ABSTRACT
The cortical and medullary thymic epithelial cell (cTEC and mTEC) lineages are essential for inducing T cell lineage commitment, T cell positive selection and the establishment of self-tolerance, but the mechanisms controlling their fetal specification and differentiation are poorly understood. Here, we show that notch signaling is required to specify and expand the mTEC lineage. Notch1 is expressed by and active in TEC progenitors. Deletion of Notch1 in TECs resulted in depletion of mTEC progenitors and dramatic reductions in mTECs during fetal stages, consistent with defects in mTEC specification and progenitor expansion. Conversely, forced notch signaling in all TECs resulted in widespread expression of mTEC progenitor markers and profound defects in TEC differentiation. In addition, lineage-tracing analysis indicated that all mTECs have a history of receiving a notch signal, consistent with notch signaling occurring in mTEC progenitors. These data provide strong evidence for a requirement for notch signaling in specification of the mTEC lineage.
KEY WORDS: Foxn1, Notch, Lineage, mTEC, Thymus
Summary: Medullary thymic epithelial cell lineage specification and progenitor formation in the fetal thymus require notch signaling via notch 1 receptors, whereas further differentiation requires notch 1 to be downregulated.
INTRODUCTION
Notch signaling is a highly conserved pathway that plays a major role in the regulation of embryonic development and controls processes such as cell fate specification, differentiation and proliferation (Kopan, 2012). Notch is a transmembrane receptor protein, of which there are four (NOTCH1-NOTCH4) in mammals. Importantly, notch ligands are also membrane-bound, ensuring that ligand–receptor interactions can only occur between adjacent cells. Binding of a ligand to the receptor triggers a proteolytic event that cleaves the intracellular domain of the receptor, allowing it to enter the nucleus and regulate the expression of downstream genes.
The thymus is the primary lymphoid organ required for T cell production. The functional component of the thymus is comprised of thymic epithelial cells (TECs), which form a unique three-dimensional network that can be broadly divided into an outer cortex and an inner medulla. T cell differentiation takes place primarily via interactions between differentiating T cells and TECs, and a complete, organized and fully functional TEC compartment is essential for production of a diverse and self-tolerant T cell repertoire. Positive selection of T cells takes place in the cortex, where thymocytes capable of recognizing self-major histocompatibility complex (MHC) molecules are selected. The cells then enter the medulla and undergo negative selection to generate self-tolerant T cells that leave the thymus and enter the periphery. Notch signaling within lymphoid progenitor cells upon entry into the thymus is required for establishing T cell fate. Lymphocyte progenitors receive a notch signal immediately upon entering the thymus, via interactions with the delta-like 4 (DLL4) ligand on TECs (Hozumi et al., 2008), that instructs them to commit to the T cell rather than alternative lineages (Pui et al., 1999). Notch signaling is also required at multiple stages during T cell development for a variety of functions, including CD4 versus CD8 lineage commitment (Maekawa et al., 2003). In addition to these crucial and well-established roles in T cell differentiation, functional evidence has begun to emerge that suggests a role for notch signaling in TECs. In addition to notch ligands, TECs also express notch receptors and pathway components (Griffith et al., 2009; Masuda et al., 2009). Gain-of-function experiments suggest that notch signaling is required to induce TEC development, particularly in the medullary lineage (Masuda et al., 2009; Goldfarb et al., 2016). These initial studies suggest that notch signaling could play important roles in the differentiation of both the lymphoid and epithelial compartments. However, definitive in vivo experiments to establish the normal roles of notch signaling in TEC development have not been performed.
All TECs have a single embryonic origin in the third pharyngeal pouch endoderm (Gordon et al., 2004), and functional studies suggest that TECs arise from a common thymic epithelial progenitor cell (TEPC) (Bennett et al., 2002; Depreter et al., 2008; Bleul et al., 2006). The precise developmental origin of TEC subsets is the subject of ongoing debate. There is evidence for both bipotent progenitors in the fetal mouse thymus (Bleul et al., 2006; Rossi et al., 2006) and for lineage-specific progenitors for cortical TECs (cTECs) (Ripen et al., 2011; Shakib et al., 2009) and medullary TECs (mTECs) (Hamazaki et al., 2007; Rodewald et al., 2001). There is also compelling evidence to suggest that a common progenitor population gives rise to mTEC lineage-specific progenitors (Hamazaki et al., 2007). Identifying key molecules involved in specification and maintenance of these different types of TEPCs will help to elucidate how and when each lineage is specified during embryonic development.
We performed a series of loss- and gain-of-function and lineage tracing experiments to investigate the specific role of NOTCH1 signaling in fetal TEC development. Our results indicate a requirement for notch signaling in the establishment and maintenance/expansion of the mTEC progenitor pool in the fetal thymus. We also provide evidence that, although all mTEC experience notch signaling, only a subset of cTECs experience active notch signaling, suggesting that notch signaling may play a previously unappreciated role in cTEC differentiation.
RESULTS
NOTCH1 activity in TEC progenitors in the fetal thymus
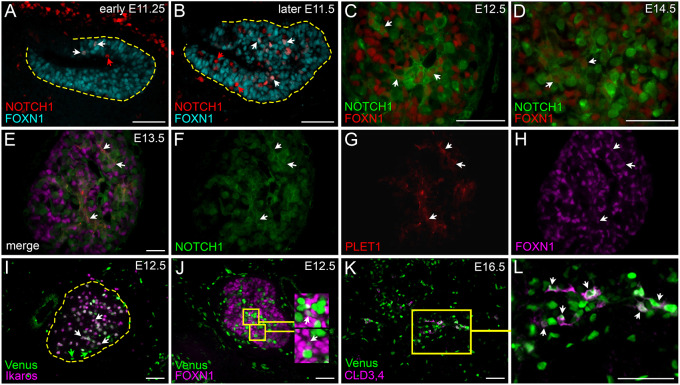
The notch receptors and their downstream targets are expressed on TECs during late fetal development (Masuda et al., 2009) [see accompanying paper (Liu et al., 2020)]. We first used immunohistochemistry (IHC) to assess NOTCH1 expression and activity in the developing thymus, as indicated by nuclear localization of cleaved NOTCH1. We first detected NOTCH1 in the nucleus in a few cells in the thymus primordium at embryonic day (E) 11.25, some of which were FOXN1+, and therefore TECs (Fig. 1A; white arrows). Thus, active NOTCH1 signaling was first detected in a few TECs around the time of initial Foxn1 expression (E11.25), and is present in a subset of TECs at later stages. More FOXN1+ cells undergoing active NOTCH1 signaling were detected in the primordium just a few hours later (Fig. 1B), and were also present at E12.5 (Fig. 1C) and E14.5 (Fig. 1D). Next, we assessed NOTCH1 expression in TEC progenitor cells (TEPC) using an antibody against PLET1, a TEPC marker (Bennett et al., 2002; Depreter et al., 2008; Ulyanchenko et al., 2016). NOTCH1+FOXN1+PLET1+ TECs were detected in the thymus at E13.5 (Fig. 1E-H), suggesting that NOTCH1 signaling plays a role in early TEPCs during fetal thymus development.
Fig. 1.
Notch1 expression and notch activity in the fetal thymus. (A,B) Immunofluorescence of E11.25 (A) and E11.5 (B) wild-type thymus for cleaved NOTCH1 (red) and FOXN1 (cyan). White arrows, co-expressing cells; red arrows, NOTCH1+;FOXN1− cells; dashed line outlines the primordium. (C,D) Immunofluorescence of E12.5 (C) and E14.5 (D) wild-type thymus for FOXN1 (red) and NOTCH1 (green). (E-H) Immunofluorescence of E13.5 wild-type thymus for NOTCH1 (green), PLET1 (red), and FOXN1 (magenta). (I) Immunofluorescence of E12.5 CBF:H2B-Venus thymus for expression of ikaros (magenta) and GFP (Venus; green). Green arrows, Venus expression in non-lymphocytes; dashed line outlines the thymus lobe. (J) Immunofluorescence of E12.5 CBF:H2B-Venus thymus for FOXN1 (magenta) and GFP (Venus; green); insets show higher magnification of double-positive cells. (K,L) Immunofluorescence of E16.5 CBF:H2B-Venus thymus for expression of Venus (green) and CLD3,4 (magenta). Box in K is the area shown in L at higher magnification. n>3 for all experiments. Scale bars: 50 µm
To further assess notch signaling in the fetal thymus, we used a CBF:H2B-Venus transgenic mouse line that expresses nuclear-localized Venus in cells undergoing active or recent notch signaling (Nowotschin et al., 2013). Flow cytometric analysis of intracellular staining with an antibody against the NOTCH1 intracellular domain (NOTCH1-IC) at E16.5 showed that although a minority of TECs were NOTCH1-IC positive, a majority of these NOTCH1-IC+ TECs were Venus-expressing cells (Fig. S1J). Furthermore, nearly all Venus+ TECs were NOTCH1-IC positive (Fig. S1J). The few Venus+ NOTCH1-IC− cells may have recently experienced NOTCH1 signaling (Nowotschin et al., 2013).
At E12.5, almost all Venus+ cells were Ikaros+ thymocytes, which are expected to be undergoing active notch signaling (Fig. 1I; white arrows). However, a few Venus+Ikaros− cells were also present at this stage (Fig. 1I; green arrows), and co-staining with FOXN1 confirmed the presence of a few Venus+ TECs (Fig. 1J). Venus+FOXN1+ TECs were more numerous at E16.5, and were also P63+ (Fig. S1A-G). Claudin-3 and claudin-4 (CLD3,4) mark mTEC progenitors (mTEPCs) in the fetal thymus at mid-gestation (Hamazaki et al., 2007); at E16.5 nearly all CLD3,4+ cells (Fig. 1K,L) and most UEA1+ mTECs (Fig. S1H,H′), which at this stage represent immature mTECs, expressed the Venus transgene, indicating they are experiencing notch signaling.
These data indicate that NOTCH1 signaling in TECs begins soon after the onset of Foxn1 expression in a subset of cells that may represent progenitors, and that by E16.5 NOTCH1 signaling may act specifically in mTEPCs.
NOTCH1 deletion in TEC results in fewer TEPCs in the fetal thymus
Because Notch1 is expressed by a subset of fetal TECs, including potential mTEPCs, we used a loss-of-function approach to determine the role of NOTCH1 signaling in TEC differentiation. We used a Notch1flox conditional allele (Yang et al., 2004) together with a Foxn1Cre strain (Gordon et al., 2007) to remove NOTCH1 function from TECs at the onset of their differentiation. Foxn1Cre;Notch1fx/fx mice had many fewer NOTCH1+ cells overall, with almost no FOXN1+NOTCH1+ cells (Fig. S1P-V), and a similar reduction in Venus+ FOXN1+ cells (Fig. S1J-O), consistent with efficient TEC-specific deletion.
