ABSTRACT
Retinoic acid (RA) signaling is essential for multiple developmental processes, including appropriate pancreas formation from the foregut endoderm. RA is also required to generate pancreatic progenitors from human pluripotent stem cells. However, the role of RA signaling during endocrine specification has not been fully explored. In this study, we demonstrate that the disruption of RA signaling within the NEUROG3-expressing endocrine progenitor population impairs mouse β cell differentiation and induces ectopic expression of crucial δ cell genes, including somatostatin. In addition, the inhibition of the RA pathway in hESC-derived pancreatic progenitors downstream of NEUROG3 induction impairs insulin expression. We further determine that RA-mediated regulation of endocrine cell differentiation occurs through Wnt pathway components. Together, these data demonstrate the importance of RA signaling in endocrine specification and identify conserved mechanisms by which RA signaling directs pancreatic endocrine cell fate.
KEY WORDS: β cell differentiation, Retinoic acid signaling, Wnt signaling, Pancreas development, Diabetes, δ cell specification
Summary: This study reveals a previously unappreciated role of RA signaling within the NEUROG3-expressing endocrine progenitor population that impairs β cell differentiation and induces ectopic expression of crucial δ cell genes.
INTRODUCTION
Cell signaling pathways are used continuously throughout development and adulthood to mediate tissue interactions and precisely control gene expression. In particular, retinoic acid (RA) signaling plays crucial roles in a wide range of developmental processes at multiple stages during embryogenesis (Ghyselinck and Duester, 2019). In vertebrates, the basic mechanisms of RA function are conserved: vitamin A is converted into RA through a series of enzymatic reactions and enters the nucleus to interact with the transcriptional effectors of the pathway, RA receptor (RAR) and retinoid X receptor (RXR), as well as co-activators and co-repressors to regulate context-specific target genes (Fig. S1A).
In the pancreas, RA signaling is necessary for the onset of pancreagenesis; previous studies have demonstrated that the inhibition of this pathway leads to pancreas agenesis (Arregi et al., 2016; Öström et al., 2008; Molotkov et al., 2005; Martín et al., 2005; Stafford and Prince, 2002). Based on these studies, exogenous RA is included in human pluripotent stem cell (hPSC) β-like cell differentiation protocols to facilitate the earliest stages of pancreas development. Although high levels of RA are crucial for initial pancreas specification, RA has been shown to subsequently inhibit endocrine differentiation in several model systems (Cardenas-Diaz et al., 2019; Pagliuca et al., 2014; Huang et al., 2014; Rezania et al., 2012; Rovira et al., 2011); therefore, most protocols use progressively lower doses of RA after pancreatic endoderm specification. Despite the reduction or exclusion of exogenous sources of RA from endocrine differentiation media at these later differentiation stages, it has been shown that a cell-autonomous source of RA might still exist (Huang et al., 2014). This is also supported by gene expression analyses in mice demonstrating the presence of many key RA pathway genes in the embryonic pancreas (Krentz et al., 2018). The function of RA signaling during vertebrate islet endocrine cell differentiation, however, has not been fully explored.
To define the role of RA signaling during pancreas endocrine development, we inhibited RA signaling specifically in endocrine progenitors by expressing the RARdn under the regulation of the Neurog3:cre allele. These studies demonstrated that RA signaling is required for both β cell specification and the inhibition of δ cell gene transcripts, including Hhex, Rbp4 and Sst. The inhibition of RA in the NEUROG3+ endocrine progenitor population resulted in reduced numbers of insulin-producing β cells, and contributed to impaired blood glucose regulation in postnatal and adult mice. Similarly, the chemical inhibition of RA specifically after NEUROG3 induction in hPSC β-like cell differentiations decreased insulin production. At the molecular level, we demonstrate that RA-mediated repression of Wnt signaling allows for proper endocrine cell differentiation. Together, these results establish the importance of RA signaling in the endocrine progenitor population for appropriate mouse and human β cell specification.
RESULTS AND DISCUSSION
The RARdn efficiently disrupts pancreas development in mice
To disrupt RA signaling in a cell-specific manner, we used the previously described dominant-negative human RAR allele that had been inserted into the R26R locus downstream of a flox-stop-flox cassette (RARdnflox; Fig. S1B; Rosselot et al., 2010). To validate that the RARdn allele functioned appropriately, we generated RARdnflox/flox; Pdx1:Cre mice to broadly inhibit RA signaling in pancreatic progenitors. Similar to previous studies (Öström et al., 2008), the disruption of RA signaling in all pancreatic progenitors led to the formation of a smaller pancreas that contained fewer β [insulin (INS)], α [glucagon (GCG)] and δ cells [somatostatin (SST)] at embryonic day (E)16.5 and E18.5 (Fig. S1C). The disruption of RA signaling using the tamoxifen-inducible Pdx1:creEsr1 allele (Gu et al., 2002) at E9.5, a slightly later stage of development after the pancreatic progenitor population has been established, also resulted in the formation of a smaller pancreas and a significant reduction in islet cluster formation, and fewer hormone-producing cells (Fig. S1D-G).
Endocrine-specific RA inhibition impairs the formation of insulin-producing β cells and causes ectopic somatostatin RNA expression
To determine whether RA signaling was also required during endocrine cell differentiation downstream of endocrine progenitor formation, we disrupted RA signaling specifically within the NEUROG3 endocrine progenitor population using RARdnflox/flox; Neurog3:cre mice. Remarkably, this disruption of RA signaling in the endocrine progenitor population resulted in significantly fewer β cells as early as E16.5, without notable changes in the other endocrine cell types (Fig. 1A,B). The timing of reduced β cell numbers, combined with no apparent changes in β cell death or proliferation, suggests the inhibition of RA signaling in endocrine progenitor cells impairs β cell differentiation (Fig. 1C,D).
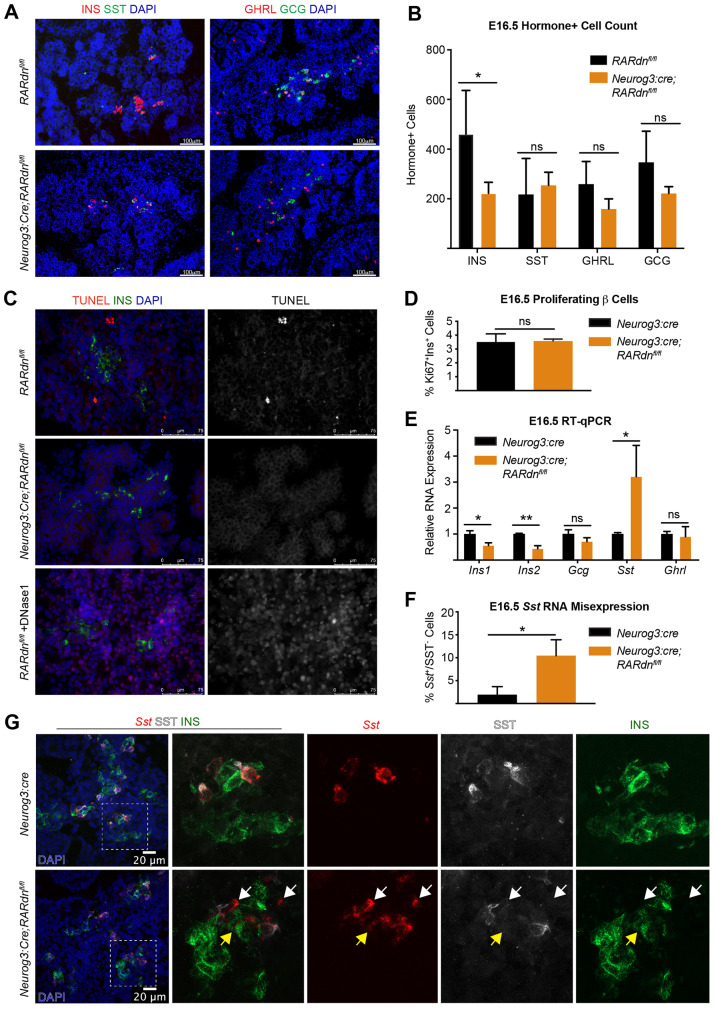
Fig. 1.
