Abstract
Base editing has the potential to improve important economic traits in agriculture and can precisely convert single nucleotides in DNA or RNA sequences into minimal double-strand DNA breaks (DSB). Adenine base editors (ABE) have recently emerged as a base editing tool for the conversion of targeted A:T to G:C, but have not yet been used in sheep. ABEmax is one of the latest versions of ABE, which consists of a catalytically-impaired nuclease and a laboratory-evolved DNA-adenosine deaminase. The Booroola fecundity (FecBB) mutation (g.A746G, p.Q249R) in the bone morphogenetic protein receptor 1B (BMPR1B) gene influences fecundity in many sheep breeds. In this study, by using ABEmax we successfully obtained lambs with defined point mutations that result in an amino acid substitution (p.Gln249Arg). The efficiency of the defined point mutations was 75% in newborn lambs, since six lambs were heterozygous at the FecBB mutation site (g.A746G, p.Q249R), and two lambs were wild-type. We did not detect off-target mutations in the eight edited lambs. Here, we report the validation of the first gene-edited sheep generated by ABE and highlight its potential to improve economically important traits in livestock.
Introduction
Clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated (Cas) 9 has been widely used to produce gene-edited animals and plants [1]. Recently, the CRISPR/Cas9 system has emerged as a simple, rapid and precise editing tool in the genomes of livestock. However, the application of this system for the introduction of defined point mutations requires the generation of double-strand DNA breaks (DSB) under the guidance of a single guide RNA (sgRNA) and homology recombination (HR) via single-stranded oligodeoxynucleotides (ssODN). Non-homologous end-joining (NHEJ) and HR are the two major DSB repair pathways [2–5]. The NHEJ pathway is active throughout the entire cell cycle and is more efficient in the repair of DSB. Thus, it is inefficient to generate animal models with defined point mutations via the HR method [6]. The base editing systems emerged thanks to the engineering of CRISPR/Cas9. Currently, two major classes of base editors exist, cytosine base editors (CBE) and adenine base editors (ABE) [7, 8].
The original CBE were developed to convert targeted C:G to T:A [7, 9] and comprised four molecules: a cytosine deaminase that catalyzes the conversion of C to U; a modified Cas9 (nCas9/dCas9) that binds target DNA; an sgRNA that directs Cas9-cytosine deaminase to the target locus; and a UGI that subverts the cellular uracil base excision repair (BER) pathway [10]. After several generations of optimization, base editor 3 (BE3) was developed and applied in a wide range of organisms, with mutation efficiencies higher than 50% in mammalian cells, mouse embryos, tripronuclear human embryos, rabbits, sheep, and goats [7, 11–16]. Since in farm animals, many of the economic traits are due to A:T to G:C substitutions, a catalytically-impaired nuclease has been fused with a laboratory evolved DNA-adenosine deaminase termed ABE to convert targeted A:T to G:C [8]. ABE catalyzes the deamination of adenine to inosine, which is treated as guanosine by the polymerase. Following DNA replication, the A:T base pairs are converted to G:C base pairs. ABE of the E. coli tRNA adenosine deaminase (TadA) have evolved through mutations, resulting in the formation of four commonly used ABE, namely ABE6.3, ABE7.8, ABE7.9, and ABE7.10 [17] with ABE7.10 being the most active base editor that shows an average editing efficiency of up to 53% [10]. The editing window of ABE7.10 targets adenosine at the protospacer adjacent motif (PAM) position 4–7 (PAM counted as 21–23), while the other three versions have slightly wider editing windows at position 4–9. However, the editing efficiency may be lower at position 4–9. To further improve the efficiency of ABE7.10, ABEmax and xCas9-ABE were generated separately. ABE7.10 was optimized through the modification of nuclear localization signals (NLS) and codon usage to obtain ABEmax. The resulting ABEmax editor can alter single nucleotide polymorphisms (SNPs) with substantially increased efficiency and low genome-wide off-target activity in a variety of mammalian cell types [18]. xCas93.7 replaced the SpCas9 of ABE7.10 to obtain xCas9-ABE, which leads to higher base editing efficiencies than ABE7.10 in HEK293T cells [19]. Although cytosine base editors often produce cell populations with mixed base editing populations, ABE do not prominently show A to non-G substitutions at the target sites. ABE also have advantages over other approaches in terms of off-target effects. In comparison with Cas9, ABE have a low off-target rate [10]. To date, the application of ABE has been validated in various organisms including human embryos, mice, rabbits, zebrafish, rice, wheat, Arabidopsis, and Brassica napus [13, 20–25].
