Abstract
Apolipoproteins govern lipoprotein metabolism and are promising biomarkers of metabolic and cardiovascular diseases. Unlike immunoassays, MS enables the quantification and phenotyping of multiple apolipoproteins. Hence, here, we aimed to develop a LC-MS/MS assay that can simultaneously quantitate 18 human apolipoproteins [A-I, A-II, A-IV, A-V, B48, B100, C-I, C-II, C-III, C-IV, D, E, F, H, J, L1, M, and (a)] and determined apoE, apoL1, and apo(a) phenotypes in human plasma and serum samples. The plasma and serum apolipoproteins were trypsin digested through an optimized procedure and peptides were extracted and analyzed by LC-MS/MS. The method was validated according to standard guidelines in samples spiked with known peptide amounts. The LC-MS/MS results were compared with those obtained with other techniques, and reproducibility, dilution effects, and stabilities were also assessed. Peptide markers were successfully selected for targeted apolipoprotein quantification and phenotyping. After optimization, the assay was validated for linearity, lower limits of quantification, accuracy (biases: –14.8% to 12.1%), intra-assay variability [coefficients of variation (CVs): 1.5–14.2%], and inter-assay repeatability (CVs: 4.1–14.3%). Bland-Altman plots indicated no major statistically significant differences between LC-MS/MS and other techniques. The LC-MS/MS results were reproducible over five repeated experiments (CVs: 1.8–13.7%), and we identified marked differences among the plasma and serum samples. The LC-MS/MS assay developed here is rapid, requires only small sampling volumes, and incurs reasonable costs, thus making it amenable for a wide range of studies of apolipoprotein metabolism. We also highlight how this assay can be implemented in laboratories.
Keywords: proteomics, lipoprotein metabolism, metabolic disease, assay development, isotopic labeling, lipid metabolism, plasma lipid, serum lipid
Apolipoproteins govern lipoprotein regulation and metabolism. They constitute a family of multifunctional proteins that structure lipoprotein particles and direct their metabolism through binding to cell-surface receptors and regulation of enzyme activities (1, 2). As apolipoproteins are involved in pro- and anti-atherosclerotic processes, they are important circulating biomarkers of metabolic dysfunction and atherosclerotic CVD (1, 3, 4).
The primary clinical application of plasma apolipoprotein measurements is the early detection of metabolic diseases leading to CVD (3–5). Plasma concentrations of apoB100 and apoA-I have been reported to predict the risk of CVD better than those of LDL-C and HDL-C (1, 3, 4). ApoC-III and lipoprotein (a) [Lp(a)] are also considered as powerful CVD risk factors (6, 7). Additionally, postprandial metabolism is important in atherogenesis, particularly in the context of obesity, insulin resistance, and diabetes. ApoB48, apoCs, and apoE have become predominant markers of CVD-related risk in this emerging field (4, 8). Furthermore, the phenotyping of the major apoE isoforms and other apolipoproteins or apolipoprotein-like proteins (1, 2, 4, 5, 9) is used to diagnose mixed dyslipidemias (10–12).
Immunoassays used for apolipoprotein quantification are associated with many drawbacks, and their standardization is limited to the major apolipoproteins (3, 4). In contrast, MS enables the analysis of multiple proteins from a small sample volume at a high throughput rate. Protocols usually involve the analysis of protein mixtures after enzymatic proteolysis, which are then analyzed by LC-MS/MS, often in combination with upstream cleaning methods to reduce sample complexity (13). Thus, LC-MS/MS allows a high level of multiplexing, enabling simultaneous quantification of several proteins (14–16) and providing further information on a patient’s metabolic profile by targeting specific polymorphisms that cannot be detected by immunoassays (3, 12, 16, 17).
However, LC-MS/MS suffers from nonnegligible between-laboratory variability stemming from the lack of harmonized protocols (3). Here, we describe a multiplexed LC-MS/MS method that can simultaneously quantitate 18 human apolipoprotein species and determine pathogenic variants resulting from CVD-associated SNPs. Each experimental step was developed, optimized, and validated in agreement with standard procedures for analytical method validations (https://clsi.org/) (18, 19), and major pitfalls were highlighted.
MATERIALS AND METHODS
Experimental design and rationale
The present multiplex LC-MS/MS assay has been developed and validated according to the Tier 2 level of guidelines for Development and Application of Targeted Mass Spectrometry Measurements of Peptides and Proteins (20). All required information relative to the guidelines is described below. The assay was validated on 160 healthy human and patient samples without any specific inclusion or exclusion criteria to get a broad range of lipid phenotypes.
Selection of peptide markers
Apolipoprotein sequences were BLAST searched using the UNIPROT tool (www.uniprot.org), and theoretical peptides were searched using ExPASy (http://web.expasy.org/peptide_mass). Peptide candidates were selected in silico to maximize the sensitivity, specificity, and stability (21). Each candidate was then experimentally sought and characterized by LC-high-resolution (HR) MS (LC-HRMS) from concentrated lipoprotein fractions after trypsin digestion (21). The most specific and detectable peptides were selected for assay sensitivity optimization.
Biological samples
Human EDTA plasma samples (80 males, 80 females; nonfasted; supplemental Table S1) and paired samples of serum (n = 160), Li-heparin (n = 72), and citrate plasma (n = 54) were provided by the French Blood Bank (Nantes, France). Samples were immediately dispatched in single-use aliquots (40 μl) and the first LC-MS/MS assay in individuals was performed within the 14 days after collection and after a single freeze/thaw cycle. Pooled samples were prepared for each matrix by mixing equal fractions from each individual sample. Ethics approval for sample collection was acquired from the institutional review board of Nantes University Hospital, and written informed consent was obtained from each subject. All studies were designed in accordance with the principles of the Declaration of Helsinki. Pooled bovine EDTA plasma (six males, six females) was purchased from Patricell Ltd. (Nottingham, UK) and used as a surrogate matrix for method validation, as bovine apolipoprotein sequences are distinct from those of humans. Samples were stored at −80°C until use.
Standard samples and quality controls
Synthetic labeled and unlabeled proteotypic peptides were provided by Thermo Scientific Biopolymers (Darmstadt, Germany). Stock solutions (1 mM) were prepared in 50% acetonitrile containing 0.1% formic acid and stored at −20°C until use. A mixed solution of unlabeled peptides was constituted and serially diluted in water to obtain seven standard solutions (Table 1). Quality control (QC) samples were prepared at three concentration levels, including lower and upper limits of quantification (LOQs). Concentrated solutions were prepared in water and then diluted 10-fold in bovine EDTA plasma (QC samples) or in water (control samples) (Table 1). Labeled peptides (i.e., containing [13C6,15N2]K, [13C6,15N4]R, or [13C6,15N]I in the C-terminal position) were used as internal standards (ISs). A mixed solution of ISs (35 μM) was prepared and added to digestion buffer (ammonium bicarbonate, 50 mM) to a final concentration of 1.75 μM.
TABLE 1.
