Abstract
Microbial interactions play an essential role in aquatic ecosystems and are of the great interest for both marine and freshwater ecologists. Recent development of new technologies and methods allowed to reveal many functional mechanisms and create new concepts. Yet, many fundamental aspects of microbial interactions have been almost exclusively studied for marine pelagic and benthic ecosystems. These studies resulted in a formulation of the Black Queen Hypothesis, a development of the phycosphere concept for pelagic communities, and a realization of microbial communication as a key mechanism for microbial interactions. In freshwater ecosystems, especially for periphyton communities, studies focus mainly on physiology, biodiversity, biological indication, and assessment, but the many aspects of microbial interactions are neglected to a large extent. Since periphyton plays a great role for aquatic nutrient cycling, provides the basis for water purification, and can be regarded as a hotspot of microbial biodiversity, we highlight that more in-depth studies on microbial interactions in periphyton are needed to improve our understanding on functioning of freshwater ecosystems. In this paper we first present an overview on recent concepts (e.g., the “Black Queen Hypothesis”) derived from state-of-the-art OMICS methods including metagenomics, metatranscriptomics, and metabolomics. We then point to the avenues how these methods can be applied for future studies on biodiversity and the ecological role of freshwater periphyton, a yet largely neglected component of many freshwater ecosystems.
Keywords: freshwater, lake periphyton, microbial interactions, Black Queen Hypothesis, OMICs tools
Introduction
The term periphyton has a long history and describes a wide range of organisms living on various submerged substrates. Perhaps the most used and popular definition of periphyton communities was formulated by Wetzel (1983): “Periphyton is a complex community of microbiota (algae, bacteria, fungi, animals, inorganic, and organic detritus) that is attached to substrata. The substrata can be inorganic or organic, living or dead.” Similarly, at present, the term “periphyton” is applied to all forms of organismic structures (except macrophytes), which are growing on submerged materials, whereby the main emphasis is set on the algal component (Håkanson and Boulion, 2002; Vadeboncoeur et al., 2006). In microbial ecology the term biofilm is widely used and means microbial attachment to any kind of substrate mediated by an extracellular matrix. In aquatic ecosystems, biofilms include a wide variety of microbial organisms including microalgae (Donlan, 2002). Thereby, the term periphyton also includes macroscopic organisms and can be used more widely. In this paper, we provide an overview on studies performed on microbial organisms attached to any kind of substrate in aquatic ecosystems, and thus will use the term periphyton for the community of micro- and macroorganisms attached to submerged surfaces (with emphasis on microalgae and their associated bacteria) as well as the term biofilm for the whole array of microorganisms (e.g., Bacteria, Protista, fungi, and microalgae, etc.). In cases where the authors of the reviewed papers have used term biofilm synonymously for the attached community of auto- and heterotrophic microorganisms, we also use this term in the context of our above given definition.
Periphyton research has a long history and resulted in numerous biodiversity studies on (i) periphyton and its application as a tool for biological indication and assessment (Schneider and Lindsrom, 2011; Vadeboncoeur et al., 2011; Kelly, 2013; Cantonati and Lowe, 2014; DeNicola and Kelly, 2014), (ii) the role of periphytic communities in primary production (Winberg, 1960, 1980; Hill et al., 1992; Håkanson and Boulion, 2002; Vadeboncoeur and Steinman, 2002; Vadeboncoeur et al., 2011, 2014), (iii) stoichiometry (Hillebrand and Sommer, 1999; Hillebrand and Kahlert, 2001; Fitter and Hillebrand, 2009), and (iv) nutrient cycling (Toet et al., 2003; Teissier et al., 2007; Kalscheur et al., 2012). Some researches had noted a cultural and ecological importance of periphyton communities and regard periphyton in multiple ways, e.g., as solar-powered biogeochemical reactors, biogenic habitats, hydraulic roughness elements, early warning systems for environmental degradation, and troves of biodiversity (Larned, 2010). Therefore, periphyton is widely used as a model object for studies on effects of abiotic factors (physical disturbances, limiting nutrients, etc.), competitive interactions, and the significance of periphyton communities in carbon and nutrient cycling in aquatic food webs (e.g., Hansson, 1992; Hillebrand and Sommer, 1999; Holomuzki et al., 2010; Larned, 2010; Mulholland and Webster, 2010). An awareness of periphytic algal communities had increased after the approval of the EU Water Framework Directive (WFD; European Union, 2000). However, the most studies on freshwater periphyton have been carried out in streams and much less has been done in lakes (Cantonati and Lowe, 2014; DeNicola and Kelly, 2014; Kahlert and Gottshalk, 2014).
Analyses of biodiversity in large lakes have shown that the majority of species, including many endemic ones, occur in the littoral and profundal zones. This pattern is mainly caused by the high substrate heterogeneity, i.e., its physical complexity provides numerous niches that allow for co-existence of a high number of species. Thus, periphyton communities contribute considerably to biodiversity in freshwater ecosystems (Vadeboncoeur et al., 2011). Recent studies have shown that, particularly in shallow lakes, periphytic primary production is relatively high (Håkanson and Boulion, 2002; Vadeboncoeur et al., 2006, 2011, 2014; Brothers et al., 2013) and in some lakes it may even contribute to more than 80–90% of the total primary production (Wyatt et al., 2014; Vesterinen et al., 2016). Besides its considerable role for primary productivity, periphyton biofilms are also “hot-spots” for numerous biogeochemical processes, including denitrification, and thus play a crucial role in nutrient remineralization and element cycling (Toet et al., 2003; Teissier et al., 2007; Kalscheur et al., 2012).
Since recently there is an increasing interest in studies on the ecological role of periphytic communities, which is related to the use of periphyton for water purification due to its binding of toxic substances (Yang et al., 2016) as well as interactions with phytoplankton, macrophytes and macrobenthos (Do Nascimento Filho et al., 2019). Absolutely new direction in biofilm studies is colonization and degradation of microplastics (Kesy et al., 2019; Amaral-Zettler et al., 2020).
Numerous papers have been published in regard to the coupling between auto- and heterotrophs in periphyton communities (Hepinstall and Fuller, 1994; Rier and Stevenson, 2002; Scott et al., 2008; Rusanov et al., 2009). These studies revealed that microbial coupling depends to a large extent on environmental variables, such as nutrient fluxes (Tank and Dodds, 2003; Carr et al., 2005; Rusanov et al., 2009), light intensity (Roeselers et al., 2007), and availability of dissolved organic carbon (DOC; Sobczak, 1996; Rier and Stevenson, 2002), etc. During the past decades some physiological aspects of interactions between auto- and heterotrophs in freshwater periphytic biofilms have been studied (Bruckner et al., 2008, 2011; Allen et al., 2015), however, many mechanistic aspects of interactions in freshwater periphyton communities are still unknown and need to be examined in more details (Allen et al., 2015).
Recently, relevant protocols describing peculiarities of material collection, preservation, and extraction for molecular studies have been developed and assessed for freshwater habitats (Zimmermann et al., 2011; Paulsen and Holmes, 2014). Yet, despite the relatively high number of biodiversity studies on freshwater periphyton, molecular data allowing for the development of the new mechanistic concepts for studying function and hence ecological role of periphyton are still missing (Tsementzi et al., 2014; Pascault et al., 2015; Lindemann et al., 2017). Some studies on microbial interactions in marine planktonic and benthic communities allowed to develop several new fundamental concepts, e.g., about chemical signaling, quorum sensing, and inter-kingdom communication (Waters and Bassler, 2005; Grossart, 2010; Amin et al., 2015; Zhou et al., 2016), the Black Queen Hypothesis (Morris et al., 2012), phycosphere (e.g., Seymour et al., 2017), microbial cross-feeding (Smith et al., 2019), and other aspects.
Our intention is to highlight the most recent concepts in microbial ecology with reference to the data obtained for marine and freshwater planktonic and benthic communities. We set a particular focus on microbial interactions to increase our understanding on the functional role of periphyton in lakes. A key question may arise, i.e., whether it is suitable to talk about freshwater periphyton when relying on data from other aquatic communities? In this paper we mainly focus on the role of algal-bacterial interactions in periphytic communities. It is well known that some algal phyla (i.e., Diatoms) can colonize the majority of aquatic habitats. Studies on algal-bacterial associations from different habitats have shown a major similarities in interactions between alga and bacteria, e.g., mutual control and selection of bacterial strains (Grossart et al., 2005; Burke et al., 2011a, b; Krohn-Molt et al., 2017; Stock et al., 2019). Since there are not many studies conducted on periphythic communities, we suppose that results and hypotheses obtained from other habitats can give us future directions for periphyton studies. We believe that application of new molecular methods—as has been done in other fields of aquatic ecology—will open up a plethora of new perspectives to studying numerous fundamental mechanisms of microbial activity and interactions in freshwater periphyton. For better guidance, in Figure 1 and Table 1 we have summarized the most prominent interactions and processes in freshwater periphyton and suggest several state of the art methods for their in-depth investigation.