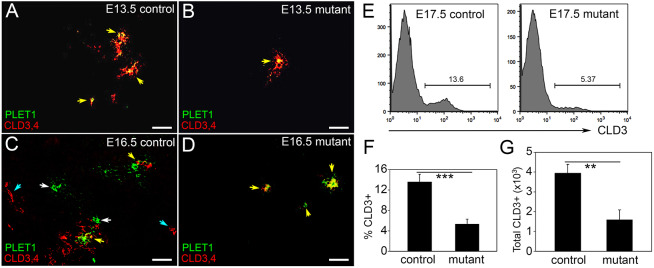
To determine the effect of loss of Notch1 on TEPC populations during fetal thymus development, we performed IHC for PLET1 (Depreter et al., 2008) and CLD3,4 (Hamazaki et al., 2007). Although CLD3,4 is a marker for the mTEPC by E15.5, it is expressed throughout the pouch at E10.5 (Hamazaki et al., 2007), and at E13.5 CLD3,4 co-localizes with PLET1 in presumed bipotent progenitors (Fig. 2A). Thus, in the control thymus, PLET1 and CLD3,4 were co-expressed at E13.5, whereas at E16.5 only a few cells co-expressed these markers (Fig. 2C; yellow arrows); most were positive for PLET1 or CLD3,4, but not both. These cells were arranged such that PLET1+CLD3,4+ double-positive (DP) cells were in clusters with PLET1+ and/or CLD3,4+ single-positive (SP) cells (Fig. 2C), rather than in homotypic clusters. This pattern is consistent with a model in which the earliest bipotent progenitor pool at E13.5 is PLET1+CLD3,4+ and expresses NOTCH1. The subset of those that experience notch signaling continue to be PLET1+CLD3,4+, commit to the mTEC lineage, then downregulate PLET1 as they differentiate. Those early progenitors that do not experience notch signaling downregulate CLD3,4 and stay PLET1+, generating a PLET1+CLD3,4− bipotent progenitor that emerges at about E15.5 (Gordon et al., 2004; Depreter et al., 2008; Ulyanchenko et al., 2016; Cook, 2010).
Fig. 2.
Notch1 deletion in TECs results in fewer TEPCs in the fetal thymus. (A-D) Immunofluorescence of E13.5 (A,B) and E16.5 (C,D) Foxn1Cre;Notch1fx/fx mutant (B,D) and control (A,C) thymi for CLD3,4 (red) and PLET1 (green). White arrows, PLET1+;CLD3,4− cells; cyan arrows, PLET1−;CLD3,4+ cells; yellow arrows, PLET1+;CLD3,4+ cells. (E) Histogram showing CLD3+ cells in Foxn1Cre;Notch1fx/fx mutant and control thymi at E17.5. (F,G) Percentage (F) and total number (G) of CLD3+ TECs in mutant and control thymi at E17.5. ***P≤0.001, **P≤0.005. n>3 for IHC, n=5 for flow cytometry. Scale bars: 50 µm.
In the Foxn1Cre;Notch1fx/fx mutant thymus at E13.5 these PLET1+CLD3,4+ clusters were rare and not always present (Fig. 2B). Not only were there fewer PLET1+ or CLD3,4+ cells overall at E16.5, but all positive cells continued to express both PLET1 and CLD3,4 (Fig. 2D). The reduction in both the percentage and number of CLD3+ cells was confirmed by flow cytometry at E17.5 (P<0.05) (Fig. 2E-G). Caspase 3 staining at E17.5 showed that the few apoptotic cells present in controls were not FOXN1+ TECs (Fig. S1P-S), and that although there was a dramatic increase in apoptotic cells in the mutants, none were FOXN1+ (Fig. S1T-W).
Together, these data show that Notch1 deletion from TECs results in fewer putative fetal TEPCs, particularly mTEPCs, as shown by fewer cells expressing PLET1 and CLD3,4. Furthermore, there were few or no PLET1−CLD3,4+ cells in the mutant thymus, suggesting a specific role for NOTCH1 in the lineage restriction of mTEPCs from a common progenitor during fetal thymus development.
TEC differentiation and organization is abnormal in Foxn1Cre;Notch1fx/fx mutants
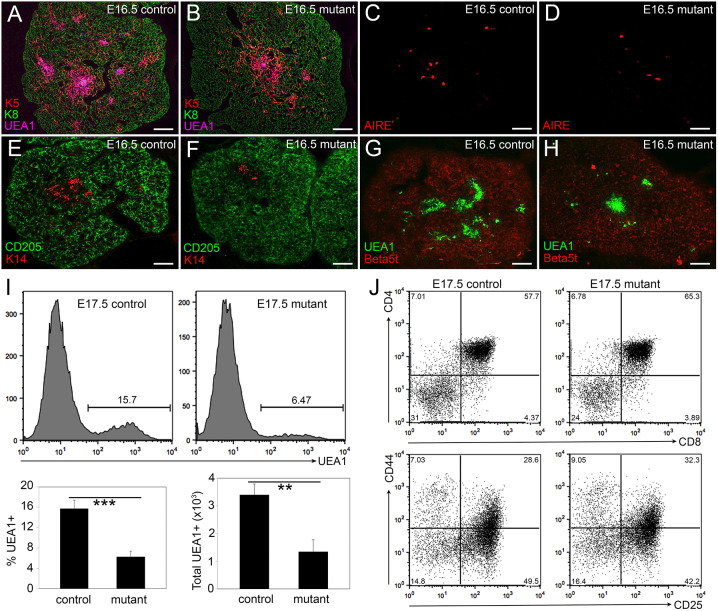
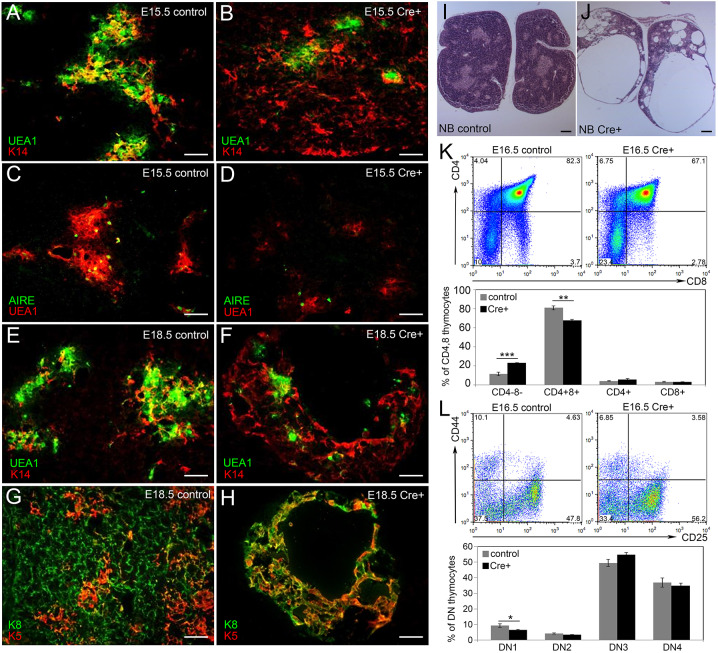
To assess TEC differentiation and function after Notch1 deletion, we performed IHC using a well-defined panel of markers that identify specific TEC subsets within the cortical and medullary compartments of the fetal thymus. We used keratin 8 (K8), CD205 and β5t to label cTECs, and keratin 5 (K5), keratin 14 (K14), AIRE and the lectin UEA1 to label mTEC subpopulations. In controls at E16.5, small distinct regions of cells positive for K5, K14 or UEA1 marked the newly expanding medulla in the developing thymus (Fig. 3A,E,G). In the Foxn1Cre;Notch1fx/fx mutant thymus, the medulla primarily consisted of one larger central region rather than several smaller islands (Fig. 3B,E,F,H). This phenotype was also seen at the newborn stage (not shown). Furthermore, there were dramatically fewer AIRE+ cells in the mutant thymus at E16.5 (Fig. 3C,D); an average of 21 cells per section for the control versus only one cell per section for the mutant, suggesting a nearly complete block in mTEC terminal differentiation. Flow cytometry confirmed the reduction in the number and frequency of mTECs in the Foxn1Cre;Notch1fx/fx mutant thymus at E17.5 (P<0.05; Fig. 3I).
Fig. 3.
Notch1 deletion from TECs affects mTEC organization and differentiation. (A,B) Immunofluorescence of E16.5 Foxn1Cre;Notch1fx/fx mutant (B) and control (A) thymus for K5 (red), K8 (green) and UEA1 (magenta). (C,D) Immunofluorescence of E16.5 Foxn1Cre;Notch1fx/fx mutant (D) and control (C) thymus for AIRE. (E,F) Immunofluorescence of E16.5 Foxn1Cre;Notch1fx/fx mutant (F) and control (E) thymus for K14 (red) and CD205 (green). (G,H) Immunofluorescence of E16.5 Foxn1Cre;Notch1fx/fx mutant (H) and control (G) thymus for UEA1 (green) and β5t (red). (I) Flow cytometry showing histogram (top), percentage (bottom left) and total number (bottom right) of UEA1+ cells in Foxn1Cre;Notch1fx/fx mutant and control thymi at E17.5. (J) Flow cytometric analysis of intrathymic thymocytes from E17.5 Foxn1Cre;Notch1fx/fx mutant and control thymi stained for CD4, CD8, CD25 and CD44. Top panels show CD4 versus CD8; bottom panels show double-negative subsets with CD44 versus CD25. ***P≤0.001, **P≤0.005. n>3 for IHC; n>5 for flow cytometry. Scale bars: 50 µm.
As total TEC numbers were similar in the E17.5 control and Foxn1Cre;Notch1fx/fx mutants (P=0.32), the reduction in mTEC frequency was correlated with a relative increase in cTEC frequency (controls, 84.26±1.65; mutants, 93.71±1.11; P=0.0001), although cTEC numbers were not significantly different (P=0.27; Fig. S2T-W). Expression of cTEC markers β5t and CD205 appeared similar to controls at E16.5 (Fig. 3E-H). Therefore, the primary defect in fetal TECs based on this analysis was in the mTEC lineage. At the newborn stage, TEC phenotypes remained consistent with those at fetal stages, including the decline in both total TEC and mTEC number and frequency, increased frequency but similar number of cTECs, and declines in PLET1+, CLD3,4+, AIRE+ and CD80+ TECs (Fig. S2).
