Abstract
The coronavirus disease 2019 (COVID-19) pandemic caused by severe acute respiratory syndrome coronavirus (SARS-CoV)-2 has resulted in the death of more than 328,000 persons worldwide in the first 5 months of 2020. Herculean efforts to rapidly design and produce vaccines and other antiviral interventions are ongoing. However, newly evolving viral mutations, the prospect of only temporary immunity, and a long path to regulatory approval pose significant challenges and call for a common, readily available, and inexpensive treatment. Strategic drug repurposing combined with rapid testing of established molecular targets could provide a pause in disease progression. SARS-CoV-2 shares extensive structural and functional conservation with SARS-CoV-1, including engagement of the same host cell receptor (angiotensin-converting enzyme 2) localized in cholesterol-rich microdomains. These lipid-enveloped viruses encounter the endosomal/lysosomal host compartment in a critical step of infection and maturation. Niemann-Pick type C (NP-C) disease is a rare monogenic neurodegenerative disease caused by deficient efflux of lipids from the late endosome/lysosome (LE/L). The NP-C disease-causing gene (NPC1) has been strongly associated with viral infection, both as a filovirus receptor (e.g., Ebola) and through LE/L lipid trafficking. This suggests that NPC1 inhibitors or NP-C disease mimetics could serve as anti-SARS-CoV-2 agents. Fortunately, there are such clinically approved molecules that elicit antiviral activity in preclinical studies, without causing NP-C disease. Inhibition of NPC1 may impair viral SARS-CoV-2 infectivity via several lipid-dependent mechanisms, which disturb the microenvironment optimum for viral infectivity. We suggest that known mechanistic information on NPC1 could be utilized to identify existing and future drugs to treat COVID-19.
Keywords: severe acute respiratory syndrome coronavirus 2, cholesterol, cholesterol trafficking, lysosomal storage disease, Niemann-Pick disease, dyslipidemias, drug repurposing, pandemic, coronavirus disease 2019, Ebola
The coronavirus disease 2019 (COVID-19) pandemic, caused by the novel severe acute respiratory syndrome coronavirus (SARS-CoV)-2 virus, has resulted in the death of over 328,000 persons worldwide in 2020 (at the time of acceptance of this article on 21 May 2020) (1). For the vast majority of infectious diseases, the implementation of a worldwide efficacious vaccination program would normally lead to their eradication. However, this outcome is dauntingly distant for COVID-19, as well as challenged by newly evolving mutations in the virus and the possible acquisition of only temporary immunity. Similarly, novel antiviral drug design, even when optimized for intervention against well-researched molecular pathways, is likely to face a prolonged path to approval. A common, readily available, and inexpensive treatment is needed.
Drug repurposing, coupled with strategic assessments of molecular mechanisms and targets, is one approach by which both containment and mitigation of this current global outbreak could be advanced. This tactic suggested several preexisting clinically approved drugs for treating COVID-19 that are currently being prescribed “off-label” or are subjects of clinical trials. Lipid enveloped viruses, such as SARS-CoV-2, enter cells by membrane fusion subsequent to acquisition by cell surface receptors. Given the localization of the host cell receptor for SARS-CoV-2 [angiotensin-converting enzyme 2 (ACE2)] to lipid-enriched membrane microdomains and the lipid-regulating activity of candidate COVID-19 therapeutics, it is surprising that, to date, the role of lipids has not been extensively investigated. Here, we describe a lipid-based strategy of antiviral drug repurposing, whereby we propose interventions in lysosomal lipid homeostasis as a treatment for COVID-19. In particular, we will emphasize the immense potential of targeting the lipid regulator defective in Niemann-Pick type C (NP-C) disease, a rare lysosomal lipid storage disorder.
THE IMPACT OF LIPIDS IN SARS-CoV-2 INFECTION AND REPLICATION
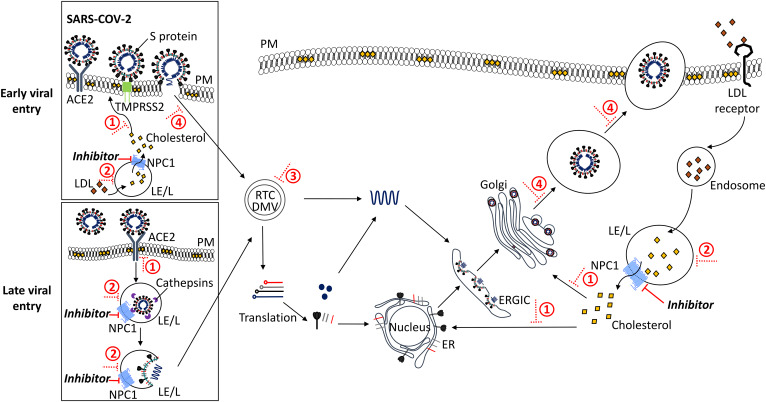
Insight into the life cycles of the majority of infectious agents is remarkably sophisticated, often informed by knowledge about the specific organism or one closely related. SARS-CoV-2 is a single-stranded RNA virus that is suspected to originate from bats and/or Malayan pangolins in exotic wildlife markets in China (2). The nucleotide sequence of SARS-CoV-2 is 82% identical to that of SARS-CoV-1, a well-studied human coronavirus that was the cause of the severe acute respiratory syndrome epidemic of 2003 (3, 4). Similar to SARS-CoV-1, the S protein of SARS-CoV-2 binds to the host cell receptor, ACE2 (5). In brief, upon binding ACE2, the type II transmembrane serine host cell protease, TMPRSS2, cleaves (“primes”) the S protein at the S1/S2 and S2′ sites, enabling viral fusion and nucleocapsid entry into the cytoplasm (Fig. 1, “early entry”) (5). Alternatively, SARS-CoV-2 is endocytosed in a clathrin- and/or caveolae-dependent manner (6, 7) and then proteolytically cleaved in the late endosome/lysosome (LE/L). This pH-dependent process is facilitated by cysteine proteases, likely cathepsin B or L, allowing for viral nucleocapsid release into the cytoplasm (5, 8) (Fig. 1, “late viral entry”). Subsequently, the virus hijacks host-cells for the formation of the replication and transcription complex, which associates with autophagosome-like intracellular membrane proliferations, such as double membrane vesicles (DMVs) (9, 10). As a consequence, the viral genomic RNA is replicated, and subgenomic RNA is transcribed and translated into structural and accessory proteins at the ER, before being transported to the ER-Golgi intermediate compartment for virion assembly. Assembled virions pass through the Golgi and then bud into vesicles before exiting the cell at the plasma membrane (PM) via exocytosis (10–15). The entry, replication, assembly, and release of the coronavirus are dependent on host factors, in which membrane lipids play a key role. We propose that lipid homeostasis presents a viral Achilles heel that could be exploited to combat both current and future pandemics.
Fig. 1.
Proposed inhibition of SARS-CoV-2 entry/replication by intervention at the NPC1 pathway. Early entry of SARS-CoV-2 into the host cell is mediated by the serine protease, TMPRSS2 (green), which proteolytically primes the viral S protein upon binding to ACE2 (dark gray) at the PM. This releases the virion into the cytoplasm. Alternatively, for late viral entry, SARS-CoV-2 is endocytosed in a clathrin- and/or caveolin-mediated manner from the PM into the host cell. The cysteine protease, cathepsin (purple), cleaves the viral S protein prior to release of the viral nucleocapsid from the LE/L into the cytoplasm. The virus hijacks host-cell machinery to form the replication and transcription complex (RTC) and DMVs where genomic RNA and subgenomic RNA (red, light gray, black, dark blue) are synthesized. Genomic RNA is replicated, and structural and accessory proteins are processed at the ER, before viral assembly at the ER-Golgi intermediate compartment (ERGIC), and bud from the Golgi into vesicles. Finally, the virus is released from the host cell by exocytosis. Under normal conditions, cholesteryl ester in LDL particles (orange) enters the cell by receptor-mediated endocytosis, is hydrolyzed to cholesterol (yellow) in the LE/L where it binds NPC1 (bright blue) and is transported to other organelles (e.g., PM, ER, ERGIC, Golgi). Inhibiting NPC1 (NPC1 inhibitors or the action of NP-C disease mimetics, solid red line) leads to lipid accumulation in the LE/L and, subsequently, 1) depletion of levels of cholesterol in the cell, affecting viral binding and priming, 2) impairment of LE/L pH and protease activity, 3) occurrence of dysregulation at LE/L-ER MCSs, and 4) impairment of endocytosis and exocytosis; overall impeding viral infectivity, replication, assembly, and release (dotted red line).
