Abstract
The computational design of transmembrane proteins with more than one membrane spanning region remains a major challenge. We report the design of transmembrane monomers, homodimers, trimers, and tetramers with 76–215 residue subunits containing 2–4 membrane spanning regions and up to 860 total residues that adopt the target oligomerization state in detergent solution. The designed proteins localize to the plasma membrane in bacteria and in mammalian cells, and magnetic tweezer unfolding experiments in the membrane indicate that they are very stable. Crystal structures of the designed dimer and tetramer--a rocket shaped structure with a wide cytoplasmic base that funnels into eight transmembrane helices--are very close to the design models. Our results pave the way for the design of multispan membrane proteins with new functions.
One Sentence Summary
We report the accurate design of transmembrane protein monomers, homodimers, trimers, and tetramers with up to 8 transmembrane spans and 860 total residues.
In recent years it has become possible to de novo design, with high accuracy, soluble protein structures ranging from 18-residue constrained peptides to megaDalton protein cages (1). There have also been advances in membrane protein design, as illustrated by an elegant zinc-transporting transmembrane peptide tetramer named Rocker (2), and an engineered ion conducting oligomer based on the C-terminal transmembrane segment (TMs) of the E. coli polysaccharide transporter Wza (3). Both are single membrane-spanning synthesized peptides with fewer than 36 residues. It has also been possible to design and confirm the transmembrane topology of multipass membrane proteins using simple sequence hydrophobicity and charge based models (4), but the extent to which the transmembrane helices pack with each other is not clear. Design of structurally defined multipass membrane proteins has remained a major challenge because of the difficulty in specifying structure within the membrane and in experimentally determining membrane protein structures generally; crystal structures of the full designed oligomeric states of Rocker and the Wza derived channel have not yet been determined, and to date there are no crystal structures of de novo designed multipass membrane proteins.
A major challenge for membrane protein design stems from the similarity of the membrane environment to protein hydrophobic cores. In the design of soluble proteins, the secondary structure and overall topology can be specified by the pattern of hydrophobic and hydrophilic residues, with the former inside the protein and the latter outside facing solvent. This core design principle cannot be used for membrane proteins, as the apolar environment of the hydrocarbon core of the lipid bilayer requires that outward facing residues in the membrane also be nonpolar. Buried hydrogen bonds between polar sidechains have been demonstrated to play an important role in the association of helical peptides within the membrane, overcoming the degeneracy in the non-polar interactions (5–7).
We reasoned that a recently developed method for designing buried hydrogen bond networks (8) could allow specification of the packing interactions of transmembrane helices in multipass transmembrane proteins. We first explored the design of helical transmembrane proteins with four TMs -- dimers of 76- to-104 residue hairpins or a single chain dimer of 156 residues -- with hydrophobic spanning regions ranging from 21 to 35 Å (Fig. 1A and Fig. 2A), repurposing the Ser and Gln containing hydrogen bond networks in a designed soluble four-helix dimer with C2 symmetry (2L4HC2_23, (Protein Data Bank (PDB) ID: 5J0K)) (8) to provide structural specificity. Four-helix bundles of different lengths with backbone geometries capable of hosting these networks were produced using parametric generating equations (9), residues comprising the hydrogen bond networks and neighboring packing residues were introduced, and the remainder of the sequence was optimized using Rosetta Monte Carlo (10) design calculations to obtain low energy sequences. Connecting loops between the helices were built using Rosetta. To specify the orientation of the designs (11) in the membrane when expressed in cells, at the designed lipid-water boundary on the extracellular/periplasmic side we incorporated a ring of amphipathic aromatic residues and at the lipid-water boundary on the cytoplasmic side, a ring of positively charged residues (Fig. 1A and Fig. 2A). Between these two rings, the surface residues are exposed to the hydrophobic membrane environment; these positions in Rosetta sequence design calculations were restricted to hydrophobic amino acids [see supplementary materials]. Consistent with the design, TMHMM predicts that the dimer designs contain 2 TMs and the single chain design (scTMHC2), 4TMs (Fig. S1). On average, for each residue ~68% of the sidechain surface area is buried in the designs, which could provide substantial van der Waals stabilization (12).