Proteotypic peptides used for apolipoprotein quantification
| Protein | Molecular Mass (kDa) | Proteotypic Peptide | Concentration Range (μM) | LQC (μM) | MQC (μM) | HQC (μM) |
| ApoA-I | 28.1 | ATEHLSTLSEK | 1–100 | 1 | 25 | 100 |
| ApoA-II | 9.3 | SPELQAEAK | 0.5–50 | 0.5 | 10 | 50 |
| ApoA-IV | 43.4 | SELTQQLNALFQDK | 0.5–50 | 0.5 | 10 | 50 |
| ApoA-V | 38.9 | VQELQEQLR | 0.01–1 | 0.01 | 0.25 | 1 |
| ApoB48 | 240.8 | LSQLQTYMI | 0.01–1 | 0.01 | 0.25 | 1 |
| ApoB100 | 512.9 | ATGVLYDYVNK | 0.1–10 | 0.1 | 2.5 | 10 |
| ApoC-I | 6.6 | TPDVSSALDK | 0.05–5 | 0.05 | 1 | 5 |
| ApoC-II | 8.2 | TAAQNLYEK | 0.25–25 | 0.25 | 5 | 25 |
| ApoC-III | 8.8 | GWVTDGFSSLK | 0.25–25 | 0.25 | 5 | 25 |
| ApoC-IV | 11.5 | ELLETVVNR | 0.05–5 | 0.05 | 1 | 5 |
| ApoD | 19.3 | VLNQELR | 0.1–10 | 0.1 | 2.5 | 10 |
| ApoE | 34.2 | LGPLVEQGR | 0.1–10 | 0.1 | 2.5 | 10 |
| ApoE2 | 34.2 | CLAVYQAGAR | 0.1–10 | 0.1 | 2.5 | 10 |
| ApoE4 | 34.2 | LGADMEDVR | 0.1–10 | 0.1 | 2.5 | 10 |
| ApoE2/E3 | 34.2 | LGADMEDVCGR | 0.1–10 | 0.1 | 2.5 | 10 |
| ApoE3/E4 | 34.2 | LAVYQAGAR | 0.1–10 | 0.1 | 2.5 | 10 |
| ApoF | 17.4 | SGVQQLIQYYQDQK | 0.05–5 | 0.05 | 1 | 5 |
| ApoH | 36.3 | ATVVYQGER | 0.1–10 | 0.1 | 2.5 | 10 |
| ApoJ | 50.1 | ELDESLQVAER | 0.1–10 | 0.1 | 2.5 | 10 |
| ApoL1 | 41.1 | VAQELEEK | 0.05–5 | 0.05 | 1 | 5 |
| ApoL1 (G0) | 41.1 | LNILNNNYK | 0.05–5 | 0.05 | 1 | 5 |
| ApoL1 (G1) | 41.1 | LNMLNNNYK | 0.05–5 | 0.05 | 1 | 5 |
| ApoL1 (G2) | 41.1 | LNILNNK | 0.05–5 | 0.05 | 1 | 5 |
| ApoM | 21.3 | AFLLTPR | 0.1–10 | 0.1 | 2.5 | 10 |
| Apo(a) | 240–800 | LFLEPTQADIALLK | 0.005–0.5 | 0.005 | 0.1 | 0.5 |
| Apo(a) Kr-IV2 | 12.5 | GTYSTTVTGR | 0.25–25 | 0.25 | 5 | 25 |
Molecular masses were adopted from UniProt (https://www.uniprot.org/) (signal peptides excluded from the calculation). Kr-IV2, apo(a) kringle IV2; LQC, low-concentration QC; MQC, middle-concentration QC; HQC, high-concentration QC.
General procedure for sample preparation
Samples were prepared with the ProteinWorks™ eXpress kit (Waters, Milford, MA), according to the manufacturer’s instructions (supplemental Table S2). Samples (40 μl) were incubated for 10 min at 80°C in digestion buffer containing ISs (100 μl) and RapidGest detergent solution (7 mg/ml, 10 μl), reduced for 20 min at 60°C with dithiothreitol (70 mM, 20 μl), alkylated for 30 min at room temperature in the dark with iodoacetamide (142 mM, 30 μl), and digested overnight at 37°C (∼16 h) with trypsin (7 mg/ml, 30 μl). Enzymatic digestion was stopped with 20% trifluoroacetic acid (TFA; 5 μl). After 15 min at 45°C, the precipitate was removed by centrifugation (15 min, 10°C, 10,000 g), and supernatants were cleaned on 30 mg Oasis HLB cartridges (Waters), which were conditioned (100% methanol; 1 ml), equilibrated (100% water; 1 ml), loaded (sample; ∼200 μl), washed (5% methanol; 1 ml), and eluted (80% methanol; 500 μl). The eluates were dried under nitrogen (45°C), reconstituted with 5% acetonitrile containing 0.1% formic acid (100 μl), and injected (10 μl) into the LC-MS/MS system. Analyses were performed on a Xevo® TQD mass spectrometer with an electrospray interface and an Acquity H-Class® UPLC™ device (Waters). The optimized LC-MS/MS and “multiple reaction monitoring” parameters are detailed in supplemental Tables S3 and S4.
Data management
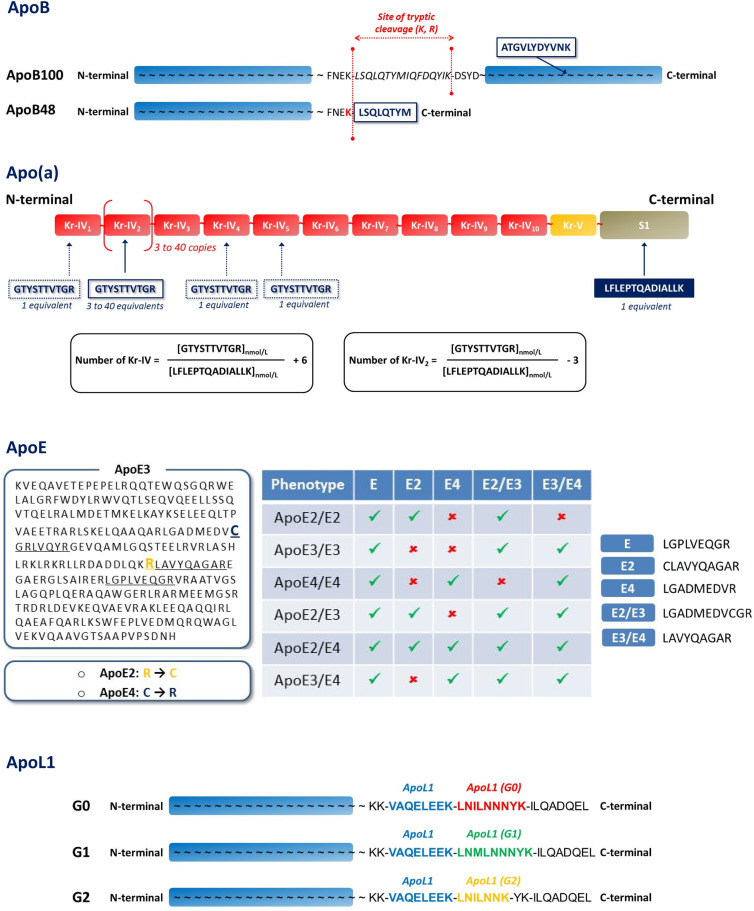
Apolipoprotein concentrations were calculated using calibration curves plotted from standard solutions and expressed in micromoles, assuming that 1 mol of peptide was equivalent to 1 mol of protein. The concentrations were then converted to standard units (milligrams per deciliter) using the molecular masses (Table 1). Unlike the apoB and apoL1 isoforms (22, 23) (Fig. 1), the apoE phenotypes and isoform concentrations (E2/E3/E4) were determined from different peptide combinations (supplemental material) (12, 24). The mean size of apo(a) was also estimated by LC-MS/MS from two proteotypic peptides (supplemental material) (25).
Fig. 1.
