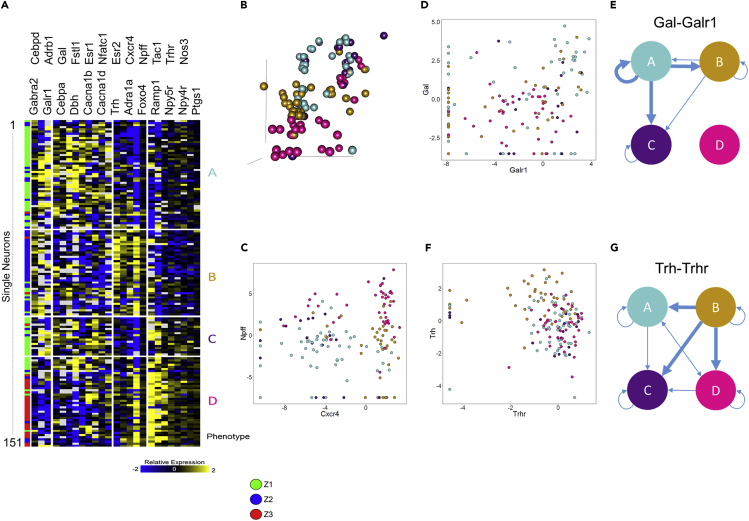
(F) Normalized qRT-PCR data showing expression of 154 genes for the 151 samples collected.
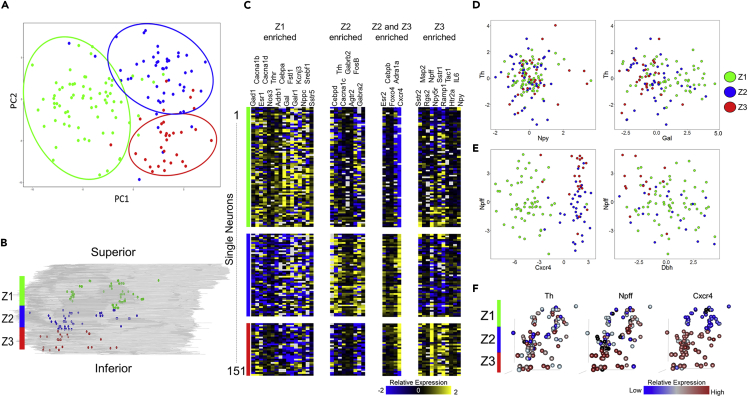
Figure 12.
Spatial Gradients of Gene Expression Profiles in the Rat ICN
(A and B) Principal-component analysis (PCA) plot of the 151 collected samples (A) show distinct separation according to their position along the z axis of the heart (B). Samples were divided into three groups along the z axis spanning from Z1 at the base of the heart to Z3 toward the apex.
(C) Pavlidis template matching using Pearson correlation with a cutoff of 0.01 was used to find genes that show specific enrichment in one or more Z groups. A cutoff of 0.001 was used to find genes enriched in both the Z2 and Z3 groups to increase specificity.
(D) Expression of tyrosine hydroxylase (Th) versus neuropeptide Y (Npy) (left) and Galanin (Gal) (right).
(E) Expression of neuropeptide FF (Npff) versus Cxcr4 (left) and Dbh (right).
(F) 3D position of collected samples colored for expression of Th, Npff, and Cxcr4. Th (F, left) and Npy (D, left) show little correlation between expression level and spatial location, whereas Galanin (D, right) appears to be upregulated in the Z1 group. Npff (F, middle and E, left) shows downregulation in the Z2 group and distinct upregulation in the Z3 group. Cxcr4 (F, right and E, left) shows very high correlation between expression level and spatial location where expression is significantly downregulated in the Z1 group and significantly upregulated in the Z2 and Z3 groups. Dbh (E, right) shows mixed expression in the Z1 and Z2 groups with low expression in the Z3 group. It can also be seen that Npff and Dbh are anticorrelated in the Z3 group. For all panels, Z1 is represented in green, Z2 in blue, and Z3 in red.
Figure 13.
Neuronal Phenotypes Contributing to Spatial Separation
(A) ANOVA was performed with a threshold of 0.001 to find genes that contribute to the separation seen between the three Z groups. Selected genes were clustered using Pearson correlation, resulting in the heatmap shown and giving rise to four different neuronal phenotypes (labeled A–D). We can see that the Z1 group was split into phenotypes A and C, with some samples from the Z1 group clustering along with the Z3 group in phenotype D. Phenotype B is composed mostly of samples residing in the Z2 group.
(B) 3D position of collected samples, colored by phenotype.
(C) Expression of Npff versus Cxcr4, colored for phenotype. We can see that the comparison of these two genes largely determines the phenotypes. Phenotype A is mostly composed of cells that have low expression of Npff and Cxcr4. Phenotype B is composed mostly of cells that have low expression of Npff and high expression of Cxcr4. Phenotype C is composed largely of cells that have high expression of Npff and low expression of Cxcr4. Phenotype D is composed largely of cells that have high expression of both Npff and Cxcr4.
(D) Expression of Galanin versus its receptor Galr1 and (E) network diagram of putative galanin-mediated connectivity between neuronal phenotypes.
(F and G) (F) Expression of thyrotropin-releasing hormone (Trh) and its receptor Trhr and (G) network diagram of putative Trh-mediated connectivity between neuronal phenotypes.
The results from these visual and quantitative analyses are presented in detail in the following section. In addition to the figures, the visualizations are also represented in movies available in the Supplemental Information.