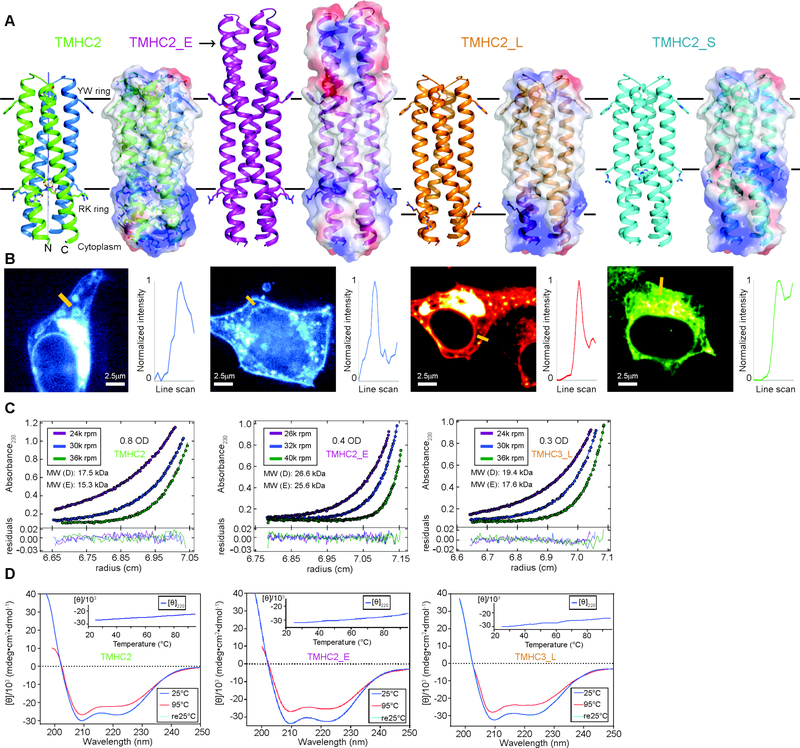
Fig. 1.
Design and characterization of proteins with four transmembrane helices. From left to right, designs and data are shown for TMHC2 (transmembrane hairpin C2), TMHC2_E (elongated), TMHC2_L (long span) and TMHC2_S (short span). (A) Design models with intra- and extra-membrane regions with different lengths. Horizontal lines demarcate the hydrophobic membrane regions. Ribbon diagrams are on left, electrostatic surfaces on right, and the neutral transmembrane regions are in gray. (B) Confocal microscopy images for HEK293T cells transfected with TMHC2 fused to mTagBFP, TMHC2_E fused to mTagBFP, TMHC2_L fused to mCherry and TMHC2_S fused to eGFP. Line scans (yellow lines in the images) across the membranes show significant increase in fluorescence across the plasma membranes for TMHC2, TMHC2_E and TMHC2_L, but less significant increase for TMHC2_S. (C) Representative analytical ultracentrifugation sedimentation-equilibrium curves at three different rotor speeds. Each data set is globally well fitted as a single ideal species in solution corresponding to the dimer molecular weight. ‘MW (D)’ and ‘MW (E)’ indicate the molecular weight of the oligomer design and that determined from experiment, respectively. (D) CD spectra and temperature melt (inset). No apparent unfolding transitions are observed up to 95°C.