Because the TEC microenvironment governs thymocyte development, we determined whether the observed TEC defects affected thymocyte populations. All stages of thymocyte differentiation require interactions with TECs. Early T cell precursors express neither CD4 nor CD8 and are termed double-negative (DN) thymocytes, which can be subdivided into four differentiation stages based on CD44 and CD25 expression. DN cells become CD4+CD8+ DP thymocytes, which undergo positive and negative selection, generating CD4+ and CD8+ SP T cells. The percentages of intrathymic DN, DP and SP cells, and of DN subsets in Foxn1Cre;Notch1fx/fx were similar to controls at E16.5 (Fig. 3J), the newborn stage and postnatal day 5 (P5; Fig. S4A,B). Further analysis of P5 thymocytes showed that maturation of αβTCR+ SP cells was decreased, based on CD69 and CD24 expression (Fig. S4C-E). γδTCR+ and FOXP3+CD4+ regulatory T cells (Treg) were also significantly decreased in newborn mutant (data not shown) and P5 (Fig. S4F-J) stages. These defects in SP and Tregs were consistent with deficiency in the mTEC compartment. However, by 2 weeks of age, the frequency and number of both cTECs and mTECs was similar to those of controls (Fig. S3).
In summary, Notch1 deletion in TECs at the onset of Foxn1 expression affects TEC organization and mTEC differentiation, but does not obviously affect T cell development in the fetal thymus. Postnatally, mTEC numbers improved, consistent with either expansion of those mTEPC that escaped early Notch1 deletion, and with redundancy with Notch2, which is also expressed widely in TECs [see accompanying paper (Liu et al., 2020); Ki et al., 2014].
Constitutive activation of notch signaling in TECs leads to an increase in TEPCs and a block in mTEC differentiation
Given that Notch1 deletion resulted in fewer TEPCs and an apparent block or reduction in fetal mTEC differentiation, we predicted that Notch1 overexpression might have the opposite effect. We therefore activated NOTCH1 signaling in all TECs from the onset of their differentiation in gain-of-function experiments using a RosaN1-IC inducible strain (Murtaugh et al., 2003) activated by the Foxn1Cre deleter strain (Gordon et al., 2007). In the RosaN1-IC mice, Cre-mediated deletion of a loxp/stop/loxp cassette results in heritable, constitutive expression of the NOTCH1 intracellular domain (N1-IC), resulting in constitutive NOTCH1-mediated signaling. We analyzed Foxn1Cre;RosaN1-IC embryos using markers of TECs, TEPCs and developing T cells.
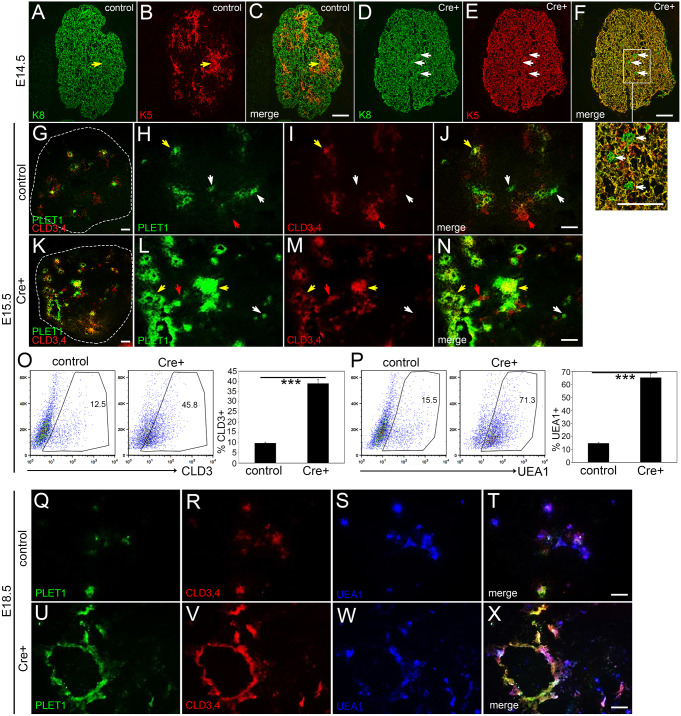
Although K5 and K8 are markers for medullary and cortical TECs, respectively, cells that co-express these markers are thought to contain a progenitor population, and are normally located at the cortico-medullary junction (Klug et al., 1998). In the control E14.5 thymus, proto-medullary areas were beginning to downregulate K8 in the center surrounded by a band of K8+K5+ cells, whereas the rest of the TECs were K5-negative, delineating the emerging cortical and medullary regions (Fig. 4A-C). However, in the Foxn1Cre;RosaN1-IC thymus at the same stage, almost all TECs were K8+K5+, with only a few single K8+ cells (Fig. 4D-F and inset). Furthermore, both PLET1+ and CLD3,4+ cells were expanded in the Foxn1Cre;RosaN1-IC thymus at E15.5 (Fig. 4G-N). Although PLET1 and CLD3,4 single-positive cells were present in the Foxn1Cre;RosaN1-IC thymus, most of these cells expressed both markers and thus probably represent mTEC-committed progenitors. Flow cytometry at E15.5 showed about a fourfold expansion in the frequency of CLD3+ cells in the Foxn1Cre;RosaN1-IC mutant thymus compared with littermate controls (P≤0.001; Fig. 4O), and the number of CLD3+ cells more than doubled in the mutant (mean=1233±387.1 (s.d.) versus 496±50.1 cells in the controls; n=3; P=0.03). Total TEC cellularity was not different between mutant and control at this stage (P=0.32). Flow cytometry for UEA1 also revealed a dramatic expansion of the medullary compartment in the Foxn1Cre;RosaN1-IC mutant thymus (P<0.05; Fig. 4P). This relative increase in progenitor-like phenotypes persisted at E18.5, by which time cysts lined with PLET1+ and CLD3,4+ cells had begun to appear (Fig. 4Q-X).
Fig. 4.
Notch1 activation in TECs causes an increase in the number of TEPCs at fetal stages. (A-F) Immunofluorescence of E14.5 Foxn1Cre;RosaN1-IC Cre+ (D-F) and control (A-C) thymus for K5 (red) and K8 (green). White arrows, K8+;K5− cells; yellow arrows, K8+;K5+ cells. (G-N) Immunofluorescence of E15.5 control (G-J) and Foxn1Cre;RosaN1-IC Cre+ (K-N) thymus for PLET1 (green) and CLD3 (red). Dashed lines in G and K outline the thymus lobe. White arrows, PLET1+;CLD3,4− cells; red arrows, PLET1−;CLD3,4+ cells; yellow arrows, PLET1+;CLD3,4+ cells (O) Flow cytometric analysis of CLD3 expression in TECs from E15.5 Foxn1Cre;RosaN1-IC Cre+ and control thymi. (P) Flow cytometric analysis of UEA1 expression in TECs from E15.5 Foxn1Cre;RosaN1-IC Cre+ and control thymi. For O,P, dot plots show one representative thymus; bar graph shows average values for three thymi. (Q-X) Immunofluorescence of E18.5 control (Q-T) and Foxn1Cre;RosaN1-IC Cre+ (U-X) thymus for PLET1 (green), CLD3 (red) and UEA1 (blue). ***P≤0.001. n>5 for IHC; n=3 for flow cytometry. Scale bars: 50 µm.
mTEC differentiation did not occur normally in the Foxn1Cre;RosaN1-IC thymus. At E15.5, instead of the normal isolated islands of K14 expression (Fig. 5A), K14 was present throughout the mutant thymus, similar to K5 (Fig. 5B). There were also fewer and smaller clusters of UEA1+ cells (Fig. 5B,D) and very few AIRE+ cells compared with controls (Fig. 5C,D), indicating a block in mTEC terminal differentiation. By E18.5, this phenotype had progressed further. Although widespread expression of K8, K5 and K14 showed that the thymus was still epithelial in nature (Fig. 5E-H), there was an almost complete absence of any recognizable organ structure at the newborn stage, as the epithelial network had essentially collapsed and the thymus was composed almost entirely of large cysts (Fig. 5I,J). Together, these data suggest that prolonged NOTCH1 signaling in TECs forces mTEC lineage commitment, but prevents differentiation, ultimately leading to a complete collapse of the TEC network.
Fig. 5.
Ectopic expression of Notch1 in all TECs blocks fetal TEC differentiation and affects T cell development. (A,B) Immunofluorescence of E15.5 Foxn1Cre;RosaN1-IC Cre+ (B) and control (A) thymus for K14 (red) and UEA1 (green). (C,D) Immunofluorescence of E15.5 Foxn1Cre;RosaN1-IC Cre+ (D) and control (C) thymus for UEA1 (red) and AIRE (green). (E,F) Immunofluorescence of E18.5 Foxn1Cre;RosaN1-IC Cre+ (F) and control (E) thymus for K14 (red) and UEA1 (green). (G,H) Immunofluorescence of E18.5 Foxn1Cre;RosaN1-IC Cre+ (H) and control (G) thymus for K5 (red) and K8 (green). (I,J) H&E staining of newborn (NB) Foxn1Cre;RosaN1-IC Cre+ (J) and control (I) thymus. (K) Flow cytometric analysis of thymocytes from E16.5 Foxn1Cre;RosaN1-IC Cre+ and control thymi stained for CD4 and CD8. (L) Flow cytometric analysis of thymocytes isolated from E16.5 Foxn1Cre;RosaN1-IC Cre+ and control thymi stained for double-negative subsets using CD44 and CD25. For K,L, dot plots show one representative thymus for each genotype; bar graph shows average values for at least 5 thymi. ***P≤0.001, **P≤0.005, *P≤0.01. n>5 for IHC; n>5 for flow cytometry. Scale bars: 50 µm.
In contrast to the loss-of-function models, thymocyte development was affected by the abnormal TEC microenvironment in the Foxn1Cre;RosaN1-IC mice. The strongest effect was on total thymocyte numbers, which were reduced in the Foxn1Cre;RosaN1-IC thymus to 1.9×106±0.51 (s.d.) compared with 12.5×106±2.12 (s.d.) thymocytes in the control (P=0.002; Fig. S5G). However, thymocyte differentiation was only mildly affected. Flow cytometry analysis of E16.5 Foxn1Cre;RosaN1-IC thymocytes revealed a slightly lower percentage of CD4+CD8+ cells (Fig. 5K) and an increase in cells at the DN3 stage (CD44−CD25+) (Fig. 5L) in E16.5 mutant thymus compared with controls, suggesting a mild block at the DN3-DN4 transition. By late fetal stages, the thymic structure had deteriorated beyond the ability to support any thymocyte development. At P6, the thymus had become a large clear cyst in Foxn1Cre;RosaN1-IC mice (Fig. S5A). Consistent with a lack of thymic T cell production, the spleen was much smaller than in wild-type controls at P6 (Fig. S5B), with both CD4 and CD8 T cells dramatically decreased (Fig. S5C-D). Surprisingly, spleen size was similar to that in controls by 4 weeks (Fig. S5B), although T cell numbers remained very low.