Endocrine-specific RA inhibition disrupts β cell development by E16.5 and increases Sst transcript expression. (A) Representative immunofluorescence images of E16.5 INS, SST, GCG and GHRL in RARdnflox/flox; Neurog3:cre mutants. (B) Quantification of A (n=4 control, n=5 mutant; statistical analysis was completed for multiple t-tests using the Holm–Sidak method to correct for multiple comparisons; *Padj<0.05 is significant). (C) TUNEL+INS staining at E16.5. DNase1(+) sample is a positive control (n=3). (D) β cell proliferation reported as a percentage of proliferating versus non-proliferating β cells (n=3, statistical analysis was performed using an unpaired parametric Student's t-test; P>0.05, not significant). (E) RT-qPCR gene expression analysis of E16.5 whole pancreata from RARdnflox/flox; Neurog3:cre mutants (n=3, statistical analysis was completed by the Benjamini, Krieger and Yekutieli two-stage step method to correct for multiple comparisons using Student's t-tests; *Q<0.05, **Q<0.005 are significant). (F) Quantification of the percentage of cells expressing Sst RNA but not SST protein (relative to all Sst RNA+ cells) in Neurog3:cre alone or RARdnflox/flox; Neurog3:cre mutants at E16.5 (n=3, statistical analysis was completed by an unpaired parametric Student's t-test; *P<0.05 is significant). Representative images can be seen in Fig. 1G. (G) Dual RNA-protein visualization using RNAscope and immunofluorescence analyses (n=3). White arrows indicate Sst RNA+ cells, SST protein–;INS protein– cells; yellow arrows indicate Sst RNA+ cells, SST protein–;INS protein+ cells. All n values represent biological replicates. Data are mean±s.d. ns, not significant.
Consistent with the reduced number of β cells in the RARdnflox/flox; Neurog3:cre mice, there was also a reduction of Ins1 and Ins2 RNA expression (Fig. 1E). Interestingly, however, there was also a significant increase in Sst RNA that did not correspond to an increase in δ cell numbers (Fig. 1E compared with Fig. 1B). To determine the explanation for discordant expression between Sst RNA and δ cell numbers, we performed RNAscope combined with immunofluorescence on E16.5 pancreatic tissue sections. This analysis verified an increase in cells expressing Sst RNA, many of which were not SST-producing δ cells (Fig. 1F,G, all arrows). Sst transcripts could be detected in other endocrine cell types, including insulin-producing β cells (Fig. 1G, yellow arrows). We also observed an additional class of Sst+ cells that did not co-express INS, SST or GCG (Fig. 1G, white arrows), which could represent a population of β cells that no longer express INS. However, the observed percentage of Sst+/SST− cells did not correlate with the more substantial decrease in β cell numbers in RA mutants (Fig. 1B,F), suggesting the β cell loss is at least partially independent of the upregulation of the δ cell transcriptional program.
Postnatal hormone expression and blood glucose homeostasis are disrupted in RA mutants
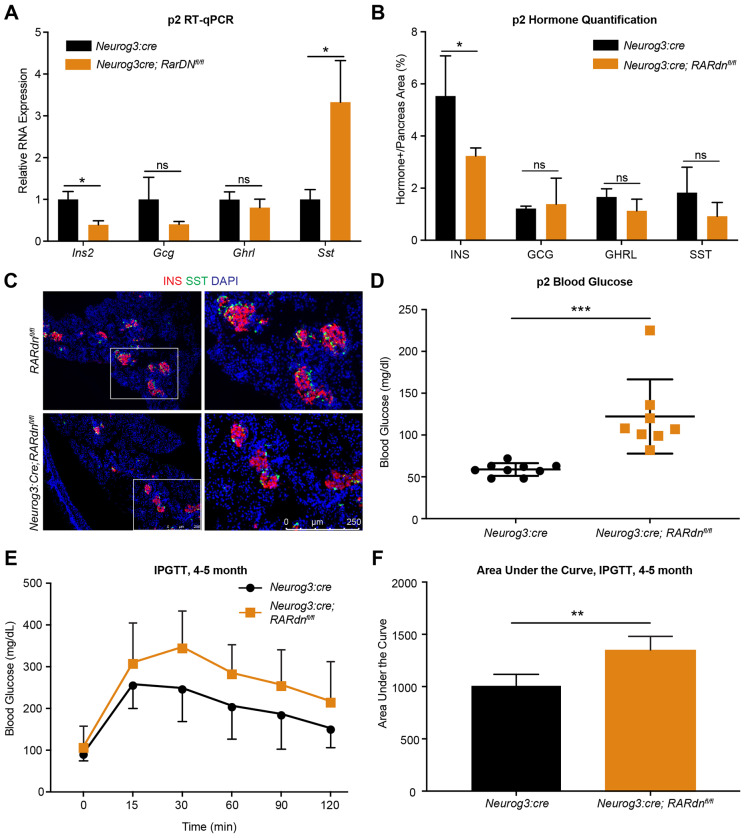
To determine whether the endocrine cell defects persisted postnatally, we assessed hormone expression in postnatal day (P)2 mice. Consistent with the defects observed at E16.5, the reduction in the number of INS+ β cells and Ins transcripts was maintained in neonates, and was again accompanied by an increase in Sst RNA expression without an impact on δ cell numbers (Fig. 2A-C). Furthermore, the RARdnflox/flox; Neurog3:cre neonatal mice were overtly hyperglycemic compared with littermate Neurog3:cre controls (Fig. 2D). Although there was a significant reduction in β cell numbers, the observed decrease is not usually sufficient to cause hyperglycemia (Bonner-Weir et al., 1983; Yasugi et al., 1976), suggesting that the remaining β cells in RARdnflox/flox; Neurog3:cre mice are dysfunctional, which could at least partially be explained by the ectopic expression of δ cell transcripts within the INS+ β cell population. This is also consistent with the glucose intolerance phenotype observed when RA signaling was specifically inhibited in adult mouse β cells (Brun et al., 2015). RARdnflox/flox; Neurog3:cre mice had a normal lifespan; however, impaired glucose homeostasis persisted into adulthood (Fig. 2E,F).
Fig. 2.