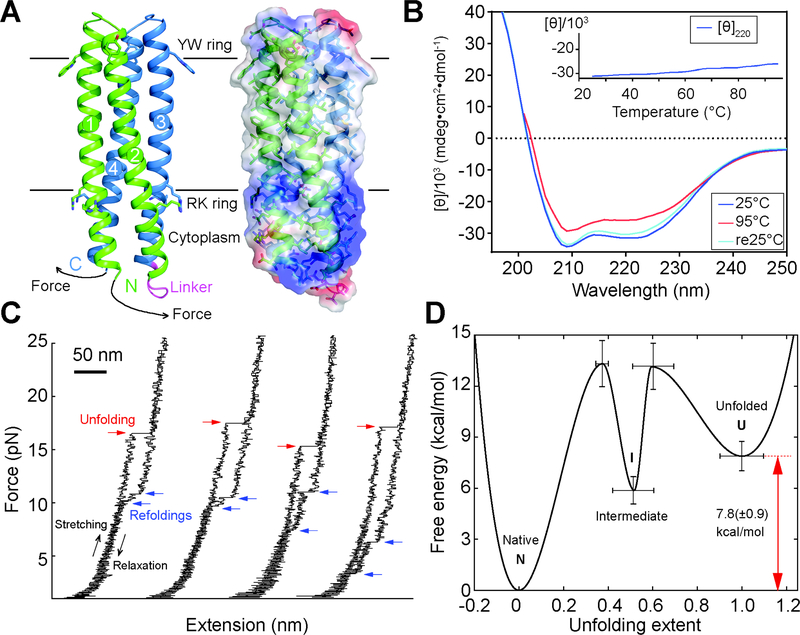
Fig. 2.
Folding stability of the 156-residue single chain TMHC2 (scTMHC2) design with four transmembrane helices. (A) Design model (left) and electrostatic surface (right) of scTMHC2. N- and C-terminal helical hairpins are colored green and blue respectively. Numbers indicate the order of the four TMs in the sequence. The linker connecting the two hairpins is colored magenta. Single-molecule forced unfolding experiments were conducted by applying mechanical tension to the N- and C-terminus of a single scTMHC2 (Fig. S5 for more details). (B) CD spectra of scTMHC2 at different temperatures. No unfolding transition is observed up to 95°C. (C) Single-molecule force-extension traces of scTMHC2. The unfolding and refolding transitions are denoted with red and blue arrows. (D) Folding energy landscape obtained from the single-molecule experiments. N, I, and U indicate the native, intermediate, and unfolded state respectively.
Synthetic genes encoding the designs were obtained and the proteins expressed in E. coli and mammalian cells using membrane protein expression vectors. The dimer design with the shortest hydrophobic span (15 residues, TMHC2_S) was poorly behaved in both E.coli and mammalian cells, but the dimer designs with longer spans TMHC2, TMHC2_E and TMHC2_L localized to the cell membrane when expressed in HEK293T cells (Fig. 1B) and in E. coli. The designed proteins were purified by extracting the E. coli membrane fraction with detergent, followed by nickel-NTA chromatography and size exclusion chromatography (SEC) with a yield of ~2 mg/L (Fig. S2A and B). The designed proteins TMHC2, TMHC2_E and TMHC2_L eluted as single peaks in SEC, and in analytical ultracentrifugation (AUC) experiments in detergent solution, the proteins sedimented as dimers consistent with the design models (Fig. 1C and Fig. S3). For the single chain scTMHC2 the major species in SEC was the monomer with a small side peak that was readily removed by purification (Fig. S2B). Circular dichroism (CD) measurements showed that the designs were alpha helical and highly thermal stable--the CD spectra at 95°C were similar to those at 25°C (Fig. 1D and Fig. 2B). TOXCAT-β–lactamase (TβL) assays (13), which couple E. coli survival to oligomerization and proper orientation of fused antibiotic resistance markers on the N and C termini, suggest that the N- and C-termini of TMHC2 are in the cytoplasm as in the design models (Fig. S4).