The bone morphogenetic protein receptor 1B (BMPRIB) gene was first identified in Booroola merino sheep, and was shown to be a major factor associated with increased ovulation rates. This gene influences follicular granulosa cell differentiation and follicular development, thus promoting ovulation [26–30]. The FecBB mutation (g.A746G, p.Q249R) in BMPR1B is highly associated with increased ovulation rate and litter size in domestic sheep breeds [28, 31–37]. In previous studies, we used the BE3 system to induce a p.R96C mutation in the sheep suppressor of cytokine signaling 2 (SOCS2) gene and nonsense mutations in the goat fibroblast growth factor 5 (FGF5) gene. The results demonstrated that CBE could be used to induce single base substitutions (C > T) in large animals with a high efficiency [15, 16]. However, to date, the application of ABE for the generation of genetically-edited large animals has not been reported. In this study, we used ABE (ABEmax) to introduce the FecBB mutation in the genome of Tan sheep, a Chinese local breed. ABEmax mRNAs and sgRNA were coinjected into ovine one-cell stage zygotes, followed by transfer of the developing embryos into surrogate ewes. Although the gene-edited lambs at the FecBB site were generated via Cas9:ssODN, the editing efficiency was low (22.7%) [33]. In the present study, by using ABE to generate founders with defined point mutations in the BMPR1B gene, we observed a much higher editing efficiency compared with the conventional ssODN approach. These results highlight the feasibility of base editors, including both CBE and ABE, to generate large animal models with targeted single nucleotide substitutions.
Methods
Animals
All experimental animals were raised at the Ningxia Tianyuan Sheep Farm, Hongsibu, Ningxia Autonomous Region, China. Water and standard food were provided ad libitum. Animals were treated according to the Guidelines for the Care and Use of Laboratory Animals at Northwest A&F University.
Design of sgRNA
The sequences targeting the g.A746G (p.Q249R) mutation in the ovine BMPR1B gene are listed in Additional file 1: Table S1. Two oligonucleotides (see Additional file 1: Table S2) were used for the in vitro transcription of sgRNAs, which were then synthesized and annealed to form double-stranded oligos. These oligos were subcloned into the pUC57-T7-gRNA vector as previously described [38]. The clones that contained the desired sequences were selected, expanded in culture, and the plasmids were extracted using a plasmid extraction kit (AP-MN-P-250G; Axygen, Union City, CA, USA). The sgRNAs were in vitro transcribed using the MEGAshortscript Kit (AM1354; Ambion, Foster City, CA, USA) and purified using the MEGAClear Kit (AM1908; Ambion). Subsequently, the ABEmax in vitro transcription vectors were used as templates to produce ABE mRNAs following previously published protocols [38].
Screening for high-efficiency ABE versions in sheep fibroblasts
Tissues from a 40-day-old sheep fetus were sectioned and cultured in DMEM medium (Gibco) containing 10% fetal bovine serum (FBS) (Gibco) and 1% penicillin–streptomycin (Gibco). After 3 to 5 days of culture, fetal fibroblasts were isolated and cultured until 70 to 90% confluence. Transfections were performed as previously reported [39]. Briefly, sheep fetal fibroblasts were respectively transfected with FecBB sgRNA (2.5 μg/μL) and ABE (ABE7.10, ABEmax and xCas9-ABE) plasmid (5 μg/μL) using Lipofectamine 3000 Reagent (Invitrogen) in 6-well culture plates. Forty-eight hours post-transfection, 0.2 μL of puromycin (10 μg/μL) was added to the medium and cells were cultured for 36 h. Then, the culture medium was replaced with puromycin-free medium to permit the complete growth of fetal fibroblasts. Genomic DNA was extracted from transfected and drug-screened fibroblasts and used for Sanger sequencing and targeted deep-sequencing. The list of primers is in Additional file 1: Table S3.
Production of mutated sheep
Ten healthy ewes (3 to 5 years old) with normal estrous cycles were selected as donors for zygote collection. The superovulation treatment of the donors was performed as previously described [40]. Briefly, an EAZI-BREED controlled internal drug release (CIDR) Sheep and Goat Device (containing 300 mg of progesterone) was inserted into the vagina of the donor ewes for 12 days and superovulation was performed 60 h prior to the removal of the CIDR Device. Each female donor was subjected to natural mating three times, the first mating was carried out 12 h after the initial estrus, and then subsequent matings were performed at 12 h intervals. Zygotes at the 1-cell stage were collected 48 h after the initial estrus by surgical operation and immediately transferred to TCM-199 medium (Gibco, Gaithersburg, MD, USA). ABEmax mRNA (25 ng μL−1) and sgRNA (10 ng μL−1) were co-injected into the cytoplasm of the collected zygotes using an Eppendorf FemtoJet system [41]. The injection pressure, compensatory pressure, and time parameter were 45 kPa, 7 kPa, and 0.1 s, respectively. Microinjections were performed on the heated stage of an Olympus ON3 micromanipulation system. Injected embryos were cultured in Quinn’s Advantage Cleavage medium (Sage Biopharma, Toronto, Canada) for 24 h and were then transferred into surrogates as previously described [39]. On average, 5.3 embryos were transferred to each recipient. The details on the number of embryos transferred to each recipient and the parents of the embryos are in Additional file 1: Table S4. Pregnancy was confirmed by observing the estrous behaviors of the surrogates at each ovulation cycle. After three estrus cycles (~ 60 days), six of the 18 recipients were pregnant. After around 150 days of pregnancy, six ewes gave birth to eight lambs; no stillbirth or dead animals were found. The eight lambs had good health conditions.