Postnatal blood glucose homeostasis is disrupted in RA mutants. (A) Gene expression analysis by RT-qPCR of P2 whole pancreata from RARdnflox/flox; Neurog3:cre mutants (n=3, statistical analysis was completed using the Benjamini, Krieger and Yekutieli two-stage step method to correct for multiple comparisons among multiple Student's t-tests; *Q<0.05 are significant). (B) Quantification of hormones in P2 pancreases (n=5, statistical analysis was completed for multiple Student's t-tests using the Holm–Sidak method to correct for multiple comparisons; *Padj<0.05 is significant). (C) Representative images of immunofluorescence at P2 for INS and SST. Scale bar applies to all panels. (D) Blood glucose at P2, reported in mg/dl. Statistical analysis was performed using an unpaired parametric Student's t-test method (***P<0.0005 is significant). (E) Glucose tolerance tests between 4 and 5 months in male RARdnfl/fl; Neurog3:cre mutants (n=4). (F) Area under the curve of the intraperitoneal glucose tolerance test (IPGTT) in E (statistical analysis was peformed using an unpaired parametric Student's t-test method; **P<0.005 is significant). All n values represent biological replicates. Data are mean±s.d.
RA signaling regulates human β cell differentiation after pancreas progenitor specification
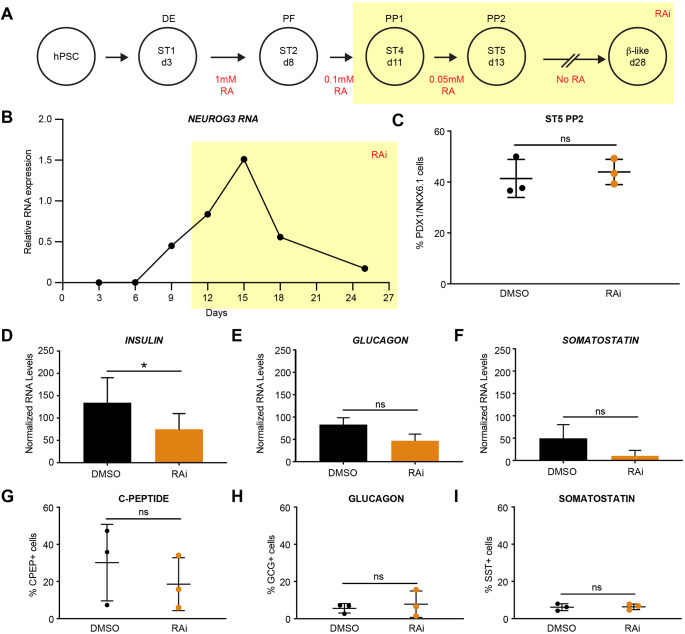
To determine whether the role of RA signaling in murine pancreatic endocrine specification was conserved during human β cell differentiation, we inhibited RA signaling at stage 4 [pancreatic progenitor stage 1 (PP1) day 11] of the human β cell differentiation protocol (Fig. 3A) (Cardenas-Diaz et al., 2019; Tiyaboonchai et al., 2017). The differentiation of human pluripotent stem cells (hPSCs) into β-like cells requires the addition of RA from stages 1 to 2 (days 3 to 8) to generate PDX1-expressing posterior foregut-like cells; as well as between stages 2 and 5 (days 8 to 13), during which time NEUROG3 begins to be expressed (Fig. 3A,B). To simulate our murine in vivo experiments, we inhibited RA signaling after NEUROG3 induction by excluding exogenous RA from the media starting at stage 4 (PP1, day 11) and adding the high-affinity pan-RAR inhibitor AGN193108 to the culture media (RAi, Fig. 3A,B) (Johnson et al., 1999). To ensure that we had not altered pancreatic progenitor differentiation, we confirmed the presence of PDX1/NKX6.1 double-positive cells at day 13, 2 days after treatment with RAi (Fig. 3C). Following an additional 2 weeks of culture in RAi-supplemented media to inhibit any potential role of autocrine RA signaling during endocrine specification, we observed a significant decrease in Ins RNA, with no statistically significant changes in either GCG or SST RNA expression (Fig. 3D-F). Analysis of endocrine cell numbers at day 28 showed no significant effect on GCG+ or SST+ cells, but there was a decrease in C-PEPTIDE+ cells, although this difference was not significant (Fig. 3G-I). These findings are in contrast to a report by Huang et al. (2014) that demonstrated that RA inhibition in human endocrine cell differentiation led to increased INS, GCG and SST. These differences might be due to the method of RA inhibition, as Huang et al. (2014) employed an ALDH inhibitor that acts upstream of RAR and does not inhibit all ALDH enzymes, raising the possibility that RA signaling could continue via other ALDH homologs (Morgan et al., 2015; Duester, 2001). Inhibition of the RA pathway at the RAR/RXR level effectively avoids potential compensatory mechanisms that could reduce the efficacy of upstream inhibitors. Overall, our results suggest that, in humans, RA signaling is important for achieving optimal INS production and, to a lesser extent, β cell differentiation (Fig. 3D,G compared with Fig. 2A,B and Fig. 1B,E).
Fig. 3.
Human β cell differentiation requires RA signaling after posterior foregut formation. (A) Summary of human β cell differentiations with RA addition in red. Highlighted area (yellow) indicates when exogenous RA was removed from the media and RA inhibitor (RAi) was added. (B) RT-qPCR gene expression analysis of NEUROG3 expression, normalized to TBP. (C) Percentage of PDX1+/NKX6.1+ cells at stage 5 (ST5) of the differentiation with and without RAi treatment (n=3 per treatment, statistical analysis was performed using an unpaired parametric Student's t-test method; P>0.05 not significant). (D-F) RT-qPCR gene expression analysis of INS, GCG and SST expression at day 28, normalized to TBP for total RNA and CHGB for endocrine cells to control for differentiation efficiency (n=3 per treatment, statistical analysis was performed using a paired parametric Student's t-test method; *P<0.05 is significant). (G-I) Percentage of cells positive for C-peptide, glucagon and somatostatin by flow cytometric analysis (n=3 per treatment, statistical analysis was performed using an unpaired parametric Student's t-test method; P>0.05, not significant). All n values represent biological replicates. ns, not significant. Data are mean±s.d.
