Abstract
Psoriasis is an immune-mediated disease, with genetic background and triggering environmental factors; however, several gaps are still present in understanding the intertwined relationship between these elements. Epigenetic mechanisms, including microRNAs (miRNAs), play an important role in the pathogenesis of psoriasis. The relationship between interleukin (IL)-17, a key cytokine in psoriasis, and these epigenetic mechanisms still needs to be elucidated. This study aimed at assessing the expression of miRNA-155, miRNA-210, and miRNA-20b in skin and sera of psoriasis patients in relation to IL-17 levels. For 20 psoriasis patients and 20 matching controls, the expression of miRNA-155, miRNA-210, and miRNA-20b was assessed using real-time polymerase chain reaction (RT-PCR), whereas IL-17/IL-17A levels were measured using quantitative enzyme-linked immunosorbent assay (ELISA) technique. MiRNA-155 expression was significantly higher in lesional skin compared to controls (P = 0.001). MiRNA-210 expression was significantly higher in both, lesional skin (P = 0.010) and sera of patients (P = 0.001) in comparison with controls. A statistically significant positive correlation was found between serum miRNA-210 expression and serum levels of IL-17/IL-17A (P = 0.010, rs = 0.562). MiRNA-20b lesional and non-lesional expression was significantly higher than controls (P < 0.001; P = 0.018). In conclusion, the expression of miRNA-155, miRNA-210, and miRNA-20b is exaggerated in psoriasis and they may be involved in disease pathogenesis. A possible relationship between miRNA-210 and IL-17 may be suggested; however, further studies are still needed to verify this relation.
Keywords: epigenetics, IL-17, miRNA-155, miRNA-20b, miRNA-210, microRNA, psoriasis
Introduction
Psoriasis, a long-lasting inflammatory cutaneous disorder, carries a multifaceted etiology that includes genetic risk factors and environmental triggers. Psoriasis imposes an immense health, social, and economic burden, particularly with the lack of curative treatment, emphasizing the necessity of exploring the genetic and molecular mechanism of psoriasis etiology.1
The central role of interleukin (IL)-17 and TNF-α in the pathogenesis of psoriasis has been constantly demonstrated. In psoriatic skin, IL-17 ligands drive an aberrant form of keratinocyte differentiation as well as an overproduction of pro-inflammatory cytokines characteristic of the psoriasis phenotype.2 Increased tissue and serum levels of TNF-α is a hallmark of psoriasis immune dysregulation which is thought to act as an upstream mediator in the IL-23/IL-17 pathway, as well as serving as a potent pro-inflammatory cytokine.3
Epigenetic networks, which include DNA methylation, chromatin modifications, and microRNA (miRNA) deregulation, might be causative elements in psoriasis.4,5 The main pathogenic event in psoriasis, aberrant keratinocyte differentiation, was demonstrated to be linked with the epigenetic state of the basal keratinocytes,6,7 and several recent research findings demonstrated that a dysregulated miRNA may be involved in the pathogenesis of psoriasis.8–10
Several inflammatory mediators are known to upregulate microRNA-155 (miRNA-155) including TNF-α. MiRNA-155 is important for Th17 cell differentiation and is required for optimum DC production of cytokines that promotes Th17 cell formation.11–13 Accordingly, IL-17/IL-17A production was found to be induced by miRNA-155.14
The hypoxia-induced microRNA-210 (miRNA-210) is involved in a variety of physiological and pathological processes.15 Upon normal CD4+ T-cell upregulation of miRNA-210, several inflammatory cytokines are induced, relevantly, IFN-γ and IL-17/IL-17A.16 The latter cytokines have been confirmed as key cytokines in psoriasis pathogenesis.17 MiRNA-210 also counteracts the functions of Treg cell through inhibiting two of its signature suppressive cytokines, namely, IL-10 and TGF-β.16
Similarly, microRNA-20b (miRNA-20b) may also regulate Th17 differentiation. In contrast to previously discussed miRNAs, overexpression of miRNA-20b seems to downregulate Th17 differentiation in vitro and in vivo,18 while its inhibition is associated with a decrease in cytokeratin markers of keratinocyte early differentiation, namely, keratin 1 (K1) and keratin 10 (K10).19
This study aimed at assessing the expression of miRNA-155, miRNA-210, and miRNA-20b in lesional and non-lesional skin as well as sera of psoriatic patients, and correlating them with IL-17 levels.
Methods
This case–control study was conducted at the Kasr ALAiny Psoriasis Unit (KAPU) of the Dermatology Department, Faculty of Medicine, Cairo University after approval from the Dermatology Research Ethical Committee (Derma REC). All participants were required to sign an informed consent to participate in this study before enrollment.