Genotyping of generated founders
Peripheral venous blood of 2 week-old lambs was sampled to extract genomic DNA. After polymerase chain reaction (PCR) amplification, Sanger sequencing was performed using the KOD-NEO-Plus enzyme (DR010A; TOYOBA, Osaka, Japan). The primers used are in Additional file 1: Table S3.
Prediction of off-target sites
The potential off-target sites with not more than three mismatches were predicted using the freely available tool Cas-OFFinder [8]. Off-target sites were searched as previously described [8]. The primers for amplifying the off-target sites and Sanger sequencing are in Additional file 1: Table S5.
Captured deep-sequencing
Target mutations were amplified using a KAPA HiFi HotStart PCR Kit (#KK2501; KAPA Biosystems, Wilmington, MA, USA) to generate deep-sequencing libraries as previously described [16]. The pool of PCR amplicons was sequenced using the MiniSeq with TruSeq HT Dual Index system (Illumina, San Diego, CA, USA).
Results and discussion
Screening for high-efficiency ABE versions in sheep fibroblasts
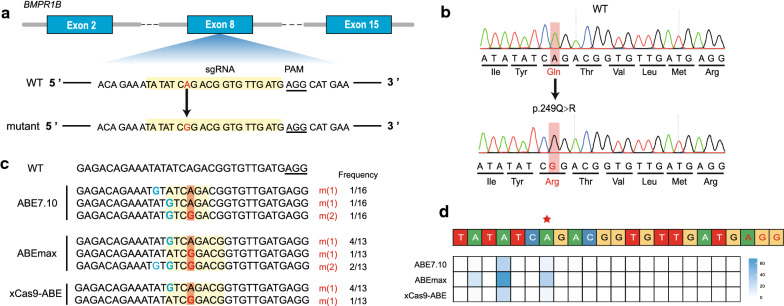
To obtain edited sheep fetal fibroblast cell lines at the target site (p.Q249R) of the BMPR1B gene, we co-transfected sgRNA and ABE (ABE7.10, ABEmax, or xCas9-ABE) plasmids into sheep fetal fibroblasts. The designed sgRNAs encompassed the target point mutation (p.Q249R) in the BMPR1B gene (Fig. 1a). DNA from transfected cells was used for Sanger sequencing, which showed overlapping peaks in the sequencing map (see Additional file 2: Figure S1). Then, we further analyzed specific genotypes using TA cloning (Fig. 1b and c). The results showed that the editing efficiency with ABE7.10, ABEmax, and xCas9-ABE plasmids was up to 3/16, 7/13 and 5/13, respectively, whereas the efficiency at the target site (p.Q249R) was 1/16, 3/13 and 1/13, respectively. Next, we carried out targeted deep-sequencing to validate the accuracy of TA cloning and obtained results that were consistent with TA cloning (Fig. 1d) and with those of other recent studies at the cellular level [11, 17, 18]. The efficiency of the ABEmax plasmid was the highest up to 53.8% (7/13), thus it was selected to produce the targeted FecBB mutation in sheep. These findings highlight how ABE can directly introduce A:T to G:C mutations into sheep fetal fibroblasts, and how it could be a more efficient approach for the generation of gene-edited sheep.
Fig. 1.
Evaluation of different ABE system mediated nucleotide substitutions of BMPR1B in sheep fibroblasts. a Schematic view of the target site in the sheep BMPR1B gene. sgRNA sequences are displayed in a yellow background. PAM sequences are underlined. The ABE-mediated nucleotide substitutions (g.A746G, p.Q249R) are highlighted in red. b Editing efficiency with ABE7.10, ABEmax, and xCas9-ABE in sheep fibroblasts. The editing window are displayed in a yellow background. The ABE-mediated nucleotide substitutions (g.A746G, p.Q249R) are highlighted in an orange background and red. Bystander mutations are indicated in blue. c Sanger sequencing chromatogram of intended mutations derived by the ABE system. d In sheep fibroblasts, editing efficiency with ABE7.10, ABEmax, and xCas9-ABE through deep sequencing in sheep fibroblasts. Bystander mutations are marked in blue. Three adenine base editors mediated nucleotide substitutions (g.A746G, p.Q249R) are highlighted in red
Generation of edited lambs
To generate lambs with a p.Q249R mutation in the BMPR1B gene, we micro-injected sgRNA and ABEmax mRNAs into the cytoplasm of 1-cell stage embryos. Ten mated Tan sheep donors were superovulated and fertilised by natural mating, producing 96 one-cell stage fertilized oocytes [38]. Of the 96 microinjected embryos, 95 were in good condition and transferred into the ampullary-isthmic junction of the oviducts in 18 recipient ewes. Finally, we obtained six pregnancies that reached a full-term gestation period (~ 150 days) and eight lambs (#25, #28, #30, #31, #34, #46, #50, and #52) were born (Table 1). Four ewes (#016, #132, #018, and #708) delivered singletons (#28, #30, #31, and #46, which were not related to each other), and two ewes (#640 and #608) delivered twins (#25 and #34, and #50 and #52, each pair being full-sibs). Details on the lambs’ parents and the recipients are in Additional file 1: Table S4.