Thus, enforced notch signaling throughout the TEC compartment during fetal development results in an abnormal TEC environment with an expanded mTEPC compartment, a major block to mTEC differentiation, and eventually causes complete collapse of the epithelial network. These data further support a role for NOTCH1 signaling in specifying the mTEPC pool during fetal development. These data also suggest that although NOTCH1 must be present for mTEPC specification, prolonged and/or excessive NOTCH1 signaling is detrimental to their differentiation.
Mosaic deletion of Notch1 shows that mTEC specification requires notch signaling
Foxn1Cre initiates Cre expression at E11.25 (Gordon et al., 2007), very similar to the timing with which mTEC specification may initiate (Nowell et al., 2011), and coincident with our expression data showing active NOTCH1 signaling in TECs in the developing thymus until E11.25 (Fig. 1A). Thus, it is possible that the few mTECs that are present in the Foxn1Cre;Notch1fx/fx mutant thymus underwent specification prior to Notch1 deletion. Because Foxn1Cre is also active throughout TEC differentiation, these cells could have deleted Notch1 after mTEC specification; but since Notch1 expression is dispensable for or even detrimental to mTEC differentiation, this later deletion would have no effect. It would, however, make it impossible for us to determine whether this scenario was correct, as we cannot determine whether Notch1 was deleted before or after mTEC specification in these mTECs.
To test this possibility, we deleted Notch1 from throughout the pharyngeal endoderm using Foxa2CreER with a single pulse of tamoxifen at E8.5 (Gordon et al., 2010), prior to the onset of Foxn1 expression (Gordon et al., 2001). We have previously shown that this single pulse of CRE activity produces a mosaic deletion in the third pharyngeal pouch (Chojnowski et al., 2016), which is ideal for testing whether Notch1-deleted cells can contribute to the mTEC lineage. Foxa2CreER;Notch1fx/fx mice had fetal thymus phenotypes consistent with those obtained using Foxn1Cre, with reductions in both mTEPCs numbers and medullary size (Figs S6,S7). Using PCR primers that selectively amplified either the undeleted or deleted allele, we performed quantitative PCR (qPCR) on sorted cTEC and mTEC populations from Cre-negative controls, Foxa2CreER;Notch1+/fx heterozygotes and Foxa2CreER;Notch1fx/fx homozygous mutants (Fig. 6A-C). As expected for mosaic deletion, all cell populations from all genotypes were positive for the undeleted allele, and the band corresponding to the deleted allele was absent from Cre-negative controls and present in all cell populations in heterozygotes (Fig. 6D). Strikingly, in Foxa2CreER;Notch1fx/fx homozygous mutants only cTEC populations had the deleted allele, which was completely absent in mTECs (Fig. 6D). These data strongly support the conclusion that specification to the mTEC lineage requires NOTCH1 signaling and are consistent with the idea that the mTECs present in the Foxn1Cre;Notch1fx/fx homozygous mutants had specified to the mTEC lineage prior to Foxn1 expression.
Fig. 6.
Notch1-deleted TECs are unable to contribute to the mTEC lineage. (A) Scheme for generating TECs with mosaic Notch1 deletion for analysis. Pregnant dams are injected at E8.5 (8 dpf), embryos are collected at E15.5 and the thymus dissected and dissociated into single cells. (B) Gating for isolation of EpCam+CD45− TECs. (C) Gating for MHCIIlo and MHCIIhi cTEC (UEA-1−) and mTEC (UEA-1+). (D) PCR of genomic DNA with primers specific for the wild-type undeleted and deleted alleles of Notch1. Genotypes and cell populations represented are indicated above each lane.
Notch signaling is required in TECs at multiple fetal stages
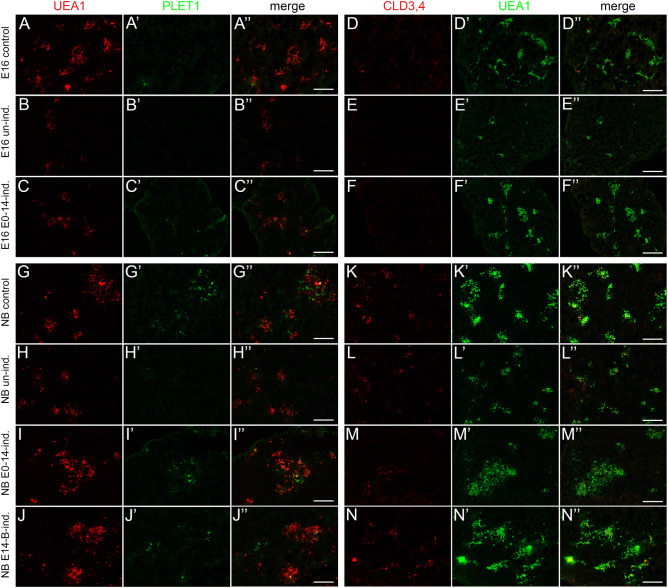
The Foxa2CreER and Foxn1Cre experiments support previous data showing that mTECs start to become specified quite early in thymus organogenesis, at around the time that Foxn1 is first expressed, and that mTEC specification is independent of Foxn1 (Nowell et al., 2011). To test the timing of Notch1 requirement in TECs across fetal development, we utilized a genetic system in which the notch pathway transcription factor RBPj is first deleted in all TECs using Foxn1Cre. Then, the capacity to respond to normal, physiological notch signals is reactivated in a temporal- and cell type-specific manner using doxycycline-controlled expression of transgenic RBPj-HA (RBPjfx/fx;Foxn1Cre;RosartTA;Teton-RBPj-HA) (Chen et al., 2019). Rbpj deletion using Foxn1Cre resulted in similar phenotypes at E16.5 and newborn stages as in Notch1 deletion, with many fewer mTECs, smaller medullary regions and near complete loss of PLET1+ and CLD3,4+ cells (‘un-induced’; Fig. 7B,E,H,L) [see accompanying paper (Liu et al., 2020)]. We then temporally activated notch signaling responsiveness in TECs by providing doxycycline from E0 to E14 (assayed at E16 and newborn) or from E14 to newborn (assayed at newborn).
Fig. 7.
Analysis of the temporal requirement for NOTCH1 signaling in fetal TEC. Labels on the left refer to the entire row; marker names across the top refer to the entire column. In each row, panels with the same letter are single color or merged versions of the same image. (A,A′,A″ and D,D′,D″) Control RBPjfx/+;Foxn1Cre;RosartTA;Teton-RBPj-HA embryos collected at E16.5 have a wild-type phenotype. (B,B′,B″ and E,E′,E″) Un-induced RBPjfx/fx;Foxn1Cre;RosartTA;Teton-RBPj-HA (RBPjind) embryos collected at E16 have a TEC-specific Rbpj null phenotype. (C,C′,C″ and F,F′,F″) RBPjind embryos injected with doxycycline daily from E0-E14 only, collected at E16. (G,G′,G″ and K,K′,K″) Control embryos collected at newborn (NB) stage. (H,H′,H″ and L,L′,L″) Un-induced RBPjind embryos collected at NB stage. (I,I′,I″ and M,M′,M″) RBPjind embryos injected with doxycycline daily from E0-E14 only, collected at NB stage. (J,J′,J″ and N,N′,N″) RBPjind embryos injected with doxycycline daily from E14-NB only, collected at NB stage. All data in this figure are quantified in Fig. 8. Scale bars: 100 µm.
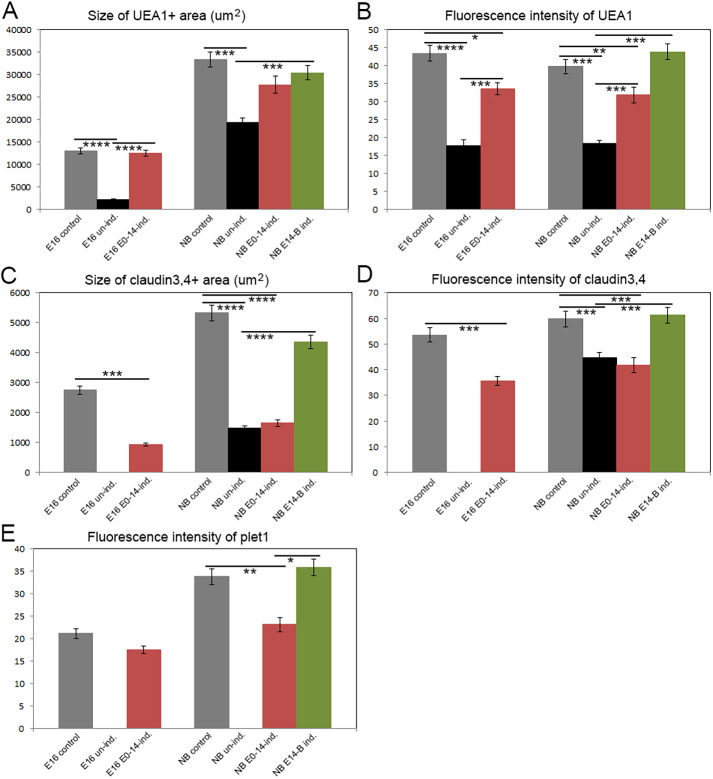
Having normal notch signaling until E14 then withdrawing doxycycline resulted in partial rescue of medullary phenotypes at both E16.5 and newborn stages (Figs 7,8). At E16, medullary area as measured by UEA-1+ cells was normal (Fig. 7F′ and Fig. 8A), although UEA-1 intensity had started to decline (Fig. 8B) and both the number and intensity of CLD3,4+ cells were less than controls (Fig. 7F and Fig. 8C,D). PLET1 staining was also similar to that in controls (Fig. 7A′,C′ and Fig. 8E). Thus, just 2 days after withdrawing notch responsiveness, mTEC markers had begun to decline. By the newborn stage, UEA-1+ area and PLET1 intensity had begun to decline, and UEA-1 intensity remained similar to that at E16.5 (Fig. 7I,I′,M′); these phenotypes were all improved relative to un-induced RBPj mutants, but the values remained less than those of controls (Fig. 8A,B,E). CLD3,4 staining remained similar to that seen at E16.5, and was now also similar to that seen for RBPj mutants, in which CLD3,4+ ‘escapers’ started to accumulate (Fig. 7M and Fig. 8C,D). Thus, notch signaling prior to E14 appears to be sufficient to establish an mTEC pool, but it fails to either expand or be maintained properly after doxycycline withdrawal and removal of notch signaling.
Fig. 8.
Quantification of TEC marker expression in temporal requirement experiments. See Fig. 7 for images. (A,B) Size and fluorescence intensity of UEA1+ areas. (C,D) Size and fluorescence intensity of CLD3,4+ areas. (E) Fluorescence intensity of PLET1+ cells. All quantification was performed using ImageJ (NIH). ****P≤0.0001, ***P≤0.001, **P≤0.005, *P≤0.01. n>3.