Wnt signaling is derepressed in RARdnflox/flox; Neurog3:cre mutants during murine endocrine specification
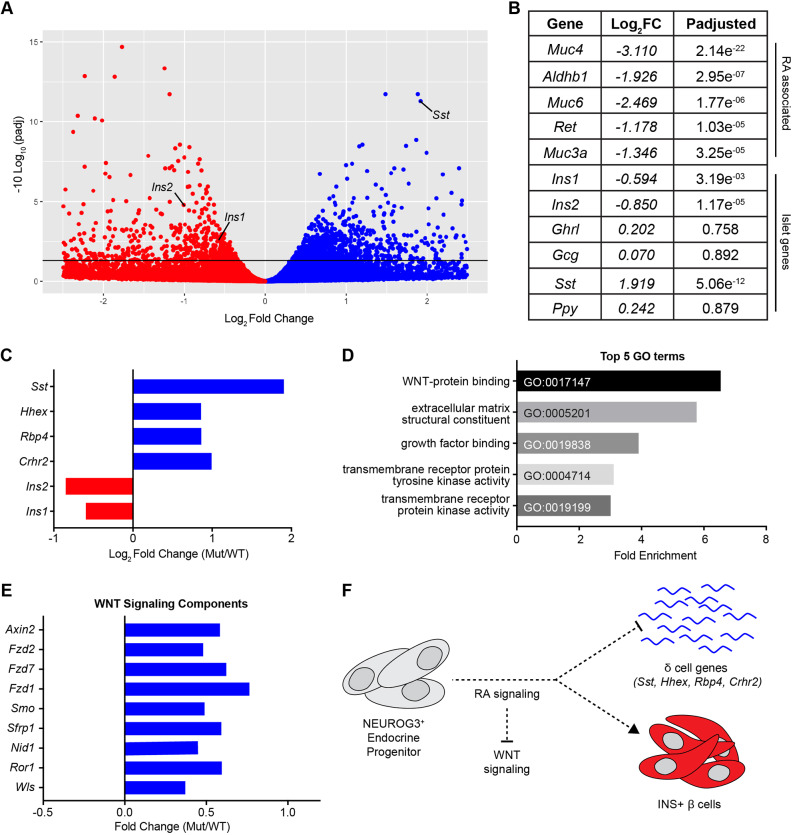
To identify pathways regulated by RA signaling in the endocrine progenitor lineage, we performed transcriptome analysis of whole-mouse pancreata at E16.5 and found 1392 significantly changed genes (Padj≤0.05) in RARdnflox/flox; Neurog3:cre mutant mice, including Ins1, Ins2 and Sst (Fig. 4A,B, Table S1). In addition to the increase in Sst transcript, we also identified several other upregulated δ cell genes, including Hhex, Rbp4 and Crhr2 (Fig. 4C) (DiGruccio et al., 2016), suggesting that RA signaling in endocrine progenitors is necessary to repress ectopic expression of the δ cell transcriptional program. Although there were also changes in several RA-associated genes, classic RA targets, such as HoxA1 (Marshall et al., 1996) and Cyp26A1 (Loudig et al., 2000), that have been described in other developmental contexts, were not significantly altered (Fig. 4A,B, Table S1). This is probably because of the absence of these genes in the NEUROG3+ endocrine progenitor population (Krentz et al., 2018), and demonstrates that RA targets are highly context dependent. Interestingly, we did observe a modest reduction (P=0.057, Padj=0.253) in Mnx1, a known RA-regulated pancreatic development factor (Dalgin et al., 2011). Endocrine deletion of Mnx1 resulted in reduced β cells and increased δ and α cells (Pan et al., 2015). Although we did not observe an α cell phenotype in RARdnflox/flox; Neurog3:cre mutants, this may be because of the lesser reduction of Mnx1 expression.
Fig. 4.
RA signaling inhibits the Wnt pathway to promote β cell specification and inhibit the δ cell transcriptional program. (A) Volcano plot of differentially expressed genes from whole-transcriptome analysis by RNA-seq of RARdnflox/flox; Neurog3:cre mutants (n=3 biological replicates). (B) A selection of genes from A with Log2 fold change and P-adjusted value (Padj≤0.05 is significant). (C) A selection of upregulated δ cell genes compared with Ins1/2 (Crhr2, Padj=0.053). (D) Significantly upregulated GO terms via PANTHER analysis of genes reported in A and Table S1. (E) Significantly enriched Wnt signaling components (GO:0017147). (F) Model demonstrating that during endocrine differentiation, RA signaling represses Wnt to promote β cell differentiation and repress δ-cell genes.
To better define the molecular mechanisms by which RA signaling in the murine endocrine lineage affects the development of hormone-producing cells, we completed a gene ontology (GO) term analysis (Mi et al., 2019) of significantly changed genes at E16.5. These GO terms revealed that Wnt signaling components were significantly affected by the inhibition of RA (Fig. 4D), including nine Wnt-associated genes that were all upregulated (Fig. 4E, GO:0017147). Bioinformatic analysis identified putative RARα-binding sites in the promoter elements of genes from this group, Ror1 and Smo, the latter hinting at cross talk with the Shh pathway. In both cases, these predicted binding sites demonstrate potential direct regulation by RA signaling (Table S2). The identification of tissue-specific enhancers will be necessary to uncover a direct link between RA and the remaining dysregulated Wnt signaling components, which will probably involve both direct and indirect regulation by RA and/or Wnt pathway components. As expected, this analysis also identified RARα-binding sites in the promoter-proximal regions of several dysregulated RA-associated genes, including Muc4, Aldh1b1 and Ret, thus validating the analysis. The dysregulated δ cell genes, however, were not enriched for RARα-binding sites in their promoter regions; however, there are several developmental studies indicating that Hhex is indirectly regulated by Wnt/β-Catenin (Rankin et al., 2011; McLin et al., 2007).
The upregulation of Wnt pathway components by RA inhibition reveals that, under normal conditions, RA functions to repress Wnt signaling, directly and/or indirectly, to promote appropriate β cell differentiation and inhibit δ-cell gene transcription (Fig. 4F). This RA-Wnt interaction has been shown to be an important mediator of development in several other biological contexts (Kumar and Duester, 2010; Bonney et al., 2018, 2016; Osei-Sarfo and Gudas, 2014; Roa et al., 2019), and importantly is consistent with recent studies suggesting that the inhibition of endogenous Wnt is necessary for improved stem cell-derived β cell maturation in human cells (Vethe et al., 2019; Sharon et al., 2019). Our findings suggest that the addition of RA, even in small amounts, to the human β cell differentiation protocol during endocrine progenitor specification could inhibit Wnt signaling to promote more robust generation of β cells, and that RA-mediated Wnt repression is necessary to repress δ cell genes in non-δ endocrine cells. Interestingly, we did not observe an increase in SST transcript expression in the human β cell differentiation platform. This could be because of the fact that the in vitro hPSC model has been engineered to optimally generate β cells, rather than the other endocrine cells. Alternatively, the unperturbed human SST RNA levels could suggest that the disruption of β cell differentiation and the upregulation of the δ cell transcriptional program are separable events, as indicated by the lack of direct correlation between the two phenotypes in the RARdnflox/flox; Neurog3:cre mutants (Fig. 1B,F,G), with only the RA regulation of β cell differentiation being conserved in humans. Finally, it remains possible that this discrepancy between mice and humans could be attributed to the artificial nature of the in vitro β cell differentiation system.
The ability to generate β cells in vitro from hPSCs has greatly improved during the last decade; however, despite significant progress, direct differentiation of functionally mature human β cells in culture remains a challenge. As these differentiation protocols have been extensively informed by developmental studies in rodent models, we examined RA signaling, a crucial mediator of early pancreas formation, in the differentiation of pancreatic endocrine cells in both mice and in the differentiation of human β-like cells. Inhibition of the RA pathway in NEUROG3+ mouse and human pancreatic endocrine progenitors resulted in defective β cell production and Ins expression, phenotypes that are at least partially due to the derepression of the Wnt signaling pathway. This study also identified a novel role for RA signaling in the repression of δ cell gene expression. Taken together, these data demonstrate a conserved and previously unappreciated role of RA signaling during pancreatic endocrine development. These results will inform β cell differentiation conditions to facilitate the generation of functionally mature human β cells in vitro, and advance our understanding of pancreatic endocrine development.