FIGURE 1.
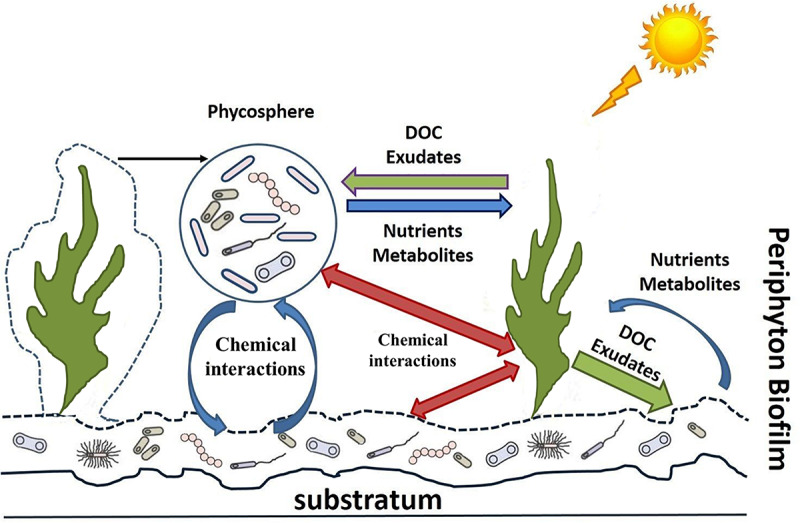
Microbial interactions in periphyton.
TABLE 1.
Overview of methods which can be used for investigation of major functional periphyton attributes.
| Type of interacting organism | Function | Methods of investigation | Achievements |
| Autotrophs | DOC excretion | Wet oxidation, High temperature combustion, etc. | Data for ecosystem modeling |
| Synthesis and excretion of exudates | Various biochemical methods, Metatranscriptomic and metabolomic analyses, Co-culturing experiments | Functional response, Metabolic interconnectedness | |
| Metabolic reactions | Combination of various OMICs tools | Functional response, Metabolic interconnectedness | |
| Chemical signaling and response | Number of chemical and biochemical methods, Combination of OMICs tools, Co-culturing experiments | Intra- and interspecies interactions | |
| Uptake of nutrients and bacterial metabolites (e.g., vitamins) | Metatranscriptomic and metabolomic analysis | Niche partioning, auxotrophy, Metabolic interconnectedness with bacteria | |
| Response on bacterial exudates and signaling molecules | Metatranscriptomic and metabolomic analysis | Alga-bacteria interactions | |
| Heterotrophs | Loss-of-function mutations | Metagenomic and metatranscriptomic analyses | Adaptive gene loss, Cross-feeding |
| Chemical signaling and response | Combination of OMICs methods, Co-culturing experiments | Alga-bacteria interactions, Interspecific interactions | |
| Metabolic reactions | Combination of OMICs methods | Cross-feeding, Interspecies interactions | |
| Biogeochemical reactions | Metatranscriptomic analysis, Various chemical and biochemical methods | Importance of community in biogeochemical cycles | |
| Response on autotrophic exudates and metabolites | Co-culturing experiments, Combination of OMICs methods | Algal-bacteria interactions |
Interactions Between Auto- and Heterotrophs in Aquatic Microbial Communities and Concepts Proposed
Principles and mechanisms of microbial interactions have been studied intensively for planktonic communities and cultures. Based on recent studies on microbial interactions in plankton, Grossart (2010) proposed the concept of an intertwined microbial network between free-living and surface-attached (particle-associated) bacteria, where surface associated bacteria may play a considerable role for nutrient cycling and biomass production. Later studies of free-living and particle-associated bacteria revealed that the connectivity between these communities increased along nutrient gradients (Hu et al., 2020). Also, it was shown that interactions in particle-associated community are stronger, than in free-living (Zhang et al., 2016; Hu et al., 2020). Microbial interactions mediated by chemical substances, e.g., allelopathy, seem to be a widespread strategy in both planktonic and benthic communities (Leflaive and Ten-Hage, 2007, 2009). So far, this concept has been frequently studied, in particular for interactions of pelagic bacteria with other organisms in pelagic environments, yet, only some studies aimed to decipher the connection of autotrophic periphyton with pelagic communities (Do Nascimento Filho et al., 2019; Dos Santos et al., 2020).
Unlike phytoplankton, biofilms represent specific organismic communities embedded in an extracellular polysaccharide matrix providing a favorable environment for short-distance microbial interactions (Jüttner, 1999; Battin et al., 2007). Studies on microbial biofilms and the main ecological concepts have been previously reviewed by Battin et al. (2007) indicating that biofilms should be viewed as complex microbial landscapes representing interconnected components of the larger landscape they colonize. Battin et al. (2007) mainly addressed the bacterial component of microbial biofilms. Later, in stream ecosystems, biofilms have been called “jungles” providing numerous ecological niches for a high number of organisms such as algae, protozoa, nematodes and even some insects, which are “among the top consumers in the stream biofilms” (Battin et al., 2016). At the same time, both local environment and biotic interactions act as drivers for local community composition (Rendueles and Ghigo, 2015). We suppose that aside of various close interactions between microbial organisms in biofilms, periphyton communities form an intertwined network with free-living bacteria (Grossart, 2010) and dead organic matter (Rusanov et al., 2009) in the surrounding environment.
Initially, in aquatic ecology, the “phycosphere” concept includes interactions between auto- and heterotrophic microorganisms, analogous to the rhizosphere concept in soils. Thereby, the phycosphere depicts an immediate region surrounding an algal cell, chain, or colony and represents a zone “in which bacterial growth is stimulated by the extracellular products of the alga” (Bell and Mitchell, 1972). Later studies on plankton confirmed that phytoplankton is surrounded by a diffusive boundary layer, in which excreted phytoplankton molecules accumulate in concentrations, high enough to be detected by bacteria allowing for intense communication and interaction with the algae (Karp-Boss et al., 1996; Jonsson et al., 2009; Amin et al., 2012). In the review of Ramanan et al. (2016), the phycosphere is compared with “an oasis for heterotrophic bacteria, constituting high concentrations of excreted organic carbon in the vast oligotrophic surroundings of both the ocean and inland waters.” At the same time, the authors point out that the phycosphere has been greatly neglected as a distinctive habitat for bacteria (Ramanan et al., 2015, 2016). Extending the phycosphere concept to the algal component of periphyton and taking into account that each species in these communities is in close contact with each other, the question arises: Do we regard the phycosphere of periphytic algae as a separate, specific ecological habitat occupied by distinct heterotrophic microbial communities? According to the concept proposed by Grossart (2010) this microbial community is interconnected by chemical means with its own members and with the surrounding periphytic and planktonic communities. Moreover, since in periphyton all organisms are in the most intimate contact, it leads to tight interactions among them. There are two main types of interactions: competitive and cooperative (Freilich et al., 2011). The cooperative concept lately was developed via the Black Queen Hypothesis, which proposes that cooperation can be a result of coevolution of coexisting bacteria and adaptive gene loss (Morris et al., 2012). This concept is discussed in more detail below.
Analysis of bacterial strains, isolated from more than 42 micro- and macroalgal species from marine and freshwater environments, revealed that bacterial species and strains having similar metabolic functions colonize similar algal taxa or groups (Burke et al., 2011a, b; Goecke et al., 2013). These metabolic functions imply the ability to degrade the complex polymers (such as polysaccharides), avoid competition, and to form mutualistic relationships with algae (Ramanan et al., 2015). In turn, this requires sharing the metabolic potentials of the interacting organisms and a complex signaling system (Ramanan et al., 2016).
Taking into account both the specificity of bacterial strains and the ability of algae to synthetize a high spectrum of active substances (Bazes et al., 2006; Bruckner et al., 2011), we may propose that algae controlling their phycosphere and attributed heterotrophic communities may represent key species or group of species defining the fate and function of microbial periphyton communities. In turn, bacterial communities influence many vital functions and the growth of algal hosts (Amin et al., 2012, 2015). Recent studies on diatom Cylindrotheca closterium and bacterial strains have shown a high specificity of bacteria and their algal hosts. Moreover, co-cultivation of this diatom with bacterial strains from a “foreign” algal host showed effects ranging from neutralism to antagonism with suppressing of algal growth (Stock et al., 2019). These results confirm the high specificity between algal host and its associated bacteria and suggest that algae can function as key-species for their associated bacterial communities.