NPC1 FUNCTIONS AS A LIPID REGULATOR AND FILOVIRUS RECEPTOR
NP-C disease is a rare (∼500 cases worldwide) and invariably fatal genetic disorder whereby lipid accumulation in the LE/L impairs organelle function leading to a cascade of visceral and neurodegenerative symptoms including dementia. Mutations in the NPC1 gene, encoding a 13 transmembrane domain protein that traverses the limiting membrane of the LE/L, account for 95% of NP-C cases. The remainder of cases are due to mutations in the NPC2 gene encoding a lysosomal lumenal protein (16). NPC1 is comprised of numerous functional domains including: the N-terminal domain (25–264 amino acids), which binds cholesterol; the second luminal loop (371–615 amino acids) where NPC2 binds; a sterol-sensing domain (SSD) (616–791 amino acids); and lastly, a cysteine-rich domain (855–1,098 amino acids) (17). Exogenous cholesteryl ester, in the form of LDLs, binds to the LDLR and is endocytosed into the LE/L, where it is hydrolyzed by acid lipase to free cholesterol. In normal cells, acquisition of cholesterol by NPC2 proteins precedes its “hand-off” to the NPC1 protein and its export from the organelle (18, 19). In situations where NPC protein activity is diminished, the ensuing sequestration of cholesterol and alterations in cellular lipid homeostasis cause a secondary accumulation of numerous bioactive lipids including various sphingolipids, phospholipids, gangliosides, and glycolipids (20). These disruptions in lipid metabolism promote extensive dysfunction of the lysosome including changes in pH, calcium/ion gradient, and lysosomal protease status (21, 22), thereby potentially deterring an invading virus.
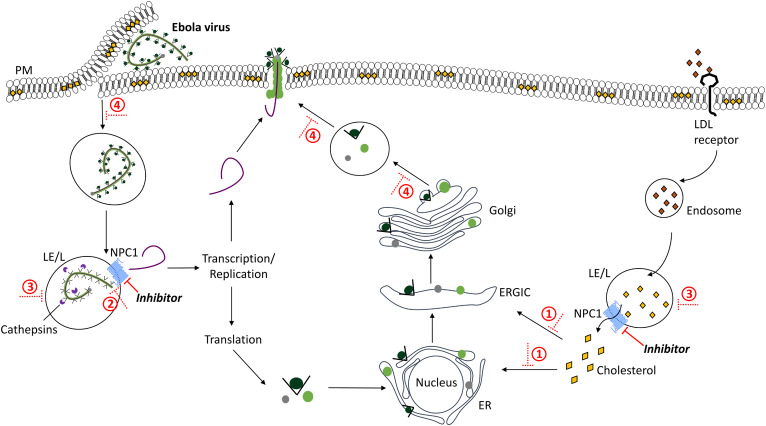
In a startling example of how research into rare diseases can extend into a larger arena, numerous studies suggest that ablation of the NPC1 pathway with mutations in the NPC1 protein confers resistance and/or lowers severity of infection in vitro and in vivo to filovirus [e.g., Ebola virus (EBOV)], retrovirus [e.g., human immunodeficiency virus (HIV)], and togavirus [e.g., Chikungunya virus (CHIKV)] (23–26). With respect to the infection of cells by EBOV, there is a clear requisite for a physical molecular interaction between the proteolytically activated EBOV glycoprotein (EBOV-GP) and NPC1 (27, 28). In EBOV infection, the EBOV-GP is trimmed by cathepsin B/L before binding to NPC1 at the second luminal loop prior to nucleocapsid release from the LE/L and subsequent viral replication and exit (27, 29, 30) (Fig. 2). Lipid transport, particularly in the context of variation in NPC1, also impacts EBOV infectivity (31), although the absence of an impact of NPC2 mutations suggests that it may be a secondary role for this virus. Similarly, the role of NPC2 in HIV viral biogenesis contradicts the impact of NPC1 mutations and inhibitors (32).
Fig. 2.
NPC1, a lipid transporter, is also a receptor for filovirus entry and replication. EBOV enters the host cell via macropinocytosis leading to activation of the EBOV-GP by host cysteine proteases, cathepsin B/L (purple), at the LE/L in a pH-dependent manner. The primed EBOV-GP binds to the second luminal loop of NPC1 and releases the virion into the cytoplasm. Upon release, the virion is replicated and transcribed, where viral structural and accessory proteins (black, dark green, light green, gray) are packed into the ER, and released from the Golgi, where the virus assembles at the PM before release from the cell via exocytosis. Under normal conditions, cholesteryl ester in LDL particles (orange) enters the cell by receptor-mediated endocytosis, is hydrolyzed to cholesterol (yellow) in the LE/L where it binds NPC1 (bright blue) and is transported to other organelles (e.g., PM, ER, ERGIC, Golgi). NPC1 inhibitors or NP-C disease mimetics 1) deplete levels of cholesterol in the cell, 2) hinder the interaction between NPC1 and the EBOV-GP, 3) impair LE/L pH and protease activity, and 4) impair endocytosis and exocytosis, hindering viral infectivity, replication, assembly, and release.
HOW DOES A RARE NEURODEGENERATIVE DISEASE CAUSED BY DEFECTIVE LIPID TRANSPORT IDENTIFY CANDIDATE COVID-19 TREATMENTS?
With respect to coronavirus infectivity, the localization of the ACE2 and TMPRSS2 proteins to cholesterol- and sphingolipid-rich microdomains in the PM is a prerequisite for efficient viral entry (33). When intracellular cholesterol movement, for example by inactivation of NPC1, is blocked at the LE/L, cholesterol levels at the PM and ER are depleted (34–36). Consistently, type I feline coronavirus (FCoV) infectivity was reduced by treatment with methyl-β-cyclodextrin, a drug that depletes cell membrane cholesterol, and virion load was restored by cholesterol supplementation (37). Similarly, inhibiting NPC1 depletes cholesterol content at cholesterol- and sphingolipid-rich microdomains, thus possibly disturbing the distribution of host cell receptors and proteases in the PM, and subsequently reducing binding/priming of viral S protein for SARS-CoV-2 entry. Cholesterol depletion may also hinder late viral entry because binding to the host cell receptor leads to endocytosis in a clathrin- and/or caveolae-dependent manner, a process that takes place in cholesterol-rich membrane domains (6, 7). Additionally, in late stage entry, where the S protein of SARS-CoV-2 is primed by cathepsin B/L in the LE/L, this proteolytic activity is impaired by cholesterol accumulation due to NPC1 deficiency and restored upon clearance of the accumulated cholesterol (38). Cholesterol accumulation due to NPC1 deficiency also affects the LE/L pH, thus decreasing the efficiency of cathepsin priming of the viral S protein. It has also been suggested that cathepsin inhibition itself leads to cholesterol accumulation (39). Therefore, inhibiting NPC1 enables a multistep blockade of viral entry through the PM or LE/L into host cells and thus represents an effective means to inhibit SARS-CoV-2 infectivity.
Inhibition of NPC1 may also impact viral replication and budding. For example, DMVs have cholesterol-enriched membranes formed from extrusions of the ER, which are the likely sites where viral replication centers are assembled (15). The nonstructural proteins of the virus target the host cell factors to induce intracellular membrane rearrangement that leads to formation of DMVs (40). Although the precise mechanism of DMV formation in a coronavirus-infected cell is not completely understood, nonvesicular lipid transport via membrane contact sites (MCSs) is proposed to play a crucial role in replication of positive strand RNA viruses (41). As NPC1 regulates the MCSs between the LE/L and the ER (42), inhibiting NPC1 reduces cholesterol supply to DMVs and impairs hepatitis C virus (HCV) replication (43). Similarly, NPC1 could interfere with DMV formation for SARS-CoV-2. Finally, the newly assembled and matured virus that buds from the Golgi undergoes exocytosis from the host cell at cholesterol- and sphingolipid-rich microdomains (44). Thus, lipid homeostasis clearly plays a crucial role for DMV formation and also vesicle membrane formation for both endocytosis and exocytosis in coronaviruses (15, 44).
Therefore, inhibition or loss of NPC1, thereby blocking cholesterol transport from the LE/L, may impair viral SARS-CoV-2 infectivity at multiple stages including viral entry, replication, and exit: 1) depletion of lipids at the PM and ER, particularly from cholesterol- and sphingolipid-rich microdomains; 2) accumulation of lipids at the LE/L; 3) dysregulation of LE/L-ER MCSs; and 4) alteration of endocytosis (entry) and exocytosis (exit); which together disturb the cellular microenvironment optimum for viral infectivity and replication (Fig. 1).