Sample size calculation
Our main outcome variables are the level of miRNA-155, miRNA-210, and miRNA-20b in psoriasis patients’ lesional and non-lesional skin, which were statistically compared by Wilcoxon signed-rank tests. Since this is the first study to be conducted on these parameters, a clinically significant effect size d = 0.7 was assumed, considered to be a moderate to large effect size as per Cohen’s 1988 criteria. A statistical power analysis was performed for sample size estimation; with a two-tailed alpha error probability set at 0.05, the projected sample size needed for the aforementioned effect size (d = 0.7) is approximately N = 20 patients to be able to reject the null hypothesis that this difference is zero with probability (power) 0.8. For reference to normal skin, an equally sized sample of healthy controls was selected. The sample size calculation was done using G*Power 3.1.9.2.
Patients’ group
A total of 20 consecutively recruited adult psoriasis vulgaris patients of both sexes, not receiving any relevant systemic or topical treatment for at least 4 weeks before initiation of our study, were recruited. A detailed history was obtained from all recruited patients.
Clinical assessment was performed using the extent of disease (%) as measured by the rule of 9,20 as well as the assessment of disease severity using Psoriasis Area and Severity Index (PASI score).21,22
Five milliliters of venous blood was collected from each patient in sterile plastic tubes, which were left 30 min to clot. After separation of serum samples by centrifugation at 3000 r/min for 15 min, the sera were stored at −80°C for further analysis.
Two punch biopsies (4 mm) were obtained from lesional as well as non-lesional skin of all recruited patients. Each sample was cut into two halves and stored in empty test tubes at −80°C.
Control group
A total of 20 healthy subjects undergoing abdominoplasty, breast reduction, and brachioplasty operations were recruited as a control group after signing informed consent. Pre-operative blood samples, as well as skin biopsies from excess skin after surgery, were obtained. All recruits were above 18 years with no history of chronic dermatological or systemic disease or a history of malnutrition.
Quantitation of IL-17/IL-17A in serum and tissue homogenate supernatant using enzyme-linked immunosorbent assay
Each skin biopsy was weighed and homogenized in 300 ul phosphate buffer saline then centrifuged at 4000 xg for 10 min. then the supernatant was used for determination of, IL-17/IL-17A concentrations in serum and supernatant using Human, IL-17/IL-17A enzyme-linked immunosorbent assay (ELISA) kit provided from Quantikine (R&D Systems, Inc., Minneapolis, MN, USA).
Fold change of miRNA-155, miRNA-210, and miRNA-20b in tissue and serum using real-time polymerase chain reaction
RNA was extracted from tissue and serum using (Qiagen, Valencia, CA, USA). RNA samples were subjected to RNA quantitation and purity assessment using the NanoDrop® (ND)-1000 spectrophotometer (NanoDrop Technologies, Inc., Wilmington, DE, USA). Reverse transcription was carried out on extracted RNA in a final volume of 20-μL RT reactions using the miScript II RT kit (Qiagen, Valencia, CA, USA). Quantitative real-time polymerase chain reaction (qPCR) was used for the detection of mature miRNAs. After the PCR cycles, melting curve analyses were performed to validate the specific generation of the expected PCR product. As there is no known control miRNA in serum, SNORD was used as an endogenous control. The expression level of miR-155, miR-210, and miR-20b was evaluated using the ΔCt method. The cycle threshold (Ct) value is the number of qPCR cycles required for the fluorescent signal to cross a specified threshold. ΔCt was calculated by subtracting the Ct values of SNORD 68 from those of target miRNAs. ΔΔCt was calculated by subtracting the ΔCt of the control samples from the ΔCt of the disease samples. The fold change in miR-155, miR-210, and miR-20b expression was calculated by the equation 2−ΔΔCt.
Statistical methods
Data were statistically described in terms of mean ± standard deviation (±SD), median and range, or frequencies (number of cases) and percentages when appropriate. A comparison of numerical variables between the study groups was done using the Mann–Whitney U test for independent samples. Within-group comparison of numerical variables was done using Freidman’s test with the Wilcoxon signed-rank test for paired (matched) samples as post hoc multiple two-group comparisons after applying Bonferroni adjustment for multiple comparisons. For comparing categorical data, chi-square (χ2) test was performed. The exact test was used instead when the expected frequency is less than 5. Correlation between various variables was done using the Spearman rank correlation equation for non-normal variables/non-linear monotonic relation. The r value was used for Pearson’s correlation coefficient for continuous variables, while r value was used for Spearman’s correlation coefficient for variables that have been converted into ranked scores, where one or more of the variables were not continuous. P values of less than 0.05 were considered statistically significant. All statistical calculations were done using computer program SPSS (Statistical Package for the Social Sciences; SPSS Inc., Chicago, IL, USA) release 15 for Microsoft Windows (2006).
Results
Clinical data of patients and controls
A total of 20 psoriasis patients (15 males (75%) and 5 females (25%)), whose ages ranged from 21 to 66 years (mean 41.80 ± SD 12.25 years) and 20 healthy subjects who served as controls, were involved in this case–control study. There was no significant difference between both groups regarding age (P = 0.694) and sex (P = 0.723) as summarized in Table 1.