We more quantitatively characterized the folding stability of scTMHC2 using single-molecule forced unfolding experiments (14, 15) (Fig. 2). The designed protein reconstituted in a bicelle was covalently attached to a magnetic bead and a glass surface through its N- and C-termini (Fig. 2A and Fig. S5). The distance between the bead and the surface was determined as a function of the applied mechanical tension. In unfolding experiments with the force slowly increasing (~0.5 pN/s), unfolding transitions were observed at ~18 pN and, upon force de-ramping, refolding transitions were observed at ~9 pN (80.1% of the recorded unfolding traces had one step unfolding transitions and 84.6% of the refolding transitions had two steps; Fig. 2C, Fig. S6 and Fig. S7). Consistent with the internal symmetry of the single-chain homodimer design (Fig. 2A and Fig. S5), the two refolding step sizes were very similar (Fig. S8). This unfolding and refolding asymmetry is consistent with a three-state free energy landscape: a native dimer state (N), an intermediate state containing only one hairpin (I), and an unfolded state (U) (Fig. S9). During unfolding at high force, only the barrier between the native and intermediate states is observed, while at the lower forces where refolding occurs, both energy barriers become prominent (Fig. S9). The transition rates between the folded, intermediate and unfolded states were determined using the Bell model (16), yielding the relative free energies of the states and the associated barrier heights (14) (Fig. 2D and Fig. S10). The overall thermodynamic stability of scTMHC2 is 7.8(±0.9) kcal/mol - on a per transmembrane helix basis, more stable than the naturally occurring helical membrane proteins studied thus far (folding free energy per helix for scTMHC2 is 2.0(±0.2) kcal/(mol⋅helix) compared to 0.7–0.9 kcal/(mol⋅helix) for GlpG (14, 17) and 1.6–1.8 kcal/(mol⋅helix) for bacteriorhodopsin) (18).
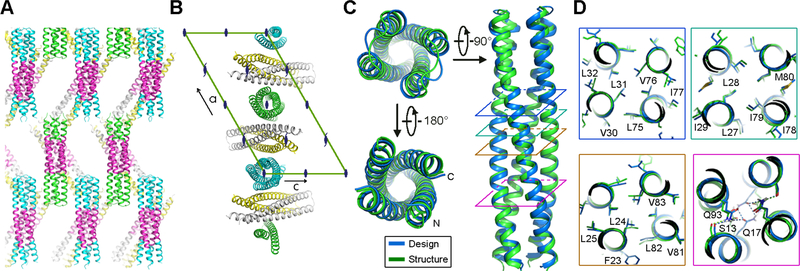
We carried out crystal screens in different detergents for each of the designs, and obtained crystals of the design with the most extensive cytoplasmic region, TMHC2_E, in n-nonyl-β-D-glucopyranoside (NG). The crystals diffracted to 2.95 Å resolution, and we solved the structure by molecular replacement with the design model. As anticipated, the extended soluble region mediates the crystal lattice packing; there are large solvent channels around the designed TMs likely due to the surrounding disordered detergent molecules (Fig. 3A). Each asymmetric unit contains four helical hairpins, two are paired in a dimer while the other two form two C2 dimers through crystallographic symmetry with two monomers in adjacent asymmetric units; the C2 axis in the design is perfectly aligned with the crystallographic two fold (Fig. 3B). The conformations of the dimers in the three biological units are nearly identical with very small differences due to crystal packing (Cα root-mean-square deviations (RMSDs): 0.60–0.84 Å) (Fig. S11). Both the overall structure and the core sidechain packing are almost identical in the crystal structure and the design model with a Cα RMSD of 0.7 Å over the core residues (Fig. 3C). Two of the three buried hydrogen bonding residues within the membrane have conformations that almost exactly match the design model (S13 and Q93), but Q17 adopts a different rotamer with the side-chain nitrogen donating a hydrogen bond to the main-chain carbonyl oxygen (Fig. 3D).
Fig. 3.
Crystal structure of the designed transmembrane dimer TMHC2_E. (A and B) Crystal lattice packing. (A) The extended soluble region mediates a large portion of the crystal lattice packing. The four helical hairpins in the asymmetric unit are colored green, gray, yellow and cyan, respectively. The TMs, in magenta, forms layers in the crystal separating the soluble regions. (B) The C2 axis of the design aligns with the crystallographic two fold. Two monomers (gray and yellow) are paired in a dimer while the other two (green and cyan) form two C2 dimers with two crystallographic adjacent monomers. The space group diagram (C121) is shown in the background. (C) Superposition of the TMHC2_E crystal structure and design model (RMSD = 0.7 Å over the core Cα atoms). (D) The side-chain packing arrangements at layers (colored squares in panel C) at different depths in the membrane are almost identical to the design model.