MATERIALS AND METHODS
Animal models
Mice were maintained under protocol 00045 as approved by the University of Colorado Denver Institutional Animal Care and Use Committee. All animals were bred on a mixed C57Bl6/129SV genetic background and group housed by sex with up to five siblings in each cage with constant access to food and water at room temperature (22°C). Cages were changed once every 2 weeks and regularly monitored for virus and parasite infection, which were never present during this study. Euthanasia was performed by CO2 inhalation and by cervical dislocation after asphyxiation as a secondary method of euthanasia. For timed matings, the identification of a vaginal plug in the morning was defined as E0.5. The male was removed from the cage and the female was monitored to ensure pregnancy until sacrifice. All mice and embryos were genotyped with primers listed in Table S4 using standard PCR with Go Taq DNA Polymerase Mastermix (Promega). All mice used are available from Jackson Laboratories: Tg(Neurog3-cre)C1Able/J (005667; RRID:IMSR_JAX:005667), B6.FVB-Tg(Pdx1-cre)6Tuv/J (014647 RRID:IMSR_JAX:014647); Gt(ROSA)26Sortm1(RARA*)Soc/HsvJ (029812 RRID:IMSR_JAX:029812) and Tg(Pdx1-cre/Esr1*)#Dam/J (024968 RRID:IMSR_JAX:024968).
hPSC culture
All hPSC studies were performed using the authenticated H1 human embryonic stem cell (hESCs) line (which is routinely checked for contamination) (Thomson et al., 1998). hPSC cell lines were cultured on 0.1% gelatin and irradiated mouse embryonic fibroblast feeder cells in Dulbecco's Modified Eagle's Medium (DMEM)/F12 supplemented with 2 mM of glutamine, and 15% KnockOut Serum Replacement [1× non-essential amino acids, penicillin/streptomycin, 0.1 mM β-mercaptoethanol and 10 ng/ml of basic fibroblast growth factor (bFGF)]. This hPSC medium was changed every day. Cells were passaged when they reached 80% confluence, approximately every 4 days, using TrypLE at a 1:6 ratio. In all hPSC cultures, 5 μM Rho-associated protein kinase (ROCK) inhibitor Y-27632 (Selleck Chemicals, S1049) was only added into the culture media for ∼18 h when passaging or thawing hPSCs.
Embryoid body generation
hPSCs were incubated with Accutase solution for 7 min at 37°C, transferred to a 50 ml Falcon tube and washed twice using 40 ml of DMEM-F12. Then, 5.5 million cells were resuspended in 5 ml of hPSC medium with 1 μM of ROCK inhibitor and plated in one well of an ultra-low attachment six-well cell culture plate. The plate was placed on a 100 rpm orbital shaker inside a 37°C incubator with 5% CO2. The cells form embryoid bodies overnight and were fed for 2 days using hPSC medium. After 2 days, the medium was removed and replaced with pancreatic differentiation medium for day 0.
Pancreatic differentiation from hPSCs
Pancreas differentiation was initiated on the hPSC embryoid bodies with day 0 media containing Roswell Park Memorial Institute growth media (RPMI) supplemented with 3 μM Chir99021 and 100 μg/ml activin A. On day 1, the media were changed to RPMI with 100 μg/ml activin A, 0.3 μM Chir99021 and 5 μg/ml bFGF. On day 2, the media were changed to serum-free defined medium with 100 μg/ml activin A. From days 3 to 5, cells were fed with DMEM-F12 containing 0.25 mM ascorbic acid, 50 ng/ml FGF7 and 1.25 μM IWP2. Day 6-8 media contained DMEM high glucose (5 g/l) supplemented with 1:100 B27 without RA, 1× GlutaMAX, 0.25 mM ascorbic acid, 1:200 ITS-X, 50 ng/ml FGF7, 0.5 μM SANT-1, 1 μM RA, 100 nM LDN-193189 and 500 nM phorbol. The media for days 9 to 11 consisted of DMEM high glucose (5 g/l) supplemented with 1:100 B27 without RA, 1× GlutaMAX, 0.25 mM ascorbic acid, 1:200 ITS-X, 2 ng/ml FGF7, 0.5 μM SANT-1, 0.1 μM RA, 200 nM LDN-193189 and 250 nM phorbol. From days 11 to 13, the media were changed to MCDB131 supplemented with 20 mM glucose, 2% fetal bovine serum (FBS), 1× GlutaMAX, 1:200 ITS-X, 10 μg/ml heparin, 10 μM zinc sulfate, 0.5 μM SANT-1, 0.05 μM RA, 200 nM LDN-193189, 1 μM T3 and 10 μM ALK5i II. For experiments with the pan-RAR inhibitor, 2 μM of AGN193108 was added to the media from day 11 onwards. Cells were harvested on day 13 for flow cytometry analysis and RNA collection using 0.25% Trypsin for 5 min. From day 13 to day 28, cells were fed every other day with media that contained MCDB131 with 20 mM glucose, 2% FBS, 1× GlutaMAX, 1:200 ITS-X, 10 μg/ml heparin, 10 μM zinc sulfate, 200 nM LDN-193189, 1 μM T3, 10 μM ALK5i II and 100 nM GSIS XX. Cells were harvested on day 28 (β-like stage) for flow cytometry analysis and RNA collection.
Immunofluorescence
Tissues were fixed in 4% paraformaldehyde (PFA) for 4 h at 4°C, washed in ice-cold 1× PBS, and incubated in 30% sucrose overnight at 4°C. The next day, samples were incubated in 50% optimum cutting temperature compound (OCT) (in sucrose) for 15 min, 100% OCT for 15 min, and frozen on dry ice. Blocks were sectioned (10-12 μm) for immunofluorescence. All antibodies used in this study are listed in Table S3. In brief, samples were blocked in 2% normal donkey serum for 30 min and incubated with primary antibodies overnight at 4°C. The next day, they were washed in PBS-T and incubated in secondary antibodies for 1-3 h, washed in PBS-T, incubated in DAPI for 10 min, washed in PBS-T and mounted using hard-set Vectashield. For prolonged storage for more than 2 weeks, nail polish was used to seal the edges of coverslips and slides were stored at 4°C. For TUNEL staining, the TMR red in situ Cell Death Detection Kit (Millipore, 12156792910) was used following the manufacturer's instructions, prior to antibody staining for insulin.
Morphometric analysis
For protein quantification, the pancreas was completely sectioned and every 10th slide was stained and quantified (five slides for E16.5 and ten slides for P2). All E16.5 embryos were sectioned without further dissection of the pancreas. At P2, the pancreas, stomach, duodenum and intestines were isolated. Images were obtained at 20× magnification using a Leica DM5500 B microscope. For both stages, DAPI-positive pancreas area was measured and was not significantly different between control and mutant samples. At E16.5, individual hormone-positive cells were counted manually using FIJI or Adobe Photoshop counting tools. P2 samples were quantified by hormone-positive area/DAPI-positive area using a Matlab program to detect positively stained pixels in each fluorescent channel. Antibody information can be found in Table S3. All quantification was performed blinded to genotype.