Table 1.
Summary of the sheep obtained with the targeted point mutations via ABEmax
| Donor sheep | 10 |
| Collected embryos | 96 |
| ABEmax-sgRNA | |
| Injected embryos | 96 |
| Transferred embryos | 95 |
| Recipient sheep | 18 |
| Pregnant recipients | 6 |
| Newborns | 8 |
| Expected defined substitution | 6 |
| Un-defined substitution | 2 |
ABEmax: the latest version of adenine base editors; sgRNA: single guide RNA
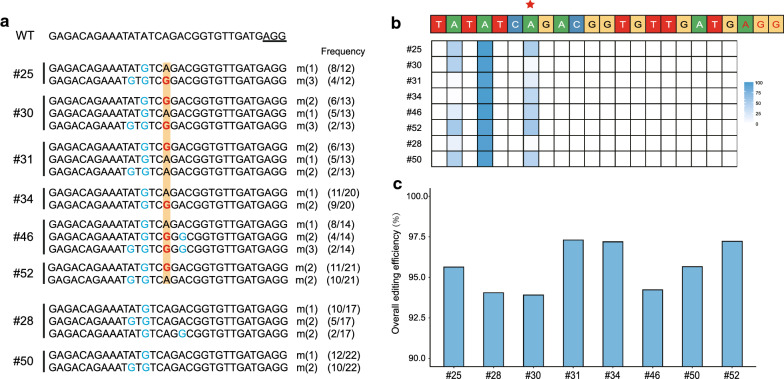
We extracted genomic DNA from the blood of these eight lambs and amplified it by PCR-based Sanger sequencing (see Additional file 1: Table S3). Sanger sequencing primarily showed that these eight lambs displayed editing events within the base editing window of ABEmax, but only six of them (#25, #30, #31, #34, #46, and #52) had the specific edit that conferred the desired Q249R substitution (see Additional file 3: Figure S2). To analyze the specific genotypes of each edited lamb, we performed TA cloning to validate single nucleotide substitutions (Fig. 2a). Sanger sequencing confirmed that lambs #25, #30, #31, #34, #46, and #52 were heterozygous mutants at the FecBB mutation site. The results of TA cloning showed that all the founders had bystander mutations. Three bystander mutations (all are A to G) were detected: from 5′ to PAM direction; A12 (Tyr to Cys) in seven founders, A14 (Ile to Met) in eight founders, and A19 (Thr to Ala) in two founders. In addition, we performed targeted deep sequencing and the results were consistent with those of TA cloning (Fig. 2b and c). All the generated founders were mosaic. This mosaic state is the result of the direct injection of the ABEmax mRNA and sgRNA in one-cell stage embryos. The translation of the injected mRNA occurs at different time points during the division of the embryo, which generates different genotypes in the same individual. In addition, recent studies showed that the non-specificity of ABEmax leads to multiple gene editing outcomes in different tissues of the generated founders [13, 42]. The mosaic state of the founders in the present study is also consistent with our previous gene-editing results [15, 16]. The potential influence of bystander mutations on fertility will be investigated further using the lambing data of the offspring. Efforts have been made to minimize the bystander effects of ABE and improve the DNA specificity [17, 18, 43]. In this study, six out of eight founders were generated with the defined point substitution (Table 1). The efficiency of single base substitutions was significantly higher than in our previous Cas9:ssODN studies on goats (24%) [44] and sheep (22.7%) [33]. In spite of the high efficiency of ABEmax, the wider base editing window at position 2-9 (PAM counted as 21–23) converts all “A” to “G” bases [17]. A reduction of the deaminase activity and the search for new Cas9 proteins are required in order to make ABEmax more precise and increase its range of use. In the future, we anticipate the development of new base editors that show improved targeted DNA specificity and reduce bystanders in the editing window.
Fig. 2.