We used a similar approach to design a transmembrane trimer with six membrane spanning helices (TMHC3) based on the 5L6HC3_1 scaffold (PDB ID: 5IZS) (8). Guided by the results with the C2 designs, we chose a hydrophobic span of ~30 Å (20 residues) (Fig. 4A). The design was expressed in E. coli and purified to homogeneity, eluting on a gel filtration column as a single homogeneous species (Fig. S2C). CD measurements showed that TMHC3 was highly thermostable with the alpha helical structure preserved at 95°C (Fig. 4B). AUC experiments showed that TMHC3 is a trimer in detergent solution consistent with the design (Fig. 4C and Fig. S12A).
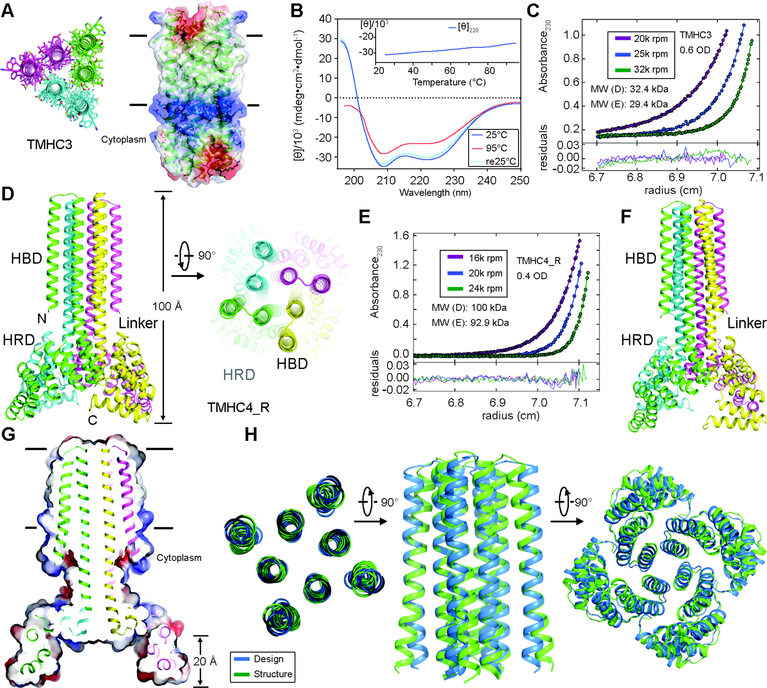
Fig. 4.
Stability and structural characterization of designs with six and eight membrane spanning helices. (A) Model of designed transmembrane trimer TMHC3 with six transmembrane helices. Stick representation from periplasmic side (left) and lateral surface view (right) are shown. (B) Circular dichroism characterization of TMHC3; the design is stable up to 95°C. (C) Representative analytical ultracentrifugation sedimentation-equilibrium curves at three different rotor speeds for TMHC3. The data fit to a single ideal species in solution with molecular weight close to that of the designed trimer. (D) Model of designed transmembrane tetramer TMHC4_R with eight transmembrane helices. The four protomers are colored green, yellow, magenta and cyan, respectively. (E) Analytical ultracentrifugation sedimentation-equilibrium curves at three different rotor speeds for TMHC4_R fit well to a single species with a measured molecular weight of ~94 kDa. (F) Crystal structure of TMHC4_R. The overall tetramer structures are very similar to the design model, with a helical bundle body and helical repeat fins. The outer helices of the transmembrane hairpins tilt off the axis by ~10°. (G) Cross section through the TMHC4_R crystal structure and electrostatic surface; the HRD forms a bowl at the base of the overall structure with a depth of ~20 Å. The transmembrane region is indicated in lines. (H) Three views of the backbone superposition of TMHC4_R crystal structure and design model.