Until recently, the methods for studying periphyton have not allowed us to dig into functional responses and interactions among individual members of periphyton communities. However, latest applications of metagenomics and metatranscriptomics together with metabolic reconstructions revealed many details of microbial interactions and their functional responses (Lindemann et al., 2017).
For example, synthesis of allelopathic substances, e.g., antibiotics and other inhibitors, by members of the biofilm community plays an essential role in determining microbial diversity and structure (Lenski and Riley, 2002; Burmolle et al., 2006). Studies on biofilms of different origin and bacterial associations with eukaryotic hosts, e.g., human pathogens (Harriott and Noverr, 2009; Lopes et al., 2012; Korgaonkar et al., 2013), as well as aquatic microbial and epiphytic communities (Burmolle et al., 2006; Allen et al., 2015; Kouzuma and Watanabe, 2015; Dittami et al., 2016), have revealed both synergistic and antagonistic inter-species interactions. It is well-known that bacteria are able to stimulate growth, morphogenesis, and zoosporogenesis of algae (Marshall et al., 2006; Goecke et al., 2013) as well as the synthesis of algal extracellular compounds (Bruckner et al., 2011). Bacteria serve as an ultimate source of cobalamin for vitamin B dependent algae (Cole, 1982; Kazamia et al., 2012) and promote algal growth via excretion of substances with hormonal nature (De-Bashan et al., 2008). In turn, DOC, excreted by algae serve as nutrients for alga-associated heterotrophic microbial community (Kazamia et al., 2012; Wyatt et al., 2014). Associations with bacteria can greatly increase the ability of algae to survive in unfavorable conditions. For example, studies on cell cultures of the brown alga Ectocarpus had shown that presence of specific associated bacteria increase the tolerance of the algal host to low salinity (Dittami et al., 2016). Besides synergistic interactions, a multispecies community inevitably faces competitive antagonistic interactions among species (Freilich et al., 2011; Allen et al., 2015; Yuan and Meng, 2020). Last, studies have shown that in the absence of carbon limitation, heterotrophs can out-compete algae for nutrients and thus suppress algal growth (Wyatt et al., 2019). Studies on freshwater periphyton revealed a frequent presence of allelopathic compounds within microbial biofilms (Jüttner and Wu, 2000; Leflaive et al., 2008) and the capability of many benthic species to produce various inhibiting substances with a highly variable chemical nature (Hagmann and Jüttner, 1996; Hirata et al., 2003; Pouvreau et al., 2007; Vanelslander et al., 2012). These substances include antibiotics and enzymes, which can be transported to competitors with extracellular vesicles and affect cellular differentiation, protein synthesis, and inhibit cell-to-cell signaling (Stubbendieck and Straight, 2016). Studies on macro- and microalgae indicate that several specific algal compounds such as fatty acids including α-linolenic (18:3 n–3) and eicosapentaenoic acids (22:5 n–3) have a strong antibacterial activity as well (Debois et al., 2010; Smith et al., 2010; Allen et al., 2015). Inhibiting the growth of both bacteria and other algae may control and shape the associated microbial communities (Allen et al., 2015).
Thus it is not surprising that studies on various microbial biofilms reveal that cell-to-cell chemical signaling plays a crucial role in a shaping of the biofilm community, e.g., by controlling processes such as invasion and colonization (Ben Jacob et al., 2004; Burmolle et al., 2006; Battin et al., 2007). Since many signaling molecules are small, they can diffuse freely through the highly hydrated biofilm matrix, depending on their hydrophobicity (Lovett et al., 2005). However, the extracellular polymeric substances have a high chemical heterogeneity creating environmental gradients which may serve as signal filters (Neu et al., 2001) or fluid channels (Espinosa-Urgel, 2003). At the same time, flow of water may dilute the pool of signaling molecules or transfer them to the more distant biofilm areas (Battin et al., 2007) resulting in chemical interactions with the surrounding environment.
Enriched cultures of planktonic bacterial communities (Garcia et al., 2015) have been studied via metatranscriptome analysis and revealed the many details of chemical interactions which may also occur in periphyton communities. However, peculiarities of periphytic communities lead to some technical difficulties. The tight relationship between bacteria and algal host (Koedooder et al., 2019), for example, render it difficult to obtain axenic cultures. Although the data on associated bacteria of green microalgae have shown that only a few species of bacteria have tight relationship with their algal host (Krohn-Molt et al., 2017), the high functional interconnectedness in periphytic communities likely result in the presence of a high number of associated, yet, non-culturable bacteria. This may result in methodical difficulties in selecting the right experimental bacterial strains, experimental design, and correct data processing. A possible way to solve these difficulties could be the joint usage of metagenomics, single cell genomics, when single the cell is isolated from the algal community together with extended microbiome and metatranscriptomic analyses (Krohn-Molt et al., 2013, 2017). Since recently, data sets of metagenomes of natural populations (i.e., biofilms) have increased significantly (Feio et al., 2020) facilitating improved data processing.
Yet, fundamental aspects of inter-species interactions among bacteria as well as among pro- and eukaryotic organisms in periphyton are still poorly understood, and their studies require the use of new methods and concepts including state-of-the-art OMICs approaches, specifically designed for complex and intertwined periphyton communities.
“Black Queen Hypothesis”: Can We Apply it to Freshwater Periphyton?
Recent applications of new molecular methods for studies on bacterial interactions of free-living lineages of oceanic plankton have resulted in the proposition of the so-called “Black Queen Hypothesis” (BQH). It suggests that some species of free-living bacteria are dependent on functions of co-occurring bacteria due to loss of a number of genes during reductive genome evolution. The hypothesis of cross-feeding assigns co-existing species as “helpers” and “beneficiaries.” The scenarios of co-existence in microbial communities are diverse and include a number of strategies (Elias and Banin, 2012). Recently bacterial cross-feeding assigned to 4 different types, depending on the strategy of interacting bacteria (Smith et al., 2019). They include interactions between self-sufficient organisms as well as auxotrophy, with and without gene loss. Thereby, the interacting strains cross-feed on metabolites of each other and gain benefit directly or indirectly providing benefit via reduction of the required energy costs (Smith et al., 2019).
Unlike many theories of coevolution, e.g., the Red Queen Hypothesis (Van Valen, 1973), the BQH proposes that relationships between helpers and beneficiaries doesn’t necessarily arise from direct interactions, but beneficiaries can simply stop a costly function that is provided by their helpers (loss-of-function mutation; Morris et al., 2012). This new paradigm caused an intense discussion in the scientific community and suggests that bacteria often form interdependent cooperative interactions in communities and develop a clear genomic signature via complementary in loss of shared diffusible functions (Sachs and Hollowell, 2012).
Metatranscriptomic studies on a mixed freshwater model culture, containing previously uncultured bacteria, have revealed a metabolic interconnectedness and niche partitioning between bacterial community members. It implies that every species in microbial communities may “play their own violin in a common concerto” by performing functions vital for the respective co-existing species (Garcia et al., 2015). Experiments with mixed algal-bacterial co-cultures have shown that in periphyton heterotrophic bacteria play a crucial ecological role and that algae-only assemblages develop slower and are less stable than in the presence of bacteria (Roeselers et al., 2007; Lubarsky et al., 2010; Kouzuma and Watanabe, 2015; Koedooder et al., 2019). Therefore, it is difficult to obtain representative axenic cultures of periphytic algae. This functional interconnectedness between photo- and heterotrophic consortia in algal-bacterial associations and, particularly, in periphyton is consistent with the above portrayed BQH (Morris et al., 2012). Yet, such interconnectedness raises the following questions: is this functional interrelationship obligatory and a result of (i) gene loss described by the BQH (when organism with lost-of-function mutation relies on self-sufficient organisms)? (ii) the proposed cross-feeding scenarios? or (iii) phenotypic plasticity caused by the present environmental conditions (Smith et al., 2019)?
Loss-of-function mutations are predicted to be the most widespread among intimately interacting bacteria, e.g., biofilm and rhizosphere consortia. Models suggest that within these communities deletions might often occur in a complementary fashion among various interacting bacteria (Morris et al., 2012; Sachs and Hollowell, 2012). Yuan and Meng (2020) modeled the development of biofilms in a bioreactor and provided an overview of prevailing interactions. They found a difference in interactions in mobile (flocks) vs. those in surface-attached biofilms. In flocks, the prevailing interactions were more competitive corresponding more with the Red Queen Hypothesis. In surface-attached biofilms, however, the relationships were less competitive and more cooperative. However, as the authors note themselves, their experiment had lasted for a too short time to demonstrate the ecological effects of possible loss-of-function mutations (Yuan and Meng, 2020).