REPURPOSING NPC1 INHIBITORS FOR ANTIVIRAL TREATMENT
Pharmacological inhibition of NPC1 represents a valuable much-used tool in the laboratory to mimic the cellular lipidosis associated with NP-C disease. The cationic amphiphilic drug U18666A was initially developed to inhibit cholesterol biosynthesis and subsequently was observed to induce lipid accumulation in the LE/L (45). More recently, it was determined that U18666A inhibits NPC1 by binding to the SSD (46); the ability of this drug to disrupt lysosomal cholesterol homeostasis has prompted numerous investigations into its antiviral properties. The infectivity of a surprising variety of enveloped viruses, such as EBOV, influenza A virus (IAV), HCV, CHIKV, Zika virus (ZIKV), Dengue virus (DENV), West Nile virus (WNV), and HIV (25, 26, 46–48), was strikingly compromised by U18666A treatment. Interestingly, the antiviral activity of U18666A against EBOV is not a simple competition for a shared ligand binding site because the EBOV-GP binds to the second luminal loop of NPC1 and not to the SSD (46). With respect to coronaviruses, U18666A strongly inhibited S protein-driven entry of SARS-CoV-1, Middle East respiratory syndrome-related coronavirus (MERS-CoV), and FCoV (49, 50). The antiviral activity of U18666A against FCoV has been extensively characterized in vitro and in vivo. In an in vitro study, Takano et al. (49) elegantly demonstrated that NPC1 plays an important role in FCoV infectivity. Antiviral activity was achieved with U18666A treatment and subsequently reversed with vorinostat, a histone deacetylase inhibitor that increases NPC1 expression and rescues lipid accumulation in vitro (51, 52). This result was then translated in vivo in FCoV-infected cats where severity of infection was suppressed relative to control, albeit the sample size was small (53).
The precise antiviral mechanism of U18666A remains to be determined although the most parsimonious explanation would reflect its general impact on sterol homeostasis. The concentration of U18666A required for antiviral activity can suggest insight into whether the antiviral activity is due to a primary effect of lipid accumulation or a secondary effect of lysosomal dysfunction caused by lipid accumulation. Relevant to the current pandemic, U18666A reduced entry of the SARS-CoV-1 S protein by 80% compared with control, at a concentration that induced lipid accumulation without increasing LE/L pH (50, 54). In contrast, a concentration known to increase LE/L pH was required to inhibit IAV (50, 55). Additionally, entry of the viral S protein of four types of human coronavirus was significantly inhibited by another pharmacological agent (clomiphene) that induces lipid accumulation without increasing LE/L pH (50, 56), suggesting that coronavirus entry may be highly dependent on host cell cholesterol transport.
We propose to repurpose clinically approved therapeutics that inhibit NPC1 and/or perturb lipid homeostasis in a similar fashion to U18666A (which did not receive regulatory approval). Antiviral activity has been shown for several readily available drugs that inhibit NPC1 and are commonly used as antidepressant, anti-inflammatory, anticancer, or antifungal agents. These drugs have the shared ability to bind NPC1 at the SSD (46, 57–60) (Table 1).
TABLE 1.
Candidate anti-SARS-CoV-2 therapeutics based on NPC1 inhibition and NP-C disease mimetic mechanisms
| Generic Name (CAS Number) | Brand Name | Clinical Contraindications/Side Effects | Conventional Use | Drug Mechanism | Antiviral Activity | References |
| NPC1 inhibitors | ||||||
| U18666Aa (3039-71-2) | U18666A | Not applicable | Laboratory use | Inhibits NPC1 | EBOV, IAV, HCV, CHIKV, ZIKV, DENV, WNV, HIV, SARS-CoV-1, MERS-CoV, type I FCoV | (25–28, 46–50, 53) |
| Imipraminea (50-49-7) | Tofranil, Janimine, Melipramine | Mood disorders, nausea, angle-closure glaucoma | Antidepressant | Inhibits NPC1, inhibits TPC2 | EBOV, CHIKV, ZIKV, DENV, WNV | (24, 26, 60, 61) |
| Cepharanthinea (481-49-2) | Cepharantin | Well tolerated | Anti-inflammatory, anticancer | Inhibits NPC1, inhibits TPC2 | HIV, Pangolin coronavirus | (58, 63–66, 117) |
| Itraconazole (84625-61-6) | Onmel, Sporanox, Tolsura | Heart failure, QT prolongation | Antifungal | Inhibits NPC1 | IAV, DENV, PeV-A3, EV, HCV, type I FCoV | (57, 67–73) |
| Posaconazole (171228-49-2) | Noxafil, Posanol | Heart failure, QT prolongation | Antifungal | Inhibits NPC1 | IAV, DENV, ZIKV, YFV, PeV-A3, EV, HCV, type I FCoV | (59, 67, 69–72) |
| NP-C disease mimetics | ||||||
| Chloroquinea,b (54-05-7) | Aralen | Heart failure, QT prolongation | Antimalarial | Lipid accumulation in the LE/L, increases pH | CHIKV, ZIKV, EBOV, HCV, HIV, IAV, EV, SARS-CoV-1, MERS-CoV, SARS-CoV-2 | (74, 80, 81, 83–87, 118) |
| Hydroxychloroquinea,b (118-42-3) | Plaquenil | Heart failure, QT prolongation | Antimalarial | Lipid accumulation in the LE/L, increases pH | HIV, SARS-CoV-1, SARS-CoV-2 | (79, 80, 82) |
| Azithromycin a,b (83905-01-5) | Zithromax, Sumamed | Heart failure, QT prolongation, liver dysfunction | Antibacterial | Lipid accumulation in the LE/L, increases pH | ZIKV, RV | (75, 90, 91) |
| Chlorpromazine hydrochloridea (69-09-0) | Sonazine, Chloractil | Dementia in older patients, mood disorders | Antipsychotic, antidepressant | Lipid accumulation in the LE/L, inhibits clathrin-mediated endocytosis | SARS-CoV-1, MERS-CoV, WNV, IV, HPEV-1, JEV, HPyV-2 | (6, 76, 84, 86, 93–98) |
| Haloperidola (52-86-8) | Haldol, Aloperidin | Dementia in older patients, mood disorders | Antipsychotic, antiemetic | Lipid accumulation in the LE/L | (78) | |
| Amiodaronea (1951-25-3) | Cordarone | Lung, liver damage, arrhythmia | Antiarrhythmic | Lipid accumulation in the LE/L, alters late compartments of the endocytic pathway | SARS-CoV-1, EBOV, HCV | (77, 100–102) |
Approved drugs that inhibit NPC1 or mimic NP-C disease that have previously shown antiviral activity. Drug specifications from https://pubchem.ncbi.nlm.nih.gov/. Contraindications and side effects are from Medline https://medlineplus.gov/drugreactions.html. Parechovirus A3 (PeV-A3); yellow fever virus (YFV); rhinovirus (RV); influenza virus (IV); human parechovirus 1 (HPEV-1); Japanese encephalitis virus (JEV); human polyomavirus 2 (HPyV-2).
Cationic amphiphile.
Currently in COVID-19 clinical trial.
Imipramine, a cationic amphiphile and an approved tricyclic antidepressant, inhibits the NPC pathway, induces lysosomal lipid accumulation, and elicits antiviral activity against several single-stranded RNA viruses. Infectivity of ZIKV, DENV, WNV, CHIKV, and EBOV was reduced upon imipramine treatment in human cell lines (24, 26). Given the promising antiviral activity of U18666A and imipramine against EBOV in vitro, Herbert et al. (24) replicated these results in vivo in an EBOV mouse model where both imipramine and U18666A significantly reduced viral replication in the liver, spleen, and serum; however, there were only moderate increases in survival in imipramine-treated mice. Imipramine has further been reported to inhibit TPC2, a two-pore channel protein required for SARS-CoV-2 entry via endocytosis (61, 62). Similar to imipramine, cepharanthine has been shown to inhibit TPC2 (63). Cepharanthine, a known NPC1 inhibitor, is an isoquinoline-containing cationic amphiphile and an approved anti-inflammatory and anticancer therapy that has also shown antiviral activity (58). Cepharanthine exhibited complete inhibition of HIV and most notably the pangolin coronavirus, which has high sequence similarity to SARS-CoV-2, while additionally sharing the same host cell receptor (ACE2) (2, 64, 65). Cepharanthine was identified to have antiviral activity against SARS-CoV-2 in vitro (66).
Further, the triazoles itraconazole and posaconazole inhibit NPC1 and are approved antifungals that elicit antiviral activity against numerous viruses in vitro [e.g., IAV, DENV, ZIKV, yellow fever virus, parechovirus A3, enterovirus (EV), and HCV] including type I FCoV (59, 67–72). Promisingly, itraconazole-treated mice infected with IAV decreased viral load in the lungs and tracheae, and improved survival relative to control (67). Further, an in silico target-based virtual ligand analysis identified itraconazole as an inhibitor of SARS-CoV-2 RNA-dependent RNA polymerase and thus a possible treatment for SARS-CoV-2; in vitro and in vivo work is required to test this hypothesis (73).
