Abstract
The yeast protein Mpo1 belongs to a protein family that is widely conserved in bacteria, fungi, protozoa, and plants, and is the only protein of this family whose function has so far been elucidated. Mpo1 is an Fe2+-dependent dioxygenase that catalyzes the α-oxidation reaction of 2-hydroxy (2-OH) long-chain FAs (LCFAs) produced in the degradation pathway of the long-chain base phytosphingosine. However, several biochemical characteristics of Mpo1, such as its catalytic residues, membrane topology, and substrate specificity, remain unclear. Here, we report that yeast Mpo1 contains two transmembrane domains and that both its N- and C-terminal regions are exposed to the cytosol. Mutational analyses revealed that three histidine residues conserved in the Mpo1 family are especially important for Mpo1 activity, suggesting that they may be responsible for the formation of coordinate bonds with Fe2+. We found that, in addition to activity toward 2-OH LCFAs, Mpo1 also exhibits activity toward 2-OH very-long-chain FAs derived from the FA moiety of sphingolipids. These results indicate that Mpo1 is involved in the metabolism of long-chain to very-long-chain 2-OH FAs produced in different pathways. We noted that the growth of mpo1Δ cells is delayed upon carbon deprivation, suggesting that the Mpo1-mediated conversion of 2-OH FAs to nonhydroxy FAs is important for utilizing 2-OH FAs as a carbon source under carbon starvation. Our findings help to elucidate the as yet unknown functions and activities of other Mpo1 family members.
Keywords: ceramides, fatty acid/oxidation, 2-hydroxy fatty acid, iron, lipids, α-oxidation, sphingolipids
FAs are major components of lipids. Most of the FAs in living organisms are nonhydroxylated. However, 2-hydroxy (2-OH) FAs also exist in a wide range of organisms from bacteria to fungi, plants, and animals, although generally in low quantities (1–5). Eukaryotic membranes are composed of glycerophospholipids, sphingolipids, and sterols. Of these, 2-OH FAs are specifically present in sphingolipids. Most sphingolipids have a hydrophobic backbone ceramide consisting of a FA and a long-chain base (LCB). In mammals, sphingolipids containing 2-OH FAs are present in specific tissues, such as brain, intestine, kidney, and reproductive tissues (3). In the brain, 2-OH FAs are abundant in the form of myelin sphingolipids (such as galactosylceramide and sulfatide), which are important for the maintenance of myelin (6–10). In plants, sphingolipids containing 2-OH FAs play an important role in the defense against infection by pathogens (11).
The main pathway of 2-OH FA production in mammals is a direct oxidation of the C2 position of FAs by FA 2-hydroxylase, which is encoded by FA2H (3, 12). Mutations in FA2H cause hereditary spastic paraplegia type 35 (SPG35), which is accompanied by myelin degeneration (6, 8, 10). Fa2h knockout mice show late-onset myelin and axon degeneration and deficits in spatial learning and memory (7, 9). Sphingolipids containing 2-OH FAs with carbon-chain lengths of C18–C24 are absent in the brains of these mice (7). FAs are classified into long-chain FAs (LCFAs; C11–C20) and very-long-chain FAs (VLCFAs; ≥C21) according to chain length (13, 14). The FA in yeast sphingolipids is predominantly a C26:0 VLCFA, most of which is 2-hydroxylated (1, 15, 16). In yeast, Scs7 (a homolog of FA2H) is responsible for 2-hydroxylation of the VLCFA moiety of ceramide (1).
In addition to the direct oxidation of FAs, 2-OH FA is also generated in the degradation pathway of the LCB phytosphingosine (PHS) (17, 18). PHS is present in the epidermis, small intestine, and kidney in mammals (19–21) and is a major LCB in yeast (15, 16). LCBs commonly have hydroxyl groups at C1 and C3 and an amino group at C2. The simplest LCB is dihydrosphingosine. The most abundant LCB in mammals is sphingosine, which has a trans double bond between C4 and C5. PHS has a hydroxyl group at C4. LCBs undergo three common reactions in their degradation pathway: phosphorylation at C1, cleavage between the C2 and C3 positions, and oxidation (5, 17, 18, 22, 23). Through these reactions LCBs are converted to two-carbon shortened FAs, and PHS is converted to 2-OH FAs (17, 18). Because 2-OH FAs cannot be used for glycerolipid synthesis or β-oxidation, they need to be converted to nonhydroxy FAs. This conversion is achieved by α-oxidation, where 2-OH FAs are metabolized to nonhydroxy FAs with one less carbon (making them odd-chain nonhydroxy FAs) (17, 18). The α-oxidation reactions differ between mammals and yeast. Mammalian α-oxidation involves three steps (CoA addition, long-chain aldehyde production by cleavage between the C1 and C2 positions, and oxidation to FAs), whereas yeast α-oxidation is conducted in one step (17, 18, 24). Accordingly, the proteins involved in α-oxidation differ between mammals and yeast. We previously identified MPO1 as the gene functioning in the degradation pathway of PHS in yeast (17). It subsequently became clear that Mpo1 is involved in the α-oxidation of 2-OH FAs as an Fe2+-dependent dioxygenase (EC 1.14.18.12), which directly converts 2-OH LCFAs to nonhydroxy FAs with one less carbon (24).
Mpo1 homologs are present in bacteria, fungi, protozoa, and plants, but not in animals. Of the members of Mpo1 family [previously known as domain of unknown function number 962 (DUF962)], Mpo1 is the only protein that has been analyzed. However, there remain many unsolved problems regarding the structural and enzymatic characterization of Mpo1. For instance, it is an ER protein that is predicted to have some transmembrane domains, but the exact number of these domains and its membrane topology are unknown; the Mpo1-catalyzed α-oxidation reaction requires Fe2+, but the region and amino acid residues involved in Fe2+ binding are unclear; and Mpo1 exhibits activity toward 2-OH LCFAs derived from PHS (24), but it is unclear whether Mpo1 is involved in α-oxidation of 2-OH VLCFAs derived from the FA portion of sphingolipids. Although the 2-OH C26 VLCFA is abundant in yeast sphingolipids, its degradation process is unclear, including whether it is subject to α-oxidation by Mpo1. Furthermore, the physiological significance of 2-OH FA degradation is also unknown. In the present study, we revealed the following unknown aspects of Mpo1: membrane topology, the residues important for its activity, its substrate specificity, and its role in the carbon starvation response. Our findings give clues to the elucidation of the functions and activities not only of Mpo1, but also of other members of the Mpo1 family.
MATERIALS AND METHODS
Yeast strains and media
The budding yeast Saccharomyces cerevisiae was used in this study. The strains we used were BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) and its derivatives 4378 (mpo1Δ::KanMX4), 261 (pxp1Δ::KanMX), and KSKY4 (BY4741, pxp1Δ::KanMX mpo1Δ::NatNT2) (25, 26). KSKY4 cells were constructed by introducing mpo1Δ::NatNT2 into strain 261. Cells without plasmid were grown in synthetic complete (SC) medium (2% d-glucose, 0.5% casamino acids, and 0.67% yeast nitrogen base without amino acids) supplemented with 20 mg/l adenine, 20 mg/l uracil, and 20 mg/l tryptophan. Cells harboring the pAKNF316 (URA3 marker) plasmid or its derivatives were grown in the above medium without uracil (SC medium supplemented with 20 mg/l adenine and 20 mg/l tryptophan). Cells bearing the plasmid encoding MPO1 fused with the enhanced GFP (EGFP) gene, under control of the MET25 promoter, were induced by removing methionine from the medium. For this purpose, medium lacking uracil and methionine (2% d-glucose, amino acid supplements without methionine, 20 mg/l adenine, and 0.67% yeast nitrogen base) was used. The following media were used to examine growth under carbon or nitrogen starvation conditions. SC medium, SC medium without a carbon source (SC−C; SC medium without glucose), and synthetic dextrose medium without a nitrogen source (SD−N; 2% d-glucose and 0.17% yeast nitrogen base without amino acids or ammonium sulfate). All yeasts were cultured at 30°C with rotation.