One of the last stochastic-spatial models, proposed by Stump et al. (2018), shows that in free-living, rather instable systems self-sufficient species (which produce all required resources) have an advantage in comparison to non-producers or those who produce only one resource from the number of resources required. Spatial structure promotes an increase in mutualistic relationships vs. self-sufficiency which is in agreement with data from natural systems. According to this model, the neighbor uncertainty is an important factor increasing the cost of loss-of-functions mutations and gives more advantage to self-sufficiency vs. mutualism according to the BQH. However, the authors also note that their model doesn’t include chemical interactions between specimens, which take place in natural systems and may substantially decrease the effect of neighbor uncertainty. When taking into account chemical signaling and chemotactic motility, the advantage can be shifted more to the side of non-producers and co-existing one-producers (Stump et al., 2018). However, in the case of biofilms and algal-bacterial interactions, microbial communities face selection according to substrate type and ability to co-exist with a specific algal host and other microbes (Goecke et al., 2013; Stock et al., 2019; Yuan and Meng, 2020). This decreased effect of neighbor uncertainty can result in strengthening mutualistic relationships via the BQH. Also, studies on diatom-bacterial interactions reveal a high species-specific interdependence of the algal host and bacteria (Grossart et al., 2005; Stock et al., 2019). Moreover, bacteria can also influence the diatom community composition themselves by promoting growth of one species and suppressing growth of others (Koedooder et al., 2019).
Thus, communities of periphyton and diatom-bacteria have a great potential for detailed evaluation of mechanisms of cross-feeding and loss-of-function mutations.
New Methods, New Concepts
For a long time, difficulties in methodology have greatly limited studies on microbial communities and algal-bacterial associations, in particular since more than 99% of bacteria from natural habitats still remain undetectable by traditional microbiological methods and are not amenable for cultivation (Curtis, 2002; Zarraonaindia et al., 2013; Solden et al., 2016). As a result, the majority of bacterial diversity has been missed in physiological studies for understanding the ecological role of heterotrophic bacteria in aquatic ecosystems (Pace, 1997). Implementation of molecular methods to ecology allowed to improve our knowledge on microbial communities, their diversity, succession, ecological functions, and physiological responses (Jackson et al., 2001; Fechner et al., 2010; Goecke et al., 2013). During the past two decades, OMICs methods have been developed and successfully applied in aquatic ecology research. Metagenomics allows to retrieve (almost complete) genome sequences from complex microbial assemblages by avoiding cultivation biases (Moran, 2009) and thus provides a useful method to address the functional potential of a given periphyton organism. However, with periphyton and other aquatic biofilms the use of molecular methods meets some difficulties. First, periphytic communities are very heterogeneous often resulting in very variable results. Second, due to the presence of extracellular DNA as well as dead and inactive cells, DNA extracted from the sample may result in an overestimation of the true diversity (Stal et al., 2019). Another way to obtain DNA from environmental samples is single cell genomics, i.e., genomic DNA is extracted from a single, mechanically isolated cell. However due to biases in amplification, it results incomplete genomes. Thus, the best result can be achieved with the joint use of these complimentary methods (Solden et al., 2016). In addition, these methods can be complemented with metaproteomics and metabolomics. Metaproteomics can be defined as “the large-scale characterization of the entire protein content of the environmental microbiota at a given point in time” (Wilmes and Bond, 2004) aiming to uncover the response of any community through expressed proteins, protein paths, and networks (Courant et al., 2014; Wilmes et al., 2015). In contrast, metabolomics investigates the complete pool of low molecular weight compounds (metabolites) in microbial communities and their changes in response to external stressors or interactions (Lankadurai et al., 2013). Both methods can be also utilized for quantification.
In contrast, metatranscriptomics reveals the genes expressed at specific environmental conditions at the moment of sampling and thus hint on specific physiological functions (Moran, 2009; Moran et al., 2013). Metatranscriptomics represents an OMICs method, which allows to disentangle the specific response of microbial species to different environmental factors including organismic interactions. As recently shown for pelagic systems, this methodology opens new perspectives in studies on interactions between algae and bacteria in freshwater periphyton, especially because it reveals the specific response of individual species at the level of gene products. Consequently, metatranscriptomics deals with a subset of genes responsible for important functions, such as nutrient metabolism and organismic interactions (Warnecke and Hess, 2009; Alexander et al., 2015). Most of such metatranscriptomic studies, however, have been conducted with plankton communities, mainly in marine and coastal environments (Krohn-Molt et al., 2013, 2017; Alexander et al., 2015; Amin et al., 2015; Garcia et al., 2015). In the case of benthic communities this method meets some challenges. In environmental samples, transcripts are low in abundance relative to the total rRNA pool. At the same time these samples (and periphytic communities as well) are rich of inhibiting compounds. These obstacles, however, can be overcome by rigorous extraction to remove RNAase enzymes, use of polyA-enrichment (if Eukaryota are targeted), and removing the rRNA with special reagents (Kuske et al., 2015). Only recently, relevant protocols became available (Zimmermann et al., 2011; Krohn-Molt et al., 2013, 2017; Paulsen and Holmes, 2014; Hovde et al., 2015), which can also be applied for freshwater periphyton. We thus believe that the application of metatranscriptomics in combination with metagenomics and single-cell genomics, which can provide the evidence of gene loss, as well as other OMICs approaches are suitable for periphyton studies and capable to reveal the basic mechanisms of chemical signaling and functional interconnectedness, both are crucial for co-existence and interactions of periphytic communities.
Conclusion
Periphyton and its microbial interactions (Figure 1) within a diverse microbial community are of great importance for whole lake primary production, nutrient cycling, and food-web structure, particularly in shallow lakes. Development and application of molecular methods in aquatic microbial ecology provide new perspectives revealing microbial diversity and the underlying mechanisms of alga-bacteria interactions, which are crucial for a mechanistic understanding of the specific ecological role of freshwater periphyton. Metatranscriptomic analysis, combined with co-culturing experiments (when a few species are cultivated together), holds the promise to reveal the mechanisms of cell-to-cell communication, signaling, niche portioning, and functional response of every interacting species in the community (Table 1). Metatranscriptome analysis together with metagenomics and other OMIC’s methods corroborated the concepts of chemical signaling in algal-bacterial interactions (Amin et al., 2015) and resulted in the proposition of the Black Queen Hypothesis in pelagic or marine benthic systems. Since in periphyton all organisms are in close interaction, these communities are very promising for further studies on species co-evolution, e.g., in the view of various hypotheses such as the Red vs. the Black Queen Hypotheses. Recent findings allow to complement and expand existing concepts of freshwater periphyton communities as interconnected microbial networks inside of diverse microbial landscapes. Development of the new concepts, based on microbial genome evolution and chemical signaling, are promising to improve our knowledge on microbial interactions and their ecological role, in particular in freshwater periphyton communities. Combination of metagenomic and metatranscriptomic techniques with more quantitative tools such as metaproteomics and metabolomics is required to uncover the ways and mechanisms of microbial interactions. In turn, a better understanding of these mechanisms enables us to predict better the ecological consequences of environmental changes (natural or man-made). The knowledge of these mechanisms in lake
periphyton is crucial for effective management and conservation of freshwater resources in a changing world.
Author Contributions
YG analyzed the literature, concepts, wrote large parts of the text, and discussed it with the coauthor. H-PG initiated the view on periphyton from the point of microbial interactions and application of the newest concepts and methods, discussed and edited the manuscript. Both authors revised the manuscript. Both authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This work was supported by State research topic of ZIN RAS No. AAAA-A19-119020690091-0. H-PG was supported by the BMBF-BIBS II project (01LC1501G1). We acknowledge the support of the Deutsche Forschungsgemeinschaft and Open Access Publishing Fund of University of Potsdam.