We sought to delineate the distribution, extent, and location of neurons on the heart in part by the use of 3D graphical models of the ICN in cardiac context both in figures and by rotating the model (Video S1). Examining the 3D model framework of the ICN from a posterior point of view shows the entire population as compact and localized to a region on the superior and posterior aspects of both left and right atria, extending inferiorly to the atrioventricular boundary or the coronary sulcus at the boundary with the ventricles, as seen in Figure 2A (also see Video S2). The additional panels, Figures 2B–2F, are enlarged images of select regions of the heart to highlight the anatomical features of ICN, in each of which various contours outlining specific features of cardiac anatomy are dimmed. This provides a better appreciation of the ICN distribution, shape, extent, and location in relation to specific cardiac structures. With all contours illuminated the zoomed-in ICN appears to comprise three or more large clusters in which neurons extend rostral to caudal almost continuously (Figure 2B). At its most superior extent, portions of the ICN lie adjacent, semi-surrounding, and posterior to the pulmonary artery and superior vena cava, on the right side of the heart (Figure 2C). As neurons extend inferior to these levels they distribute in the shape of “a comma” and extend laterally to the left. Neurons at these levels are situated on both atria and within the interatrial septum (IAS) (Figure 2D). Further inferior the ICN neurons can be seen to distribute in relation to the contours of the pulmonary veins in Figure 2E, extending on the posterior surface of the left atrium to the boundary with the left ventricle. In summary, the 3D reconstruction illustrates how the most prominent clusters of neurons are positioned on the left atrium and/or across both atria as in Figure 2F. Most neurons appear to be located on the posterior surface of the left atrium. Other neurons, more superiorly located, are located within the IAS and on the right atrium. This again shows that the ICN is associated with the posterior wall of both left and right atria as well as the IAS (Ai et al., 2007, Cheng et al., 1999, Cheng et al., 2017). Using visual inspection for grouping, there are three large clusters of ICN neurons associated with the atria denoted as #1, #2, and #3. From this perspective, clusters #1 and #2 appear to be principally on the epicardial surface of the left atrium, whereas cluster #3, located adjacent to the borders of the superior vena cava, left atrium, and right atrium, appears to be largely on the right atrium.
Cardiac features have been contoured and colored accordingly, with mapped neurons marked in yellow. Partial projection of the neurons at the base of the heart are also shown.
The ICN distributes on the posterior atria. To position, in higher resolution, the neurons associated with posterior cardiac structures, we used the virtual section “partial projection” tool from the Tissue Mapper software. Using partial projection, a user-defined subset of the 3D heart model and neurons in any plane and thickness can be displayed, more than single sections and less than the full 3D model. Using this tool we made a virtual section that displays only neurons in a 3-mm sagittal section oriented superior-inferior, centered on the posterior cardiac surface, as seen in Figure 3. This shows a more restricted portion of the heart structure while presenting a more coherent view than single sections can provide. At these levels the majority of neurons appear inferior to the base of the heart, extending to the coronary sulcus. This thick section and point of view is focused on the posterior surface of the heart. This has the advantage of providing a broader context than single sections and also a more focused examination than the full 3D heart context. In Figure 3 this partial projection of the ICN localizes a compact population of neuronal clusters with a superior-inferior orientation, distributed on both atria and in the IAS, but preferentially on the left atrium.
By rotating the 3D model, we found that a superior view, i.e., looking down at the base of the heart, provided the ability to distinguish neurons associated with the right versus the left atrium. Figure 4A takes a superior view of the 3D heart and ICN, viewing the base of the heart from a viewpoint that highlights the profile of the IAS (also see Video S3). In particular, the distribution of neurons in the left and right atria can be observed in relation to the IAS, with neurons associated with both atria, but more so to the left atrium, with a much smaller population within the IAS itself. In Figure 4A, the entire population of the ICN is seen from a superior viewpoint allowing appreciation of the full distribution of neurons on the left and right atria.
As in rat the ICN is often associated with the hilum on the base of the heart we examined only those neurons with that specific neuroanatomical distribution and localization. In Figures 4B′ and 4B we aim to acquire and then only visualize neurons plausibly located on the base of the heart. As described in previous sections, using “partial projection,” a user-defined subset of the 3D heart model and neurons in any plane and thickness can be displayed, more than single sections and less than the full 3D model. Using the partial projection tool, we made a virtual section that displays only neurons at the base of the heart (as seen in Figure 4B). These ICN neurons are on the epicardial aspect of the base associated with the major pulmonary vessels there at the cardiac hilum. These neurons appear to be on the right atrium and in the IAS.
Our results show that the majority of neurons are positioned more inferiorly to the base of the heart, extending caudally to the boundary with the ventricles as seen in Figures 2 and 3. To visualize their distribution and location across the two atria we again took a superior view of the heart to observe left-right neuronal distribution of neurons, but used the Tissue Mapper software “partial projection” tool (to “cut off the base/hilum), and observe only neurons inferior to the base of the heart (Figure 5, Video S4). This graphic shows those ICN neurons as a transverse or axial visualization of a 3-mm-thick slab of tissue. Neuron clusters are abundant across both atria, but in contrast to the base of the heart neurons are more abundant on the left atrium.
The 3D reference models of the heart and ICN are derived from whole-mount stained histological tissues (i.e., whole rodent hearts). It is also informative to look at these single tissue sections and appreciate the position of the neurons in them. The “native” section images in Figure 6 are from the data acquisition pipeline in Figure 1A. These six tissue section images are at 0.5 μm (x-y) resolution and allow appreciation of the cellular histology for discriminating neurons. As shown at six different levels of the heart (Figure 6) individual neurons could be identified and their locations determined in relation to the heart chambers and other structures. This heart was sectioned in a near-sagittal plane, and Figures 6A–6F move sequentially from the right to the left side of the heart. The image volume is available online through the SPARC Data Portal. Figure 6A is close to the right side limit to the ICN, and Figure 6F is close to the left limit. The numbers in each panel indicate the right-left position of the section. Thus, Figure 6A is 1,285 μm from the right heart edge and Figure 6F is 4,020 μm farther left. In these tissue sections, neurons form clusters on the surface of the walls of the right and left atria. As seen in Figures 2 and 4 these clusters continue across many sections, so that when the maps are combined (stacked) across sections, these neurons form somewhat continuous structures in the 3D model. These structures may represent previously identified cardiac ganglionated plexuses (Cheng et al., 1999, Cheng et al., 2004).
The qualitative inspection of ICN clustering finds areas of high and low neuronal packing density, whereas a more quantitative and objective approach to these observations involves use of “partitioning around medoids (PAM)” algorithm to assay packing density or clustering (Kaufman and Rousseeuw, 1987). These clusters can be viewed in Tissue Mapper in 3D cardiac context (Figure 7A; Video S5) and without cardiac features, as the ICN alone, in a posterior point of view (Figure 7B) and in Figures 7C–7E in rotated points of view to highlight the shapes of the clusters. These clusters can be represented as surface plots, as in Figure 7F, where the height of the surface contour represents the neuronal packing density.