Table 1.
Clinical characteristics of patient group.
| Clinical variable | Results |
|---|---|
| Extent of disease (%) | |
| Range | 7–80 |
| Median | 30 |
| PASI | |
| Range | 5.6–56.8 |
| Median | 19 |
| Course (no. %) | |
| Progressive | 7 (35%) |
| Remissions and exacerbations | 13 (65%) |
| Duration (months) | |
| Range | 8–360 |
| Mean ± SD | 122.55 ± 106.91 |
| Family history (no. %) | |
| Negative | 16 (80%) |
| Positive | 4 (20%) |
| Hypertension (no. %) | |
| Positive | 3 (15%) |
| Negative | 17 (85%) |
| Diabetes mellitus | |
| Positive | 2 (10%) |
| Negative | 18 (90%) |
PASI: Psoriasis Area and Severity Index; SD: standard deviation.
Tissue expression of the miRNAs
Fold increase in miRNA-155, miRNA-210, and miRNA-20b level was significantly higher in lesional skin than non-lesional and control skin. Non-lesional miRNA-20b fold increase was also significantly higher than the control skin (Table 2).
Table 2.
MiRNA fold change of studied groups.
| MiRNA | Psoriatic skin miRNA fold
change |
Patients serum miRNA fold
change |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| 155 |
210 |
20b |
155 | 210 | 20b | ||||
| Lesional | Non-lesional | Lesional | Non-lesional | Lesional | Non-lesional | ||||
| Range Median |
0.8–5 1.4 |
0–2.7 0.91 |
0–11.4 2.30 |
0–6.4 1.06 |
1.1–15.8 3.18 |
0.2–8.1 1.36 |
0.2–1.9 1.18 |
0.5–25.3 4.88 |
0.2–31.5 1.95 |
|
P
value Controls |
0.001* | 0.001* | 0.001* | – | – | – | |||
| 0.001* | 0.511 | 0.010* | 0.198 | 0.001* | 0.018* | 0.050 | 0.001* | 0.056 | |
significant p value <0.05.
All studied miRNAs showed a significant positive correlation between their lesional and non-lesional levels (P < 0.001).
Lesional miRNA-210 positively correlated with extent of disease (P = 0.027, r = 0.495) and both its lesional and non-lesional expression correlated positively with PASI score (P = 0.006, r = 0.596 and P = 0.004, r = 0.610, respectively).
A statistically significant positive correlation was detected between non-lesional expression of miRNA-20b and PASI score (P = 0.011, r = 0.588).
Serum expression of miRNAs
All studied miRNA serum levels were higher in patients compared to controls; however, this increase was only significant for miRNA-210 (Table 2).
IL-17/IL-17A
Lesional IL-17/IL-17A levels were significantly higher than non-lesional and control skin, while non-lesional IL-17/IL-17A was significantly higher than control skin. Similarly, the serum IL-17/IL-17A level was significantly higher in patients than in controls (Table 3).
Table 3.
IL-17 levels in patients and controls.
| Lesional (pg/g) | Non-lesional (pg/g) | Serum | |
|---|---|---|---|
| Patients | |||
| Range | 1553–20,829 | 302–854 | 20.7–35.1 |
| Median | 5357.4 | 601.17 | 24.81 |
| Controls | |||
| Range | 118.1–497.6 | 10.7–23.7 | |
| Median | 243.97 | 17.87 | |
| P value | <0.001* | <0.001* | |
significant p value <0.05.
Correlation between miRNA-155, miRNA-210, miRNA-20b, and IL-17/IL-17A in patients
In lesional skin, a statistically significant positive correlation was detected between miRNA-155 and miRNA-20b (P = 0.020, rs = 0.514), whereas in the serum samples, a statistically significant positive correlation was detected between miRNA-210 and serum IL-17/IL-17A (P = 0.010, rs = 0.562).
Another statistically significant positive correlations were present between lesional miRNA-20b expression and its serum expression (P = 0.046, rs = 0.451) (Figure 1).
Figure 1.
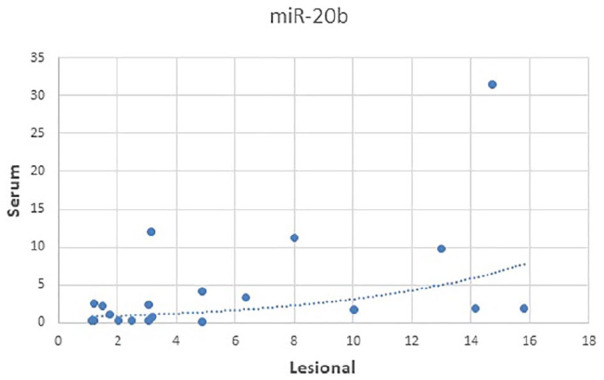
Correlation between lesional and serum expression of miRNA-20b in psoriasis patients.