References
- Alexander H., Jenkins B. D., Rynearson T. A., Dyhrman S. T. (2015). Metatranscriptome analyses indicate resource partitioning between diatoms in the field. Proc. Natl. Acad. Sci. U.S.A. 112 E2182–E2190. 10.1073/pnas.1421993112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen J. L., Ten-Hage L., Leflaive J. (2015). Impairment of benthic diatom adhesion and photosynthetic activity by allelopathic compounds from a green alga: involvement of free fatty acids? Environ. Sci. Pol. Res. 22 13669–13680. 10.1007/s11356-014-3873-9 [DOI] [PubMed] [Google Scholar]
- Amaral-Zettler L. A., Zettler E. R., Mincer T. J. (2020). Ecology of the plastisphere. Nat. Rev. Microbiol. 18 139–151. 10.1038/s41579-019-0308-0 [DOI] [PubMed] [Google Scholar]
- Amin S. A., Hmelo L. R., van Tol H. M., Durham B. P., Carlson L. T., Heal K. R., et al. (2015). Interaction and signaling between a cosmopolitan phytoplankton and associated bacteria. Nature 522 98–101. 10.1038/nature14488 [DOI] [PubMed] [Google Scholar]
- Amin S. A., Parker M. S., Armbrust E. V. (2012). Interactions between diatoms and bacteria. Microbiol. Mol. Biol. Rev. 76 667–684. 10.1128/MMBR.00007-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battin T. J., Besemer K., Bengtsson M. M., Romani A. M., Packmann A. I. (2016). The ecology and biogeochemistry of stream biofilms. Nat. Rev. Microbiol. 254 251–263. 10.1038/nrmicro.2016.15 [DOI] [PubMed] [Google Scholar]
- Battin T. J., Sloan W. T., Kjelleberg S., Daims H., Head I. M., Curtis T. P., et al. (2007). Microbial landscapes: new paths to biofilm research. Nat. Rev. Microbiol. 5 76–81. 10.1038/nrmicro1556 [DOI] [PubMed] [Google Scholar]
- Bazes A., Silkina A., Defera D., Bernède-Bauduinb C., Quéménera E., Braud J.-P., et al. (2006). Active substances from Ceramium botryocarpum used as antifouling products in aquaculture. Aquaculture 258 664–674. 10.1016/j.aquaculture.2006.04.017 [DOI] [Google Scholar]
- Bell W., Mitchell R. (1972). Chemotactic and growth responses of marine bacteria to algal extracellular products. Biol. Bull. 143 265–277. 10.2307/1540052 [DOI] [Google Scholar]
- Ben Jacob E., Becker I., Shapira Y., Levine H. (2004). Bacterial linguistic communication and social intelligence. Trends Microbiol. 12 366–372. 10.1016/j.tim.2004.06.006 [DOI] [PubMed] [Google Scholar]
- Brothers S. M., Hilt S., Attermeyer K., Grossart H. P., Kosten S., Lischke B., et al. (2013). A regime shift from macrophyte to phytoplankton dominance enhances carbon burial in a shallow, eutrophic lake. Ecosphere 4:137 10.1890/ES13-00247.1 [DOI] [Google Scholar]
- Bruckner C. G., Bahulikar R., Rahalkar M., Schink B., Kroth P. B. (2008). Bacteria associated with benthic diatoms from Lake Constance: phylogeny and influences on diatom growth and secretion of extracellular polymeric substances. Appl. Environ. Microbiol. 74 7740–7749. 10.1128/AEM.01399-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruckner C. G., Rehm C., Grossart H.-P., Kroth P. G. (2011). Growth and release of extracellular organic compounds by benthic diatoms depend on interactions with bacteria. Environ. Microbiol. 13 1052–1063. 10.1111/j.1462-2920.2010.02411.x [DOI] [PubMed] [Google Scholar]
- Burke C., Steinberg P., Rusch D., Kjelleberg S., Thomas T. (2011a). Bacterial community assembly based on functional genes rather than species. Proc. Natl. Acad. Sci. U.S.A. 108 14288–14293. 10.1073/pnas.1101591108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke C., Thomas T., Lewis M., Steinberg P., Kjelleberg S. (2011b). Composition, uniqueness and variability of the epiphytic bacterial community of the green alga Ulva australis. ISME J. 5 590–600. 10.1038/ismej.2010.164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burmolle M., Webb J. S., Rao D., Hansen L. H., Sorensev S. J., Kjelleberg S. (2006). Enhanced biofilm formation and increased resistance to antimicrobial agents and bacterial invasion are caused by synergistic interactions in multispecies biofilms. Appl. Environ. Microbiol. 72 3916–3923. 10.1128/AEM.03022-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantonati M., Lowe R. L. (2014). Lake benthic algae: toward an understanding of their ecology. Freshw. Sci. 33 475–486. 10.1086/676140 [DOI] [Google Scholar]
- Carr G. M., Morin A., Chambers P. A. (2005). Bacteria and algae in stream periphyton along a nutrient gradient. Freshw. Biol. 50 1337–1350. 10.1111/j.1365-2427.2005.01401.x [DOI] [Google Scholar]
- Cole J. J. (1982). Interactions between bacteria and algae in aquatic ecosystems. Annu. Rev. Ecol. Syst. 13 291–314. 10.1146/annurev.es.13.110182.001451 [DOI] [Google Scholar]
- Courant F., Antignac J.-P., Dervilly-Pinel G., Le Bizec B. (2014). Basics of mass spectrometry based metabolomics. Proteomics 14 2369–2388. 10.1002/pmic.201400255 [DOI] [PubMed] [Google Scholar]
- Curtis T. P. (2002). Estimating prokaryotic diversity and its limits. Proc. Natl. Acad. Sci. U.S.A. 99 10494–10499. 10.1073/pnas.142680199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De-Bashan L. E., Antoun H., Bashan Y. (2008). Involvement of indole-3- acetic acid produced by the growth-promoting bacterium Azospirillum spp. in promoting growth of Chlorella vulgaris. J. Phycol. 44 938–947. 10.1111/j.1529-8817.2008.00533.x [DOI] [PubMed] [Google Scholar]
- Debois A. P., Walton M., Smith V. J. (2010). Differential antibacterial activities of fusiform and oval morphotypes of Phaeodactylum tricornutum (Bacillariophyceae). J. Mar. Biol. Assoc. U.K. 90 769–774. 10.1017/s0025315409991366 [DOI] [Google Scholar]
- DeNicola D. M., Kelly M. G. (2014). Role of periphyton in ecological assessments of lakes. Freshw. Sci. 33 619–638. 10.1086/676117 [DOI] [Google Scholar]
- Dittami S. M., Duboscq-Bidot L., Perennou M., Gobet A., Corre E., Boyen C., et al. (2016). Host–microbe interactions as a driver of acclimation to salinity gradients in brown algal cultures. ISME J. 10 51–63. 10.1038/ismej.2015.104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do Nascimento Filho S. L., De França E. J., De Melo J., Do Nascimento Moura A. (2019). Interactions between benthic microalgae, nutrients and benthic macroinvertebrates in reservoirs from the semi-arid Neotropical region. Fundam. Appl. Limnol. 19 237–254. 10.1127/fal/2019/1180 [DOI] [Google Scholar]
- Donlan R. M. (2002). Biofilms: microbial Life on Surfaces. Emerg. Infect. Dis. 8 881–890. 10.3201/eid0809.020063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dos Santos T. R., Carolina Castilho M. C., Henry R., Ferragut C. (2020). Relationship between epipelon, epiphyton and phytoplankton in two limnological phases in a shallow tropical reservoir with high Nymphaea coverage. Hydrobiologia 847 1121–1137. 10.1007/s10750-019-04172-2 [DOI] [Google Scholar]
- Elias S., Banin E. (2012). Multi-species biofilms: living with friendly neighbors. FEMS Microbiol. Rev. 36 990–1004. 10.1111/j.1574-6976.2012.00325.x [DOI] [PubMed] [Google Scholar]
- Espinosa-Urgel M. (2003). Resident parking only: rhamnolipids maintain fluid channels in biofilms. J. Bacteriol. 185 699–700. 10.1128/JB.185.3.699-700.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- European Union (2000). Directive 2000/60/EC of the european parliament and of the council of 23 October 2000 establishing a framework for Community action in the field of water policy. Off. J. Eur. Commun. Ser. L 327 1–73. [Google Scholar]
- Fechner L. C., Vincent-Hubert F., Gaubert P., Bouchez T., Gourlay-France C., Tusseau-Vuillemin M.-H. (2010). Combined eukaryotic and bacterial community fingerprinting of natural freshwater biofilms using automated ribosomal intergenic spacer analysis. FEMS Microbiol. Ecol. 74 542–553. 10.1111/j.1574-6941.2010.00968.x [DOI] [PubMed] [Google Scholar]