Visualization of neuronal clusters based on packing density by use of partitioning around medoids (PAM) algorithm. Clusters are shown as a flat map projection as well as their mapped locations within the ICN.
It is possible that, like the neurons in the brain, these clusters of neurons may be somewhat variable in precise spatial distribution from animal to animal, but identifiable as they are consistently present at the coarse-grain anatomical locations relative to notable cardiac features. By analyzing the connectivity and molecular phenotype of individual neurons from these clusters, it may be possible to identify functional groupings within the ICN. They may have a consistent connectome of functionally specific projections and may also have distinct molecular phenotypes. The representation of the anatomy of molecular phenotypes of the ICN requires the data acquisition pipeline described in Figure 1B. Figures 8 and 9 show rat heart sections acquired in this pipeline. Figure 8 presents overviews of regions containing ICN neurons from which laser capture microdissection (LCM) samples were acquired, and Figure 9 the detailed neuronal groupings from which single neurons were sampled.
Combining 3D Anatomical Mapping of the ICN with Laser Capture Microdissection of Single Neurons for Molecular Phenotype Analysis
The sections in Figures 8 and 9 are single sections from a female rat heart in which we mapped the 3D organization of the ICN. The heart was sectioned in the transverse plane going rostro-caudally between the base and apex. The tissue sections shown are at regular intervals at levels where ICN neurons are present in the heart. At each level the position of captured neurons is indicated in yellow. These distributions in two dimensions are like what is seen in Figures 2, 3, 4, and 5 in 3D or stacks, with relatively continuous distributions of neurons that tend to clump in groups consistent with being described as ganglionated plexuses. Distinct clusters of widely separated cell groups are associated with both atria. Sections are numbered from the base, and neurons were found in sections 39 through 470.
The neurons marked with yellow dots in Figure 9 indicate the single cells mapped within the ICN. An effort was made to select cells randomly but broadly representative across all sections. Neurons that were lifted from the tissue by performing LCM at Z1 levels are marked green (Figures 9B and 9C), and those from Z2 levels are marked in blue (Figure 9D). Such microdissected neurons (n = 151) were used to perform single-cell transcriptomics. The molecular phenotype of each neuron is associated with the position of each cell in the heart's 3D coordinate framework. The genes expressed and the cell types of these isolated neurons combined back into the specific distributions in 3D context generated by stacking the serial images. This quantitative display can then be used for the visualization of laser capture neurons and linked to their molecular phenotype datasets. Figures 10A and 10B use the “data-driven” ICN quantitative clustering as was done in Figure 7. However, this ICN was “undersampled” for mapping and is not for comparison to Figure 7. Rather it is to provide context for the clustering and distribution of the ICN neurons lifted for molecular profiling in Figures 10C and 10D (Video S6).
Neurons are colored based off of clusters identified through PAM algorithm as described.
Molecular Analysis of Single Neurons in the 3D ICN Context
We obtained a high-throughput transcriptomic dataset containing 23,254 data points from 151 samples of single neurons (Figure 11). Each sample was assayed for the expression of 154 genes selected as associated with neuromodulation and cardiac function. We analyzed this transcriptomics data using principal-component analysis (PCA) to identify the structured variation that, if present, could organize the neurons into subgroups based on the variability in the gene expression profiles (Figure 12). We mapped the three major neuron groups that arose from PCA results to their spatial location in the three-dimensional coordinates of ICN. Interestingly, the superior-inferior position (base to apex direction) accounted for the most robust clustering along the first two principal components PC1 and PC2 that account for the top two dominant sources of variation in the gene expression data (Figure 12A). Samples were divided into three Z position groups (Z1, Z2, Z3) where Z1 is closest to the base, moving down to Z3 near the inferior aspect of the atria (Figure 12). Examining the gene expression profiles of samples within the Z groups, distinct sets of genes were distinctly enriched in each of the Z1, Z2, and Z3 positional groups, with a small subset being enriched in both Z2 and Z3 (Figure 12C). Notably, the neurons within each of the Z groups were isolated from multiple ganglia (Figure 11), suggesting that the heterogeneity of molecular phenotypes is more constrained than the spatial distribution of these neurons.
We also analyzed the distribution of select molecular profiles in a pairwise fashion to assess the alignment of conventionally described cell types in the ICN. For example, a pairwise comparison of Th and Npy expression demonstrated that whereas several neurons could be classified as Th only or Npy only, a subset of neurons were both Th and Npy positive. These Th+ Npy+ neurons were distributed throughout ICN without any apparent bias toward a narrow spatial location (Figure 12D, left). These results are consistent with results from immunohistochemistry where NPY-positive cells and TH-positive cells were observed as broadly distributed in the ICN (Richardson et al., 2003), but did not have a correlated expression as was suggested in other results (Crick et al., 1994). Examination of Galanin along with Th, however, shows a more organized pattern with higher expression of Galanin in the Z1 position groups, but with little correlation with Th generally (Figure 12D, right). Interestingly, we found that certain genes showed a very strong spatial localization bias, and examining them in a pairwise manner reveals further co-localization patterns. For example, Cxcr4 and Npff were highly correlated with spatial location. Cxcr4 expression was almost exclusively high in the Z2 and Z3 positional groups, whereas Npff expression was the highest in Z3, was the lowest in Z2, and was mid-range in Z1 (Figure 12D, left). Cxcr4 and Npff were co-expressed in the neurons in the Z3 positional group. By contrast, the neurons in the Z3 positional group with high expression levels of Npff showed distinctively low expression levels of Dbh, with little correlation between the two genes in Z1 and Z2 neurons (Figure 12D, right). Cxcr4 is also known as the NPY3 receptor, suggesting that these neurons may be responsive to NPY+ sympathetic fibers projecting to the heart from the stellate ganglion. Npff has been shown to increase heart rate when acting on receptors in the rat heart, working synergistically with adrenergic signaling pathways (Allard et al., 1995). Using the mapping techniques discussed in the previous sections, we can also visualize the expression patterns of these genes within the context of their three-dimensional localization, making it possible for us to delve deeper into the relationship between their expression patterns and locations within the rat ICN (Figure 12F).