To explore our capability to design membrane proteins with more complex topologies, we designed a C4 tetramer with a two ring helical bundle membrane spanning region composed of 8 TMs and an extended bowl shaped cytoplasmic domain formed by repeating structures emanating away from the symmetry axis (Fig. 4D). The design has an overall rocket shape with a height of ~100 Å and can be divided into three regions: the helical bundle domain (HBD), the helical repeat domain (HRD), and the helical linker between the two. The central HBD was derived from the soluble design 5L8HC4_6 (8) and the bowl from a designed helical repeat protein homo-oligomer (tpr1C4_2) (19). Helical linkers were built using RosettaRemodel (20) - a 9-residue junction was found to yield the correct helical register (Fig. S13). Following Rosetta sequence design calculations, a gene encoding the lowest energy design, TMHC4_R, was synthesized. The protein was expressed in E. coli and purified using nickel affinity and gel filtration chromatography; the final yield was ~3 mg/L and the purified protein chromatographed as a monodisperse peak in SEC (Fig. S2C). CD experiments showed that the design was alpha-helical and thermostable up to 95°C (Fig. S12B). AUC measurements showed that TMHC4_R is a tetramer in detergent solution, consistent with the design model (Fig. 4E and Fig. S12C). After a systematic effort to screen detergents for crystallization, we obtained crystals in a combination of n-Decyl-β-D-Maltopyranoside (DM) and NG in the P4 space group that diffracted to 3.9 Å resolution. We solved the crystal structure by molecular replacement using the design model (Rwork/Rfree = 0.29/0.32 with unambiguous electron density) (Table S1 and Fig. S14). The crystal lattice packing is primarily between the extended cytoplasmic domains; there may be minor detergent-mediated interactions between the transmembrane and helical repeat (HR) domains as well (Fig. S15).
Although the resolution is insufficient for evaluating the details of the side-chain packing, it does allow backbone-level comparisons. There are four TMHC4_R monomers in one asymmetric unit, with nearly identical structures (Cα RMSDs between 0.2 and 0.6 Å) (Fig. S16A). The Cα RMSDs between the structure and design model are 1.2–1.8 Å for the monomer transmembrane helices, 0.3–0.4 Å for the linkers, 1.1–1.5 Å for the HR domains, and 3.3–3.6 Å for the overall structure (Fig. S16B). As in the case of the C2 design, the C4 symmetry axis of the design coincides with the crystallographic axes of the crystal lattice (Fig. S16C). The four tetramer structures on the crystal C4 axes have overall structures very similar to each other and to the design model (Fig. 4, F and G, and Fig. S16A); the tetrameric transmembrane domain, HR domain, and overall tetramer structure have Cα RMSDs to the design model of 1.3–1.5 Å, 3.3–3.8 Å and 3.3–3.8 Å, respectively (Fig. 4H and Fig. S16D, left panel). The deviation in the HR domain may result from crystal packing interactions between the termini; the Cα RMSDs over the first 162 residues are 2.2–2.3 Å (Fig. S16D, right panel). The main deviation from the design model is a tilting of the outer helices of transmembrane hairpins from the axis by ~10° (Fig. 4, F and G).
The agreement between the crystal structures of TMHC2_E and TMHC4_R with the design models demonstrates that transmembrane homo-oligomers containing multiple membrane spanning regions and extensive extracellular domains can now be accurately designed. For future work, the general approach of first designing and characterizing hydrogen bond network containing soluble versions of the desired transmembrane structures, and then converting to integral membrane proteins by redesigning the membrane exposed residues, could be quite robust. Single-molecule forced unfolding and thermal denaturation experiments show that the designed proteins are highly stable. While the designs lack the classic small residue packing in the core thought to be an important driver of membrane protein folding (21–24), like natural membrane proteins they bury more surface area than typical soluble proteins, thereby maximizing van der Waals packing contributions (12). The range of the design features--variable transmembrane and extracellular helix lengths and twists, extensive soluble domains and diverse oligomeric states--are significant steps towards the complexity of natural transmembrane proteins with multiple membrane spanning regions and extra membrane domains that play important roles in ligand/substrate recognition and structure stabilization as in the ATP binding cassette (ABC) transporters, ion channels, ryanodine receptor and gamma-secretase (25, 26). The capability to accurately designing complex multipass transmembrane proteins that can be expressed in cells opens the door to design of a new generation of multipass membrane protein structures and functions.