Discussion
miRNA regulation of gene expression at the post-transcriptional level is considered an important epigenetic mechanism that seems to be involved in the pathogenesis of psoriasis.5 Multiple pieces of research have revealed the role of Th17 cells as important regulators of psoriatic skin inflammation where IL-17/IL-17A induces the production of various chemokines and cytokines by keratinocytes, thereby bridging the innate and adaptive immune systems to sustain a chronic inflammation.23,24 The relation between miRNAs and Th 17 cells is occasionally reported in the literature and may be involved in psoriasis.14
In this study, our results revealed a significant fold increase in psoriasis lesional miRNA-155, 210, and 20b. Except for miRNA-20b, these miRNAs’ fold increase was not statistically significant in non-lesional skin. The expression of the examined miRNAs was significantly higher in lesional skin compared to non-lesional skin. Moreover, the serum fold increase of these miRNAs was higher in patients than controls, but the difference was only statistically significant for miRNA-210 (Table 2). As expected, lesional, non-lesional, and serum IL-17/IL-17A levels were significantly higher in patients when compared to controls (Table 3).
MiRNA-155 is upregulated by TNF-α and is an important factor for Th17 cell differentiation.11,12 It directly inhibiting SOCS1, a negative regulator of the JAK/ STAT signaling pathway, thus enhancing Th17 cell function and differentiation. Accordingly, IL-17/IL-17A production was found to be induced by miRNA-155.14 Moreover, miRNA-155 is required for optimum DC production of cytokines that promote Th17 cell formation.13
In this work, miRNA-155 showed 1.4-fold upregulation in psoriatic skin, which was in concordance with the studies of Zibert et al.8 in which miRNA-155 showed 1.24-fold elevation, and Xia and Zhang25 in which showed 2.7- fold upregulation in psoriatic skin. Moreover, miR-155 was reported to be upregulated in dermal mesenchymal stem cells (MSCs) of psoriasis patients.26 As described by Xu et al.,10 miRNA-155 expression in MSCs is induced by inflammatory cytokines IFN-γ, TNF-α, or IL-1, which are all upregulated in psoriasis.
Interestingly, miRNA-155 was also shown to negatively regulate the MSC iNOS expression by inhibiting its target gene TAB2. iNOS is an inflammation-induced enzyme that plays an important role in NO synthesis, inflammatory cytokine secretion, and angiogenesis. Inhibition of iNOS expression in MSCs leads to a decrease in the immunosuppressive capacity of these cells which cannot sufficiently suppress the inflammatory response in psoriatic lesions.10
The increased expression of miRNA-155 in lesional psoriatic skin in comparison with non-lesional skin we report here is in concordance with the finding of Løvendorf and Skov27 of upregulated miRNA-155 in psoriasis plaque dermal inflammatory cell infiltrates vs normal psoriatic reticular dermis. On the other hand, although previous investigators reported an increased miRNA-155 expression detected in psoriatic patients’ peripheral blood mononuclear cells (PBMCs) that was correlated with PASI,28 we did not detect such correlation.
These findings support the role of miRNA-155 in the pathogenesis of psoriasis, and it can be hypothesized that inflammatory cytokines involved in psoriasis pathogenesis as IFN-γ, TNF-α, or IL-1 increase the expression of miRNA155, which in turn augments the inflammatory process in psoriasis by upregulating IL-17/IL-17A production and rendering the immune-suppressing dermal MSCs dysfunctional. In addition, miRNA-155 is capable of directly increasing TNF-α levels,29 one of the major cytokines involved in psoriasis.30 Although the expression of miRNA-155 was not correlated with the levels of IL-17/IL-17A in psoriasis patients, this may be explained by the fact that Th17 are not the only cells capable of IL-17/IL-17A production, in addition to the different pathways that may influence IL-17/IL-17A production in psoriasis.
Furthermore, the data regarding psoriasis being not as common in less polluted rural areas than polluted urban communities31,32 and that miRNA-155 is upregulated by pollution,33,34 may be a link between environmental factors and psoriasis, and certainly deserves further investigations.
MiRNA-210 is a member of the hypoxia-induced miRNA that is known to upregulate angiogenesis and inhibit apoptosis.35 Hypoxia-inducible factor 1α (HIF-1α) is vital for Th17 differentiation36 and was found to be overexpressed in psoriatic skin37,38 and serum.39 HIF-1α was found to drive miRNA-210 overexpression.40 Together with the ability of miRNA-210 to induce crucial psoriasis mediators like IL-17/IL-17A as well as to attenuate the immune regulatory functions of Tregs16 makes this miRNA an ideal target for therapeutic targeting in psoriasis.
Our study demonstrated elevated expression of miRNA-210 in psoriatic skin and serum, thus providing further support of the role of miRNA-210 in the pathogenesis of psoriasis. Furthermore, Zhao et al.16 demonstrated the upregulation of miRNA-210 in PBMCs of psoriasis patients compared to healthy controls.