In contrast, restoration of notch signaling responsiveness beginning at E14 and continuing until birth substantially restored medullary phenotypes at the newborn stage. UEA-1, CLD3,4 and PLET1 intensities were all similar to controls and significantly increased relative to both uninjected and E0-E14 injected samples (Fig. 7J,J′,M,M′ and Fig. 8A,B,D,E). Only the number of CLD3,4+ cells (measured as area) remained below that of controls, although significantly improved relative to uninduced and E0-E14 injected samples (Fig. 8C). Furthermore, in both E0-E14 and E14-newborn samples, staining for CLD3,4 and PLET1 was largely non-overlapping, similar to that in controls (Fig. S8) and distinct from the maintenance of overlapping staining seen in E16.5 Foxn1Cre;Notch1fx/fx mutants (Fig. 2), demonstrating that progression from PLET1+CLD3,4+ to expression of only one or the other marker is NOTCH1-dependent.
Together, these data suggest that notch signaling is required not only for initial mTEC lineage specification, but also for maintenance and/or expansion of the mTEPCs throughout fetal stages. These data are also consistent with the possibility that mTEPCs can continue to be specified at later fetal stages.
Lineage analysis of active notch signaling in the fetal thymus
We used two NOTCH1 activity-trap mouse lines to trace the lineage of TECs experiencing relatively high (N1IP::CreLO) or lower (N1IP::CreHI) levels of NOTCH1 activation (Liu et al., 2015). In these two strains, the NOTCH1 intracellular domain was replaced with Cre, such that NOTCH1 signaling triggers proteolytic cleavage and Cre is able to move to the nucleus. We used these two strains to activate a CAG-tdTomato reporter (Madisen et al., 2010) and permanently label cells receiving a NOTCH1 signal and their progeny. Co-staining the resulting fetal thymi with TEC markers allowed us to identify all TECs that arise from N1IP::Cre;tdTomato+ cells through ontogeny. Interestingly, we observed different patterns of NOTCH1 signaling lineage history in the fetal thymus using these two lineage reporter lines (Figs 9,10).
Fig. 9.
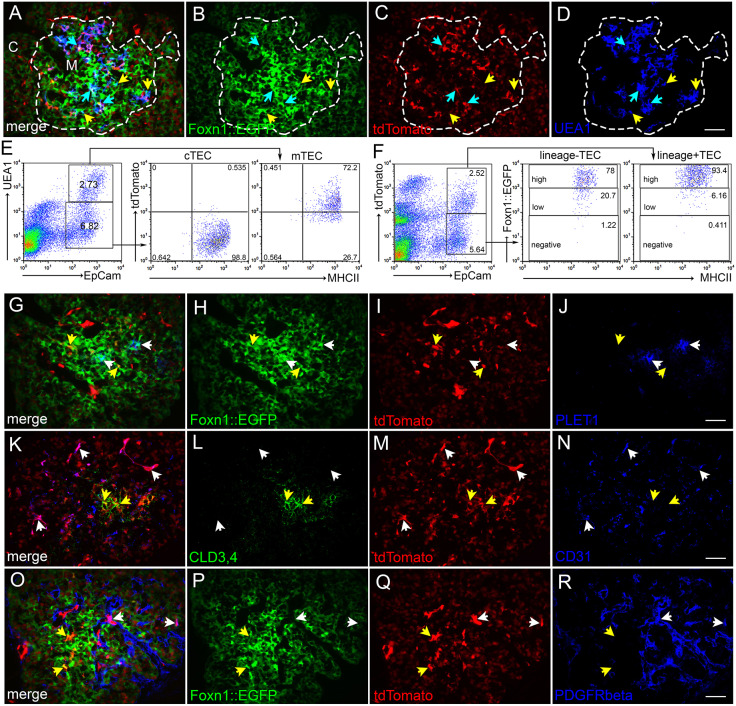
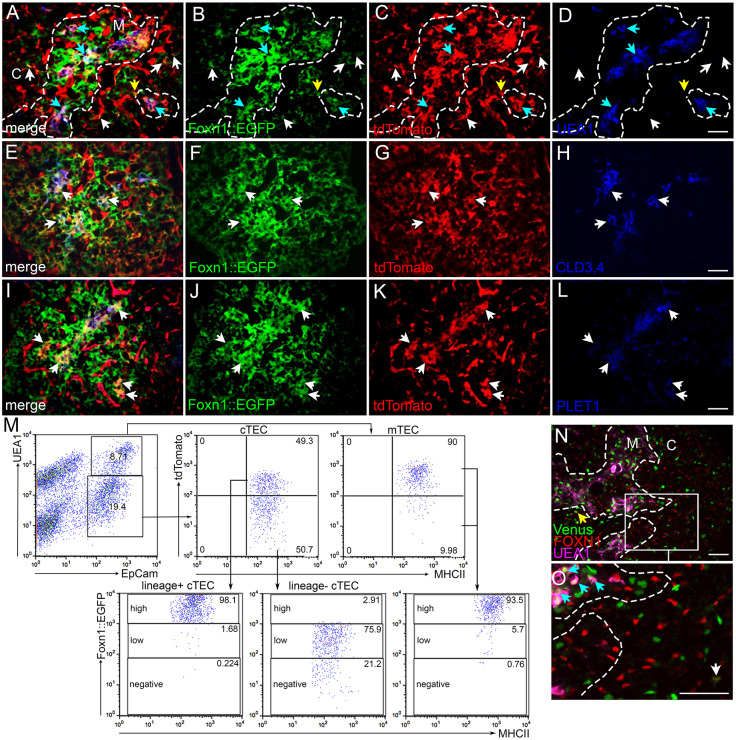
NOTCH1 signaling lineage tracing in TEPCs: N1IP::CreLO;tdTomato. (A-D) Immunofluorescence of E14.5 N1IP::CreLO;tdTomato;FOXN1::EGFP thymus for expression of FOXN1::EGFP (green; B), tdTomato (red; C) and UEA1 (blue; D). Dashed line outlines medulla. Cyan arrows, GFP+;tdTomato+;UEA1+ cells; yellow arrows, GFP+;tdTomato−;UEA1+ cells. (E) Flow cytometric analysis of newborn N1IP::CreLO;tdTomato thymus stained for EpCam, UEA1 and MHCII, showing percentage of UEA1+;MHCIIhi mTECs and UEA1−;MHCIIhi cTECs that express the N1IP::CreLO;tdTomato reporter. (F) Flow cytometric analysis of newborn N1IP::CreLO;tdTomato;FOXN1::EGFP thymus stained for EpCam and MHCII showing FOXN1::EGFP levels in the EpCam+;N1IP::CreLO;tdTomato+ and EpCam+;N1IP::CreLO;tdTomato− TEC populations. (G-J) Immunofluorescence of E14.5 N1IP::CreLO;tdTomato;FOXN1::EGFP thymus for FOXN1::EGFP (green; H), tdTomato (red; I) and PLET1 (blue; J). White arrows, GFP+;tdTomato−;PLET1+ TEPCs; yellow arrows, GFP+;tdTomato+;PLET1− TECs. (K-N) Immunofluorescence of E14.5 N1IP::CreLO;tdTomato thymus for CLD3,4 (green; L), tdTomato (red; M) and CD31 (blue; N). White arrows, CLD3,4−;tdTomato−;CD31+ endothelial cells; yellow arrows, CLD3,4+;tdTomato+;CD31− mTEPCs. (O-R) Immunofluorescence of E14.5 N1IP::CreLO;tdTomato; FOXN1::EGFP thymus for FOXN1::EGFP (green; P), tdTomato (red; Q) and PDGFR-β (blue; R). White arrows, GFP−;tdTomato+;PDGFR-β+ pericytes; yellow arrows, GFP+;tdTomato+;PDGFR-β− TECs. C, cortex. M, medulla. n>3 for IHC; n>5 for flow cytometry. Scale bars: 50 µm.
Fig. 10.
NOTCH1 signaling lineage tracing in TEPCs: N1IP::CreHI;tdTomato. (A-D) Immunofluorescence of E14.5 N1IP::CreHI;tdTomato;FOXN1::EGFP thymus for expression of FOXN1::EGFP (green; B), tdTomato (red; C) and UEA1 (blue; D). White arrows, GFP+;tdTomato+;UEA1− cTECs; yellow arrow, GFP+;tdTomato+;UEA1− cell at the cortico-medullary junction; cyan arrows, GFP+;tdTomato+;UEA1+ mTECs. Dashed line outlines medulla. (E-H) Immunofluorescence of E14.5 N1IP::CreHI;tdTomato;FOXN1::EGFP thymus for FOXN1::EGFP (green; I), tdTomato (red; J) and Cld3,4 (blue; K). Arrows, GFP+;tdTomato+;CLD3,4+ cells. (I-L) Immunofluorescence of E14.5 N1IP::CreHI;tdTomato;FOXN1::EGFP thymus for, FOXN1::EGFP (green; M), tdTomato (red; N) and PLET1 (blue; O). Arrows, GFP+;tdTomato+;PLET1+ cells. (M) Flow cytometric analysis of newborn N1IP::CreHI;tdTomato;FOXN1::EGFP thymus stained for EpCam, UEA1 and MHCII showing percentage of UEA1+ mTECs and UEA1− cTECs that express the N1IP::CreHI;tdTomato reporter. Lower plots show FOXN1::EGFP expression levels in UEA1−;tdTomato+ cTECs and UEA1−;tdTomato− cTECs. (N,O) Immunofluorescence of E16.5 CBF:H2B-Venus thymus for, FOXN1 (red) and UEA1 (magenta). Yellow arrow, Venus+; FOXN1+; UEA1− TECs at the cortico-medullary junction; white arrow, Venus+; FOXN1+; UEA1− TECs in the cortex; cyan arrows, Venus+; FOXN1+; UEA1+ mTECs. Box in N is zoomed area in O. Dashed line outlines medulla. C, cortex. M, medulla. n>3 for IHC; n>5 for flow cytometry. Scale bars: 50 µm.