Plasmids
The pAKNF316 vector (CEN) is designed to express a protein tagged with 3×FLAG at the N terminus under the control of the glycerol-3-phosphate dehydrogenase gene (TDH3) promoter (27). The pNK46 plasmid encoding 3×FLAG-MPO1 is a derivative of the pAKNF316 vector (18). The pMT12 (3×FLAG-MPO1 Y15A), pMT13 (3×FLAG-MPO1 H19A), pMT14 (3×FLAG-MPO1 H28A), pMT15 (3×FLAG-MPO1 H115A), pMT16 (3×FLAG-MPO1 E119A), pMT17 (3×FLAG-MPO1 P123A), pKSK32 (3×FLAG-MPO1 Y15H), pKSK33 (3×FLAG-MPO1 H19Y), pKSK34 (3×FLAG-MPO1 H28Y), pKSK35 (3×FLAG-MPO1 H115Y), and pKSK36 (3×FLAG-MPO1 E119D) plasmids were constructed using a QuikChange site-directed mutagenesis kit (Agilent Technologies, Santa Clara, CA) with the pNK46 plasmid and appropriate primers. The pMT66 (N terminus), pMT67 (Lys47), pMT72 (Leu69), pMT68 (Val93), pMT69 (Arg121), and pMT70 (C terminus) plasmids encode 3×FLAG-MPO1 fused with N-glycosylation reporter at the indicated positions. These are derivatives of the pNK46 plasmid. For their construction, the BamHI site was first introduced into each position using a QuikChange site-directed mutagenesis kit. The resulting plasmids were digested with BamHI and ligated with the N-glycosylation reporter fragment, which had been prepared by digesting the pMT50 plasmid encoding amino acid residues 40–221 of the invertase Suc2, using BamHI.
The pUG36 vector (URA3 marker; CEN; GenBank accession number, AF298791.1; a gift from Dr. J. H. Hegemann, University of Duesseldorf) is designed to express a protein fused with EGFP at the N terminus, under control of the MET25 promoter. The pNK47 (EGFP-MPO1), pKSK17 (EGFP-MPO1 Y15A), pKSK18 (EGFP-MPO1 H19A), pKSK19 (EGFP-MPO1 H28A), pKSK20 (EGFP-MPO1 H115A), pKSK21 (EGFP-MPO1 E119A), and pKSK22 (EGFP-MPO1 P123A) plasmids were constructed by cloning the WT or respective mutant MPO1 gene into the pUG36 vector.
Protein deglycosylation
Deglycosylation of Mpo1 carrying the N-glycosylation reporter was performed using endoglycosidase H (New England Biolabs, Beverly, MA), according to the manufacturer’s instructions.
Immunoblotting
Immunoblotting was performed as described previously (28). Rabbit anti-FLAG antibody (1:1,000 dilution) (18) and rabbit anti-Pgk1 polyclonal antibody (1:500 dilution), which was raised against a peptide (NH2-CDANTKTVTDKEGIPAG-COOH), were used as the primary antibodies. A horseradish peroxidase-conjugated anti-rabbit IgG F(ab′)2 fragment (1:7,500 dilution; GE Healthcare Life Sciences, Little Chalfont, UK) was used as the secondary antibody. Immunodetection was performed using the Pierce Western blotting substrate (Thermo Fisher Scientific, Waltham, MA).
PHS labeling assay
The PHS labeling assay using [11,12-3H]PHS was performed as described previously (17).
In vitro 2-OH FA α-oxidation assays
The membrane fraction of the yeast cells was prepared by ultracentrifugation after the cells had been crushed with zirconia beads, as described previously (24). A proteoliposome containing 3×FLAG-Mpo1 was prepared using the ProteoLiposome Expression Kit (CellFree Sciences, Ehime, Japan), essentially as described previously (24). Briefly, 3×FLAG-Mpo1 was translated by the cell-free system using wheat germ lysate from the mRNA transcribed from the pZKN29 plasmid (24) in the presence of liposomes. The pEU-E01-T1R1 plasmid included in the kit was used as a control. Liposomes were prepared as follows. A lipid solution [2 mg PC (1-palmitolyl-2-oleyl-sn-glycero-3-phosphocholine; Avanti Polar Lipids, Alabaster, AL) and 20 μg 2-OH C24:0 FA (2-OH C24:0-COOH; Cayman Chemical, Ann Arbor, MI) in 200 μl chloroform] was dried in a glass tube. The dried lipids were suspended in 80 μl of the translation buffer SUB-AMIX SGC, incubated at room temperature for 1 h, mixed vigorously for 5 min, and sonicated. The membrane fraction (10 μg) was incubated with FeCl2 and 2-OH FA [2-OH C16:0-COOH (Wako Pure Chemical Industries, Osaka, Japan) or 2-OH C14:0-COOH (Merck, Darmstadt, Germany)] at 37°C. The proteoliposomes (3.86 μl) were incubated with 200 μM FeCl2 at 37°C. After the reactions, the lipids were extracted by successive addition and mixing with 3.75 vol of chloroform/methanol (1:2, v/v), 1.25 vol of chloroform, and 1.25 vol of water. After centrifugation (9,000 g, room temperature, 1 min), the organic phase was recovered and dried. The reaction product FAs were derivatized to N-(4-aminomethylphenyl)pyridine (AMPP) using an AMP+ MS kit (Cayman Chemical) according to the manufacturer’s instructions and quantified by LC/MS/MS as described below.
LC/MS/MS analyses
To quantify PCs, the lipids were extracted from the yeast and purified by TLC as described previously (24). During lipid extraction, 1 nmol C12:0/C12:0 PC (Avanti Polar Lipids) was added as an internal standard. To quantify 2-OH ceramides, the lipids were extracted from the yeast by incubating cell pellets containing ∼1.2 × 108 cells with 1 ml of ethanol/water/diethyl ether/pyridine/15 M ammonia (15:15:5:1:0.018, v/v/v/v/v) at 60°C for 30 min. After centrifugation (2,000 g, room temperature, 1 min), the supernatant was recovered. The extracted lipids were then subjected to successive addition and mixing of 1,013 μl chloroform/methanol/5 M HCl (30:50:1, v/v/v), 525 μl of chloroform, and 525 μl of 1% KCl. Phases were separated by centrifugation (2,000 g, room temperature, 3 min), and lipids were recovered from the organic phase, dried, and dissolved in 200 μl of chloroform/methanol (1:2, v/v). A portion of the samples (5 μl) was subjected to LC/MS/MS analysis. 2-OH C24:0 PHS-ceramide [N-(2′-(R)-OH lignoceroyl)-D-ribo-PHS; Avanti Polar Lipids] was used as the external standard.
The PCs and ceramides extracted from yeast cells and the FAs derivatized to AMPP after the in vitro assay were resolved by UPLC on a reverse-phase column (Acquity UPLC CSH C18 column; particle size, 1.7 μm; column size, 2.1 × 100 mm; Waters, Milford, MA) at 55°C and detected using an electrospray ionization tandem triple quadrupole spectrometer (Xevo TQ-S; Waters) in MRM mode. LC conditions for the PCs and ceramides were as follows. The flow rate was 0.4 ml/min in the binary gradient system using a mobile phase A [acetonitrile/water (3:2, v/v) containing 10 mM ammonium formate] and a mobile phase B [2-propanol/acetonitrile (9:1, v/v) containing 10 mM ammonium formate]. The elution gradient steps were as follows: 0 min, 40% B; 0–18 min, gradient to 100% B; 18–23 min, 100% B; 23–23.1 min, gradient to 40% B; 23.1–25 min, 40% B. LC conditions for the FAs derivatized to AMPP were as previously described (24). 2-OH ceramides were detected by selecting the appropriate m/z values for the quadrupole mass filters Q1 (2-OH C24:0 PHS-ceramide, 666.3 and 684.3; 2-OH C24:0 PHS-ceramide, 680.3 and 698.3; 2-OH C26:0 PHS-ceramide, 694.3 and 712.3; for [M-H2O+H]+ and [M+H]+ ions, respectively) and Q3 (300.2), with the collision energy set at 35 eV. The Q1 and Q3 values and collision energies for PCs and FAs derivatized to AMPP were as previously described (24). Data were analyzed and quantified using MassLynx software (Waters).