The significant positive correlation between serum expression of miRNA-210 and IL-17/IL-17A levels in psoriatic patients (P = 0.010, r = 0.562) provides further support for the important role of this miRNA in stimulating IL-17/IL-17A production and focuses the light on the possible interaction between miRNA-210 and Th17 cells in the pathogenesis of psoriasis. Moreover, the positive correlation between lesional miRNA-210 with both PASI and body surface area (BSA) affected by psoriasis suggests an important relationship between this miRNA and the disease activity and may encourage further investigations to explore the possibility of using this miRNA as one of the markers of psoriasis severity.
Interestingly, although non-lesional miRNA-210 expression was not significantly higher (P = 0.198) than normal control skin, it positively correlated with PASI (P = 0.004, r = 0.610) as well as lesional miRNA-210 (P < 0.001, r = 0.884). Moreover, miRNA-210 expression was significantly higher in patients’ sera compared to controls (P = 0.001). Such observations may suggest a direct participation of miRNA-210 in the development and severity of psoriasis.
Again, environmental factors may be linked to psoriasis through miRNA-210, as several authors have demonstrated dysregulation of this miRNA with arsenic intake.41,42 Furthermore, because of the role of miRNA-210 in cardiovascular diseases,43 this encourages investigations of this miRNA for links between cardiovascular diseases and psoriasis.
MiRNA-20b is a member of the miR-106a-363 cluster, which are members of miR-17 family44 that are frequently overexpressed in various human neoplasias.45–47 Lei et al.48 showed that miRNA-20b regulates vascular endothelial growth factor (VEGF), which is mediated by HIF-1α and STAT3.49
In experimental autoimmune encephalomyelitis (EAE), when miRNA-20b was overexpressed, a decrease in Th17 differentiation by targeting RORγt and STAT3 was observed with a resultant decrease in disease severity.18 However, in our study, miRNA-20b was overexpressed, did not correlate with IL-17/IL-17A, and the non-lesional levels correlated with the PASI score. Thus, it does not seem that miRNA-20b plays a protective role in the setting of psoriasis. Conversely, it has been demonstrated that miR-20b binds to the 3′-UTR of phosphatase and tensin homolog (PTEN) and suppresses its translation, and therefore decreases PTEN protein.50–52 PTEN has been reported to be downregulated in psoriatic lesions,53,54 explaining the excessive proliferation and resistance to apoptosis in psoriatic epidermis.54 The upregulation of p63 in the psoriatic epidermis may augment miRNA-20b expression which acts downstream of p63.19,55
It had been previously demonstrated that miRNA expression in plasma and serum appears to reflect the extrusion of miRNAs from distant tissues or organs or disease pathways.56–59 In this study, this was the case only for miRNA-20b which showed such relation between its serum and lesional levels (P = 0.046, rs = 0.451) (Figure 1). Lesional miRNA-20b also correlated positively with its non-lesional levels (P < 0.001, rs = 0.709). Thus, it seems that both tissue and serum miRNA20-b in psoriasis patients may share similar mechanisms of regulation.
Both lesional and serum levels of miRNA-155 and miRNA-20b correlated positively in our patients (P = 0.020, rs = 0.514 and P = 0.013, rs = 0.544, respectively), and serum miRNA-155 also correlated positively with lesional miRNA-20b expression (P = 0.027, rs = 0.493). This implies that there may be common factors responsible for the upregulation of these miRNAs. It would be interesting to investigate whether inflammatory mediators known to upregulate miRNA-155, like TNF-α, may also upregulate miRNA-20b.
MiRNA-155 and 210 levels were insignificantly higher in non-lesional compared to normal control skin but were significantly higher in lesional skin compared to non-lesional and control skin. These findings may support the hypotheses of Naldi60 and Zhang et al.61 that epigenetic mechanisms, including miRNAs expression, may explain the discordance of psoriasis development among certain portions of the skin within the same patient.
These data suggest the possible involvement of the miRNAs examined in the pathogenesis of psoriasis; however, the data are limited by the small sample size. Another limitation would be the lack of follow-up for the measured proteins after therapeutic intervention, which would have possibly shed more light on the interactions of the examined miRNAs and IL-17/IL-17A axis in the context of psoriasis.
Conclusion
This study highlights the possible contribution of miRNA-155, 210, and 20b in the pathogenesis of psoriasis. These results need further verification in large-scale studies to determine the degree of involvement of these miRNAs in psoriasis, which also seem to be involved in comorbidities associated with psoriasis. The significance of identifying environmental factors associated with miRNAs regulation in psoriasis cannot be underscored enough as it may provide possible means to limit disease progression, exacerbations, and severity.
Footnotes
Declaration of conflicting interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Ethical approval: Ethical approval for this study was obtained from Dermatology Research Ethical Committee (15 September 2017, Serial No. 10/2015).
