ABSTRACT
Cell ablation is a powerful method for elucidating the contributions of individual cell populations to embryonic development and tissue regeneration. Targeted cell loss in whole organisms has been typically achieved through expression of a cytotoxic or prodrug-activating gene product in the cell type of interest. This approach depends on the availability of tissue-specific promoters, and it does not allow further spatial selectivity within the promoter-defined region(s). To address this limitation, we have used the light-inducible GAVPO transactivator in combination with two genetically encoded cell-ablation technologies: the nitroreductase/nitrofuran system and a cytotoxic variant of the M2 ion channel. Our studies establish ablative methods that provide the tissue specificity afforded by cis-regulatory elements and the conditionality of optogenetics. Our studies also demonstrate differences between the nitroreductase and M2 systems that influence their efficacies for specific applications. Using this integrative approach, we have ablated cells in zebrafish embryos with both spatial and temporal control.
KEY WORDS: Optogenetics, Nitroreductase, M2 ion channel, Cell ablation, Neural injury, Zebrafish
Summary: Combining the light-inducible GAVPO transactivator with a cytotoxic channel enables robust cell ablation in zebrafish embryos with spatial and temporal precision.
INTRODUCTION
Tissue development, homeostasis and regeneration rely on functional networks composed of diverse cell types. To probe these mechanisms, several genetically encoded technologies have been developed to achieve cell ablation with spatiotemporal control, including cytotoxic proteins (Chelur and Chalfie, 2007; Kurita et al., 2003; Slanchev et al., 2005; Smith et al., 2002, 2007) and enzymes that can covert prodrugs into cytotoxic molecules (Bridgewater et al., 1995; Chu et al., 2007; Curado et al., 2007; Jung et al., 2007; Pisharath et al., 2007; Springer and Niculescu-Duvaz, 2000). Both of these approaches can be combined with tissue-specific promoters to spatially restrict ablation; however, cis-regulatory sequences that are exclusive to the targeted cells are not always available, resulting in collateral damage to other tissues. For example, nitroreductase and nitrofuran-based toxins have been used to ablate motor neurons in zebrafish larvae (Ohnmacht et al., 2016), providing an alternative to mechanically induced spinal cord lesions (Becker and Becker, 2001; Briona et al., 2015; Mokalled et al., 2016; Reimer et al., 2009; Yu et al., 2011). Although this chemical-genetic technique was able to elicit regenerative responses in the central nervous system (CNS), the promoter used for these studies also led to nitroreductase expression in the heart and pancreas. As result, metronidazole treatment also induced heart damage and edema (Ohnmacht et al., 2016).
Light-inducible technologies have the potential to achieve targeted cell ablation in a more precise manner. Toward that goal, chromophores that generate reactive oxygen species (ROS) upon exposure to light have been applied in various systems (Buckley et al., 2017; Bulina et al., 2006; Makhijani et al., 2017; Qi et al., 2012; Sarkisyan et al., 2015; Xu and Chisholm, 2016). However, these methods can require sustained irradiation for efficient cell ablation, limiting their ability to target specific cell populations in dynamic systems, and ROS sensitivity also varies between cell types (Williams et al., 2013). We reasoned that light-inducible expression of cytotoxic proteins or toxin-activating enzymes might represent a more general and versatile approach. In particular, we envisioned this could be achieved using a photoactivatable transcription factor that can drive the expression of exogenous transgenes. One candidate construct for this strategy is a synthetic transcription factor, GAVPO, that was originally developed for light-inducible gene expression in mammalian systems (Wang et al., 2012). The GAVPO transactivator contains a Gal4 DNA-binding domain, a variant of the VIVID light-oxygen-voltage (LOV) domain and the p65 transcriptional activation domain. Upon exposure to blue light, the LOV domain undergoes a photo-reductive reaction that forms a covalent adduct between its flavin adenine dinucleotide chromophore and a neighboring cysteine residue. The adduct stabilizes a conformational state that promotes LOV domain self-association, generating a transcriptionally active GAVPO dimer. Light-activated GAVPO then drives the expression of upstream activating sequence (UAS)-controlled genes until it reverts to the inactive monomer with a half-life of ∼2 h (Wang et al., 2012).
In this report, we investigate the efficacy of light-inducible cell ablation in zebrafish using two genetically encoded cell-ablation technologies and the GAVPO transactivator. We demonstrate that the cytotoxic ion channel variant M2H37A (Lam et al., 2010; Smith et al., 2002, 2007) acts through non-apoptotic pathways to kill neurons and other cell types with greater efficacy and faster kinetics than the nitroreductase/nitrofuran system. We further establish GAVPO as an effective tool for achieving photoactivatable gene transcription in zebrafish, using both embryos injected with GAVPO mRNA and stable transgenic lines with neuronal expression of the transactivator. Finally, we show that integrating the GAVPO and M2H37A technologies enables targeted cell death in zebrafish embryos and larvae. We anticipate that optogenetic cell ablation will be a versatile approach for studying developmental and regenerative biology.
RESULTS
Genetically encoded neuron ablation in zebrafish
To develop conditional models of CNS damage, we pursued neuronal ablation through two genetically encoded technologies: bacterial nitroreductase (NTR) and viral ion channel M2. The Escherichia coli gene, nsfB, encodes a flavoprotein that can reduce a variety of nitroaromatic compounds (Zenno et al., 1996). Among the substrates of this nitroreductase (NTR) is the prodrug metronidazole, which is reductively converted into a DNA interstrand crosslinking agent that triggers cell death. M2 normally functions as a proton-selective channel during influenza infection (Shimbo et al., 1996; Wharton et al., 1994), and a single point mutation in the transmembrane domain (H37A) converts M2 into a constitutively active, non-selective cation channel (Gandhi et al., 1999; Wang et al., 1995). This loss of specificity renders M2H37A cytotoxic (Lam et al., 2010; Le Tissier et al., 2005; Smith et al., 2002, 2007). The availability of M2-targeting antiviral drugs, such as rimantadine and amantadine (Intharathep et al., 2008; Schnell and Chou, 2008), also enables pharmacological control of M2H37A function.
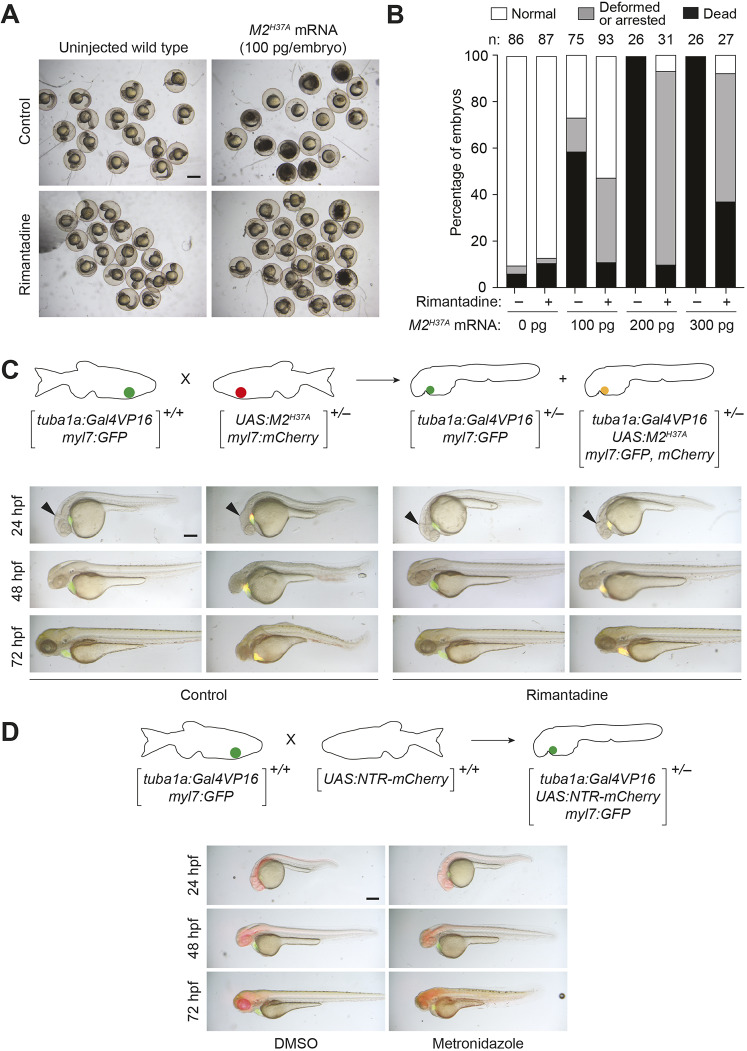
Exogenous expression of NTR in combination with metronidazole has been widely used to selectively ablate cells in zebrafish (Curado et al., 2007, 2008; Pisharath and Parsons, 2009; Pisharath et al., 2007). However, the M2H37A channel has not been applied previously in this model organism. We therefore injected zebrafish embryos with varying amounts of M2H37A mRNA and monitored their embryonic development. Toxicity was initially observed at the blastula stages, and the embryos were unable to proceed through gastrulation (Fig. 1A,B). Injecting increasing amounts of M2H37A mRNA rapidly killed 100% of the embryos. Culturing the M2H37A-expressing embryos in the presence of 100 µg/ml rimantadine enabled some to develop normally until 24 h post fertilization (hpf). However, the channel blocker was unable to fully protect against channel-mediated toxicity, demonstrating the need to control M2H37A expression.
Fig. 1.