Analysis of the N1IP::CreLO;tdTomato reporter (Fig. 9) at E14.5 identified only those cells that had themselves or their progenitors experienced a high level of NOTCH1 signaling prior to or at that stage. To assess TECs positive for this marker, we used both the tdTomato reporter and FOXN1::GFP to identify TECs (N1IP::CreLO;tdTomato;FOXN1::EGFP) (see Fig. S9 for gating controls used for these two markers). At E14.5, a subset of medullary TECs marked by UEA1 staining were lineage-positive (Fig. 9A-D, blue arrows), although a substantial fraction of mTECs were lineage-negative (Fig. 9A-D, yellow arrows). Consistent with this result, flow cytometry showed that about 75% of MHCIIhi;UEA1+ mTECs expressed the N1IP::CreLO;tdTomato reporter at the newborn stage (Fig. 9E, right panel), whereas fewer than 1% of MHCIIhi;UEA1− cTECs had experienced high levels of NOTCH1 activity (Fig. 9E, middle panel). Almost all lineage-positive TECs (N1IP::CreLOtdTomato+EpCAM+) and lineage-negative TECs (N1IP::CreLO;tdTomato−) were FOXN1::EGFP+MHCII+ (Fig. 9F), confirming the TEC identity of the cells. In terms of progenitors, CLD3,4+ cells expressed the N1IP::CreLO;tdTomato reporter (Fig. 9L,M, yellow arrows), whereas PLET1+ cells did not (Fig. 9H-J, white arrows). These data are consistent with our CBF:H2B-Venus reporter data (Fig. 1K,L) showing that the mTEPC pool is undergoing active notch signaling; these data specifically show that CLD3,4+ cells have experienced a high level of NOTCH1 signal. Lineage-positive non-TEC cells (N1IP::CreLOtdTomato+ cells negative for TEC markers) were vascular-associated, as indicated by co-expression with CD31 (Fig. 9K-N, white arrows) and PDGFR-β (Fig. 9O-R, white arrows).
Next, we assessed the expression pattern of the N1IP::CreHI;tdTomato reporter, which reports a broader range of NOTCH1 signaling, in the thymus at E14.5 (Fig. 10). Almost all UEA1+ mTECs expressed the N1IP::CreHI;tdTomato reporter at E14.5, as shown by IHC (Fig. 10C,D, cyan arrows), and at the newborn stage using flow cytometric analysis (Fig. 10M). Consistent with our other expression, signaling and lineage results, all CLD3,4+ cells were N1IP::CreHI;tdTomato+ (Fig. 10E-H, arrows). However, in contrast to the results from the N1IP::CreLO reporter, most or all PLET1+ cells were also positive for this reporter (Fig. 10I-L, arrows). These results support a model in which all TEPCs have experienced at least low levels of NOTCH1 signaling, whereas those receiving a high level of signaling commit to the mTEC fate.
Analysis of the N1IP::CreHI;tdTomato reporter in cTECs showed that some lineage-positive FOXN1::GFP+ TECs could also be detected in the cortex (Fig. 10A-D, white arrows). Flow cytometry revealed that about half of all cTECs (EpCam+UEA1−) were tdTomato+ at the newborn stage (Fig. 10M). This finding reveals a previously unidentified split in the cTEC population, based on history of NOTCH1 signaling. Essentially all (>98%) of the NOTCH1 lineage-positive cTECs (EpCam+UEA1−N1IP::CreHItdTomato+) were FOXN1::EGFPhi (Fig. 10M). However, none of the NOTCH1 lineage-negative cTECs expressed a high level of FOXN1::EGFP (Fig. 10M). These FOXN1::EGFP low cells also had lower MHCII surface levels than the FOXN1::EGFP high cells (MFI 256±18.73 (s.d.) versus 360±31.53; P=0.008). Thus, the expression levels of FOXN1 and MHCII are correlated in these cell populations, consistent with previous studies, and the lower levels are also consistent with a less differentiated phenotype.
Finally, to assess the level of current or recent notch signaling as opposed to a history of signaling, we analyzed CBF:H2B-Venus expression at E16.5. Although a substantial fraction of mTECs and all CLD3,4+ mTECs were Venus+FOXN1+, there were only rare Venus+FOXN1+ cells in the cortex (Fig. 10N,O). This result suggests the existence of two distinct populations of cells within the lineage-negative cTECs and suggests that the notch lineage-positive cTECs arise from a relatively small population of cTECs undergoing active notch signaling.
In summary, we have generated a fate map of NOTCH1 signaling during TEC ontogeny using two NOTCH1 activity-trap mouse lines. Our data reveal that all mTECs, but only a subset of cTECs, have experienced NOTCH1 signaling during fetal thymus development.
DISCUSSION
TECs represent the major functional component of the thymus, yet the mechanisms controlling their differentiation during fetal development remain largely unknown, particularly in terms of lineage specification and progenitor cell maintenance. In the current study, we provide evidence that NOTCH1 signaling is required to specify the lineage-restricted mTEPC pool in the fetal thymus. We show that all mTEPCs in the fetal thymus exhibit active NOTCH1 signaling from early in organogenesis and have a lineage history of high levels of notch signaling. Ablation of Notch1 in TECs results in fewer TEPCs and causes a block in specification of mTEPCs, as Notch1-null TECs are unable to contribute to the mTEC lineage after mosaic deletion. In contrast, NOTCH1 activation in TECs results in expansion of the TEPC pool, but then subsequent mTEC differentiation is blocked. These data indicate that notch signaling is required for specification of mTEPCs and promotes their expansion, but that notch signaling must cease for mTEC differentiation to mature phenotypes to occur. The fact the removal of notch signaling in TECs after E14 results in progressive loss of the mTEC population also suggests that notch signaling is required for maintenance of mTEPCs or for their proliferation.
A parallel study in the Blackburn laboratory (University of Edinburgh) targeting Rbpj and thus globally affecting notch signaling came to a similar conclusion (Liu et al., 2020).The similarity in phenotypes from targeting Rbpj and Notch1 suggests that at fetal stages NOTCH1 is the major mediator of notch signaling in TECs. Interestingly, Rbpj deletion continued to show reduced mTECs at 2 weeks postnatal, in contrast to our results where only Notch1 was deleted, even though we used the same Cre driver. This difference suggests that fetal TECs primarily rely on NOTCH1 signaling, whereas NOTCH1 and NOTCH2 may have redundant function in TECs at postnatal stages. However, Notch2 and Notch 3 are also expressed in fetal TEC in a pattern similar to Notch1 (see Liu et al., 2020) and it is unclear why they are unable to provide redundant function at that stage.
The developmental origins of separate cortical and medullary TEC lineages and the existence and identity of bipotent TEPCs remain controversial. Whether they arise from a common bipotent progenitor or individual lineage-restricted progenitors is still uncertain, with evidence for both (Bleul et al., 2006; Rossi et al., 2006; Ripen et al., 2011; Shakib et al., 2009; Hamazaki et al., 2007; Rodewald et al., 2001) Furthermore, it is still unclear exactly when and how the fetal and adult TEPC populations arise and what their relationships might be. Our data do not definitively prove either the bipotent or the individual lineage-restricted progenitor model, but do provide clear indications of how different lineages are related, and show that mTECs and cTECs require different signals for specification.
We identify NOTCH1 as a key molecule required for the establishment and expansion of the mTEPC pool. Our functional studies revealed that NOTCH1 pathway inhibition or activation both affected the mTEPC pool in the fetal thymus. Our data are consistent with a model in which NOTCH1 signaling acts on an early fetal bipotent progenitor, that is PLET1+CLD3,4+, which gives rise to a PLET1−CLD3,4+ mTEC-specific TEPC pool that has experienced high levels of notch signaling, sometime between E13.5 and E16.5. Whether this PLET1+CLD3,4+ TEPC also gives rise to the cortical lineage is not clear, as other lineage studies have suggested that all TECs arise from a progenitor expressing cortical markers (Ripen et al., 2011; Shakib et al., 2009). However, it is clear that cTECs do not all experience notch signaling, at least not at levels we can detect with our lineage reporters, and that cTECs in general can develop in the absence of notch signaling. In either case, our data indicate that a bipotent progenitor would itself probably not experience notch signaling, although its immediate daughter cells could.
We propose a model in which NOTCH1 signaling is required to generate the mTEPC pool during fetal thymus development (Fig. 11). Lineage restriction of these cells occurs according to whether or not the bipotent progenitor itself or its daughter cells, experience high levels of NOTCH1 signaling. In this model, all TECs arise from a common PLET1+CLD3,4+ bipotent progenitor cell present in the early fetal thymus, although it is also formally possible that this early TEPC population contains separate cortical and medullary progenitors. Regardless, those cells that do receive a NOTCH1 signal become PLET1−CLD3,4+ mTEC lineage-restricted TEPCs; those that do not receive a notch signal downregulate the genes encoding CLD3,4 (Cldn3 and Cldn4) to establish a PLET1+CLD3,4− TEPC population by about E15.5. cTECs develop from TEPCs that have never seen a notch signal, either by default or under the influence of a second unknown signal. Thus, when Notch1 is deleted from TECs (as in the Foxn1Cre;Notch1fx/fx and Foxg1Cre;Notch1fx/fx models presented here) the PLET1+CLD3,4+ TEPCs fail to downregulate Plet1 and the mTEPC lineage-restricted pool is not generated; they also fail to downregulate Cldn3 and Cldn4 to establish the PLET1+CLD3,4− TEPC population. Our data also show that Notch1 must be downregulated after mTEPCs are specified for differentiation of the PLET1−CLD3,4+ cells into more mature mTECs, consistent with previous reports (Goldfarb et al., 2016). Thus, in our Foxn1Cre;RosaN1-IC overexpression model, prolonged NOTCH1 signaling prevents mTEC differentiation.
Fig. 11.

Model for the role of NOTCH1 signaling during fetal TEC development. In this model, all fetal TECs derive from a common PLET1+;CLD3,4+ progenitor pool at E13.5 that develops into a PLET1+;CLD3,4− bipotent progenitor (TEPC) by mid-gestation. This PLET1+;CLD3,4− becomes lineage-restricted into either mTEPCs or cTEPCs. Those that experience low levels of notch signaling downregulate PLET1 and upregulate CLD3,4, committing to the mTEC lineage; these mTEPCs then experience high levels of notch signaling to drive initial expansion and differentiation. Notch1 expression must then be downregulated in those cells for mTEC differentiation to result in functional AIRE+ mTECs. The progeny of PLET1+ TEPCs that do not receive a NOTCH1 signal commit to the cTEC lineage. At some point during their differentiation, a separate exposure to low notch signaling results in upregulation of Foxn1, presumably leading to cTEC maturation. It is also possible that the cTEC lineage splits into two different functional populations depending on exposure to low level notch signaling (dotted arrow); in the absence of more cTEC markers and functional information, these two possibilities cannot be distinguished.
Our fate mapping lineage analysis showed that only half of fetal cTECs have experienced NOTCH1 signaling, and that these cTECs have uniformly higher levels of Foxn1 and MHCII expression than those that are notch lineage-negative. These data indicate that notch signaling may also play a role in later cTEC differentiation that is distinct from the mTEC role, uncovering a previously unidentified diversity within cTECs based on having experienced notch signaling or not (Fig. 9). Compared with mTECs, little is known about the cTEC lineage and its development during ontogeny. As these two lineage-negative and lineage-positive populations also differ in their levels of Foxn1 and MHCII expression, it is reasonable to conclude that these populations may be distinct either in their level of maturity or their function. Although we did not detect an obvious change in cTECs in our Notch1 deletion model, the relative lack of cTEC markers means that we have little power to do so based on known markers. As a result, we can only speculate at this point what the relationship between these two cTEC subsets may be. As the lineage-positive cTECs cannot give rise to lineage-negative cTECs due to the nature of our reporters, either the lineage-negative cTECs must give rise to lineage-positive cTECs upon experiencing notch signaling or the two populations arise independently. Regardless, this result indicates that low level notch signaling acts on cTECs, and opens new avenues of investigation into cTEC differentiation.