Funding: The author(s) received no financial support for the research, authorship, and/or publication of this article.
Informed consent: Written informed consent was obtained from all subjects before the study.
ORCID iD: Mohamed El-Komy  https://orcid.org/0000-0002-9653-2938
https://orcid.org/0000-0002-9653-2938
References
- 1. Zhou F, Shen C, Xu J, et al. (2016) Epigenome wide association data implicates DNA methylation mediated genetic risk in psoriasis. Clinical Epigenetics 8: 131–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Martin DA, Towne JE, Kricorian G, et al. (2013) The emerging role of IL-17 in the pathogenesis of psoriasis: Preclinical and clinical findings. Journal of Investigative Dermatology 133(1): 17–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zaba LC, Suárez-Fariñas M, Fuentes-Duculan J, et al. (2009) Effective treatment of psoriasis with etanercept is linked to suppression of IL-17 signaling, not immediate response TNF genes. The Journal of Allergy and Clinical Immunology 124(5): 1022, 10.e1–e395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lu Q. (2013) The critical importance of epigenetics in autoimmunity. Journal of Autoimmunity 41: 1–5. [DOI] [PubMed] [Google Scholar]
- 5. Chandra A, Ray A, Senapati S, et al. (2015) Genetic and epigenetic basis of psoriasis pathogenesis. Molecular Immunology 64(2): 313–323. [DOI] [PubMed] [Google Scholar]
- 6. Rishi V, Bhattacharya P, Chatterjee R, et al. (2010) CpG methylation of half-CRE sequences creates C/EBP alpha binding sites that activate some tissue-specific genes. Proceedings of the National Academy of Sciences of the United States of America 107(47): 20311–20316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gudjonsson JE, Krueger G. (2012) A role for epigenetics in psoriasis: Methylatedcytosine-guanine sites differentiate lesional from nonlesional skin and from normal skin. Journal of Investigative Dermatology 132(3 Pt.1): 506–508. [DOI] [PubMed] [Google Scholar]
- 8. Zibert JR, Løvendorf MB, Litman T, et al. (2010) MicroRNAs and potential target interactions in psoriasis. Journal of Dermatological Science 58(3): 177–185. [DOI] [PubMed] [Google Scholar]
- 9. Meisgen F, Xu N, Wei T, et al. (2012) miR-21 is up-regulated in psoriasis and suppresses T cell apoptosis. Experimental Dermatology 21(4): 312–314. [DOI] [PubMed] [Google Scholar]
- 10. Xu N, Meisgen F, Butler LM, et al. (2013) MicroRNA-31 is overexpressed in psoriasis and modulates inflammatory cytokine and chemokine production in keratinocytes via targeting serine/threonine kinase 40. Journal of Immunology 190(2): 678–688. [DOI] [PubMed] [Google Scholar]
- 11. O’Connell RM, Taganov KD, Boldin MP, et al. (2007) MicroRNA-155 is induced during the macrophage inflammatory response. Proceedings of the National Academy of Sciences of the United States of America 104(5): 1604–1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ruggiero T, Trabucchi M, De Santa F, et al. (2009) LPS induces KH-type splicing regulatory protein-dependent processing of microRNA-155 precursors in macrophages. The FASEB Journal 23(9): 2898–2908. [DOI] [PubMed] [Google Scholar]
- 13. O’Connell RM, Kahn D, Gibson WS, et al. (2010) MicroRNA 155 promotes autoimmune inflammation by enhancing inflammatory T cell development. Immunity 33(4): 607–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yao R, Ma YL, Liang W, et al. (2012) MicroRNA 155 modulates Treg and Th17 cells differentiation and Th17 cell function by targeting SOCS1. PLoS ONE 7(10): e46082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ivan M, Harris AL, Martelli F, et al. (2008) Hypoxia response and microRNAs: No longer two separate worlds. Journal of Cellular and Molecular Medicine 12(5A): 1426–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhao M, Wang LT, Liang GP, et al. (2014) Up-regulation of microRNA 210 induces immune dysfunction via targeting FOXP3 in CD4(+) T cells of psoriasis vulgaris. Journal of Clinical Immunology 150(1): 22–30. [DOI] [PubMed] [Google Scholar]
- 17. Quaglino P, Bergallo M, Ponti R, et al. (2011) Th1, Th2, Th17 and regulatory T cell pattern in psoriatic patients: Modulation of cytokines and gene targets induced by etanercept treatment and correlation with clinical response. Dermatology 223(1): 57–67. [DOI] [PubMed] [Google Scholar]
- 18. Zhu E, Wang X, Zheng B, et al. (2014) miR-20b suppresses Th17 differentiation and the pathogenesis of experimental autoimmune encephalomyelitis by targeting RORγt and STAT3. Journal of Immunology 192(12): 5599–5609. [DOI] [PubMed] [Google Scholar]