REPURPOSING NP-C DISEASE MIMETICS AS CANDIDATE COVID-19 THERAPEUTICS
Targeting the cascade of metabolic effects that arise from lipid accumulation (independent of NPC1 inhibition) could also be an effective therapy to treat COVID-19, given the dependence of SARS-CoV-2 on host lipids and organelles for infection and replication. Interestingly, several clinically approved drugs, including antiviral drugs, mimic NP-C disease with lipid accumulation in the LE/L that further disrupts cellular lipid homeostasis (74–79). These include chloroquine and its derivative, hydroxychloroquine, as well as azithromycin, chlorpromazine, and amiodarone. Chloroquine was initially approved as an antimalarial agent and later approved to treat rheumatoid arthritis due to its anti-inflammatory properties. This quinolone-containing cationic amphiphile has shown antiviral activity against numerous viral infections in vitro (e.g., CHIKV, ZIKV, EBOV, HCV, HIV, IAV, EV, SARS-CoV-1, MERS-CoV, and SARS-CoV-2) (80–84). In a SARS-CoV-1 replication mouse model, chloroquine did not reduce viral titer in the lung (85). Recent work (non-peer reviewed) from the same laboratory suggests that chloroquine treatment reduced weight loss and protected against inflammation in the lungs in a SARS-CoV-1 mouse model (MA15); however; there was no difference in viral titer in the lungs (86). In FCoV-infected cats, chloroquine showed a better clinical outcome relative to control cats, albeit this was achieved independently of antiviral efficacy and was possibly a consequence of the anti-inflammatory activity of chloroquine (87). Chloroquine is a weak base that increases the pH of the LE/L, thereby impeding membrane fusion and function of lysosomal proteases (e.g., cathepsin B/L) required for viral entry/replication (7). As depicted in Fig. 3, chloroquine treatment produces a comparable accumulation of cholesterol to that observed in NPC1-deficient cells. In SARS-CoV-1 infection, chloroquine interferes with the glycosylation of ACE2, which negatively affects receptor binding and thus viral entry (81). Hydroxychloroquine, a less toxic derivative of chloroquine, has the same antiviral properties as chloroquine; both compounds inhibited pre- and post-entry steps in SARS-CoV-2 infection in cell culture (82, 83, 88). In a large retrospective study (1,376 COVID-19 patients), no benefit was identified in hydroxychloroquine-treated patients; however, treated patients were more severely ill at baseline than the control group (89). Azithromycin, a cationic amphiphile and an approved antibacterial, also leads to an increase in lysosomal pH and an accumulation of lipids. Azithromycin has previously shown antiviral activity in vitro against the ZIKV and rhinovirus (90, 91), and has been tested in combination with hydroxychloroquine in COVID-19 patients with inconclusive results (92). Approval of the use of these compounds awaits randomized controlled clinical trials. Nevertheless, these drugs have been used to treat severely ill patients in the early months of the 2019/2020 pandemic (92).
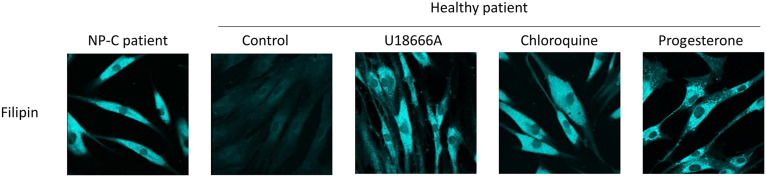
Fig. 3.
Representative cationic amphiphile drugs inhibit intracellular transport of cholesterol and mimic NP-C disease. Healthy patient fibroblasts were treated overnight with either the NPC1 inhibitor U18666A (5 μM) or NP-C disease mimetics chloroquine (10 μM) or progesterone (10 μM) and stained with filipin to visualize unesterified cholesterol. Images were obtained using a laser scanning spectral confocal microscope (Leica TCS SP-2). In all cases, U18666A, chloroquine, and progesterone phenocopy the biochemical hallmark of lysosomal cholesterol accumulation in NP-C patient fibroblasts. Cholesterol accumulation impairs lysosomal function, which may mediate the antiviral activity of the interventions discussed in this review.
Chlorpromazine hydrochloride (chlorpromazine), a cationic amphiphile approved as an antipsychotic and antidepressant, inhibited infectivity in vitro of SARS-CoV-1, MERS-CoV, WNV, influenza virus, human parechovirus 1, Japanese encephalitis virus, and human polyomavirus 2 by specifically inhibiting the formation of clathrin-coated pits required for clathrin-dependent endocytosis at the PM (6, 84, 93–98). It is suspected that SARS-CoV-2 may undergo clathrin-dependent endocytosis upon entry into the host cell. Having identified chlorpromazine as a hit in an earlier in vitro drug library screen against SARS-CoV-1 and MERS-CoV (98), Weston et al. (86) (non-peer reviewed) then tested its antiviral activity in SARS-CoV-2, where chlorpromazine inhibited SARS-CoV-2 entry. These promising in vitro results were translated when chlorpromazine-treated mice showed reduced weight loss and reduced signs of SARS-CoV-1 infection relative to control; however, there was no difference in viral titer in the lungs (86).
Haloperidol, a cationic amphiphile and approved antipsychotic and antiemetic drug, was identified as a candidate antiviral therapy against SARS-CoV-2 (99). Haloperidol may hinder viral entry and replication by inhibiting cholesterol trafficking from the LE/L (78). Lastly, amiodarone, a cationic amphiphile and antiarrhythmic, has been observed to inhibit infectivity of SARS-CoV-1, EBOV, and HCV in vitro (100–102) and, in the case of SARS-CoV-1, prevent transit of the virus through the endosomal pathway.
GENETIC PROCLIVITY TO VIRAL INFECTION AND DISEASE PROGRESSION
In addition to the anticipated preponderance of COVID-19 disease in the aged, there are clear and unexplained gender and ethnic biases to disease severity. In the context of any human infection, multiple factors impact the clinical outcome of a disease including comorbidities, the pathogen genome, and the genome of the host. Heterozygote advantage, whereby carriers of polymorphisms at a locus with two alleles have a higher fitness than either homozygote has been advanced as a mechanism by which deleterious genetic variants are maintained in human populations. The most established example of this balancing selection arises in hemoglobinopathies where carriers of sickle cell anemia or thalassemia exhibit resistance to Plasmodium falciparum and thus malaria (103). At the biochemical level, this likely reflects accelerated phagocytosis of infected sickled or thalassemic cells. Given the role of NPC1 and the lysosome in an abundance of viral life cycles, including EBOV and coronaviruses, we propose that heterozygous genetic variation at NPC1 or indeed any of the approximately 50 loci that in the homozygous state cause a lysosomal storage disorder, may account for divergent clinical outcomes and prognosis for enveloped viruses, including SARS-CoV-2.
An important animal experiment provided proof of concept to this premise. Homozygous recessive (Npc1−/−) and, importantly, Npc1+/− (heterozygous) mice were resistant to viral infection and protected from EBOV disease (24). Similarly, naturally occurring variants in human NPC1 were observed to impair EBOV infectivity in vitro (31). Human NPC1 carriers (heterozygotes) are generally healthy and may even be relatively resistant to atherosclerosis (104). Nevertheless, gene dosage was observed to quantitatively impact lipid homeostasis in fibroblasts (105, 106). Whether persons with half-normal levels of the NPC1 protein (i.e., heterozygotes) have altered susceptibility to SARS-CoV-2 remains to be determined.
Intriguingly, for unknown reasons, males appear to be more susceptible to COVID-19 than females, a pattern previously observed in SARS-CoV-1 and MERS-CoV (107–110). Progesterone, a female sex hormone and an approved contraceptive, inhibits lysosome acidification and the release of cholesterol from the lysosome, thereby phenocopying NP-C disease (Fig. 3) (111). Could this contribute to the greater burden of disease in males than females? In mice infected with IAV, progesterone-treated mice displayed limited inflammation and aided pulmonary repair; however, treatment did not increase resistance or inhibit viral replication (112). In contrast, progesterone increases the risk of HIV and herpes simplex virus 2 by enhancing viral replication (113); therefore, progesterone functions differently depending on the viral family. Interestingly, the majority (87%) of pregnant COVID-19-positive women at the time of delivery, when progesterone levels are high, were asymptomatic (114). We hypothesize that in COVID-19, progesterone may function in a protective manner: this hypothesis should be further investigated.
CONCLUSIONS
Rapid and efficacious repurposing of any of the established globally prescribed therapeutics that are also NPC1 inhibitors or NP-C disease mimetics has the potential to contribute to global relief from the health, human, and economic consequences of this pandemic, while the march toward an effective vaccine proceeds. We envisage that these candidates could be administered in three dosages: 1) a prophylactic dosage to provide a competitive edge for the immune system upon initial infection, 2) a mitigating dosage to permit immune response while keeping the disease at bay, and 3) a curative dosage to enable timely treatment of significantly infected patients. If variants of SARS-CoV-2 develop, or if acquired immunity proves to be temporary, prophylaxis and treatment will become more important considering the time required for vaccine development. Additionally, NP-C disease studies might uncover more loosely connected compounds that can be valuable to investigate in preparation for the future viral pandemics undoubtedly to come.