M2H37A channel variant is toxic to zebrafish embryos and ablates neurons more rapidly than NTR. (A) Wild-type zygotes were injected with the designated amount of M2H37A mRNA and cultured in the absence or presence of 100 μg/ml rimantadine. Representative bright-field micrographs of 24 hpf embryos from two independent experiments are shown. Scale bar: 500 μm. (B) Phenotypic distributions of embryos injected with the designated amount of M2H37A mRNA. The embryos were scored at 24 hpf, and the number of embryos (n) per condition from two or three independent experiments is indicated. Statistical analyses: χ2=389, d.f.=14, P<0.00001. (C) Comparison of Tg(tuba1a:Gal4VP16;myl7:GFP) and Tg(tuba1a:Gal4VP16; UAS:M2H37A;myl7:gfp,mCherry) embryos cultured in the absence or presence of 100 µg/ml rimantadine. Neuronal expression of M2H37A induces loss of the midbrain-hindbrain boundary (arrowheads) at 24 hpf and increasingly severe CNS deficits as development continues. Treatment with rimantadine starting at 10 hpf rescues these neuronal defects in Tg(tuba1a:Gal4VP16;UAS:M2H37A;myl7:gfp,mCherry) embryos. Representative bright-field and epifluorescence micrographs (overlays) from three independent experiments are shown. (D) Comparison of Tg(tuba1a:Gal4VP16;UAS:NTR-mCherry;myl7:GFP) embryos cultured with 5 mM metronidazole or an equivalent amount of DMSO, starting at 10 hpf. Metronidazole-treated embryos first exhibit CNS defects at 48 hpf. Representative bright-field and epifluorescence micrographs (overlays) from two independent experiments are shown. All embryos are shown in lateral view, anterior left. Scale bars: 250 µm. Statistics for the observed phenotypes are in Table S1.
We next examined whether M2H37A can be used to ablate zebrafish neurons by establishing a system for Gal4/UAS-dependent expression in these cells. We generated heterozygous Tg(UAS:M2H37A;myl7:mCherry) zebrafish, using five tandem UAS sequences to minimize basal M2H37A expression (Ma et al., 2013), and crossed them with a homozygous Tg(tuba1a:Gal4VP16;myl7:GFP) line, which restricts Gal4VP16 expression to the developing nervous system (Goldman et al., 2001). We then selected progeny that contained one copy of each transgene, which could be confirmed by the presence of both GFP and mCherry fluorescence in the heart (driven by the myl7 promoter of each construct) (Fig. 1C). The double transgenic embryos expressed M2H37A in neural tissues (Fig. S1), and they exhibited CNS deficits that appeared as early as 24 hpf and progressed over time (Fig. 1C). Rimantadine rescued these defects, confirming the role of M2H37A channel activity in neuronal loss (Fig. 1C).
To compare the ablative efficacies of the M2H37A and NTR/metronidazole systems, we also crossed homozygous Tg(UAS:NTR-mCherry) (Goldman et al., 2001) and homozygous Tg(tuba1a:Gal4VP16;myl7:GFP) zebrafish, and cultured the resulting embryos in medium supplemented with DMSO or 5 mM metronidazole starting at bud stage (10 hpf). In contrast to zebrafish with neuronal M2H37A expression, metronidazole treatment of the NTR-expressing progeny did not induce CNS defects until 48 hpf, suggesting the two ablative technologies kill neural cells with different kinetics and possibly through distinct mechanisms (Fig. 1D). We next used TUNEL staining to detect apoptosis in M2H37A- and NTR-expressing embryos (Fig. S2A). Although we observed overt developmental defects as early as 24 hpf in M2H37A-expressing embryos, significant TUNEL staining was not detected until 32 hpf, and TUNEL-positive cells were distributed in regions within and near the CNS. Because TUNEL staining detects DNA fragmentation that occurs later in the apoptotic process, we also stained the embryos for activated caspase 3, the initiator caspase for apoptosis. Similar to our TUNEL results, we did not observe activated caspase 3 until 32 hpf (Fig. S2B). In comparison, NTR-expressing embryos treated with 5 mM metronidazole first exhibited phenotypic defects at 48 hpf, which coincided with robust TUNEL staining localized to the CNS (Fig. S2A). Thus, M2H37A appears to kill zebrafish neurons through necrosis, which, when continuously expressed in the developing CNS, subsequently causes the apoptosis of surrounding cells. This progressive mechanism of tissue loss is similar to that associated with CNS damage (Oyinbo, 2011), potentially making M2H37A a useful tool for studying neural injury and recovery.
Light-inducible cell ablation using transient GAVPO expression
Using cytotoxic proteins to ablate specific cell populations, including regions within a tissue of interest, requires spatiotemporal control of their expression or function. To accomplish this goal, we pursued light-dependent NTR and M2H37A expression in zebrafish embryos and larvae. A number of strategies for photoactivatable gene expression have been described, including GAVPO (Fig. 2A), EL222, an EL222-derived construct (TAEL) and two-hybrid-like systems based on CYR2-CIB1 or PhyB-PIF3 heterodimerization (Liu et al., 2012; Motta-Mena et al., 2014; Reade et al., 2017; Shimizu-Sato et al., 2002). Of these technologies, EL222, TAEL and CYR2-CIB1have been applied previously in zebrafish, but they are limited by the minutes-scale lifetimes of their light-activated states. The PhyB-PIF3 system requires co-administration of phycocyanobilin, an exogenous chromophore that cannot penetrate zebrafish embryos (Buckley et al., 2016). As GAVPO uses endogenous flavin and has a light-state half-life of 2 h, we envisioned that this optogenetic tool could afford transcriptional control with transient illumination, enabling genetically encoded ablation of dynamic cell populations.
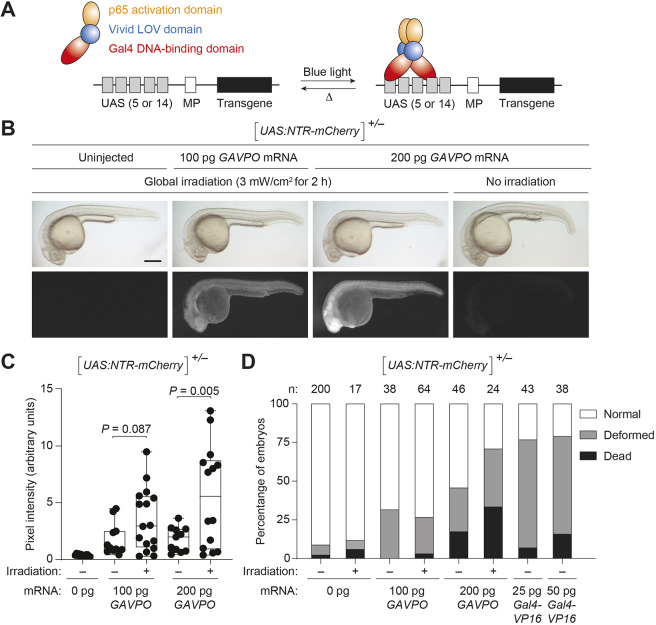
Fig. 2.
GAVPO conveys light-inducible gene expression in zebrafish. (A) Mechanism of GAVPO-dependent transcription of a transgene with an upstream activating sequence (UAS) and minimal promoter (MP). (B) Tg(UAS:NTR-mCherry) zygotes were injected with the designated amounts of GAVPO mRNA, irradiated with a blue LED lamp at 6 hpf, and then imaged at 24 hpf. Representative bright-field and epifluorescence micrographs from two to four independent experiments are shown. GAVPO induces NTR-mCherry expression in a light- and concentration-dependent manner. Scale bar: 250 µm. (C) Quantification of mCherry fluorescence in Tg(UAS:NTR-mCherry) zygotes were injected with the designated amounts of GAVPO mRNA, as described in B. The box extends from the 25th to 75th percentiles. The whiskers extend to the minimum and maximum values. The horizontal line indicates the average value. (D) The designated amounts of GAVPO or Gal4VP16 mRNA were injected into Tg(UAS:NTR-mCherry) zygotes, and the embryos were either cultured in the dark or globally exposed to blue LED light (3 mW/cm2) from 6 to 8 hpf. Developmental phenotypes were then scored at 24 hpf, using the indicated number of embryos (n) per condition from two to four independent experiments. Statistical analyses: χ2=190, d.f.=14, P<0.00001. Statistics for the observed phenotypes are in Table S2.
We assessed GAVPO activity in Tg(UAS:NTR-mCherry) zebrafish by injecting transgenic zygotes with various amounts of GAVPO mRNA (Fig. 2B). The embryos were then raised in the dark until 6 hpf, after which half were exposed to blue light (470 nm) from a light-emitting diode (LED) lamp for 2 h. NTR-mCherry levels were subsequently assessed by fluorescence microscopy at 24 hpf. GAVPO expression induced ubiquitous NTR-mCherry expression in a light- and dose-dependent manner (Fig. 2B,C), and the majority of animals developed normally when the GAVPO mRNA was injected at a dose of 100 pg/embryo (Fig. 2D). At 200 pg doses, GAVPO mRNA led to embryonic deformity or lethality. In comparison, Gal4VP16 mRNA caused developmental defects even when only 25 pg was injected per embryo, corroborating previous reports of Gal4VP16 toxicity (Köster and Fraser, 2001; Scott et al., 2007) (Fig. 2D). Taken together, these findings indicate that the GAVPO system can be used to convey light-dependent gene expression in multiple zebrafish cell types and may provide a less-toxic alternative to the currently available Gal4 driver lines.