Notch signaling functions via cell-cell contact, therefore the NOTCH1 signal that TECs experience must be triggered by ligands expressed on adjacent cells. But what are these cells? What cells express the ligand(s) and what are the ligands? The cells could be other TECs, thymocytes, endothelial cells and/or neural crest-derived mesenchymal cells. It has been suggested that thymocytes are at least one source of ligand, and that an interaction between these two cell types is required for TEC development (Masuda et al., 2009). In the current study, we first observed active NOTCH1 signaling in FOXN1+ cells at early E11.5, which is coincident with the first wave of lymphocyte entry to the primordium (Harman et al., 2003), although it is clear in our data that TECs are not adjacent to thymocytes when undergoing notch signaling. Of note, at this early stage there are few cellular sources of notch ligands, and the most likely source based on our expression data is other fetal TECs, which express multiple notch ligands, including jagged 1 and delta-like 4 (Griffith et al., 2009; Masuda et al., 2009; Ki et al., 2014; Harman et al., 2003; Liu et al., 2020). Whether the specific ligands and their cellular source change during ontogeny or have functional consequences for TEC biology remains to be determined.
MATERIALS AND METHODS
Mice
At the University of Georgia
Notch1flox (stock no. 006951), RosaN1-IC (Stock No. 008159), CBF:H2B-Venus (stock no. 020942) and CAG-tdTomato (stock no. 007909) mice were obtained from The Jackson Laboratories (Bar Harbor, ME). N1IP::CreHI and N1IP::CreLO strains were a gift from Dr Raphael Kopan (Cincinnati Children's Hospital Medical Center, Cincinnati, OH, USA) (Liu et al., 2015). FOXN1::EGFP (enhanced green fluorescent protein) mice were a gift from Dr Thomas Boehm (Max Planck Institute of Immunobiology, Freiburg, Germany) (Terszowski et al., 2006). Foxn1Cre and Foxa2Cre strains have been described elsewhere (Gordon et al., 2007; Hébert and McConnell, 2000). All colonies were maintained on a majority C57BL6/J genetic background. At noon on the day of detecting, a vaginal plug was designated embryonic day 0.5 (E0.5) and confirmed by morphological features.
All mice and embryos were genotyped by PCR using DNA extracted from tail tissue. EGFP primer sequences were fwd, 5′-GTTCATCTGCACCACCGGC-3′ and rev, 5′-TTGTGCCCCAGGATGTTG C-3′. Primer sequences for Notch1flox, RosaN1-IC, CBF:H2B-Venus, CAG-tdTomato, Foxn1Cre (Foxn1ex9cre, stock no. 018448), and Foxg1Cre (stock no. 006084) strains are available from The Jackson Laboratories (Bar Harbor, ME). In all cases, Cre-negative animals or embryos were used as littermate controls. Values of n for all experiments are shown in the figure legends.
All experiments involving animals were performed with approval from the UGA Institutional Animal Care and Use Committee.
At the University of Toronto
RBPj-inducible (RBPjind or RBPjfx/fx;RosartTA;Teton-RBPj-HA) mice, described elsewhere (Chen et al., 2019), were bred to Foxn1Cre mice (RBPjfx/fx;Foxn1Cre;RosartTA;Teton-RBPj-HA) and maintained in the Comparative Research Facility of the Sunnybrook Research Institute under specific pathogen-free conditions. All animal procedures were approved by the Sunnybrook Research Institute Animal Care Committee and performed in accordance with the committee's ethical standards. For induction of notch responsiveness, pregnant mice were given 1 mg/ml doxycycline (Sigma-Aldrich) in drinking water supplemented with 5% Splenda ad libitum.
Immunofluorescence and histology
For cryosectioning, mouse embryos were snap-frozen in liquid nitrogen and stored at −80°C. Tissues were sectioned at 8 µm and fixed in ice-cold acetone for 2 min. Tissues were rinsed with phosphate buffered saline (PBS), blocked with 10% donkey serum in PBS for 30 min at room temperature, then incubated with appropriate primary antibodies overnight at 4°C. The following antibodies were used: anti-cleaved NOTCH1 (Cell Signaling Technologies, 4147, 1:200), anti-NOTCH1 (Origene, EP1238Y, 1:200), anti-FOXN1 (Santa Cruz, G-20, 1:200), anti-CD31 (BD, MEC13.3, 1:100), anti-PDGFR-β (R&D Systems, AF1042, 1:50), anti-ikaros (Santa Cruz, M-20, 1:200), anti-GFP (Abcam, ab13970, 1:200), anti-PLET1 (rat supernatant from cell line ID4-20), anti-claudin 3 (Life Technologies, 34-1700, 1:200), anti-claudin 4 (Life Technologies, 36-4800, 1:200), anti-β5t (MBL, PD021, 1:200), anti-CD205 (BioLegend, 138202, 1:200), anti-AIRE (Santa Cruz, M-300, 1:200), anti-K5 (Covance, AF138, 1:1000), anti-K8 (rat supernatant, Troma1), anti-K14 (Covance, AF64, 1:1000) or UEA1 lectin (Vector Labs, X0922, 1:400). Secondary detection was performed with donkey anti-primary species. For N1IP::Cre;tdTomato;Foxn1::EGFP observation, tissues were fixed in 4% paraformaldehyde (PFA) in PBS for 5 min at 4°C, washed with PBS followed by 10% sucrose in PBS for 1 h, then 30% sucrose in PBS overnight. Tissues were embedded in OCT and stored at −80°C until sectioning. Sections were examined by fluorescent microscopy using a Zeiss Axioplan 2 microscope (Thornwood, NY).
For paraffin sectioning, tissues were collected and fixed in 4% PFA for 2-3 h. Tissues were dehydrated through an ethanol series (70, 80, 90, 96 and 100%) and embedded in paraffin wax using standard procedures. Sections (8 µm) were cut and rinsed in xylene before rehydration through a reverse ethanol series. Antigen retrieval was performed by boiling slides in 10 mM sodium citrate buffer, pH 6, for 30 min. Sections were stained using appropriate primary and secondary antibodies as described above, and imaged using fluorescence microscopy.
Hematoxylin and eosin staining was performed on paraffin sections using standard procedures; sections were then imaged on a Zeiss Axioplan microscope.
Flow cytometry
For TEC analysis, fetal or newborn stage thymi were dissected and digested in 1 mg/ml collagenase/dispase (Roche, Basel, Switzerland) and passed through a 100 μm mesh to remove debris. Thymi were processed individually before genotyping. PE-Cy7 conjugated anti-CD45 (BioLegend, 30-F11, 1:150) and APC-conjugated anti-EpCam (BioLegend, G8.8, 1:150) were used to isolate TEC populations. Ly51-PE (BioLegend, 6C3, 1:150), CD80-PE (BioLegend, 16-10A1, 1:150), PLET1, AIRE, UEA1 lectin, anti- claudin 3 and anti-MHCII (M5/114.15.2, BioLegend, 1:150) were used in the TEC analysis. Cells were refixed in 1% PFA in PBS and analyzed using a CyAn ADP Flow Cytometer (Beckman Coulter, Miami, FL). Data were collected using a four-decade log amplifier and stored in list mode for subsequent analysis using FlowJo Software (Tree Star, Ashland, OR).
Thymocytes were harvested from individual fetal or newborn stage thymi and suspended in FACS buffer (PBS containing 2% fetal bovine serum). Thymi were processed individually before genotyping. Cells were incubated with conjugated monoclonal antibodies CD4-FITC (BioLegend, GK1.5, 1:150), CD8-PE (BioLegend, 53-6.7, 1:150), CD25-APC (BD, PC61, 1:150), CD44-PerCP (BioLegend, IM-7, 1:150), TCRβ-PE/Cy7 (BioLegend, H57-597, 1:150), TCRγδ-APC (BioLegend, GL3, 1:150), CD69-APC (BioLegend, H1.2F3, 1:150), CD24-FITC (BioLegend, M1/69, 1:150) or FOXP3-PE (FJK-16s, eBioscience) at 4°C for 30 min, washed and fixed with 1% PFA (EM Sciences, Ft. Washington, PA) before analysis on a CyAn ADP Flow Cytometer (Beckman Coulter, Miami, FL). Data were collected using a four-decade log amplifier and were stored in list mode for subsequent analysis using FlowJo Software.
Cell isolation and genomic PCR
E15.5 thymi were harvested and processed individually to generate a single cell suspension (as described above). TEC populations were isolated based on staining with PE-Cy7-conjugated anti-CD45, APC-conjugated anti-EpCam, UEA1 lectin and anti-MHCII as described in the text. DNA was purified from sorted cell populations using QIAamp DNA Mini kit (QIAGEN). PCR was performed using the following primer sequences: fwd-1 (undeleted allele), 5′-TACTTAGAGCGGGGCAGAGA-3′; fwd-2 (deleted allele), 5′-CTGAGGCCTAGAGCCTTGAA-3′; rev (both deleted and undeleted alleles), 5′-ACTCCGACACCCAATACCTG-3′.
Statistics
Data are presented as mean±s.d. N was at least three for each genotype in each experiment, as indicated in the text and/or figure legends. Comparisons between two groups were made using Student's t-test, multiple comparisons used ANOVA with a Tukey's post-test. P<0.05 or Q<0.05 was considered significant.
Supplementary Material
Acknowledgements
We thank E. Richie for providing the K8 and PLET1 antibodies, and C. C. Blackburn and E. Richie for reading the manuscript and helpful discussions. We thank J. Nelson in the Center for Tropical and Emerging Global Diseases Flow Cytometry Facility at the University of Georgia for flow cytometry and cell sorting technical support.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: J.L., J.C.Z.-P., N.R.M.; Methodology: J.C.Z.-P.; Validation: J.L., J.G., E.L.Y.C., L.W.; Formal analysis: J.L., J.G., N.R.M.; Investigation: J.L., J.G., E.L.Y.C., L.W.; Resources: J.C.Z.-P.; Writing - original draft: J.L., N.R.M.; Writing - review & editing: J.L., J.G., E.L.Y.C., J.C.Z.-P., N.R.M.; Visualization: J.L., J.G., L.W.; Supervision: J.C.Z.-P., N.R.M.; Project administration: J.L., N.R.M.; Funding acquisition: N.R.M.