- Feio M. J., Filipe A. F., Garcia-Raventós A., Alba Ardura A., Calapez R. A., Ana Maria Pujante A. M., et al. (2020). Advances in the use of molecular tools in ecological and biodiversity assessment of aquatic ecosystems. Limnetica 39 419–440. 10.23818/limn.39.27 [DOI] [Google Scholar]
- Fitter A., Hillebrand H. (2009). Microbial food web structure affects bottom-up effects and elemental stoichiometry in periphyton assemblages. Limnol. Oceanogr. 54 2183–2200. 10.4319/lo.2009.54.6.2183 [DOI] [Google Scholar]
- Freilich S., Zarecki R., Eilam O., Shtifman Segal E., Henry C. S., Kupiec M., et al. (2011). Competitive and cooperative metabolic interactions in bacterial communities. Nat. Commu. 2:589. 10.1038/ncomms1597 [DOI] [PubMed] [Google Scholar]
- Garcia S. L., Buck M., McMahon K. D., Grossart H.-P., Eiler A., Warnecke F. (2015). Auxotrophy and intrapopulation complementary in the ‘interactome’ of a cultivated freshwater model community. Mol. Ecol. 24 4449–4459. 10.1111/mec.13319 [DOI] [PubMed] [Google Scholar]
- Goecke F., Thiel V., Wiese J., Labes A., Imhoff J. F. (2013). Algae as an important environment for bacteria – phylogenetic relationships among new bacterial species isolated from algae. Phycology 52 14–24. 10.2216/12-24.1 [DOI] [Google Scholar]
- Grossart H. P. (2010). Ecological consequences of bacterioplankton lifestyles: changes in concepts are needed. Environ. Microb. Rep. 2 706–714. 10.1111/j.1758-2229.2010.00179.x [DOI] [PubMed] [Google Scholar]
- Grossart H.-P., Levold F., Allgaier M., Simon M., Brinkhof T. (2005). Marine diatom species harbour distinct bacterial communities. Environ. Microbiol. 7 860–873. 10.1111/j.1462-2920.2005.00759.x [DOI] [PubMed] [Google Scholar]
- Hagmann L., Jüttner F. (1996). Fischerellin A, a novel photosystem-IIinhibiting allelochemical of the cyanobacterium Fischerella muscicola with antifungal and herbicidal activity. Tetrahedron Lett. 37 6529–6542. [Google Scholar]
- Håkanson L., Boulion V. V. (2002). The lake Foodweb – Modelling Predation And Abiotoc/Biotic Interactions. Leiden: Backhuys Publishers. [Google Scholar]
- Hansson L.-A. (1992). Factors regulating periphytic algal biomass. Limnol. Oceanogr. 37 322–328. 10.4319/lo.1992.37.2.0322 [DOI] [Google Scholar]
- Harriott M. M., Noverr M. C. (2009). Candida albicans and Staphylococcus aureus form polymicrobial biofilms: effects on antimicrobial resistance. Antimicrob. Agents Chemother. 53 3914–3922. 10.1128/AAC.00657-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepinstall J. A., Fuller R. L. (1994). Periphyton reactions to different light and nutrient levels and the response of bacteria to these manipulations. Arch. Hydrobiol. 131 161–173. [Google Scholar]
- Hill W. R., Boston H. L., Steinman A. D. (1992). Grazers and nutrients simultaneously limit lotic primary productivity. Canad. J. Fish. Aquat. Sci. 49 504–512. 10.1139/f92-059 [DOI] [Google Scholar]
- Hillebrand H., Kahlert M. (2001). Effect of grazing and nutrient supply on periphyton biomass and nutrient stoichiometry in habitats of different productivity. Limnol. Oceanogr. 46 1881–1898. 10.4319/lo.2001.46.8.1881 [DOI] [Google Scholar]
- Hillebrand H., Sommer U. (1999). The nutrient stoichiometry of benthic microalgal growth: Redfield proportions are optimal. Limnol. Oceanogr. 44 440–446. 10.4319/lo.1999.44.2.0440 [DOI] [Google Scholar]
- Hirata K., Yoshitomi S., Dwi S., Iwabe O., Mahakhant A., Polchai J., et al. (2003). Bioactivities of nostocine A produced by a freshwater cyanobacterium Nostoc spongiaeforme TISTR 8169. J. Biosci. Bioeng. 95 512–517. 10.1016/s1389-1723(03)80053-1 [DOI] [PubMed] [Google Scholar]
- Holomuzki J. R., Feminella J. W., Power M. E. (2010). Biotic interactions in freshwater benthic habitats. J. N. Am. Benthol. Soc. 29 220–244. 10.1899/08-044.1 [DOI] [Google Scholar]
- Hovde B. T., Deodato C. R., Hunsperger H. M., Ryken S. A., Yost W., Jha R. K., et al. (2015). Genome sequence and transcriptome analyses of Chrysochromulina tobin: metabolic tools for enhanced algal fitness in the prominent order prymnesiales (Haptophyceae). PLoS Genet 11:e1005469. 10.1371/journal.pgen.1005469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Xie G., Jiang X., Shao K., Tang X., Gao G. (2020). The relationships between the free-living and particle-attached bacterial communities in response to elevated eutrophication. Front. Microbiol. 11:423. 10.3389/fmicb.2020.00423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson C. R., Churchill P. F., Roden E. E. (2001). Successional changes in bacterial assemblage structure during epilithic biofilm development. Ecology 82 555–566. 10.1890/0012-9658(2001)082[0555:scibas]2.0.co;2 [DOI] [Google Scholar]
- Jonsson P. R., Pavia H., Toth G. (2009). Formation of harmful algal blooms cannot be explained by allelopathic interactions. Proc. Natl Acad. Sci. U.S.A. 106 11177–11182. 10.1073/pnas.0900964106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jüttner F. (1999). “Allelochemical control of natural photoautotrophic biofilms,” in Biofilms in Aquatic Environment, eds Keevil C. W., Godfree A., Holt D., Dow C. (Cambridge: Royal Society of Chemistry; ), 43–50. [Google Scholar]
- Jüttner F., Wu J.-T. (2000). Evidence of allelochemical activity in subtropical cyanobacterial biofilms of Taiwan. Arch. Hydrobiol. 147 505–517. 10.1127/archiv-hydrobiol/147/2000/505 [DOI] [Google Scholar]
- Kahlert M., Gottshalk S. (2014). Differences in benthic diatom assemblages between streams and lakes in Sweden and implications for ecological assessment. Freshw. Sci. 33 655–669. 10.1086/675727 [DOI] [Google Scholar]
- Kalscheur K. N., Rojas M., Peterson C. G., Kelly J. J., Gray K. A. (2012). Algal exudates and stream organic matter influence the structure and function of denitrifying bacterial communities. Microb. Ecol. 64 881–892. 10.1007/s00248-012-0091-1 [DOI] [PubMed] [Google Scholar]
- Karp-Boss L., Boss E., Jumars P. A. (1996). Nutrient fluxes to planktonic osmotrophs in the presence of fluid motion. Oceanogr. Mar. Biol. Annu. Rev. 34 71–107. [Google Scholar]
- Kazamia E., Czesnick H., Nguyen T. T. V., Croft M. T., Sherwood E., Sasso S., et al. (2012). Mutualistic interactions between vitamin B12-dependent algae and heterotrophic bacteria exhibit regulation. Environ. Microbiol. 14 1466–1476. 10.1111/j.1462-2920.2012.02733.x [DOI] [PubMed] [Google Scholar]
- Kelly M. G. (2013). Data rich, information poor? ten years of the water framework directive in Europe. Eur. J. Phycol. 48 437–450. 10.1080/09670262.2013.852694 [DOI] [Google Scholar]
- Kesy K., Oberbeckmann S., Kreikemeyer B., Labrenz M. (2019). Spatial environmental heterogeneity determines young biofilm assemblages on microplastics in baltic sea mesocosms. Front. Microbiol. 10:1665. 10.3389/fmicb.2019.01665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koedooder C., Stock W., Willems A., Mangelinck S., De Troch M., Vyverman W., et al. (2019). Diatom-bacteria interactions modulate the composition and productivity of benthic diatom biofilms. Front. Microbiol. 10:1255. 10.3389/fmicb.2019.01255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korgaonkar A., Trivedi U., Rumbaugh K. P., Whiteley M. (2013). Community surveillance enhances Pseudomonas aeruginosa virulence during polymicrobial infection. PNAS 110 1059–1064. 10.1073/pnas.1214550110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzuma A., Watanabe K. (2015). Exploring the potential of algae/bacteria interactions. Curr. Opin. Biotech. 33 125–129. 10.1016/j.copbio.2015.02.007 [DOI] [PubMed] [Google Scholar]