We next examined whether GAVPO can be coupled with targeted illumination to achieve both spatial and temporal control of gene expression. We injected heterozygous Tg(UAS:NTR-mCherry) zygotes with GAVPO mRNA and irradiated the embryonic shield at 6 hpf for 20 s, using an epifluorescence microscope equipped with a 470/40 nm filter, a 20× objective and an iris diaphragm. Zebrafish fate-mapping studies have shown that cells within this dorsal organizing center give rise to the notochord, head mesoderm and hatching gland (Kimmel et al., 1990; Shih and Fraser, 1996), and we observed red fluorescence in these tissues at later developmental stages (Fig. 3A). To determine whether the expressed NTR-mCherry can impart nitrofuran sensitivity, we treated a cohort of the shield-irradiated embryos with either 5 mM metronidazole or DMSO vehicle alone from 10 to 32 hpf. We then fixed and immunostained the embryos for activated caspase 3. Although basal levels of caspase 3 activation increased with metronidazole treatment, we did not observe any evidence of GAVPO/NTR-mediated apoptosis in the notochord (Fig. S2C). Thus, even levels of NTR-mCherry expression that can be readily detected by fluorescence microscopy are not sufficient to convey nitrofuran-induced apoptosis within this time frame.
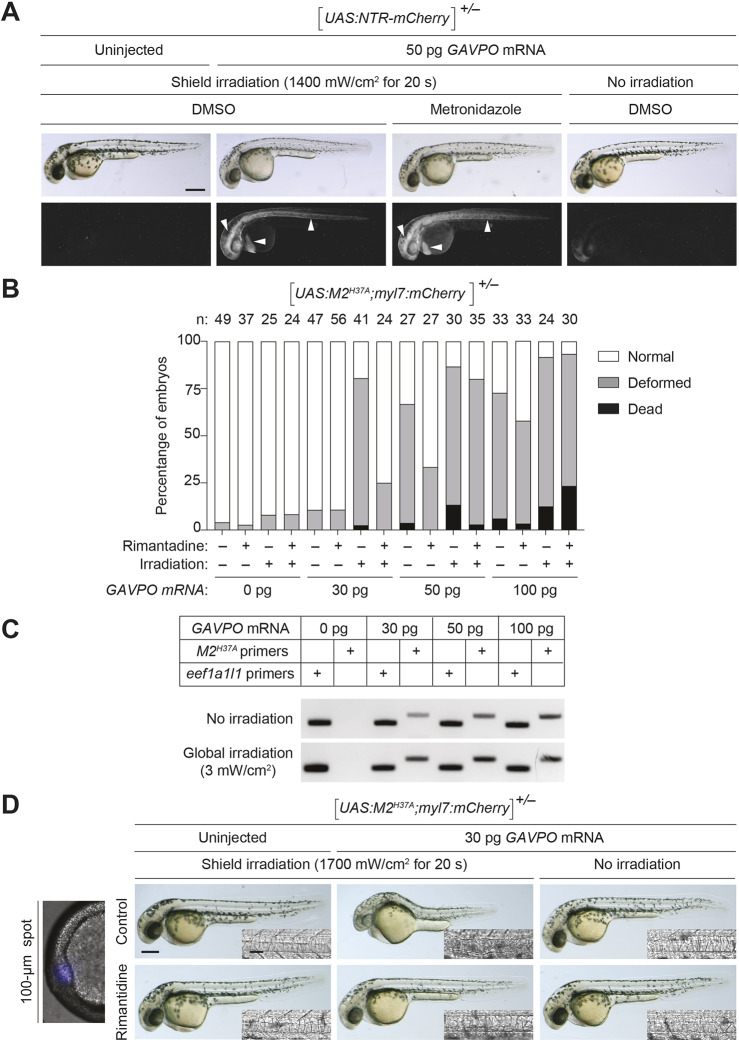
Fig. 3.
GAVPO-mediated toxicity in UAS:NTR-mCherry and UAS:M2H37A zebrafish lines. (A) Tg(UAS:NTR-mCherry) zygotes were injected with the designated amounts of GAVPO mRNA, and their embryonic shields were irradiated at 6 hpf using an epifluorescence microscope equipped with a 470/40 nm filter. An iris diaphragm was used to restrict illumination to a 100 µm diameter region. The 10 hpf embryos were dechorionated and treated with DMSO or 5 mM metronidazole, and imaged at 32 hpf. Representative bright-field and epifluorescence micrographs from two independent experiments are shown. Shield-irradiation of the GAVPO-expressing embryos induces NTR-mCherry expression in the notochord, head mesoderm and hatching gland (arrowheads). (B) The designated amounts of GAVPO mRNA were injected into Tg(UAS:UAS:M2H37A; myl7:mCherry) zygotes, and the embryos were either cultured in the dark or globally exposed to blue LED light (3 mW/cm2) from 6 to 8 hpf and then cultured in the absence or presence of 100 μg/ml rimantadine. Developmental phenotypes were scored at 24 hpf, using the indicated number of embryos (n) per condition from two to four independent experiments. Statistical analyses: χ2=301, d.f.=30, P<0.00001. (C) RT-PCR analyses of M2H37A expression in Tg(UAS:UAS:M2H37A;myl7:mCherry) embryos injected with GAVPO mRNA and treated as described above. The expression of the translation elongation factor eef1a1l1 was used as a normalization control. The amplification reaction was performed using a standard PCR with 40 cycles for M2 and 35 cycles for eef1a1l1 amplification. (D) Tg(UAS:M2H37A;myl7:mCherry) zygotes were injected with the designated amounts of GAVPO mRNA, cultured in the absence or presence of 100 µg/ml rimantadine, shield-irradiated at 6 hpf as described in A, and imaged at 30 hpf. Representative bright-field and differential interference contrast (DIC) micrographs from three independent experiments are shown. Shield irradiation of the GAVPO-expressing embryos disrupts development of the notochord (inset), head and hatching gland. All embryos are shown in lateral view, anterior left. Scale bars: 250 µm in A,D; 50 µm in D (insets). Statistics for the observed phenotypes are in Table S3.
As our studies with Tg(tuba1a:Gal4VP16;myl7:GFP) zebrafish indicated that M2H37A can ablate neurons with faster kinetics than the NTR/metronidazole system, we investigated whether the GAVPO transactivator could be used to express cytotoxic levels of this constitutively active channel. We injected varying amounts of GAVPO mRNA into heterozygous Tg(UAS:M2H37A;myl7:mCherry) zygotes, establishing 30 pg/embryo as the maximum dose that permits normal embryonic development in this transgenic line under dark-state conditions (Fig. 3B). Higher concentrations of GAVPO mRNA caused patterning defects, and RT-PCR analyses confirmed that this coincides with increased light-independent M2H37A expression (Fig. 3C). We then injected Tg(UAS:M2H37A;myl7:mCherry) zygotes with 30 pg/embryo of GAVPO mRNA, and focally irradiated the shield with blue light at 6 hpf for 20 s using the epifluorescence microscope. The resulting embryos exhibited deformities of the notochord and head when imaged at 30 hpf (Fig. 3D), matching the transgene expression we observed in Tg(UAS:NTR-mCherry) zebrafish that had been injected with GAVPO mRNA and shield irradiated (Fig. 3A). In addition, culturing the focally irradiated embryos in the presence of the M2 blocker rimantadine, prevented these developmental defects.
Light-inducible neuron ablation through tissue-specific GAVPO expression
Having established the ability of transiently expressed GAVPO to convey light-inducible cell ablation, we explored the efficacy of transgenic lines that stably express GAVPO in a neuron-specific manner. To target GAVPO expression to post-mitotic neurons, we generated zebrafish carrying GAVPO under control of the neuronal promoter elavl3 (also known as HuC) (Kim et al., 1996; Park et al., 2000a,b). Multiple founders were screened for single insertions of the GAVPO expression construct and raised to generate two stable lines. When crossed to homozygous Tg(UAS:NTR-mCherry) zebrafish, both lines yielded embryos that exhibited photoinducible, neuron-specific mCherry fluorescence in the expected Mendelian ratios (Fig. S3A). One of the heterozygous lines exhibited more robust mCherry fluorescence in the developing spinal cord, and we incrossed this line to generate fertile, homozygous Tg(elavl3:GAVPO) zebrafish. GAVPO expression in this line recapitulated endogenous elavl3 expression (Fig. S3B).