Selection of proteotypic peptides for apolipoprotein polymorphisms. A: Proteotypic peptides used to distinguish apoB48 and apoB100. B: Proteotypic peptides used to assess the mean polymorphic size of apo(a) [i.e., Kringle IV repeats (Kr-IV)]. C: Identification of apoE phenotypes by selective combinations of apoE proteotypic peptides. D: Identification of apoL1 isoforms by specific proteotypic peptides.
Optimization of sample preparation
Solid-phase extraction (SPE) was optimized from QC samples treated as described above. The wash and elution conditions were optimized with increasing levels of methanol or acetonitrile in water, with or without additives (0.1% TFA or 0.5% NH4OH) and were added directly after sample loading (n = 8 per condition). Several acetonitrile/water mixtures containing 0.1% formic acid were tested for sample reconstitution after drying (n = 8 per condition). Sample volumes, incubation times, and temperatures were optimized using a pooled human EDTA plasma sample to improve proteolysis (n = 8 per condition). Control samples were used to assess recoveries, matrix effects, and peptide stabilities throughout the experiments.
Method validation
The LC-MS/MS assay was validated via four independent experiments consisting of two calibration curves and 18 QCs (three concentration levels, n = 6). The assay linearity was illustrated by R2 coefficients calculated from calibration curves by linear regression analysis (1/x weighting, origin excluded). The intra-assay and inter-assay imprecisions were expressed by the coefficients of variation (CVs) obtained at each QC level. The accuracy was expressed by the mean bias between the theoretical and measured concentrations at each QC level. Targeted lower LOQs were validated with signal-to-noise ratios greater than 10. Limits of acceptance were set at ±15% for accuracy and variability. To assess matrix effects and carry-over, the pooled human EDTA plasma was diluted in bovine EDTA plasma (1:0, 1:1, 1:3, and 1:7; v:v). Thirty replicates per dilution level were then trypsin digested, randomized, and injected into the LC-MS/MS system. Matrix effects were determined by comparing individual measurements in EDTA plasma versus those in serum, Li-heparin plasma, and citrate plasma.
Cross-validations
Individual EDTA plasma concentrations of apolipoproteins obtained by LC-MS/MS (n = 160) were compared with those obtained in a blinded fashion by standardized immuno-turbidimetry, sandwich ELISA, LC-HRMS, and Western blot (supplemental material). Three replicates of each QC sample and six replicates of the plasma pool were included in each LC-MS/MS experiment and randomly injected throughout the assay to ascertain the quality of our results as well as the proteolysis efficiency. Spearman correlations were calculated; and Bland-Altman plots were generated to compare our LC-MS/MS approach with the other methods (26).
Statistical analyses
Graphics and analyses were achieved with GraphPad Prism software (version 6.0, GraphPad Software Inc., La Jolla, CA). The D’Agostino-Pearson test was used to estimate data distribution and select the most appropriate statistical test for data comparisons (significance at P < 0.05). Network analysis based on Spearman correlation analysis was also performed. The Spearman’s rank correlation coefficient was computed (R software) and displayed using the “corrplot package.”
RESULTS
Selection of peptide markers
In silico investigations led to the identification of numerous peptide candidates per target apolipoprotein. The most specific and detectable peptides were selected to optimize the assay sensitivity and specificity by LC-HRMS experiments (not shown). Peptide candidates were primarily detected as doubly charged precursor ions, except for apoA-I (a triply charged ion). After MS/MS fragmentation, each precursor ion yielded several specific and singly charged “y” or “b” product ions (except for apoA-I, doubly charged y ions), ascertaining thereby the peptide sequences (supplemental Fig. S1). The most intense and specific peptides were selected and synthetized for the manual optimization of the multiple reaction monitoring transitions used for LC-MS/MS analyses (supplemental Tables S4–S6).
Optimization of sample preparation
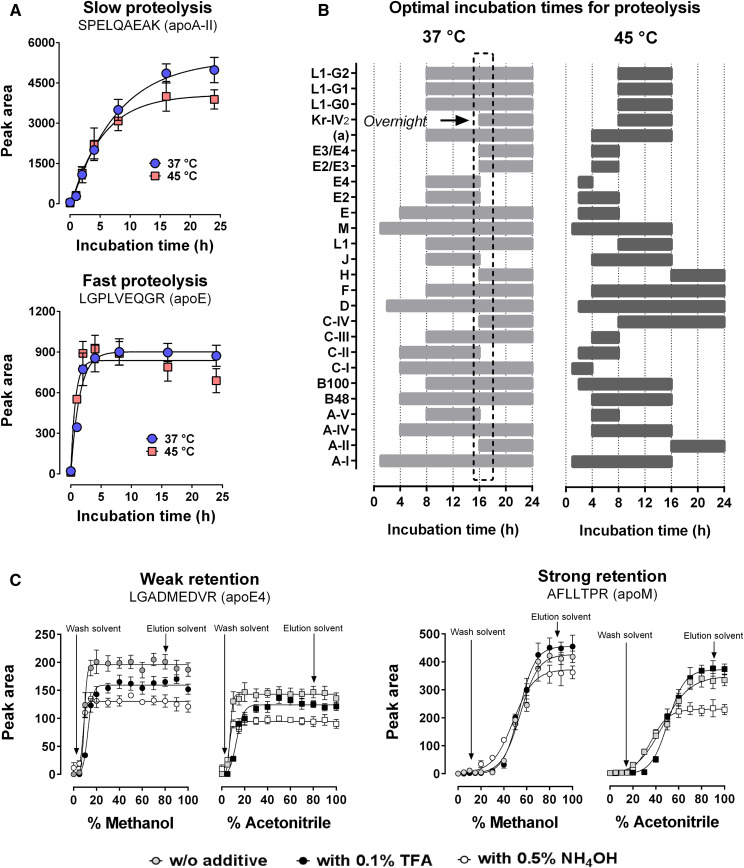
Trypsin proteolysis optimization showed contrasting digestion profiles between apolipoproteins (Fig. 2A). Unlike the manufacturer’s recommendations (2 h, 45°C), the optimal condition was reached with an overnight trypsin incubation at 37°C (Fig. 2B). All other recommendations were confirmed by testing four incubation times on the pooled human EDTA plasma (not shown). SPE was also optimized on bovine EDTA plasma spiked with known amounts of synthetic peptides (QC). Methanol was slightly more efficient than acetonitrile for peptide recovery, while acidic (TFA) and basic (NH4OH) additives did not improve extraction (Fig. 2C). The wash and elution solvents were 5% and 80% methanol, respectively. After drying, several mixtures were tested for sample reconstitution to obtain the optimal signal intensity (supplemental Fig. S2). The most optimal mixture was 5% acetonitrile containing 0.1% formic acid. Higher concentrations of acetonitrile led to peak distortions for some polar peptides. Despite SPE, the plasma peptide detection was strongly reduced by 15–85%, compared with aqueous controls. These matrix effects were corrected after IS normalization and ranged from −9% to +12% (supplemental Fig. S3). Different plasma volumes (10, 20, 30, 40, and 50 μl) were also assessed, and 40 μl was found to be more suitable for assay sensitivity and proteolysis efficiency (not shown).
Fig. 2.
Optimization of sample preparation for apolipoprotein quantification. Trypsin incubation times were optimized using a mixture of human EDTA plasma samples. Solid-phase extraction protocols were optimized using bovine plasma samples spiked with a solution of synthetic peptides (MQC). A: Representative examples of peptides generated from trypsin digestion that require slow and fast proteolysis times. B: Summary of optimal incubation times for all proteotypic peptides of apolipoproteins in human plasma. The optimal digestion time is indicated by the black box. C: Representative examples of proteotypic peptides that were weakly or strongly retained in the cartridges (C18, reversed phase) during solid-phase extraction. Values are presented as the mean ± standard deviation (n = 8).