Insight and understanding of NP-C disease has enabled us to deduce a set of compounds to intervene in COVID-19 and explain the potential of these drugs on a mechanistic level. It is no surprise that many of the drugs proposed here have appeared as hits in SARS-CoV-2 antiviral drug screening dragnets. Interestingly, many of these drugs (Table 1) are cationic amphiphilic and lysosomotropic drugs that disrupt the normal functions of the LE/L and induce lipid accumulation. The detection of low serum cholesterol levels in COVID-19 patients (115) may be in agreement with our hypothesis that SARS-CoV-2 hijacks host cholesterol, but this requires further investigation of lipid profiles pre- and post-infection. Given the role of the NPC1-dependent LE/L lipid pathway in coronavirus infections, we urge that the known mechanistic information on NPC1 be utilized within the context of COVID-19 and associated candidate therapeutics. Symptoms associated with homozygosity at these loci are unlikely to arise from use of the NPC1 inhibitors and mimetics discussed here, based on the previous use of these compounds for decades and the short treatment time to overcome infection. Although the compounds are well tolerated, they are not without side effects in some individuals (Table 1) and should not be administered in the absence of a clinical trial. The range of studies described here implicate pharmacological reductions in NPC1, and thus lysosomal function, as an achievable, appropriate, and safe target for antivirals, both for the current and forthcoming pandemics.
Acknowledgments
The authors thank Paul Atkinson for comments on earlier versions of this article. During the review of this article, Ballout et al. (116) also reviewed the connections between NP-C disease and COVID-19.
Footnotes
Abbreviations:
- ACE2
- angiotensin-converting enzyme 2
- CHIKV
- Chikungunya virus
- COVID-19
- coronavirus disease 2019
- DENV
- Dengue virus
- DMV
- double membrane vesicle
- EBOV
- Ebola virus
- EBOV-GP
- Ebola virus glycoprotein
- EV
- enterovirus
- FCoV
- feline coronavirus
- HCV
- hepatitis C virus
- HIV
- human immunodeficiency virus
- IAV
- influenza A virus
- LE/L
- late endosome/lysosome
- MCS
- membrane contact site
- MERS-CoV
- Middle East respiratory syndrome-related coronavirus
- NP-C
- Niemann-Pick type C
- PM
- plasma membrane
- SARS-CoV
- severe acute respiratory syndrome coronavirus
- SSD
- sterol-sensing domain
- WNV
- West Nile virus
- ZIKV
- Zika virus
This work was supported by the Ara Parseghian Medical Research Foundation (S.L.S., A.B.M.), Dana’s Angels Research Trust and Actelion Pharmaceuticals Ltd. (S.L.S.), National Institutes of Health Grants DK54320 and GM1129465 (S.L.S.), and Wellington Medical Research Foundation and Research for Life (A.B.M.). A.B.M. was supported as a Peter Pentchev Research Fellow of the National Niemann-Pick Disease Foundation, a Senior Fellow in Biomedical Sciences of the Charles H. Revson Foundation, and a National Institutes of Health Postdoctoral Fellow in Arteriosclerosis (Grant T32 HL07343). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The authors declare that they have no conflicts of interest with the contents of this article.
REFERENCES
- 1.Dong E., Du H., and Gardner L.. 2020. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect. Dis. 20: 533–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lam T. T., Shum M. H., Zhu H. C., Tong Y. G., Ni X. B., Liao Y. S., Wei W., Cheung W. Y., Li W. J., Li L. F., et al. Identifying SARS-CoV-2 related coronaviruses in Malayan pangolins. Nature. Epub ahead of print. March 26, 2020; doi:10.1038/s41586-020-2169-0. [DOI] [PubMed] [Google Scholar]
- 3.Chan J. F., Kok K. H., Zhu Z., Chu H., To K. K., Yuan S., and Yuen K. Y.. 2020. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microbes Infect. 9: 221–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xu J., Zhao S., Teng T., Abdalla A. E., Zhu W., Xie L., Wang Y., and Guo X.. 2020. Systematic comparison of two animal-to-human transmitted human coronaviruses: SARS-CoV-2 and SARS-CoV. Viruses. 12: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., Schiergens T. S., Herrler G., Wu N. H., Nitsche A., et al. 2020. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 181: 271–280.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Inoue Y., Tanaka N., Tanaka Y., Inoue S., Morita K., Zhuang M., Hattori T., and Sugamura K.. 2007. Clathrin-dependent entry of severe acute respiratory syndrome coronavirus into target cells expressing ACE2 with the cytoplasmic tail deleted. J. Virol. 81: 8722–8729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang N., and Shen H-M.. 2020. Targeting the endocytic pathway and autophagy process as a novel therapeutic strategy in COVID-19. Int. J. Biol. Sci. 16: 1724–1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang Z. Y., Huang Y., Ganesh L., Leung K., Kong W. P., Schwartz O., Subbarao K., and Nabel G. J.. 2004. pH-dependent entry of severe acute respiratory syndrome coronavirus is mediated by the spike glycoprotein and enhanced by dendritic cell transfer through DC-SIGN. J. Virol. 78: 5642–5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reid C. R., Airo A. M., and Hobman T. C.. 2015. The virus-host interplay: biogenesis of +RNA replication complexes. Viruses. 7: 4385–4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Wilde A. H., Snijder E. J., Kikkert M., and van Hemert M. J.. 2018. Host factors in coronavirus replication. Curr. Top. Microbiol. Immunol. 419: 1–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ujike M., and Taguchi F.. 2015. Incorporation of spike and membrane glycoproteins into coronavirus virions. Viruses. 7: 1700–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Song Z., Xu Y., Bao L., Zhang L., Yu P., Qu Y., Zhu H., Zhao W., Han Y., and Qin C.. 2019. From SARS to MERS, thrusting coronaviruses into the spotlight. Viruses. 11: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lim Y. X., Ng Y. L., Tam J. P., and Liu D. X.. 2016. Human coronaviruses: a review of virus-host interactions. Diseases. 4: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen Y., Liu Q., and Guo D.. 2020. Emerging coronaviruses: genome structure, replication, and pathogenesis. J. Med. Virol. 92: 418–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fung T. S., and Liu D. X.. 2014. Coronavirus infection, ER stress, apoptosis and innate immunity. Front. Microbiol. 5: 296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vanier M. T. 2010. Niemann-Pick disease type C. Orphanet J. Rare Dis. 5: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hammond N., Munkacsi A. B., and Sturley S. L.. 2019. The complexity of a monogenic neurodegenerative disease: More than two decades of therapeutic driven research into Niemann-Pick type C disease. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 1864: 1109–1123. [DOI] [PubMed] [Google Scholar]
- 18.Infante R. E., Wang M. L., Radhakrishnan A., Kwon H. J., Brown M. S., and Goldstein J. L.. 2008. NPC2 facilitates bidirectional transfer of cholesterol between NPC1 and lipid bilayers, a step in cholesterol egress from lysosomes. Proc. Natl. Acad. Sci. USA. 105: 15287–15292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kwon H. J., Abi-Mosleh L., Wang M. L., Deisenhofer J., Goldstein J. L., Brown M. S., and Infante R. E.. 2009. Structure of N-terminal domain of NPC1 reveals distinct subdomains for binding and transfer of cholesterol. Cell. 137: 1213–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walkley S. U., and Vanier M. T.. 2009. Secondary lipid accumulation in lysosomal disease. Biochim. Biophys. Acta. 1793: 726–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu E. A., and Lieberman A. P.. 2019. The intersection of lysosomal and endoplasmic reticulum calcium with autophagy defects in lysosomal diseases. Neurosci. Lett. 697: 10–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Elrick M. J., Yu T., Chung C., and Lieberman A. P.. 2012. Impaired proteolysis underlies autophagic dysfunction in Niemann-Pick type C disease. Hum. Mol. Genet. 21: 4876–4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rajakumar T., Munkacsi A. B., and Sturley S. L.. 2017. Exacerbating and reversing lysosomal storage diseases: from yeast to humans. Microb. Cell. 4: 278–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Herbert A. S., Davidson C., Kuehne A. I., Bakken R., Braigen S. Z., Gunn K. E., Whelan S. P., Brummelkamp T. R., Twenhafel N. A., Chandran K., et al. 2015. Niemann-Pick C1 is essential for ebolavirus replication and pathogenesis in vivo. MBio. 6: e00565-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tang Y., Leao I. C., Coleman E. M., Broughton R. S., and Hildreth J. E.. 2009. Deficiency of Niemann-Pick type C-1 protein impairs release of human immunodeficiency virus type 1 and results in Gag accumulation in late endosomal/lysosomal compartments. J. Virol. 