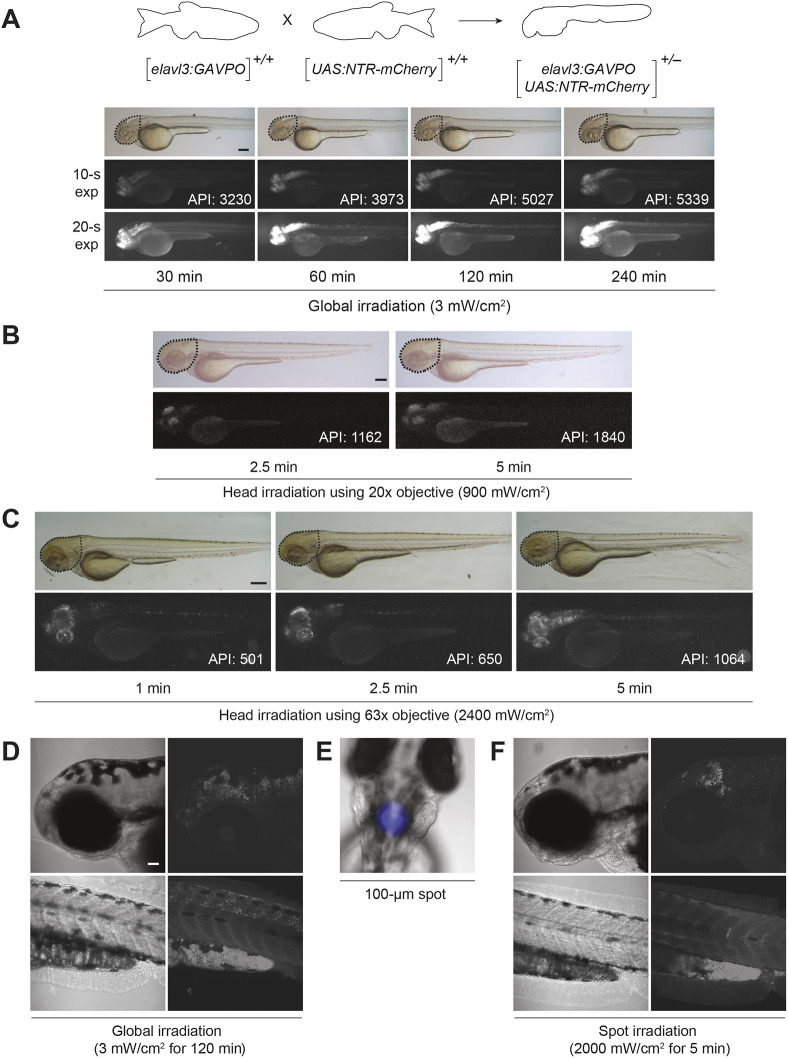
We next tested the ability of the elavl3:GAVPO transgene to convey light-inducible gene expression in neurons. We crossed homozygous Tg(elavl3:GAVPO) and homozygous Tg(UAS:NTR-mCherry) zebrafish to obtain embryos that contained one copy of each transgene. The double transgenic embryos were irradiated at different developmental stages for 2 h using the blue LED lamp and imaged 8 h post irradiation (hpi). We found that the mCherry fluorescence coincided with GAVPO expression at 36 and 48 hpf (Fig. S3B,C), and correlated with the duration and intensity of blue-light illumination (Fig. 4A-C, Fig. S4). Focal irradiation using the epifluorescence microscope induced mCherry expression in the targeted neurons within the zebrafish brain, whereas global irradiation concurrently induced mCherry expression in the spinal cord (Fig. 4D-F). We also examined the sensitivity of the GAVPO transactivator to ambient light by exposing the double transgenic embryos to a white LED lamp (intensity at 470 nm=1.8 mW/cm2). These embryos exhibited substantially lower levels of mCherry expression in comparison with those irradiated with blue light for the same duration (Fig. S4C).
Fig. 4.
Light-inducible, neuron-specific gene expression using the GAVPO system. (A) Tg(elavl3:GAVPO;UAS:NTR-mCherry) embryos were irradiated for varying durations, using a blue LED lamp and starting at 48 hpf. The embryos were then imaged 8 h later, and representative bright-field and epifluorescence micrographs (with short or long exposure times) are shown. NTR-mCherry was expressed in neural tissues, at levels that increased with the length of irradiation. Dashed outlines indicate the region used to determine the average pixel intensity (API) for each micrograph. (B,C) The heads of 48 hpf Tg(elavl3:GAVPO;UAS:NTR-mCherry) embryos were irradiated using an epifluorescence microscope equipped with a 470/40 nm filter and either a 20× (B) or 63× (C) objective. The embryos were imaged 8 h later, and representative bright-field and epifluorescence micrographs are shown. NTR-mCherry expression was observed in anterior neural tissues, at levels that increased with the duration and intensity of irradiation. Dashed outlines indicate the region used to determine the average pixel intensity (API) for each micrograph. (D-F) Tg(elavl3:GAVPO;UAS:NTR-mCherry) embryos were illuminated either globally using a blue LED lamp (D) or within a 100 µm diameter region in the head using an epifluorescence microscope equipped with a 470/40 nm filter, 20× objective and iris diaphragm (E,F). Representative bright-field and confocal fluorescence micrographs from two independent experiments are shown, and total sample sizes are indicated. LED illumination induced NTR-mCherry expression throughout the head, whereas spot-irradiated embryos exhibited mCherry fluorescence in only a localized anterior region. Embryo orientations: A-D,F, lateral view and anterior left; E, dorsal view, anterior up. Scale bars: 200 µm in A; 300 µm in B,C; 50 µm in D-F. Statistics for the observed phenotypes are in Table S4.
We then crossed homozygous Tg(UAS:M2H37A;myl7:mCherry) zebrafish with the homozygous Tg(elavl3:GAVPO) line and exposed their Tg(elavl3:GAVPO;UAS:M2H37A;myl7:mCherry) progeny to blue light at 48 hpf for varying lengths of time. As determined by whole-mount immunostaining, M2H37A levels correlated with the duration of blue-light irradiation (Fig. S5). Rimantadine treatment led to higher M2H37A expression levels, likely owing to its ability to inhibit the cytotoxic channel and promote the survival of M2H37A-expressing neurons.
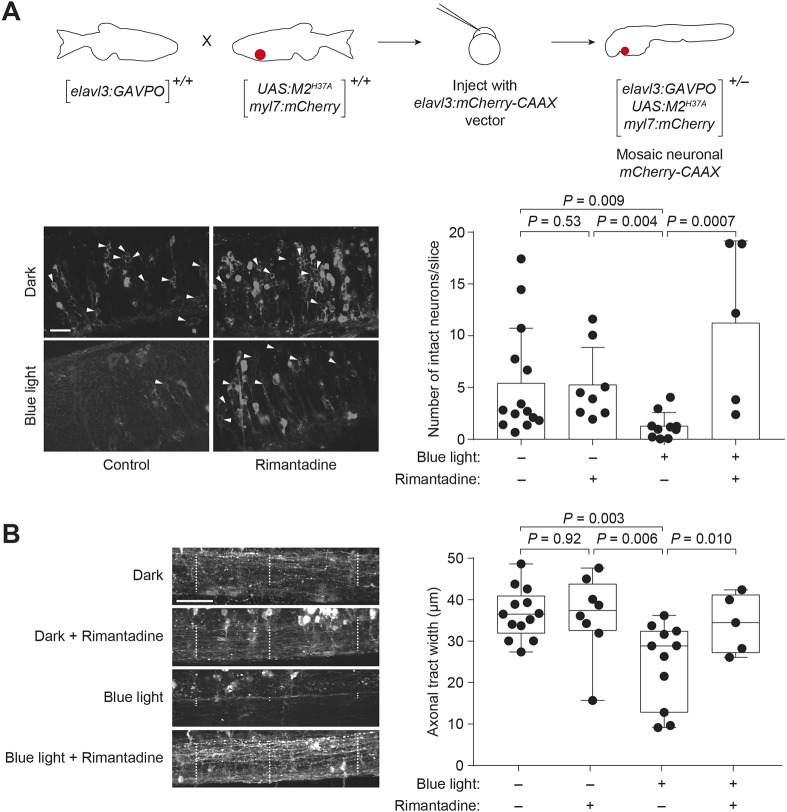
Finally, we sought to confirm that Tg(elavl3:GAVPO;UAS:M2H37A;myl7:mCherry) zebrafish exhibit light-dependent neuronal loss. As M2H37A-induced cell death is not associated with caspase 3 activation or DNA fragmentation that can be detected by TUNEL staining, we monitored neuronal ablation by optical microscopy. We injected Tg(elavl3:GAVPO;UAS:M2H37A;myl7:mCherry) zygotes with an elavl3:mCherry-CAAX construct to label post-mitotic neurons in a mosaic manner (Fig. 5A), facilitating their visualization and quantification. We raised the embryos to 48 hpf, globally irradiated a subset with the blue LED lamp for 8 h, and imaged the zebrafish at 12 hpi. The irradiated animals had significantly fewer intact neurons in the hindbrain than those maintained in the dark or cultured in rimantadine-containing medium after exposure to blue light (Fig. 5A). The spinal cords of irradiated Tg(elavl3:GAVPO;UAS:M2H37A;myl7:mCherry) embryos also exhibited reduced axon bundle widths (Fig. 5B). In contrast to the Gal4VP16-driver line with high levels of constitutive neural M2H37A expression (Fig. 1C), we did not observe gross abnormalities in the irradiated transgenic animals and caspase 3 staining of these embryos had limited apoptosis (Fig. S5B). Thus, the effects of transient M2H37A expression can be restricted to the targeted tissues.
Fig. 5.
Light-inducible neural ablation using the GAVPO/M2H37A system. (A) Heterozygous Tg(elavl3:GAVPO;UAS:M2H37A;myl7:mCherry) embryos were injected with an elavl3:mCherry-CAAX construct to label neurons in a mosaic manner. The injected embryos were globally irradiated with a blue LED lamp at 48 hpf for 8 h, fixed 12 h later and then imaged using a confocal microscope. Representative micrographs from a hindbrain slice just above the otic vesicle are shown. Irradiated embryos have fewer intact neurons (arrowheads) than those cultured in the dark or treated with 100 µg/ml rimantadine during blue-light illumination. Neurons were counted in ImageJ in a blinded manner, and the average number of neurons per slice±s.d. for each experimental condition is shown. (B) Representative micrographs from a maximal intensity projection of the spinal cord just above the cloaca, with axonal tract widths in selected regions indicated by the dashed lines. The average axonal tract width±s.d. for each experimental condition are shown. Values for individual fish are shown. The box extends from the 25th to 75th percentiles. The whiskers extend to the minimum and maximum values. The horizontal line indicates the average value. Embryo orientations: A,B, lateral view, anterior left. Scale bars: 25 µm.