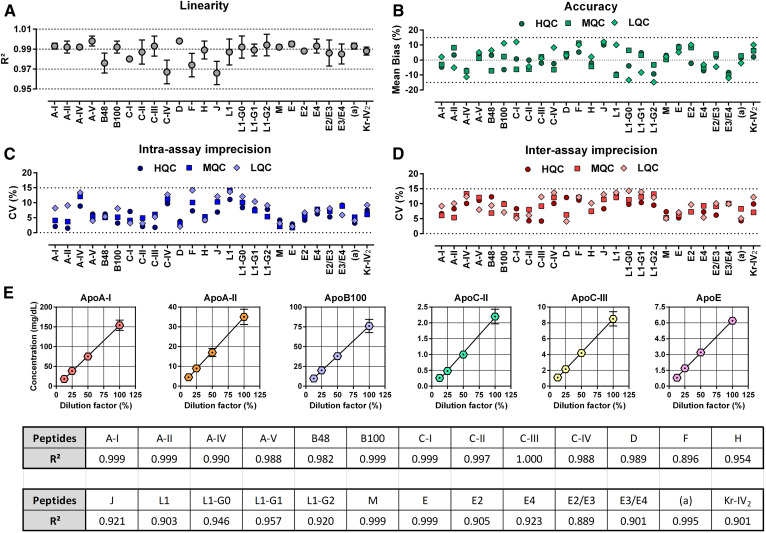
Reliability of the assay
After calibration curve analysis (n = 8), the mean R2 value ranged from 0.967 ± 0.012 to 0.998 ± 0.001 (linear regression, 1/x weighted, origin excluded), and the CVs never exceeded 14.5% over the concentration range tested (Fig. 3A, supplemental Table S7). For the QCs, the mean absolute bias did not deviate by more than ±14.8%, compared with the expected concentrations (Fig. 3B). Intra- and inter-assay CVs never exceeded 14.2% and 14.3%, respectively (Fig. 3C, D; supplemental Table S8). Moreover, the signal-to-noise ratios measured at the lower LOQs were greater than 10 for all target peptides (supplemental Fig. S4). The assay variability was determined by using several aliquots of pooled human EDTA plasma diluted in bovine plasma at four dilution levels (n = 30 per dilution level). Samples were randomly injected into the LC-MS/MS instrument to estimate the carry-over between injections (Fig. 3E). As expected, the concentrations of peptides generated by trypsin proteolysis were linearly decreased according to the dilutions (R2 range: 0.889–1.000).
Fig. 3.
LC-MS/MS validation, repeatability, and effect of matrix dilution. The analytical validation was performed over four distinct experiments. Synthetic peptides were spiked into EDTA bovine plasma and then submitted to the entire experimental process. Repeatability was evaluated using pooled human EDTA plasma serially diluted in bovine EDTA plasma. A: Assay linearity was illustrated by the R2 coefficient (mean ± standard deviation, n = 8) calculated from calibration curves by linear regression analysis (origin excluded, 1/x weighting). B: Assay accuracy was expressed by the mean bias between the theoretical and the measured concentration (n = 6 × 4 per level). Intra-assay (C) and inter-assay (D) imprecisions were expressed by the CVs obtained at each QC level (n = 6 × 4 per level). E: Repeatability of the assay and matrix effects (mean ± standard deviation, n = 30 per dilution level, randomly injected). Dotted lines indicate our limits of acceptance (±15%) in panels B, C, and D. LQC, low-concentration QC; MQC, middle-concentration QC; HQC, high-concentration QC.
Comparison with other methods
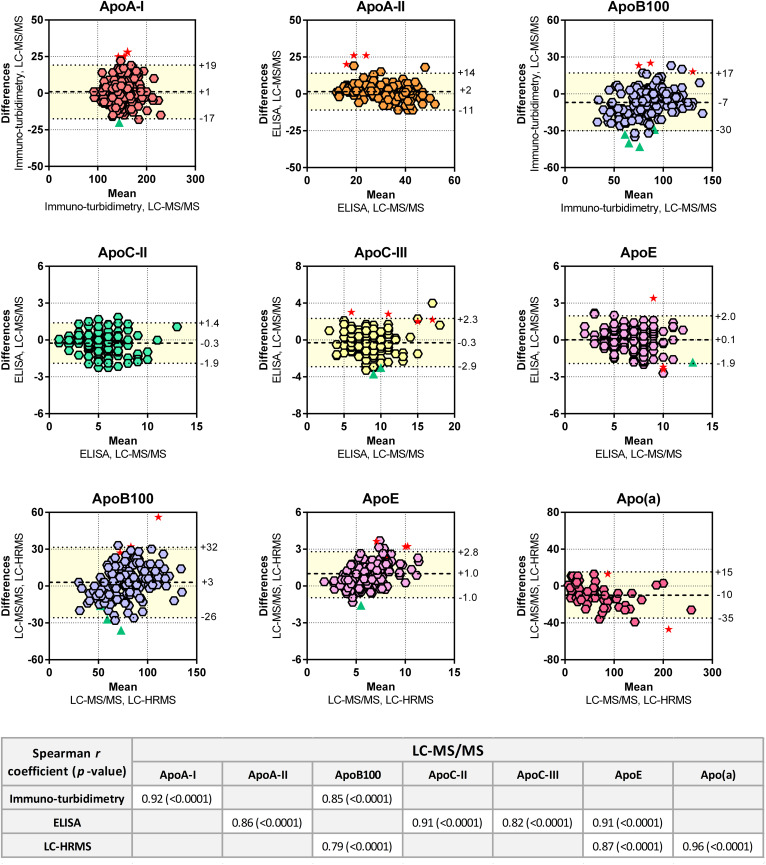
The apolipoprotein concentrations measured by LC-MS/MS in individual EDTA plasma samples were similar to those obtained by immuno-turbidimetry, ELISA, and LC-HRMS (Fig. 4). The LC-MS/MS data were also strongly correlated with those obtained by the other techniques. The Bland-Altman plots highlighted nonnegligible concentration differences between the analytical approaches, with deviations of up to 50%. The most marked differences in apolipoprotein concentrations were observed between LC-MS/MS and LC-HRMS measurements, suggesting that matrix effects strongly impacted the MS measurements. LC-HRMS (Orbitrap®) experiments were also limited compared with those performed by LC-MS/MS (triple quadrupole) because of HRMS lower level of multiplexing ability related to its lower dynamic range. Only a few outliers were identified from the Bland-Altman plots (<5%), and most of them were related to the initial sample quality (i.e., hemolyzed or opalescent appearance, Fig. 4). This observation was not systematic because 12 hemolyzed and 23 opalescent plasma samples were identified.
Fig. 4.
Cross-validation of the LC-MS/MS assay with other techniques. Bland-Altman plots and Spearman correlations were generated to test the similarity of various methods (n = 160). For the Bland-Altman plots, the difference of values (y axis) obtained from two methods was plotted according to the average value obtained by two methods. The mean difference and the limits of agreement (colored area), corresponding to the 95% confidence level (i.e., mean ± 1.96 × standard deviation), are represented. Red stars indicate hemolyzed plasma samples. Green triangles indicate opalescent plasma samples. Spearman correlations between LC-MS/MS and other methods are indicated.