Supplementary Material
Acknowledgments
This work was supported by the Howard Hughes Medical Institute (D.B.) and the National Institutes of Health (R01GM063919 to J.U.B.). P.L. was supported by the Raymond and Beverly Sackler fellowship. D.M. was supported by the Basic Science Research Program through the National Research Foundation of Korea funded by the Ministry of Education (NRF-2016R1A6A3A03007871). We thank J. Sumida for analytical ultracentrifugation support; A. Kang for crystallization support; D. Ma and Z. Wang for crystallography support; the staff at the Advanced Light Source and P. Huang, Y. Hsia, A. Ford, L. Stewart, C. Xu and many other members of the Baker lab for helpful discussions. This work was facilitated by the Hyak supercomputer at the University of Washington. Coordinates and structure files have been deposited to the Protein Data Bank with accession codes: 6B87 (TMHC2_E), 6B85 (TMHC4_R). Author contributions: P.L. and D.B. designed the research and P.L., D.M., F.D., K.Y.W, J.U.B. and D.B. wrote the manuscript. P.L. and D.B. carried out design calculations and developed the membrane protein design method. P.L. purified and characterized the designed proteins. D.M. and J.U.B. designed, performed and analyzed single-molecule forced unfolding experiments. K.Y.W. and M.D.V. performed mammalian cell localization experiment. P.L. crystallized the designed proteins. P.L. collected and analyzed crystallographic data with help from W.X.. F.D. solved structures with help from P.L.. S.E.B., C.Z., J.F., G.U., and W.S. contributed the soluble scaffolds. V.K.M. wrote the amino acid composition based energy term. All authors discussed results and commented on the manuscript.
References and Notes
- 1.Huang P-S, Boyken SE, Baker D, The coming of age of de novo protein design. Nature. 537, 320–327 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Joh NH et al. , De novo design of a transmembrane Zn2 -transporting four-helix bundle. Science. 346, 1520–1524 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mahendran KR et al. , A monodisperse transmembrane α-helical peptide barrel. Nat. Chem. 9, 411–419 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Whitley P, Nilsson I, von Heijne G, De novo design of integral membrane proteins. Nat. Struct. Mol. Biol. 1, 858–862 (1994). [DOI] [PubMed] [Google Scholar]
- 5.Choma C, Gratkowski H, Lear JD, DeGrado WF, Asparagine-mediated self-association of a model transmembrane helix. Nat. Struct. Biol. 7, 161–166 (2000). [DOI] [PubMed] [Google Scholar]
- 6.Zhou FX, Cocco MJ, Russ WP, Brunger AT, Engelman DM, Interhelical hydrogen bonding drives strong interactions in membrane proteins. Nat. Struct. Biol. 7, 154–160 (2000). [DOI] [PubMed] [Google Scholar]
- 7.Tatko CD, Nanda V, Lear JD, Degrado WF, Polar networks control oligomeric assembly in membranes. J. Am. Chem. Soc. 128, 4170–4171 (2006). [DOI] [PubMed] [Google Scholar]
- 8.Boyken SE et al. , De novo design of protein homo-oligomers with modular hydrogen-bond network-mediated specificity. Science. 352, 680–687 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang P-S et al. , High thermodynamic stability of parametrically designed helical bundles. Science. 346, 481–485 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rohl CA, Strauss CEM, Misura KMS, Baker D, in Methods in Enzymology (2004), pp. 66–93. [DOI] [PubMed] [Google Scholar]
- 11.von Heijne G, Membrane-protein topology. Nat. Rev. Mol. Cell Biol. 7, 909–918 (2006). [DOI] [PubMed] [Google Scholar]
- 12.Oberai A, Joh NH, Pettit FK, Bowie JU, Structural imperatives impose diverse evolutionary constraints on helical membrane proteins. Proc. Natl. Acad. Sci. U. S. A. 106, 17747–17750 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elazar A et al. , Mutational scanning reveals the determinants of protein insertion and association energetics in the plasma membrane. Elife. 5 (2016), doi: 10.7554/eLife.12125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Min D, Jefferson RE, Bowie JU, Yoon T-Y, Mapping the energy landscape for second-stage folding of a single membrane protein. Nat. Chem. Biol. 11, 981–987 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Min D, Arbing MA, Jefferson RE, Bowie JU, A simple DNA handle attachment method for single molecule mechanical manipulation experiments. Protein Sci. 25, 1535–1544 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bell G, Models for the specific adhesion of cells to cells. Science. 200, 618–627 (1978). [DOI] [PubMed] [Google Scholar]