DISCUSSION
Our studies demonstrate that GAVPO can be used to achieve optogenetic control of gene expression in zebrafish. Although it has previously been reported that the GAVPO mRNA disrupts zebrafish development and reduces embryo viability (Reade et al., 2017), we readily generated viable, fertile transgenic lines, indicating that the photoactivatable transactivator is not inherently toxic. The efficacy of GAVPO in zebrafish opens the door to new experimental approaches for this model organism, as this VIVID LOV domain-based system has certain advantages over other optogenetic approaches. For example, the homodimeric GAVPO transactivator can be introduced into organisms as a single construct, whereas CYR2/CIB1- and PhyB/PIF-based technologies are heterodimeric systems (Buckley et al., 2016; Liu et al., 2012). The photoactivated state of GAVPO is also long-lived, with a mean lifetime that is more than two orders of magnitude greater than that of EL222 and its derivatives (τ∼2 h versus 30 s) (Motta-Mena et al., 2014; Wang et al., 2012). As a result, sustained GAVPO activity can be achieved with a single pulse of blue light, allowing transcriptional control of specific cell populations in developing embryos and other dynamic living systems. The reliance of GAVPO on endogenous flavin chromophores and its compatibility with the myriad of existing Gal4/UAS systems are other major strengths.
These attributes highlight the versatility of GAVPO as an optogenetic tool and, in principle, its functionality could be enhanced through modifications of its LOV domain. For example, the light-state lifetime of the VIVID LOV domain can be increased up to tenfold by mutating the flavin-binding site (Zoltowski et al., 2009). Introducing these residue changes into the GAVPO transactivator could prolong transcriptional responses to a single illumination pulse. We also observed that GAVPO mRNA was more toxic to Tg(UAS:M2H37A;myl7:mCherry) embryos than their Tg(UAS:NTR-mCherry) counterparts, even when the mRNA-injected zebrafish were cultured in the dark. As M2H37A is constitutively ablative and NTR is not, we interpret these results as evidence of dark-state GAVPO activity. Consistent with this model, rimantadine attenuates GAVPO mRNA-dependent toxicity in Tg(UAS:M2H37A;myl7:mCherry) embryos. We therefore anticipate that basal activation of UAS-driven transgenes will increase with GAVPO concentration, and strong promoters may constrain the dynamic range of this photoactivatable transactivator. This limitation could be overcome by using alternative cis-regulatory sequences or by screening multiple transgenic lines, or, alternatively, GAVPO variants with lower dark-state dimer affinities could be developed.
Our studies also establish M2H37A expression as an effective strategy for ablating specific cell populations in whole organisms. In addition to providing the first demonstration that M2H37A is cytotoxic in zebrafish, we reveal important differences between this non-selective cation channel and the NTR/metronidazole system. M2H37A expression kills cells with greater efficacy than NTR-mediated prodrug activation, as evidenced by their respective activities in the zebrafish CNS and axial mesoderm. M2H37A and NTR/metronidazole also ablate cells through distinct mechanisms. M2H37A appears to initially trigger neuronal necrosis, and surrounding cells can subsequently undergo apoptosis, at least when the cytotoxic channel is continuously expressed in the targeted tissue (e.g. using a CNS Gal4VP16 driver). In comparison, NTR-mediated activation of metronidazole induces programmed cell death. These differences make M2H37A a promising tool for studying injury, a process that involves both necrosis and apoptosis. For example, previous studies have shown that spinal cord injuries lead to sustained apoptosis that can last for weeks (Crowe et al., 1997; Oyinbo, 2011). M2H37A could be particularly advantageous for investigating molecular and cellular responses to CNS damage, as commonly used models of spinal cord injury rely on mechanical perturbations that can concurrently damage muscle and skin (Becker et al., 1997; Bhatt et al., 2004). The sensitivity of M2H37A to rimantadine also enables pharmacological tuning of this cytotoxic channel, allowing one to minimize basal activity and maximize inducibility.
Finally, by combining the GAVPO and M2H37A, we have achieved optical control of cell ablation in a whole organism. In comparison with other genetically encoded methods for cell ablation, the GAVPO/M2H37A system can eliminate cells of interest without requiring tissue-specific promoters. When GAVPO is expressed ubiquitously, cell targeting can be guided by morphological cues alone, as illustrated by our ablation of progenitors within the zebrafish shield. Moreover, optically inducing M2H37A expression within the shield did not appear to cause neighboring cells to apoptose, contrasting the non-cell-autonomous effects we observed with Gal4VP16-driven M2H37A expression in the developing CNS. Thus, it may be possible to use the GAVPO/M2H37A system to elucidate the developmental fates of other embryonic cell populations. Alternatively, cis-regulatory sequences can be used to restrict GAVPO expression to certain tissues, enabling the ablation of specific cell populations within these promoter-defined regions. The latter approach could be used to generate tissue injuries in a spatially defined manner. Based on these capabilities, we anticipate that the GAVPO/M2H37A system will be a versatile tool for deconstructing developmental and regenerative mechanisms in whole organisms.
MATERIALS AND METHODS
Zebrafish husbandry
Adult zebrafish [wild-type AB strain and Tg(UAS-E1b:NTR-mCherry) (Davison et al., 2007); 3-18 months] were obtained from the Zebrafish International Resource Center. The transgenic line Tg(tuba1a:Gal4VP16;myl7:GFP) (Goldman et al., 2001) was a generous gift from Philippe Mourrain (Stanford University, CA, USA). All zebrafish lines were raised according to standard protocols. Embryos were obtained through natural matings and cultured at either 28.5°C or 32°C in E3 medium. The culture medium was also supplemented with 0.003% (w/v) PTU to prevent pigmentation. All animal procedures were approved by the Administrative Panel on Laboratory Animal Care at Stanford University (protocol 10511) and the University of Wyoming (protocol 2018017KM00326-02).
Plasmids
To generate GAVPO mRNA, the GAVPO-coding region with an upstream Kozak site was PCR amplified from pGAVPO plasmid (a gift from Yi Yang, East China University of Science and Technology, Shanghai, China) using the following primers containing BamHI and XbaI sites (underlined): 5-GAATGGATCCGCCACCATGAAGCTACTGTC-3′ and 5′-GAACTCTAGAGTGTACATTACTTGTCATCATCGTC-3′. The resulting PCR product was digested and ligated into pCS2+ to generate pCS2-GAVPO. The insertion was sequenced to confirm ligation fidelity. pCS2-M2H37A (a gift from Tim Mohun, Francis Crick Institute, London, UK) and pCS2-GAVPO constructs were linearized with SacII, and their corresponding mRNAs were synthesized using in vitro SP6-dependent runoff transcription (Invitrogen).
To generate the Tol2-elavl3:GAVPO plasmid, the GAVPO-coding region with an upstream Kozak sequence was PCR amplified using the following primers: 5′-CCACCTGCAGATAATTGTTTAAACCACTCCGCCACCATGAAG-3′ and 5′-AGTAAAACGACGGCCAGGATCCACCGGTCTGCTATTACTTGTCATCATCGTC-3′. Tol2-elavl3:GCaMP6s plasmid (Misha Ahrens; Addgene 59531) was digested with AgeI, and Tol2-elavl3:GAVPO was assembled from the resulting vector and GAVPO PCR product using Gibson Assembly Master Mix (NEB). The entire plasmid was sequenced to confirm ligation fidelity.
To generate the Tol2-5xUAS-TATA:M2H37A;myl7:mCherry construct, the M2-coding region was PCR amplified using the following primers, which contain HindIII and ApaI sites (underlined): 5′-GAACAAGCTTGCCACCATGAGTCTTCTAACCG-3′ and 5′-GAATGGGCCCTTACTCCAGCTCTATGTTGAC-3′. This PCR product was then digested and ligated into pU5 (a gift from Yi Yang) to generate pU5-M2H37A. The 5xUAS-TATA:M2H37A region was then PCR amplified using the following primers: 5′-ACACAGGCCAGATGTGGGCCCGTACTTGGAGCGGCCGCA-3′ and 5′-GTCTGGATCATCATCGATGCGGCCGCAAGCCATAGAGCCCACCG-3′. pBH (a gift from Michael Nonet; Washington University, St Louis, MO, USA) was digested with ApaI and NotI, and the Tol2-5xUAS-TATA:M2H37A;myl7:mCherry plasmid was assembled from the digested pBH and 5xUAS-TATA:M2H37A PCR product using Gibson Assembly Master Mix (NEB). The entire plasmid was sequenced to confirm ligation fidelity.
To generate the Tol2-elavl3:mCherry-CAAX plasmid, the mCherry- and CAAX-coding regions with an upstream Kozak sequence were PCR amplified using the following primers: mCherry, 5′-TATATTTTCCACCTGCAGATAATTACCGGTGCCACCATGGTGAGCAAG-3′ and 5′-TAAAACGACGGCCAGGATCCACGCGTCTATTACATAATTACACACTTTGTCTTTGACTTC-3′; CAAX, 5′-TATATTTTCCACCTGCAGATAATTACCG GTTCCGCCACCATGGTGAGC-3′ and 5′-AGTAAAACGACGGCCAGGATCCACACGCGTCTATTACATAATTACACACTTTGTCTTTGA CTTCTTTTTCTTCTTTTTAC-3′. Tol2-elavl3:GCaMP6s was digested with AgeI, and Tol2-elavl3:mCherry-CAAX was assembled from the digested plasmid and PCR products using Gibson Assembly Master Mix (NEB). The insert was sequenced to confirm ligation fidelity.