Apolipoprotein concentrations
The circulating apolipoprotein concentrations are provided in Table 2. Several plasma apolipoprotein concentrations were slightly but significantly different between males and females. To ensure assay repeatability, EDTA plasma measurements were repeated over five distinct experiments. The calculated CVs ranged from 1.8% to 13.7% and were within acceptable limits for MS experiments. Of note, intra- and inter-assay CVs calculated from the QC samples and the plasma pool never exceeded 15%, validating the reproducibility of the measurements and the quality of the preparation and analytical runs. The apolipoprotein concentrations were significantly over- or underestimated in serum, Li-heparin, and citrate plasma, in comparison to EDTA plasma. The plasma apolipoprotein concentrations were also compared with other biochemical data (supplemental Fig. S5). As expected, strong and significant correlations were found between apolipoproteins and lipids, strengthening the reliability of LC-MS/MS for providing relevant measurements (e.g., apoA-I versus HDL-C, apoB100 versus LDL-C, etc.).
TABLE 2.
Apolipoprotein concentrations in nonfasted individuals and matrix effect assessment
| Apolipoprotein | EDTA Plasma (n = 160) | Serum (n = 160) | Li-Heparin Plasma (n = 72) | Citrate Plasma (n = 54) | |||
| Male (n = 80) | Female (n = 80) | a | CV (%) | ||||
| b | |||||||
| ApoA-I | 145 [126–160] | 154 [133–178] | 0.0016 | 5.6 | +5.4 | +1.2 | −2.3 |
| ApoA-II | 33 [26–39] | 34 [28–39] | 0.5868 | 8.1 | +37.2c | +29.4c | +17.3d |
| ApoA-IV | 9 [8–11] | 9 [11–13] | 0.0002 | 10.1 | −8.8c | −16.3c | −18.4c |
| ApoA-V | 0.02 [0.01–0.06] | 0.02 [0.01–0.04] | 0.0452 | 11.3 | +34.2c | +30.1c | +16.9c |
| ApoB48 | 1.4 [1.3–1.6] | 1.7 [1.5–1.9] | 0.0001 | 9.8 | +12.1e | +9.5e | +11.5d |
| ApoB100 | 86 [68–100] | 74 [55–96] | 0.1059 | 4.8 | +18.3c | +14.6c | +9.4d |
| ApoC-I | 3.9 [3.1–4.9] | 3.4 [2.6–4.0] | 0.0024 | 2.4 | +16.2b | +7.6e | +2.1 |
| ApoC-II | 5.5 [4.1–6.9] | 4.4 [3.4–5.9] | 0.0009 | 4.2 | +8.3c | −5.6d | −16.4c |
| ApoC-III | 8.0 [6.6–10.2] | 7.5 [6.4–8.9] | 0.0045 | 5.2 | +7.9 | +2.1 | −3.2 |
| ApoC-IV | 0.09 [0.07–0.17] | 0.04 [0.03–0.12] | 0.0001 | 12.5 | −14.5c | −21.3c | −18.9c |
| ApoD | 3.8 [3.2–4.3] | 3.1 [2.5–3.5] | 0.0001 | 2.7 | +8.8c | +2.1 | −1.4 |
| ApoE | 6.4 [5.1–7.8] | 6.3 [5.1–7.8] | 0.4685 | 7.3 | +58.7c | +47.3c | +43.2c |
| ApoF | 0.38 [0.16–0.67] | 0.44 [0.24–0.73] | 0.1284 | 13.7 | +5.4 | +7.1 | +3.5 |
| ApoH | 5.5 [4.7–6.3] | 5.2 [4.6–5.9] | 0.3832 | 4.8 | +62.4c | +59.3c | +44.7c |
| ApoJ | 11 [9–14] | 11 [10–13] | 0.4713 | 9.4 | −4.3c | +2.1 | −6.1c |
| ApoL1 | 1.0 [0.9–1.2] | 1.3 [1.1–1.8] | 0.0001 | 11.8 | −6.3d | +1.2 | +4.2 |
| ApoM | 2.5 [2.1–2.9] | 2.4 [2.0–2.7] | 0.1566 | 1.8 | −5.4c | −2.3e | −1.8 |
| Apo(a) (nM) | 22 [12–63] | 39 [16–77] | 0.1085 | 3.8 | +34.2c | +21.4c | +15.3c |
Values are medians [25th to 75th percentiles]. Apolipoprotein concentrations are in milligrams per deciliter, unless otherwise specified. EDTA plasma samples were assayed five times. CVs were calculated over five experiments.
Mann-Whitney test or unpaired t-test.
Wilcoxon matched-pairs signed rank test or paired t-test.
P < 0.001.
P < 0.01.
P < 0.05.
Apolipoprotein polymorphisms
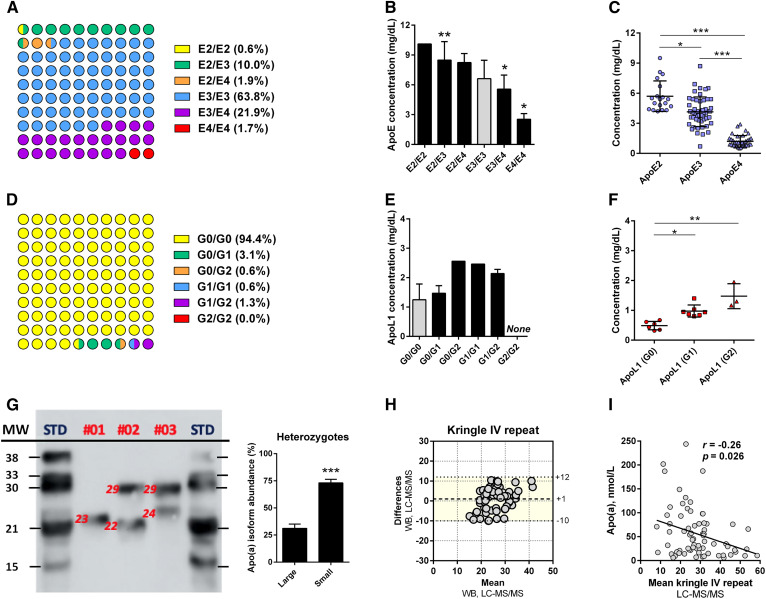
An expected distribution of apoE phenotypes with a predominance of apoE3 carriers was observed (Fig. 5A). The total apoE plasma concentrations were significantly different between the apoE phenotypes, with apoE2 and apoE4 carriers exhibiting the highest and the lowest concentrations, respectively (Fig. 5B). We also confirmed that the apoE2 levels were higher than apoE3 levels in E2/E3 heterozygote carriers, whereas apoE4 levels were lower than apoE3 levels in E2/E4 heterozygote carriers (Fig. 5C). The apoL1 phenotypes were also determined and showed a sharp predominance of the most common nonrisk G0/G0 phenotype (Fig. 5D). We did not find any significant difference in the total apoL1 concentrations according to the phenotype (Fig. 5E). However, the apoL1 isoforms were differently expressed in heterozygotes (G0 < G1 < G2) (Fig. 5F). In addition, the apo(a) polymorphic sizes were estimated by LC-MS/MS. Western blot experiments revealed that 65% of patients displayed two detectable apo(a) isoforms and showed that the smallest isoform was usually the most abundant (Fig. 5G). Unlike Western blot, LC-MS/MS was not able to discriminate both isoforms in heterozygotes. However, the Bland-Altman plot showed that LC-MS/MS can reliably estimate the average size of apo(a) (Fig. 5H). LC-MS/MS analysis also confirmed the slight but significant negative correlation between the apo(a) plasma concentration and the average of apo(a) polymorphic size (Fig. 5I). Finally, LC-MS/MS was able to clearly discriminate intestinal apoB48 from hepatic apoB100, despite strong sequence homologies (Table 2).