RESULTS
Mpo1 is a membrane protein with two transmembrane domains and N and C termini exposed to the cytosol
Although we had previously revealed that Mpo1 is an ER protein (17), the number and regions of its transmembrane domains and its membrane topology remained unknown. Our hydrophobicity plot revealed that Mpo1 has two highly hydrophobic regions (H2 and H3) and three moderately hydrophobic regions (H1, H4, and H5) (Fig. 1A). To determine its membrane topology, we performed an N-glycosylation reporter assay. In this assay, an N-glycosylation cassette containing multiple N-glycosylation sites is inserted into certain regions of the target protein. Because N-linked sugar chains are added to proteins only on the luminal side of the ER, the orientation (cytosolic or luminal side of the ER) of the region into which the cassette was inserted can be determined by examining the N-glycosylation status of the cassette (29, 30). We inserted an N-glycosylation cassette into the middle of each hydrophilic region that was flanked by two hydrophobic regions (after Lys47, Leu69, Val93, or Arg121), as well as at the N terminus and the C terminus of Mpo1 (Fig. 1A). These inserts were expressed as 3×FLAG-tagged proteins in the mpo1Δ cells and detected by immunoblotting. Of the six proteins with an N-linked glycosylation cassette insert, only the one with the cassette inserted after Leu69 exhibited slower mobility on SDS-PAGE than expected for its molecular weight (Fig. 1B). This mobility shift was canceled upon treatment with endoglycosidase H, which removes the N-linked sugar chain. These results indicate that a hydrophilic region containing Leu69 is present in the ER lumen. The fact that the other inserts were not N-glycosylated suggests that they and their adjacent hydrophilic regions are oriented to the cytosolic side of the ER. From these results, we concluded that Mpo1 contains two transmembrane domains and that both its N and its C termini face the cytosol (Fig. 1C).
Fig. 1.
Mpo1 is an integral membrane protein with two transmembrane domains. A: A hydrophobicity plot of Mpo1 was generated using MacVector software (MacVector, Apex, NC) with Manavalan hydrophobicity values (40) and a window size of 11. Arrows indicate the sites of N-glycosylation reporter cassette insertion. H, hydrophobic region; term, terminus. B: Yeast cells 4378 (mpo1Δ) bearing the pMT66 (3×FLAG-MPO1 with N-glycosylation reporter cassette inserted at the N terminus), pMT67 (the cassette after Lys47), pMT72 (after Leu69), pMT68 (after Val93), pMT69 (after Arg121), or pMT70 (at the C terminus) plasmid were grown to early log-phase. Total cell lysates were prepared, treated with or without endoglycosidase H (Endo H), separated by SDS-PAGE, and subjected to immunoblotting with anti-FLAG antibody. C: Membrane topology model of Mpo1. Mpo1 spans the ER membrane twice via its H2 and H3 regions, and its N and C termini are oriented to the cytosol. The H1, H4, and H5 regions of Mpo1 may be peripherally associated with the membrane. The positions of the Mpo1 motif (YXXXH....H....HXXXE) as well as the cassette insert sites (arrows) are indicated.
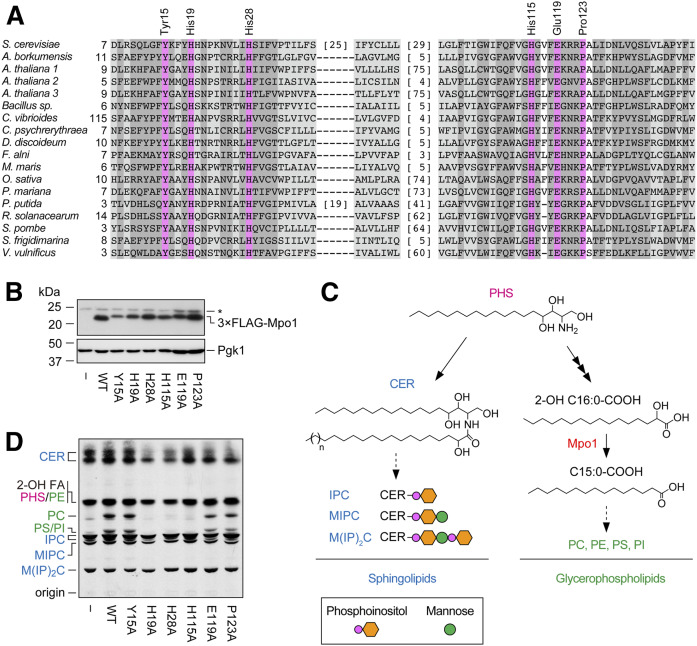
Three histidine residues are important for the activity of Mpo1
Mpo1 is an Fe2+-dependent dioxygenase (24), but the amino acid residues important for its activity, and in particular those involved in the binding of Fe2+, had remained unclear. To identify these, the amino acid sequences of 18 bacterial, fungal, protozoan, and plant Mpo1 family members were aligned. Six amino acid residues (Tyr15, His19, His28, His115, Glu119, and Pro123) were highly conserved (Fig. 2A). To examine their importance for Mpo1 activity, each amino acid residue was substituted to Ala, and the resulting mutants (Y15A, H19A, H28A, H115A, E119A, and P123A) were expressed in the mpo1Δ cells. Although the expression levels of the Y15A mutant were slightly lower than those of the WT protein, the other mutants were detected at almost the same levels as the WT (Fig. 2B).
Fig. 2.
His19, His28, and His115 residues are important for Mpo1 function. A: Amino acid sequence alignment of Mpo1 family members. Amino acid sequences of S. cerevisiae Mpo1 (GenBank accession number, CAA96710) and Mpo1 homologs [Alcanivorax borkumensis (CAL16258); Arabidopsis thaliana (AAL06870, AAM63163, and AAG52360); Bacillus sp. (AGX03529.1); Caulobacter vibrioides (AAK25125); Colwellia psychrerythraea (AAZ26546); Dictyostelium discoideum (EAL69507); Frankia alni (CAJ60547.1); Maricaulis maris (ABI65291); Oryza sativa (AAK98685); Picea mariana (AAC32136); Pseudomonas putida (AFK68339); Ralstonia solanacearum (CAD17957); Schizosaccharomyces pombe (CAB11031); Shewanella frigidimarina (ABI71109); and Vibrio vulnificus (AAO07035)] were aligned using the BLAST tool (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Pink highlights represent amino acid residues that are identical across all members, and the amino acid numbers of these conserved amino acid residues in the yeast S. cerevisiae are indicated above the alignment. Dark gray and light gray highlights represent amino acid residues with strong and weak similarity, respectively. B, D: Yeast cells 4378 (mpo1Δ) bearing the pAKNF316 (vector), pNK46 (3×FLAG-MPO1), pMT12 (3×FLAG-MPO1 Y15A), pMT13 (3×FLAG-MPO1 H19A), pMT14 (3×FLAG-MPO1 H28A), pMT15 (3×FLAG-MPO1 H115A), pMT16 (3×FLAG-MPO1 E119A), or pMT17 (3×FLAG-MPO1 P123A) plasmid were to early log-phase. B: Total cell lysates were prepared, separated by SDS-PAGE, and subjected to immunoblotting with anti-FLAG or anti-Pgk1 (loading control) antibody. The asterisk indicates nonspecific bands. C: PHS metabolic pathway. PHS is metabolized to complex sphingolipids [inositol phosphorylceramide (IPC), mannosylinositol phosphorylceramide (MIPC), and mannosyldiinositol phosphorylceramide (M(IP)2C)] via ceramide (CER). Alternatively, in the degradation pathway, C18 PHS is metabolized to 2-OH C16:0-COOH, which is then converted to C15:0-COOH by Mpo1-catalyzed α-oxidation and incorporated into glycerophospholipids (PC, PE, PS, and PI). D: Cells were labeled with 0.016 μCi [3H]PHS at 30°C for 3 h. Lipids were extracted, separated by normal-phase TLC, and detected by autoradiography.