Detection of ABEmax-mediated nucleotide substitutions in founder animals. a Genotypes of target sites by TA cloning in all founder animals. Bystander mutations are highlighted in blue. The ABEmax-mediated nucleotide substitutions (g.A746G, p.Q249R) are highlighted in red. b Genotypes of target sites through deep sequencing in eight founder animals. Bystander mutations are marked in blue. The ABEmax-mediated nucleotide substitutions (g.A746G, p.Q249R) are highlighted in red. c Mutation rate at the targeted region in eight founder animals
Analysis of off-target mutations in edited animals
To evaluate the off-target effects of ABE, we used Sanger sequencing. Five off-target sites (OT1–OT5) were predicted (see Additional file 1: Table S6) using the Cas-OFFinder program [8]. PCR products from eight gene-edited founders and single gene-edited fetal fibroblasts were sequenced and no off-target events were detected (see Additional file 4: Figure S3 and Additional file 5: Figure S4). Thus, compared to CBE [7, 13, 45–47], ABE produce almost no off-target events [21–24, 42, 48]. These results highlight the accuracy and potential applications of ABE for gene therapy. Although we used the Cas-OFFinder program that was developed for Cas9, the off-target effects of Cas9 and ABE are likely to differ, which means that independent off-target assessments will be required in future studies [10, 49].
Conclusions
In summary, we provide the first report on the application of ABE in large animals to generate sheep with targeted amino acid substitutions. Although we are unable to demonstrate the phenotypes in gene-modified animals at this stage, the results provide an alternative approach to improve animal production, and contribute to the validation of key SNPs that underlie agriculturally important traits.
Supplementary information
Additional file 1. Table S1. sgRNA of target sites. Table S2. Oligonucleotides for generating transcription of sgRNA expression vectors. Table S3. Primers for genotyping and amplifying Cas9/sgRNA targeted BMPR1B fragment. Table S4. Detailed summary of the lambs generated with ABEmax-mediated base editing. Table S5. Primers for genotyping and amplifying predicted off-target site fragments. Table S6. List of predicted off-target sites.
Additional file 2: Figure S1. Overlapping peaks in sequencing maps of the DNA from transfected cells.
Additional file 3: Figure S2. Sanger sequencing maps of the DNA from the eight founder animals.
Additional file 4: Figure S3. Detection of potential off-targeted sites by Sanger sequencing in founder animals. Five potential off-targeted sites (OT1–OT5) were predicted by Cas-OFFinder. Sanger sequencing was used to determine substitution at predicted target sites for the eight founder animals.
Additional file 5: Figure S4. Detection of potential off-targeted sites by Sanger sequencing in sheep fibroblasts. Five potential off-targeted sites (OT1–OT5) were predicted by Cas-OFFinder. Sanger sequencing was used to determine substitutions at predicted target sites in sheep fibroblasts.
Acknowledgements
The authors would like to thank their colleagues in Prof. Chen Yulin’s laboratory, specifically, Yunrui Mao and Yangbin Xu, who assisted with herd management, surgical operations and offspring care.
Authors’ contributions
SZ, YD, YL, GL, XW, BM, and YC conceived the study. SZ, JL, BZ, CZ, PK, YW, HY, XH, XZ and BM performed the experiments. YD, CL, SH and XW analyzed the data set. QK and YW provided samples. SZ, XW, XH and BP wrote the article. All authors read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (31772571), Local Grants (NXTS2018-001, 2018KJXX-009), and Local Road Research Board (2017NY-072). XW is a Tang Scholar at Northwest A&F University.
Availability of data and materials
All relevant results are within this paper and its additional files. The raw targeted deep sequencing data is available at NCBI SRA database under the BioProject ID: PRJNA562971.
Ethics approval and consent to participate
The present study was approved by the Animal Care and Use Committee of Northwest A&F University (Approval ID: 2014ZX08008002).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Shiwei Zhou and Yige Ding contributed equally to this work
Contributor Information
Shiwei Zhou, Email: zhoushiwei66@163.com.
Yige Ding, Email: yigeding5923@163.com.
Jiao Liu, Email: liujiaodk@163.com.
Yao Liu, Email: liuyao306@163.com.
Xiaoe Zhao, Email: zhxiaoe126@126.com.
Guanwei Li, Email: liguanweitx@163.com.
Chenguang Zhang, Email: zhangchenguang1027@163.com.
Chao Li, Email: lichao_xinong@163.com.
Ying Wang, Email: nd2013wang@163.com.
Peter Kalds, Email: peterkalds@nwafu.edu.cn.
Yawei Gao, Email: gyw1993310@qq.com.
Bo Zong, Email: 1842237949@qq.com.
Xiaoyu Huang, Email: 724678049@qq.com.
Shuhong Huang, Email: huangshuhongzjx@163.com.
Honghao Yu, Email: geneyhh@126.com.
Qifang Kou, Email: 13995004618@163.com.
Bjoern Petersen, Email: bjoern.petersen@fli.de.
Xingxu Huang, Email: huangxx@shanghaitech.edu.cn.
Xiaolong Wang, Email: xiaolongwang@nwafu.edu.cn.
Baohua Ma, Email: mabh@nwafu.edu.cn.
Yulin Chen, Email: chenyulin@nwafu.edu.cn.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12711-020-00554-6.