We evaluated the gene expression data for differences between the three Z positional groups using a one-way ANOVA with 85 degrees of freedom. Hierarchical clustering of samples using the genes with ANOVA p < 0.001 elucidated four subtypes of neurons (Figure 13A–D), with subtypes A and C being found primarily in the Z1 group, B in the Z2 group, and D in the Z3 group along with a number of samples from the Z1 group (Figures 13A and 13B). We analyzed the molecular patterns that underlie the spatially organized neuronal subtypes to identify correlated modules of gene expression and the combinations of modules that distinguish each neuronal subtype. For example, we see that comparing Npff to Cxcr4 expression largely delineates the different phenotypes. We saw that Npff and Cxcr4 greatly correlate with spatial position of the Z groupings, where the Z1 group shows a range of expression of Npff. When examining phenotypes, however, we see that phenotype A is largely composed of cells that have low expression of both Npff and Cxcr4, whereas phenotype C largely accounts for the cells that show high expression in Npff and low expression of Cxcr4. We also examined the distributions of neuropeptide and corresponding receptor gene expression across the four identified phenotypes. For example, expression patterns between galanin and its receptor, Galr1, show that most cells in the phenotype A express high levels of galanin, with a large subset also expressing high levels of Galr1, indicating that cells in phenotype A largely drive galanin signaling both through autocrine signaling and acting as the source for paracrine signaling, whereas cells from phenotypes A, B, and C act as the target for paracrine signaling. Most cells in phenotype D showed low expression for both Gal and Galr1 (Figures 13D and 13E). Examining expression patterns between thyrotropin-releasing hormone (Trh) and its receptor Trhr shows very different cell-cell signaling connectivity patterns when compared with those for galanin. Although a small set of cells from all four phenotypic groups show coexpression of both Trh and Trhr, the most noticeable pattern is that phenotype B has consistently high levels of Trh, but not Trhr, indicating that cells in phenotype B primarily act as the source for paracrine signaling between phenotype B and the other three identified phenotypes.
Discussion
Recapitulation of the Major Findings and Their Significance
In this article, we present a method to identify individual neurons in the cardiac ICN and comprehensively record the neurons' relative spatial positions in the heart at micron-scale resolution. Combined with the ability to extract these neurons for downstream applications, we can now understand the gene expression of individual neurons in the ICN in their spatial context. Together, we present an integrated pipeline for cardiac molecular phenotype data acquisition. Here, we also demonstrate the use of KESM (McCormick, 2002), on cardiac tissue for the purposes of making a cardiac atlas at a micron-level resolution. This process and approach can be expanded to other organ systems for the microscopic mapping of neuronal structures.
All these results were made possible by a team science approach involving four groups each providing distinct but necessary technology, as required by the SPARC Program. The approach demonstrated here should be applicable to other organs with intrinsic nervous systems. This approach can also be extended to human hearts in health and in heart failure, seeking neuromodulators involved in regulating heart function. Understanding the cellular and molecular processes governing innervation and the functional control of the myocardium in health and disease is the mechanistic basis for the development of neuraxial therapies to prevent sudden cardiac death and arrhythmias (Fukuda et al., 2015). The anatomical and molecular data are all archived in the DAT-CORE Portal of the SPARC Program for public access and use (Tables S1 and S2 and Figure S1). The inspiration for our approach comes from the Allen Institute Brain Atlas, which has been at the forefront of brain sectioning and 3D reconstruction (Lein et al., 2007). Creating a reference framework and mapping anatomical positions of neurons and molecular phenotypes is an ongoing and very active field in brain research, and its extension to the cardiac brain, ICN, benefits from it. Our contribution also resembles reports that provide a data scaffold to enable adding new studies, such as Hsu and Bhandawat (2016).
We find the rat ICN to be a bounded, relatively compact population within the context of the larger 3D anatomy of the heart. It is present on the hilum on the base of the heart, on both the left and right sides, as well as in the IAS. The majority of neurons are on the posterior or dorsal surface of both atria, but more prominently on the left side, extending to the coronary sulcus separating the atria from the ventricles. Within the ICN, neurons tend to cluster and to some extent appear to be continuously distributed in the superior-inferior direction. Quantitative analysis of neuronal packing density suggests several distinct anatomical groupings. The molecular phenotypes of these individual neurons are variable and diverse and reveal many previously unknown neuromodulatory regulators to be present. Spatial analysis suggests that many of these genes have specific gradients of expression, some of which is specifically associated with the neuronal packing density clusters, suggesting functional specificity by molecular phenotype and location within ICN.
Anatomical Mapping of Intrinsic Cardiac Neurons in the 3D Reconstructed Whole Heart
While qualitative and gross anatomical descriptions of the anatomy of the ICN have been presented, we here bring forth, at a cellular level, a comprehensive atlas, and 3D reconstruction of the ICN in the rat. This work represents a quantum leap in a long history of attempts to understand the anatomical substrate upon which the neuronal control of cardiac function is built. There has not yet been an integrative effort to generate a comprehensive digitized neurocardiac anatomical atlas for any species, or any effort to generate a histological foundation for molecular as well as functional mapping at the single-neuron level for the whole heart of any species.
Prior studies have attempted to map the intrinsic cardiac ganglia, the autonomic afferent and efferent nerve innervation of these cardiac ganglia, and cardiac myocardium. Commonly, immunohistochemical or immunofluorescent staining of a neuronal marker (acetylcholinesterase, tyrosine hydroxylase, choline acetyltransferase, PGP9.5, CGRP, SP) and a fluorescent tracer were used to examine sections, whole mounts, or whole hearts with microscopic or macroscopic organ imaging. Mapping efforts of ICN and cardiac nerve innervation in small animals have included mouse (Ai et al., 2007, Hoard et al., 2008, Li et al., 2010, Li et al., 2014, Rysevaite et al., 2011a, Rysevaite et al., 2011b), rat (Ai et al., 2007, Cheng et al., 1999, Cheng et al., 2004, Cheng and Powley, 2000, Pauza et al., 2000, Richardson et al., 2003), guinea pig (Hardwick et al., 2014, Pauza et al., 2000, Steele et al., 1994), and rabbit (Saburkina et al., 2014, Pauziene et al., 2016). Mapping of ICN was also performed in the hearts of larger animals and humans: dogs (Cardinal et al., 2009, Pauza et al., 2000, Singh et al., 2013, Xi et al., 1991), pigs (Arora et al., 2003, Pauza et al., 2014, Nakamura et al., 2016), and humans (Pauza et al., 2000, Petraitiene et al., 2014, Singh et al., 1996, Singh et al., 2013). The ICN in all these species is characterized and summarized by Wake and Brack (2016). With these efforts, either restricted anatomical regions in tissue sections or flat whole mounts were mapped at the microscopic level or large gross anatomical regions were mapped macroscopically.