Funding
This study was supported by the National Institutes of Health (R21 AI107465 to N.R.M.) and a Canadian Institutes of Health Research grant (FND-154332 to J.C.Z.-P.). E.L.Y.C. was supported by an Ontario Graduate Scholarship and J.C.Z.-P. is supported by a Canada Research Chair in Developmental Immunology. Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.178988.supplemental
References
- Bennett A. R., Farley A., Blair N. F., Gordon J., Sharp L. and Blackburn C. C. (2002). Identification and characterization of thymic epithelial progenitor cells. Immunity 16, 803-814. 10.1016/S1074-7613(02)00321-7 [DOI] [PubMed] [Google Scholar]
- Bleul C. C., Corbeaux T., Reuter A., Fisch P., Mönting J. S. and Boehm T. (2006). Formation of a functional thymus initiated by a postnatal epithelial progenitor cell. Nature 441, 992-996. 10.1038/nature04850 [DOI] [PubMed] [Google Scholar]
- Chen E. L. Y., Thompson P. K. and Zuniga-Pflucker J. C. (2019). RBPJ-dependent Notch signaling initiates the T cell program in a subset of thymus-seeding progenitors. Nat. Immunol. 20, 1456-1468. 10.1038/s41590-019-0518-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chojnowski J. L., Trau H. A., Masuda K. and Manley N. R. (2016). Temporal and spatial requirements for Hoxa3 in mouse embryonic development. Dev. Biol. 415, 33-45. 10.1016/j.ydbio.2016.05.010 [DOI] [PubMed] [Google Scholar]
- Cook A. M. (2010). Proliferation and lineage potential in fetal thymic epithelial progenitor cells. Ph.D. thesis, University of Edinburgh. http://hdl.handle.net/1842/4650 [Google Scholar]
- Depreter M. G. L., Blair N. F., Gaskell T. L., Nowell C. S., Davern K., Pagliocca A., Stenhouse F. H., Farley A. M., Fraser A., Vrana J. et al. (2008). Identification of Plet-1 as a specific marker of early thymic epithelial progenitor cells. Proc. Natl. Acad. Sci. USA 105, 961-966. 10.1073/pnas.0711170105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldfarb Y., Kadouri N., Levi B., Sela A., Herzig Y., Cohen R. N., Hollenberg A. N. and Abramson J. (2016). HDAC3 is a master regulator of mTEC development. Cell Rep. 15, 651-665. 10.1016/j.celrep.2016.03.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon J., Bennett A. R., Blackburn C. C. and Manley N. R. (2001). Gcm2 and Foxn1 mark early parathyroid- and thymus-specific domains in the developing third pharyngeal pouch. Mech. Dev. 103, 141-143. 10.1016/S0925-4773(01)00333-1 [DOI] [PubMed] [Google Scholar]
- Gordon J., Wilson V. A., Blair N. F., Sheridan J., Farley A., Wilson L., Manley N. R. and Blackburn C. C. (2004). Functional evidence for a single endodermal origin for the thymic epithelium. Nat. Immunol. 5, 546-553. 10.1038/ni1064 [DOI] [PubMed] [Google Scholar]
- Gordon J., Xiao S., Hughes B., Su D. M., Navarre S. P., Condie B. G. and Manley N. R. (2007). Specific expression of lacZ and cre recombinase in fetal thymic epithelial cells by multiplex gene targeting at the Foxn1 locus. BMC Dev. Biol. 7, 69 10.1186/1471-213X-7-69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon J., Patel S. R., Mishina Y. and Manley N. R. (2010). Evidence for an early role for BMP4 signaling in thymus and parathyroid morphogenesis. Dev. Biol. 339, 141-154. 10.1016/j.ydbio.2009.12.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith A. V., Fallahi M., Nakase H., Gosink M., Young B. and Petrie H. T. (2009). Spatial mapping of thymic stromal microenvironments reveals unique features influencing T lymphoid differentiation. Immunity 31, 999-1009. 10.1016/j.immuni.2009.09.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamazaki Y., Fujita H., Kobayashi T., Choi Y., Scott H. S., Matsumoto M. and Minato N. (2007). Medullary thymic epithelial cells expressing Aire represent a unique lineage derived from cells expressing claudin. Nat. Immunol. 8, 304-311. 10.1038/ni1438 [DOI] [PubMed] [Google Scholar]
- Harman B. C., Jenkinson E. J. and Anderson G. (2003). Entry into the thymic microenvironment triggers Notch activation in the earliest migrant T cell progenitors. J. Immunol. 170, 1299-1303. 10.4049/jimmunol.170.3.1299 [DOI] [PubMed] [Google Scholar]
- Hébert J. M. and McConnell S. K. (2000). Targeting of cre to the Foxg1 (BF-1) locus mediates loxP recombination in the telencephalon and other developing head structures. Dev. Biol. 222, 296-306. 10.1006/dbio.2000.9732 [DOI] [PubMed] [Google Scholar]
- Hozumi K., Mailhos C., Negishi N., Hirano K.-, Yahata T., Ando K., Zuklys S., Holländer G. A., Shima D. T. and Habu S. (2008). Delta-like 4 is indispensable in thymic environment specific for T cell development. J. Exp. Med. 205, 2507-2513. 10.1084/jem.20080134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ki S., Park D., Selden H. J., Seita J., Chung H., Kim J., Iyer V. R. and Ehrlich L. I. R. (2014). Global transcriptional profiling reveals distinct functions of thymic stromal subsets and age-related changes during thymic involution. Cell Rep. 9, 402-415. 10.1016/j.celrep.2014.08.070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klug D. B., Carter C., Crouch E., Roop D., Conti C. J. and Richie E. R. (1998). Interdependence of cortical thymic epithelial cell differentiation and T-lineage commitment. Proc. Natl. Acad. Sci. USA 95, 11822-11827. 10.1073/pnas.95.20.11822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopan R. (2012). Notch signaling. Cold Spring Harbor Perspect. Biol. 4, a011213 10.1101/cshperspect.a011213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Brunskill E., Boyle S., Chen S., Turkoz M., Guo Y., Grant R. and Kopan R. (2015). Second-generation Notch1 activity-trap mouse line (N1IP::CreHI) provides a more comprehensive map of cells experiencing Notch1 activity. Development 142, 1193-1202. 10.1242/dev.119529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, D., Kousa, A. I., O'Neill, K. E., Rouse, P., Popis, M., Farley, A. M., Tomlinson, S. R., Ulyanchenko, S., Guillemot, F., Seymour, P. A. et al (2020). Canonical Notch signaling controls the early thymic epithelial progenitor cell state and emergence of the medullary epithelial lineage in fetal thymus development. Development 147, dev178582. 10.1242/dev.178582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L., Zwingman T. A., Sunkin S. M., Oh S. W., Zariwala H. A., Gu H., Ng L. L., Palmiter R. D., Hawrylycz M. J., Jones A. R. et al. (2010). A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 13, 133-140. 10.1038/nn.2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa Y., Tsukumo S.-, Chiba S., Hirai H., Hayashi Y., Okada H., Kishihara K. and Yasutomo K. (2003). Delta1-Notch3 interactions bias the functional differentiation of activated CD4+ T cells. Immunity 19, 549-559. 10.1016/S1074-7613(03)00270-X [DOI] [PubMed] [Google Scholar]
- Masuda K., Germeraad W. T. V., Satoh R., Itoi M., Ikawa T., Minato N., Katsura Y., Van Ewijk W. and Kawamoto H. (2009). Notch activation in thymic epithelial cells induces development of thymic microenvironments. Mol. Immunol. 46, 1756-1767. 10.1016/j.molimm.2009.01.015 [DOI] [PubMed] [Google Scholar]
- Murtaugh L. C., Stanger B. Z., Kwan K. M. and Melton D. A. (2003). Notch signaling controls multiple steps of pancreatic differentiation. Proc. Natl. Acad. Sci. USA 100, 14920-14925. 10.1073/pnas.2436557100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowell C. S., Bredenkamp N., Tetélin S., Jin X., Tischner C., Vaidya H., Sheridan J. M., Stenhouse F. H., Heussen R., Smith A. J. H. et al. (2011). Foxn1 regulates lineage progression in cortical and medullary thymic epithelial cells but is dispensable for medullary sublineage divergence. PLoS Genet. 7, e1002348 10.1371/journal.pgen.1002348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowotschin S., Xenopoulos P., Schrode N. and Hadjantonakis A.-K. (2013). A bright single-cell resolution live imaging reporter of Notch signaling in the mouse. BMC Dev. Biol. 13, 15 10.1186/1471-213X-13-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pui J. C., Allman D., Xu L., Derocco S., Karnell F. G., Bakkour S., Lee J. Y., Kadesch T., Hardy R. R., Aster J. C. et al. (1999). Notch1 expression in early lymphopoiesis influences B versus T lineage determination. Immunity 11, 299-308. 10.1016/S1074-7613(00)80105-3 [DOI] [PubMed] [Google Scholar]
- Ripen A. M., Nitta T., Murata S., Tanaka K. and Takahama Y. (2011). Ontogeny of thymic cortical epithelial cells expressing the thymoproteasome subunit β5t. Eur. J. Immunol. 41, 1278-1287. 10.1002/eji.201041375 [DOI] [PubMed] [Google Scholar]
- Rodewald H.-R., Paul S., Haller C., Bluethmann H. and Blum C. (2001). Thymus medulla consisting of epithelial islets each derived from a single progenitor. Nature 414, 763-768. 10.1038/414763a [DOI] [PubMed] [Google Scholar]
- Rossi S. W., Jenkinson W. E., Anderson G. and Jenkinson E. J. (2006). Clonal analysis reveals a common progenitor for thymic cortical and medullary epithelium. Nature 441, 988-991. 10.1038/nature04813 [DOI] [PubMed] [Google Scholar]
- Shakib S., Desanti G. E., Jenkinson W. E., Parnell S. M., Jenkinson E. J. and Anderson G. (2009). Checkpoints in the development of thymic cortical epithelial cells. J. Immunol. 182, 130-137. 10.4049/jimmunol.182.1.130 [DOI] [PubMed] [Google Scholar]
- Terszowski G., Müller S. M., Bleul C. C., Blum C., Schirmbeck R., Reimann J., Pasquier L. D., Amagai T., Boehm T. and Rodewald H. R. (2006). Evidence for a functional second thymus in mice. Science 312, 284-287. 10.1126/science.1123497 [DOI] [PubMed] [Google Scholar]
- Ulyanchenko S., O'Neill K. E., Medley T., Farley A. M., Vaidya H. J., Cook A. M., Blair N. F. and Blackburn C. C. (2016). Identification of a bipotent epithelial progenitor population in the adult thymus. Cell Rep. 14, 2819-2832. 10.1016/j.celrep.2016.02.080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Klein R., Tian X., Cheng H.-T., Kopan R. and Shen J. (2004). Notch activation induces apoptosis in neural progenitor cells through a p53-dependent pathway. Dev. Biol. 269, 81-94. 10.1016/j.ydbio.2004.01.014 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.