Fig. 5.
LC-MS/MS analysis of apoE, apoL1, and apo(a) polymorphisms. A: Distribution of the apoE phenotype was determined by LC-MS/MS and LC-HRMS (100% agreement). B, C: Influence of the apoE phenotype on the total apoE plasma concentration and the plasma concentrations of the apoE isoforms in heterozygous patients (E2/E3, E2/E4, and E3/E4). D: Distribution of the apoL1 phenotype as determined by LC-MS/MS. E, F: Influence of the apoL1 phenotype on the total apoL1 plasma concentration and the plasma concentrations of the apoL1 isoforms in heterozygous patients (G0/G1, G0/G2, and G1/G2). G: Determination of apo(a) polymorphic sizes [kringle IV (KrIV) repeats] by Western blot. H: Bland-Altman plots were generated to test the similarity of LC-MS/MS and Western blot for the determination of the apo(a) polymorphic size (n = 71, LC-MS/MS detectable values). The mean difference and the limits of agreement (colored area), corresponding to the 95% confidence level (i.e., mean ± 1.96 × standard deviation), are represented. I: Influence of the apo(a) polymorphic size on the apo(a) plasma concentration (Spearman’s test). *P < 0.05, **P < 0.01, ***P < 0.001 (Kruskal-Wallis test). For B and E, the plasma concentrations were compared with the healthy phenotypes (i.e., E3/E3 or G0/G0).
Peptide and sample stabilities
Because some peptides could be not highly stable during storage, we assessed the stability of stock solutions by using new freshly synthetized peptides. Stock solutions of the labeled and unlabeled synthetic peptides were found stable for 18 months at −20°C. The peptides were also stable in bovine plasma throughout sample preparation processes. Digested samples were stable for 5 days under refrigeration (10°C) and for 18 months at −20°C (not shown). Furthermore, the stability of the biological samples was assessed on pooled plasma/serum samples (supplemental Table S9). Unlike serum, Li-heparin, and citrate plasma, the apolipoproteins were stable for 12 months at −80°C and after five freeze/thaw cycles (−80°C). They were also stable for 24 h at room temperature, 1 week at 4°C, and 3 months at −20°C. It is of note that oxidized forms of peptides carrying a methionine residue (+16 and/or +32 mass unit shift) were systematically detected after long-term storage or after repeated freeze/thaw cycles (>3). This suggested that the use of oxidized synthetic peptides is warranted for evaluating and quantifying the possible degradation of samples during storage.
DISCUSSION
LC-MS/MS enables measurements of multiple apolipoproteins in a single run but requires specific sample manipulations prior to analysis. Hence, the development of harmonized protocols is required to minimize between-laboratory variability and standardize such assays. Herein, we developed a high-throughput MS-based protocol for large-scale profiling of human circulating apolipoproteins. All steps of sample preparation were optimized from a commercially available kit before being validated according to standard guidelines. The method was then compared with other techniques and was shown to be reliable for apolipoprotein quantification and major SNP determination at very competitive costs, compared with traditional assays.
Lipoprotein metabolism abnormalities are important in metabolic diseases leading to CVD (1, 3, 4). Because apolipoproteins direct lipoprotein metabolism, their measurements can be used to improve CVD risk prediction primarily based on traditional plasma lipid testing (3). Beyond apoA-I and apoB (1, 16, 27), strong associations have been reported between CVD risk and apoC-II, apoC-III, and apoE plasma levels (7, 10, 28). The risk of developing cardiovascular or metabolic diseases also has been associated with other apolipoproteins (2, 5, 28–30), whose functions are less understood and deserve further investigations (3, 31, 32). Moreover, the detection of disease-specific polymorphisms could increase the specificity of CVD risk assessment, as some isoforms display altered functionalities (12, 16, 24, 33, 34).
Whereas current immunoassays are unavailable for some apolipoproteins and lack the capacity for multiplexing, MS enables the analysis of multiple molecules from a single sample preparation (3). MS can also detect and quantify apolipoprotein variants resulting from a single amino acid mutation (22, 24). Hence, MS constitutes a powerful tool for the large-scale profiling of apolipoproteins (14–16, 35, 36). However, MS protocols involve multiple steps: enzymatic digestion of complex samples to transform proteins into peptides (13); SPE to reduce sample complexity; and LC-MS/MS to separate and detect signature peptides. Thus, MS-based assays suffer from great heterogeneity stemming from the lack of harmonized protocols including the applied sampling material, proteolysis conditions, cleaning methods, and proteolytic peptides (3).
The selection of peptide markers is critical to accurately quantify proteins by LC-MS/MS. They must be stable, sensitive, efficiently released from proteolysis, and not interfere with nontargeted proteins. Our peptide candidates satisfy these criteria, and most of them have already been validated (12, 14–16, 22, 23, 36, 37). Nevertheless, despite careful method validations, the differences noticed in peptide selection could contribute to some extent to between-laboratory variability. Our set of peptide markers was successfully validated in terms of linearity, specificity, low LOQ, precision, accuracy, and short- and long-term stabilities (18, 19). However, due to the high specificity, a single subtle modification of a peptide sequence, such as a single amino acid substitution or the oxidation of a methionine residue, will substantially alter the mass of the wild-type or native peptide and, as a result, preclude proper detection by MS. Here, we selected one peptide per target apolipoprotein to screen a large set of species, but the selection of two to three peptide markers per apolipoprotein (when possible) should be warranted for improving between-laboratory reproducibility and detecting any deviating responses (3).
Another source of variability is the proteolysis yield, which leads to an optimal release of peptide markers and is related to the selected material, reagents, experimental conditions, and choice of signature peptides (14–16). To maximize the between-laboratory reproducibility, we used a commercial kit specifically dedicated for proteomics. All manufacturers’ instructions were validated and/or optimized to improve assay efficiency on our set of peptides, and we used internal controls (QCs and plasma pools) for validating each analytical batch. However, the use of certified reference standards is required for clinical implementation of apolipoprotein measurements and for ascertaining proteolysis efficiency.
Despite using SPE, the peptide markers were strongly impacted by matrix effects, causing sharp and variable signal reductions. This was corrected by using dedicated ISs enriched with stable isotopes. Our investigations confirmed that the applied sampling material and anticoagulant significantly influenced the results (15), possibly due to matrix effects, different proteolysis yields, or sample stability. EDTA plasma was selected as the reference material, but further studies are needed prior to the implementation of standardized protocols. Importantly, the measurement of plasma apolipoproteins may be clinically less relevant than lipoprotein subclass-specific analysis (3). Matrix effects were strongly reduced in lipoprotein subclasses isolated by ultracentrifugation, gel filtration, or phosphotungstic acid precipitation of apoB-containing particles (not shown). While ultracentrifugation or gel filtration processes would complicate protocol harmonization, precipitation could be easily standardized even if analyses are limited to apoA-I-containing (HDL) or apoB-containing (non-HDL) particles.