References
- 1.Sander JD, Keith Joung J. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol. 2014;32:347–355. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ceccaldi R, Rondinelli B, D’Andrea AD. Repair pathway choices and consequences at the double-strand break. Trends Cell Biol. 2016;26:52–64. doi: 10.1016/j.tcb.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chu VT, Weber T, Wefers B, Wurst W, Sander S, Rajewsky K, et al. Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nat Biotechnol. 2015;33:543–548. doi: 10.1038/nbt.3198. [DOI] [PubMed] [Google Scholar]
- 4.Paquet D, Kwart D, Chen A, Sproul A, Jacob S, Teo S, et al. Efficient introduction of specific homozygous and heterozygous mutations using CRISPR/Cas9. Nature. 2016;533:125–129. doi: 10.1038/nature17664. [DOI] [PubMed] [Google Scholar]
- 5.Sakuma T, Nakade S, Sakane Y, Suzuki KT, Yamamoto T. MMEJ-assisted gene knock-in using TALENs and CRISPR-Cas9 with the PITCh systems. Nat Protoc. 2016;11:118–133. doi: 10.1038/nprot.2015.140. [DOI] [PubMed] [Google Scholar]
- 6.Chiruvella KK, Lang Z, Wilson TE. Repair of double-strand breaks by end joining. Cold Spring Harb Perspect Biol. 2013;5:a012757. doi: 10.1101/cshperspect.a012757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016;533:420–424. doi: 10.1038/nature17946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gaudelli NM, Komor AC, Rees HA, Packer MS, Badran AH, Bryson DI, et al. Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature. 2017;551:464–471. doi: 10.1038/nature24644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nishida K, Arazoe T, Yachie N, Banno S, Kakimoto M, Tabata M, et al. Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science. 2016;353:aaf8729. doi: 10.1126/science.aaf8729. [DOI] [PubMed] [Google Scholar]
- 10.Molla KA, Yang Y. CRISPR/Cas-mediated base editing: technical considerations and practical applications. Trends Biotechnol. 2019;37:1121–1142. doi: 10.1016/j.tibtech.2019.03.008. [DOI] [PubMed] [Google Scholar]
- 11.Komor AC, Zhao KT, Packer MS, Gaudelli NM, Waterbury AL, Koblan LW, et al. Improved base excision repair inhibition and bacteriophage Mu Gam protein yields C:G-to-T: A base editors with higher efficiency and product purity. Sci Adv. 2017;3:eaa04774. doi: 10.1126/sciadv.aao4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim K, Ryu SM, Kim ST, Baek G, Kim D, Lim K, et al. Highly efficient RNA-guided base editing in mouse embryos. Nat Biotechnol. 2017;35:435–437. doi: 10.1038/nbt.3816. [DOI] [PubMed] [Google Scholar]
- 13.Liu Z, Chen M, Chen S, Deng J, Song Y, Lai L, et al. Highly efficient RNA-guided base editing in rabbit. Nat Commun. 2018;9:2717. doi: 10.1038/s41467-018-05232-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zeng Y, Li J, Li G, Huang S, Yu W, Zhang Y, et al. Correction of the Marfan syndrome pathogenic FBN1 mutation by base editing in human cells and heterozygous embryos. Mol Ther. 2018;26:2631–2637. doi: 10.1016/j.ymthe.2018.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou S, Cai B, He C, Wang Y, Ding Q, Liu J, et al. Programmable base editing of the sheep genome revealed no genome-wide off-target mutations. Front Genet. 2019;10:215. doi: 10.3389/fgene.2019.00215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li G, Zhou S, Li C, Cai B, Yu H, Ma B, et al. Base pair editing in goat: nonsense codon introgression into FGF5 results in longer hair. FEBS J. 2019;286:4675–4692. doi: 10.1111/febs.14983. [DOI] [PubMed] [Google Scholar]
- 17.Koblan LW, Doman JL, Wilson C, Levy JM, Tay T, Newby GA, et al. Improving cytidine and adenine base editors by expression optimization and ancestral reconstruction. Nat Biotechnol. 2018;36:843–846. doi: 10.1038/nbt.4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu JH, Miller SM, Geurts MH, Tang W, Chen L, Sun N, et al. Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature. 2018;556:57–63. doi: 10.1038/nature26155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li G, Liu X, Huang S, Zeng Y, Yang G, Lu Z, et al. Efficient generation of pathogenic A-to-G mutations in human tripronuclear embryos via ABE-mediated base editing. Mol Ther Nucleic Acids. 2019;17:289–296. doi: 10.1016/j.omtn.2019.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qin W, Lu X, Liu Y, Bai H, Li S, Lin S. Precise A•T to G•C base editing in the zebrafish genome. BMC Biol. 2018;16:139. doi: 10.1186/s12915-018-0609-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Z, Lu Z, Yang G, Huang S, Li G, Feng S, et al. Efficient generation of mouse models of human diseases via ABE- and BE-mediated base editing. Nat Commun. 2018;9:2338. doi: 10.1038/s41467-018-04768-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hua K, Tao X, Yuan F, Wang D, Zhu JK. Precise A•T to G•C base editing in the rice genome. Molec Plant. 2018;11:627–630. doi: 10.1016/j.molp.2018.02.007. [DOI] [PubMed] [Google Scholar]