RNA extraction and RT-qPCR in mice
Whole pancreata were collected and total RNA extractions were completed using the RNA Easy Mini Kit and eluted in 30 μl RNAse-free H2O. RNA (200 ng) was used to generate cDNA via the iScript cDNA Synthesis Kit (Bio-Rad). The cDNA was then diluted in RNAse free H2O to 1 ng/μl, and 4 ng cDNA was used in each qPCR reaction using the SsoAdvanced Universal Probes Supermix (Bio-Rad) with TaqMan probes (Table S3) and run on the Bio-Rad CFX96 Real-Time PCR Detection System. All expression levels were normalized to Actb and quantified using the 2−ΔΔCt method; control samples were averaged and set to 1 to determine gene expression changes in mutants.
RNA-seq
RNA was extracted as described above and quality was checked by obtaining RNA integrity numbers (RINs) using the eukaryote Total RNA Nano Kit for the Agilent 2100 Bioanalyzer. Samples with RINs >8.0 were subjected to RNA-seq using the NovaSeq 6000 for paired-end sequencing (2×150), from poly-A selected total RNA, by the University of Colorado Cancer Center Genomics and Microarray Core Facility. Reads were quality checked using FastQC and subjected to Cutadapt to trim adapters. These reads were then aligned to Mm10 using HISAT2 (Kim et al., 2015). Aligned reads were mapped to genes using Ensembl (Mus_musculus.GRCm38.95.gtf) and HTseq-count. Files were converted from SAM to BAM format and aligned to the genome using SAMtools (Li et al., 2009). Differential gene expression was examined using the DESeq2 package (Love et al., 2014) in RStudio. Gene ontology enrichment analysis was carried out using Panther (Mi et al., 2019).
Putative RARα binding site identification
Transcript locations and sequences of promoter regions defined as 0-500 bp upstream of the transcriptional start site for differentially expressed genes were gathered using the GenomicFeatures package in RStudio (Lawrence et al., 2013). The RStudio package Biostrings (https://bioconductor.org/packages/release/bioc/html/Biostrings.html) was used to search for the JASPER Rara_1 PB0053.1 position weight matrix (PWM) in these promoter proximal sequences. Only motif matches with a quantitative PWM score that was ≥80% of the maximum score were kept for consideration (Wasserman and Sandelin, 2004).
Glucose tolerance tests
Mice were fasted overnight (∼16 h) and blood glucose was measured from tail vein blood for a 0 min time point just before a 2 mg D-glucose/gram mouse mass intraperitoneal injection. Blood glucose was monitored at 15, 30, 45, 60, 90 and 120 min after injection. All blood glucose measurements were completed using the Contour 7151H blood glucose monitor with Contour 7097C blood glucose strips.
RNAscope
RNAscope was performed largely as described in ACD biosciences RNAscope Fluorescent Multiplex Kit Quick Guide (acdbio.com/documents/product-documents). Previously fixed and sectioned E16.5 samples were postfixed at 4°C overnight in 4% PFA. The next morning, slides were washed in deionized water and dehydrated using a series of increasing concentrations of ethanol and baked at 60°C for 10 min before being treated with hydrogen peroxide for 10 min. Antigen retrieval was carried out by boiling sections in target retrieval reagent for 10 s, which were then allowed to cool to room temperature for 5 min. Slides were washed with deionized water, subjected to mild protease digestion for 20 min in a humidified chamber, washed again in deionised water and incubated in prewarmed probes (Table S3) for 2 h at 40°C. Slides were washed with 1× wash buffer and placed in 5× SSC overnight. The next morning, slides were again washed in wash buffer and incubated in AMP1, AMPI2 and AMPI3 solutions for 30 min, 30 min and 15 min, respectively, at 40°C, with washes in wash buffer in between each incubation. To develop signal, we used Opal dyes (Table S3) diluted in multiplex tyramide signal amplification buffer for only the channels we visualized in each experiment. Each channel was developed sequentially by incubating samples in HRP-C1/2/3 for 15 min at 40°C, washed and then incubated in the pre-diluted Opal dye for 30 min at 40°C. Slides were then washed in wash buffer and incubated in HRP blocker for 15 min at 40°C. This solution was removed and slides were again cleaned in wash buffer, then in PBS-T and subjected to antibody staining as described previously, but mounted using ProLong gold mounting media in place of Vectashield. All samples were imaged at 63× on a Zeiss LSM800 confocal microscope.
Flow cytometry
Single cell suspensions were prepared by treating cells with 0.25% Trypsin/EDTA for 3 to 5 min. For intracellular staining, cells were fixed with 1.6% PFA (Electron Microscopy Sciences) for 30 min at 37°C. Cells were washed, permeabilized and stained with 1× saponin buffer (BioLegend). Primary antibodies were diluted to the appropriate concentrations in 100 μl of saponin buffer and cells were stained for 30 min at room temperature. Samples were washed twice using 100 μl saponin buffer and incubated for 30 min using the appropriate secondary antibody. Following staining, cells were resuspended in fluorescence-activated cell sorting buffer (DPBS with 0.1% bovine serum albumin and 0.1% sodium azide). All samples were run on a FACSCanto II or CytoFLEX flow cytometer (Becton Dickinson) and analyzed using the FlowJo (Treestar) software program.
RNA extraction and RT-qPCR in human cells
Cells were lysed using the lysis buffer provided with the PureLink RNA Micro Kit (Invitrogen, 12183-016) and stored at −80°C. To harvest RNA, samples were thawed at 4°C and RNA was extracted using the PureLink RNA Micro Kit following the manufacturer's instructions. RNAse free water (14 μl) was used to resuspend the isolated RNA. cDNA was produced using the SuperScript III First-Strand Synthesis System kit (Invitrogen). qPCR was carried out using a LightCycler 480 II with SYBR Select Master Mix (Invitrogen). For all experiments, TBP (Veazey and Golding, 2011) was used as a housekeeping gene to determine relative gene expression levels.
Quantification and statistical analysis
Graphs and statistical analyses were generated using GraphPad Prism 8. Statistical analyses include n values (biological replicates), statistical tests and significance based on either P-value/P-adjusted value/q-value, which are detailed in the figure legends. For mouse analyses, each n value represents a different animal. Analyses were performed with littermate controls, when available. Gender was not assessed for any embryonic studies but all adult mouse glucose tolerance tests were completed on male mice. The E16.5 RNA-seq experiments were analyzed by the DESeq2 platform that reports both P-value and P-adjusted, with the latter being used as a significance cutoff at Padj≤0.05 as defined by the Benjamini-Hochberg procedure, which corrects for multiple hypothesis testing.