It is of note that MS requires an initial expensive investment in both apparatus acquisition and maintenance (especially high-resolution systems). However, its high ability for multiplexing (especially triple quadrupole systems) allows for the reduction of sample volumes (<50 μl vs. several aliquots of up to 1 ml), experimental times (∼400 samples × 18 markers can be performed per week per instrument), and costs (by at least 10-fold), compared with traditional immunoassays. Therefore, LC-MS/MS enables large-scale profiling of plasma apolipoproteins in large cohorts, which is essential for improving the sensitivity of studies regarding the determination of novel CVD risk markers (1). Another advantage of LC-MS/MS for apolipoprotein metabolism studies is its ability to conduct metabolic flux analyses with stable isotope-labeled tracers. The labeled tracer (e.g., 2H3-leucine) is perfused or injected into patients, and blood samples are collected. Its incorporation within apolipoproteins is then measured over time by LC-MS/MS, and the production and catabolic rates are deduced from kinetic curves (9, 21, 24, 25). All peptide markers selected here carry at least one leucine residue (except apoH), which makes it possible to conduct such studies. In addition, apolipoproteins also exhibit proteoforms arising from posttranslational modifications that could dramatically affect their functionalities (17). Some studies have unraveled the presence and alteration of such modifications for apolipoproteins in diabetes and CVD (38–40). However, such proteoforms are numerous and present at low stoichiometric levels. Therefore, their study often requires specific enrichment methods prior to LC-MS/MS analyses (41) and remains a huge challenge.
In conclusion, MS not only allows the large-scale profiling of circulating apolipoproteins in human cohorts but also enables further research investigations that traditional immunoassays cannot achieve. The attractive costs and the reduced experimental times associated with multiplexed LC-MS/MS assays make this technique of interest for clinical diagnostics. Nevertheless, LC-MS/MS in clinical laboratories still requires a high level of standardization, including: 1) automated sample preparation; 2) the selection of appropriate sampling materials (e.g., EDTA plasma); 3) the harmonized selection of peptide markers; and 4) the use of certified reference standards.
Data availability
All data are contained within the article or in the supplemental material section. All chromatograms used for quantification are available in the supplemental material.
Supplementary Material
Footnotes
Abbreviations:
- CV
- coefficient of variation
- IS
- internal standard
- LC-HRMS
- LC-high-resolution MS
- LOQ
- limit of quantification
- QC
- quality control
- SPE
- solid-phase extraction
- TFA
- trifluoroacetic acid
This work was supported by grants from the Fondation de France and by the French national Cholesterol Personalized Innovation (CHOPIN) project, funded by the Agence Nationale de la Recherche (ANR-16-RHUS-0007) and coordinated by the Centre Hospitalo-Universitaire of Nantes. V.B. received scholarships from the Région Réunion and the European Union (European Regional Development Fund INTERREG V). Additional financial support was provided by the Biogenouest Corsaire core facility. The authors declare that they have no conflicts of interest with the contents of this article.
The online version of this article (available at https://www.jlr.org) contains a supplement.
REFERENCES
- 1.Dominiczak M. H., and Caslake M. J.. 2011. Apolipoproteins: metabolic role and clinical biochemistry applications. Ann. Clin. Biochem. 48: 498–515. [DOI] [PubMed] [Google Scholar]
- 2.Perdomo G., and Henry Dong H.. 2009. Apolipoprotein D in lipid metabolism and its functional implication in atherosclerosis and aging. Aging (Albany NY). 1: 17–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van den Broek I., Sobhani K., and Van Eyk J. E.. 2017. Advances in quantifying apolipoproteins using LC-MS/MS technology: implications for the clinic. Expert Rev. Proteomics. 14: 869–880. [DOI] [PubMed] [Google Scholar]
- 4.Renee Ruhaak L., van der Laarse A., and Cobbaert C. M.. 2019. Apolipoprotein profiling as a personalized approach to the diagnosis and treatment of dyslipidaemia. Ann. Clin. Biochem. 56: 338–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brahimaj A., Ligthart S., Ikram M. A., Hofman A., Franco O. H., Sijbrands E. J. G., Kavousi M., and Dehghan A.. 2017. Serum levels of apolipoproteins and incident type 2 diabetes: a prospective cohort study. Diabetes Care. 40: 346–351. [DOI] [PubMed] [Google Scholar]
- 6.Tsimikas S. 2017. A test in context: lipoprotein(a): diagnosis, prognosis, controversies, and emerging therapies. J. Am. Coll. Cardiol. 69: 692–711. [DOI] [PubMed] [Google Scholar]
- 7.Chan D. C., Chen M. M., Ooi E. M. M., and Watts G. F.. 2008. An ABC of apolipoprotein C-III: a clinically useful new cardiovascular risk factor? Int. J. Clin. Pract. 62: 799–809. [DOI] [PubMed] [Google Scholar]
- 8.Smith D., Watts G. F., Dane-Stewart C., and Mamo J. C.. 1999. Post-prandial chylomicron response may be predicted by a single measurement of plasma apolipoprotein B48 in the fasting state. Eur. J. Clin. Invest. 29: 204–209. [DOI] [PubMed] [Google Scholar]
- 9.Croyal M., Billon-Crossouard S., Goulitquer S., Aguesse A., León L., Fall F., Chétiveaux M., Moyon T., Blanchard V., Ouguerram K., et al. . 2018. Stable isotope kinetic study of apoM (apolipoprotein M). Arterioscler. Thromb. Vasc. Biol. 38: 255–261. [DOI] [PubMed] [Google Scholar]
- 10.Huang Y. 2010. Mechanisms linking apolipoprotein E isoforms with cardiovascular and neurological diseases. Curr. Opin. Lipidol. 21: 337–345. [DOI] [PubMed] [Google Scholar]
- 11.Blanchard V., Croyal M., Khantalin I., Ramin-Mangata S., Chemello K., Nativel B., Blom D. J., Marais A. D., and Lambert G.. 2019. Reduced lipoprotein(a) associated with the apolipoprotein E2 genotype confers cardiovascular protection in familial hypercholesterolemia. JACC Basic Transl. Sci. 4: 425–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martínez-Morillo E., Nielsen H. M., Batruch I., Drabovich A. P., Begcevic I., Lopez M. F., Minthon L., Bu G., Mattsson N., Portelius E., et al. . 2014. Assessment of peptide chemical modifications on the development of an accurate and precise multiplex selected reaction monitoring assay for apolipoprotein e isoforms. J. Proteome Res. 13: 1077–1087. [DOI] [PubMed] [Google Scholar]
- 13.Giansanti P., Tsiatsiani L., Low T. Y., and Heck A. J. R.. 2016. Six alternative proteases for mass spectrometry-based proteomics beyond trypsin. Nat. Protoc. 11: 993–1006. [DOI] [PubMed] [Google Scholar]
- 14.Ceglarek U., Dittrich J., Becker S., Baumann F., Kortz L., and Thiery J.. 2013. Quantification of seven apolipoproteins in human plasma by proteotypic peptides using fast LC-MS/MS. Proteomics Clin. Appl. 7: 794–801. [DOI] [PubMed] [Google Scholar]
- 15.Dittrich J., Adam M., Maas H., Hecht M., Reinicke M., Ruhaak L. R., Cobbaert C., Engel C., Wirkner K., Löffler M., et al. . 2018. Targeted on-line SPE-LC-MS/MS assay for the quantitation of 12 apolipoproteins from human blood. Proteomics. 18: doi:10.1002/pmic.201700279. [DOI] [PubMed] [Google Scholar]