- 19. Wu N, Sulpice E, Obeid P, et al. (2012) The miR-17 family links p63 protein to MAPK signaling to promote the onset of human keratinocyte differentiation. PLoS ONE 7(9): e45761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Livingston EH, Lee S. (2000) Percentage of burned body surface area determination in obese and nonobese patients. Journal of Surgical Research 91(2): 106–110. [DOI] [PubMed] [Google Scholar]
- 21. Wittkowski KM, Krueger JG, Leonardi C, et al. (2011) Clinical symptoms of skin, nails, and joints manifest independently in patients with concomitant psoriasis and psoriatic arthritis. PLoS ONE 6(6): e20279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ports WC, Khan S, Lan S, et al. (2013) A randomized phase 2a efficacy and safety trial of the topical Janus kinase inhibitor tofacitinib in the treatment of chronic plaque psoriasis. British Journal of Dermatology 169(1): 137–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Adami S, Cavani A, Rossi F, et al. (2014) The role of interleukin-17A in psoriatic disease. Biodrugs 28(6): 487–497. [DOI] [PubMed] [Google Scholar]
- 24. Lynde CW, Poulin Y, Vender R, et al. (2014) Interleukin 17A: Toward a new understanding of psoriasis pathogenesis. Joint American Academy of Dermatology 71(1): 141–150. [DOI] [PubMed] [Google Scholar]
- 25. Xia J, Zhang W. (2014) MicroRNAs in normal and psoriatic skin. Physiological Genomics 46(4): 113–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hou RX, Liu RF, Zhao XC, et al. (2016) Increased miR-155-5p expression in dermal mesenchymal stem cells of psoriatic patients: Comparing the microRNA expression profile by microarray. Genetics and Molecular Research 15(3): 38631. [DOI] [PubMed] [Google Scholar]
- 27. Løvendorf MB, Skov L. (2015) miRNAs in inflammatory skin diseases and their clinical implications. Expert Review of Clinical Immunology 11(4): 467–477. [DOI] [PubMed] [Google Scholar]
- 28. García-Rodríguez S, Arias-Santiago S, Blasco-Morente G, et al. (2017) Increased expression of microRNA-155 in peripheral blood mononuclear cells from psoriasis patients is related to disease activity. The Journal of the European Academy of Dermatology and Venereology 31(2): 312–322. [DOI] [PubMed] [Google Scholar]
- 29. Tili E, Michaille JJ, Cimino A, et al. (2007) Modulation of miR-155 and miR-125b levels following lipopolysaccharide/TNF-alpha stimulation and their possible roles in regulating the response to endotoxin shock. Journal of Immunology 179(8): 5082–5089. [DOI] [PubMed] [Google Scholar]
- 30. Chiricozzi A, Guttman-Yassky E, Suárez-Fariñas M, et al. (2011) Integrative responses to IL-17 and TNF-α in human keratinocytes account for key inflammatory pathogenic circuits in psoriasis. Journal of Investigative Dermatology 131(3): 677–687. [DOI] [PubMed] [Google Scholar]
- 31. Braathen LR, Botten G, Bjerkedal T. (1989) Prevalence of psoriasis in Norway. Acta Dermato-Venereologica 142: 5–8. [PubMed] [Google Scholar]
- 32. Valenzuela F, Silva P, Valdés MP, et al. (2011) Epidemiology and quality of life of patients with psoriasis in Chile. Actas Dermosifiliogr 102(10): 810–816. [DOI] [PubMed] [Google Scholar]
- 33. Levänen B, Bhakta NR, Torregrosa Paredes P, et al. (2013) Altered microRNA profiles in bronchoalveolar lavage fluid exosomes in asthmatic patients. The Journal of Allergy and Clinical Immunology 131(3): 894–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Vrijens K, Bollati V, Nawrot TS. (2015) MicroRNAs as potential signatures of environmental exposure or effect: A systematic review. Environmental Health Perspectives 123(5): 399–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hu S, Huang M, Li Z, et al. (2010) MicroRNA-210 as a novel therapy for treatment of ischemic heart disease. Circulation 122(11 Suppl): S124–S131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Korn T, Bettelli E, Oukka M, et al. (2009) IL-17 and Th17 Cells. Annual Review of Immunology 27: 485–517. [DOI] [PubMed] [Google Scholar]
- 37. Rosenberger C, Solovan C, Rosenberger AD, et al. (2007) Upregulation of hypoxia-inducible factors in normal and psoriatic skin. Journal of Investigative Dermatology 127(10): 2445–2452. [DOI] [PubMed] [Google Scholar]
- 38. Ioannou M, Sourli F, Mylonis I, et al. (2009) Increased HIF-1 alpha immunostaining in psoriasis compared to psoriasiform dermatitides. Journal of Cutaneous Pathology 36(12): 1255–1261. [DOI] [PubMed] [Google Scholar]