- Krohn-Molt I., Alawi M., Förstner K. U., Wiegandt A., Burkhardt L., Indenbirken D., et al. (2017). Insights into microalga and bacteria interactions of selected phycosphere biofilms using metagenomic, transcriptomic, and proteomic approaches. Front. Microbiol. 8:1941. 10.3389/fmicb.2017.01941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krohn-Molt I., Wemheuer B., Alawi M., Poehlein A., Güllert S., Schmeisser C., et al. (2013). Metagenome survey of a multispecies and alga-associated biofilm revealed key elements of bacterial-algal interactions in photobioreactors. Appl. Environ. Microbiol. 79 6196–6206. 10.1128/AEM.01641-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuske C. R., Hesse C. N., Challacombe J. F., Cullen D., Herr J. R., Mueller R. C., et al. (2015). Prospects and challenges for fungal metatranscriptomics of complex communities. Fungal Ecol. 14 133–137. 10.1016/j.funeco.2014.12.005 [DOI] [Google Scholar]
- Lankadurai B. P., Nagato E. G., Simpson M. J. (2013). Environmental metabolomics: an emerging approach to study organism responses to environmental stressors. Environ. Rev. 21 180–205. 10.1139/er-2013-0011 [DOI] [Google Scholar]
- Larned S. T. (2010). A prospectus for periphyton: recent and future ecological research. J. N. Am. Benthol. Soc. 29 182–206. 10.1899/08-063.1 [DOI] [Google Scholar]
- Leflaive J., Boulêtreau S., Buffan-Dubau E., Ten-Hage L. (2008). Temporal patterns in epilithic biofilms—relation with a putative allelopathic activity. Fund. Appl. Limnol. 173 121–134. 10.3354/ame01616 [DOI] [Google Scholar]
- Leflaive J., Ten-Hage L. (2007). Algal and cyanobacterial secondary metabolites in freshwaters: a comparison of allelopathic compounds and toxins. Freshwat. Biol. 52 199–214. 10.1111/j.1365-2427.2006.01689.x [DOI] [Google Scholar]
- Leflaive J., Ten-Hage L. (2009). Allelopathic interactions in benthic biofilms: effects of abiotic conditions on production of and sensitivity to allelochemicals. J. N. Am. Benthol. Soc. 28 271–280. 10.1111/1758-2229.12436 [DOI] [PubMed] [Google Scholar]
- Lenski R. E., Riley M. A. (2002). Chemical warfare from an ecological perspective. Proc. Natl. Acad. Sci. U.S.A. 99 556–558. 10.1073/pnas.022641999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindemann S. R., Mobberley J. M., Cole J. K., Markillie L. M., Taylor R. C., Huang E., et al. (2017). Predicting species-resolved macronutrient acquisition during succession in a model phototrophic biofilm using an integrated ‘Omics’ approach. Front. Microbiol. 8:1020. 10.3389/fmicb.2017.01020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes S. P., Ceri H., Azevedo F., Pereira M. O. (2012). Antibiotic resistance of mixed biofilms in cystic fibrosis: impact of emerging microorganisms on treatment of infection. Int. J. Antimicrob. Agents 40 260–263. 10.1016/j.ijantimicag.2012.04.020 [DOI] [PubMed] [Google Scholar]
- Lovett G. M., Jones C. G., Turner M. G., Weathers K. C. (2005). Ecosystem Function in Heterogeneous Landscapes. New York, NY: Springer. [Google Scholar]
- Lubarsky H. V., Hubas C., Chockolek M., Larson F., Manz W., Paterson D. M., et al. (2010). The stabilisation potential of individual and mixed assemblages of natural bacteria and microalgae. PLoS One 5:e13794. 10.1371/journal.pone.0013794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall K., Joint I., Callow M. E., Callow J. A. (2006). Effect of marine bacterial isolates on the growth and morphology of axenic plantlets of the green alga Ulva linza. Microbial. Ecol. 52 302–310. 10.1007/s00248-006-9060-x [DOI] [PubMed] [Google Scholar]
- Moran M. A. (2009). Metatranscriptomics: eavesdropping on complex microbial communities. Microbe 4 329–335. 10.1128/microbe.4.329.1 [DOI] [Google Scholar]
- Moran M. A., Satinsky B., Gifford S. M., Luo H., Rivers A., Chan L.-K., et al. (2013). Sizing up metatranscriptomics. ISME J. 7 237–243. 10.1038/ismej.2012.94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris J. J., Lenski R. E., Zinser E. R. (2012). The Black queen hypothesis: evolution of dependencies through adaptive gene loss. mBio 3:e036-12. 10.1128/mBio.00036-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulholland P. J., Webster J. R. (2010). Nutrient dynamics in streams and the role of J-NABS. J. N. Am. Benthol. Soc. 29 110–117. [Google Scholar]
- Neu T. R., Swerhone G. D. W., Lawrence J. R. (2001). Assessment of lectin-binding analysis for in situ detection of glycoconjugates in biofilm systems. Microbiology 147 299–313. 10.1099/00221287-147-2-299 [DOI] [PubMed] [Google Scholar]
- Pace N. R. (1997). A molecular view of microbial diversity and the biosphere. Science 276, 734–740. 10.1126/science.276.5313.734 [DOI] [PubMed] [Google Scholar]
- Pascault N., Loux V., Derozier S., Martin V., Debroas D., Maloufi S., et al. (2015). Technical challenges in metatranscriptomic studies applied to the bacterial communities of freshwater ecosystems. Genetica 143 157–167. 10.1007/s10709-014-9783-4 [DOI] [PubMed] [Google Scholar]
- Paulsen I. T., Holmes A. J. (2014). Environmental Microbiology Methods And Protocols. Berlin: Springer. [Google Scholar]
- Pouvreau J. B., Housson E., Le Tallec L., Morancais M., Rince Y., Fleurence J., et al. (2007). Growth inhibition of several marine diatom species induced by the shading effect and allelopathic activity of marennine, a blue-green polyphenolic pigment of the diatom Haslea ostrearia (Gaillon/Bory) Simonsen. J. Exp. Mar. Biol. Ecol. 352 212–225. 10.1016/j.jembe.2007.07.011 [DOI] [Google Scholar]
- Ramanan R., Kang Z., Kim B.-H., Cho D.-H., Jin L., Oh H.-M., et al. (2015). Phycosphere bacterial diversity in green algae reveals an apparent similarity across habitats. Algal Res. 8 140–144. 10.1016/j.algal.2015.02.003 [DOI] [Google Scholar]
- Ramanan R., Kim B.-H., Cho D.-H., Oh H.-M., Kim H.-S. (2016). Algae–bacteria interactions: Evolution, ecology and emerging applications. Biotech. Adv. 34 14–29. 10.1016/j.biotechadv.2015.12.003 [DOI] [PubMed] [Google Scholar]
- Rendueles O., Ghigo J.-M. (2015). Mechanisms of competition in biofilm communities. Microbiol. Spectr. 3:MB–0009–2014. 10.1128/microbiolspec.MB-0009-2014 [DOI] [PubMed] [Google Scholar]
- Rier S. T., Stevenson R. J. (2002). Effects of light, dissolved organic carbon, and inorganic nutrients on the relationship between algae and heterotrophic bacteria in stream periphyton. Hydrobiologia 489 179–184. 10.1023/A:1023284821485 [DOI] [Google Scholar]
- Roeselers G., Van Loosdrecht M. C., Muyzer G. (2007). Heterotrophic pioneers facilitate phototrophic biofilm development. Microb. Ecol. 54 578–585. 10.1007/s00248-007-9238-x [DOI] [PubMed] [Google Scholar]
- Rusanov A. G., Grossart H.-P., Pusch M. T. (2009). Periphytic bacterial and algal response to a hydraulic gradient under different light levels: Test of algal-bacterial the coupling in the laboratory. Fund. Appl. Limnol. 175 339–353. 10.1127/1863-9135/2009/0175-0339 [DOI] [Google Scholar]
- Sachs J. L., Hollowell A. C. (2012). The origins of cooperative bacterial communities. mBio 3:e099-12. 10.1128/mBio.00099-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider S. C., Lindsrom E.-A. (2011). The periphyton index of trophic status PIT: a new eutrophication metric based on non-diatomaceous benthic algae in Nordic rivers. Hydrobiologia 665 143–155. 10.1007/s10750-011-0614-7 [DOI] [Google Scholar]
- Scott J. T., Back J. A., Taylor J. M., King R. S. (2008). Does nutrient enrichment decouple algal–bacterial production in periphyton? J. N. Am. Benthol. Soc. 27 332–344. 10.1899/07-108.1 [DOI] [Google Scholar]
- Seymour J., Amin S., Raina J.-B., Stocker R. (2017). Zooming in on the phycosphere: the ecological interface for phytoplankton-bacteria relationships. Nat. Microbiol. 2:65. 10.1038/nmicrobiol.2017.65 [DOI] [PubMed] [Google Scholar]
- Smith N. W., Shorten P. R., Altermann E., Roy N. C., McNabb W. C. (2019). the classification and evolution of bacterial cross-feeding. Front. Ecol. Evol. 7:153 10.3389/fevo.2019.00153 [DOI] [Google Scholar]