Generation of transgenic zebrafish lines
Transgenic zebrafish were generated using Tol2-mediated transgenesis as previously described (Suster et al., 2009). For each line generated, plasmid DNA and Tol2 mRNA were premixed and co-injected into one-cell-stage embryos (12-50 pg of plasmid; 50 pg of mRNA). Fish were raised to adulthood and mated with wild-type AB fish to identify founders with germline transmission, and those yielding F2 generations with monoallelic expression were used to establish transgenic lines. Both heterozygous and homozygous Tg(elavl3:GAVPO) and homozygous Tg(UAS:M2H37A;myl7:mCherry) lines were used in subsequent studies.
GAVPO photoactivation
All experimental procedures with GAVPO-expressing zebrafish used a Wratten #29 filter (Kodak) to minimize GAVPO activation by the microscopy light sources required for embryological procedures, monitoring and imaging. For global irradiations, a plate containing E3 medium and embryos was mounted onto a mirrored surface and then fixed onto a Vortex-Genie (Scientific Industries). Dechorionated embryos were irradiated with a blue or white LED light source (TaoTronics), during which the plate was vortexed to ensure that embryos were irradiated from all sides. Light intensity received by the embryos was measured as 3 mW/cm2 (1.8 mW/cm2 for white LEDs). For individual irradiations, the embryos were either mounted in an injection tray or in 1% low-melt agarose on a slide. Each embryo was irradiated using a Leica DM4500B upright compound microscope equipped with a mercury lamp, a GFP filter (ex: 470 nm, 40 nm bandpass) and a 20×/0.5 NA or a 63× water-immersion objective. To optically target the larval head, the fully open diaphragm was used to irradiate a region spanning the forebrain and the otic vesicle. For focal irradiation, an iris diaphragm was used to limit the targeted region to a 100 μm diameter. Light intensities from the mercury lamp were measured before and after the experiment, and they ranged from 900 to 2400 mW/cm2.
Fluorescence intensity quantification
Live embryos were mounted in 1% low-melting-point agarose on a glass slide. Embryos were imaged using a Leica M205FA microscope equipped with a SPOT Flex color camera and images were captured using SPOT software (Molecular Devices) or using an Olympus SZX16 microscope equipped with an Olympus DP80 dual-chip camera. Fluorescent images were captured with DP80 in monochrome mode. Pixel intensities were quantified using ImageJ, using the polygon tool on the bright-field image to circumscribe the region of interest. The average mCherry fluorescence intensity within the same region of the corresponding fluorescent micrograph was then calculated.
Reverse transcriptase (RT)-PCR
RNA was extracted from 10-15 embryos per condition at 10 hpf using the RNeasy Micro Kit (Qiagen). Reverse transcription, using olig(dT)20 primers was performed with the SuperScript III First Strand Synthesis System (Invitrogen). The following primers were used to amplify M2 and normalized against eukaryotic translation elongation factor 1 alpha 1, like 1 (eef1a1l1), by standard PCR (55°C, 35 cycles for eef1a1l1 and 40 cycles for M2 amplification): eef1a1l1 forward, 5′-AGAAGGAAGCCGCTGAGATGG-3′; eef1a1l1 reverse, 5′-TCCGTTCTTGGAGATACCAGCC-3′; M2H37A forward, 5′-GCCGCGAGTATCATTGGGAT-3′; M2H37A reverse, 5′-TCAGGCACTCCTTCCGTAGA-3′.
Immunostaining
Embryos were fixed at the desired timepoint in 4% (w/v) paraformaldehyde (PFA) in PBS at 4°C overnight and then stored in methanol. After rehydration, embryos were permeabilized with acetone and proteinase K, immunostained and imaged with a Leica DM4500B compound microscope equipped with a Retiga-SRV or Prime cMOS camera (QImaging) or Zeiss LSM 700 and LSM 800 confocal microscopes equipped with a MA-PMT. The following antibodies were used: mouse monoclonal anti-HuC/HuD (1:100 dilution, Molecular Probes, A-21271), mouse monoclonal anti-mCherry (1:500 dilution, Abcam, ab125096), rabbit polyclonal anti-active caspase 3 (1:100 dilution, BD Biosciences, BDB559565), rabbit polyclonal anti-influenza A virus M2 (1:100 dilution, GeneTex, GTX125951), goat anti-mouse and goat anti-rabbit antibodies conjugated to Alexa Fluor 488, 555 or 594 (1:200 dilution, Roche).
TUNEL staining
For M2H37A and NTR experiments, embryos were raised in E3 medium supplemented with either 100 µg/ml rimantadine or 5 mM metronidazole, respectively. Embryos raised in E3 medium treated with an equivalent amount of DMSO vehicle were used as controls. The embryos were then dechorionated using forceps, fixed in 4% PFA at the desired timepoint, and stored in methanol. Embryos were rehydrated and permeabilized with diluted bleach solution containing 0.8% potassium hydroxide, 3% hydrogen peroxide and 0.2% Triton X-100. After bleaching, the embryos were washed with PBS supplemented with 0.2% Triton X-100 (PBS-X), dehydrated with methanol and stored overnight at −20°C. After rehydration, embryos were repeatedly washed with PBS-X and digested using proteinase K. Embryos were re-fixed in 4% PFA followed by treatment with a pre-chilled solution (2:1) of ethanol and acetic acid. Using the ApopTag Peroxidase In Situ Apoptosis Detection Kit (Millipore Sigma), the samples were incubated in equilibration buffer for 1 h and then TdT reaction buffer overnight at 37°C. Embryos were washed in diluted stop solution and PBS-X and then incubated with a blocking solution containing: 2% (w/v) blocking reagent (Roche), 0.15 M NaCl, 0.1 M maleic acid and 20% (v/v) sheep serum (Sigma #S2263). The embryos were incubated with horseradish peroxidase-conjugated anti-digoxigenin antibody overnight at 4°C and washed with PBS-X. Embryos were stained with fresh DAB mix (Metal Enhanced DAB Kit Thermo Fisher Scientific #34065), washed and fixed in 4% PFA. The embryos were stored and imaged in a solution of 90% glycerol in PBS.
In situ hybridization
Whole-mount in situ hybridization was performed according to standard protocols (Broadbent and Read, 1999). Zebrafish cDNA was prepared from RNA extracted from embryos as previously described (Payumo et al., 2015). T3-promoter containing PCR products were then amplified with the designated primers (T3 sequence underlined): elavl3, 5′-CACCTCACGCATCCTGGTAA-3′ and 5′-ATTAACCCTCACTAAAGGGATGGTCTTGAACGAGACCTGC-3′; GAVPO Probe 1, 5′-AAGAAAAACCGAAGTGCGCC-3′ and 5′-ATTAACCCTCACTAAAGGGATGTTTCATCTCGCACCGGAA-3′; GAVPO Probe 2, 5′-GCTCTGATTCTGTGCGACCT-3′ and 5′-ATTAACCCTCACTAAAGGGAATCAGCATGGGCTCAGTTGT-3′. RNA probes were in vitro transcribed from the PCR products using the MEGAscript T3 Transcription Kit (Ambion), substituting nucleotides from the kit with digoxigenin-UTP (Roche). For GAVPO staining, both probes were used simultaneously in separate tubes. Embryos were imaged using a Leica M205FA microscope equipped with a SPOT Flex color camera and images were captured using SPOT software (Molecular Devices).
Neuron labeling and counting
Heterozygous Tg(elavl3:GAVPO;UAS:M2H37A;myl7:mCherry) zygotes were co-injected with plasmid DNA encoding elavl3:mCherry-CAAX (25 pg/embryo) and Tol2 mRNA (50 pg/embryo). The resulting embryos were raised until 48 hpf, irradiated for 8 h and then fixed 12 h after irradiation in 4% PFA for 1 h at room temperature. The embryos were then mounted laterally in 1% low-melting agarose and imaged with a Zeiss LSM 700 confocal microscope equipped with a 63×/0.5 NA water-immersion objective. Z-stacks were generated from images taken at 2- to 5-μm intervals, using the following settings: 1024×1024 pixels, 8 speed, 4 averaging. Neurons were scored as intact when mCherry fluorescence was localized only to the plasma membrane and excluded from the cytoplasm. Counting was performed by an observer blinded to the experimental conditions. For spinal cord measurements, maximum intensity projections were made in ImageJ, and three measurements were made for each projection and averaged to determine the axonal tract width.
Statistical analyses
For all zebrafish experiments, at least two breeding tanks, each containing three or four males and three to five females from separate stocks, were set up to generate embryos. Embryos from each tank were randomly distributed across tested conditions, and unfertilized and developmentally abnormal embryos were removed prior to irradiation or compound treatment. No statistical methods were used to determine sample size per condition. Experimental statistics for the phenotypes reported in Figs 1-4 are provided in Tables S1-S4, respectively. χ2 analyses were conducted for the phenotypic distributions reported in Figs S1 and S4. For the cell counting and axonal tract width measurements reported in Fig. 5, the scorer was blinded to treatment conditions. Values for individual fish are plotted, and each distribution was assessed using the Shapiro-Wilk test and determined to be non-normal. A Kruskal-Wallis test with a two-stage linear step-up procedure of Benjamini, Krieger and Yekutieli was used to determine differences between all conditions. P-values were corrected for multiple comparisons testing by controlling the false discovery rate to 0.05.
Supplementary Material
Acknowledgements
We thank Shannon Linch for assistance with mating and genotyping. Imaging was supported in part by the University of Wyoming's Integrated Microscopy Core (NIH P20 GM121310).