Next, we performed [3H]PHS labeling experiments. PHS is metabolized either to sphingolipids or glycerophospholipids (Fig. 2C) (17). In the latter, which is a degradation pathway, the PHS metabolite 2-OH FAs are converted to nonhydroxy odd-chain FAs by α-oxidation, followed by incorporation into glycerophospholipids. When the mpo1Δ cells harboring the vector were labeled with [3H]PHS, PHS was metabolized only to sphingolipids (Fig. 2D, blue) and not to glycerophospholipids (Fig. 2D, green). Expression of WT Mpo1 in mpo1Δ cells restored this impaired PHS-to-glycerophospholipid metabolism. PHS was also not metabolized to glycerophospholipids in the mpo1Δ cells expressing the H19A, H28A, or H115A mutant, but it was in those expressing the Y15A, E119A, or P123A mutant. These results indicate that three histidine residues, His19, His28, and His115, are important for Mpo1 function. In several Fe2+-binding proteins, histidine residues are involved in the binding action (31–35), implying that His19, His28, and His115 are responsible for Fe2+ binding.
In the yeast S. cerevisiae, C18 PHS is the most abundant PHS species, followed by C20 PHS (30, 36). These are metabolized to C15:0 FA (C15:0-COOH) and C17:0-COOH, respectively, via the degradation pathway involving Mpo1. A portion of the C15:0-COOH thus generated is converted to C17:0-COOH via FA elongation, and some of both of these are converted to C15:1-COOH and C17:1-COOH, respectively, by desaturation (24). These odd-chain LCFAs are incorporated into glycerophospholipids after conversion to acyl-CoAs (17). To quantitatively evaluate the activity of each Mpo1 mutant, the amounts of PC, the most abundant glycerophospholipid class, were measured by LC/MS/MS. The measurement results for five of the most abundant of the PC species containing odd-chain LCFAs that are products of Mpo1 (or their derivatives) are shown in Fig. 3A: C16:1–C15:0, C16:1–C17:1, C18:1–C15:0, C18:1–C15:1, and C16:1–C17:0. The quantities of these odd-chain PC species in mpo1Δ cells harboring vector were much lower than in those expressing WT Mpo1, but were similar to those in the H19A- or H115A-expressing cells. The levels of odd-chain PCs in H28A-expressing cells were lower than in WT-expressing cells, but higher than in cells harboring vector. The quantities of odd-chain PCs in the mpo1Δ cells expressing the Y15A, E119A, or P123A mutant were similar to those of the WT Mpo1-expressing cells.
Fig. 3.
Tyr15, His19, His28, His115, and Glu119 residues are important for Mpo1 function. A, B: Yeast cells 4378 (mpo1Δ) bearing the pAKNF316 (vector), pNK46 (3×FLAG-MPO1), pMT12 (3×FLAG-MPO1 Y15A), pMT13 (3×FLAG-MPO1 H19A), pMT14 (3×FLAG-MPO1 H28A), pMT15 (3×FLAG-MPO1 H115A), pMT16 (3×FLAG-MPO1 E119A), or pMT17 (3×FLAG-MPO1 P123A) plasmid were grown to early log-phase. Cells were treated with 0.1% ethanol (A) or 10 μM 2-OH C16:0-COOH (B) for 3 h at 30°C. Lipids were extracted, and odd-chain PCs were analyzed by LC/MS/MS. The left panels represent the values for each odd-chain PC species, and the right panels show the total values for all odd-chain PCs. Values represent the mean (percent) ± SD of total PC quantities (even-chain plus odd-chain PCs) from three independent experiments. The statistical significance of differences between cells harboring the pNK46 plasmid (WT 3×FLAG-MPO1) and cells harboring another type of plasmid was analyzed (**P < 0.01, *P < 0.05; Dunnett’s test).
Next, the quantities of odd-chain PCs were measured in the presence of 10 μM 2-OH C16:0-COOH in the medium; that is, they were measured under conditions in which the substrate of Mpo1 was abundant. Upon addition of 2-OH C16:0-COOH, odd-chain PC levels in mpo1Δ cells expressing WT Mpo1 increased approximately 11-fold (compare Fig. 3B with Fig. 3A: the total percentage of odd-chain PCs is 9.8% in the former and 0.9% in the latter). However, the increase was very slight in cells harboring vector or expressing the H19A, H28A, or H115A mutants, and although the increase was greater in those expressing the Y15A or E119A mutants, it was still smaller than that in the WT-expressing cells (Fig. 3B). The difference from the WT-expressing cells was smallest in the cells expressing the E119A mutant. These results indicate that the activity of all of these mutants (H19A, H28A, H115A, Y15A, and E119A) was depressed. Although there was no observable effect of the mutation on the activity of cells expressing the Y15A or E119A mutant at low concentrations of the substrate (physiological conditions), the effects became apparent under an increased substrate concentration (addition of 2-OH C16:0-COOH to the medium). Together with the results shown in Fig. 2D, these findings indicate that the order of the effect on Mpo1 activity of the mutations is H19A = H115A > H28A > Y15A > E119A > P123A.
The amino acid sequence important for Mpo1 activity (YXXXH.....H.....HXXXE; hereafter we refer to this as the Mpo1 motif) resembles the Fe2+-binding motifs in class I diiron-oxo proteins (motif 1, H....HXXXE; motif 2, HXXXH.....HXXXXD) (31, 33). To investigate the functional relationship between the Mpo1 motif and those of class I diiron-oxo proteins, the tyrosine (Tyr15) and glutamate (Glu119) residues of Mpo1 were replaced with the corresponding amino acid residues from the latter (histidine and aspartate, respectively). The resulting Y15H and E119D mutants were expressed in mpo1Δ cells, and their activity was examined by [3H]PHS labeling. The activity and expression levels of the two mutants were comparable to the WT protein (Fig. 4A, B). Tyrosine can potentially form a coordinate bond with Fe2+, as can histidine. Both glutamate and aspartate are acidic amino acids. We speculated that these similarities would make it possible to replace the amino acid residues in the Mpo1 motif with the corresponding amino acid residues from the class I diiron-oxo proteins.
Fig. 4.
The histidine residues of the Mpo1 motif cannot be substituted with tyrosine residues. A, B: Yeast cells 4378 (mpo1Δ) bearing the pAKNF316 (vector), pNK46 (3×FLAG-MPO1), pKSK32 (3×FLAG-MPO1 Y15H), pKSK33 (3×FLAG-MPO1 H19Y), pKSK34 (3×FLAG-MPO1 H28Y), pKSK35 (3×FLAG-MPO1 H115Y), or pKSK36 (3×FLAG-MPO1 E119D) plasmid were grown to early log-phase. A: Total cell lysates were prepared, separated by SDS-PAGE, and subjected to immunoblotting with anti-FLAG or anti-Pgk1 (loading control) antibody. B: Cells were labeled with 0.016 μCi [3H]PHS at 30°C for 3 h. Lipids were extracted, separated by normal-phase TLC, and detected by autoradiography. CER, ceramide; IPC, inositol phosphorylceramide; MIPC, mannosylinositol phosphorylceramide; M(IP)2C, mannosyldiinositol phosphorylceramide.
We also created mutants in which each of the histidine residues in the Mpo1 motif was replaced with a tyrosine residue (H19Y, H28Y, and H115Y). None of these mutants exhibited any activity (Fig. 4B), similar to the alanine mutants (H19A, H28A, and H115A; Fig. 2D). Thus, tyrosine residues cannot substitute for the function of the histidine residues in Mpo1, probably due to the differences in the orientation and/or position of the coordinate bond ligands.