- Smith V. J., Desbois A. P., Dyrynda E. A. (2010). Conventional and unconventional antimicrobials from fish, marine invertebrates and micro-algae. Mar. Drugs 8 1213–1262. 10.3390/md8041213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobczak W. V. (1996). Epilithic bacterial responses to variations in algal biomass and labile dissolved organic carbon in water. J. North. Am. Benthol. Soc. 15 143–154. 10.2307/1467944 [DOI] [Google Scholar]
- Solden L., Lloyd K., Wrighton K. (2016). The bright side of microbial dark matter: lessons learned from the uncultivated majority. Curr. Opin. Microbiol. 31 217–226. 10.1016/j.mib.2016.04.020 [DOI] [PubMed] [Google Scholar]
- Stal L. J., Bolhuis H., Cretoiu M. S. (2019). Phototrophic marine benthic microbiomes: the ecophysiology of these biological entities. Environ. Microbiol. 21 1529–1551. 10.1111/1462-2920.14494 [DOI] [PubMed] [Google Scholar]
- Stock W., Blommaert L., De Troch M., Mangelinckx S., Willems A., Vyverman W., et al. (2019). Host specificity in diatom–bacteria interactions alleviates antagonistic effects. FEMS Microbiol. Ecol. 95:fiz171. 10.1093/femsec/fiz171 [DOI] [PubMed] [Google Scholar]
- Stubbendieck R. M., Straight P. D. (2016). Multifaceted interfaces of bacterial competition. J. Bacteriol. 198 2145–2155. 10.1128/JB.00275-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stump S. M., Johnson E. C., Sun Z., Klausmeier C. A. (2018). How spatial structure and neighbor uncertainty promote mutualists and weaken black queen effects. J. Theor. Biol. 446 33–60. 10.1016/j.jtbi.2018.02.031 [DOI] [PubMed] [Google Scholar]
- Tank J. L., Dodds W. K. (2003). Nutrient limitation of epilithic and epixylic biofilms in 10 North American streams. Freshw. Biol. 48 1031–1049. 10.1046/j.1365-2427.2003.01067.x [DOI] [Google Scholar]
- Teissier S., Torre M., Delmas F., Garabetian F. (2007). Detailing biogeochemical N budgets in riverine epilithic biofilms. J. N. Am. Benthol. Soc. 26 178–190. 10.1899/0887-3593(2007)26[178:dbnbir]2.0.co;2 [DOI] [Google Scholar]
- Toet S., Huibers L., Van Logtestijn R. S. P., Verhoeven J. T. A. (2003). Denitrification in the periphyton associated with plant shoots and in the sediment of a wetland system supplied with sewage treatment plant effluent. Hydrobiologia 501 29–44. 10.1023/A:1026299017464 [DOI] [Google Scholar]
- Tsementzi D., Poretsky R., Rodriguez-R L. M., Luo C., Konstantinidis K. T. (2014). Evaluation of metatranscriptomic protocols and application to the study of freshwater microbial communities. Environ. Microbiol. Rep. 6 640–655. 10.1111/1758-2229.12180 [DOI] [PubMed] [Google Scholar]
- Vadeboncoeur Y., Devlin S. P., McIntyre P. B., Vander Zanden M. J. (2014). Is there light after depth? Distribution of periphyton chlorophyll and productivity in lake littoral zones. Freshw. Sci. 33 524–536. 10.1086/676315 [DOI] [Google Scholar]
- Vadeboncoeur Y., Kalff J., Christoffersen K., Jeppesen E. (2006). Substratum as a driver of variation in periphyton chlorophyll and productivity in lakes. J. N. Am. Benthol. Soc. 25 379–392. 10.1899/0887-3593(2006)25[379:saadov]2.0.co;2 [DOI] [Google Scholar]
- Vadeboncoeur Y., McIntyre P. B., Vander Zanden M. J. (2011). Borders of biodiversity: life at the Edge of the World’s Large Lakes. Bioscience 61 526–537. 10.1525/bio.2011.61.7.7 [DOI] [Google Scholar]
- Vadeboncoeur Y., Steinman A. D. (2002). Periphyton function in lake ecosystems. Sci. World J. 2 1449–1468. 10.1100/tsw.2002.294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Valen L. (1973). A new evolutionary law. Evol. Theory 1 1–30. [Google Scholar]
- Vanelslander B., Paul C., Grueneberg J., Prince E. K., Gillard J., Sabbe K., et al. (2012). Daily bursts of biogenic cyanogen bromide (BrCN) control biofilm formation around a marine benthic diatom. Proc. Natl. Acad. Sci. U.S.A. 109 2412–2417. 10.1073/pnas.1108062109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vesterinen J., Devlin S. P., Syvaranta J., Jones R., I (2016). Accounting for littoral primary production by periphyton shifts a highly humic boreal lake towards net autotrophy. Freshw. Biol. 61 265–276. 10.1111/fwb.12700 [DOI] [Google Scholar]
- Warnecke F., Hess M. (2009). A perspective: metatranscriptomics as a tool for the discovery of novel biocatalysts. J. Biotechnol. 142 91–95. 10.1016/j.jbiotec.2009.03.022 [DOI] [PubMed] [Google Scholar]
- Waters C. M., Bassler B. L. (2005). Quorum sensing: cell-to cell communication in bacteria. Annu. Rev. Cell Dev. Biol. 21 319–346. 10.1146/annurev.cellbio.21.012704.131001 [DOI] [PubMed] [Google Scholar]
- Wetzel R. (ed.) (1983). “Periphyton of freshwater ecosystems,” in Proceedings of the First International Workshop on Periphyton of Freshwater Ecosystems Held, Vaxjo. [Google Scholar]
- Wilmes P., Bond P. L. (2004). The application of two-dimensional polyacrylamide gel electrophoresis and downstream analyses to a mixed community of prokaryotic microorganisms. Environ. Microbiol. 6 911–920. 10.1111/j.1462-2920.2004.00687.x [DOI] [PubMed] [Google Scholar]
- Wilmes P., Heintz-Buschart A., Bond P. L. (2015). A decade of metaproteomics: where we stand and what the future holds. Proteomics 15 3409–3417. 10.1002/pmic.201500183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winberg G. G. (1960). Primary Production Of The Waterbodies. Minsk: Academy of Sciences of BSSR. [Google Scholar]
- Winberg G. G. (1980). “General characteristics of freshwater ecosystems based on Soviet IBP studies,” in Functioning of Freshwater Ecosystems, eds Le Cren E. D., McConnell R. H. (Cambridge: Cambridge University Press; ), 481–491. [Google Scholar]
- Wyatt K. H., Seballos R. C., Shoemaker M. N., Brown S. P., Chandra S., Kuehn K. A., et al. (2019). Resource constraints highlight complex microbial interactions during lake biofilm development. J. Ecol. 107 2737–2746. 10.1111/1365-2745.13223 [DOI] [Google Scholar]
- Wyatt K. H., Tellez E., Woodke R. L., Bidner R. J., Davison I. R. (2014). Effects of nutrient limitation on the release and use of dissolved organic carbon from benthic algae in Lake Michigan. Freshw. Sci. 33 557–567. 10.1086/675453 [DOI] [Google Scholar]
- Yang J., Tang C., Wang F., Wua Y. (2016). Co-contamination of Cu and Cd in paddy fields: using periphyton to entrap heavy metals. J. Hazard. Mater. 304 150–158. 10.1016/j.jhazmat.2015.10.051 [DOI] [PubMed] [Google Scholar]
- Yuan S., Meng F. (2020). Ecological insights into the underlying evolutionary patterns of biofilm formation from biological wastewater treatment systems: red or black queen hypothesis? Biotechnol. Bioeng. 117 1–11. 10.1002/bit.27289 [DOI] [PubMed] [Google Scholar]
- Zarraonaindia I., Smith D. P., Gilbert J. A. (2013). Beyond the genome: community-level analysis of the microbial world. Biol. Philos. 28 261–282. 10.1007/s10539-012-9357-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Xiao W., Jiao N. (2016). Linking biochemical properties of particles to particle attachedand free-living bacterial community structure along the particle density gradient from freshwater to open ocean. J. Geophys. Res. Biogeosci. 121 2261–2274. 10.1002/2016JG003390 [DOI] [Google Scholar]
- Zhou J., Lyu Y., Richlen M. L., Anderson D. M., Cai Z. (2016). Quorum sensing is a language of chemical signals and plays an ecological role in algal-bacterial interactions. Crit. Rev. Plant Sci. 35 81–105. 10.1080/07352689.2016.1172461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann J., Jahn R., Gemeinholzer B. (2011). Barcoding diatoms: evaluation of the V4 subregion on the 18S rRNA gene, including new primers and protocols. Organ. Divers. Evol. 11 173–192. 10.1007/s13127-011-0050-6 [DOI] [Google Scholar]