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: K.M., J.K.C.; Validation: K.M., P.C., P.A.P., M.A.A.; Formal analysis: K.M., P.C., J.K.C.; Investigation: K.M., P.C., P.A.P., M.A.A.; Writing - original draft: K.M., J.K.C.; Writing - review & editing: K.M., P.C., P.A.P., M.A.A., J.K.C.; Visualization: K.M., P.C., J.K.C.; Supervision: K.M., J.K.C.; Project administration: K.M., J.K.C.; Funding acquisition: K.M., J.K.C.
Funding
We gratefully acknowledge financial support from the National Institutes of Health (R35 GM127030 to J.K.C.), from a Craig H. Neilsen Foundation Postdoctoral Fellowship (313804 to K.M.), from a Stanford School of Medicine Dean's Postdoctoral Fellowship (P.C.) and from the Stanford ChEM-H Undergraduate Scholars Program (P.A.P.). Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.183640.supplemental
Peer review history
The peer review history is available online at https://dev.biologists.org/lookup/doi/10.1242/dev.183640.reviewer-comments.pdf
References
- Becker T. and Becker C. G. (2001). Regenerating descending axons preferentially reroute to the gray matter in the presence of a general macrophage/microglial reaction caudal to a spinal transection in adult zebrafish. J. Comp. Neurol. 433, 131-147. 10.1002/cne.1131 [DOI] [PubMed] [Google Scholar]
- Becker T., Wullimann M. F., Becker C. G., Bernhardt R. R. and Schachner M. (1997). Axonal regrowth after spinal cord transection in adult zebrafish. J. Comp. Neurol. 377, 577-595. 10.1002/(SICI)1096-9861(19970127)377:4<577::AID-CNE8>3.0.CO;2-# [DOI] [PubMed] [Google Scholar]
- Bhatt D. H., Otto S. J., Depoister B. and Fetcho J. R. (2004). Cyclic AMP-induced repair of zebrafish spinal circuits. Science 305, 254-258. 10.1126/science.1098439 [DOI] [PubMed] [Google Scholar]
- Bridgewater J. A., Springer C. J., Knox R. J., Minton N. P., Michael N. P. and Collins M. K. (1995). Expression of the bacterial nitroreductase enzyme in mammalian cells renders them selectively sensitive to killing by the prodrug CB1954. Eur. J. Cancer 31A, 2362-2370. 10.1016/0959-8049(95)00436-X [DOI] [PubMed] [Google Scholar]
- Briona L. K., Poulain F. E., Mosimann C. and Dorsky R. I. (2015). Wnt/ß-catenin signaling is required for radial glial neurogenesis following spinal cord injury. Dev. Biol. 403, 15-21. 10.1016/j.ydbio.2015.03.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broadbent J. and Read E. M. (1999). Wholemount in situ hybridization of Xenopus and zebrafish embryos. In Molecular Methods in Developmental Biology: Xenopus and Zebrafish (ed. Guille M.), pp. 57-67. Totowa, NJ: Humana Press. [DOI] [PubMed] [Google Scholar]
- Buckley C. E., Moore R. E., Reade A., Goldberg A. R., Weiner O. D. and Clarke J. D. W. (2016). Reversible optogenetic control of subcellular protein localization in a live vertebrate embryo. Dev. Cell 36, 117-126. 10.1016/j.devcel.2015.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley C., Carvalho M. T., Young L. K., Rider S. A., McFadden C., Berlage C., Verdon R. F., Taylor J. M., Girkin J. M. and Mullins J. J. (2017). Precise spatio-temporal control of rapid optogenetic cell ablation with mem-KillerRed in Zebrafish. Sci. Rep. 7, 5096 10.1038/s41598-017-05028-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulina M. E., Chudakov D. M., Britanova O. V., Yanushevich Y. G., Staroverov D. B., Chepurnykh T. V., Merzlyak E. M., Shkrob M. A., Lukyanov S. and Lukyanov K. A. (2006). A genetically encoded photosensitizer. Nat. Biotechnol. 24, 95-99. 10.1038/nbt1175 [DOI] [PubMed] [Google Scholar]
- Chelur D. S. and Chalfie M. (2007). Targeted cell killing by reconstituted caspases. Proc. Natl. Acad. Sci. USA 104, 2283-2288. 10.1073/pnas.0610877104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu Q. D., Sun L., Li J., Byrnes K., Chervenak D., DeBenedetti A., Mathis J. M. and Li B. D. L. (2007). Rat adenocarcinoma cell line infected with an adenovirus carrying a novel herpes-simplex virus-thymidine kinase suicide gene construct dies by apoptosis upon treatment with ganciclovir. J. Surg. Res. 143, 189-194. 10.1016/j.jss.2006.12.007 [DOI] [PubMed] [Google Scholar]
- Crowe M. J., Bresnahan J. C., Shuman S. L., Masters J. N. and Beattie M. S. (1997). Apoptosis and delayed degeneration after spinal cord injury in rats and monkeys. Nat. Med. 3, 73-76. 10.1038/nm0197-73 [DOI] [PubMed] [Google Scholar]
- Curado S., Anderson R. M., Jungblut B., Mumm J., Schroeter E. and Stainier D. Y. R. (2007). Conditional targeted cell ablation in zebrafish: a new tool for regeneration studies. Dev. Dyn. 236, 1025-1035. 10.1002/dvdy.21100 [DOI] [PubMed] [Google Scholar]
- Curado S., Stainier D. Y. R. and Anderson R. M. (2008). Nitroreductase-mediated cell/tissue ablation in zebrafish: a spatially and temporally controlled ablation method with applications in developmental and regeneration studies. Nat. Protoc. 3, 948-954. 10.1038/nprot.2008.58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davison J. M., Akitake C. M., Goll M. G., Rhee J. M., Gosse N., Baier H., Halpern M. E., Leach S. D. and Parsons M. J. (2007). Transactivation from Gal4-VP16 transgenic insertions for tissue-specific cell labeling and ablation in zebrafish. Dev. Biol. 304, 811-824. 10.1016/j.ydbio.2007.01.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi C. S., Shuck K., Lear J. D., Dieckmann G. R., DeGrado W. F., Lamb R. A. and Pinto L. H. (1999). Cu(II) inhibition of the proton translocation machinery of the influenza A virus M2 protein. J. Biol. Chem. 274, 5474-5482. 10.1074/jbc.274.9.5474 [DOI] [PubMed] [Google Scholar]
- Goldman D., Hankin M., Li Z., Dai X. and Ding J. (2001). Transgenic zebrafish for studying nervous system development and regeneration. Transgenic Res. 10, 21-33. 10.1023/A:1008998832552 [DOI] [PubMed] [Google Scholar]
- Intharathep P., Laohpongspaisan C., Rungrotmongkol T., Loisruangsin A., Malaisree M., Decha P., Aruksakunwong O., Chuenpennit K., Kaiyawet N., Sompornpisut P. et al. (2008). How amantadine and rimantadine inhibit proton transport in the M2 protein channel. J. Mol. Graph. Model. 27, 342-348. 10.1016/j.jmgm.2008.06.002 [DOI] [PubMed] [Google Scholar]
- Jung J., Hackett N. R., Pergolizzi R. G., Pierre-Destine L., Krause A. and Crystal R. G. (2007). Ablation of tumor-derived stem cells transplanted to the central nervous system by genetic modification of embryonic stem cells with a suicide gene. Hum. Gene Ther. 18, 1182-1192. 10.1089/hum.2007.078 [DOI] [PubMed] [Google Scholar]
- Kim C.-H., Ueshima E., Muraoka O., Tanaka H., Yeo S.-Y., Huh T.-L. and Miki N. (1996). Zebrafish elav/HuC homologue as a very early neuronal marker. Neurosci. Lett. 216, 109-112. 10.1016/0304-3940(96)13021-4 [DOI] [PubMed] [Google Scholar]
- Kimmel C. B., Warga R. M. and Schilling T. F. (1990). Origin and organization of the zebrafish fate map. Development 108, 581-594. [DOI] [PubMed] [Google Scholar]
- Köster R. W. and Fraser S. E. (2001). Tracing transgene expression in living zebrafish embryos. Dev. Biol. 233, 329-346. 10.1006/dbio.2001.0242 [DOI] [PubMed] [Google Scholar]
- Kurita R., Sagara H., Aoki Y., Link B. A., Arai K. and Watanabe S. (2003). Suppression of lens growth by alphaA-crystallin promoter-driven expression of diphtheria toxin results in disruption of retinal cell organization in zebrafish. Dev. Biol. 255, 113-127. 10.1016/S0012-1606(02)00079-9 [DOI] [PubMed] [Google Scholar]
- Lam V. K., Tokusumi T., Cerabona D. and Schulz R. A. (2010). Specific cell ablation in Drosophila using the toxic viral protein M2(H37A). Fly 4, 338-343. 10.4161/fly.4.4.13114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Tissier P. R., Carmignac D. F., Lilley S., Sesay A. K., Phelps C. J., Houston P., Mathers K., Magoulas C., Ogden D. and Robinson I. C. A. F. (2005). Hypothalamic growth hormone-releasing hormone (GHRH) deficiency: targeted ablation of GHRH neurons in mice using a viral ion channel transgene. Mol. Endocrinol. 19, 1251-1262. 10.1210/me.2004-0223 [DOI] [PubMed] [Google Scholar]
- Liu H., Gomez G., Lin S., Lin S. and Lin C. (2012). Optogenetic control of transcription in zebrafish. PLoS ONE 7, e50738 10.1371/journal.pone.0050738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z., Du Z., Chen X., Wang X. and Yang Y. (2013). Fine tuning the LightOn light-switchable transgene expression system. Biochem. Biophys. Res. Comm. 440, 419-423. 10.1016/j.bbrc.2013.09.092 [DOI] [PubMed] [Google Scholar]