To exclude the possibility that the cause of the impaired PHS/2-OH FA metabolism in the Mpo1 mutant-expressing cells was due to mislocalization, the intracellular localization of each alanine mutant was examined using EGFP-fusion. All Mpo1 mutants exhibited a double ring structure characteristic of yeast ER, which is composed of perinuclear ER and cortical ER, as does the WT Mpo1 (Fig. 5). Therefore, none of the mutations affected the intracellular localization of Mpo1.
Fig. 5.
Mpo1 mutants are localized in the ER. Yeast cells 4378 (mpo1Δ) bearing the pUG36 (EGFP), pNK47 (EGFP-MPO1), pKSK17 (EGFP-MPO1 Y15A), pKSK18 (EGFP-MPO1 H19A), pKSK19 (EGFP-MPO1 H28A), pKSK20 (EGFP-MPO1 H115A), pKSK21 (EGFP-MPO1 E119A), or pKSK22 (EGFP-MPO1 P123A) plasmid were grown in medium lacking uracil. Expression of EGFP-MPO1 was induced by transferring the cells to the medium lacking uracil and methionine at 30°C for 3 h. The cells were observed under a DM5000B fluorescence microscope (Leica Microsystems, Wetzlar, Germany). Scale bar, 5 μm.
Next, to investigate the possibility that the cause of the impaired PHS/2-OH FA metabolism in the Mpo1 mutant-expressing cells was decreased Mpo1 activity, the enzyme activity of each alanine mutant was examined by in vitro assay. The membrane fraction of the mpo1Δ cells expressing each Mpo1 mutant was prepared and incubated with 1 mM 2-OH C16:0-COOH in the presence of 1 mM Fe2+, and the amount of C15:0-COOH produced was determined by LC/MS/MS analysis. The α-oxidation activity in cells expressing WT Mpo1 was approximately five and one-half times higher than that in cells harboring vector (Fig. 6A). Those expressing the Y15A, H19A, H28A, or H115A mutants exhibited very low levels of activity, similar to the cells harboring vector. The activity in cells expressing the E119A mutant was only slightly lower than that in the WT cells, and in cells expressing the P123A mutant, it was comparable to that of the WT cells.
Fig. 6.
Tyr15, His19, His28, His115, and Glu119 residues are important for Mpo1 activity. A–C: Membrane fractions (10 μg) prepared from yeast cells 4378 (mpo1Δ) bearing the pAKNF316 (vector; A), pNK46 [3×FLAG-MPO1 (A–C)], pMT12 [3×FLAG-MPO1 Y15A (A, C)], pMT13 [3×FLAG-MPO1 H19A (A, C)], pMT14 [3×FLAG-MPO1 H28A (A, C)], pMT15 [3×FLAG-MPO1 H115A (A, C)], pMT16 [3×FLAG-MPO1 E119A (A, C)], or pMT17 [3×FLAG-MPO1 P123A (A)] plasmid were incubated with 1 mM 2-OH FA [(2-OH C16:0-COOH (A); 2-OH C14:0-COOH (B, C)] in the presence of Fe2+ [1 mM (A); indicated concentrations (B, C)] at 37°C for 3 h (A) or 4.5 h (B, C). Lipids were extracted, and the odd-chain FAs produced (C15:0-COOH (A); C13:0-COOH (B, C)] were derivatized to AMPP, followed by quantification by LC/MS/MS. Values represent the mean ± SD from three independent experiments (A, B). The statistical significance of differences between cells harboring pNK46 plasmid (WT 3×FLAG-MPO1) and cells harboring another type of plasmid was analyzed (A) (**P < 0.01, *P < 0.05; Dunnett’s test).
WT Mpo1 was active not only toward 2-OH C16:0-COOH but also toward 2-OH C14:0-COOH, producing C13:0-COOH (Fig. 6B). When the α-oxidation activity of Mpo1 toward 2-OH C14:0-COOH was measured using different concentrations of the Fe2+, the activity reached a plateau at an Fe2+ concentration of 20 μM. The KD value of Mpo1 for Fe2+ was calculated to be 9.7 μM. However, no activity was observed for the Y15A, H19A, H28A, or H115A mutants, even when the Fe2+ concentration was increased to 1 mM (Fig. 6C), suggesting that the affinity of these mutants for Fe2+ was much lower and that their KD values were above 1 mM.
Pxp1 is not involved in the α-oxidation of 2-OH LCFAs in yeast
When 2-OH C16:0-COOH was added to the mpo1Δ cells (and mpo1Δ cells harboring vector), the levels of odd-chain PCs were increased slightly but significantly (Figs. 3 and 7) (24). This indicates that a minor alternative α-oxidation pathway exists in yeast. Mammals do not have an Mpo1 homolog, and α-oxidation of 2-OH FAs is carried out in three steps (CoA addition, cleavage, and oxidation) (18). The 2-OH acyl-CoA lyases (HACL1 and HACL2) catalyze the rate-limiting step (cleavage reaction) in this α-oxidation (18, 37). Yeast contains a homolog of HACL1 and HACL2, Pxp1, although its function is unknown. To investigate the possibility that Pxp1 is responsible for the minor alternative α-oxidation pathway, odd-chain PC levels in mpo1Δ pxp1Δ double-mutant cells treated with ethanol or 2-OH C16:0-COOH were compared with those in mpo1Δ cells. The increases in odd-chain PC levels resulting from the addition of 2-OH C16:0-COOH were similar between mpo1Δ cells and mpo1Δ pxp1Δ cells (Fig. 7). Therefore, Pxp1 is not involved in the α-oxidation of 2-OH LCFAs in yeast.
Fig. 7.
Pxp1 is not involved in the α-oxidation of 2-OH FA. Yeast cells BY4741 (WT), 4378 (mpo1Δ), 261 (pxp1Δ), and KSKY4 (pxp1Δ mpo1Δ) were grown to early log-phase. Cells were treated with 0.1% ethanol or 10 μM 2-OH C16:0-COOH for 3 h at 30°C. Lipids were extracted, and odd-chain PCs were analyzed by LC/MS/MS. Values represent the mean ± SD summed percentages of the quantities of PCs containing C15 and C17 FAs out of the total quantity of PCs in three independent experiments. The statistical significance of differences was analyzed (**P < 0.01, *P < 0.05; Tukey’s test).
Mpo1 is active toward 2-OH VLCFAs
In yeast, 2-OH FAs are generated not only via the PHS degradation pathway but also through the oxidation of the C2 position of FAs in ceramide by the sphingolipid 2-hydroxylase Scs7 (1, 17). The 2-OH FAs produced by degradation of PHS are LCFAs (mainly C16 and C18) and are mostly incorporated into glycerophospholipids after being converted to nonhydroxy odd-chain LCFAs by Mpo1 (Fig. 3) (24). The substrates of Scs7 are thought to be VLCFAs (mainly C26) in ceramides (1, 38), although the possibility remains that free VLCFAs are substrates of Scs7. So far, the metabolism of 2-OH VLCFAs in sphingolipids is completely unknown. We examined the possibility that Mpo1 converts 2-OH C26:0-COOH into C25:0-COOH after the release of 2-OH C26:0-COOH from sphingolipids by inositol phosphosphingolipid phospholipase C and ceramidase. The levels of ceramide containing 2-OH C26:0-COOH were comparable between WT and mpo1Δ cells (Fig. 8A), but those of ceramide containing 2-OH C25:0-COOH were approximately 38-fold lower in mpo1Δ than in WT cells (Fig. 8B). The latter is predicted to be produced from the Mpo1 product C25:0-COOH by CoA addition, amide bond formation with LCB (ceramide production), and 2-hydroxylation (Fig. 8C). We then performed an in vitro assay using purified Mpo1 and 2-OH C24:0-COOH as the 2-OH VLCFA substrate. Mpo1 indeed exhibited activity toward 2-OH C24:0-COOH (Fig. 8D). These results indicate that Mpo1 is involved not only in α-oxidation of 2-OH LCFAs, which are the metabolites of PHS, but also in α-oxidation of 2-OH VLCFAs produced by the Scs7-mediated pathway (Fig. 8E).