Although these studies demonstrate the location of the ICN, the extrinsic cardiac innervation by afferent and autonomic nerves, and the intrinsic cardiac innervation, they cannot provide an anatomical substrate for integrating functional and molecular data with anatomically identified ICN neurons in their precise locations in the whole heart. In contrast, our work provides a model to precisely integrate anatomical and molecular data in the 3D digitally reconstructed whole heart with high resolution at the micron scale.
Recent technological advances in tissue clearing techniques make the heart transparent (Chung et al., 2013). Following tissue clearing, immunohistochemistry and various microscopy techniques can be applied to map ICN and extrinsic and intrinsic cardiac nerves. Confocal and two-photon imaging requires laser scanning, which is slow and hence practical for only small regions. ICN and extrinsic and intrinsic cardiac nerves may be beautifully imaged with light sheet microscopy, as demonstrated on the murine heart (Ding et al., 2018). This approach can present an ability to create an unbiased view of the myocardium at the single-cell level. However, there appears to be a trade-off between the physical width of the lightsheet (which can determine the axial resolution) and the size of the volume that can be imaged (Richardson and Lichtman, 2015). Hence, lightsheet microscopy with the highest resolution is limited to tissues that are a few hundred micrometers thick. In addition, the electrophoretic process in tissue clearing is not optimal for preservation of charged molecules, including DNA and RNA. Thus, our approach provides a much more comprehensive, reliable, and precise model to integrate anatomical, functional, and molecular data.
Molecular Heterogeneity of Intrinsic Cardiac Neurons: Functional Significance
The current understanding of ICN is that they modulate cardiac physiological functions of chronotropy, dromotropy, inotropy, and lusitropy. Several anatomical studies have hinted at the complexity of cardiac ganglia organization both in the cell body locations (Hasan, 2013, Hoover et al., 2009, Li et al., 2014, Pauziene et al., 2016, Rysevaite et al., 2011a, Rysevaite et al., 2011b, Saburkina et al., 2014) and the complexity of afferent and efferent projections to and from the ICN (Cheng et al., 2004, Cheng et al., 1999, Cheng and Powley, 2000, Lin et al., 2014, Li et al., 2014, Li et al., 2010). Although some work has hinted at the vast heterogeneity of the ICN from a functional perspective (Beaumont et al., 2013), there has not yet been a systematic assay capable of analyzing the molecular substrates that underlies this functional heterogeneity. For example, from a population perspective, it is exactly this type of heterogeneity that may determine the differences between patients who respond to vagal stimulation and those who do not. Furthermore, determination of the deficits in the ICN that drive cardiovascular pathology can lead to new avenues of therapy not only for patients with heart failure but also with an eye toward preventative approaches, as the transition from health to disease is better understood from the perspective of the ICN (Herring, 2015, Longpré et al., 2014). This work elucidates the transcriptional heterogeneity of anatomically positioned ICN in the rat heart, providing anatomical and molecular substrates for functional heterogeneity of cardiac ICNs.
The distinct molecular phenotypes that are defined along the base-to-apex axis have not been previously described. That such an organizational pattern exists may be a result of the embryological development of the intrinsic cardiac ganglia, predominantly from the migration of neural crest cells that differentiate into neurons. The migrating neural crest cells enter the heart at the base and spread out within the atria toward the apex (Fukiishi and Morriss-Kay, 1992, Hildreth et al., 2008). As in the brain, neurons in the heart likely follow chemical gradients to determine the direction and extent of migration, offering a possible explanation for why molecular phenotypes can be defined along the z axis and opening the door for further work to determine which molecules make up these gradients. That cardiac conduction tissues run generally from base to apex may also suggest a functional implication for the separation of neurons in this fashion. In mouse hearts, not all intrinsic cardiac ganglia originate from migrating neural crest cells, especially those that influence nodal pacemakers (Hildreth et al., 2008). Further development of more comprehensive profiling of single neurons within their anatomical context will provide important information as to the role of both neural crest and non-neural crest neurons in the heart.
Data Analysis and Integration Framework
To piece together the relationship between 3D histological reconstructions and single-cell molecular data, data storage and annotation must be carefully considered. The use of an anatomical ontology not only provides a means by which neurons can be anatomically referred to but also makes it so that data from different hearts can be more readily compared. The generation of such data storage and annotation is not a trivial matter and requires collaboration between technicians, researchers, software developers, and data storage specialists. Although purely mathematical and statistical methods for interpreting these data are crucial, the complexity quickly gets to a point where such approaches are challenging, making the ability to qualitatively interpret the data essential. To this end, MBF Bioscience has designed custom software, TissueMaker and Tissue Mapper, allowing for visualization of molecular and anatomical patterns. All processed and raw data are available via the SPARC Data Portal (https://data.sparc.science). The data structure adheres to the SPARC data format and is curated by the SPARC data curation team to align the data to community ontologies, to ensure data integrity, and to extract the Minimal Information Standard metadata.
The standardized data will enable mapping onto a 3D scaffold via a common coordinate framework to be used as a reference atlas for comparison across animals as well as across species and experimental conditions. These efforts are presently ongoing in coordination with the MAPCORE group of the SPARC program.
Future Directions for Scalability and Extension
We have established approaches that will now support scaling to acquire several male and female ICN providing a 3D framework as a foundational data resource, useful for developing detailed anatomical neural circuit/connectomic maps for neural control of the heart and showing 3D distribution/gradients of molecular phenotypes. This granularity in the understanding of cardiac neurons has the potential to unlock a new generation of cardiac therapeutics to treat or prevent all forms of cardiac pathology. To achieve the desired integration of different data types, the data from other approaches are visualized within the ICN of the 3D framework. The demonstration of cross-species application of the approach supports scaling to organisms with larger and more complex hearts such as pig and human, and to extension to other organs.