Supplementary Material
Acknowledgements
We thank members of the Sussel lab for helpful feedback on this project, specifically Dr Michelle Guney and Alexandra Theis for critical readings of the manuscript; and Helen Moses and Amira Sheikh for technical assistance. We also thank Dr Julie Siegenthaler (University of Colorado Denver) for generously providing us with the RARdnflox mice. We are grateful for the technical assistance provided for many specialized techniques and analyses presented here: RNA-seq was conducted at the University of Colorado Denver Microarray and Genomics Core at the Cancer Center by Katrina Diener; RNA-sequencing analysis was completed with substantial help from Dr Ruth Singer and the University of Colorado Denver RNA Biosciences Initiative; and RNAscope experiments were performed with the assistance of Dr Laura Hudish.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: D.S.L., C.R., S.X., L.S.; Methodology: D.S.L., S.K., C.R., S.X., P.G., L.S.; Software: O.S., D.G.; Formal analysis: D.S.L., S.K., D.S., E.P.B., E.A.; Investigation: D.S.L., S.K., C.R., D.S., E.P.B., E.A., S.X.; Resources: O.S., C.M., P.G., L.S.; Data curation: D.S.L., O.S., D.G.; Writing - original draft: D.S.L., L.S.; Writing - review & editing: D.S.L., S.K., C.R., P.G., L.S.; Visualization: D.S.L., S.K., P.G., L.S.; Supervision: D.G., C.M., P.G., L.S.; Funding acquisition: D.S.L., P.G., L.S.
Funding
This work was supported by the American Diabetes Association (1-18-PDF-107 to D.S.L.) and the National Institute of Diabetes and Digestive and Kidney Diseases at the National Institutes of Health (R01 DK118155 to P.G. and L.S.; R01 DK082590 to L.S.). Deposited in PMC for release after 12 months.
Data availability
Sequencing data have been deposited in GEO under accession number GSE144953. The MatLab program used for analyses of hormone expression at P2 is available upon request.
Supplementary information
Supplementary information available online at https://dev.biologists.org/lookup/doi/10.1242/dev.189977.supplemental.
Peer review history
The peer review history is available online at https://dev.biologists.org/lookup/doi/10.1242/dev.189977.reviewer-comments.pdf.
References
- Arregi I., Climent M., Iliev D., Strasser J., Gouignard N., Johansson J. K., Singh T., Mazur M., Semb H., Artner I. et al. (2016). Retinol dehydrogenase-10 regulates pancreas organogenesis and endocrine cell differentiation via paracrine retinoic acid signaling. Endocrinology 157, 4615-4631. 10.1210/en.2016-1745 [DOI] [PubMed] [Google Scholar]
- Bonner-Weir S., Trent D. F. and Weir G. C. (1983). Partial pancreatectomy in the rat and subsequent defect in glucose-induced insulin release. J. Clin. Invest. 71, 1544-1553. 10.1172/JCI110910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonney S., Harrison-Uy S., Mishra S., Macpherson A. M., Choe Y., Li D., Jaminet S.-C., Fruttiger M., Pleasure S. J. and Siegenthaler J. A. (2016). Diverse functions of retinoic acid in brain vascular development. J. Neurosci. 36, 7786-7801. 10.1523/JNEUROSCI.3952-15.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonney S., Dennison B. J. C., Wendlandt M. and Siegenthaler J. A. (2018). Retinoic acid regulates endothelial beta-catenin expression and pericyte numbers in the developing brain vasculature. Front. Cell Neurosci. 12, 476 10.3389/fncel.2018.00476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brun P.-J., Grijalva A., Rausch R., Watson E., Yuen J. J., Das B. C., Shudo K., Kagechika H., Leibel R. L. and Blaner W. S. (2015). Retinoic acid receptor signaling is required to maintain glucose-stimulated insulin secretion and β-cell mass. FASEB J. 29, 671-683. 10.1096/fj.14-256743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardenas-Diaz F. L., Osorio-Quintero C., Diaz-Miranda M. A., Kishore S., Leavens K., Jobaliya C., Stanescu D., Ortiz-Gonzalez X., Yoon C., Chen C. S. et al. (2019). Modeling monogenic diabetes using human ESCs reveals developmental and metabolic deficiencies caused by mutations in HNF1A. Cell Stem Cell 25, 273-289.e5. 10.1016/j.stem.2019.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalgin G., Ward A. B., Hao L. T., Beattie C. E., Nechiporuk A. and Prince V. E. (2011). Zebrafish mnx1 controls cell fate choice in the developing endocrine pancreas. Development 138, 4597-4608. 10.1242/dev.067736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Digruccio M. R., Mawla A. M., Donaldson C. J., Noguchi G. M., Vaughan J., Cowing-Zitron C., Van Der Meulen T. and Huising M. O. (2016). Comprehensive alpha, beta and delta cell transcriptomes reveal that ghrelin selectively activates delta cells and promotes somatostatin release from pancreatic islets. Mol. Metab. 5, 449-458. 10.1016/j.molmet.2016.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duester G. (2001). Genetic dissection of retinoid dehydrogenases. Chem-Biol. Interact. 130-132, 469-480. 10.1016/S0009-2797(00)00292-1 [DOI] [PubMed] [Google Scholar]
- Ghyselinck N. B. and Duester G. (2019). Retinoic acid signaling pathways. Development 146, dev167502 10.1242/dev.167502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu G., Dubauskaite J. and Melton D. A. (2002). Direct evidence for the pancreatic lineage: NGN3+ cells are islet progenitors and are distinct from duct progenitors. Development 129, 2447-2457. [DOI] [PubMed] [Google Scholar]
- Huang W., Wang G., Delaspre F., Vitery M. C., Beer R. L. and Parsons M. J. (2014). Retinoic acid plays an evolutionarily conserved and biphasic role in pancreas development. Dev. Biol. 394, 83-93. 10.1016/j.ydbio.2014.07.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson A. T., Wang L., Gillett S. J. and Chandraratna R. A. (1999). High affinity retinoic acid receptor antagonists: analogs of AGN 193109. Bioorg. Med. Chem. Lett. 9, 573-576. 10.1016/s0960-894x(99)00047-5 [DOI] [PubMed] [Google Scholar]
- Kim D., Langmead B. and Salzberg S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357-360. 10.1038/nmeth.3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krentz N. A. J., Lee M. Y. Y., Xu E. E., Sproul S. L. J., Maslova A., Sasaki S. and Lynn F. C. (2018). Single-cell transcriptome profiling of mouse and hESC-derived pancreatic progenitors. Stem Cell Rep. 11, 1551-1564. 10.1016/j.stemcr.2018.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S. and Duester G. (2010). Retinoic acid signaling in perioptic mesenchyme represses Wnt signaling via induction of Pitx2 and Dkk2. Dev. Biol. 340, 67-74. 10.1016/j.ydbio.2010.01.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence M., Huber W., Pagès H., Aboyoun P., Carlson M., Gentleman R., Morgan M. T. and Carey V. J. (2013). Software for computing and annotating genomic ranges. PLoS Comput. Biol. 9, e1003118 10.1371/journal.pcbi.1003118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. and 1000 Genome Project Data Processing Subgroup (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078-2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loudig O., Babichuk C., White J., Abu-Abed S., Mueller C. and Petkovich M. (2000). Cytochrome P450RAI(CYP26) promoter: a distinct composite retinoic acid response element underlies the complex regulation of retinoic acid metabolism. Mol. Endocrinol. 14, 1483-1497. 10.1210/mend.14.9.0518 [DOI] [PubMed] [Google Scholar]