Fig. 8.
Mpo1 exhibits activity toward 2-OH VLCFAs. A, B: BY4741 (WT) and 4378 (mpo1Δ) cells were grown to early log-phase. Lipids were extracted and ceramides containing PHS and 2-OH FAs [C26:0 (A); C25:0 (B)] were analyzed by LC/MS/MS. Values represent the mean ± SD from three independent experiments (**P < 0.01; Student’s t-test). C: Model of the metabolic pathway of 2-OH C26:0-ceramide. 2-OH C26:0-ceramide is degraded to 2-OH C26:0-COOH by the ceramidase Ypc1 or Ydc1. The 2-OH C26:0-COOH thus generated is then converted to C25:0-COOH by Mpo1-catalyzed α-oxidation. After conversion to C25:0-CoA by acyl-CoA synthetases, C25:0-COOH is incorporated into ceramide, followed by 2-hydroxylation by Scs7. In yeast, there are seven acyl-CoA synthetases (Faa1–4, Fat1 and Fat2, and Acs1). Ceramide synthase is composed of Lag1, Lac1, and Lip1. D: The mRNAs transcribed from pEU-E01-T1R1 (control) and pZKN29 (3×FLAG-MPO1) plasmids were incubated with wheat germ extract at 15°C for 20 h in the presence of PC-based liposomes containing 2-OH C24:0-COOH to produce Mpo1-containing proteoliposomes. The generated proteoliposomes were incubated at 37°C for 6 h in the presence of 1 mM Fe2+. Lipids were extracted, and the reaction product C23:0-COOH was derivatized to AMPP, followed by quantification by LC/MS/MS. Values represent the mean ± SD from three independent experiments (**P < 0.01; Student’s t-test). E: 2-OH FA metabolic pathway involving Mpo1. The structural formula represents the major yeast sphingolipid containing C18 PHS and 2-OH C26:0-COOH, with R representing a polar head group. Mpo1 is involved not only in the α-oxidation of 2-OH C16:0-COOH produced by the PHS metabolic pathway, but also in that of 2-OH C26:0-COOH generated by removal from sphingolipids. C15:0-COOH is used for glycerophospholipid production, whereas C25:0-COOH is utilized for sphingolipid synthesis. CER, ceramide.
Mpo1 is important for carbon utilization from 2-OH FAs under carbon starvation conditions
Although we revealed that Mpo1 is involved in the metabolism of a variety of chain lengths of 2-OH FAs, the physiological significance of Mpo1 remains largely unknown. Deletion of the MPO1 gene did not affect growth under nutrient-rich growth conditions (24) (Fig. 9A). We examined the effects of MPO1 deficiency on growth under starvation conditions. The growth of mpo1Δ cells was slower than that of WT cells under carbon-deprived conditions (Fig. 9B). On the other hand, the growth of mpo1Δ cells was similar to that of WT cells under nitrogen starvation conditions (Fig. 9C). Addition of nonhydroxy FAs (C16:0-COOH), but not 2-OH FAs (2-OH C16:0-COOH), to the carbon-deprived medium restored the growth of mpo1Δ cells (Fig. 9D, E). These results suggest that yeast utilizes 2-OH FAs as a carbon source under carbon-starved conditions, whereas this utilization is abolished in mpo1Δ cells due to impaired α-oxidation of 2-OH FAs.
Fig. 9.
α-Oxidation of 2-OH FAs by Mpo1 is important for growth under carbon-starved conditions. A–E: BY4741 (WT) and 4378 (mpo1Δ) cells were grown in SC medium to early-log phase [absorbance at 600 nm (A600) = ∼1.0]. Cells were washed with SC medium (A), SC–C [carbon-deprived (–C) medium (B, D, E)], or synthetic dextrose (SD–N) [nitrogen-deprived (–N) medium (C)] twice, and suspended in each medium. Cells were cultured at 30°C for 1, 2, and 3 days in the absence (A–C) or presence of 20 μM C16:0-COOH (D) or 2-OH C16:0-COOH (E), and A600 was used as an indicator of cell density. The values are relative to the A600 values at the start of growth after washing and represent the mean ± SD from three independent experiments (**P < 0.01; Student’s t-test).
DISCUSSION
Mpo1 is an Fe2+-dependent dioxygenase, and oxygen bound to Fe2+ is involved in the α-oxidation of 2-OH FAs (24). However, the amino acid residues involved in Fe2+ binding were unclear. In the present study, we have revealed that three histidine residues (His19, His28, and His115) are particularly important for Mpo1 activity (Figs.2–4, 6). The KD value of WT Mpo1 toward Fe2+ was ∼10 μM (Fig. 6B), but no activity was detected for the mutants in which one of these histidine residues had been substituted (H19A, H28A, and H115A), even in the presence of 1 mM Fe2+. In class I diiron-oxo proteins, each of the two histidine-containing motifs (motif 1, H....HXXXE; motif 2, HXXXH....HXXXXD) forms a coordinate bond with Fe2+ (31, 33). On the other hand, the amino acid sequence that we have revealed here, which is important for Mpo1 activity, is YXXXH....H....HXXXE (the Mpo1 motif). In this sequence, the tyrosine and glutamate residues can be replaced by histidine and aspartate residues, respectively (Fig. 4B). The H....HXXXE portion of the Mpo1 motif is identical to motif 1 of class I diiron-oxo proteins, and the combination of YXXXH and HXXXE in the Mpo1 motif is similar to motif 2. At present, it is not clear which residues in the Mpo1 motif actually form the coordinate bonds with Fe2+. However, we speculate that the His19 and His115 residues, which are indispensable for Mpo1 activity in any of our assays, are at least involved in the bond formation process.
Depressed activity of the H19A, H28A, and H115A mutants was observed in our [3H]PHS labeling experiment (Fig. 2D) and our measurements of odd-chain PC levels (Fig. 3A). In contrast, the activity of the Y15A and E119A mutants was not depressed in those experiments, although this was the case to some extent when 2-OH C16:0-COOH was added to the culture medium (Fig. 3B), and also in the in vitro assay (Fig. 6A). In the first two in vivo experiments, the Mpo1-mediated reaction was carried out under conditions that mimicked physiological conditions in the living organism, with low levels of endogenous substrates. Under these conditions, a slight depression of enzyme activity was difficult to detect. On the other hand, the latter two experiments were conducted under comparatively unnatural conditions: the in vivo experiment was conducted in the presence of high levels of substrate, and the in vitro conditions were not necessarily optimized for the enzyme, enabling us to detect the depression of its activity with high sensitivity. Our results indicate that Tyr15 and Glu119 also have some role in the enzyme activity of Mpo1.
Because Mpo1 is not present in mammals, 2-OH FAs (carbon chain length = n) are metabolized to nonhydroxylated FAs (n–1) through a different pathway than in yeast. Mpo1 converts 2-OH FAs (n) to nonhydroxy FAs (n–1) in one step (24). In contrast, in mammals, 2-OH FAs (n) are first converted to 2-OH acyl-CoAs, then cleaved to nonhydroxy fatty aldehydes (n–1), and finally converted to nonhydroxy FAs (n–1) (18, 37). The second cleavage step is catalyzed by 2-OH acyl-CoA lyases HACL1 and HACL2, with HACL2 being the predominant enzyme (18). Although the homolog of these HACLs exists in yeast (Pxp1), it is not involved in 2-OH FA metabolism (Fig. 7). Mpo1 and HACLs differ in both their substrates (2-OH FAs vs. 2-OH acyl-CoAs) and their cofactors (Fe2+ vs. thiamine pyrophosphate) (18, 24, 37). Consistent with the fact that HACL2 activity is not dependent on Fe2+, histidine residues conserved in the Mpo1 family are absent in HACLs.