Limitations of the Study
-
•
In the anatomical representations there is some interobserver variability in the manual segmenting. The borders of these anatomic structures depend to some extent on subjective interpretation.
-
•
The single-neuron LCM molecular data collected provide limited, preliminary findings on gradients, heterogeneity, or anatomic patterning. They rest on reliable, replicable multiplexed RT-qPCR, providing substantial but less-than-ideal numbers of samples and depth of gene expression data.
-
•
At this time, single-cell RNA sequencing (RNA-seq) from laser captured cells (regardless of laser capture method) falls short of producing reliable and replicable data. Other single-cell RNA-seq technologies, such as 10×, drop-seq, nuc-seq, and others, necessarily forgo anatomic specificity. As of this writing complex transcriptomic profiling techniques like MERFISH are in their proof-of-concept phases and were not accessible for this work.
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, James Schwaber (james.schwaber@jefferson.edu).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
Knife-edge scanning microscopy and 3D reconstructed rat heart data: https://doi.org/10.26275/WOX9-GQZP.
Laser capture microdissection imaging and molecular profile data: https://doi.org/10.26275/XMYX-RNM9.
Knife-edge scanning microscopy (KESM): Strateos, San Francisco, CA: https://www.strateos.com/kesm/
Tissue Mapper, Tissue Maker, Biolucida: MBF Bioscience, Williston, VT: https://www.mbfbioscience.com/tissue-mapper, https://www.mbfbioscience.com/tissuemaker, https://www.mbfbioscience.com/biolucida.
Original data have been deposited to SPARC DAT-CORE Data Portal: https://sparc.science/data?type=dataset&q=heart.
MAPCORE group of the SPARC program: https://mapcore-documentation.readthedocs.io/en/latest/RatHeart.html.
SciCrunch database: https://scicrunch.org/.
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
We would like to thank Dr. Mahyar Osanlouy of the Auckland Bioengineering Institute for ongoing efforts in the process of fitting segmented data to the heart scaffold. Financial support for this work was provided by the National Institutes of Health under the Stimulating Peripheral Activity to Relieve Conditions (SPARC) program, Grant OT2 OD023848 subaward, NHLBI Grant U01HL133360 to J.S.S. and R.V., NIH R15 1R15HL137143-01A1, and NIH 1 U01 NS113867-01 to J.C. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Author Contributions
Conceptualization, J.S.S., R.V., Z.C., S.T., and J.G.; Methodology, J.S.S., R.V., S.T., N.F., T.H., S.E., Z.C., and J.G.; Software, S.T. and M.H.; Validation, Z.C., C.L., J.C., and L.E.; Formal Analysis, J.G., S.A., A.M., and R.V.; Investigation, J.G., S.A., C.L., S.R., and J.C.; Resources, S.T., M.H., N.F., T.H., and S.E.; Data Curation, J.S.S., A.M., and R.V.; Writing – Original Draft, J.S.S. and J.G.; Writing – Review and Editing, J.S.S., J.G., and R.V.; Visualization, C.L., A.M., S.R., S.T., M.H., and S.E.; Supervision, J.S.S., R.V., and Z.C.; Project Administration, J.S.S.; Funding Acquisition, J.S.S. and R.V.
Declaration of Interests
S.T. and M.H. are paid employees of MBF Bioscience (Williston, VT). S.T. and M.H. are also funded by the NIH Common Fund award, OT3OD025349 to Dr. Shivkumar at University of California Los Angeles (subaward to J.S.S., R.V., J.C.), to create multi-scale, multi-organ, multi-species SPARC map management as a part of SPARC Portal. The software development efforts described in this manuscript preceded integration with SPARC DRC. Owing to intellectual property right restrictions, we cannot provide the Tissue Mapper, Tissue Maker or Biolucida Converter source code or its documentation at this time.
Strateos and MBF Bioscience are commercial entities, and the authors affiliated with them are company employees. The remaining authors declare that no competing interests exist.
Published: June 26, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.isci.2020.101140.
Contributor Information
Zixi (Jack) Cheng, Email: zixi.cheng@ucf.edu.
Rajanikanth Vadigepalli, Email: rajanikanth.vadigepalli@jefferson.edu.
James S. Schwaber, Email: james.schwaber@jefferson.edu.
Supplemental Information
This file also details the metadata such as the X, Y, Z coordinates, slide number, and cell count per sample
References
- Ai J., Gozal D., Li L., Wead W.B., Chapleau M.W., Wurster R., Yang B., Li H., Liu R., Cheng Z. Degeneration of vagal efferent axons and terminals in cardiac ganglia of aged rats. J. Comp. Neurol. 2007;504:74–88. doi: 10.1002/cne.21431. [DOI] [PubMed] [Google Scholar]
- Allard M., Labrouche S., Nosjean A., Laguzzi R. Mechanisms underlying the cardiovascular responses to peripheral administration of NPFF in the rat. J. Pharmacol. Exp. Ther. 1995;274:577–583. [PubMed] [Google Scholar]
- Armour J.A., Murphy D.A., Yuan B.X., Macdonald S., Hopkins D.A. Gross and microscopic anatomy of the human intrinsic cardiac nervous system. Anat. Rec. 1997;247:289–298. doi: 10.1002/(SICI)1097-0185(199702)247:2<289::AID-AR15>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Arora R.C., Cardinal R., Smith F.M., Ardell J.L., Dell’Italia L.J., Armour J.A. Intrinsic cardiac nervous system in tachycardia induced heart failure. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2003;285:R1212–R1223. doi: 10.1152/ajpregu.00131.2003. [DOI] [PubMed] [Google Scholar]
- Beaumont E., Salavatian S., Southerland E.M., Vinet A., Jacquemet V., Armour J.A., Ardell J.L. Network interactions within the canine intrinsic cardiac nervous system: implications for reflex control of regional cardiac function. J. Physiol. (Lond) 2013;591:4515–4533. doi: 10.1113/jphysiol.2013.259382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardinal R., Pagé P., Vermeulen M., Ardell J.L., Armour J.A. Spatially divergent cardiac responses to nicotinic stimulation of ganglionated plexus neurons in the canine heart. Auton. Neurosci. 2009;145:55–62. doi: 10.1016/j.autneu.2008.11.007. [DOI] [PubMed] [Google Scholar]