83: 7982–7995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wichit S., Hamel R., Bernard E., Talignani L., Diop F., Ferraris P., Liegeois F., Ekchariyawat P., Luplertlop N., Surasombatpattana P., et al. 2017. Imipramine inhibits Chikungunya virus replication in human skin fibroblasts through interference with intracellular cholesterol trafficking. Sci. Rep. 7: 3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carette J. E., Raaben M., Wong A. C., Herbert A. S., Obernosterer G., Mulherkar N., Kuehne A. I., Kranzusch P. J., Griffin A. M., Ruthel G., et al. 2011. Ebola virus entry requires the cholesterol transporter Niemann-Pick C1. Nature. 477: 340–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Côte M., Misasi J., Ren T., Bruchez A., Lee K., Filone C. M., Hensley L., Li Q., Ory D., Chandran K., et al. 2011. Small molecule inhibitors reveal Niemann-Pick C1 is essential for Ebola virus infection. Nature. 477: 344–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang H., Shi Y., Song J., Qi J., Lu G., Yan J., and Gao G. F.. 2016. Ebola viral glycoprotein bound to its endosomal receptor Niemann-Pick C1. Cell. 164: 258–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gong X., Qian H., Zhou X., Wu J., Wan T., Cao P., Huang W., Zhao X., Wang X., Wang P., et al. 2016. Structural insights into the Niemann-Pick C1 (NPC1)-mediated cholesterol transfer and Ebola infection. Cell. 165: 1467–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kondoh T., Letko M., Munster V. J., Manzoor R., Maruyama J., Furuyama W., Miyamoto H., Shigeno A., Fujikura D., Takadate Y., et al. 2018. Single-nucleotide polymorphisms in human NPC1 influence filovirus entry into cells. J. Infect. Dis. 218: S397–S402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Coleman E. M., Walker T. N., and Hildreth J. E.. 2012. Loss of Niemann-Pick type C proteins 1 and 2 greatly enhances HIV infectivity and is associated with accumulation of HIV Gag and cholesterol in late endosomes/lysosomes. Virol. J. 9: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Glende J., Schwegmann-Wessels C., Al-Falah M., Pfefferle S., Qu X., Deng H., Drosten C., Naim H. Y., and Herrler G.. 2008. Importance of cholesterol-rich membrane microdomains in the interaction of the S protein of SARS-coronavirus with the cellular receptor angiotensin-converting enzyme 2. Virology. 381: 215–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wojtanik K. M., and Liscum L.. 2003. The transport of low density lipoprotein-derived cholesterol to the plasma membrane is defective in NPC1 cells. J. Biol. Chem. 278: 14850–14856. [DOI] [PubMed] [Google Scholar]
- 35.Lusa S., Blom T. S., Eskelinen E. L., Kuismanen E., Mansson J. E., Simons K., and Ikonen E.. 2001. Depletion of rafts in late endocytic membranes is controlled by NPC1-dependent recycling of cholesterol to the plasma membrane. J. Cell Sci. 114: 1893–1900. [DOI] [PubMed] [Google Scholar]
- 36.Underwood K. W., Jacobs N. L., Howley A., and Liscum L.. 1998. Evidence for a cholesterol transport pathway from lysosomes to endoplasmic reticulum that is independent of the plasma membrane. J. Biol. Chem. 273: 4266–4274. [DOI] [PubMed] [Google Scholar]
- 37.Takano T., Satomi Y., Oyama Y., Doki T., and Hohdatsu T.. 2016. Differential effect of cholesterol on type I and II feline coronavirus infection. Arch. Virol. 161: 125–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Elrick M. J., and Lieberman A. P.. 2013. Autophagic dysfunction in a lysosomal storage disorder due to impaired proteolysis. Autophagy. 9: 234–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cermak S., Kosicek M., Mladenovic-Djordjevic A., Smiljanic K., Kanazir S., and Hecimovic S.. 2016. Loss of cathepsin B and L leads to lysosomal dysfunction, NPC-like cholesterol sequestration and accumulation of the key Alzheimer’s proteins. PLoS One. 11: e0167428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Angelini M. M., Akhlaghpour M., Neuman B. W., and Buchmeier M. J.. 2013. Severe acute respiratory syndrome coronavirus nonstructural proteins 3, 4, and 6 induce double-membrane vesicles. MBio. 4: e00524-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Z., He G., Filipowicz N. A., Randall G., Belov G. A., Kopek B. G., and Wang X.. 2019. Host lipids in positive-strand RNA virus genome replication. Front. Microbiol. 10: 286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Höglinger D., Burgoyne T., Sanchez-Heras E., Hartwig P., Colaco A., Newton J., Futter C. E., Spiegel S., Platt F. M., and Eden E. R.. 2019. NPC1 regulates ER contacts with endocytic organelles to mediate cholesterol egress. Nat. Commun. 10: 4276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stoeck I. K., Lee J. Y., Tabata K., Romero-Brey I., Paul D., Schult P., Lohmann V., Kaderali L., and Bartenschlager R.. 2017. Hepatitis C virus replication depends on endosomal cholesterol homeostasis. J. Virol. 92: e01196-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bukrinsky M. I., Mukhamedova N., and Sviridov D.. 2020. Lipid rafts and pathogens: the art of deception and exploitation. J. Lipid Res. 61: 601–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cenedella R. J. 2009. Cholesterol synthesis inhibitor U18666A and the role of sterol metabolism and trafficking in numerous pathophysiological processes. Lipids. 44: 477–487. [DOI] [PubMed] [Google Scholar]
- 46.Lu F., Liang Q., Abi-Mosleh L., Das A., De Brabander J. K., Goldstein J. L., and Brown M. S.. 2015. Identification of NPC1 as the target of U18666A, an inhibitor of lysosomal cholesterol export and Ebola infection. eLife. 4: e12177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Eckert N., Wrensch F., Gartner S., Palanisamy N., Goedecke U., Jager N., Pohlmann S., and Winkler M.. 2014. Influenza A virus encoding secreted Gaussia luciferase as useful tool to analyze viral replication and its inhibition by antiviral compounds and cellular proteins. PLoS One. 9: e97695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Elgner F., Ren H., Medvedev R., Ploen D., Himmelsbach K., Boller K., and Hildt E.. 2016. The intracellular cholesterol transport inhibitor U18666A inhibits the exosome-dependent release of mature Hepatitis C virus. J. Virol. 90: 11181–11196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Takano T., Endoh M., Fukatsu H., Sakurada H., Doki T., and Hohdatsu T.. 2017. The cholesterol transport inhibitor U18666A inhibits type I feline coronavirus infection. Antiviral Res. 145: 96–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wrensch F., Winkler M., and Pohlmann S.. 2014. IFITM proteins inhibit entry driven by the MERS-coronavirus spike protein: evidence for cholesterol-independent mechanisms. Viruses. 6: 3683–3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pipalia N. H., Cosner C. C., Huang A., Chatterjee A., Bourbon P., Farley N., Helquist P., Wiest O., and Maxfield F. R.. 2011. Histone deacetylase inhibitor treatment dramatically reduces cholesterol accumulation in Niemann-Pick type C1 mutant human fibroblasts. Proc. Natl. Acad. Sci. USA. 108: 5620–5625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Munkacsi A. B., Chen F. W., Brinkman M. A., Higaki K., Gutierrez G. D., Chaudhari J., Layer J. V., Tong A., Bard M., Boone C., et al. 2011. An “exacerbate-reverse” strategy in yeast identifies histone deacetylase inhibition as a correction for cholesterol and sphingolipid transport defects in human Niemann-Pick type C disease. J. Biol. Chem. 286: 23842–23851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Doki T., Tarusawa T., Hohdatsu T., and Takano T.. 2020. In vivo antiviral effects of U18666A against type I feline infectious peritonitis virus. Pathogens. 