To date, 3,062 Mpo1 homologs have been found in a wide range of species (1,986 species) from bacteria, fungi, protozoa, and plants (http://pfam.xfam.org/family/PF06127#tabview=tab1). The function of yeast Mpo1 (2-OH FA dioxygenase) was the first function to be elucidated from this family (17, 24). Sequence alignment of representative Mpo1 family members revealed that the Mpo1 family contains three highly conserved regions consisting of 32, 7, and 40 amino acid residues [collectively called the Mpo1 domain (previously the DUF962 domain) hereafter], respectively (Fig. 2A). Mpo1 (S. cerevisiae) has gaps of 25 and 29 amino acids separating these highly conserved regions. We also revealed that Mpo1 spans the ER membrane via the highly hydrophobic regions H2 and H3. These regions correspond to the gaps between the highly conserved regions in the Mpo1 family. On the other hand, in some Mpo1 family members (e.g., Alcanivorax borkumensis and Bacillus sp.), the three highly conserved regions (Mpo1 domain) are nearly contiguous, with almost no gaps between them, suggesting that they do not have transmembrane domains. Considering the high sequence conservation in this family of enzymes, it is highly probable that all family members are dioxygenases, like Mpo1. However, the substrates of the soluble family members may not be lipids (2-OH FAs) but rather hydrophilic 2-OH-containing molecules. In addition, some Mpo1 family members contain domains such as cytochrome P450-like, protein kinase, or peptidase (http://pfam.xfam.org/family/PF06127#tabview=tab1), in addition to the Mpo1 domain, suggesting that this family has a variety of cellular functions.
The regions other than the transmembrane domains of Mpo1 are oriented toward the cytosol; in other words, the active sites of Mpo1 exist in the cytosol. This means that 2-OH FAs, which are the substrates of Mpo1, exist in the cytosolic leaflet of the ER membrane with their carboxy group exposed to the cytosol, and receive oxygen attack from the cytosol. The first irreversible reaction in the degradation pathway of LCBs is catalyzed by LCB 1-phosphate lyase. The active site of this enzyme is also oriented to the cytosolic side of the ER membrane (39). Therefore, the metabolism of LCBs proceeds in the cytosolic leaflet of the ER membrane.
Mpo1 is involved not only in 2-OH LCFA metabolism in the PHS degradation pathway, but also in the metabolism of 2-OH VLCFAs (Fig. 8B). Furthermore, in our in vitro assay, Mpo1 exhibited activity toward the 2-OH LCFAs 2-OH C16:0-COOH (24) (Fig. 6A) and 2-OH C14:0-COOH (Fig. 6B), as well as toward the 2-OH VLCFA 2-OH C24:0-COOH (Fig. 8D). These results indicate that Mpo1 is active toward 2-OH FAs with a wide range of chain lengths and produced via different pathways (Fig. 8E).
Deletion of MPO1 caused growth retardation under carbon-starved conditions (Fig. 9B). The cells recovered from the growth retardation after addition of nonhydroxy FA to the medium (Fig. 9D). On the other hand, deletion of MPO1 did not affect growth under nitrogen starvation conditions (Fig. 9C). These results suggest that Mpo1 is not involved in the induction of autophagy, but rather functions to supply carbon sources from 2-OH FAs. Both 2-OH FAs and PHS, a precursor of 2-OH FAs, are components of sphingolipids. Although little is known about sphingolipid metabolism under carbon-starved conditions, our results suggest that sphingolipids are degraded and utilized as a carbon source to produce energy. 2-OH FAs cannot be used as substrates for β-oxidation. Therefore, they must be converted to nonhydroxy FAs by Mpo1 to become substrates.
In conclusion, here, we have revealed the membrane topology, substrate specificity, amino acid residues important for activity, and physiological role of Mpo1. Of the Mpo1 family members, only Mpo1 itself has been analyzed. However, our findings give clues as to the enzymatic characterization and roles of the entire Mpo1 family.
Data availability
All data are contained within the article.
Acknowledgments
The authors thank Dr. J. H. Hegemann (University of Düsseldorf) for the pUG36 vector.
Footnotes
Abbreviations:
- AMPP
- N-(4-aminomethylphenyl)pyridine
- EGFP
- enhanced GFP
- LCB
- long-chain base
- LCFA
- long-chain FA
- 2-OH
- 2-hydroxy
- PHS
- phytosphingosine
- SC
- synthetic complete
- VLCFA
- very-long-chain FA
This work was supported by the Advanced Research and Development Programs for Medical Innovation (AMED-CREST), Japan Agency for Medical Research and Development (AMED) Grant JP20gm0910002h0006 (to A.K.), and Japan Society for the Promotion of Science (JSPS) KAKENHI Grant JP18H03976 (to A.K.). The authors declare that they have no conflicts of interest with the contents of this article.
REFERENCES
- 1.Haak D., Gable K., Beeler T., and Dunn T.. 1997. Hydroxylation of Saccharomyces cerevisiae ceramides requires Sur2p and Scs7p. J. Biol. Chem. 272: 29704–29710. [DOI] [PubMed] [Google Scholar]
- 2.Ring M. W., Schwar G., and Bode H. B.. 2009. Biosynthesis of 2-hydroxy and iso-even fatty acids is connected to sphingolipid formation in myxobacteria. ChemBioChem. 10: 2003–2010. [DOI] [PubMed] [Google Scholar]
- 3.Hama H. 2010. Fatty acid 2-hydroxylation in mammalian sphingolipid biology. Biochim. Biophys. Acta. 1801: 405–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Markham J. E., Lynch D. V., Napier J. A., Dunn T. M., and Cahoon E. B.. 2013. Plant sphingolipids: function follows form. Curr. Opin. Plant Biol. 16: 350–357. [DOI] [PubMed] [Google Scholar]
- 5.Kihara A. 2016. Synthesis and degradation pathways, functions, and pathology of ceramides and epidermal acylceramides. Prog. Lipid Res. 63: 50–69. [DOI] [PubMed] [Google Scholar]
- 6.Edvardson S., Hama H., Shaag A., Gomori J. M., Berger I., Soffer D., Korman S. H., Taustein I., Saada A., and Elpeleg O.. 2008. Mutations in the fatty acid 2-hydroxylase gene are associated with leukodystrophy with spastic paraparesis and dystonia. Am. J. Hum. Genet. 83: 643–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zöller I., Meixner M., Hartmann D., Bussow H., Meyer R., Gieselmann V., and Eckhardt M.. 2008. Absence of 2-hydroxylated sphingolipids is compatible with normal neural development but causes late-onset axon and myelin sheath degeneration. J. Neurosci. 28: 9741–9754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dick K. J., Eckhardt M., Paisan-Ruiz C., Alshehhi A. A., Proukakis C., Sibtain N. A., Maier H., Sharifi R., Patton M. A., Bashir W., et al. 2010. Mutation of FA2H underlies a complicated form of hereditary spastic paraplegia (SPG35). Hum. Mutat. 31: E1251–E1260. [DOI] [PubMed] [Google Scholar]
- 9.Potter K. A., Kern M. J., Fullbright G., Bielawski J., Scherer S. S., Yum S. W., Li J. J., Cheng H., Han X., Venkata J. K., et al. 2011. Central nervous system dysfunction in a mouse model of FA2H deficiency. Glia. 59: 1009–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rattay T. W., Lindig T., Baets J., Smets K., Deconinck T., Sohn A. S., Hortnagel K., Eckstein K. N., Wiethoff S., Reichbauer J., et al. 2019. FAHN/SPG35: a narrow phenotypic spectrum across disease classifications. Brain. 142: 1561–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nagano M., Ishikawa T., Fujiwara M., Fukao Y., Kawano Y., Kawai-Yamada M., and Shimamoto K.. 2016. Plasma membrane microdomains are essential for Rac1-RbohB/H-mediated immunity in rice. Plant Cell. 28: 1966–1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hamberg M., Ponce de Leon I., Rodriguez M. J., and Castresana C.. 2005. α-Dioxygenase. Biochem. Biophys. Res. Commun. 338: 169–174. [DOI] [PubMed] [Google Scholar]