- 23.Ryu SM, Koo T, Kim K, Lim K, Baek G, Kim ST, et al. Adenine base editing in mouse embryos and an adult mouse model of Duchenne muscular dystrophy. Nat Biotechnol. 2018;36:536–539. doi: 10.1038/nbt.4148. [DOI] [PubMed] [Google Scholar]
- 24.Kang BC, Yun JY, Kim ST, Shin Y, Ryu J, Choi M, et al. Precision genome engineering through adenine base editing in plants. Nat Plants. 2018;4:427–431. doi: 10.1038/s41477-018-0178-x. [DOI] [PubMed] [Google Scholar]
- 25.Mulsant P, Lecerf F, Fabre S, Schibler L, Monget P, Lanneluc I, et al. Mutation in bone morphogenetic protein receptor-IB is associated with increased ovulation rate in Booroola Merino ewes. Proc Natl Acad Sci USA. 2001;98:5104–5109. doi: 10.1073/pnas.091577598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Montgomery GW, Galloway SM, Davis GH, McNatty KP. Genes controlling ovulation rate in sheep. Reproduction. 2001;121:843–852. [PubMed] [Google Scholar]
- 27.Wilson T, Wu XY, Juengel JL, Ross IK, Lumsden JM, Lord EA, et al. Highly prolific Booroola sheep have a mutation in the intracellular kinase domain of bone morphogenetic protein IB receptor (ALK-6) that is expressed in both oocytes and granulosa cells. Biol Reprod. 2001;64:1225–1235. doi: 10.1095/biolreprod64.4.1225. [DOI] [PubMed] [Google Scholar]
- 28.Reader KL, Haydon LJ, Littlejohn RP, Juengel JL, McNatty KP. Booroola BMPR1B mutation alters early follicular development and oocyte ultrastructure in sheep. Reprod Fertil Dev. 2012;24:353–361. doi: 10.1071/RD11095. [DOI] [PubMed] [Google Scholar]
- 29.Davis GH, Balakrishnan L, Ross IK, Wilson T, Galloway SM, Lumsden BM, et al. Investigation of the Booroola (FecB) and Inverdale (FecX(I)) mutations in 21 prolific breeds and strains of sheep sampled in 13 countries. Anim Reprod Sci. 2006;92:87–96. doi: 10.1016/j.anireprosci.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 30.Tian XE, Sun HX, Wang YJ. Genetic polymorphism of BMPR-IB gene and effect on litter size in three sheep breeds. J Northwest AF Univ. 2009;37:31–36. [Google Scholar]
- 31.Chu MX, Liu ZH, Jiao CL, He YQ, Fang L, Ye SC, et al. Mutations in BMPR-IB and BMP-15 genes are associated with litter size in Small Tailed Han sheep (Ovis aries) J Anim Sci. 2007;85:598–603. doi: 10.2527/jas.2006-324. [DOI] [PubMed] [Google Scholar]
- 32.Zhou S, Yu H, Zhao X, Cai B, Ding Q, Huang Y, et al. Generation of gene-edited sheep with a defined Booroola fecundity gene (FecBB) mutation in bone morphogenetic protein receptor type 1B (BMPR1B) via clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated Cas9. Reprod Fertil Dev. 2018;30:1616–1621. doi: 10.1071/RD18086. [DOI] [PubMed] [Google Scholar]
- 33.Mahdavi M, Nanekarani S, Hosseini SD. Mutation in BMPR-IB gene is associated with litter size in Iranian Kalehkoohi sheep. Anim Reprod Sci. 2014;147:93–98. doi: 10.1016/j.anireprosci.2014.04.003. [DOI] [PubMed] [Google Scholar]
- 34.Roy J, Polley S, De S, Mukherjee A, Batabyal S, Pan S, et al. Polymorphism of fecundity genes (FecB, FecX, and FecG) in the Indian Bonpala sheep. Anim Biotechnol. 2011;22:151–162. doi: 10.1080/10495398.2011.589239. [DOI] [PubMed] [Google Scholar]
- 35.Kumar S, Mishra AK, Kolte AP, Dash SK, Karim SA. Screening for Booroola (FecB) and Galway (FecXG) mutations in Indian sheep. Small Ruminant Res. 2008;80:57–61. [Google Scholar]
- 36.Yue YJ, Yang BH, Xia L, Liu JB, Feng RL. Simultaneous identification of FecB and FecX G mutations in Chinese sheep using high resolution melting analysis. J Appl Anim Res. 2011;39:164–168. [Google Scholar]