- 17.Guo R et al. , Steric trapping reveals a cooperativity network in the intramembrane protease GlpG. Nat. Chem. Biol. 12, 353–360 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chang Y-C, Bowie JU, Measuring membrane protein stability under native conditions. Proc. Natl. Acad. Sci. U. S. A. 111, 219–224 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fallas JA et al. , Computational design of self-assembling cyclic protein homo-oligomers. Nat. Chem. 9, 353–360 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang P-S et al. , RosettaRemodel: A Generalized Framework for Flexible Backbone Protein Design. PLoS One. 6, e24109 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang S-Q et al. , The membrane- and soluble-protein helix-helix interactome: similar geometry via different interactions. Structure. 23, 527–541 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walters RFS, DeGrado WF, Helix-packing motifs in membrane proteins. Proc. Natl. Acad. Sci. U. S. A. 103, 13658–13663 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feng X, Barth P, A topological and conformational stability alphabet for multipass membrane proteins. Nat. Chem. Biol. 12, 167–173 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meruelo AD, Han SK, Kim S, Bowie JU, Structural differences between thermophilic and mesophilic membrane proteins. Protein Sci. 21, 1746–1753 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shi Y, A Glimpse of Structural Biology through X-Ray Crystallography. Cell. 159, 995–1014 (2014). [DOI] [PubMed] [Google Scholar]
- 26.Fernandez-Leiro R, Scheres SHW, Unravelling biological macromolecules with cryo-electron microscopy. Nature. 537, 339–346 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.von Heijne G, Membrane protein structure prediction. J. Mol. Biol. 225, 487–494 (1992). [DOI] [PubMed] [Google Scholar]
- 28.Alford RF et al. , An Integrated Framework Advancing Membrane Protein Modeling and Design. PLoS Comput. Biol. 11, e1004398 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yarov-Yarovoy V, Schonbrun J, Baker D, Multipass membrane protein structure prediction using Rosetta. Proteins. 62, 1010–1025 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rocklin GJ et al. , Global analysis of protein folding using massively parallel design, synthesis, and testing. Science. 357, 168–175 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.The PyMOL Molecular Graphics System, version 1.7.4 (Schrödinger, LLC; ). [Google Scholar]
- 32.Otwinowski Z, Minor W, Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997). [DOI] [PubMed] [Google Scholar]
- 33.Collaborative Computational Project, Number 4, The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 (1994). [DOI] [PubMed] [Google Scholar]
- 34.Adams PD et al. , PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.DiMaio F et al. , Improved low-resolution crystallographic refinement with Phenix and Rosetta. Nat. Methods. 10, 1102–1104 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Song Y et al. , High-resolution comparative modeling with RosettaCM. Structure. 21, 1735–1742 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lebowitz J, Lewis MS, Schuck P, Modern analytical ultracentrifugation in protein science: a tutorial review. Protein Sci. 11, 2067–2079 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schuck P, On the analysis of protein self-association by sedimentation velocity analytical ultracentrifugation. Anal. Biochem. 320, 104–124 (2003). [DOI] [PubMed] [Google Scholar]
- 39.Hanggi P, Talkner P & Borkovec M, Reaction-Rate Theory: Fifty Years after Kramers. Rev. Mod. Phys. 62, 251–341 (1990). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.