9: 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shoemaker C. J., Schornberg K. L., Delos S. E., Scully C., Pajouhesh H., Olinger G. G., Johansen L. M., and White J. M.. 2013. Multiple cationic amphiphiles induce a Niemann-Pick C phenotype and inhibit Ebola virus entry and infection. PLoS One. 8: e56265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lafourcade C., Sobo K., Kieffer-Jaquinod S., Garin J., and van der Goot F. G.. 2008. Regulation of the V-ATPase along the endocytic pathway occurs through reversible subunit association and membrane localization. PLoS One. 3: e2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Johansen L. M., Brannan J. M., Delos S. E., Shoemaker C. J., Stossel A., Lear C., Hoffstrom B. G., Dewald L. E., Schornberg K. L., Scully C., et al. 2013. FDA-approved selective estrogen receptor modulators inhibit Ebola virus infection. Sci. Transl. Med. 5: 190ra79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Head S. A., Shi W. Q., Yang E. J., Nacev B. A., Hong S. Y., Pasunooti K. K., Li R. J., Shim J. S., and Liu J. O.. 2017. Simultaneous targeting of NPC1 and VDAC1 by itraconazole leads to synergistic inhibition of mTOR signaling and angiogenesis. ACS Chem. Biol. 12: 174–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lyu J., Yang E. J., Head S. A., Ai N., Zhang B., Wu C., Li R. J., Liu Y., Yang C., Dang Y., et al. 2017. Pharmacological blockade of cholesterol trafficking by cepharanthine in endothelial cells suppresses angiogenesis and tumor growth. Cancer Lett. 409: 91–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Trinh M. N., Lu F., Li X., Das A., Liang Q., De Brabander J. K., Brown M. S., and Goldstein J. L.. 2017. Triazoles inhibit cholesterol export from lysosomes by binding to NPC1. Proc. Natl. Acad. Sci. USA. 114: 89–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Underwood K. W., Andemariam B., McWilliams G. L., and Liscum L.. 1996. Quantitative analysis of hydrophobic amine inhibition of intracellular cholesterol transport. J. Lipid Res. 37: 1556–1568. [PubMed] [Google Scholar]
- 61.Zhang X., Chen W., Li P., Calvo R., Southall N., Hu X., Bryant-Genevier M., Feng X., Geng Q., Gao C., et al. 2019. Agonist-specific voltage-dependent gating of lysosomal two-pore Na(+) channels. eLife. 8: e51423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ou X., Liu Y., Lei X., Li P., Mi D., Ren L., Guo L., Guo R., Chen T., Hu J., et al. 2020. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 11: 1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Weber C., and Opatz T.. 2019. Chapter one - bisbenzylisoquinoline alkaloids. In The Alkaloids: Chemistry and Biology. H-J. Knölker, editor. Academic Press, Amsterdam. 1–114. [DOI] [PubMed] [Google Scholar]
- 64.Fan H. H., Wang L. Q., Liu W. L., An X. P., Liu Z. D., He X. Q., Song L. H., and Tong Y. G.. 2020. Repurposing of clinically approved drugs for treatment of coronavirus disease 2019 in a 2019-novel coronavirus (2019-nCoV) related coronavirus model. Chin. Med. J. (Engl.). 133: 1051–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Matsuda K., Hattori S., Komizu Y., Kariya R., Ueoka R., and Okada S.. 2014. Cepharanthine inhibited HIV-1 cell-cell transmission and cell-free infection via modification of cell membrane fluidity. Bioorg. Med. Chem. Lett. 24: 2115–2117. [DOI] [PubMed] [Google Scholar]
- 66.Jeon S., Ko M., Lee J., Choi I., Byun S. Y., Park S., Shum D., and Kim S.. Identification of antiviral drug candidates against SARS-CoV-2 from FDA-approved drugs. Antimicrob. Agents Chemother. Epub ahead of print. May 4, 2020; doi:10.1128/AAC.00819-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schloer S., Goretzko J., Kuhnl A., Brunotte L., Ludwig S., and Rescher U.. 2019. The clinically licensed antifungal drug itraconazole inhibits influenza virus in vitro and in vivo. Emerg. Microbes Infect. 8: 80–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Takano T., Akiyama M., Doki T., and Hohdatsu T.. 2019. Antiviral activity of itraconazole against type I feline coronavirus infection. Vet. Res. 50: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Takano T., Wakayama Y., and Doki T.. 2019. Endocytic pathway of feline coronavirus for cell entry: Differences in serotype-dependent viral entry pathway. Pathogens. 8: 300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Meutiawati F., Bezemer B., Strating J., Overheul G. J., Zusinaite E., van Kuppeveld F. J. M., van Cleef K. W. R., and van Rij R. P.. 2018. Posaconazole inhibits dengue virus replication by targeting oxysterol-binding protein. Antiviral Res. 157: 68–79. [DOI] [PubMed] [Google Scholar]
- 71.Rhoden E., Nix W. A., Weldon W. C., and Selvarangan R.. 2018. Antifungal azoles itraconazole and posaconazole exhibit potent in vitro antiviral activity against clinical isolates of parechovirus A3 (Picornaviridae). Antiviral Res. 149: 75–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Strating J. R., van der Linden L., Albulescu L., Bigay J., Arita M., Delang L., Leyssen P., van der Schaar H. M., Lanke K. H., Thibaut H. J., et al. 2015. Itraconazole inhibits enterovirus replication by targeting the oxysterol-binding protein. Cell Rep. 10: 600–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wu C., Liu Y., Yang Y., Zhang P., Zhong W., Wang Y., Wang Q., Xu Y., Li M., Li X., et al. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm. Sin. B. Epub ahead of print. February 27, 2020; doi:10.1016/j.apsb.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Porpaczy Z., Tomasek J. J., and Freeman D. A.. 1997. Internalized plasma membrane cholesterol passes through an endosome compartment that is distinct from the acid vesicle-lysosome compartment. Exp. Cell Res. 234: 217–224. [DOI] [PubMed] [Google Scholar]
- 75.Nujić K., Banjanac M., Munic V., Polancec D., and Erakovic Haber V.. 2012. Impairment of lysosomal functions by azithromycin and chloroquine contributes to anti-inflammatory phenotype. Cell. Immunol. 279: 78–86. [DOI] [PubMed] [Google Scholar]
- 76.Wehrmann Z. T., Hulett T. W., Huegel K. L., Vaughan K. T., Wiest O., Helquist P., and Goodson H.. 2012. Quantitative comparison of the efficacy of various compounds in lowering intracellular cholesterol levels in Niemann-Pick type C fibroblasts. PLoS One. 7: e48561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Piccoli E., Nadai M., Caretta C. M., Bergonzini V., Del Vecchio C., Ha H. R., Bigler L., Dal Zoppo D., Faggin E., Pettenazzo A., et al. 2011. Amiodarone impairs trafficking through late endosomes inducing a Niemann-Pick C-like phenotype. Biochem. Pharmacol. 82: 1234–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Canfrán-Duque A., Barrio L. C., Lerma M., de la Pena G., Serna J., Pastor O., Lasuncion M. A., and Busto R.. 2016. First-generation antipsychotic haloperidol alters the functionality of the late endosomal/lysosomal compartment in vitro. Int. J. Mol. Sci. 17: 404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Salata C., Calistri A., Parolin C., Baritussio A., and Palu G.. 2017. Antiviral activity of cationic amphiphilic drugs. Expert Rev. Anti Infect. Ther. 15: 483–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.D’Alessandro S., Scaccabarozzi D., Signorini L., Perego F., Ilboudo D. P., Ferrante P., and Delbue S.. 2020. The use of antimalarial drugs against viral infection. Microorganisms. 8: 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Vincent M. J., Bergeron E., Benjannet S., Erickson B. R., Rollin P. E., Ksiazek T. G., Seidah N. G., and Nichol S. T.. 2005. Chloroquine is a potent inhibitor of SARS coronavirus infection and spread. Virol. J. 2: 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Liu J., Cao R., Xu M., Wang X., Zhang H., Hu H., Li Y., Hu Z., Zhong W., and Wang M.. 2020. Hydroxychloroquine, a less toxic derivative of chloroquine, is effective in inhibiting SARS-CoV-2 infection in vitro. Cell Discov. 6: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M., Shi Z., Hu Z., Zhong W., and Xiao G.. 2020. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 30: 269–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.de Wilde A. H., Jochmans D., Posthuma C. C., Zevenhoven-Dobbe J. C., van Nieuwkoop S., Bestebroer T. M., van den Hoogen B. G., Neyts J., and Snijder E. J.. 2014. Screening of an FDA-approved compound library identifies four small-molecule inhibitors of Middle East respiratory syndrome coronavirus replication in cell culture. Antimicrob. Agents Chemother. 58: 4875–4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Barnard D. L., Day C. W., Bailey K., Heiner M., Montgomery R., Lauridsen L., Chan P. K., and Sidwell R. W.. 2006. Evaluation of immunomodulators, interferons and known in vitro SARS-CoV inhibitors for inhibition of SARS-CoV replication in BALB/c mice. Antivir. Chem. Chemother. 17: 275–284. [DOI] [PubMed] [Google Scholar]
- 86.Weston S., Coleman C. M., Haupt R., Logue J., Matthews K., and Frieman M. B.. 2020. Broad anti-coronaviral activity of FDA approved drugs against SARS-CoV-2 in vitro and SARS-CoV in vivo. bioRxiv 10.1101/2020.03.25.008482. [DOI] [PMC free article] [PubMed]