- Love M. I., Huber W. and Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall H., Morrison A., Studer M., Pöpperl H. and Krumlauf R. (1996). Retinoids and Hox genes. FASEB J. 10, 969-978. 10.1096/fasebj.10.9.8801179 [DOI] [PubMed] [Google Scholar]
- Martín M., Gallego-Llamas J., Ribes V., Kedinger M., Niederreither K., Chambon P., Dollé P. and Gradwohl G. (2005). Dorsal pancreas agenesis in retinoic acid-deficient Raldh2 mutant mice. Dev. Biol. 284, 399-411. 10.1016/j.ydbio.2005.05.035 [DOI] [PubMed] [Google Scholar]
- Mclin V. A., Rankin S. A. and Zorn A. M. (2007). Repression of Wnt/β-catenin signaling in the anterior endoderm is essential for liver and pancreas development. Development 134, 2207-2217. 10.1242/dev.001230 [DOI] [PubMed] [Google Scholar]
- Mi H., Muruganujan A., Huang X., Ebert D., Mills C., Guo X. and Thomas P. D. (2019). Protocol Update for large-scale genome and gene function analysis with the PANTHER classification system (v.14.0). Nat. Protoc. 14, 703-721. 10.1038/s41596-019-0128-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molotkov A., Molotkova N. and Duester G. (2005). Retinoic acid generated by Raldh2 in mesoderm is required for mouse dorsal endodermal pancreas development. Dev. Dyn. 232, 950-957. 10.1002/dvdy.20256 [DOI] [PubMed] [Google Scholar]
- Morgan C. A., Parajuli B., Buchman C. D., Dria K. and Hurley T. D. (2015). N,N-diethylaminobenzaldehyde (DEAB) as a substrate and mechanism-based inhibitor for human ALDH isoenzymes. Chem. Biol. Interact. 234, 18-28. 10.1016/j.cbi.2014.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osei-Sarfo K. and Gudas L. J. (2014). Retinoic acid suppresses the canonical Wnt signaling pathway in embryonic stem cells and activates the noncanonical Wnt signaling pathway. Stem Cells 32, 2061-2071. 10.1002/stem.1706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Öström M., Loffler K. A., Edfalk S., Selander L., Dahl U., Ricordi C., Jeon J., Correa-Medina M., Diez J. and Edlund H. (2008). Retinoic acid promotes the generation of pancreatic endocrine progenitor cells and their further differentiation into β-cells. PLoS ONE 3, e2841 10.1371/journal.pone.0002841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagliuca F. W., Millman J. R., Gürtler M., Segel M., Van Dervort A., Ryu J. H., Peterson Q. P., Greiner D. and Melton D. A. (2014). Generation of functional human pancreatic β cells in vitro. Cell 159, 428-439. 10.1016/j.cell.2014.09.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan F. C., Brissova M., Powers A. C., Pfaff S. and Wright C. V. E. (2015). Inactivating the permanent neonatal diabetes gene Mnx1 switches insulin-producing β-cells to a δ-like fate and reveals a facultative proliferative capacity in aged β-cells. Development 142, 3637-3648. 10.1242/dev.126011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rankin S. A., Kormish J., Kofron M., Jegga A. and Zorn A. M. (2011). A gene regulatory network controlling hhex transcription in the anterior endoderm of the organizer. Dev. Biol. 351, 297-310. 10.1016/j.ydbio.2010.11.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezania A., Bruin J. E., Riedel M. J., Mojibian M., Asadi A., Xu J., Gauvin R., Narayan K., Karanu F., O'neil J. J. et al. (2012). Maturation of human embryonic stem cell-derived pancreatic progenitors into functional islets capable of treating pre-existing diabetes in mice. Diabetes 61, 2016-2029. 10.2337/db11-1711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roa L. A., Bloemen M., Carels C. E. L., Wagener F. A. and Von Den Hoff J. W. (2019). Retinoic acid disrupts osteogenesis in pre-osteoblasts by down-regulating WNT signaling. Int. J. Biochem. Cell Biol. 116, 105597 10.1016/j.biocel.2019.105597 [DOI] [PubMed] [Google Scholar]
- Rosselot C., Spraggon L., Chia I., Batourina E., Riccio P., Lu B., Niederreither K., Dollé P., Duester G., Chambon P. et al. (2010). Non-cell-autonomous retinoid signaling is crucial for renal development. Development 137, 283-292. 10.1242/dev.040287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rovira M., Huang W., Yusuff S., Shim J. S., Ferrante A. A., Liu J. O. and Parsons M. J. (2011). Chemical screen identifies FDA-approved drugs and target pathways that induce precocious pancreatic endocrine differentiation. Proc. Natl. Acad. Sci. USA 108, 19264-19269. 10.1073/pnas.1113081108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon N., Vanderhooft J., Straubhaar J., Mueller J., Chawla R., Zhou Q., Engquist E. N., Trapnell C., Gifford D. K. and Melton D. A. (2019). Wnt signaling separates the progenitor and endocrine compartments during pancreas development. Cell Rep. 27, 2281-2291.e5. 10.1016/j.celrep.2019.04.083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stafford D. and Prince V. E. (2002). Retinoic acid signaling is required for a critical early step in zebrafish pancreatic development. Curr. Biol. 12, 1215-1220. 10.1016/S0960-9822(02)00929-6 [DOI] [PubMed] [Google Scholar]
- Thomson J. A., Itskovitz-Eldor J., Shapiro S. S., Waknitz M. A., Swiergiel J. J., Marshall V. S. and Jones J. M. (1998). Embryonic stem cell lines derived from human blastocysts. Science 282, 1145-1147. 10.1126/science.282.5391.1145 [DOI] [PubMed] [Google Scholar]
- Tiyaboonchai A., Cardenas-Diaz F. L., Ying L., Maguire J. A., Sim X., Jobaliya C., Gagne A. L., Kishore S., Stanescu D. E., Hughes N. et al. (2017). GATA6 plays an important role in the induction of human definitive endoderm, development of the pancreas, and functionality of pancreatic β cells. Stem Cell Rep. 8, 589-604. 10.1016/j.stemcr.2016.12.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veazey K. J. and Golding M. C. (2011). Selection of stable reference genes for quantitative rt-PCR comparisons of mouse embryonic and extra-embryonic stem cells. PLoS One 6, e27592 10.1371/journal.pone.0027592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vethe H., Ghila L., Berle M., Hoareau L., Haaland O. A., Scholz H., Paulo J. A., Chera S. and Raeder H. (2019). The effect of Wnt pathway modulators on human iPSC-derived pancreatic beta cell maturation. Front. Endocrinol. 10, 293 10.3389/fendo.2019.00293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasserman W. W. and Sandelin A. (2004). Applied bioinformatics for the identification of regulatory elements. Nat. Rev. Genet. 5, 276-287. 10.1038/nrg1315 [DOI] [PubMed] [Google Scholar]
- Yasugi H., Mizumoto R., Sakurai H. and Honjo I. (1976). Changes in carbohydrate metabolism and endocrine function of remnant pancreas after major pancreatic resection. Am. J. Surg. 132, 577-580. 10.1016/0002-9610(76)90346-9 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.