- Makhijani K., To T.-L., Ruiz-González R., Lafaye C., Royant A. and Shu X. (2017). Precision optogenetic tool for selective single- and multiple-cell ablation in a live animal model system. Cell Chem. Biol. 24, 110-119. 10.1016/j.chembiol.2016.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mokalled M. H., Patra C., Dickson A. L., Endo T., Stainier D. Y. R. and Poss K. D. (2016). Injury-induced ctgfa directs glial bridging and spinal cord regeneration in zebrafish. Science 354, 630-634. 10.1126/science.aaf2679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motta-Mena L. B., Reade A., Mallory M. J., Glantz S., Weiner O. D., Lynch K. W. and Gardner K. H. (2014). An optogenetic gene expression system with rapid activation and deactivation kinetics. Nat. Chem. Biol. 10, 196-202. 10.1038/nchembio.1430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnmacht J., Yang Y., Maurer G. W., Barreiro-Iglesias A., Tsarouchas T. M., Wehner D., Sieger D., Becker C. G. and Becker T. (2016). Spinal motor neurons are regenerated after mechanical lesion and genetic ablation in larval zebrafish. Development 143, 1464-1474. 10.1242/dev.129155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyinbo C. A. (2011). Secondary injury mechanisms in traumatic spinal cord injury: a nugget of this multiply cascade. Acta Neurobiol. Exp. (Wars) 71, 281-299. [DOI] [PubMed] [Google Scholar]
- Park H.-C., Hong S.-K., Kim H.-S., Kim S.-H., Yoon E.-J., Kim C.-H., Miki N. and Huh T.-L. (2000a). Structural comparison of zebrafish Elav/Hu and their differential expressions during neurogenesis. Neurosci. Lett. 279, 81-84. 10.1016/S0304-3940(99)00940-4 [DOI] [PubMed] [Google Scholar]
- Park H.-C., Kim C.-H., Bae Y.-K., Yeo S.-Y., Kim S.-H., Hong S.-K., Shin J., Yoo K.-W., Hibi M., Hirano T. et al. (2000b). Analysis of upstream elements in the HuC promoter leads to the establishment of transgenic zebrafish with fluorescent neurons. Dev. Biol. 227, 279-293. 10.1006/dbio.2000.9898 [DOI] [PubMed] [Google Scholar]
- Payumo A. Y., Walker W. J., McQuade L. E., Yamazoe S. and Chen J. K. (2015). Optochemical dissection of T-box gene-dependent medial floor plate development. ACS Chem. Biol. 10, 1466-1475. 10.1021/cb5010178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisharath H. and Parsons M. J. (2009). Nitroreductase-mediated cell ablation in transgenic zebrafish embryos. Methods Mol. Biol. 546, 133-143. 10.1007/978-1-60327-977-2_9 [DOI] [PubMed] [Google Scholar]
- Pisharath H., Rhee J. M., Swanson M. A., Leach S. D. and Parsons M. J. (2007). Targeted ablation of beta cells in the embryonic zebrafish pancreas using E. coli nitroreductase. Mech. Dev. 124, 218-229. 10.1016/j.mod.2006.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y. B., Garren E. J., Shu X., Tsien R. Y. and Jin Y. (2012). Photo-inducible cell ablation in Caenorhabditis elegans using the genetically encoded singlet oxygen generating protein miniSOG. Proc. Natl. Acad. Sci. USA 109, 7499-7504. 10.1073/pnas.1204096109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reade A., Motta-Mena L. B., Gardner K. H., Stainier D. Y., Weiner O. D. and Woo S. (2017). TAEL: a zebrafish-optimized optogenetic gene expression system with fine spatial and temporal control. Development 144, 345-355. 10.1242/dev.139238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimer M. M., Kuscha V., Wyatt C., Sorensen I., Frank R. E., Knuwer M., Becker T. and Becker C. G. (2009). Sonic hedgehog is a polarized signal for motor neuron regeneration in adult zebrafish. J. Neurosci. 29, 15073-15082. 10.1523/JNEUROSCI.4748-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkisyan K. S., Zlobovskaya O. A., Gorbachev D. A., Bozhanova N. G., Sharonov G. V., Staroverov D. B., Egorov E. S., Ryabova A. V., Solntsev K. M., Mishin A. S. et al. (2015). KillerOrange, a genetically encoded photosensitizer activated by blue and green light. PLoS ONE 10, e0145287 10.1371/journal.pone.0145287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell J. R. and Chou J. J. (2008). Structure and mechanism of the M2 proton channel of influenza A virus. Nature 451, 591-595. 10.1038/nature06531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott E. K., Mason L., Arrenberg A. B., Ziv L., Gosse N. J., Xiao T., Chi N. C., Asakawa K., Kawakami K. and Baier H. (2007). Targeting neural circuitry in zebrafish using GAL4 enhancer trapping. Nat. Methods 4, 323-326. 10.1038/nmeth1033 [DOI] [PubMed] [Google Scholar]
- Shih J. and Fraser S. E. (1996). Characterizing the zebrafish organizer: microsurgical analysis at the early-shield stage. Development 122, 1313-1322. [DOI] [PubMed] [Google Scholar]
- Shimbo K., Brassard D. L., Lamb R. A. and Pinto L. H. (1996). Ion selectivity and activation of the M2 ion channel of influenza virus. Biophys. J. 70, 1335-1346. 10.1016/S0006-3495(96)79690-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu-Sato S., Huq E., Tepperman J. M. and Quail P. H. (2002). A light-switchable gene promoter system. Nat. Biotechnol. 20, 1041-1044. 10.1038/nbt734 [DOI] [PubMed] [Google Scholar]
- Slanchev K., Stebler J., de la Cueva-Mendez G. and Raz E. (2005). Development without germ cells: the role of the germ line in zebrafish sex differentiation. Proc. Natl. Acad. Sci. USA 102, 4074-4079. 10.1073/pnas.0407475102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C. A., Graham C. M., Mathers K., Skinner A., Hay A. J., Schroeder C. and Thomas D. B. (2002). Conditional ablation of T-cell development by a novel viral ion channel transgene. Immunology 105, 306-313. 10.1046/j.0019-2805.2002.01376.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S. J., Kotecha S., Towers N. and Mohun T. J. (2007). Targeted cell-ablation in Xenopus embryos using the conditional, toxic viral protein M2(H37A). Dev. Dyn. 236, 2159-2171. 10.1002/dvdy.21233 [DOI] [PubMed] [Google Scholar]
- Springer C. J. and Niculescu-Duvaz I. (2000). Approaches to gene-directed enzyme prodrug therapy (GDEPT). Adv. Exp. Med. Biol. 465, 403-409. 10.1007/0-306-46817-4_35 [DOI] [PubMed] [Google Scholar]
- Suster M. L., Kikuta H., Urasaki A., Asakawa K. and Kawakami K. (2009). Transgenesis in zebrafish with the tol2 transposon system. Methods Mol. Biol. 561, 41-63. 10.1007/978-1-60327-019-9_3 [DOI] [PubMed] [Google Scholar]
- Wang C., Lamb R. A. and Pinto L. H. (1995). Activation of the M2 ion channel of influenza virus: a role for the transmembrane domain histidine residue. Biophys. J. 69, 1363-1371. 10.1016/S0006-3495(95)80003-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Chen X. and Yang Y. (2012). Spatiotemporal control of gene expression by a light-switchable transgene system. Nat. Methods 9, 266-269. 10.1038/nmeth.1892 [DOI] [PubMed] [Google Scholar]
- Wharton S. A., Belshe R. B., Skehel J. J. and Hay A. J. (1994). Role of virion M2 protein in influenza virus uncoating: specific reduction in the rate of membrane fusion between virus and liposomes by amantadine. J. Gen. Virol. 75, 945-948. 10.1099/0022-1317-75-4-945 [DOI] [PubMed] [Google Scholar]
- Williams D. C., El Bejjani R., Ramirez P. M., Coakley S., Kim S. A., Lee H., Wen Q., Samuel A., Lu H., Hilliard M. A. et al. (2013). Rapid and permanent neuronal inactivation in vivo via subcellular generation of reactive oxygen with the use of KillerRed. Cell Rep. 5, 553-563. 10.1016/j.celrep.2013.09.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S. and Chisholm A. D. (2016). Highly efficient optogenetic cell ablation in C. elegans using membrane-targeted miniSOG. Sci. Rep. 6, 21271 10.1038/srep21271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y.-M., Cristofanilli M., Valiveti A., Ma L., Yoo M., Morellini F. and Schachner M. (2011). The extracellular matrix glycoprotein tenascin-C promotes locomotor recovery after spinal cord injury in adult zebrafish. Neuroscience 183, 238-250. 10.1016/j.neuroscience.2011.03.043 [DOI] [PubMed] [Google Scholar]
- Zenno S., Koike H., Tanokura M. and Saigo K. (1996). Gene cloning, purification, and characterization of NfsB, a minor oxygen-insensitive nitroreductase from Escherichia coli, similar in biochemical properties to FRase I, the major flavin reductase in Vibrio fischeri. J. Biochem. 120, 736-744. 10.1093/oxfordjournals.jbchem.a021473 [DOI] [PubMed] [Google Scholar]
- Zoltowski B. D., Vaccaro B. and Crane B. R. (2009). Mechanism-based tuning of a LOV domain photoreceptor. Nat. Chem. Biol. 5, 827-834. 10.1038/nchembio.210 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.