- 37.Shen B, Zhang J, Wu H, Wang J, Ma K, Li Z, et al. Generation of gene-modified mice via Cas9/RNA-mediated gene targeting. Cell Res. 2013;23:720–723. doi: 10.1038/cr.2013.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang X, Niu Y, Zhou J, Yu H, Kou Q, Lei A, et al. Multiplex gene editing via CRISPR/Cas9 exhibits desirable muscle hypertrophy without detectable off-target effects in sheep. Sci Rep. 2016;6:32271. doi: 10.1038/srep32271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang X, Yu H, Lei A, Zhou J, Zeng W, Zhu H, et al. Generation of gene-modified goats targeting MSTN and FGF5 via zygote injection of CRISPR/Cas9 system. Sci Rep. 2015;5:13878. doi: 10.1038/srep13878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang Y, Ding Y, Liu Y, Zhou S, Ding Q, Yan H, et al. Optimisation of the clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9: single-guide RNA (sgRNA) delivery system in a goat model. Reprod Fertil Dev. 2019;31:1533–1537. doi: 10.1071/RD18485. [DOI] [PubMed] [Google Scholar]
- 41.Bae S, Park J, Kim JS. Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics. 2014;30:1473–1475. doi: 10.1093/bioinformatics/btu048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ma Y, Yu L, Zhang X, Xin C, Huang S, Bai L, et al. Highly efficient and precise base editing by engineered dCas9-guide tRNA adenosine deaminase in rats. Cell Discov. 2018;4:39. doi: 10.1038/s41421-018-0047-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang L, Zhang X, Wang L, Yin S, Zhu B, Xie L, et al. Increasing targeting scope of adenosine base editors in mouse and rat embryos through fusion of TadA deaminase with Cas9 variants. Protein Cell. 2018;9:814–819. doi: 10.1007/s13238-018-0568-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Niu Y, Zhao X, Zhou J, Li Y, Huang Y, Cai B, et al. Efficient generation of goats with defined point mutation (I397V) in GDF9 through CRISPR/Cas9. Reprod Fertil Dev. 2018;30:307–312. doi: 10.1071/RD17068. [DOI] [PubMed] [Google Scholar]
- 45.Kim YB, Komor AC, Levy JM, Packer MS, Zhao KT, Liu DR. Increasing the genome-targeting scope and precision of base editing with engineered Cas9-cytidine deaminase fusions. Nat Biotechnol. 2017;35:371–376. doi: 10.1038/nbt.3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee HK, Willi M, Miller SM, Kim S, Liu C, Liu DR, et al. Targeting fidelity of adenine and cytosine base editors in mouse embryos. Nat Commun. 2018;9:4804. doi: 10.1038/s41467-018-07322-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gapinske M, Luu A, Winter J, Wood WS, Kostan KA, Shiva N, et al. CRISPR-SKIP: programmable gene splicing with single base editors. Genome Biol. 2018;19:107. doi: 10.1186/s13059-018-1482-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yan F, Kuang Y, Ren B, Wang J, Zhang D, Lin H, et al. Highly efficient A.T to G.C base editing by Cas9n-guided tRNA adenosine deaminase in rice. Mol Plant. 2018;11:631–634. doi: 10.1016/j.molp.2018.02.008. [DOI] [PubMed] [Google Scholar]
- 49.Kim D, Lim K, Kim ST, Yoon SH, Kim K, Ryu SM, et al. Genome-wide target specificities of CRISPR RNA-guided programmable deaminases. Nat Biotechnol. 2017;35:475–480. doi: 10.1038/nbt.3852. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Table S1. sgRNA of target sites. Table S2. Oligonucleotides for generating transcription of sgRNA expression vectors. Table S3. Primers for genotyping and amplifying Cas9/sgRNA targeted BMPR1B fragment. Table S4. Detailed summary of the lambs generated with ABEmax-mediated base editing. Table S5. Primers for genotyping and amplifying predicted off-target site fragments. Table S6. List of predicted off-target sites.
Additional file 2: Figure S1. Overlapping peaks in sequencing maps of the DNA from transfected cells.
Additional file 3: Figure S2. Sanger sequencing maps of the DNA from the eight founder animals.
Additional file 4: Figure S3. Detection of potential off-targeted sites by Sanger sequencing in founder animals. Five potential off-targeted sites (OT1–OT5) were predicted by Cas-OFFinder. Sanger sequencing was used to determine substitution at predicted target sites for the eight founder animals.
Additional file 5: Figure S4. Detection of potential off-targeted sites by Sanger sequencing in sheep fibroblasts. Five potential off-targeted sites (OT1–OT5) were predicted by Cas-OFFinder. Sanger sequencing was used to determine substitutions at predicted target sites in sheep fibroblasts.
Data Availability Statement
All relevant results are within this paper and its additional files. The raw targeted deep sequencing data is available at NCBI SRA database under the BioProject ID: PRJNA562971.