- 16.van den Broek I., Romijn F. P. H. T. M., Nouta J., van der Laarse A., Drijfhout J. W., Smit N. P. M., van der Burgt Y. E. M., and Cobbaert C. M.. 2016. Automated multiplex LC-MS/MS assay for quantifying serum apolipoproteins A-I, B, C-I, C-II, C-III, and E with qualitative apolipoprotein E phenotyping. Clin. Chem. 62: 188–197. [DOI] [PubMed] [Google Scholar]
- 17.Nedelkov D. 2017. Mass spectrometric studies of apolipoprotein proteoforms and their role in lipid metabolism and type 2 diabetes. Proteomes. 5: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pum J. 2019. A practical guide to validation and verification of analytical methods in the clinical laboratory. Adv. Clin. Chem. 90: 215–281. [DOI] [PubMed] [Google Scholar]
- 19.Allinson J. L. 2018. Clinical biomarker validation. Bioanalysis. 10: 957–968. [DOI] [PubMed] [Google Scholar]
- 20.Carr S. A., Abbatiello S. E., Ackermann B. L., Borchers C., Domon B., Deutsch E. W., Grant R. P., Hoofnagle A. N., Hüttenhain R., Koomen J. M., et al. . 2014. Targeted peptide measurements in biology and medicine: best practices for mass spectrometry-based assay development using a fit-for-purpose approach. Mol. Cell. Proteomics. 13: 907–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Croyal M., Fall F., Ferchaud-Roucher V., Chétiveaux M., Zaïr Y., Ouguerram K., Krempf M., and Nobécourt E.. 2016. Multiplexed peptide analysis for kinetic measurements of major human apolipoproteins by LC/MS/MS. J. Lipid Res. 57: 509–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou H., Hoek M., Yi P., Rohm R. J., Mahsut A., Brown P., Saunders J., Chmielowski R. A., Ren N., Shuster D., et al. . 2013. Rapid detection and quantification of apolipoprotein L1 genetic variants and total levels in plasma by ultra-performance liquid chromatography/tandem mass spectrometry. Rapid Commun. Mass Spectrom. 27: 2639–2647. [DOI] [PubMed] [Google Scholar]
- 23.Pan Y., Zhou H., Mahsut A., Rohm R. J., Berejnaia O., Price O., Chen Y., Castro-Perez J., Lassman M. E., McLaren D., et al. . 2014. Static and turnover kinetic measurement of protein biomarkers involved in triglyceride metabolism including apoB48 and apoA5 by LC/MS/MS. J. Lipid Res. 55: 1179–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blanchard V., Ramin-Mangata S., Billon-Crossouard S., Aguesse A., Durand M., Chemello K., Nativel B., Flet L., Chétiveaux M., Jacobi D., et al. . 2018. Kinetics of plasma apolipoprotein E isoforms by LC-MS/MS: a pilot study. J. Lipid Res. 59: 892–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Croyal M., Ouguerram K., Passard M., Ferchaud-Roucher V., Chétiveaux M., Billon-Crossouard S., de Gouville A-C., Lambert G., Krempf M., and Nobécourt E.. 2015. Effects of extended-release nicotinic acid on apolipoprotein (a) kinetics in hypertriglyceridemic patients. Arterioscler. Thromb. Vasc. Biol. 35: 2042–2047. [DOI] [PubMed] [Google Scholar]
- 26.Bland J. M., and Altman D. G.. 1986. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet. 1: 307–310. [PubMed] [Google Scholar]
- 27.Marcovina S., and Packard C. J.. 2006. Measurement and meaning of apolipoprotein AI and apolipoprotein B plasma levels. J. Intern. Med. 259: 437–446. [DOI] [PubMed] [Google Scholar]
- 28.Pechlaner R., Tsimikas S., Yin X., Willeit P., Baig F., Santer P., Oberhollenzer F., Egger G., Witztum J. L., Alexander V. J., et al. . 2017. Very-low-density lipoprotein-associated apolipoproteins predict cardiovascular events and are lowered by inhibition of APOC-III. J. Am. Coll. Cardiol. 69: 789–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tavernier G., Caspar-Bauguil S., and Viguerie N.. 2020. Apolipoprotein M: new connections with diet, adipose tissue and metabolic syndrome. Curr. Opin. Lipidol. 31: 8–14. [DOI] [PubMed] [Google Scholar]
- 30.Bick A. G., Akwo E., Robinson-Cohen C., Lee K., Lynch J., Assimes T. L., DuVall S., Edwards T., Fang H., Freiberg S. M., et al. . 2019. Association of APOL1 risk alleles with cardiovascular disease in blacks in the million veteran program. Circulation. 140: 1031–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tailleux A., Duriez P., Fruchart J-C., and Clavey V.. 2002. Apolipoprotein A-II, HDL metabolism and atherosclerosis. Atherosclerosis. 164: 1–13. [DOI] [PubMed] [Google Scholar]
- 32.Wang F., Kohan A. B., Lo C-M., Liu M., Howles P., and Tso P.. 2015. Apolipoprotein A-IV: a protein intimately involved in metabolism. J. Lipid Res. 56: 1403–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Genovese G., Friedman D. J., Ross M. D., Lecordier L., Uzureau P., Freedman B. I., Bowden D. W., Langefeld C. D., Oleksyk T. K., Uscinski Knob A. L., et al. . 2010. Association of trypanolytic ApoL1 variants with kidney disease in African Americans. Science. 329: 841–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Erqou S., Thompson A., Di Angelantonio E., Saleheen D., Kaptoge S., Marcovina S., and Danesh J.. 2010. Apolipoprotein(a) isoforms and the risk of vascular disease: systematic review of 40 studies involving 58,000 participants. J. Am. Coll. Cardiol. 55: 2160–2167. [DOI] [PubMed] [Google Scholar]
- 35.Agger S. A., Marney L. C., and Hoofnagle A. N.. 2010. Simultaneous quantification of apolipoprotein A-I and apolipoprotein B by liquid-chromatography-multiple- reaction-monitoring mass spectrometry. Clin. Chem. 56: 1804–1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.von Zychlinski A., Williams M., McCormick S., and Kleffmann T.. 2014. Absolute quantification of apolipoproteins and associated proteins on human plasma lipoproteins. J. Proteomics. 106: 181–190. [DOI] [PubMed] [Google Scholar]
- 37.Lassman M. E., McLaughlin T. M., Zhou H., Pan Y., Marcovina S. M., Laterza O., and Roddy T. P.. 2014. Simultaneous quantitation and size characterization of apolipoprotein(a) by ultra-performance liquid chromatography/mass spectrometry. Rapid Commun. Mass Spectrom. 28: 1101–1106. [DOI] [PubMed] [Google Scholar]
- 38.Koska J., Yassine H., Trenchevska O., Sinari S., Schwenke D. C., Yen F. T., Billheimer D., Nelson R. W., Nedelkov D., and Reaven P. D.. 2016. Disialylated apolipoprotein C-III proteoform is associated with improved lipids in prediabetes and type 2 diabetes. J. Lipid Res. 57: 894–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Azizkhanian I., Trenchevska O., Bashawri Y., Hu J., Koska J., Reaven P. D., Nelson R. W., Nedelkov D., and Yassine H. N.. 2016. Posttranslational modifications of apolipoprotein A-II proteoforms in type 2 diabetes. J. Clin. Lipidol. 10: 808–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yassine H. N., Jackson A. M., Reaven P. D., Nedelkov D., Nelson R. W., Lau S. S., and Borchers C. H.. 2014. The application of multiple reaction monitoring to assess Apo A-I methionine oxidations in diabetes and cardiovascular disease. Transl. Proteom. 4–5: 18–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chatterjee B., and Thakur S. S.. 2018. Investigation of post-translational modifications in type 2 diabetes. Clin. Proteomics. 15: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained within the article or in the supplemental material section. All chromatograms used for quantification are available in the supplemental material.