- 39. Vasilopoulos Y, Sourli F, Zafiriou E, et al. (2013) High serum levels of HIF-1α in psoriatic patients correlate with an over-expression of IL-6. Cytokine 62(1): 38–39. [DOI] [PubMed] [Google Scholar]
- 40. Dang K, Myers KA. (2015) The role of hypoxia-induced miR-210 in cancer progression. International Journal of Molecular Sciences 16(3): 6353–6372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Meng XZ, Zheng TS, Chen X, et al. (2011) microRNA expression alteration after arsenic trioxide treatment in HepG-2 cells. Journal of Gastroenterology and Hepatology 26(1): 186–193. [DOI] [PubMed] [Google Scholar]
- 42. Wang J, Cui Q. (2012) Specific roles of microRNAs in their interactions with environmental factors. Journal of Nucleic Acids 2012: 978384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Greco S, Gaetano C, Martelli F. (2014) HypoxamiR regulation and function in ischemic cardiovascular diseases. Antioxidants & Redox Signaling 21(8): 1202–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tanzer A, Stadler PF. (2004) Molecular evolution of a microRNA cluster. Journal of Molecular Biology 339(2): 327–335. [DOI] [PubMed] [Google Scholar]
- 45. Matsubara H, Takeuchi T, Nishikawa E, et al. (2007) Apoptosis induction by antisense oligonucleotides against miR-17-5p and miR-20a in lung cancers overexpressing miR-17-92. Oncogene 26(41): 6099–6105. [DOI] [PubMed] [Google Scholar]
- 46. Ventura A, Young AG, Winslow MM, et al. (2008) Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell 132(5): 875–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Inomata M, Tagawa H, Guo YM, et al. (2009) MicroRNA-17-92 down-regulates expression of distinct targets in different B-cell lymphoma subtypes. Blood 113(2): 396–402. [DOI] [PubMed] [Google Scholar]
- 48. Lei Z, Li B, Yang Z, et al. (2009) Regulation of HIF-1α and VEGF by miR-20b tunes tumor cells to adapt to the alteration of oxygen concentration. PLoS ONE 4(10): e7629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Cascio S, D’Andrea A, Ferla R, et al. (2010) miR-20b modulates VEGF expression by targeting HIF-1 alpha and STAT3 in MCF-7 breast cancer cells. Journal of Cellular Physiology 224(1): 242–249. [DOI] [PubMed] [Google Scholar]
- 50. Li J, Yen C, Liaw D, et al. (1997) PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science 275(5308): 1943–1947. [DOI] [PubMed] [Google Scholar]
- 51. Zhou W, Shi G, Zhang Q, et al. (2014) MicroRNA-20b promotes cell growth of breast cancer cells partly via targeting phosphatase and tensin homologue (PTEN). Cell and Bioscience 4(1): 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Chalhoub N, Baker SJ. (2009) PTEN and the PI3-kinase pathway in cancer. Annual Review of Pathology 4: 127–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Li Y, Man X, You L, et al. (2014) Downregulation of PTEN expression in psoriatic lesions. International Journal of Dermatology 53(7): 855–860. [DOI] [PubMed] [Google Scholar]
- 54. Hong KK, Gwak MJ, Song J, et al. (2016) Nuclear factor-κB pathway activation and phosphatase and tensin homolog downregulation in psoriasis. British Journal of Dermatology 174(2): 433–435. [DOI] [PubMed] [Google Scholar]
- 55. Shen CS, Tsuda T, Fushiki S, et al. (2005) The expression of p63 during epidermal remodeling in psoriasis. The Journal of Dermatology 32(4): 236–242. [DOI] [PubMed] [Google Scholar]
- 56. Mitchell PS, Parkin RK, Kroh EM, et al. (2008) Circulating microRNAs as stable blood-based markers for cancer detection. Proceedings of the National Academy of Sciences of the United States of America 105(30): 10513–10518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wang K, Zhang S, Marzolf B, et al. (2009) Circulating microRNAs, potential biomarkers for drug-induced liver injury. Proceedings of the National Academy of Sciences of the United States of America 106(11): 4402–4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Tsujiura M, Ichikawa D, Komatsu S, et al. (2010) Circulating microRNAs in plasma of patients with gastric cancers. British Journal of Cancer 102(7): 1174–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zahm AM, Thayu M, Hand NJ, et al. (2011) Circulating microRNA is a biomarker of pediatric Crohn disease. Journal of Pediatric Gastroenterology and Nutrition 53(1): 26–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Naldi L. (2013) Risk factors for psoriasis. Current Dermatology Reports 2: 58–65. [Google Scholar]
- 61. Zhang P, Zhao M, Liang G, et al. (2013) Whole-genome DNA methylation in skin lesions from patients with psoriasis vulgaris. Journal of Autoimmunity 41: 17–24. [DOI] [PubMed] [Google Scholar]