- Cheng Y.-F., Chang Y.-T., Chen W.-H., Shih H.-C., Chen Y.-H., Shyu B.-C., Chen C.-C. Cardioprotection induced in a mouse model of neuropathic pain via anterior nucleus of paraventricular thalamus. Nat. Commun. 2017;8:826. doi: 10.1038/s41467-017-00891-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z., Powley T.L. Nucleus ambiguus projections to cardiac ganglia of rat atria: an anterograde tracing study. J. Comp. Neurol. 2000;424:588–606. [PubMed] [Google Scholar]
- Cheng Z., Powley T.L., Schwaber J.S., Doyle F.J. Projections of the dorsal motor nucleus of the vagus to cardiac ganglia of rat atria: an anterograde tracing study. J. Comp. Neurol. 1999;410:320–341. [PubMed] [Google Scholar]
- Cheng Z., Zhang H., Guo S.Z., Wurster R., Gozal D. Differential control over postganglionic neurons in rat cardiac ganglia by NA and DmnX neurons: anatomical evidence. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2004;286:R625–R633. doi: 10.1152/ajpregu.00143.2003. [DOI] [PubMed] [Google Scholar]
- Chung K., Wallace J., Kim S.Y., Kalyanasundaram S., Andalman A.S., Davidson T.J., Mirzabekov J.J., Zalocusky K.A., Mattis J., Denisin A.K. Structural and molecular interrogation of intact biological systems. Nature. 2013;497:332–337. doi: 10.1038/nature12107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crick S.J., Wharton J., Sheppard M.N., Royston D., Yacoub M.H., Anderson R.H., Polak J.M. Innervation of the human cardiac conduction system. A quantitative immunohistochemical and histochemical study. Circulation. 1994;89:1697–1708. doi: 10.1161/01.cir.89.4.1697. [DOI] [PubMed] [Google Scholar]
- Ding Y., Bailey Z., Messerschmidt V., Nie J., Bryant R., Rugonyi S., Fei P., Lee J., Hsiai T.K. Light-sheet fluorescence microscopy for the study of the murine heart. J. Vis. Exp. 2018 doi: 10.3791/57769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukiishi Y., Morriss-Kay G.M. Migration of cranial neural crest cells to the pharyngeal arches and heart in rat embryos. Cell Tissue Res. 1992;268:1–8. doi: 10.1007/BF00338048. [DOI] [PubMed] [Google Scholar]
- Fukuda K, Kanazawa H, Aizawa Y, Ardell J.L., Shivkumar K. Cardiac innervation and sudden cardiac death. Circ. Res. 2015;116:2005–2019. doi: 10.1161/CIRCRESAHA.116.304679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardwick J.C., Ryan S.E., Beaumont E., Ardell J.L., Southerland E.M. Dynamic remodeling of the Guinea pig intrinsic cardiac plexus induced by chronic myocardial infarction. Auton. Neurosci. 2014;181:4–12. doi: 10.1016/j.autneu.2013.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan W. Autonomic cardiac innervation: development and adult plasticity. Organogenesis. 2013;9:176–193. doi: 10.4161/org.24892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herring N. Autonomic control of the heart: going beyond the classical neurotransmitters. Exp. Physiol. 2015;100:354–358. doi: 10.1113/expphysiol.2014.080184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildreth V., Webb S., Bradshaw L., Brown N.A., Anderson R.H., Henderson D.J. Cells migrating from the neural crest contribute to the innervation of the venous pole of the heart. J. Anat. 2008;212:1–11. doi: 10.1111/j.1469-7580.2007.00833.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoard J.L., Hoover D.B., Mabe A.M., Blakely R.D., Feng N., Paolocci N. Cholinergic neurons of mouse intrinsic cardiac ganglia contain noradrenergic enzymes, norepinephrine transporters, and the neurotrophin receptors tropomyosin-related kinase A and p75. Neuroscience. 2008;156:129–142. doi: 10.1016/j.neuroscience.2008.06.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoover D.B., Isaacs E.R., Jacques F., Hoard J.L., Pagé P., Armour J.A. Localization of multiple neurotransmitters in surgically derived specimens of human atrial ganglia. Neuroscience. 2009;164:1170–1179. doi: 10.1016/j.neuroscience.2009.09.001. [DOI] [PubMed] [Google Scholar]
- Hsu C.T., Bhandawat V. Organization of descending neurons in Drosophila melanogaster. Sci. Rep. 2016;6:20259. doi: 10.1038/srep20259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman L., Rousseeuw P.J. Clustering by means of medoids. In: Dodge Y., editor. Statistical Data Analysis Based on the L1-Norm and Related Methods. North Holland Publishing Company; 1987. pp. 405–416. [Google Scholar]
- Lein E.S., Hawrylycz M.J., Ao N., Ayres M., Bensinger A., Bernard A., Boe A.F., Boguski M.S., Brockway K.S., Byrnes E.J. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- Lin M., Hatcher J.T., Wurster R.D., Chen Q.-H., Cheng Z.J. Characteristics of single large-conductance Ca2+-activated K+ channels and their regulation of action potentials and excitability in parasympathetic cardiac motoneurons in the nucleus ambiguus. Am. J. Physiol. Cell Physiol. 2014;306:C152–C166. doi: 10.1152/ajpcell.00423.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Hatcher J.T., Hoover D.B., Gu H., Wurster R.D., Cheng Z.J. Distribution and morphology of calcitonin gene-related peptide and substance P immunoreactive axons in the whole-mount atria of mice. Auton. Neurosci. 2014;181:37–48. doi: 10.1016/j.autneu.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Huang C., Ai J., Yan B., Gu H., Ma Z., Li A.Y., Xinyan S., Harden S.W., Hatcher J.T. Structural remodeling of vagal afferent innervation of aortic arch and nucleus ambiguus (NA) projections to cardiac ganglia in a transgenic mouse model of type 1 diabetes (OVE26) J. Comp. Neurol. 2010;518:2771–2793. doi: 10.1002/cne.22363. [DOI] [PubMed] [Google Scholar]