- 87.Takano T., Katoh Y., Doki T., and Hohdatsu T.. 2013. Effect of chloroquine on feline infectious peritonitis virus infection in vitro and in vivo. Antiviral Res. 99: 100–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Liu C., Zhou Q., Li Y., Garner L. V., Watkins S. P., Carter L. J., Smoot J., Gregg A. C., Daniels A. D., Jervey S., et al. 2020. Research and development on therapeutic agents and vaccines for COVID-19 and related human coronavirus diseases. ACS Cent. Sci. 6: 315–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Geleris J., Sun Y., Platt J., Zucker J., Baldwin M., Hripcsak G., Labella A., Manson D., Kubin C., Barr R. G., et al. Observational study of hydroxychloroquine in hospitalized patients with COVID-19. N. Engl. J. Med. Epub ahead of print. May 7, 2020; doi:10.1056/NEJMoa2012410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Iannetta M., Ippolito G., and Nicastri E.. 2017. Azithromycin shows anti-Zika virus activity in human glial cells. Antimicrob. Agents Chemother. 61: e01152-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gielen V., Johnston S. L., and Edwards M. R.. 2010. Azithromycin induces anti-viral responses in bronchial epithelial cells. Eur. Respir. J. 36: 646–654. [DOI] [PubMed] [Google Scholar]
- 92.Meyerowitz E. A., Vannier A. G. L., Friesen M. G. N., Schoenfeld S., Gelfand J. A., Callahan M. V., Kim A. Y., Reeves P. M., and Poznansky M. C.. 2020. Rethinking the role of hydroxychloroquine in the treatment of COVID-19. FASEB J. 34: 6027–6037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Krizanová O., Ciampor F., and Veber P.. 1982. Influence of chlorpromazine on the replication of influenza virus in chick embryo cells. Acta Virol. 26: 209–216. [PubMed] [Google Scholar]
- 94.Chu J. J., and Ng M. L.. 2004. Infectious entry of West Nile virus occurs through a clathrin-mediated endocytic pathway. J. Virol. 78: 10543–10555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Joki-Korpela P., Marjomaki V., Krogerus C., Heino J., and Hyypia T.. 2001. Entry of human parechovirus 1. J. Virol. 75: 1958–1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Nawa M., Takasaki T., Yamada K. I., Kurane I., and Akatsuka T.. 2003. Interference in Japanese encephalitis virus infection of Vero cells by a cationic amphiphilic drug, chlorpromazine. J. Gen. Virol. 84: 1737–1741. [DOI] [PubMed] [Google Scholar]
- 97.Pho M. T., Ashok A., and Atwood W. J.. 2000. JC virus enters human glial cells by clathrin-dependent receptor-mediated endocytosis. J. Virol. 74: 2288–2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Dyall J., Coleman C. M., Hart B. J., Venkataraman T., Holbrook M. R., Kindrachuk J., Johnson R. F., Olinger G. G. Jr., Jahrling P. B., Laidlaw M., et al. 2014. Repurposing of clinically developed drugs for treatment of Middle East respiratory syndrome coronavirus infection. Antimicrob. Agents Chemother. 58: 4885–4893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gordon D. E., Jang G. M., Bouhaddou M., Xu J., Obernier K., O’Meara M. J., Guo J. Z., Swaney D. L., Tummino T. A., Hüttenhain R., et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. Epub ahead of print. April 30, 2020; doi:10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Stadler K., Ha H. R., Ciminale V., Spirli C., Saletti G., Schiavon M., Bruttomesso D., Bigler L., Follath F., Pettenazzo A., et al. 2008. Amiodarone alters late endosomes and inhibits SARS coronavirus infection at a post-endosomal level. Am. J. Respir. Cell Mol. Biol. 39: 142–149. [DOI] [PubMed] [Google Scholar]
- 101.Gehring G., Rohrmann K., Atenchong N., Mittler E., Becker S., Dahlmann F., Pohlmann S., Vondran F. W., David S., Manns M. P., et al. 2014. The clinically approved drugs amiodarone, dronedarone and verapamil inhibit filovirus cell entry. J. Antimicrob. Chemother. 69: 2123–2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Cheng Y. L., Lan K. H., Lee W. P., Tseng S. H., Hung L. R., Lin H. C., Lee F. Y., Lee S. D., and Lan K. H.. 2013. Amiodarone inhibits the entry and assembly steps of hepatitis C virus life cycle. Clin. Sci. (Lond.). 125: 439–448. [DOI] [PubMed] [Google Scholar]
- 103.Allison A. C. 1964. Polymoprhism and natural selection in human populations. Cold Spring Harb. Symp. Quant. Biol. 29: 137–149. [DOI] [PubMed] [Google Scholar]
- 104.Zhang J. R., Coleman T., Langmade S. J., Scherrer D. E., Lane L., Lanier M. H., Feng C., Sands M. S., Schaffer J. E., Semenkovich C. F., et al. 2008. Niemann-Pick C1 protects against atherosclerosis in mice via regulation of macrophage intracellular cholesterol trafficking. J. Clin. Invest. 118: 2281–2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kruth H. S., Comly M. E., Butler J. D., Vanier M. T., Fink J. K., Wenger D. A., Patel S., and Pentchev P. G.. 1986. Type C Niemann-Pick disease. Abnormal metabolism of low density lipoprotein in homozygous and heterozygous fibroblasts. J. Biol. Chem. 261: 16769–16774. [PubMed] [Google Scholar]
- 106.Geberhiwot T., Moro A., Dardis A., Ramaswami U., Sirrs S., Marfa M. P., Vanier M. T., Walterfang M., Bolton S., Dawson C., et al. 2018. Consensus clinical management guidelines for Niemann-Pick disease type C. Orphanet J. Rare Dis. 13: 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Karlberg J., Chong D. S., and Lai W. Y.. 2004. Do men have a higher case fatality rate of severe acute respiratory syndrome than women do? Am. J. Epidemiol. 159: 229–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Grasselli G., Zangrillo A., Zanella A., Antonelli M., Cabrini L., Castelli A., Cereda D., Coluccello A., Foti G., Fumagalli R., et al. Baseline characteristics and outcomes of 1591 patients infected with SARS-CoV-2 admitted to ICUs of the Lombardy Region, Italy. JAMA. Epub ahead of print. April 6, 2020; doi:10.1001/jama.2020.5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Garg S., Kim L., Whitaker M., O’Halloran A., Cummings C., Holstein R., Prill M., Chai S. J., Kirley P. D., Alden N. B., et al. 2020. Hospitalization rates and characteristics of patients hospitalized with laboratory-confirmed coronavirus disease 2019 - COVID-NET, 14 States, March 1–30, 2020. MMWR Morb. Mortal. Wkly. Rep. 69: 458–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Jansen A., Chiew M., Konings F., Lee C. K., and Ailan L.. 2015. Sex matters - a preliminary analysis of Middle East respiratory syndrome in the Republic of Korea, 2015. Western Pac. Surveill. Response J. 6: 68–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Butler J. D., Blanchette-Mackie J., Goldin E., O’Neill R. R., Carstea G., Roff C. F., Patterson M. C., Patel S., Comly M. E., Cooney A., et al. 1992. Progesterone blocks cholesterol translocation from lysosomes. J. Biol. Chem. 267: 23797–23805. [PubMed] [Google Scholar]
- 112.Hall O. J., Limjunyawong N., Vermillion M. S., Robinson D. P., Wohlgemuth N., Pekosz A., Mitzner W., and Klein S. L.. 2016. Progesterone-based therapy protects against influenza by promoting lung repair and recovery in females. PLoS Pathog. 12: e1005840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ragupathy V., Xue W., Tan J., Devadas K., Gao Y., and Hewlett I.. 2016. Progesterone augments cell susceptibility to HIV-1 and HIV-1/HSV-2 co-infections. J. Mol. Endocrinol. 57: 185–199. [DOI] [PubMed] [Google Scholar]
- 114.Sutton D., Fuchs K., D’Alton M., and Goffman D.. Universal screening for SARS-CoV-2 in women admitted for delivery. N. Engl. J. Med. Epub ahead of print. May 28, 2020; doi:10.1056/NEJMc2009316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hu X., Chen D., Wu L., He G., and Ye W.. 2020. Low serum cholesterol level among patients with COVID-19 infection in Wenzhou, China. Lancet Preprints 10.2139/ssrn.3544826. [Google Scholar]
- 116.Ballout R. A., Sviridov D., Bukrinsky M. I., and Remaley A. T.. The lysosome: a potential juncture between SARS-CoV-2 infectivity and Niemann-Pick disease type C, with therapeutic implications. FASEB J. Epub ahead of print. May 5, 2020; doi:10.1096/fj.202000654R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Tang Z. H., Cao W. X., Guo X., Dai X. Y., Lu J. H., Chen X., Zhu H., and Lu J. J.. 2018. Identification of a novel autophagic inhibitor cepharanthine to enhance the anti-cancer property of dacomitinib in non-small cell lung cancer. Cancer Lett. 412: 1–9. [DOI] [PubMed] [Google Scholar]
- 118.Mauthe M., Orhon I., Rocchi C., Zhou X., Luhr M., Hijlkema K. J., Coppes R. P., Engedal N., Mari M., and Reggiori F.. 2018. Chloroquine inhibits autophagic flux by decreasing autophagosome-lysosome fusion. Autophagy. 14: 1435–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]