- 13.Kihara A. 2012. Very long-chain fatty acids: elongation, physiology and related disorders. J. Biochem. 152: 387–395. [DOI] [PubMed] [Google Scholar]
- 14.Sassa T., and Kihara A.. 2014. Metabolism of very long-chain fatty acids: genes and pathophysiology. Biomol. Ther. (Seoul). 22: 83–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dickson R. C. 2008. Thematic review series: sphingolipids. New insights into sphingolipid metabolism and function in budding yeast. J. Lipid Res. 49: 909–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ejsing C. S., Sampaio J. L., Surendranath V., Duchoslav E., Ekroos K., Klemm R. W., Simons K., and Shevchenko A.. 2009. Global analysis of the yeast lipidome by quantitative shotgun mass spectrometry. Proc. Natl. Acad. Sci. USA. 106: 2136–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kondo N., Ohno Y., Yamagata M., Obara T., Seki N., Kitamura T., Naganuma T., and Kihara A.. 2014. Identification of the phytosphingosine metabolic pathway leading to odd-numbered fatty acids. Nat. Commun. 5: 5338. [DOI] [PubMed] [Google Scholar]
- 18.Kitamura T., Seki N., and Kihara A.. 2017. Phytosphingosine degradation pathway includes fatty acid α-oxidation reactions in the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA. 114: E2616–E2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iwamori M., Costello C., and Moser H. W.. 1979. Analysis and quantitation of free ceramide containing nonhydroxy and 2-hydroxy fatty acids, and phytosphingosine by high-performance liquid chromatography. J. Lipid Res. 20: 86–96. [PubMed] [Google Scholar]
- 20.Nishimura K. 1987. Phytosphingosine is a characteristic component of the glycolipids in the vertebrate intestine. Comp. Biochem. Physiol. B. 86: 149–154. [DOI] [PubMed] [Google Scholar]
- 21.Wartewig S., and Neubert R. H.. 2007. Properties of ceramides and their impact on the stratum corneum structure: a review. Part 1: ceramides. Skin Pharmacol. Physiol. 20: 220–229. [DOI] [PubMed] [Google Scholar]
- 22.Nakahara K., Ohkuni A., Kitamura T., Abe K., Naganuma T., Ohno Y., Zoeller R. A., and Kihara A.. 2012. The Sjögren-Larsson syndrome gene encodes a hexadecenal dehydrogenase of the sphingosine 1-phosphate degradation pathway. Mol. Cell. 46: 461–471. [DOI] [PubMed] [Google Scholar]
- 23.Ohkuni A., Ohno Y., and Kihara A.. 2013. Identification of acyl-CoA synthetases involved in the mammalian sphingosine 1-phosphate metabolic pathway. Biochem. Biophys. Res. Commun. 442: 195–201. [DOI] [PubMed] [Google Scholar]
- 24.Seki N., Mori K., Kitamura T., Miyamoto M., and Kihara A.. 2019. Yeast Mpo1 is a novel dioxygenase that catalyzes the α-oxidation of a 2-hydroxy fatty acid in an Fe2+-dependent manner. Mol. Cell. Biol. 39: e00428-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brachmann C. B., Davies A., Cost G. J., Caputo E., Li J., Hieter P., and Boeke J. D.. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 14: 115–132. [DOI] [PubMed] [Google Scholar]
- 26.Winzeler E. A., Shoemaker D. D., Astromoff A., Liang H., Anderson K., Andre B., Bangham R., Benito R., Boeke J. D., Bussey H., et al. 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 285: 901–906. [DOI] [PubMed] [Google Scholar]
- 27.Narita T., Naganuma T., Sase Y., and Kihara A.. 2016. Long-chain bases of sphingolipids are transported into cells via the acyl-CoA synthetases. Sci. Rep. 6: 25469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kitamura T., Takagi S., Naganuma T., and Kihara A.. 2015. Mouse aldehyde dehydrogenase ALDH3B2 is localized to lipid droplets via two C-terminal tryptophan residues and lipid modification. Biochem. J. 465: 79–87. [DOI] [PubMed] [Google Scholar]
- 29.Kim H., Melen K., and von Heijne G.. 2003. Topology models for 37 Saccharomyces cerevisiae membrane proteins based on C-terminal reporter fusions and predictions. J. Biol. Chem. 278: 10208–10213. [DOI] [PubMed] [Google Scholar]
- 30.Kihara A., Sakuraba H., Ikeda M., Denpoh A., and Igarashi Y.. 2008. Membrane topology and essential amino acid residues of Phs1, a 3-hydroxyacyl-CoA dehydratase involved in very long-chain fatty acid elongation. J. Biol. Chem. 283: 11199–11209. [DOI] [PubMed] [Google Scholar]
- 31.Sheriff S., Hendrickson W. A., and Smith J. L.. 1987. Structure of myohemerythrin in the azidomet state at 1.7/1.3 Å resolution. J. Mol. Biol. 197: 273–296. [DOI] [PubMed] [Google Scholar]
- 32.Shanklin J., Whittle E., and Fox B. G.. 1994. Eight histidine residues are catalytically essential in a membrane-associated iron enzyme, stearoyl-CoA desaturase, and are conserved in alkane hydroxylase and xylene monooxygenase. Biochemistry. 33: 12787–12794. [DOI] [PubMed] [Google Scholar]
- 33.Fox B. G., Shanklin J., Ai J., Loehr T. M., and Sanders-Loehr J.. 1994. Resonance Raman evidence for an Fe-O-Fe center in stearoyl-ACP desaturase. Primary sequence identity with other diiron-oxo proteins. Biochemistry. 33: 12776–12786. [DOI] [PubMed] [Google Scholar]
- 34.Holmes M. A., Le Trong I., Turley S., Sieker L. C., and Stenkamp R. E.. 1991. Structures of deoxy and oxy hemerythrin at 2.0 Å resolution. J. Mol. Biol. 218: 583–593. [DOI] [PubMed] [Google Scholar]
- 35.Leitgeb S., and Nidetzky B.. 2008. Structural and functional comparison of 2-His-1-carboxylate and 3-His metallocentres in non-haem iron(II)-dependent enzymes. Biochem. Soc. Trans. 36: 1180–1186. [DOI] [PubMed] [Google Scholar]
- 36.Dickson R. C., Nagiec E. E., Skrzypek M., Tillman P., Wells G. B., and Lester R. L.. 1997. Sphingolipids are potential heat stress signals in Saccharomyces. J. Biol. Chem. 272: 30196–30200. [DOI] [PubMed] [Google Scholar]
- 37.Foulon V., Sniekers M., Huysmans E., Asselberghs S., Mahieu V., Mannaerts G. P., Van Veldhoven P. P., and Casteels M.. 2005. Breakdown of 2-hydroxylated straight chain fatty acids via peroxisomal 2-hydroxyphytanoyl-CoA lyase: a revised pathway for the α-oxidation of straight chain fatty acids. J. Biol. Chem. 280: 9802–9812. [DOI] [PubMed] [Google Scholar]
- 38.Zhu G., Koszelak-Rosenblum M., Connelly S. M., Dumont M. E., and Malkowski M. G.. 2015. The crystal structure of an integral membrane fatty acid α-hydroxylase. J. Biol. Chem. 290: 29820–29833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ikeda M., Kihara A., and Igarashi Y.. 2004. Sphingosine-1-phosphate lyase SPL is an endoplasmic reticulum-resident, integral membrane protein with the pyridoxal 5′-phosphate binding domain exposed to the cytosol. Biochem. Biophys. Res. Commun. 325: 338–343. [DOI] [PubMed] [Google Scholar]
- 40.Manavalan P., and Ponnuswamy P. K.. 1978. Hydrophobic character of amino acid residues in globular proteins. Nature. 275: 673–674. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data are contained within the article.