- Longpré J.-P., Salavatian S., Beaumont E., Armour J.A., Ardell J.L., Jacquemet V. Measure of synchrony in the activity of intrinsic cardiac neurons. Physiol. Meas. 2014;35:549–566. doi: 10.1088/0967-3334/35/4/549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick D.A. Cortical and subcortical generators of normal and abnormal rhythmicity. Int. Rev. Neurobiol. 2002;49:99–114. doi: 10.1016/s0074-7742(02)49009-5. [DOI] [PubMed] [Google Scholar]
- Nakamura K., Ajijola O.A., Aliotta E., Armour J.A., Ardell J.L., Shivkumar K. Pathological effects of chronic myocardial infarction on peripheral neurons mediating cardiac neurotransmission. Auton. Neurosci. 2016;197:34–40. doi: 10.1016/j.autneu.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauza D.H., Rysevaite K., Inokaitis H., Jokubauskas M., Pauza A.G., Brack K.E., Pauziene N. Innervation of sinoatrial nodal cardiomyocytes in mouse. A combined approach using immunofluorescent and electron microscopy. J. Mol. Cell. Cardiol. 2014;75:188–197. doi: 10.1016/j.yjmcc.2014.07.016. [DOI] [PubMed] [Google Scholar]
- Pauza D.H., Skripka V., Pauziene N., Stropus R. Morphology, distribution, and variability of the epicardiac neural ganglionated subplexuses in the human heart. Anat. Rec. 2000;259:353–382. doi: 10.1002/1097-0185(20000801)259:4<353::AID-AR10>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Pauziene N., Alaburda P., Rysevaite-Kyguoliene K., Pauza A.G., Inokaitis H., Masaityte A., Rudokaite G., Saburkina I., Plisiene J., Pauza D.H. Innervation of the rabbit cardiac ventricles. J. Anat. 2016;228:26–46. doi: 10.1111/joa.12400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petraitiene V., Pauza D.H., Benetis R. Distribution of adrenergic and cholinergic nerve fibres within intrinsic nerves at the level of the human heart hilum. Eur. J. Cardiothorac. Surg. 2014;45:1097–1105. doi: 10.1093/ejcts/ezt575. [DOI] [PubMed] [Google Scholar]
- Richardson D.S., Lichtman J.W. Clarifying tissue clearing. Cell. 2015;162:246–257. doi: 10.1016/j.cell.2015.06.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson R.J., Grkovic I., Anderson C.R. Immunohistochemical analysis of intracardiac ganglia of the rat heart. Cell Tissue Res. 2003;314:337–350. doi: 10.1007/s00441-003-0805-2. [DOI] [PubMed] [Google Scholar]
- Rysevaite K., Saburkina I., Pauziene N., Noujaim S.F., Jalife J., Pauza D.H. Morphologic pattern of the intrinsic ganglionated nerve plexus in mouse heart. Heart Rhythm. 2011;8:448–454. doi: 10.1016/j.hrthm.2010.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rysevaite K., Saburkina I., Pauziene N., Vaitkevicius R., Noujaim S.F., Jalife J., Pauza D.H. Immunohistochemical characterization of the intrinsic cardiac neural plexus in whole-mount mouse heart preparations. Heart Rhythm. 2011;8:731–738. doi: 10.1016/j.hrthm.2011.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saburkina I., Gukauskiene L., Rysevaite K., Brack K.E., Pauza A.G., Pauziene N., Pauza D.H. Morphological pattern of intrinsic nerve plexus distributed on the rabbit heart and interatrial septum. J. Anat. 2014;224:583–593. doi: 10.1111/joa.12166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S., Johnson P.I., Lee R.E., Orfei E., Lonchyna V.A., Sullivan H.J., Montoya A., Tran H., Wehrmacher W.H., Wurster R.D. Topography of cardiac ganglia in the adult human heart. J. Thorac. Cardiovasc. Surg. 1996;112:943–953. doi: 10.1016/S0022-5223(96)70094-6. [DOI] [PubMed] [Google Scholar]
- Singh S., Sayers S, Walter JS, Thomas D, Dieter RS, Nee LM, Wurster RD. Hypertrophy of neurons within cardiac ganglia in human, canine, and rat heart failure: the potential role of nerve growth factor. J. Am. Heart Assoc. 2013;2:e000210. doi: 10.1161/JAHA.113.000210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele P.A., Gibbins I.L., Morris J.L., Mayer B. Multiple populations of neuropeptide-containing intrinsic neurons in the Guinea-pig heart. Neuroscience. 1994;62:241–250. doi: 10.1016/0306-4522(94)90327-1. [DOI] [PubMed] [Google Scholar]
- Wake E., Brack K. Characterization of the intrinsic cardiac nervous system. Auton. Neurosci. 2016;199:3–16. doi: 10.1016/j.autneu.2016.08.006. [DOI] [PubMed] [Google Scholar]
- Xi X., Randall W.C., Wurster R.D. Morphology of intracellularly labeled canine intracardiac ganglion cells. J. Comp. Neurol. 1991;314:396–402. doi: 10.1002/cne.903140213. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Cardiac features have been contoured and colored accordingly, with mapped neurons marked in yellow. Partial projection of the neurons at the base of the heart are also shown.
Visualization of neuronal clusters based on packing density by use of partitioning around medoids (PAM) algorithm. Clusters are shown as a flat map projection as well as their mapped locations within the ICN.
Neurons are colored based off of clusters identified through PAM algorithm as described.
This file also details the metadata such as the X, Y, Z coordinates, slide number, and cell count per sample
Data Availability Statement
Knife-edge scanning microscopy and 3D reconstructed rat heart data: https://doi.org/10.26275/WOX9-GQZP.
Laser capture microdissection imaging and molecular profile data: https://doi.org/10.26275/XMYX-RNM9.
Knife-edge scanning microscopy (KESM): Strateos, San Francisco, CA: https://www.strateos.com/kesm/
Tissue Mapper, Tissue Maker, Biolucida: MBF Bioscience, Williston, VT: https://www.mbfbioscience.com/tissue-mapper, https://www.mbfbioscience.com/tissuemaker, https://www.mbfbioscience.com/biolucida.
Original data have been deposited to SPARC DAT-CORE Data Portal: https://sparc.science/data?type=dataset&q=heart.
MAPCORE group of the SPARC program: https://mapcore-documentation.readthedocs.io/en/latest/RatHeart.html.
SciCrunch database: https://scicrunch.org/.